Abstract
The involvement of cyclic nucleotide gated ion channels (CNGCs) in the signal transduction of animal light and odorant perception is well documented. Although plant CNGCs have recently been revealed to mediate multiple stress responses and developmental pathways, studies that aim to elucidate their structural and regulatory properties are still very much in their infancy. The structure–function relationship of plant CNGCs was investigated here by using the chimeric Arabidopsis AtCNGC11/12 gene that induces multiple defence responses in the Arabidopsis mutant constitutive expresser of PR genes 22 (cpr22) for the identification of functionally essential residues. A genetic screen for mutants that suppress cpr22-conferred phenotypes identified over 20 novel mutant alleles in AtCNGC11/12. One of these mutants, suppressor S58 possesses a single amino acid substitution, arginine 557 to cysteine, in the αC-helix of the cyclic nucleotide-binding domain (CNBD). The suppressor S58 lost all cpr22 related phenotypes, such as spontaneous cell death formation under ambient temperature conditions. However, these phenotypes were recovered at 16 °C suggesting that the stability of channel function is affected by temperature. In silico modelling and site-directed mutagenesis analyses suggest that arginine 557 in the αC-helix of the CNBD is important for channel regulation, but not for basic function. Furthermore, another suppressor mutant, S136 that lacks the entire αC-helix due to a premature stop codon, lost channel function completely. Our data presented here indicate that the αC-helix is functionally important in plant CNGCs.
Keywords: Calcium ion channel, CNGC, cpr22, cyclic nucleotide gated ion channel, environment, programmed cell death, temperature
Introduction
For plant survival, cationic nutrients play essential roles in a wide variety of physiological aspects during growth and development. They also play important roles for signal transduction. For example, Ca2+ and K+ fluxes have been reported to be important for abiotic stress as well as biotic stress responses. Uptake and distribution of cationic nutrients mainly relies on membrane-localized cation transporter proteins. Based on genomic sequence data, Arabidopsis contains over 150 cation transport proteins (Mäser et al., 2001). Among them cyclic nucleotide-gated ion channels (CNGCs) form a large gene family consisting of 20 members that have been implicated in defence responses, development, and ion homeostasis in plants (Talke et al., 2003; Kaplan et al., 2007; Chin et al., 2009).
CNGCs were first discovered in retinal photoreceptors and olfactory sensory neurons and so far, six CNGC channel genes have been found in mammalian genomes (Zagotta and Siegelbaum, 1996; Zufall et al., 1994).
The structure of CNGCs is similar to that of the voltage-gated outward rectifying K+-selective ion channel (Shaker) proteins, including a cytoplasmic N-terminus, six membrane spanning regions (S1–S6), a pore domain located between S5 and S6, and a cytoplasmic C-terminus (Zagotta and Siegelbaum, 1996). However, CNGCs are opened by the direct binding of cyclic nucleotides (CN), such as cAMP and cGMP (Fesenko et al., 1985). The CN-binding domain (CNBD) of CNGCs is located at the cytoplasmic C-terminus and exhibits significant sequence similarity to that of protein kinase A, protein kinase G, and the catabolite activator protein of E. coli (Bridges et al., 2005). The cytoplasmic C-terminus contains a CNBD that is connected to the pore domain by a C-linker region. Important functional features of CNGCs were extensively studied in animal systems and it has been suggested that the subunit composition of the respective channel complex is an important determinant for functional features such as ligand sensitivity, selectivity, and gating. (Kaupp and Seifert, 2002).
The first plant CNGC, HvCBT1 (Hordeum vulgare calmodulin (CaM)-binding transporter), was identified as a CaM-binding protein in barley (Schuurink et al., 1998). Subsequently, several CNGCs were identified from Arabidopsis and Nicotiana tabacum (Arazi et al., 1999; Köhler et al., 1999). A precise analysis of the CaM binding site has been reported using the tobacco CNGC, NtCBP4 as well as Arabidopsis AtCNGC1 and AtCNGC2 and it has been suggested to be located at the αC-helix of the CNBD in these plant CNGCs (Arazi et al., 2000; Köhler and Neuhaus, 2000). However, only a handful of studies on the structure–function analysis of plant CNGCs have so far been published (Hua et al., 2003; Bridges et al., 2005; Kaplan et al., 2007; Baxter et al., 2008).
The Arabidopsis mutant constitutive expresser of PR genes 22 (cpr22), which contains a novel chimeric CNGC, AtCNGC11/12, shows environmentally sensitive defence responses, such as heightened salicylic acid (SA) accumulation and a hypersensitive response (HR)-like programmed cell death (Yoshioka et al., 2001; Moeder and Yoshioka, 2009). It has been reported that the expression of AtCNGC11/12 and its channel activity is attributable for the cpr22 phenotype (Yoshioka et al., 2006; Baxter et al., 2008). Interestingly, all SA dependent phenotypes, are suppressed under high humidity conditions and enhanced by low humidity (Yoshioka et al., 2001). This type of environmental sensitivity has been reported for various pathogen resistance mutants as well as on defence responses in wild type plants (Moeder and Yoshioka, 2009).
The importance of the interaction between the C-linker domain and the CNBD domain for basic channel function was reported previously by Baxter et al. (2008). In addition, it was shown that intragenic mutants of cpr22 are useful tools to study the structure–function relationship of CNGCs (Baxter et al., 2008). The use of intragenic mutants of cpr22 were used here for the structure–function study and to reveal the importance of the αC-helix of the CNBD for stable channel function in planta.
Materials and methods
Plant growth conditions
Arabidopsis thaliana plants were grown on Pro-Mix soil (Premier Horticulture Inc., Red Hill, PA, USA) in a growth chamber under ambient humidity as described by Silva et al. (1999). Nicotiana benthamiana plants were grown on the same soil in a greenhouse under a 14:10 h light:dark regimen at 25 °C (day) and 20 °C (night).
Suppressor screening and identification of the S58 mutant
Suppressor no. 58 (S58) was identified in the same screen that was described in Baxter et al. (2008).
Trypan blue staining
Leaf samples were taken from 3–4-week-old plants grown on soil. Trypan blue staining was performed as described previously (Yoshioka et al., 2001).
RNA extraction, RT-PCR, and Northern hybridization
Small-scale RNA extraction was carried out using the TRIzol reagent (Invitrogen, Carlsbad, MO, CA), according to the manufacturer's instructions. Reverse transcriptase (RT)-PCR was performed using cDNA generated by SuperScript™ II Reverse Transcriptase (Invitrogen, Carlsbad, MO, CA) according to the manufacturer's instructions. For the detection of CNGC gene expression in N. benthamiana, CNGC, and actin gene expression in yeast, and β-tubulin gene expression in Arabidopsis, the same sets of primers described by Baxter et al. (2008) were used. For the detection of PR-1 by RT-PCR, PR-1F: 5′-GCTCTTGTAGGTGCTCTTGTT-3′ and PR-1R; 5′-CAGCTCTTATTTGTATTATTT-3′ were used. Northern hybridization of the PR-1 gene was performed as previously described (Yoshioka et al., 2006). For the RT-PCR, 25 cycles for each analysis were applied.
Pathogen infection
Infection with Hyaloperonospora arabidopsidis isolate Emwa1 was performed as described previously (Yoshioka et al., 2001).
Plasmid construction
The yeast expression vector plasmid pYES2-empty vector (Invitrogen), pYES2-AtCNGC12, pYES2-AtCNGC11/12, and pYES2-AtCNGC11/12:E519K (S73) were constructed as previously described (Yoshioka et al., 2006; Baxter et al., 2008). For pYES2-AtCNGC11/12:R557C (S58), total RNA was extracted from S58 and cDNA was generated as described above. cDNA of AtCNGC11/12:R557C was then amplified by RT-PCR using primers: BamH-ATG3: 5′-GGGATCCCATGAATCTTCAGAGGAGAAA-3′ and cDNA14-R1: 5′-CACTATGCTTCAGCCTTTGC-3′, and then cloned into pGEM-T Easy (Promega, Madison, WI). In the case of pYES2-AtCNGC11/12:Q543X (S136), AtCNGC11/12 cDNA was used as template for PCR using primers: BamH-ATG3: 5′-GGGATCCCATGAATCTTCAGAGGAGAAA-3′ and Reverse 136 stop mutation R: 5′-TTATCTTTGAAAGACATTTAA-3′. Both cDNA clones were then subcloned into pYES2 at the BamHI and NotI restriction enzyme sites. AtCNGC:R557C (S58) cDNA minus stop was generated by RT-PCR using forward 5′-CTCTAGACATGAATCTTCAGAGGAGAAA-3′ and reverse 5′-AGTCTAGATGCTTCAGCCTTTGC-3′ primers and subsequently cloned into pGEM-T easy (Promega, Madison, WI). This clone was then subcloned into the XbaI site in pMBP3 (Yoshioka et al., 2006; Urquhart et al., 2007). For site-directed mutagenesis, AtCNGC:R557C (S58) cDNA was excised using XbaI and subsequently subcloned into pBluescript (Stratagene, La Jolla, CA). The point mutation R557I (C1627T) was introduced into AtCNGC11/12 cDNA in pBluescript (Baxter et al., 2008) using the QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The AtCNGC11/12:R557I cDNA was then subcloned into pMBP3 for transient assays in N. benthamiana. All constructed plasmids were sequenced for fidelity.
Agrobacterium-mediated transient expression
Agrobacterium-mediated transient expression in N. benthamiana was performed as described by Urquhart et al. (2007). The expression of these genes was confirmed by RT-PCR (see RNA extraction and RT-PCR).
Functional complementation in yeast
K+ yeast mutant complementation:
The K+-uptake deficient yeast mutant strain CY162 (MAT α, ura3-52, trk1,2) was transformed with the empty plasmid pYES2, pYES2-AtCNGC12, pYES2-AtCNGC11/12, pYES2-AtCNGC11/12:R557C, pYES2-AtCNGC12:R557I and pYES2-AtCNGC11/12:Q543X following the lithium acetate transformation protocol (Ausubel et al., 1987). Yeast transformants were grown in synthetic minimal media supplemented with 0.1 mM KCI (Leng et al., 1999) and hygromycin (5 mg l−1) (Mercier et al., 2004) to suppress background growth. Growth rates were monitored by determining the OD600 of the growing yeast cultures at 20 h and 40 h.
Ca2+ yeast mutant complementation:
A wild-type strain of S. cerevisiae W303-1A (Wallis et al., 1989) and the Ca2+ channel mutant (cch1::TRP1 null mutant) strain K927 (Locke et al., 2000) were provided by Dr H Iida (Tokyo Gakugei University). K927 was transformed with pYES2 empty vector, pYES2-AtCNGC12, pYES2-AtCNGC11/12, pYES2-AtCNGC11/12:R557C, pYES2-AtCNGC12:R557I, and pYES2-AtCNGC11/12:Q543X following the lithium acetate transformation protocol (Ausubel et al., 1987).
To test for complementation of the cch1 mutation, yeast transformants were grown to logarithmic phase in synthetic minimal media and were diluted to 106 cells ml−1 and exposed to 20 μM α-mating factor in modified synthetic minimal media containing 100 μM CaCl2 (Muller et al., 2001). 100 μl aliquots of cells were harvested by centrifugation and resuspended in 10 mg ml−1 Trypan blue solution at 4, 8, and 12 h. Yeast viability was measured by assessing the ratio of stained to unstained cells under brightfield microscopy. A minimum of 200 cells were counted for each transformant.
Ca2+ uptake was measured by the method described by Kurusu et al. (2004) with slight modifications. K927 transformants were grown to logarithmic phase in synthetic minimal media and diluted to 107 cells ml−1. Cultures were then preincubated for 120 min at 30 °C in 10 mM MES-TRIS buffer (pH 6.0) containing 100 mM glucose. 45CaCl2 was added to a final concentration of 72 kBq ml−1. 100 μl aliquots were harvested at 10, 20, and 30 min by centrifugation and washed five times with washing solution (20 mM MgCl2 0.1 mM LaCl3). Radioactivity in yeast cells were measured using a liquid scintillation counter. All experiments have been conducted with three biological repeats with three technical repeats.
Green fluorescence protein (GFP) visualization by confocal microscopy
Agrobacterium-mediated transient expression in N. benthamiana was performed as described in Urquhart et al. (2007) at 16 °C and 22 °C. Plants were shifted to 16 °C 12 h after infiltration, and protein stability in either condition was confirmed by GFP expression at 30 h. Small sections of the infiltrated area were excised and used for confocal microscopy. Confocal fluorescence images were acquired using a Leica TCS SP5 confocal system with AOBS® (HCS PL APO CS 40× immersion oil objective; NA, 1.25) with the AOTF for the argon laser (488 nm) set at 35% and the detection window at 500–600 nm (Leica Microsystems Inc., Wetzlar, Germany).
Computational modelling and sequence alignment
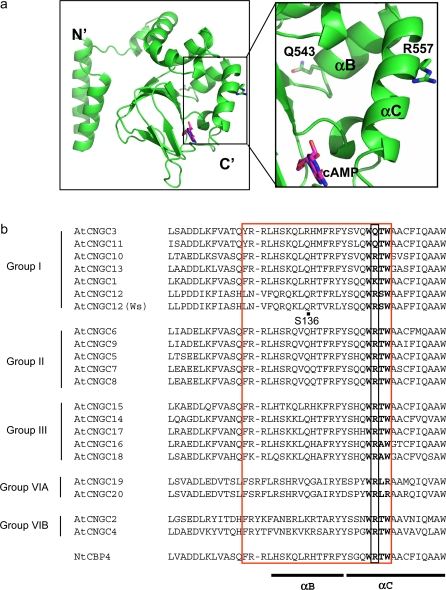
The tertiary structure modelling of AtCNGC11/12 was conducted as described previously (Baxter et al., 2008) using the crystallized structure of the cytoplasmic C-terminus of invertebrate CNGC, SpIH (Flynn et al., 2007, PDB no. 2PTM). The protein fold recognition server (Phyre; Kelley and Sternberg, 2009) was used to model the protein coordinates with estimated precision of 100%. All the images were generated using PyMOL (DeLano, 2002).
The sequence alignments of the CNBD amino acid sequences of all 20 Arabidopsis CNGCs were aligned using ClustalW (Thompson et al., 1994). The accession numbers of all protein sequences used for the alignment are indicated in the figure legends.
Results
The intragenic suppressor S58 loses cpr22 related phenotypes under ambient conditions
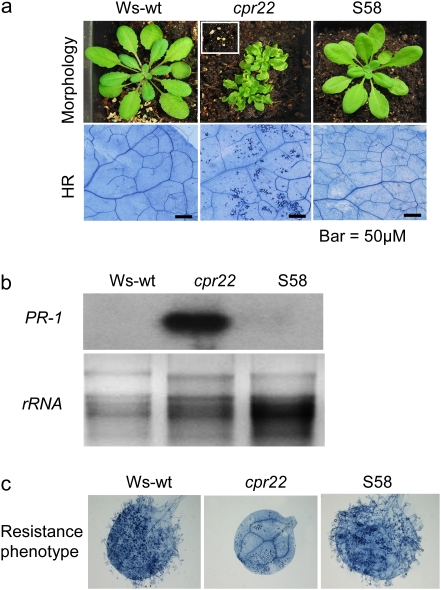
The suppressor screen of cpr22 was reported previously (Baxter et al., 2008). Through this screen more than 20 novel mutant alleles in AtCNGC11/12 have been discovered so far. One of these mutants, suppressor S58 was found to be morphologically identical to the wild type (wt) plants (Fig. 1a). The homozygosity of the cpr22 allele in S58 was confirmed by PCR-based marker analysis (data not shown). In contrast to homozygous cpr22, S58 is neither lethal, nor shows HR-like spontaneous lesion formation and constitutive PR-1 gene expression (Fig. 1a, b). Pathogen resistance was evaluated using the oomycete pathogen, Hyaloperonospora arabidopsidis, isolate Emwa1, which is virulent for the ecotype Wassilewskija (Ws, the background ecotype of cpr22). cpr22 showed enhanced resistance to Emwa1 (Yoshioka et al., 2006). As predicted, suppressor S58 lost cpr22-mediated enhanced resistance to this pathogen (Fig. 1c; see Supplementary Table S1 at JXB online). Taken together; it is concluded that suppressor S58 lost all tested cpr22-related phenotypes under ambient temperature condition (22 °C). Sequence analysis of the cpr22 gene (AtCNGC11/12) revealed one nucleotide substitution, C to T in the cyclic nucleotide binding domain (CNBD) that caused an amino acid substitution, Arginine 557 to Cysteine (R557C) in S58.
Fig. 1.
Characterization of the suppressor mutant S58. (a) Morphological phenotypes and spontaneous HR cell death formation of wilde type (Ws-wt), cpr22, and suppressor 58 (S58). A cpr22 homozygous plant is shown in the white square. Approximately 4-week-old plants were used. (b) Northern blot analysis for PR-1 gene expression in Ws-wt, cpr22, and S58. The samples were taken from approximately 4-week-old plants. Ethidium bromide staining of ribosomal RNA (rRNA; lower panel) served as a loading control. (c) Growth of Hyaloperonospora arabidopsidis, isolate Emwa1 in Ws-wt, cpr22, and S58. Plants were infected by spraying a conidiospore suspension of 106 ml−1 on 7-d-old plants. The Trypan blue analysis 8 d after infection was done to visualize pathogen growth.
The genetic nature of S58 was evaluated by backcrossing with cpr22 homozygous plants. As shown in Supplementary Table S2 at JXB online, all B1 (backcross, 1st generation) plants showed cpr22 heterozygous-like phenotypes, and the following self-pollinated B2 generation showed a segregation of 1 (wild type like):2 (cpr22 heterozygous like):1 (lethal). This indicated the semi-dominant nature of this mutation supporting the idea that aforementioned intragenic mutation R557C is the cause of the phenotype suppression. RT-PCR in homozygous cpr22 and S58 mutants confirmed that there is no significant difference in expression of AtCNGC11/12 and AtCNGC11/12:R557C (data not shown).
The S58 mutation (R557C) does not affect channel function in yeast
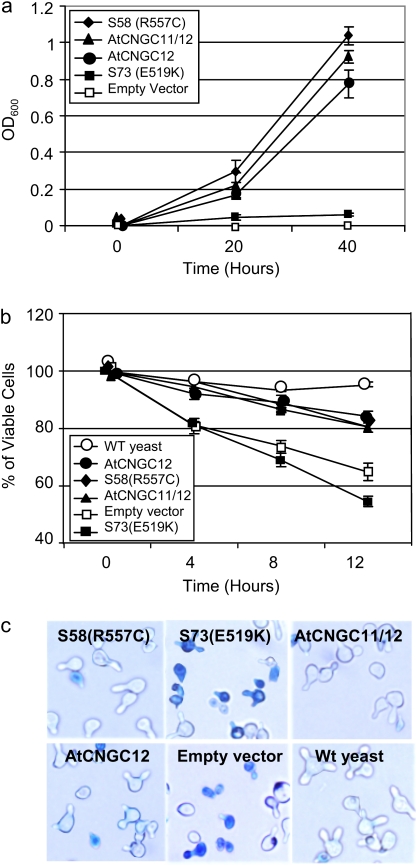
Growth enhancement of mutant yeast has been previously demonstrated upon expression of various plant CNGCs (Köhler et al., 1999; Leng et al., 1999; Ali et al., 2006). More recently, it was suggested that AtCNGC11, 12, and AtCNGC11/12 can function as K+ channels as well as Ca2+ channels when expressed in the same systems (Yoshioka et al., 2006; Urquhart et al., 2007). Therefore, it is possible that the loss of cpr22-related phenotypes in S58 can be attributed to either a loss in basic ion channel function, or a loss in the constitutive active character of AtCNGC11/12 while still maintaining basic channel function. To address this point, a heterologous expression system using K+ and Ca2+ uptake-deficient yeast mutants was used to evaluate the channel function of AtCNGC11/12:R557C. The trk1,trk2 K+ uptake-deficient yeast mutant, CY162 (Leng et al., 1999), transformed with AtCNGC11/12 or AtCNGC11/12:R557C was tested in low external K+ and in the presence of the cationic antibiotic hygromycin. As shown in Fig. 2a, AtCNGC11/12 and AtCNGC12 were able to complement the trk1,trk2 phenotype compared to the empty vector control. Interestingly, the mutants carrying AtCNGC11/12:R557C were also able to complement the trk1,trk2 phenotype with the same efficiency as AtCNGC11/12, whereas another CNBD mutant, S73 (E519K), which affects ion channel function (Baxter et al., 2008) did not, suggesting that the R557C mutation does not affect its basic ion channel function. To explore if this is also the case for Ca2+ channel function, a similar complementation analysis was conducted using the Ca2+-uptake yeast mutant strain K927 (cchl::TRP1; Locke et al., 2000). CCH1 has been implicated in Ca2+ influx in response to mating pheromones (Fischer et al., 1997). Previously, it has been shown that Ca2+ is important for cell death induction by the expression of AtCNGC11/12 (Urquhart et al., 2007). Therefore, it is possible that only Ca2+ channel function is affected by the S58 mutation. However as shown in Fig. 2c by Trypan blue staining, AtCNGC11/12, AtCNGC12, and AtCNGC11/12:R557C rescued this yeast phenotype to comparable levels, indicating that AtCNGC11/12:R557C is functional as a Ca2+ ion channel. These data were quantitatively confirmed in Fig. 2b. In addition, 45Ca2+ uptake analysis was conducted using the same yeast mutant strain. Although the 45Ca2+ uptake did not reach the same level as wild-type yeast, K927 mutant yeast with AtCNGC11/12 or AtCNGC11/12:R557C had a higher uptake rate of 45Ca2+ than K927 carrying the empty vector (see Supplementary Fig. S1 at JXB online). The same result was observed in four independent experiments. No significant difference between AtCNGC11/12 and AtCNGC11/12:R557C was observed, suggesting that the R557 (S58) mutation does not impair the function of AtCNGC11/12. This result is consistent with the mating pheromone analyses using the same yeast mutant strain (Fig. 2b). Taken together, it was concluded that the mutation R557C does not affect basic ion channel function but rather affects the regulation of AtCNGC11/12.
Fig. 2.
Yeast complementation analyses. (a) AtCNGC11/12, AtCNGC12, and S58 (AtCNGC11/12:R557C) complemented the K+-uptake deficient mutant CY162, whereas S73 and the empty vector did not. Data are the average of three biological repeats ±SE. Student's t test shows significant differences between the empty vector/S73 and AtCNGC11/12, AtCNGC12, or S58 at 20 h and 40 h (P <0.05). Experiments have been performed more than three times with similar results. (b) AtCNGC11/12, AtCNGC12, and S58 (AtCNGC11/12:R557C) complemented the Ca2+-uptake deficient mutant K927, whereas S73 and empty vector did not. Data are the average of three biological repeats ±SE. Student's t test shows significant differences between the empty vector and AtCNGC11/12, AtCNGC12, or S58 at 4, 8, and 12 h (P <0.05).The experiment has been repeated more than three times with comparable results. (c) Yeast viability analysis by Trypan blue staining. AtCNGC11/12, AtCNGC12, and S58 (AtCNGC11/12:R557C) rescued the cell death phenotype of the Ca2+-uptake deficient mutant K927, whereas S73 and empty vector did not. (This figure is available in colour at JXB online.)
AtCNGC11/12:R557C recovers the activity to induce cell death under low temperature
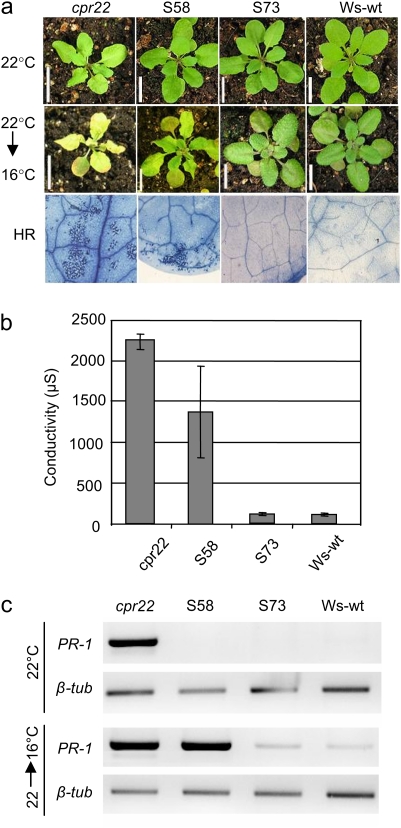
As mentioned above, cpr22-related phenotypes show temperature sensitivity. Relatively high temperatures (>28 °C) can suppress its phenotypes and on the other hand relatively lower temperature (<16 °C) enhanced them (Mosher et al., 2010). Since the R557C mutation seems to suppress the constitutive active character of AtCNGC11/12 but not basic ion channel function, the question was asked if lower temperature can restore this constitutive active character (channel activity). Strikingly, a temperature shift from 22 °C to 16 °C induced cpr22-like morphology, such as curly leaves 4 d after shift in S58. Chlorosis on leaves, which indicates cell death development, could also be observed (Fig. 3a). Trypan blue staining as well as electrolyte leakage analyses further confirmed cell death development in S58 (Fig. 3a, b). PR-1 gene expression also recovered after the shift from 22 °C to 16 °C (Fig. 3c), further supporting the temperature sensitivity of this mutant.
Fig. 3.
Temperature sensitivity of cpr22-related phenotypes in cpr22, S58, S73, and Ws-wt plants after a shift from 22 °C to 16 °C. (a) S58 displayed cpr22-morphology after temperature shift. cpr22 showed enhancement of HR cell death and S58 induced HR cell death and cpr22-related phenotypes after the temperature shift. No cell death induction was observed in another intragenic suppressor, S73 and Ws-wt under both conditions. Photographs were taken 7 d after the shift. (b) Quantitative analysis of cell death by electrolyte leakage in cpr22, S58, S73, and Ws-wt. Samples were taken 7 d after the shift. (c) RT-PCR analysis of PR-1 gene expression in cpr22, S58, S73, and Ws-wt. Temperature shift induced PR-1 gene expression in S58, whereas no significant change was observed in S73 and Ws-wt. β-tubulin served as a loading control. Samples were taken 7 d after the shift.
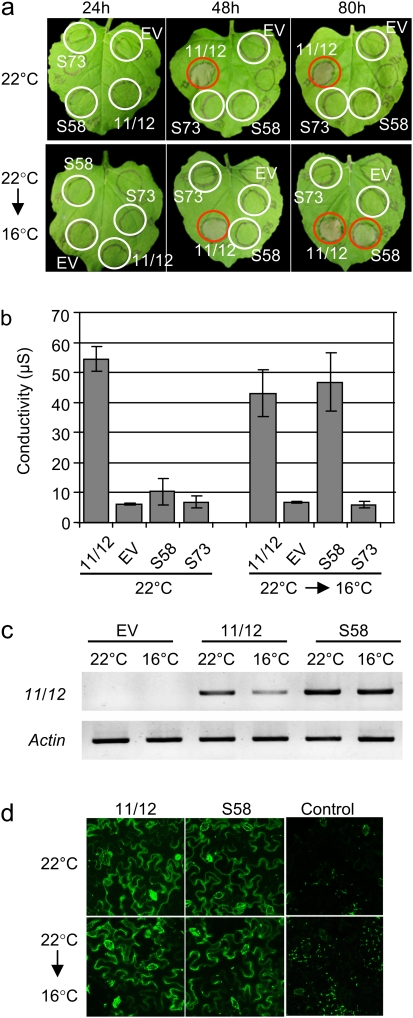
To characterize the temperature sensitivity of S58 further (AtCNGC11/12:R557C), Agrobacterium-mediated transient expression analysis was conducted in Nicotiana benthamiana. This is an established system to analyse HR cell death development and has been used to show that AtCNGC11/12 induces cell death in a synchronized manner (Urquhart et al., 2007). As shown in Fig. 4a, cell death was induced by the transient expression of AtCNGC11/12 but not AtCNGC11/12:R557C at 22 °C (ambient temperature) indicating the inactivity of AtCNGC11/12:R557C in planta under this condition. On the other hand, AtCNGC11/12:R557C recovered cell death induction in plants that were shifted from 22 °C to 16 °C. This was also confirmed quantitatively by electrolyte leakage analysis (Fig. 4b). Transcript and protein levels were not significantly altered by the temperature shift as depicted by RT-PCR (Fig. 4c) and GFP fluorescence (Fig. 4d). Note that while we have observed enhanced cell death in cpr22 when it was shifted from 22 °C to16 °C (Fig. 3a), there is no significant difference between 22 °C and 16 °C when AtCNGC11/12 was transiently expressed in N. benthamiana. This is probably due to the sampling timing. The samples for Fig. 4b were taken at 80 h after Agrobacterium infiltration. At this time point, cell death development induced by transient expression of AtCNGC11/12 is almost completed and also it is very strong and uniform compared to the mutant itself due to the constitutive CaMV35S promoter.
Fig. 4.
Temperature sensitivity of cell death induction by transient expression of AtCNGC11/12, empty vector, S58 (AtCNGC11/12:R557C), and S73 (AtCNGC11/12:E519K) in Nicotiana benthamiana. (a) Induction of cell death in N. benthamiana 24, 48, and 80 h after Agrobacterium infiltration, either shifted from 22 °C to 16 °C at 12 h after Agrobacterium infiltration (lower panels) or not shifted (upper panels). Cell death induction was observed in the leaf area expressing S58, but not empty vector (EV) or S73 after the temperature shift. Cell death induced by AtCNGC11/12 was enhanced by temperature shift. Red circles indicate HR development. (b) Quantitative analysis of cell death in N. benthamiana by electrolyte leakage of leaf discs. S58 expression induced cell death after the temperature shift, but not empty vector (EV) or S73. Samples were taken 80 h after Agrobacterium infiltration. (c) RT-PCR analysis of leaf discs from N. benthamiana leaves expressing AtCNGC11/12, S58 or empty vector (EV). The temperature shift did not significantly affect gene expression of AtCNGC11/12 or AtCNGC11/12:R557C in N. benthamiana leaf discs. actin served as a loading control. Samples were taken 24 h after Agrobacterium- infiltration (12 h after the shift). (d) The expression of AtCNGC11/12:GFP and AtCNGC11/12:R557C:GFP was not altered by the temperature shift. The samples were taken 30 h after Agrobacterium infiltration. The fluorescence of the GFP-fusion proteins was monitored by confocal microscopy.
3D computational modelling of AtCNGC:R557C
In order to predict the role of R557 in channel structure, a computational analysis of the three-dimensional structure of the cytoplasmic C-terminal region was conducted. Previously, the crystal structure of the cytoplasmic C-terminal region of a hyperpolarization-activated cyclic nucleotide-modulated channel, HCN2, has been used as a template (Zagotta et al., 2003, PDB ID: 1Q50, Baxter et al., 2008). The recently published crystal structure of another HCN, SpIH was used here (crystallized with cAMP; Flynn et al., 2007; PDB ID: 2PTM) that possesses higher structural similarity than HCN2 to AtCNGC12 (H Abdel-Hamid and D Shahinas, unpublished data). As shown in Fig. 5a, R557 is located in the middle of the αC-helix in the CNBD. The importance of this αC-helix for cNMP binding has been reported in animal CNGCs (Rehmann et al., 2007). According to the crystal structure of the CNBD of HCN2 in the presence of cAMP, cAMP binds in the anti-configuration between the β-roll and the αC-helix of the CNBD. In our model, the side chain of R557 faces away from the interior of the cNMP binding pocket formed by the β-roll of the CNBD and therefore, likely does not bind to cNMPs directly (Fig. 5a). Hydrophobic interactions between the αC-helix and the base of cNMPs were also postulated to stabilize cNMP binding (Rehmann et al., 2007), further suggesting that the hydrophilic R557 does not directly interact with cNMPs.
Fig. 5.
The location of R557 and Q543 in tertiary structure and amino acid sequence alignment. (a) Ribbon diagram of the cytoplasmic C-terminal region of AtCNGC11/12 (AtCNGC12) (left panel) and close-up of the indicated area of the left panel (right panel). R557 is located in the αC-helix and Q543 is located in the αB-helix of the CNBD. cAMP is indicated by pink colour. (b) Alignment of the area of R557 with 20 Arabidopsis CNGCs and tobacco NtCBP4. NCBI:AF079872, AtCNGC3:CAB40128, AtCNGC11:AAD20357, AtCNGC10:AAF73128, AtCNGC13:AAL27505, AtCNGC1:AAK43954, AtCNGC12:AAd23055, AtCNGC12 (Ws ecotype):EU541495, AtCNGC6:AAC63666, AtCNGC9:CAB79774, AtCNGC5:T52573, AtCNGC7:AAG12561, AtCNGC8:NP_173408, AtCNGC15:AAD29827, AtCNGC14:AAD23886, AtCNGC17:CAB81029, ATCNGC16:CAB41138, AtCNGC18:CAC01886, AtCNGC19:BAB02061, AtCNGC20:BAB02062, AtCNGC2:CAC01740, AtCNGC4:T52574. The black box indicates the position of R557. The red box indicates the CaM binding domain and bold characters highlight the critical four amino acids for the CaM binding suggested by Arazi et al. (2000). The location of Q543 (S136) is indicated by a black dot.
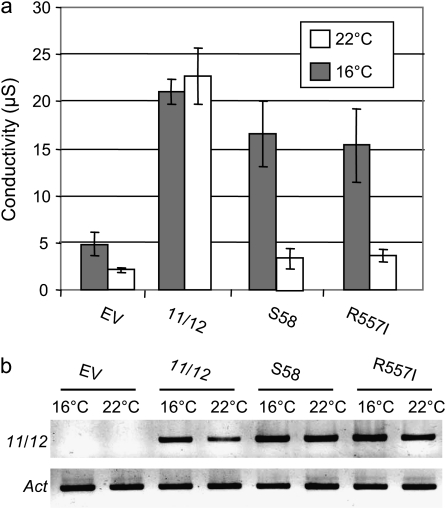
A sequence comparison revealed that R557 is conserved in 17 out of 20 Arabidopsis CNGCs (Fig. 5b). Only three CNGCs, AtCNGC1, 3, and 11 have a lysine (K) or glutamine (Q) that mediate ionic interactions similarly to arginine (R) in this position. All of them are hydrophilic (hydropathy index: –4.5). Cysteine is slightly polar due to its –SH group but does not mediate any ionic interactions (hydropathy index: +2.5). To address whether a more hydrophobic residue with less polarity than cysteine such as isoleucine (hydropathy index: +4.5) can completely disrupt channel function, AtCNGC11/12:R557I was created by site-directed mutagenesis and it was transiently expressed in N. benthamiana to assess cell death induction. As shown in Fig. 6a, cell death is not induced by AtCNGC11/12:R557I at 22 °C, but is recovered at 16 ° C similar to AtCNGC11/12:R557C. The expression of all constructs was confirmed by RT-PCR (Fig. 6b). Therefore, even by altering R557 to isoleucine complete suppression of channel function at 16 °C could not be obtained.
Fig. 6.
AtCNGC11/12:R557I expression induces cell death similarly to S58 in Nicotiana benthamiana at lower temperature. Quantitative analysis of cell death in N. benthamiana was assessed by electrolyte leakage of leaf discs. AtCNGC11/12:R557I expression induced cell death to the same degree as AtCNGC11/12:R557C (S58) after the temperature shift from 22 °C to 16 °C but not at 22 °C. (b) RT-PCR analysis of leaf discs from N. benthamiana leaves expressing AtCNGC11/12, S58 (AtCNGC11/12:R557C), AtCNGC11/12:R557I or empty vector (EV). The temperature shift did not significantly affect gene expression of AtCNGC11/12, AtCNGC11/12:R557C or AtCNGC11/12:R557I in N. benthamiana leaf discs. Actin (Act) served as a loading control.
The αC and αB helices in the CNBD are crucial for channel function
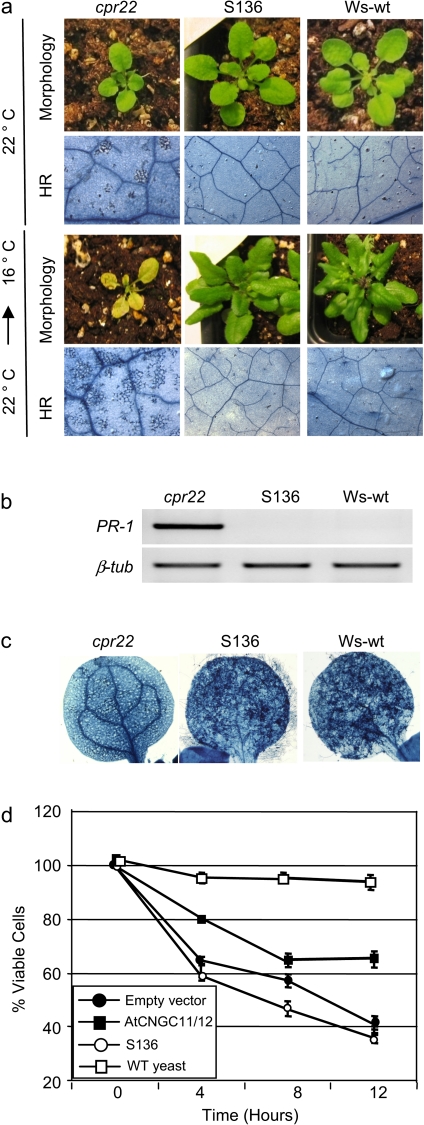
The αC-helix in the CNBD was suggested to play an important role for animal CNGC function/channel gating (Rehmann et al., 2007). However, so far there is almost no report about the importance of the αC-helix in plant CNGCs except one report showing that the CaM binding domain is located in the αC-helix (Arazi et al., 2000). To study the role of the αC-helix in channel function further, another intragenic suppressor mutant, S136, was used that has a premature stop codon at Q543 (C to T point mutation) in the CNBD. Our structural model revealed Q543 to be located at the middle of the αB helix, indicating that S136 does not have the entire αC-helix and only a partial αB helix (Fig. 5a). S136 did not display cpr22 phenotypes under 22 °C or 16 °C (Fig. 7a). It also lost constitutive expression of PR-1 and enhanced pathogen resistance (Fig. 7b, c). Furthermore, heterologous expression in the previously mentioned trk1,trk2 K+ and cchl Ca2+ yeast mutants revealed that AtCNGC11/12:Q543X does not have channel function (Fig. 7d; data not shown). Taken together, these data suggest that the αC- helix and possibly the αB-helix of the CNBD is functionally essential for CNGC function in plants.
Fig. 7.
Characterization of the premature stop codon mutant, S136. (a) Morphological and cell death phenotypes of Ws-wt, cpr22 and S136 with and without temperature shift. S136, unlike S58 does not induce cell death after a shift from 22 °C to 16 °C. Samples were taken 7 d after the shift. (b) RT-PCR analysis for PR-1 gene expression in Ws-wt, cpr22, and S136. The samples were taken from approximately 4-week-old plants. β-tublin (β-tub) served as a loading control. (c) Growth of Hyaloperonospora arabidopsidis, isolate Emwa1 in Ws-wt, cpr22, and S136. Plants were infected by spraying a conidiospore suspension of 106 ml−1 on 7-d-old plants. The Trypan blue analysis 8 d after infection was done to visualize pathogen growth. (d) Yeast complementation analysis using the Ca2+-uptake deficient mutant K927. Only AtCNGC11/12 but not S136 (AtCNGC11/12: Q543X) rescued the K927 phenotype. Data are the average of three biological repeats ±SE. Student's t test shows a significant difference between AtCNGC11/12 and both empty vector and S136 at 12 h (P <0.05).The experiment has been repeated more than three times with similar results.
Discussion
Through this study it has been found that R557 in the αC-helix of the CNBD plays an important role in stable channel regulation. Substitution of R557C in the Arabidopsis mutant cpr22 impaired cell death formation and other cpr22-related phenotypes suggesting a disruption of channel function. However, the cpr22-related phenotypes as well as cell death caused by transient expression of AtCNGC11/12:R557C could still be observed under slightly lower temperatures (16 °C) suggesting an alteration in channel regulation rather than channel function. Yeast complementation analysis demonstrated that AtCNGC11/12:R557C maintains its basic channel function for both K+ and Ca2+ conductance, further supporting this notion. R557 is highly conserved in the CNGC family and all residues in this position share similar polarity and hydrophobicity. It has been examined if a change to an even more hydrophobic and less polar residue (R557I) would have a stronger effect that completely disrupts basic channel function. However, this channel remained to be conditional just like AtCNGC11/12:R557C, further strengthening the notion that R557 is important for the regulation of channel activity rather than basic channel function.
In animal systems, it has been reported that cNMPs bind within the pocket formed by the αC-helix and the β-barrel composed of the eight β sheets in the CNBD (Weber and Steitz, 1987; Rehmann et al., 2007). The αC-helix was suggested to function as the lid of this pocket that stabilizes the cNMP binding by forming hydrophobic interactions with the bound cNMP (Rehmann et al., 2007). However, because R557 is hydrophilic, it does not seem to participate directly in cNMP binding. Our computational modelling also supported this notion.
Regarding the role of the αC-helix in plant CNGCs, a 19–20 amino acid sequence of this region was suggested to be the CaM binding domain in AtCNGC1 and AtCNGC2 by Köhler and Neuhaus (2000) using yeast two hybrid analysis. Arazi et al. (2000) biochemically demonstrated that a 23 amino acid sequence overlapping with this 19–20 amino acids is the CaM binding domain in the tobacco CNGC, NtCBP4. Furthermore, they reported that the four additional amino acids (W R T/S W) which are located just outside of the 19–20 amino acid sequence are crucial for efficient binding. As shown in the alignment in Fig. 5b, R557 is located in this crucial sequence (indicated by bold characters). Considering the fact that R557C altered its regulation, not the basic channel function itself, it can be hypothesized that R557C displays an alteration in its binding affinity to CaM which causes an alteration to the regulation of its function. Considering the importance of CaM in regulating this channel group, our finding is significant. Biochemical studies that aim to identify the CaM interactions with AtCNGC11/12 (AtCNGC12) are underway.
The experimental results involving of suppressor S136 was unexpected, since the deletion of the part of the CNBD that includes the CaM binding site has been reported to enhance the AtCNGC1 channel function in yeast (Ali et al., 2006). Ali et al. (2006) demonstrated that two deletion constructs of AtCNGC1 showed enhanced K+ channel function in a yeast complementation analysis. They hypothesized that the binding of the negative regulator CaM does not occur in these deletion constructs thereby promoting channel activity. Based on our computational modelling, the deletion of S136 is positioned between these two constructs (data not shown). However, we did not see any channel function using both K+ and Ca2+ yeast mutants in the case of S136, suggesting the importance of the αC-helix and possibly the αB-helix for its basic channel function. This discrepancy could be due either to differences in regulatory properties of different AtCNGC subunits or to unknown structural changes caused by the different deletion positions used in both studies. As of now, the answer remains unknown and further analyses will be required to understand these differences.
In this study, it has been demonstrated that R557 in the αC-helix of the CNBD of CNGCs plays an important role for channel regulation and that the αC-helix and possibly αB-helix are crucial for plant CNGC channel function. It has been proposed that the αC-helix binds to both cNMP and CaM as a mechanism to regulate CNGC channel gating (Arazi et al., 2000). Our data here demonstrates the importance of the αC-helix in a plant CNGC for the first time. Further precise structural and biochemical studies to elucidate the role of the αC-helix are underway.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Ca2+-uptake analysis using Calcium-45 AtCNGC11/12 and S58 (AtCNGC11/12:R557C) partially complemented the Ca2+- uptake deficient mutant K927, whereas the empty vector did not.
Supplementary Material
Acknowledgments
This project was supported by a Discovery Grant from NSERC (Natural Science and Engineering Research Council of Canada), CFI (Canadian Foundation for Innovation), and ORF (Ontario Research Fund) to KY, and a graduate student fellowship from Egyptian government to HA.
We thank Dr Leon Kochian at Cornell University for providing the CY162 yeast mutant strain. We also thank Dr Hidetoshi Iida at Tokyo Gakugei University and Dr Kyle W Cunningham at Johns Hopkins University for providing the K927 yeast mutant strain as well as for technical advice. For their patient technical assistance, we thank Dr Ali Rashid at the University of Connecticut, Dr Kazuyuki Kuchitsu, and Dr Takamitsu Kurusu at the Tokyo University of Science. We also would like to thank Mr William Urquhart for his assistance for plasmid construction.
References
- Ali R, Zielinski RE, Berkowitz GA. Expression of plant cyclic nucleotide-gated cation channels in yeast. Journal of Experimental Botany. 2006;57:125–138. doi: 10.1093/jxb/erj012. [DOI] [PubMed] [Google Scholar]
- Arazi T, Sunkar R, Kaplan B, Fromm H. A tobacco plasma membrane calmodulin-binding transporter confers Ni2+ tolerance and Pb2+ hypersensitivity in transgenic plants. The Plant Journal. 1999;20:171–182. doi: 10.1046/j.1365-313x.1999.00588.x. [DOI] [PubMed] [Google Scholar]
- Arazi T, Kaplan B, Fromm H. A high-affinity calmodulin-binding site in tobacco plasma-membrane channel protein coincides with a characteristic element of cyclic nucleotide-binding domains. Plant Molecular Biology. 2000;42:591–601. doi: 10.1023/a:1006345302589. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Bent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York: John Wiley and Sons; 1987. [Google Scholar]
- Baxter J, Moeder W, Urquhart W, Shahinas D, Chin K, Christendat D, Kang HG, Angelova M, Kato N, Yoshioka K. Identification of a functionally essential amino acid for Arabidopsis cyclic nucleotide gated ion channels using the chimeric AtCNGC11/12 gene. The Plant Journal. 2008;56:457–469. doi: 10.1111/j.1365-313X.2008.03619.x. [DOI] [PubMed] [Google Scholar]
- Bridges D, Fraser ME, Moorhead GBG. Cyclic nucleotide bindning proteins in the Arabidopsis thaliana and Oryza sativa genomes. BMC Bioinformatics. 2005;11:6. doi: 10.1186/1471-2105-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, Moeder W, Yoshioka K. Biological roles of cyclic-nucleotide-gated ion channels in plants: what we know and don't know about this 20 member ion channel family. Botany. 2009;87:668–677. [Google Scholar]
- DeLano WL. The PyMOL molecular graphics system. Palo Alto, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- Fesenko EE, Kolesnikov SS, Lyubarsky AL. Induction by cyclic GMP of cationic conductance in plasma membrane of retinal rod outer segment. Nature. 1985;313:310–313. doi: 10.1038/313310a0. [DOI] [PubMed] [Google Scholar]
- Fischer M, Schnell N, Chattaway J, Davies P, Dixon G, Sanders D. The Saccharomyces cerevisiae CCH1 gene is involved in calcium influx and mating. FEBS Letters. 1997;419:259–262. doi: 10.1016/s0014-5793(97)01466-x. [DOI] [PubMed] [Google Scholar]
- Flynn GE, Black KD, Islas LD, Sankaran B, Zagotta WN. Structure and rearrangements in the carboxy-terminal region of SpIH channels. Structure. 2007;15:671–682. doi: 10.1016/j.str.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua BG, Mercier RW, Zielinski RE, Berkowitz GA. Functional interaction of calmodulin with a plant cyclic nucleotide gated cation channel. Plant Physiology and Biochemistry. 2003;41:945–954. [Google Scholar]
- Kaplan B, Sherman T, Fromm H. Cyclic nucleotide-gated channels in plants. FEBS Letters. 2007;581:2237–2246. doi: 10.1016/j.febslet.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Kaupp UB, Seifert R. Cyclic nucleotide-gated ion channels. Physiological Reviews. 2002;82:769–824. doi: 10.1152/physrev.00008.2002. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJE. Protein structure prediction on the web: a case study using the Phyre server. Nature Protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Köhler C, Merkle T, Neuhaus G. Characterization of a novel gene family of putative cyclic nucleotide- and calmodulin-regulated ion channels in Arabidopsis thaliana. The Plant Journal. 1999;18:97–104. doi: 10.1046/j.1365-313x.1999.00422.x. [DOI] [PubMed] [Google Scholar]
- Köhler C, Neuhaus G. Characterisation of calmodulin binding to cyclic nucleotide-gated ion channels from Arabidopsis thaliana. FEBS Letters. 2000;4710:133–136. doi: 10.1016/s0014-5793(00)01383-1. [DOI] [PubMed] [Google Scholar]
- Kurusu T, Sakurai Y, Miyao A, Hirochika H, Kuchitsu K. Identification of a putative voltage-gated Ca2+ permeable channel (OsTPC1) involved in Ca2+ influx and regulation of growth and development in rice. Plant and Cell Physiology. 2004;45:693–702. doi: 10.1093/pcp/pch082. [DOI] [PubMed] [Google Scholar]
- Leng Q, Mercier RW, Yao W, Berkowitz GA. Cloning and first functional characterization of a plant cyclic nucleotide-gated cation channel. Plant Physiology. 1999;121:753–761. doi: 10.1104/pp.121.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke EG, Bonilla M, Liang L, Takita Y, Cunningham KW. A homolog of voltage-gated Ca2+ channels stimulated by depletion of secretory Ca2+ in yeast. Molecular Cell Biology. 2000;29:6686–6694. doi: 10.1128/mcb.20.18.6686-6694.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser P, Thomine S, Schroeder JI, et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiology. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier RW, Rabinowitz NM, Gaxiola RA, Ali R, Berkowitz GA. Use of hygromycin hypersensitivity of a K+ uptake yeast mutant as a functional assay of plant cyclic nucleotide gated cation channels. Plant Physiology and Biochemistry. 2004;42:529–536. doi: 10.1016/j.plaphy.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Moeder W, Yoshioka K. Environmental sensitivity in pathogen resistant Arabidopsis mutants. In: Yoshioka K, Shinozaki K, editors. Signal crosstalk in plant stress responses. Ames, IA: Wiley-Blackwell; 2009. pp. 113–135. [Google Scholar]
- Mosher S, Moeder W, Nishimura N, Jikumaru Y, Joo SH, Urquhart W, Klessig DF, Kim SK, Nambara E, Yoshioka K. The lesion mimic mutant cpr22 shows alterations in abscisic acid signaling and abscisic acid insensitivity in a salicylic acid-dependent manner. Plant Physiology. 2010;152:1–13. doi: 10.1104/pp.109.152603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller EM, Locke EG, Cunningham KW. Differential regulation of two Ca2+ influx systems by pheromone signaling in Saccharomyces cerevisiae. Genetics. 2001;159:1527–1538. doi: 10.1093/genetics/159.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmann H, Wittinghofer A, Bos JL. Capturing cyclic nucleotides in action: snapshots from crystallographic studies. Nature Reviews Molecular Cell Biology. 2007;8:63–73. doi: 10.1038/nrm2082. [DOI] [PubMed] [Google Scholar]
- Schuurink RC, Shartzer SF, Fath A, Jones RL. Characterization of a calmodulin-binding transporter from the plasma membrane of barley aleurone. Proceedings of the National Academy of Sciences, USA. 1998;95:1944–1949. doi: 10.1073/pnas.95.4.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva H, Yoshioka K, Dooner HK, Klessig DF. Characterization of a new Arabidopsis mutant exhibiting enhanced disease resistance. Molecular Plant–Microbe Interactions. 1999;12:1053–1063. doi: 10.1094/MPMI.1999.12.12.1053. [DOI] [PubMed] [Google Scholar]
- Talke IN, Blaudez D, Maathuis FJM, Sanders D. CNGCs: prime targets of plant cyclic nucleotide signaling? Trends in Plant Science. 2003;8:286–293. doi: 10.1016/S1360-1385(03)00099-2. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart W, Gunawardena AHLAN, Moeder W, Ali R, Berkowitz GA, Yoshioka K. The chimeric cyclic nucleotide-gated ion channel ATCNGC11/12 constitutively induces programmed cell death in a Ca2+ dependent manner. Plant Molecular Biology. 2007;65:747–761. doi: 10.1007/s11103-007-9239-7. [DOI] [PubMed] [Google Scholar]
- Wallis JW, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- Weber IT, Steitz TA. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 Å resolution. Journal of Molecular Biology. 1987;198:311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]
- Yoshioka K, Kachroo P, Tsui F, Sharma SB, Shah J, Klessig DF. Environmentally-sensitive, SA-dependent defence response in the cpr22 mutant of Arabidopsis. The Plant Journal. 2001;26:447–459. doi: 10.1046/j.1365-313x.2001.2641039.x. [DOI] [PubMed] [Google Scholar]
- Yoshioka K, Moeder W, Kang HG, Kachroo P, Masmoudi K, Berkowitz G, Klessig DF. The chimeric Arabidopsis CYCLIC NUCLEOTIDE-GATED ION CHANNEL11/12 activates multiple pathogen resistance responses. The Plant Cell. 2006;18:747–763. doi: 10.1105/tpc.105.038786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta WN, Siegelbaum SA. Structure and function of cyclic nucleotide-gated channels. Annual Review of Neuroscience. 1996;19:235–263. doi: 10.1146/annurev.ne.19.030196.001315. [DOI] [PubMed] [Google Scholar]
- Zagotta WN, Olivier NB, Black KD, Young EC, Olson R, Gouaux E. Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature. 2003;425:200–205. doi: 10.1038/nature01922. [DOI] [PubMed] [Google Scholar]
- Zufall F, Firestein S, Shepherd GM. Cyclic nucleotide-gated ion channels and sensory transduction in olfactory receptor neurons. Annual Review of Biophysics and Biomolecure Structure. 1994;23:577–607. doi: 10.1146/annurev.bb.23.060194.003045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.