Abstract
Under hypoxia, tumor cells produce a secretion that modulates their microenvironment to facilitate tumor angiogenesis and metastasis. Here, we observed that hypoxic or reoxygenated A431 carcinoma cells exhibited enhanced angiogenic and metastatic potential such as reduced cell-cell and cell-extracellular matrix adhesion, increased invasiveness, and production of a secretion with increased chorioallantoic membrane angiogenic activity. Consistent with these observations, quantitative proteomics revealed that under hypoxia the tumor cells secreted proteins involved in angiogenesis, focal adhesion, extracellular matrix-receptor interaction, and immune cell recruitment. Unexpectedly, the secreted proteins were predominantly cytoplasmic and membrane proteins. Ultracentrifugation at 100,000 × g precipitated 54% of the secreted proteins and enriched for many exosome-associated proteins such as the tetraspanins and Alix and also proteins with the potential to facilitate angiogenesis and metastasis. Two tetraspanins, CD9 and CD81, co-immunoprecipitated. Together, these data suggested that tumor cells secrete proteins and exosomes with the potential to modulate their microenvironment and facilitate angiogenesis and metastasis.
Cancer is the leading cause of mortality, causing one in eight deaths worldwide with 90% of these deaths attributable to metastases (1). Generally, a primary non-metastatic cancer begins at a localized focus and is resectable with good prognosis, but once metastasized, it is usually unresectable, and controlling its spread with radio- and chemotherapy remains ineffective (2). In fact, prognoses of highly metastatic cancers have not improved in the last century (3).
To sustain growth and survival in their hostile microenvironment, rapidly growing tumors have to overcome hypoxia (Hx)1 and a lack of nutrients through either angiogenesis to ensure an adequate supply of oxygen and nutrients or metastasis to a more conducive microenvironment. Therefore, therapeutic intervention targeting tumor angiogenesis or metastasis represents a viable strategy for regulating tumor growth. Indeed, antitumor angiogenesis drugs such as anti-VEGF therapy have proven to be clinically efficacious (4). However, the therapeutic efficacy of such treatments is generally short lived as tumors are proficient at adopting alternative pathways to circumvent the therapeutic block. For example, prolonged anti-VEGF treatment on tumors is known to select for the tumor cells that recruit alternative angiogenesis signaling pathways involving fibroblast growth factor, platelet-derived growth factor (PDGF), and angiopoietins (5). Therefore, to develop effective therapeutics, a comprehensive understanding of the complex processes that are central to metastasis and angiogenesis would likely reveal more robust and less redundant therapeutic targets. Because emerging evidence implicates Hx as a key inducer of angiogenesis and metastasis in tumors (6) and because extracellular signals emanating from the tumor cells will be necessary in modulating the extracellular matrix (ECM) to facilitate the cell migration during tumor development (7), we focused on elucidating the secretome (8) of tumor cells in their adaptation to Hx.
A431 squamous carcinoma cells have been used as a model to study the oxidative stress- or EGFR-mediated angiogenesis and tumor growth (9, 10) and in a xenograft model for metastasis (11). Here, A431 cells were used to investigate the effects of Hx and hypoxia/reoxygenation (Reox) stresses on metastasis and angiogenic potential. We observed that under Hx the tumor cells exhibited reduced adhesion to their neighboring cells or ECM accompanied by enhanced invasiveness into Matrigel. We also noted that secretion from the hypoxic A431 cells was more efficient at inducing angiogenesis in the chorioallantoic membrane (CAM) assay. These observations suggest that Hx and/or Reox potentiated the angiogenic and metastatic phenotype in A431 cells, possibly through the secretion of proangiogenic and prometastatic factors. To test this hypothesis, we used mass spectrometry-based and cytokine array proteomics approaches to perform high throughput elucidation of the secretome of A431 tumor cells. High throughput proteomics analysis by mass spectrometry has been applied successfully to uncover potential cancer biomarkers as well as elucidate the tumorigenic mechanism (12, 13). Here, we utilized quantitative proteomics in analyzing the tumor secretome and delineating the dynamic changes in the secretome during Hx and Reox with a specific focus on signals that are potentially useful to the survival of a tumor in a hostile tumor microenvironment.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
All reagents were purchased from Sigma unless otherwise specified. Antibodies to extracellular and cytoplasmic domains of EGFR were purchased from Affinity BioReagents (Golden, CO). Anti-E-cadherin and HIF-1α antibody were purchased from BD Pharmingen. Anti-KLK6 antibody was from Abcam (Cambridge, UK). Park7, actin, and tubulin were from Santa Cruz Biotechnology (Santa Cruz, CA). The human cytokine antibody array and receptor tyrosine kinase phosphorylation array were from RayBiotech, Inc. (Norcross, GA). The iTRAQ reagent kit was from Applied Biosystems (Foster City, CA).
Cell Culture and Hypoxia/Reoxygenation Treatment
The A431 squamous carcinoma, MDA-MB-231 breast adenocarcinoma, A549 non-small cell lung (NSCL), H1299 NSCL, and HFF-1 foreskin fibroblast cells were purchased from ATCC. A431, MDA-MB-231, and HFF-1 cells were cultured in DMEM with 10% FBS. A549 and H1299 cells were cultured in RPMI 1640 medium with 10% FBS. For all experiments, 2 × 106 cells of each cell line were seeded on 10-cm dishes in DMEM or RPMI 1640 medium with 10% FBS and incubated under 5% CO2 at 37 °C. On reaching 30–40% confluency, the cells were adapted to serum deprivation by incubating in Minimum Essential Medium α with 0.5% FBS for 12 h. The cells were washed five times with 1× PBS and three times with serum-free medium before incubating in serum-free medium. The cells were exposed to Nx (21% O2, 5% CO2) for 48 h or Hx (<0.1% O2, 5% CO2) for 48 or 72 h. For Reox stress, the cells were exposed to Hx for 48 h, changed to a fresh medium, and then exposed to Nx (21% O2, 5% CO2) for an additional 24 h.
Cell Viability and Cell Cycle Analysis
Cell viability was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. Cell cycle analysis was performed by staining cells with propidium iodide followed by flow cytometry.
Immunofluorescence Analysis and Immunoblotting
To visualize filamentous actin (F-actin) and E-cadherin, immunofluorescent staining with FITC-phalloidin and anti-E-cadherin antibody was performed and viewed under a Zeiss LSM 510 laser scanning confocal microscope (Carl Zeiss, Inc., Oberkochen, Germany). Cells were seeded into 20 × 20-mm coverslips, treated for the indicated conditions, fixed by 4% paraformaldehyde, and then permeabilized with 0.05% Triton X-100 in PBS for 20 min. The cells were incubated with goat serum to block nonspecific binding sites before incubation with monoclonal anti-E-cadherin (BD Pharmingen; 1:100) for 1 h, washed, and then incubated with FITC-conjugated secondary antibody for 1 h. For visualization of filamentous actin, fixed cells were stained with 500 ng/ml phalloidin (Sigma) in PBS containing 1% goat serum for 15 min. After that, cells were washed, mounted, and examined by confocal microscope. The procedures described above were carried out at room temperature. Immunoblotting was performed after SDS-PAGE by probing with specific antibodies against EGFR, KLK-6, CDH-1, Park7, HIF-1α, actin, and α-tubulin and visualization by ECL.
Adhesion and Transwell Invasion Assay
24-well plates were coated with 40 μg/ml type I collagen in DMEM. Uncoated wells were incubated with 0.5% BSA in DMEM to serve as negative control. Cells were treated under Nx, Hx, or Reox conditions; harvested; resuspended in serum-free medium; and seeded at 2.5 × 105 live cells/well in a 24-well plate. Cells were incubated for 0.5, 2, 12, or 24 h at 37 °C. After incubation, the detached cells were removed by serum-free medium, and attached cells were fixed with 95% ethanol for 10 min and stained with 0.1% crystal violet for 25 min. The plate was immersed in fresh tap water to remove excess dye and then allowed to air dry for 5–10 min. The adherent cells were visualized under light microscopy at 200× magnification. 200 μl of 0.5% Triton X-100 was then added to each well to solubilize the cells overnight at room temperature. The absorbance was measured at A595 using a microplate reader (Tecan Magellan, Männedorf, Switzerland).
The cell invasion assay was performed using 24-well Transwell plates containing 8-μm-pore size polycarbonate filters (Corning Costar, Cambridge, MA). Transwell inserts were coated with 100 μl of Matrigel (diluted 1:2 in α-minimum Eagle's medium; BD Biosciences) and allowed to solidify for 1 h at 37 °C. 4 × 104 cells were seeded in serum-free medium in each Transwell and incubated for the indicated times. The Transwells were then removed, and cells that had not migrated through the filter and had remained inside each Transwell were removed by wiping with a cotton swab. Cells that had migrated through the filter of each Transwell and adhered to the other side of the filter of the Transwell were stained with the crystal violet for image capture and counted under a light microscope.
CAM Assay
Shells of 4-day fertilized eggs (Chew's Agriculture, Singapore) were scored equatorially with a cutoff wheel, and the contents were gently transferred into sterilized 80-mm2 plastic weigh boats and incubated ex ovo in an egg incubator (Brinsea, North Somerset, UK) at 37.7 °C at 76% humidity under a home-made plastic shield to prevent the albumen from drying out. For each onplant, two nylon mesh layers were embedded into 3 mg/ml collagen mixed with conditioned medium (CM). Collagen onplants were placed randomly on CAMs of 10-day-old chick embryos. Distinct blood vessels appearing in the onplants above the plane of the lower mesh were identified.
Sample Preparation, iTRAQ Labeling, and LC-MS
CM was collected and concentrated by centrifugation with Amicon ultrafiltration units (5-kDa molecular weight cutoff; Millipore, Billerica, MA). The secreted proteins were labeled with the isobaric tags as followed: the Nx-treated sample was labeled with 114; the sample under Hx for 48 or 72 h was labeled with 115 or 117, respectively; and the Reox-treated sample was labeled with 116 isobaric tag. Protein digestion and labeling with iTRAQ reagents were performed according to the supplier's instruction. The labeled sample was fractionated using the Shimadzu Prominence UFLC system (Kyoto, Japan) connected to a PolySULFOETHYL A column (PolyLC, Columbia, MD; 4.6 × 200 mm, 5-μm particle size, 200-Å pore size). Fractionation was performed with a 60-min linear gradient of 0–500 mm KCl (10 mm KH2PO4, pH 3.0, 25% acetonitrile) at a flow rate of 1 ml/min. A total of 40 fractions were collected and desalted using Sep-Pak C18 cartridges (Waters). Each fraction was dried down and redissolved in 100 μl of 0.1% formic acid for MS analysis. All fractions were analyzed using a QSTAR Elite mass spectrophotometer coupled with an on-line Tempo nano-MDLC system. For each analysis, 10 μl of peptide mixture was injected and separated on a home-packed nanobored C18 column with a picofrit nanospray tip (75-μm inner diameter × 15 cm, 5-μm particles) (New Objectives, Woburn, MA). The separation was performed at a constant flow rate of 300 nl/min with a 120-min gradient. The mass spectrometer was set to perform data acquisition in the positive ion mode with a selected mass range of 300–2000 m/z. Peptides with 2+ to 4+ charge states were selected for MS/MS. The three most abundant peptides above a five-count threshold were selected for MS/MS, and each selected target ion was dynamically excluded for 30 s with ±30 mDa mass tolerance. Smart information-dependent acquisition was activated with automatic collision energy and automatic MS/MS accumulation. The fragment intensity multiplier was set to 20, and the maximum accumulation time was 2 s. The peak areas of the iTRAQ reporter ions reflect the relative abundances of the proteins in the samples (14, 15).
Database Searching and Criteria
Protein identification and peptide quantification were performed using ProteinPilot software version 3.0 (Applied Biosystems) by searching the combined raw data from the three runs of two independent biological samples against the concatenated “target” (International Protein Index (IPI) human database, version 3.34, including 69,164 sequences and 29,064,824 residues) and “decoy” (the reverse amino acid sequence for false discovery rate (FDR) estimation) databases. The Paragon and Pro Group algorithms in the ProteinPilot software were used for peptide identification and isoform-specific quantification. User-defined parameters were as follows: (i) sample type, iTRAQ fourplex (peptide-labeled); (ii) cysteine alkylation, methyl methanethiosulfonate; (iii) digestion, trypsin; (iv) instrument, QSTAR Elite ESI; (v) special factors, none; (vi) species, none; (vii) specify processing, quantitate and bias correction; (viii) ID focus, biological modifications; and (ix) search effort, thorough.
To minimize false positive results, a strict cutoff for protein identification was applied with the unused ProtScore ≥2, which corresponds to a confidence limit of 99%, and at least two peptides with 95% confidence were considered for protein identification as listed in supplemental Table S1. With this filter, the corresponding FDR was calculated from the decoy database. The resulting data set was autobias-corrected to get rid of any variations imparted due to unequal mixing during combining different labeled samples. For iTRAQ quantitation, the peptide for quantification was automatically selected by the Pro Group algorithm (at least one peptide with 99% confidence) to calculate the reporter peak area, error factor (EF), and p value. The proteins for further analysis were filtered with EF <2, p value <0.05, and 1.3-fold changes (>1.30 or <0.77) relative to the normoxic control unless the proteins were known to be of interest from our functional assay. The list of proteins used is shown in Tables I and II.
Table I. Differentially regulated proteins classified into focal adhesion or ECM components quantified by iTRAQ.
48N, 48-h normoxia; 48H, 48-h hypoxia; 72H, 72-h hypoxia; 48H-24R, 48-h hypoxia followed by 24-h normoxia. These proteins met the criteria, i.e. unused ProtScore > 2.0, p value < 0.05 and EF < 2. Specific information for selected protein list is supplied in supplemental Table S1.
| No. | Accession no. | Gene symbol | 48H | 72H | 48H-24R | Molecular function |
|---|---|---|---|---|---|---|
| 1 | IPI00028911.1 | DAG1 | 0.42 | 0.42 | 0.41 | Adhesion molecule |
| 2 | IPI00807403.1 | ALCAM | 0.61 | 0.85 | 0.74 | Adhesion molecule |
| 3 | IPI00032313.1 | S100A4 | 1.98 | 2.47 | 1.18 | ECM/MMP regulation |
| 4 | IPI00645849.1 | ECM1 | 0.22 | 0.23 | 0.31 | Adhesion molecule |
| 5 | IPI00002406.2 | BCAM | 0.61 | 1.40 | 0.72 | Adhesion molecule |
| 6 | IPI00001754.1 | F11R | 1.16 | 1.65 | 1.14 | Adhesion molecule |
| 7 | IPI00018274.1 | EGFR | 0.19 | 0.23 | 0.39 | Cadherin signaling |
| 8 | IPI00744692.1 | TALDO1 | 1.33 | 1.35 | 1.28 | Cadherin signaling |
| 9 | IPI00744889.1 | CDH1 | 1.70 | 1.79 | 1.33 | Cadherin signaling |
| 10 | IPI00216951.2 | DSG2 | 0.59 | 0.74 | 0.74 | Cadherin signaling |
| 11 | IPI00215948.4 | CTNNA1 | 1.21 | 1.32 | 1.26 | Cadherin signaling |
| 12 | IPI00008215.1 | ME1 | 1.18 | 1.28 | 1.25 | Cadherin signaling |
| 13 | IPI00645614.2 | CDH3 | 0.76 | 0.79 | 0.58 | Cadherin signaling |
| 14 | IPI00007257.4 | CLSTN1 | 0.22 | 0.25 | 0.46 | Cell adhesion molecule |
| 15 | IPI00018219.1 | TGFBI | 0.30 | 0.37 | 0.48 | Cell adhesion molecule |
| 16 | IPI00005563.1 | TINAGL1 | 0.26 | 0.28 | 0.39 | Cell adhesion molecule |
| 17 | IPI00024284.4 | HSPG2 | 0.27 | 0.27 | 0.34 | ECM |
| 18 | IPI00296099.6 | THBS1 | 0.31 | 0.33 | 0.87 | ECM |
| 19 | IPI00030255.1 | PLOD3 | 0.83 | 0.84 | 1.13 | ECM |
| 20 | IPI00783665.2 | LAMA5 | 0.22 | 0.23 | 0.24 | ECM, integrin signaling |
| 21 | IPI00298281.3 | LAMC1 | 0.31 | 0.30 | 0.33 | ECM, integrin signaling |
| 22 | IPI00377045.3 | LAMA3 | 0.46 | 0.44 | 0.52 | ECM, integrin signaling |
| 23 | IPI00333541.6 | FLNA | 1.28 | 1.47 | 1.21 | Integrin signaling |
| 24 | IPI00382696.1 | FLNB | 1.28 | 1.16 | 1.21 | Integrin signaling |
| 25 | IPI00307162.2 | VCL | 1.30 | 1.39 | 1.17 | Integrin signaling |
| 26 | IPI00645194.1 | ITGB1 | 1.22 | 1.36 | 1.19 | Integrin signaling |
| 27 | IPI00220846.1 | ITGB4 | 0.90 | 0.90 | 0.90 | Integrin signaling |
| 28 | IPI00301058.5 | VASP | 1.33 | 1.32 | 1.51 | Integrin signaling |
| 29 | IPI00642154.2 | PTPRF | 0.94 | 1.11 | 1.57 | Other receptor |
| 30 | IPI00479252.1 | PTPRS | 0.53 | 1.45 | 0.55 | Other receptor |
| 31 | IPI00644648.2 | PTPRK | 0.64 | 0.68 | 0.61 | Other receptor |
| 32 | IPI00023673.1 | LGALS3BP | 0.21 | 0.25 | 0.39 | Other signaling molecule |
| 33 | IPI00465431.7 | LGALS3 | 1.77 | 1.10 | 0.78 | Other signaling molecule |
Table II. Differentially regulated proteins involved in cell cycle, cell proliferation, or protease quantified by iTRAQ.
48N, 48-h normoxia; 48H, 48-h hypoxia; 72H, 72-h hypoxia; 48H-24R, 48-h hypoxia followed by 24-h normoxia. These proteins met the criteria, i.e. unused ProtScore > 2.0, p value < 0.05, and EF < 2. Specific information for selected protein list is supplied in supplemental Table S1.
| No. | Accession no. | Gene symbol | 48H | 72H | 48H-24R | Molecular function |
|---|---|---|---|---|---|---|
| Cell proliferation and cell cycle control | ||||||
| 1 | IPI00020956.1 | HDGF | 1.38 | 1.28 | 1.49 | Cell proliferation |
| 2 | IPI00444386.2 | IGFBP3 | 0.44 | 0.50 | 1.69 | Cell proliferation |
| 3 | IPI00029235.1 | IGFBP6 | 0.40 | 0.42 | 0.45 | Cell proliferation |
| 4 | IPI00018219.1 | TGFBI | 0.30 | 0.37 | 0.48 | Cell proliferation |
| 5 | IPI00030557.2 | CTH | 1.54 | 1.3 | 1.66 | Cell proliferation |
| 6 | IPI00009342.1 | IQGAP1 | 1.41 | 1.27 | 1.34 | Cell cycle |
| 7 | IPI00014177.3 | SEPT2 | 1.11 | 0.69 | 1.37 | Cell cycle |
| 8 | IPI00784614.1 | SEPT9 | 1.34 | 1.24 | 1.20 | Cell cycle |
| 9 | IPI00411765.3 | SFN | 1.70 | 1.48 | 1.15 | Cell cycle |
| 10 | IPI00021536.1 | CALML5 | 1.33 | 1.35 | 1.15 | Cell proliferation, cell cycle |
| 11 | IPI00018274.1 | EGFR | 0.19 | 0.23 | 0.39 | Cell proliferation, cell cycle |
| 12 | IPI00016179.1 | S100A13 | 1.20 | 1.31 | 0.99 | Cell proliferation, cell cycle |
| Proteases and inhibitors | ||||||
| 1 | IPI00011229.1 | CTSD | 0.2003 | 0.2911 | 0.5735 | Aspartic protease |
| 2 | IPI00011285.1 | CAPN1 | 1.3355 | 1.3443 | 0.9401 | Cysteine protease |
| 3 | IPI00640357.1 | USP14 | 0.9752 | 1.169 | 0.8557 | Cysteine protease |
| 4 | IPI00177728.3 | CNDP2 | 1.36 | 1.3975 | 1.2423 | Metalloprotease |
| 5 | IPI00023845.1 | KLK6 | 0.2277 | 0.2445 | 0.4102 | Serine protease |
| 6 | IPI00032325.1 | CSTA | 1.913 | 1.6308 | 1.804 | Serine protease |
| 7 | IPI00021828.1 | CSTB | 1.437 | 1.507 | 1.1018 | Serine protease |
| 8 | IPI00023728.1 | GGH | 0.2121 | 0.2881 | 0.3213 | Other hydrolase |
| 9 | IPI00843928.1 | NRD1 | 1.37 | 1.1605 | 1.583 | Other hydrolase |
| 10 | IPI00642211.3 | RNPEP | 0.9363 | 1.2044 | 0.9604 | Other hydrolase |
| 11 | IPI00642739.1 | TIMP1 | 0.2503 | 0.2418 | 0.4891 | Metalloprotease inhibitor |
| 12 | IPI00027166.1 | TIMP2 | 0.2663 | 0.3119 | 1.362 | Metalloprotease inhibitor |
| 13 | IPI00027444.1 | SERPINB1 | 1.1652 | 1.2671 | 1.2161 | Serine protease inhibitor |
| 14 | IPI00472082.2 | SERPINB5 | 1.5669 | 1.7318 | 1.497 | Serine protease inhibitor |
| 15 | IPI00749398.1 | SERPINB6 | 1.1875 | 0.9935 | 1.3079 | Serine protease inhibitor |
| 16 | IPI00007118.1 | SERPINE1 | 0.597 | 0.6351 | 0.4845 | Serine protease inhibitor |
| 17 | IPI00032140.4 | SERPINH1 | 0.9549 | 1.946 | 0.7934 | Serine protease inhibitor |
Human Cytokine Antibody Array
Cytokine concentrations in CM were quantified by using human cytokine antibody arrays (G series 2000, Ray Biotech, Inc., Norcross, GA) according to the manufacturer's instructions. The cytokine array was processed and quantified by using the Axon GenePix 4000B microarray scanner and GenePix Pro Accuity 4.0 microarray informatics software (Molecular Devices). The positive controls on each slide were used to normalize the intensity of the individual spots being compared on different slides. The complete list of cytokines in the arrays is summarized in supplemental Table S2.
Isolation and Characterization of Exosomes
Exosomes were isolated as described previously (16). Briefly, culture supernatant was collected and subjected to two successive centrifugations to remove cells and debris: 500 × g for 10 min and 2000 × g for 10 min. The resultant supernatant was filtered with a 0.22-μm pore filter followed by ultracentrifugation at 100,000 × g for 2 h using an SW41 rotor (Beckman Coulter Instruments, Fullerton, CA). The ultracentrifuged pellet was washed by resuspension in PBS before ultracentrifugation at 100,000 × g for 1 h. The washed exosomal pellet was resuspended in PBS and analyzed by Western blot or LC-MS/MS (supplemental Table S3).
To verify the existence of exosomes in the cell culture medium, we performed CD81 immunoprecipitation. Dynabead M-280 sheep anti-mouse IgG (Invitrogen) was washed with 0.1% BSA in PBS before incubation with mouse anti-human CD81 antibody for 2 h with gentle shaking at room temperature. The Dynabeads were washed twice and incubated with CM with gentle shaking for 2 h at room temperature. The supernatant was then collected, and the Dynabeads were gently washed twice with PBS. The supernatant and the Dynabeads were denatured, resolved by 4–12% SDS-PAGE, and analyzed by Western blotting as described above.
Statistical Analysis
Experimental data are presented as the mean of each condition ± S.E. One-way analysis of variance was performed for comparing more than two groups, and paired Student's t test was performed when comparing two conditions (SPSS for Windows® version 12.0). Statistical significance was accepted at p < 0.05 (*) and p < 0.01 (**).
RESULTS
Hypoxia and Reoxygenation Resulted in Loss of Cell Adhesiveness
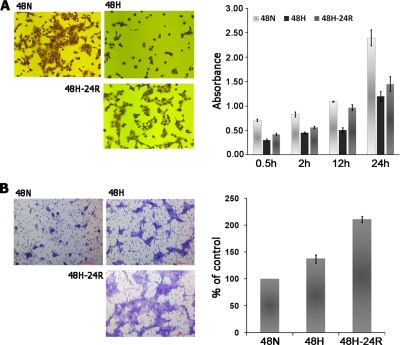
To migrate and intravasate during metastasis, tumor cells must first lose their cell-cell and cell-ECM adhesions (17, 18) and then reorganize their cytoskeleton to facilitate the cellular movement (19). To determine whether Hx or Reox facilitates metastasis through enhancing a loss of adhesiveness to ECM in tumor cells, we assessed the adhesiveness of A431 cells to collagen-coated culture dishes after exposure to Hx and Reox stresses. We observed that after 30 min of seeding onto collagen-coated culture dishes the reduced adhesiveness of Hx-treated cells relative to controls became apparent, and this disparity continued to increase for up to 24 h. The Reox-treated cells exhibited similar suppressed adhesiveness although to a lesser extent (Fig. 1A). In addition, this phenotype was accompanied by the modulation of several key proteins known to be important in cellular adhesion such as E-cadherin, α-catenin, vinculin, and Snail.
Fig. 1.
A, adhesion of A431 cells to collagen-coated culture dishes after pretreatment with either 48-h Hx or Reox. Adhesiveness was determined by seeding pretreated cells into the culture dishes and incubating the dishes for the indicated duration followed by removal of non-adherent cells. The remaining adherent cells were stained with crystal violet, and quantitation was obtained via solubilization of the dye and measuring the absorption at 595 nm. B, chemoinvasion assay of A431 cells under Hx or Reox. Cells were seeded on a porous membrane coated with a Matrigel (pseudo-ECM) layer, which assayed the ability of the cells to degrade the Matrigel and migrate to the other side of the membrane. Unmigrated cells were removed, and the remainder were stained with crystal violet. Representative images were taken at random, and quantitation was performed through cell counting in a blinded fashion. 48N, 48-h normoxia; 48H, 48-h hypoxia; 48H-24R, 48-h hypoxia followed by 24 h normoxia. Error bars denote S.D.
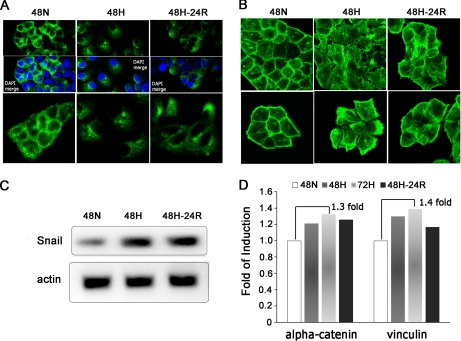
Immunostaining of the cells revealed that during Hx and Reox E-cadherin in the plasma membrane and cytoplasm clustered into foci (Fig. 2A). Loss of E-cadherin-mediated adhesion was further validated via RT-PCR for Snail, a negative E-cadherin transcription regulator (20). Snail mRNA levels were elevated under both Hx and Reox (Fig. 2C). There were also concomitant increases in secretion of α-catenin and vinculin, key stabilizers of the E-cadherin complex (21, 22), under Hx and Reox by quantitative proteomics analysis (Fig. 2D).
Fig. 2.
Hx-induced morphological changes, delocalization of E-cad, and possible dissociation of E-cad-α-catenin complexes. A, immunostaining of E-cad to visualize its localization and expression levels under the indicated conditions. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). E-cadherin appears to be localized to the cell periphery under Nx (left panel), but this distribution was disrupted under Hx (middle panel) and Reox (right panel) accompanied with a concomitant drop in signal intensity. Magnification, ×63. B, visualization of F-actin via FITC-phalloidin staining. Hx and Reox appear to induce stress fiber and ruffle formation as well as an observable increase in cell size. Magnification, ×63. C, analysis of human Snail transcript levels through RT-PCR. Snail was found to be similarly up-regulated under Hx and Reox. Actin levels served as control. D, secreted levels of α-catenin and vinculin quantitated by iTRAQ analysis were found to be elevated under Hx and Reox. 48N, 48-h normoxia; 48H, 48-h hypoxia; 72H, 72-h hypoxia; 48H-24R, 48-h hypoxia followed by 24-h normoxia.
To determine whether, in addition to the reduced adhesion to ECM, Hx and Reox stresses also cause cytoskeletal reorganization in tumor cells, Hx- and Reox-treated A431 cells were stained with phalloidin to visualize the distribution of the F-actin. We observed increased stress fiber formation and membrane ruffling (23) in both Hx- and Reox-treated A431 cells (Fig. 2B) with the effect of Hx being more severe. Furthermore, both forms of cellular stress resulted in obvious morphological changes accompanied by an increase in the cell size and detachment of hypoxic cells from each other.
Hypoxia and Reoxygenation Increased Invasiveness of A431 Cells across Reconstituted ECM Layer
One of the rate-limiting steps in the metastatic cascade is the invasion of the basement membrane, which distinguishes malignant from benign tumors (24). To determine whether Hx and Reox increased the metastatic potential of the tumor cells by increasing their invasiveness, we used a chemoinvasion assay to estimate the ability of A431 cells to degrade a pseudo-ECM layer and migrate through the ECM layer. Hx and Reox enhanced invasiveness by 1.4-fold and over 2-fold of controls, respectively (Fig. 1B). This observation suggested that hypoxic A431 cells produced and/or activated proteolytic enzymes to degrade the ECM to increase their invasiveness.
Conditioned Medium Collected from Hypoxia-treated Cells or Reoxygenated Cells Induces Angiogenesis in Vivo
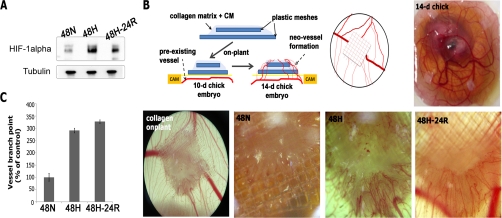
One of the most prominent effects of hypoxia is the induction of tumor angiogenesis (25). To determine whether A431 cells increase secretion of angiogenic factors under Hx and Reox conditions, we tested serum-free medium conditioned by A431 cells under Nx, Hx, or Reox for angiogenic potential. The CM was collected, concentrated, and assayed for angiogenic potential using the CAM angiogenesis assay in which chick embryos were cultured in vitro until cessation of the developmental angiogenesis (Day 10). A collagen gel containing concentrated CM was placed at random spots on the CAM. Induced angiogenesis was quantified by counting the number of branch points on Day 14. Relative to CM from untreated cells, there was a 2.9- and 3.3-fold elevation in induced angiogenesis by Hx CM and Reox CM, respectively (Fig. 3, B and C). Therefore, hypoxia enhanced the angiogenic potential of A431 cellular secretion and provided a mechanism to facilitate tumor angiogenesis in a growing tumor with a hypoxic core.
Fig. 3.
Analysis of HIF-1α levels and angiogenic potentials. A, immunoblotting against HIF-1α revealed its relative elevation under Hx and the modest decline after Reox. Tubulin was used as loading control. B, collagen onplants, with or without CM, supported by two layers of nylon mesh were randomly placed on the CAM of 10-day-old chick embryos ex ovo. Induced angiogenesis was quantitated by taking random representative images of onplants and counting the number of branch points and extent of branching of blood vessels with the aid of a dissecting microscope in a blinded fashion. C, the angiogenic index is the percentage of branch points with newly formed blood vessels relative to controls. CM from both Hx and Reox induced a marked elevation in angiogenesis of 290 and 330%, respectively. 48N, 48-h normoxia; 48H, 48-h hypoxia; 48H-24R, 48-h hypoxia followed by 24-h normoxia; WB, Western blot; d, day.
Quantitative Analysis Revealed Significant Alterations to Tumor Secretome
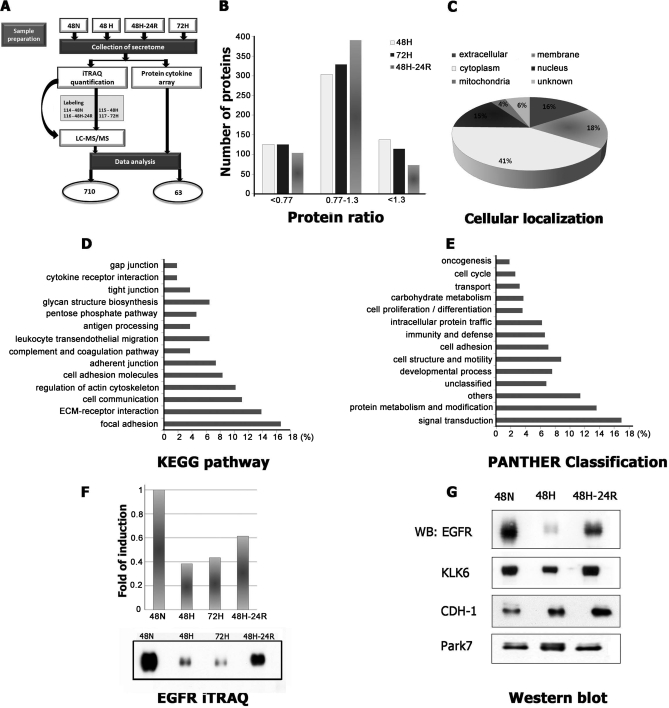
Our above observations suggested that Hx modulated cell-cell and cell-ECM adhesiveness, ECM proteolytic activity, and angiogenic potential of the cellular secretion of A431 cells such that the cells were poised to migrate and invade or to induce infiltration of angiogenic cells to vascularize the tumor. As these activities are largely extracellular, they were probably mediated by cellular secretion into extracellular spaces. To elucidate the molecular mechanisms mediating these extracellular activities, we first profiled the proteins secreted by A431 cells under Nx and Hx to determine whether proteins secreted under hypoxic condition could promote the observed hypoxia-induced cellular activities. The secreted proteins were profiled using iTRAQ-based quantitative proteomics (see “Experimental Procedures” and Fig. 4A). Proteins in the CM were extracted, labeled with an isobaric mass tag (48-h Nx, 114; 48-h Hx, 115; 48-h Reox, 116; 72-h Hx, 117), and analyzed by two-dimensional LC-MS/MS. To reduce analytical variation and false positive results while increasing sensitivity and reproducibility, iTRAQ analysis was performed on two independent biological samples (26). The combined data were searched with the concatenated target and decoy IPI human databases. The following stringent criteria were used for the identification and quantification of proteins in the iTRAQ experiments: (i) at least two unique peptides with >95% confidence, (ii) unused ProtScore ≥2 corresponding to ≥99% confidence, and (iii) FDR estimated to be 0.0%. An iTRAQ ratio with EF <2 and p value <0.05 was used unless the proteins were known to be of interest from our functional assay (listed in supplemental Table S1). Relative quantities of >1.3 or <0.77 (1/1.3) were considered to be up- or down-regulated, respectively, with ∼42% of the proteins secreted under Hx falling into either category. Using a combination of annotations from GeneCards, TMHMM, and SecretomeP, we observed that ∼32% of proteins were categorized into membrane or extracellular region. Further functional segregation of the secretome subset was performed utilizing the KEGG pathway or PANTHER database (27, 28). To corroborate the results obtained by the iTRAQ approach, we analyzed the levels of four differentially expressed proteins, EGFR, CDH1, KLK6, and Park7, in Nx, Hx, and Reox secretomes by Western blot. Western blot analysis confirmed the results produced by iTRAQ analysis, and consistency in quantitation was verified by cytokine array, although the -fold changes obtained by the three approaches were slightly different from each other (Fig. 4, F and G).
Fig. 4.
Summary of iTRAQ quantification data from A431 secretome. A, schematic overview of our quantitative analysis of the A431 secretome. B, distribution of the 710 proteins identified by mass spectrometry based on their quantities relative to the control. C, distribution of the identified proteins based on their cellular localization. Predictions were obtained from gene ontology annotation (GeneCards), computational prediction of transmembrane α helix (TMHMM), and prediction of non-classical protein secretion (SecretomeP 2.0). Ontological distribution of secreted and membrane proteins was based on the KEGG pathway database (D) and PANTHER database (E). F, comparative analysis of protein expression levels using iTRAQ and Western blot for EGFR. G, selected proteins EGFR, CDH1, KLK6, and Park7 were further analyzed by Western blot under Nx, Hx, and Reox for validation of iTRAQ data. 48N, 48-h normoxia; 48H, 48-h hypoxia; 48H-24R, 48-h hypoxia followed by 24-h normoxia; WB, Western blot.
Functional Cluster Analysis of Differentially Regulated Proteins under Hypoxia and Reoxygenation
Based on the ontological and then functional clustering analysis of the secreted proteins or secretome, we observed that under hypoxia the highest occurring molecular functions in the secretomes were associated with the cellular adhesion events: focal adhesion (16.7%), ECM-receptor interaction (13.9%), cell communication (11.1%), cell adhesion molecules (8.3%), and adherent junction (7.4%) (Fig. 4D). To determine whether there was a link between gain of metastatic potential by hypoxic cells and the adhesion-associated secreted factors, we focused on specific components in the ECM or focal adhesion, dynamic structures that regulate the cell-cell or cell-ECM adhesions (Table I) (29).
We observed a decline in the secretion of the ECM components such as DAG1 (dystroglycan), a laminin-binding glycoprotein involved in linking the extracellular matrix to the cytoskeleton (30); laminins (LAMA3, LAMA5, and LAMC1); THBS1 (thrombospondin-1), a potent antiangiogenic molecule and mediator of cell adhesion (31); and HSPG2 (heparan sulfate proteoglycan 2), a major component of the basement membranes and extracellular matrix (32). Up-regulated proteins include filamins (FLNA and FLNB), actin cross-linkers that anchor membrane proteins to the actin cytoskeleton (33); F11 receptor, an important regulator of tight junction assemblies (34); plectin-1, a cross-linker of actin, microtubules, and intermediate filaments (35); VASP (vasodilator-stimulated phosphoprotein), involved in invasive migration of cancer cells (36); S100A4, a metastasis promoter involved in up-regulation of matrix metalloproteases (MMPs) and down-regulation of tissue inhibitors of matrix metalloprotease (TIMPs) (37); VCL (vinculin), involved in anchoring F-actin to the membrane (38); and LGALS3 (lectin, galactoside-binding, soluble-3), a protein that cross-links Mgat-5 to the cell surface. Mgat-5 was found to be the rate-limiting protein for cytokine signaling and consequently for epithelial-mesenchymal transition, mobility, and metastasis (39).
Next, we focused on ECM proteolytic activity as hypoxic A431 cells were observed to exhibit increased invasiveness through ECM (Fig. 1B). Several proteins classified into the protease family were identified as highly up-regulated in the Hx or Reox secretome (Table II) including CAPN1 (calpain 1), CSTA (cathepsin A), and CSTB (cathepsin B). CAPN1, a Ca2+-dependent cysteine protease, was found to be involved in hypoxia-mediated cell death in a caspase-dependent manner (40). CSTA and CSTB, cysteine proteases, were known to be related to cancer development (41). Taken together, we could detect down-regulation of protease inhibitors including TIMP1 and TIMP2, major inhibitors of MMPs, which would be expected to be involved in cancer progression by degrading and remodeling of ECM (42).
The ontological analysis of the secretome revealed that many biological processes were modulated under Hx or Reox condition. To map these secreted proteins to the phenotypic changes that we observed above, we focused on the cell cycle control and proliferation as reduced cell cycle arrest was a prominent cellular phenotype under Hx (Fig. 5, A and B). Several components of the cell cycle regulation and proliferation were significantly up- or down-regulated under Hx (Table II). Up-regulated proteins include stratifin (SFN), a negative cell cycle regulator that causes G2 arrest (43); cystathionine γ-lyase (CTH), a negative regulator of cell proliferation and cell cycle via modulation of ERK (extracellular signal-regulated kinase) 1/2 phosphorylation (44); and hepatoma-derived growth factor (HDGF), involved in the anchorage-independent growth, cell invasion, and formation of the neovasculature of NSCL cancer (45). It is interesting to note that most of the growth factors and their binding proteins including IGFBPs, fibroblast growth factor-binding protein, and TGF-β were down-regulated under Hx (Table II). Such changes could account for the reduced cell proliferation and cell cycle arrest under Hx and Reox conditions. Several proteins known to facilitate biological functions that are pertinent to reduced cell-cell and cell-ECM adhesion, increased invasiveness, and increased chorioallantoic membrane angiogenesis are examined in detail under the “Discussion.”
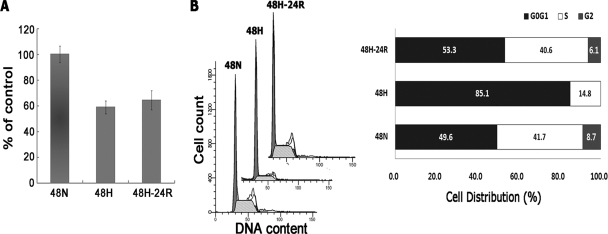
Fig. 5.
Effects of Hx and Reox on A431 cell proliferation and cell cycle arrest. A, cells were subjected to the indicated conditions, and proliferation was subsequently analyzed via the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. Cell proliferation was found to be suppressed by both Hx and Reox treatments but to a lesser extent in the latter case. B, induction of cell cycle arrest in G0G1 phase by Hx. Cells were treated with the fluorescent dye propidium iodide, and measurements of DNA content were performed via flow cytometry. The distribution of cells based on cell cycle stage is shown. Hx induced severe arrest in G0G1 phase that was released by Reox, evidenced by cells entering both S and G2 phases. 48N, 48-h normoxia; 48H, 48-h hypoxia; 48H-24R, 48-h hypoxia followed by 24-h normoxia.
Cytokine Protein Array Analysis Unveiled Changes in Extracellular Signaling Components
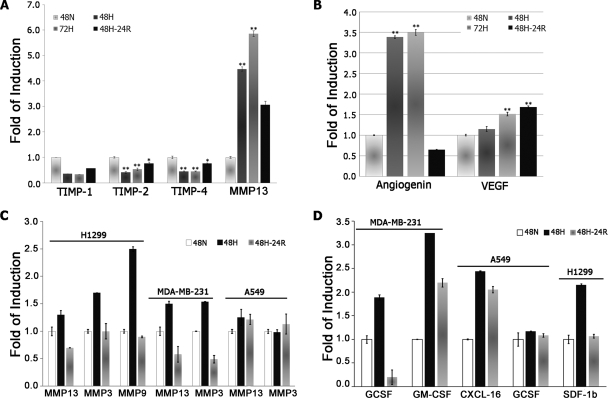
The cytokine profile for each of the conditioned media was performed using two independent biological samples on protein arrays printed with 174 anti-cytokine antibodies in replicate. Using internal positive and negative controls in setting the intensity gates netted a total of 63 cytokines (supplemental Table S2). Of these, 10 (interleukin-3 (IL-3), MMP-13, angiogenin (ANG), growth-related oncogene (GRO/GRO-α), granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-1α, VEGF, and PDGF-BB) were significantly increased (>1.5-fold), whereas three (IGF-II (insulin-like growth factor-II), TIMP-2, and TIMP-4) were down-regulated (Table III and Fig. 6). Intriguingly, hematopoietic stimulators GM-CSF and PDGF-BB showed enhanced secretion under extended 72-h Hx.
Table III. Potential angiogenic and antiangiogenic cytokines differentially regulated under Hx and Reox conditions.
48N, 48-h normoxia; 48H, 48-h hypoxia; 72H, 72-h hypoxia; 48H-24R, 48-h hypoxia followed by 24-h normoxia. Selected cytokine list was subjected to one-way analysis of variance and showed statistical difference by a p value of less than 0.05. Intensity and S.D. of selected cytokines is supplied in supplemental Table S2.
| No. | Protein | -Fold induction |
|||
|---|---|---|---|---|---|
| 48N | 48H | 72H | 48H-24R | ||
| Increased cytokines under hypoxia | |||||
| 1 | IL-3 | 1.0 | 8.5 | 3.0 | 8.1 |
| 2 | MMP-13 | 1.0 | 4.5 | 5.9 | 3.1 |
| 3 | Angiogenin | 1.0 | 3.4 | 3.5 | 0.6 |
| 4 | IL-1α | 1.0 | 2.0 | 4.3 | 1.7 |
| 5 | GRO | 1.0 | 1.8 | 5.2 | 1.5 |
| GRO-α | 1.0 | 2.1 | 2.6 | 2.2 | |
| 6 | PDGF-BB | 1.0 | 1.0 | 5.4 | 2.0 |
| 7 | VEGF | 1.0 | 1.2 | 1.5 | 1.7 |
| 8 | G-CSF | 1.0 | 2.0 | 2.3 | 1.7 |
| 9 | GM-CSF | 1.0 | 0.5 | 1.8 | 0.6 |
| Decreased cytokines under hypoxia | |||||
| 1 | IGF-II | 1.0 | 0.5 | 0.8 | 0.9 |
| 2 | TIMP-4 | 1.0 | 0.4 | 0.4 | 0.8 |
| 3 | TIMP-2 | 1.0 | 0.4 | 0.5 | 0.8 |
Fig. 6.
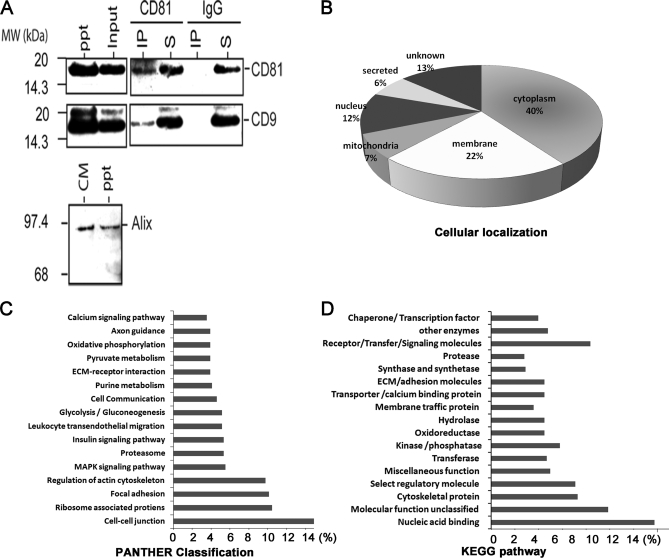
A, co-immunoprecipitation of CD81 and CD9. After immunoprecipitation of CM with anti-CD81 or mouse IgG, the immunoprecipitate (IP) and supernatant (S) were analyzed by Western blot hybridization using antibodies against CD81 and CD9. Western blotting using the 200,000 × g precipitate (ppt; exosome fraction) showed that the size and density of the CD81-CD9 complex in the CM were consistent with that of an exosome. A Western blot analysis of Alix using the CM and exosome fraction (ppt) is shown. B–D, summary of LC-MS data from the A431 exosome. B, distribution of the identified proteins based on their cellular localization. Predictions were obtained from gene ontology annotation (GeneCards), computational prediction of transmembrane α helix (TMHMM), and prediction of non-classical protein secretion (SecretomeP 2.0). The ontological distribution of secreted and membrane proteins was based on the KEGG pathway database (C) and PANTHER database (D). MAPK, mitogen-activated protein kinase.
Cytoplasmic Composition of Secretome Implicated the Presence of Exosome
Analysis of the secretome indicated that 40% of the proteins are cytoplasmic proteins or are not known to be secreted or membrane-bound proteins (supplemental Table S1). Among these proteins, we observed the presence of numerous exosome-associated proteins such as CD9 tetraspanin protein; Rab proteins (Ras superfamily of small GTP-binding proteins), e.g. RAB1B and RAB7A; annexins, e.g. ANXA1, ANXA2, and ANXA5; MHC class 1; glyceraldehyde-3-phosphate dehydrogenase; PKM2; EEF1A1; HSP90; actin; and tubulin (Table IV) (46). We hypothesized that the high proportion of cytoplasmic proteins in the secretome was due to the presence of exosomes in the secretion. Exosomes are bilipid membrane vesicles containing both RNA and proteins (46), and cancer cells have been shown to secrete exosomes into their extracellular microenvironment (47–50). To confirm the presence of exosomes in the secretion of A431 cells, we ultracentrifuged the CM for enrichment of the presumptive exosomes. The ultracentrifuged pellet from the CM was analyzed by LC-MS/MS and Western blot. Exosome-associated proteins (supplemental Table S3) such as glyceraldehyde-3-phosphate dehydrogenase, pyruvate kinase, EEF1A1, HSP70, HSP90, actin, tubulin, Rab proteins, annexins, and MHC class 1 (46) were clearly detected in the ultracentrifuged pellet by LC-MS/MS. The proteins in this ultracentrifuged pellet contained 54% of the proteins found in the CM, which also included many cytosolic proteins as listed in supplemental Table S3. Ontological and functional clustering analysis of the proteins in the ultracentrifuged pellet also revealed that the highest occurring molecular functions and biological processes were those important in metastasis such as focal adhesion, ECM-receptor interaction, cell adhesion molecules, and cell-cell junction (Fig. 6). To further confirm the presence of exosome in the secretion of A431 cells, the CM and ultracentrifuged pellet were analyzed for the presence of Alix, a protein frequently found in exosomes (51). The presence of Alix in both the CM and the ultracentrifuged pellet was consistent with the presence of exosomes in the secretion of A431 cells. Next, we determined whether the immunoprecipitation of CD81, one of the tetraspanin proteins frequently found in exosomes, would also immunoprecipitate other exosome-associated tetraspanin proteins such as CD9 (51). As shown in Fig. 6A, CD9 co-immunoprecipitated with CD81, suggesting that they were both physically associated with each other, possibly on an exosome in the CM. As this CD9-CD81 complex was also detected in a 100,000 × g precipitate, the size and density of this complex were consistent with that of an exosome.
Table IV. Proteins identified by LC-MS/MS in A431 cell-derived exosomes.
Specific information of proteins identified in exosomes is supplied in supplemental Table S3.
| Accession no. | Gene symbols |
|---|---|
| Exosome marker proteins | |
| IPI00000736 | TSPAN15 |
| IPI00027438 | FLOT1 |
| IPI00789008 | FLOT2 |
| IPI00008494 | ICAM1 |
| IPI00000190 | CD81 |
| IPI00215997 | CD9 |
| Antigens | |
| IPI00152540 | CD109 |
| IPI00410488 | CD276 |
| IPI00297160 | CD44 |
| IPI00000059 | CD58 |
| IPI00011302 | CD59 |
| IPI00002252 | CD68 |
| IPI00299412 | CD97 |
| IPI00745220 | HLA-A A-25 α chain precursor |
| IPI00472855 | HLA-A A-30 α chain precursor |
| IPI00472825 | HLA-A A-32 α chain precursor |
| IPI00472013 | HLA-A A-33 α chain precursor |
| IPI00743503 | HLA-A A-34 α chain precursor |
| IPI00472882 | HLA-A A-68 α chain precursor |
| IPI00472112 | HLA-A A-11 α chain precursor |
| IPI00647457 | HLA-A class I, A |
| IPI00872948 | HLA-A MHC class I antigen |
| IPI00472921 | HLA-A A-29 α chain precursor |
| IPI00472652 | HLA-C B-37 α chain precursor |
| IPI00746105 | HLA-C B-40 α chain precursor |
| IPI00472676 | HLA-C B-42 α chain precursor |
| IPI00472767 | HLA-C B-55 α chain precursor |
| IPI00472434 | HLA-C B-57 α chain precursor |
| IPI00472867 | HLA-C B-67 α chain precursor |
| IPI00004657 | HLA-C B-7 α chain precursor |
| IPI00472943 | HLA-C B-73 α chain precursor |
| IPI00472753 | HLA-C B-81 α chain precursor |
| IPI00026650 | HLA-C Cw-1 α chain precursor |
| IPI00743716 | HLA-C Cw-12 α chain precursor |
| IPI00473006 | HLA-C Cw-17 α chain precursor |
| IPI00472605 | HLA-C Cw-2 α chain precursor |
| IPI00642409 | HLA-C Cw-7 α chain precursor |
| IPI00816057 | HLA-C MHC class I antigen |
| Antigens | |
| IPI00015988 | HLA-G α chain G precursor |
| IPI00004672 | HLA-H α chain H precursor |
| IPI00152441 | HM13 |
| Ras-related GTP-binding proteins | |
| IPI00016513 | RAB10 |
| IPI00020436 | RAB11B |
| IPI00016373 | RAB13 |
| IPI00291928 | RAB14 |
| IPI00383449 | RAB15 |
| IPI00014577 | RAB18 |
| IPI00005719 | RAB1A |
| IPI00008964 | RAB1B |
| IPI00007755 | RAB21 |
| IPI00007756 | RAB22A |
| IPI00031169 | RAB2A |
| IPI00102896 | RAB2B |
| IPI00021475 | RAB33B |
| IPI00328180 | RAB34 |
| IPI00300096 | RAB35 |
| IPI00001618 | RAB39 |
| IPI00448725 | RAB4B |
| IPI00023510 | RAB5A |
| IPI00017344 | RAB5B |
| IPI00016339 | RAB5C |
| IPI00023526 | RAB6A |
| IPI00016342 | RAB7A |
| IPI00028481 | RAB8A |
| IPI00024282 | RAB8B |
| Annexins | |
| IPI00218918 | ANXA1 |
| IPI00414320 | ANXA11 |
| IPI00455315 | ANXA2 |
| IPI00024095 | ANXA3 |
| IPI00793199 | ANXA4 |
| IPI00329801 | ANXA5 |
| IPI00002460 | ANXA7 |
| IPI00471941 | ANXA8 |
| IPI00218835 | ANXA8L2 |
DISCUSSION
In solid tumors, a substantial fraction of the tumor cells is subjected to either chronic or acute Hx, and the emerging evidences have suggested that hypoxic cells tend to acquire metastatic character and resistance to cancer therapy (17, 52). Moreover, Reox of hypoxic cells is thought to augment invasiveness of cells as supported by the reports of increased spontaneous metastases in vivo and in clinical data (53, 54). Together these observations led us to examine the effects of Hx and Reox on several key metastatic processes and to postulate that the tumor secretome under these conditions modulates the tumor microenvironment and facilitates the acquired metastatic character. To elucidate the changes in the tumor secretome in response to Hx and Reox, we used quantitative proteomics to profile the secretome and identify candidate pathways as targets to develop strategies that will synergize with existing cancer therapies.
Hypoxia Decreased Cell Adhesion and Increased Invasiveness of A431 Cells
To metastasize, malignant cells must first detach from the dense, cross-linked type I collagen network of the ECM and migrate through the host vasculature before extravasating the vasculature and infiltrating the host tissues (55). Therefore, tumor metastasis is dependent on the ability of the tumor to degrade the surrounding ECM and to reduce the cell adhesion. A key mediator of the former is the proteolytic system of MMPs and their inhibitors, TIMPs, that plays pivotal roles in ECM remodeling (42, 56). This system regulates the tumor microenvironment via modulation of the cell-cell and cell-ECM adhesion and the levels and activities of growth factors, cytokines, and their receptors through either up-regulation or down-regulation of the relative activities of MMPs and their inhibitors, TIMPs. Thus, it is possible that our observation of increased invasion of A431 cells across ECM under Hx and Reox was due to the hyperactivation of this proteolytic system. Indeed, analysis of the secretome revealed not only a 6-fold increase in MMP-13 levels but also an increase in the known enhancers of MMP-13 including several cytokines such as S100A4 (37), IL-1, VEGF, and PDGF-BB (57) as well as the decrease in TGF-β, a potential transcriptional suppressor of MMP-13 (58) (Tables I and III). These changes in the secretion of MMP proteolytic enzymes suggest that Hx and Reox facilitate metastasis of cancer cells by increasing the secretion of MMP-13. Further investigation revealed that other cancer cell lines such as MDA-MB-231 (breast), A549, and H1299 (lung) cells but not normal human foreskin fibroblasts also responded to Hx by increasing secretion of MMPs such as MMP-3, -9, and -13 (Fig. 7C). These observations suggested that the underlying mechanism for increased tumor metastasis during Hx is partially mediated by increased secretion of the MMPs.
Fig. 7.
A, relative quantities of the secreted MMP-13-TIMP proteolytic system components, MMP-13 and TIMP-1, -2, and -4, determined by cytokine arrays. MMP-13 secretion was found to be elevated by 450, 585, and 300% by 48-h Hx, 72-h Hx, and Reox, respectively, whereas TIMP levels wersuppressed under both Hx and Reox. B, relative levels of secreted ANG and VEGF as determined by cytokine array. Both were significantly elevated under Hx, but ANG dropped precipitously to 65% of control under Reox, whereas secreted VEGF levels increased further. Statistical significance was accepted at p < 0.05 (*) and p < 0.01 (**). C, relative quantities of the secreted MMPs in MDA-MB-231, A549, and H1299 cells determined by cytokine arrays. MMP-3, -9, and -13 secretion was found to be elevated under 72-h Hx. D, relative levels of secreted chemokines related to recruitment of immune cells, G-CSF, GM-CSF, CXCL-16, and stroma-derived factor-1 (SDF-1beta), as determined by cytokine array. Secretion of these chemokines was significantly elevated under Hx. 48N, 48-h normoxia; 48H, 48-h hypoxia; 48H-24R, 48-h hypoxia followed by 24-h normoxia.
However, the actual extracellular MMP activity depends not only on its expression levels but also on its interaction with TIMPs (59). As TIMPs inhibit MMP activity, MMP activity correlates inversely with TIMP expression level (60, 61). It is therefore significant that in response to Hx tumor cells reduced the secretion of three of the four known TIMPs, TIMP-1, -2, and -4 (Fig. 7A), coordinately with increased MMP secretion to effectively increase the overall proteolytic activity and facilitate degradation of the ECM for metastatic activity. Intriguingly, TIMPs have been implicated in the inhibition of tumorigenesis, angiogenesis, and metastasis independent of MMP inhibition (62). For example, TIMPs have been associated with increased formation of stable complexes between E-cadherin and β-catenin (63). Consistent with this, we noted a delocalization and loss of E-cadherin at the cell membrane with concomitant up-regulation of Snail and acquisition of mesenchymal morphology (Fig. 2A), possibly for gain of cell motility (64). E-cadherin, an important component of cell-cell adhesion, forms adhesion complexes that are stabilized by bridges between E-cadherin adhesion complex and actin cytoskeleton through α-catenin and vinculin. E-cadherin is often lost at the invasive front of a tumor, and this loss represents an important metastatic step (21, 22). Interestingly, proteomics analysis revealed that under Hx or Reox stress the tumor increased the secretion of α-catenin, vinculin, and E-cadherin in the secretion (Fig. 2D). This suggested that E-cadherin adhesion complexes were disassembled, and their components were discarded through secretion, leading to the loss of cell adhesiveness. Moreover, the tumor cell under Hx had reduced ECM components in the secretome that together with disassembly of E-cadherin adhesion complexes were consistent with diminished cell adhesiveness.
Increased Angiogenic Potential and Self-enhancing Mechanism under Hypoxia
Angiogenesis is regarded as a metastatic prerequisite as it permits extended tumor growth and passage to distant sites. A431 cells under Hx enhanced secretion of several angiogenic factors, ANG, VEGF, IL-1α, IL-3, GRO-α, and PDGF-BB (Fig. 7 and Table III), and these factors with the exception of ANG remained elevated even after Reox. This angiogenic potential was validated through the use of an in vivo CAM assay, which demonstrated that CM from hypoxic and reoxygenated A431 cells induced a marked increase in angiogenesis over controls (Fig. 4). We further observed that secretion of some angiogenic factors, PDGF-BB and GM-CSF, increased further with prolonged Hx (PDGF-BB: 72-h Hx/48-h Hx = 5.4). This is consistent with the hypothesis that extended Hx causes tumors to activate alternative angiogenic pathways to secure their nutrient supply (4, 5), and this activation presumably facilitates development of resistance to antiangiogenic therapy (such as bevacizumab).
Additionally, cancer cells situated in the hostile microenvironments secrete potent chemoattractant to recruit immune cells, which could further assist tumor cell invasion by secreting the ECM-degrading enzymes (7). Recruited immune cells might alter the tumor microenvironment by expression of potent prometastatic and proangiogenic factors. Our results show that G-CSF and GM-CSF, potent chemokinetic molecules for tumor-associated macrophages and neutrophils (7, 65), were up-regulated in the secretome of hypoxic A431 cells, and we also discovered that several cancer cell lines including MDA-MB-231, A549, and H1299 cells also secreted chemokines such as G-CSF, GM-CSF, stroma-derived factor-1, and CXCL-16 (CXC motif ligand 16) under the Hx stress (Fig. 7D). Together, these results indicated that cancer cells could potentially modulate their microenvironment as a self-sustaining mechanism in a hostile environment by secreting the cytokines.
Tumor Cell Secretion Is Encapsulated in Exosomes
Proteomics analysis of the secretome revealed that ∼40% of proteins (Fig. 4C) were classified into cytosolic proteins, many of which were exosome-associated proteins. This unexpected discovery led us to postulate that many of the secreted proteins were encapsulated in the exosomes. Exosomes are 40–100-nm bilipid membrane vesicles that are secreted by most cell types. They are thought to mediate the cell-cell communication and facilitate biological processes such as antigen presentation, cell growth, apoptotic cellular breakdown, and malignant transformation (50, 66). We discovered that more than 50% of the proteins secreted by A431 cells under hypoxia were found to be present in the presumptive exosome preparation. Significantly, these proteins also have the potential to facilitate biological functions and processes that are important in angiogenesis and metastasis. However, the significance of these exosomes in mediating the Hx- and Reox-induced cellular phenotype in A431 cell line remains to be determined. Based on a recent review of tumor exosomes (50, 66), it is highly conceivable that these exosomes may be functional. If so, they are likely to add a new dimension to our present understanding of the tumor microenvironment and its modulation.
Conclusions
In this study, we analyzed the effects of Hx and Reox on several key metastatic prerequisites. Our findings were consistent with the reported prometastatic effects of Hx, i.e. enhanced angiogenesis, increased invasion, and loss of cell adhesiveness. As these cellular changes were highly dependent on the cellular microenvironment, we determined whether cellular secretion mediated these cellular changes through modulation of the microenvironment. Indeed, the proteins secreted by tumor cells under Hx and Reox could potentially support the Hx-induced cellular changes. For example, the increased secretion of proteases such as MMPs and cysteine proteases correlated well with the reduced adhesion and invasiveness of tumor cells under Hx. The importance of proteases in tumor biology is reflected in their use as targets for cancer therapy because of their abnormal expression (67). We also observed that tumor cells secrete many proteins such as IL-1, VEGF, PDGF-BB, TGF-β, and S100A4 with the potential to activate the MMP-TIMP proteolytic system. Lastly, analysis of the tumor secretome under Hx and Reox revealed that tumor cells may adapt to hostile microenvironments by secreting cytokines including angiogenin, VEGF, G-CSF, and GM-CSF to induce angiogenesis or acquire a self-sustaining mechanism. Taken together, our proteomics analysis of secretion by tumor cells under Hx or Reox has revealed a large number of candidate molecules in the regulation of tumor metastasis and angiogenesis. These molecules could potentially serve as therapeutic targets or disease biomarkers. Unexpectedly, we observed that more than 50% of the secreted proteins were found in presumptive exosomes. As exosomes are known to have a myriad of functional activities, our observation is likely to add another dimension to our present understanding of the tumor microenvironment and its regulation by tumor cells.
Acknowledgments
We thank Mark Featherstone, Peter Cheung, Tinnan Guo, Xin Li, Yi Zhu, Bamaprasad Dutta, and Abdul Qader Al-Aidaroos for invaluable discussions and comments on the manuscript.
* This work was supported by the Ministry of Education (ARC Grant T206B3211) and the Agency for Science, Technology and Research (BMRC Grant 08/1/22/19/575) of Singapore.
 This article contains supplemental Tables S1–S3.
This article contains supplemental Tables S1–S3.
1 The abbreviations used are:
- Hx
- hypoxia
- Reox
- hypoxia/reoxygenation
- ECM
- extracellular matrix
- CAM
- chorioallantoic membrane
- Nx
- normoxia
- CM
- conditioned medium
- E-cad
- E-cadherin
- CDH1
- E-cadherin
- KLK6
- kallikrein 6
- MMP
- matrix metalloproteinase
- ANG
- angiogenin
- GRO/GRO-α
- growth-related oncogene
- G-CSF
- granulocyte colony-stimulating factor
- GM-CSF
- granulocyte-macrophage colony-stimulating factor
- IGF-II
- insulin like growth factor-II
- TIMP
- tissue inhibitor of matrix metalloproteinase
- MHC
- major histocompatibility complex
- VEGF
- vascular endothelial growth factor
- PDGF
- platelet-derived growth factor
- EGFR
- epidermal growth factor receptor
- iTRAQ
- isobaric tag for relative and absolute quantitation
- NSCL
- non-small cell lung
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- IPI
- International Protein Index
- FDR
- false discovery rate
- EF
- error factor
- F-actin
- filamentous actin
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- IGFBP
- insulin-like growth factor-binding protein
- TGF-β
- transforming growth factor-β
- IL
- interleukin
- TMHMM
- Trans Membrane prediction using Hidden Markov Models
- PANTHER
- Protein ANalysis THrough Evolutionary Relationships.
REFERENCES
- 1.Hanahan D., Weinberg R. A. (2000) The hallmarks of cancer. Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 2.Stacker S. A., Baldwin M. E., Achen M. G. (2002) The role of tumor lymphangiogenesis in metastatic spread. FASEB J 16, 922–934 [DOI] [PubMed] [Google Scholar]
- 3.Sporn M. B. (1997) The war on cancer: a review. Ann. N.Y. Acad. Sci 833, 137–146 [DOI] [PubMed] [Google Scholar]
- 4.Ferrara N., Hillan K. J., Novotny W. (2005) Bevacizumab (Avastin), a humanized anti-VEGF monoclonal antibody for cancer therapy. Biochem. Biophys. Res. Commun 333, 328–335 [DOI] [PubMed] [Google Scholar]
- 5.Bergers G., Hanahan D. (2008) Modes of resistance to anti-angiogenic therapy. Nat. Rev. Cancer 8, 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magagnin M. G., Sergeant K., van den Beucken T., Rouschop K. M., Jutten B., Seigneuric R., Lambin P., Devreese B., Koritzinsky M., Wouters B. G. (2007) Proteomic analysis of gene expression following hypoxia and reoxygenation reveals proteins involved in the recovery from endoplasmic reticulum and oxidative stress. Radiother. Oncol 83, 340–345 [DOI] [PubMed] [Google Scholar]
- 7.Joyce J. A., Pollard J. W. (2009) Microenvironmental regulation of metastasis. Nat. Rev. Cancer 9, 239–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sze S. K., de Kleijn D. P., Lai R. C., Khia Way Tan E., Zhao H., Yeo K. S., Low T. Y., Lian Q., Lee C. N., Mitchell W., El Oakley R. M., Lim S. K. (2007) Elucidating the secretion proteome of human embryonic stem cell-derived mesenchymal stem cells. Mol. Cell Proteomics 6, 1680–1689 [DOI] [PubMed] [Google Scholar]
- 9.Juarez J. C., Manuia M., Burnett M. E., Betancourt O., Boivin B., Shaw D. E., Tonks N. K., Mazar A. P., Doñate F. (2008) Superoxide dismutase 1 (SOD1) is essential for H2O2-mediated oxidation and inactivation of phosphatases in growth factor signaling. Proc. Natl. Acad. Sci. U.S.A 105, 7147–7152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Nedawi K., Meehan B., Kerbel R. S., Allison A. C., Rak J. (2009) Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc. Natl. Acad. Sci. U.S.A 106, 3794–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao F., Li Y., O'Connor W., Zanetta L., Bassi R., Santiago A., Overholser J., Hooper A., Mignatti P., Dejana E., Hicklin D. J., Bohlen P. (2000) Monoclonal antibody to vascular endothelial-cadherin is a potent inhibitor of angiogenesis, tumor growth, and metastasis. Cancer Res 60, 6805–6810 [PubMed] [Google Scholar]
- 12.Simpson R. J., Bernhard O. K., Greening D. W., Moritz R. L. (2008) Proteomics-driven cancer biomarker discovery: looking to the future. Curr. Opin. Chem. Biol 12, 72–77 [DOI] [PubMed] [Google Scholar]
- 13.Hanash S. M., Pitteri S. J., Faca V. M. (2008) Mining the plasma proteome for cancer biomarkers. Nature 452, 571–579 [DOI] [PubMed] [Google Scholar]
- 14.Datta A., Park J. E., Li X., Zhang H., Ho Z. S., Heese K., Lim S. K., Tam J. P., Sze S. K. (2010) Phenotyping of an in vitro Model of Ischemic Penumbra by iTRAQ-based Shotgun Quantitative Proteomics. J. Proteome Res 9, 472–484 [DOI] [PubMed] [Google Scholar]
- 15.Guo T., Gan C. S., Zhang H., Zhu Y., Kon O. L., Sze S. K. (2008) Hybridization of pulsed-Q dissociation and collision-activated dissociation in linear ion trap mass spectrometer for iTRAQ quantitation. J. Proteome Res 7, 4831–4840 [DOI] [PubMed] [Google Scholar]
- 16.Conde-Vancells J., Rodriguez-Suarez E., Embade N., Gil D., Matthiesen R., Valle M., Elortza F., Lu S. C., Mato J. M., Falcon-Perez J. M. (2008) Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J. Proteome Res 7, 5157–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sullivan R., Graham C. H. (2007) Hypoxia-driven selection of the metastatic phenotype. Cancer Metastasis Rev 26, 319–331 [DOI] [PubMed] [Google Scholar]
- 18.Toffoli S., Michiels C. (2008) Intermittent hypoxia is a key regulator of cancer cell and endothelial cell interplay in tumours. FEBS J 275, 2991–3002 [DOI] [PubMed] [Google Scholar]
- 19.Olson M. F., Sahai E. (2009) The actin cytoskeleton in cancer cell motility. Clin. Exp. Metastasis 26, 273–287 [DOI] [PubMed] [Google Scholar]
- 20.Blechschmidt K., Sassen S., Schmalfeldt B., Schuster T., Höfler H., Becker K. F. (2008) The E-cadherin repressor Snail is associated with lower overall survival of ovarian cancer patients. Br. J. Cancer 98, 489–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bajpai S., Correia J., Feng Y., Figueiredo J., Sun S. X., Longmore G. D., Suriano G., Wirtz D. (2008) {alpha}-Catenin mediates initial E-cadherin-dependent cell-cell recognition and subsequent bond strengthening. Proc. Natl. Acad. Sci. U.S.A 105, 18331–18336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiss E. E., Kroemker M., Rüdiger A. H., Jockusch B. M., Rüdiger M. (1998) Vinculin is part of the cadherin-catenin junctional complex: complex formation between alpha-catenin and vinculin. J. Cell Biol 141, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huot J., Houle F., Rousseau S., Deschesnes R. G., Shah G. M., Landry J. (1998) SAPK2/p38-dependent F-actin reorganization regulates early membrane blebbing during stress-induced apoptosis. J. Cell Biol 143, 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albini A., Benelli R. (2007) The chemoinvasion assay: a method to assess tumor and endothelial cell invasion and its modulation. Nat. Protoc 2, 504–511 [DOI] [PubMed] [Google Scholar]
- 25.Liao D., Johnson R. S. (2007) Hypoxia: a key regulator of angiogenesis in cancer. Cancer Metastasis Rev 26, 281–290 [DOI] [PubMed] [Google Scholar]
- 26.Gan C. S., Chong P. K., Pham T. K., Wright P. C. (2007) Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ). J. Proteome Res 6, 821–827 [DOI] [PubMed] [Google Scholar]
- 27.Thomas P. D., Campbell M. J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. (2003) PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 13, 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang B., Kirov S., Snoddy J. (2005) WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 33, W741–W748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia N., Thodeti C. K., Hunt T. P., Xu Q., Ho M., Whitesides G. M., Westervelt R., Ingber D. E. (2008) Directional control of cell motility through focal adhesion positioning and spatial control of Rac activation. FASEB J 22, 1649–1659 [DOI] [PubMed] [Google Scholar]
- 30.Sgambato A., Migaldi M., Montanari M., Camerini A., Brancaccio A., Rossi G., Cangiano R., Losasso C., Capelli G., Trentini G. P., Cittadini A. (2003) Dystroglycan expression is frequently reduced in human breast and colon cancers and is associated with tumor progression. Am. J. Pathol 162, 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simantov R., Silverstein R. L. (2003) CD36: a critical anti-angiogenic receptor. Front. Biosci 8, s874–s882 [DOI] [PubMed] [Google Scholar]
- 32.Murdoch A. D., Dodge G. R., Cohen I., Tuan R. S., Iozzo R. V. (1992) Primary structure of the human heparan sulfate proteoglycan from basement membrane (HSPG2/perlecan). A chimeric molecule with multiple domains homologous to the low density lipoprotein receptor, laminin, neural cell adhesion molecules, and epidermal growth factor. J. Biol. Chem 267, 8544–8557 [PubMed] [Google Scholar]
- 33.Popowicz G. M., Schleicher M., Noegel A. A., Holak T. A. (2006) Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem. Sci 31, 411–419 [DOI] [PubMed] [Google Scholar]
- 34.Liu Y., Nusrat A., Schnell F. J., Reaves T. A., Walsh S., Pochet M., Parkos C. A. (2000) Human junction adhesion molecule regulates tight junction resealing in epithelia. J. Cell Sci 113, 2363–2374 [DOI] [PubMed] [Google Scholar]
- 35.Svitkina T. M., Verkhovsky A. B., Borisy G. G. (1996) Plectin sidearms mediate interaction of intermediate filaments with microtubules and other components of the cytoskeleton. J. Cell Biol 135, 991–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han G., Fan B., Zhang Y., Zhou X., Wang Y., Dong H., Wei Y., Sun S., Hu M., Zhang J., Wei L. (2008) Positive regulation of migration and invasion by vasodilator-stimulated phosphoprotein via Rac1 pathway in human breast cancer cells. Oncol. Rep 20, 929–939 [PubMed] [Google Scholar]
- 37.Sherbet G. V. (2009) Metastasis promoter S100A4 is a potentially valuable molecular target for cancer therapy. Cancer Lett 280, 15–30 [DOI] [PubMed] [Google Scholar]
- 38.Weller P. A., Ogryzko E. P., Corben E. B., Zhidkova N. I., Patel B., Price G. J., Spurr N. K., Koteliansky V. E., Critchley D. R. (1990) Complete sequence of human vinculin and assignment of the gene to chromosome 10. Proc. Natl. Acad. Sci. U.S.A 87, 5667–5671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Partridge E. A., Le Roy C., Di Guglielmo G. M., Pawling J., Cheung P., Granovsky M., Nabi I. R., Wrana J. L., Dennis J. W. (2004) Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science 306, 120–124 [DOI] [PubMed] [Google Scholar]
- 40.Kim M. J., Oh S. J., Park S. H., Kang H. J., Won M. H., Kang T. C., Hwang I. K., Park J. B., Kim J. I., Kim J., Lee J. Y. (2007) Hypoxia-induced cell death of HepG2 cells involves a necrotic cell death mediated by calpain. Apoptosis 12, 707–718 [DOI] [PubMed] [Google Scholar]
- 41.Su H., Hu N., Shih J., Hu Y., Wang Q. H., Chuang E. Y., Roth M. J., Wang C., Goldstein A. M., Ding T., Dawsey S. M., Giffen C., Emmert-Buck M. R., Taylor P. R. (2003) Gene expression analysis of esophageal squamous cell carcinoma reveals consistent molecular profiles related to a family history of upper gastrointestinal cancer. Cancer Res 63, 3872–3876 [PubMed] [Google Scholar]
- 42.Noël A., Jost M., Maquoi E. (2008) Matrix metalloproteinases at cancer tumor-host interface. Semin. Cell Dev. Biol 19, 52–60 [DOI] [PubMed] [Google Scholar]
- 43.Hermeking H. (2003) The 14-3-3 cancer connection. Nat. Rev. Cancer 3, 931–943 [DOI] [PubMed] [Google Scholar]
- 44.Yang G., Cao K., Wu L., Wang R. (2004) Cystathionine gamma-lyase overexpression inhibits cell proliferation via a H2S-dependent modulation of ERK1/2 phosphorylation and p21Cip/WAK-1. J. Biol. Chem 279, 49199–49205 [DOI] [PubMed] [Google Scholar]
- 45.Zhang J., Ren H., Yuan P., Lang W., Zhang L., Mao L. (2006) Down-regulation of hepatoma-derived growth factor inhibits anchorage-independent growth and invasion of non-small cell lung cancer cells. Cancer Res 66, 18–23 [DOI] [PubMed] [Google Scholar]
- 46.Théry C., Ostrowski M., Segura E. (2009) Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol 9, 581–593 [DOI] [PubMed] [Google Scholar]
- 47.Al-Nedawi K., Meehan B., Micallef J., Lhotak V., May L., Guha A., Rak J. (2008) Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol 10, 619–624 [DOI] [PubMed] [Google Scholar]
- 48.Simpson R. J., Jensen S. S., Lim J. W. (2008) Proteomic profiling of exosomes: current perspectives. Proteomics 8, 4083–4099 [DOI] [PubMed] [Google Scholar]
- 49.Skog J., Würdinger T., van Rijn S., Meijer D. H., Gainche L., Sena-Esteves M., Curry W. T., Jr., Carter B. S., Krichevsky A. M., Breakefield X. O. (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol 10, 1470–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schorey J. S., Bhatnagar S. (2008) Exosome function: from tumor immunology to pathogen biology. Traffic 9, 871–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olver C., Vidal M. (2007) Proteomic analysis of secreted exosomes. Subcell. Biochem 43, 99–131 [DOI] [PubMed] [Google Scholar]
- 52.Teicher B. A. (1994) Hypoxia and drug resistance. Cancer Metastasis Rev 13, 139–168 [DOI] [PubMed] [Google Scholar]
- 53.Cairns R. A., Hill R. P. (2004) Acute Hypoxia Enhances Spontaneous Lymph Node Metastasis in an Orthotopic Murine Model of Human Cervical Carcinoma. Cancer Res 64, 2054–2061 [DOI] [PubMed] [Google Scholar]
- 54.Rofstad E. K., Halsør E. F. (2002) Hypoxia-associated spontaneous pulmonary metastasis in human melanoma xenografts: involvement of microvascular hot spots induced in hypoxic foci by interleukin 8. Br. J. Cancer 86, 301–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hay E. D. (1991) Cell Biology of Extracellular Matrix, Plenum Press, New York, 371–384 [Google Scholar]
- 56.Page-McCaw A., Ewald A. J., Werb Z. (2007) Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol 8, 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Roeyen C. R., Ostendorf T., Denecke B., Bokemeyer D., Behrmann I., Strutz F., Lichenstein H. S., LaRochelle W. J., Pena C. E., Chaudhuri A., Floege J. (2006) Biological responses to PDGF-BB versus PDGF-DD in human mesangial cells. Kidney Int 69, 1393–1402 [DOI] [PubMed] [Google Scholar]
- 58.Rydziel S., Varghese S., Canalis E. (1997) Transforming growth factor beta1 inhibits collagenase 3 expression by transcriptional and post-transcriptional mechanisms in osteoblast cultures. J. Cell. Physiol 170, 145–152 [DOI] [PubMed] [Google Scholar]
- 59.Melendez-Zajgla J., Del Pozo L., Ceballos G., Maldonado V. (2008) Tissue inhibitor of Metalloproteinases-4. The road less traveled. Mol. Cancer 7, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang M., Liu Y. E., Greene J., Sheng S., Fuchs A., Rosen E. M., Shi Y. E. (1997) Inhibition of tumor growth and metastasis of human breast cancer cells transfected with tissue inhibitor of metalloproteinase 4. Oncogene 14, 2767–2774 [DOI] [PubMed] [Google Scholar]
- 61.Lichtinghagen R., Musholt P. B., Stephan C., Lein M., Kristiansen G., Hauptmann S., Rudolph B., Schnorr D., Loening S. A., Jung K. (2003) mRNA expression profile of matrix metalloproteinases and their tissue inhibitors in malignant and non-malignant prostatic tissue. Anticancer Res 23, 2617–2624 [PubMed] [Google Scholar]
- 62.Gomez D. E., Alonso D. F., Yoshiji H., Thorgeirsson U. P. (1997) Tissue inhibitors of metalloproteinases: structure, regulation and biological functions. Eur. J. Cell Biol 74, 111–122 [PubMed] [Google Scholar]
- 63.Ho A. T., Voura E. B., Soloway P. D., Watson K. L., Khokha R. (2001) MMP inhibitors augment fibroblast adhesion through stabilization of focal adhesion contacts and up-regulation of cadherin function. J. Biol. Chem 276, 40215–40224 [DOI] [PubMed] [Google Scholar]
- 64.Huber M. A., Kraut N., Beug H. (2005) Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr. Opin. Cell Biol 17, 548–558 [DOI] [PubMed] [Google Scholar]
- 65.Raman D., Baugher P. J., Thu Y. M., Richmond A. (2007) Role of chemokines in tumor growth. Cancer Lett 256, 137–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Al-Nedawi K., Meehan B., Rak J. (2009) Microvesicles: messengers and mediators of tumor progression. Cell Cycle 8, 2014–2018 [DOI] [PubMed] [Google Scholar]
- 67.Ala-aho R., Kähäri V. M. (2005) Collagenases in cancer. Biochimie 87, 273–286 [DOI] [PubMed] [Google Scholar]