Abstract
The rapid expansion of sequence data and the development of statistical approaches that embrace varying evolutionary rates among lineages have encouraged many more investigators to use DNA and protein data to time species divergences. Here, we report results from a systematic evaluation, by means of computer simulation, of the performance of two frequently used relaxed-clock methods for estimating these times and their credibility intervals (CrIs). These relaxed-clock methods allow rates to vary in a phylogeny randomly over lineages (e.g., BEAST software) and in autocorrelated fashion (e.g., MultiDivTime software). We applied these methods for analyzing sequence data sets simulated using naturally derived parameters (evolutionary rates, sequence lengths, and base substitution patterns) and assuming that clock calibrations are known without error. We find that the estimated times are, on average, close to the true times as long as the assumed model of lineage rate changes matches the actual model. The 95% CrIs also contain the true time for ≥95% of the simulated data sets. However, the use of incorrect lineage rate model reduces this frequency to 83%, indicating that the relaxed-clock methods are not robust to the violation of underlying lineage rate model. Because these rate models are rarely known a priori and are difficult to detect empirically, we suggest building composite CrIs using CrIs produced from MultiDivTime and BEAST analysis. These composite CrIs are found to contain the true time for ≥97% data sets. Our analyses also verify the usefulness of the common practice of interpreting the congruence of times inferred from different methods as a reflection of the accuracy of time estimates. Overall, our results show that simple strategies can be used to enhance our ability to estimate times and their CrIs when using the relaxed-clock methods.
Keywords: molecular clocks, lineage rate models, divergence times, credibility intervals, simulations
Introduction
Molecular clock methods are becoming indispensable for establishing the chronological dimension of the tree of life (Hedges and Kumar 2009). The exponential increase in the amount of sequence data available is reflected in the number of studies applying molecular clocks to larger data sets and increasing numbers of taxonomic groups (Benton and Ayala 2003; Kumar 2005; Donoghue and Benton 2007; Hedges and Kumar 2009). Molecular clocks are being applied not only to date species divergences where few fossils or geochemical data (e.g., biomarkers) exist but also for dating more recent events in evolution where a far larger amount of paleontological evidence exists to establish a temporal history of species (Hedges and Kumar 2003; Brocks and Pearson 2005). These molecular clock time estimates have been useful in highlighting links between species divergences and major events in Earth's evolution, patterns of parallel speciation/niche availability, and the relationship between times from fossils and molecules (e.g., Hedges et al. 1996; Tamura et al. 2004; Donoghue and Benton 2007).
Evaluation of divergence times produced by using molecular clock methods is frequently based on their comparisons with paleontological, geological, and geochemical record (e.g., Donoghue and Benton 2007; Kodner et al. 2008; Givnish et al. 2009). Molecular and nonmolecular time estimates do not always agree, and their differences have fuelled debates on possible biases inherent in both types of data and the methods of analyses (Ayala 1999; Smith and Peterson 2002; Graur and Martin 2004; Hedges and Kumar 2004; Reisz and Muller 2004; Blair and Hedges 2005; Pulquerio and Nichols 2007; Peterson et al. 2008). One reason for the observed differences between molecular- and fossil-based divergence times is that the latter often concerns the morphological modification of a descendant lineage compared with the former, which dates the genetic divergence immediately following the speciation event (e.g., Hedges et al. 1996; Steiper and Young 2008). However, it is rarely possible to resolve large differences between molecular and nonmolecular time estimates in this way. A case in point is the timing of origin of animal phyla recorded in the Cambrian explosion where molecular clock estimates for divergences are often much older than paleontological estimates (Wray et al. 1996; Smith and Peterson 2002; Hedges et al. 2004; Blair and Hedges 2005; Peterson et al. 2008).
In order to assess the utility of molecular clock estimates, many investigators compare times obtained using alternative calibrations, different software packages, and alternative taxa and gene samplings (Hedges et al. 2004; Perez-Losada et al. 2004; Ho et al. 2005; Linder et al. 2005; Hug and Roger 2007; Lepage et al. 2007; Rutschmann et al. 2007; Brown et al. 2008; Poux et al. 2008). Such investigations provide information on the robustness of estimated time to the data subsamples and evolutionary assumptions, but they do not provide a systematic evaluation of the accuracy and bias of the time estimates and associated credibility intervals (CrIs).
Different clock methods may produce disparate times due to their implicit handling of the rate heterogeneity across lineages, the number and position of calibrations, and the set of genes analyzed (Bromham and Penny 2003; Ho and Larson 2006; Pulquerio and Nichols 2007). The interactions among these factors in any empirical data set hinder attempts to systematically assess their effect on the time estimates. For example, using two empirical data sets, Hug and Roger (2007) investigated the effect of the position of a single calibration point on the estimate of divergence time of the deepest node in their phylogeny. The estimates are found to depend on the position of the calibration, the relaxed-clock method used, and the data set analyzed. In contrast, Hedges et al. (2004) have reported similar estimates across multiple methods when data set and calibration times were held constant throughout the analysis.
In the absence of the knowledge of the true divergence times, which is frequently the case in empirical studies, it is not possible to assess which combination of clock method and data subset has produced the best estimate. For this reason, computer simulations are employed to directly compare the estimates of divergence times with the simulated (true) times. For example, Bayesian and maximum likelihood methods have been reported to recover the true rate (and, thus, time) when the model they assume coincides with that used for simulating sequences. CrIs generated by taking into account different sources of uncertainty (e.g., number of genes, imprecision of the calibration, rate variation) are also found to contain the true time in 95% of the simulations (Sanderson 1997; Kishino et al. 2001; Ho et al. 2005; Kumar et al. 2005; Drummond et al. 2006). However, an assessment of the robustness of different relaxed-clock methods under autocorrelated rate (AR) and random rate (RR) changes remains unexplored, even though these methods are frequently used to estimate divergence times without knowing the actual model of evolutionary change. Furthermore, an evaluation of the effects of the number of calibrations on the time estimation is lacking.
Therefore, we have conducted a computer simulation study to examine the absolute and relative performance of molecular clock methods when the evolutionary rate varies among lineages under different models of rate change, and the phylogeny and calibration points are known perfectly. We have simulated a large number of sequence alignments based on a set of 448 naturally derived substitution rate and pattern parameters, including the evolutionary rate, sequence length, and G + C content (Rosenberg and Kumar 2003). In producing these alignments, we modeled evolutionary rates among lineages such that their change was AR (ancestor and descendant rates were correlated) or RR. We evaluated two relaxed-clock methods—the method of Thorne and Kishino (2002; MultiDivTime) and the method of Drummond and Rambaut (2007; BEAST)—as they are primarily designed to model such evolutionary rate changes among lineages in the estimation procedure. We focused on the estimation of the absolute times using single and multiple genes, where one or more true calibration points were applied. We also explored the frequency with which the CrIs reported by these two methods include the true time because statistical tests of hypotheses require their use.
Materials and Methods
We simulated gene alignments starting from naturally derived ranges of parameters. These were drawn from 448 orthologous mammalian sequences, including the number of sites (range 147–9,359 sites), the evolutionary rate (range 0.47–3.95 substitutions/site per billion years), the GC content (range 31–93%), and the transition/transversion ratio (range 2.2–26.6) (Rosenberg and Kumar 2003). DNA simulations were carried out using the SeqGen program (Rambaut and Grassly 1997) under the HKY model of nucleotide substitution (Hasegawa et al. 1985). A phylogeny consisting of 14 species with node divergence times inspired by those known for groups of mammals was used (fig. 1). Even though the naturally derived parameters and the model phylogeny were based on the mammalian taxa, we expect the simulation results to be applicable to a wide range of genes and phylogenies, because of the diversity of parameter sets considered.
FIG. 1.
The model timetree used in computer simulations. The internal nodes are labeled a–l, with node a being the ingroup root node. Extant taxa are A through M plus the outgroup (out). True times for each internal node are as follows: a: 173 Myr, b: 92 Myr, c: 90 Myr, d: 81 Myr, e: 85 Myr, f: 65 Myr, g: 74 Myr, h: 46 Myr, i: 20 Myr, j: 23 Myr, k: 10 Myr, and l: 5 Myr.
In DNA sequence simulation, evolutionary rate (and thus the amount of change) on an evolutionary lineage (branch) of the tree was generated by assuming that the rate variation was autocorrelated in ancestral and descendant lineages (AR) or varied independently (RR). In AR simulations, the mean autocorrelation was set to 1 (ν in MultiDivTime) following Thorne and Kishino (2002). In RR simulations, the randomized evolutionary rate for each branch was drawn from a uniform distribution over the interval from 0.5r to 1.5r, where r is the nominal rate for the entire gene. For short sequences and slow evolutionary rates, it is possible that identical simulated sequences are produced for closely related species. In AR simulations, this happened for only 3% (15 of the 448 genes) of the genes where at least two taxa had identical sequences. In the RR simulations, all genes had different sequences across taxa. Exclusion of time estimates from these replicates did not alter our conclusions or the results presented.
Simulated sequences were analyzed in MultiDivTime and BEAST programs (Thorne and Kishino 2002; Drummond and Rambaut 2007). We estimated branch lengths under the F84 model using the Estbranches program and generated the maximum likelihood estimates of the shape parameter of the Gamma distribution of evolutionary rates among sites and the transition/transversion ratio using the PAML program (Nei et al. 2001; Yang 2007). This procedure was inspired by the common analysis practices of biologists, especially the use of Gamma distribution to model rate variation, even though the sequences were simulated with uniform substitution rate among sites. (Results with and without Gamma distribution of rates are expected to be identical because the estimate of shape parameter was more than two for every data set and exceeded ten for >95% of the simulated data sets.)
For MultiDivTime program, the time estimation process was completed after 10,000 samplings of the Markov chain, a sampling frequency of 100, and a burn-in of 100,000. The mean of the prior distribution for the ingroup root time (rttm) was set at 173 Myr. Other parameters such as the mean of the prior distribution for the rate of evolution (rtrate) and the mean of the prior distribution for the autocorrelation parameter (brownmean) were calculated specifically for each alignment using the branch lengths information from the Estbranches program and the ingroup root prior; rtrate was given by the median of the root-to-tip branch lengths divided by rttm, whereas brownmean was obtained dividing a constant value of 1.5 by rttm, as suggested by the author. Standard deviations of these parameters (rtrateSD and brownSD) were set equal to the parameters themselves, which is a common practice.
In addition to analyzing individual gene alignment separately, we generated 100 concatenated data sets of ten genes each from AR and RR alignments. These concatenated subsets were analyzed both in a nonpartitioned (NP) and a partitioned (P) fashion. We also generated five concatenated alignments for AR and RR simulated sequences with 20, 30, 60, and 100 genes each.
BEAST analyses were conducted by using the model topology (fig. 1) under the HKY model plus gamma (four categories) and a lognormal relaxed-clock model. The number of generations necessary to reach convergence and effective sample sizes above 200 varied depending on the data set, and thus, burn-in and sampling frequency were adjusted accordingly. Even with extensive computing resources available to us, it was not possible to complete BEAST analyses for many data sets either due to excessive time required or because of the failure of BEAST calculations to converge. Finally, BEAST produced results for 68 AR and 83 RR ten-gene concatenations.
In these molecular clock analyses, we used different calibration sets to test the effect of single versus multiple as well as shallow versus deep calibrations. The single calibrations chosen were nodes b (92 Myr), h (46 Myr), i (20 Myr), and k (10 Myr) in figure 1. Pairs of calibrations used were h and k, d (81 Myr) and h, and j (23 Myr) and k. Because the uncertainty in the calibration points was not the primary topic of interest in this study, all calibrations were provided as perfectly known, which necessitated the use of ±1 Myr uncertainty in MultiDivTime and a uniform distribution of ±1 Myr around the true time in BEAST.
Results
We begin with results from the MultiDivTime analysis of individual alignments. Because MultiDivTime models autocorrelated changes in evolutionary rates over lineages, we first used alignments generated using the AR. The distributions of the estimated times (448 estimates for each node) show a strong central tendency and are generally symmetrical (fig. 2). Distributions resulting from the use of single and double calibrations are similar in shape. The standard deviation of these distributions over all nodes is 19% (12–41%) of the mean for estimates based on single calibrations, which is slightly larger than that for two calibrations where the standard deviation is 14% (7–28%) of the mean. Therefore, the use of an additional (perfect) calibration point leads to more precise estimates (smaller standard deviation), as expected. Regardless of the number of calibrations used, the dispersion of the time estimates around the true time depends on the position of the calibration relative to the estimated node. That is, smaller dispersions are associated with nodes closer to the calibration points. Nonetheless, the central tendencies are not shifted away from the true time even for the highly dispersed cases.
FIG. 2.
Distributions of single gene time estimates obtained from MultiDivTime analysis of autocorrelated sequences. Results for four nodes are shown for a subset of single and double calibrations. Vertical dotted lines mark the true time, with the arrows indicating the mean of the inferred time distributions Cal, calibration.
Performance of CrIs
MultiDivTime produces 95% CrIs that convey the level of uncertainty in the estimated times. Overall, double calibrations produce ∼20% narrower CrIs, that is, times are estimated with a greater precision using multiple calibrations (fig. 3A). Another important measure of success of a statistical estimator is the frequency at which the 95% CrIs contains the true time in all the replicates; it should be at least 95% (i.e., less than 5% failure rate). This requirement was fulfilled for most nodes in single and double calibrations, except for the three deep nodes in the phylogeny (a, c, and e; fig. 1). Their CrIs did not contain the true time in >5% replicates for some calibration sets (fig. 3B and supplementary table S1, Supplementary Material online). The node with the highest average failure rate (10%) is the deepest one (node a), probably because it is separated by a long internal branch from the rest of the tree. These results indicate that single gene time estimates and their CrIs may be misleading (conservative in rejecting the null hypothesis) even when perfect calibration times are used, and the distribution of lineage rate change is modeled correctly in the estimation procedure.
FIG. 3.
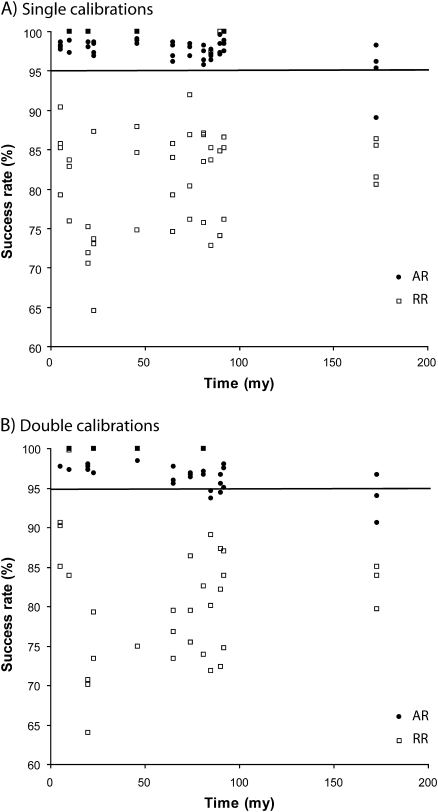
Comparison of the size of the CrIs from single and double calibrations (A) and the percent cases in which the CrI contained the true time (B). All results are for the MultiDivTime analysis of autocorrelated sequences. In panel A, all values for each node are averages over 3,136 replicates and calibration points. In panel B, for each node, there are seven success rates (percentage of replicates for which the CrI contains the true time), which correspond to seven calibration sets and 448 replicates. In some cases, less than seven results are visible because of overlapping points. The horizontal line marks the 95% threshold, which is the expected value because we constructed 95% CrIs. All success rate values below 95% are circled with letters referring to nodes in Fig. 1.
In practical data analysis, however, MultiDivTime is routinely applied even if there is no guarantee that the lineage rates are autocorrelated. Therefore, we evaluated the performance of MultiDivTime for sequence alignments generated under a RR model, where lineages could deviate from the average rate by ±50% under a uniform distribution of rates. The CrI failure rates increase significantly when single calibrations are used (up to ∼35%; fig. 4A and supplementary table S2, Supplementary Material online). The use of double calibrations does not alleviate the problem (fig. 4B and supplementary table S2, Supplementary Material online). Thus, the use of additional calibrations is not helpful when the model of lineage rate change is misspecified. The only exceptions to the results mentioned above are time estimates for nodes close to a single deep calibration point (nodes c and e), where the failure rates are 0% and 3%, respectively. This performance is likely a result of the proximity of these nodes to the deep calibration point (node b). Therefore, MultiDivTime produces biased estimates of CrIs when the underlying assumption of autocorrelated lineage rates is violated.
FIG. 4.
The relative success rates of CrIs in containing the true time when using MultiDivTime for the analysis of AR and RR simulated sequences using single (A) and double (B) calibrations . The horizontal line marks the 95% threshold.
Multigene Estimates of Species Divergence Time
In the above, we considered the CrIs produced in single gene analyses, along with the distribution of individual time estimates. We next examined how well the mean and other measures of the central values of distributions of individual time estimates over genes coincided with the true time. This is useful because multiple individual gene times have been used by many investigators to generate species divergence times (e.g., Wray et al. 1996; Kumar and Hedges 1998; Nei et al. 2001). In this case, the simple mean, mode, or geometric mean of the distribution of individual gene time estimates is used to infer the time of species divergences (e.g., Morrison 2008). Simple arithmetic means of the gene time estimates from MultiDivTime are close to the true time (±10%) for a majority of the nodes for both AR and RR simulations. However, we find that the inferred times for the shallowest nodes can depart considerably from the true time (nodes i, j, k, and l) and that the degree of departure depends on the position of the calibration. Divergence times for these nodes were overestimated in MultiDivTime up to 29% for AR and 41% for RR alignments when the calibration point was outside of the cluster formed by terminal taxa I–L.
The poor performance in timing the ages of shallow nodes cannot be remedied by using an additional calibration point, as single and double calibration analyses produced very similar results. We also examined whether the use of a geometric mean may improve the inference because the time distributions are never strictly symmetrical (e.g., Morrison 2008). The geometric means of the gene time estimates are on average 5–7% different from the true times for AR and RR, respectively, which is slightly better than the arithmetic mean. The problem of overestimation of times for the shallow nodes is reduced, but not completely resolved, by the use of geometric means (12% and 18% for AR and RR alignments compared with 17% and 28% for arithmetic mean).
Instead, the normalized difference between the estimate and true times for shallow nodes is smaller than that for the deeper nodes when at least one calibration point (in both the single or the double calibration analyses) was within the shallow node cluster (taxa I–L; supplementary figs. S1 and S2, Supplementary Material online). The largest departures from the true time are seen for the deepest node when only shallow calibrations are utilized. Therefore, the use of distant calibrations is expected to yield poor time estimates even when using a large number of genes.
In addition to the mean time estimates, confidence intervals can be obtained from the distribution of gene times (in our simulations 448 genes for each node) such that the lower and upper boundaries of the interval correspond to the 2.5th and 97.5th percentile of this distribution, respectively. We calculated these confidence intervals directly from the observed distribution of individual gene times for each node because multigene times are not always normally distributed.
These multigene confidence intervals are very wide and include the true time for all nodes in both AR and RR cases. On the contrary, the confidence intervals calculated based on the standard error of the mean (mean ±1.96 × SEM) are too conservative (e.g., Kumar and Hedges 1998), and provide overly narrow intervals that fail to include the true time for a majority of nodes in both AR and RR simulated data sets (see supplementary tables S3 and S4, Supplementary Material online).
Instead of estimating times from a distribution of individual gene estimates, most investigators now create concatenations of the gene alignments and estimate divergence times with or without retaining the information on the individual gene boundaries. In order to examine the accuracy of MultiDivTime in analyzing such data, we constructed 100 concatenations of ten randomly selected gene alignments for AR and RR simulations separately. First, we carried out the nonpartitioned analysis for the ten gene alignments, treating them as a single supergene. We again used single and double calibration points in these analyses.
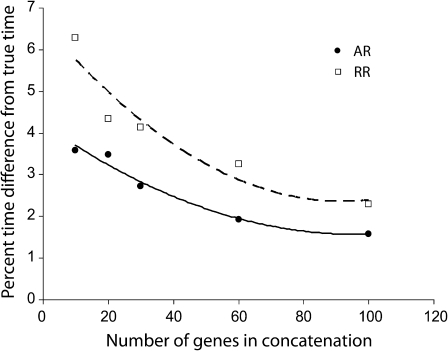
As expected, multigene alignments produce better estimates than the single gene alignments generated in both AR and RR simulations, and these concatenation time estimates have smaller dispersions around the true time (fig. 5). Central tendencies of time distributions for AR and RR are similar to each other, although RR distributions are wider (see supplementary fig. S3, Supplementary Material online). Increasingly larger numbers of concatenated genes result in a progressive improvement in the point estimate of time. For example, 30-gene concatenations simulated under AR conditions yield time estimates that were, on average, 24% closer to the true time than those from the 10-gene concatenations (fig. 6). Increasing the number of genes initially leads to a rapid increase in the accuracy of time estimates, but this increase becomes slower and plateaus after 60 genes (see also Kumar et al. 2005).
FIG. 5.
Increased accuracy of times inferred from ten-gene concatenations (thick line) compared with those from single genes (thin line). A total of 100 ten-gene concatenations and 448 single genes were analyzed. RR sequences showed patterns similar to the ARs (presented here). The percent time difference is given by ([estimated time − true time]/true time) and is estimated for each replicate independently. MultiDivTime results from single genes (thinner line) and from concatenations (bolded line) are shown for AR simulations. For a comparison, see figure 2 for the distribution of actual time estimates for the nodes and calibrations for which results are shown here.
FIG. 6.
The effect of increasing number of genes on the difference between estimated and true times. Each data point is the average percent time difference obtained for all nodes using double calibrations with MultiDivTime. Filled circles, autocorrelated simulated sequences; empty squares, RR simulated sequences. A second-order polynomial fits the data (R2 = 0.97 for AR sequences and 0.90 for RR sequences).
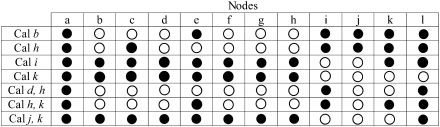
Next, we examined the frequency with which the CrIs reported by MultiDivTime for each multigene alignment contained the true time. We found that the multigene concatenations produce much narrower CrIs than those from single gene estimates; multigene CrIs are less than half as wide. For AR simulated data sets, multigene CrIs contain the true time in 95% of the cases, which is expected because the simulation and estimation models match. However, the failing 5% CrIs are not equally distributed among nodes and calibration sets. All nodes experience a failure rate above 5% under at least one calibration condition with the most extreme case being the deepest node a that fails under all calibrations used. Similarly, none of the calibrations applied gives CrIs wide enough to include the true time in a significant percentage of the simulations for all nodes. The worst cases are those using the calibration duo j,k or a single calibration i, which exhibit the highest failure rates with only 10% of the nodes experiencing failure rates less than 5% (fig. 7).
FIG. 7.
Nodes and calibration combinations yielding CrIs with success rates ≥95% (open circles) and <95% (filled circles). All analyses were conducted by using MultiDivTime on autocorrelated sequences. Cal, calibration.
However, there is no clear relationship between the position of the calibration points and the success rate of CrIs, as both shallow and deep calibrations produce many nodes with low success rates. These results are consistent with the observations of Hug and Roger (2007), who also did not find discernable correlation between time estimates and the depth of the calibration. These failures cannot be fully explained by biases that may be introduced when concatenating sequence alignments that have evolved with vastly different rates and patterns, as largely similar results are obtained when the individual gene boundaries are retained in the analyses such that the evolutionary parameters are estimated specifically for each gene (partitioned analysis). However, for one calibration condition (node k), there is significant improvement with all nodes having failure rates below 5% (see supplementary fig. S4, Supplementary Material online).
In the analysis of RR multigene concatenations (nonpartitioned analysis), the success rate is significantly worse for MultiDivTime CrIs because the divergence times show wider distributions and the CrIs are not wide enough to include the true times. Overall, more than 20% of the CrIs failed to contain the true time, with the CrIs for a larger majority of nodes (93%) failing to contain the true time in greater than 5% of data sets. Again, the partitioned analysis did not improve the situation. Instead, the failure rates became higher. CrI failure rates were greater than 5% for 98% of the nodes compared with 91% for the nonpartitioned analysis. This problem is caused by decreases in the size of the CrIs (∼10%) in the partitioned analysis; this is unexpected, as the partitioned analysis should produce wider CrIs because it involves the estimation of greater number of parameters compared with the nonpartitioned analysis.
On the other hand, an increase of the number of genes in the multigene concatenation data sets improves the time estimates, as the 30-gene RR concatenations produced estimates that were 34% closer to the true time than those from the 10-gene concatenations. Furthermore, the failure rates of CrIs decreased significantly as well (66% compared with 93% for 30-gene vs. 10-gene concatenations; supplementary fig. S5, Supplementary Material online). Therefore, it is better to use multigene alignments in relaxed-clock analyses (see references in Hedges and Kumar 2009).
The higher failure rates observed for RR simulations in MultiDivTime analysis are due to the violation of the primary assumption of ARs in the MultiDivTime software. BEAST does not make this assumption, so we tested the performance of BEAST for RR data and compared it with the performance of MultiDivTime. We expected that the use of BEAST would produce narrower time estimate distributions and decrease the failure rate of the CrIs. Indeed, the use of BEAST leads to a significant improvement (table 1 and supplementary figs. S6–S8, Supplementary Material online). In particular, 96% of the CrIs contain the true time for RR multigene concatenations compared with only 78% for MultiDivTime when comparing estimates obtained under the same conditions. Moreover, the CrIs contain the true time in 95% of the data sets for 80% of the nodes (table 1). This improved performance could be produced by either wider CrIs or CrIs more frequently distributed around the true time. We found that, on average, the width of the CrIs does not differ between BEAST and MultiDivTime, but rather, the CrIs are shifted toward the true time. Therefore, the use of BEAST that employs the correct lineage rate model (RR) leads to better results than those obtained by using MultiDivTime.
Table 1.
Percentage of Nodes with CrI Success Rate above 95% in MDT and BEAST.
| ARs (%) |
RRs (%) |
|||||
| Calibration | MDT | BEAST | cCrI | MDT | BEAST | cCrI |
| d, h | 50 | 50 | 80 | 20 | 90 | 80 |
| h, k | 60 | 20 | 90 | 0 | 70 | 90 |
| j, k | 10 | 10 | 20 | 0 | 80 | 80 |
NOTE.—MDT, MultiDivTime. CrIs are estimated using the concatenated alignments. All calibration nodes are excluded from the total number of nodes considered because they were constrained around the true time. Results from the cCrIs are also shown.
This led us to examine the performance of BEAST for the analysis of the AR data sets that violate its assumption of uncorrelated lineage rate changes. BEAST performed much more poorly for AR data compared with the RR data. For the AR data, the time distributions are wider for BEAST compared with MultiDivTime, and the BEAST CrIs contain the true time in 95% data sets only for 27% of the nodes on average, which is much smaller than that seen for MultiDivTime (table 1 and supplementary figs. S9–S11, Supplementary Material online). The reliability of CrIs by BEAST decreased despite the CrI sizes being, on average, 10% larger for AR sequences compared with RR. For BEAST estimates, like those for MultiDivTime, individual nodes with the highest failure rates are those more distant from the calibrations. Therefore, the highest percentage of nodes with unsuccessful CrIs is given by the use of shallow calibrations (j and k). Overall, BEAST produces poor results when the sequences have evolved with ARs.
Concatenated gene sets (average length ∼14,000 sites) produce better time estimates, and smaller CrIs, than single genes (average length ∼1,350 sites). In addition to the reduction in sampling variance associated with the use of more data, the rate equalization among lineages is an additional possible factor in the improved accuracy. The latter may result from an averaging effect of evolutionary rates across lineages when individual genes are concatenated because each autocorrelated gene alignment was generated independently of other genes in our simulations. This means that different sets of lineages evolved slower or faster than average in different genes, which, when concatenated, would likely reduce rate differences across lineages. An inspection of multigene concatenation phylogenies with branch lengths confirmed this effect.
This prompted us to simulate an alternative scenario where evolutionary rate deviations among segments were synchronized, which produces a long alignment with all ∼10,000 bp sites experiencing the same autocorrelation. This scenario simulates genome-wide biases in rate differences. Application of MultiDivTime to this data set represents a best case scenario: the lineage rate model used in estimation coincides with the simulations scheme. We then compared the time estimates obtained using these simulated sequences with those obtained from comparable length multigene concatenations. The individual time estimates are similar between the two results: on average, inferred times differ from the true times 4.5% and 4.9% for multigene concatenation and the genome-wide scenario (AR simulations) (see supplementary fig. S12, Supplementary Material online). However, the CrIs for the rate-synchronized simulations are >50% wider than those obtained from the ten-gene concatenations. Therefore, significant estimation variance is introduced by the need to account for autocorrelation of lineage rates.
Finally, we evaluated the general perception that similar results obtained from different clock methods are more reliable (i.e., closer to the true divergence time). We examined the similarity of time estimates for the same data set from MultiDivTime and BEAST and quantified the difference between the true and the estimated times, pooling all results where MultiDivTime and BEAST estimated times do not differ by more than 5%. For these cases, we found that the estimated times are much more similar to the true time (32% in AR and 27% in RR data sets) compared with those for all other replicates. Furthermore, their CrIs are more likely to include the true time. This provides initial support for the common practice of arguing for higher reliability of estimates if multiple ones produce similar results (e.g., Hedges et al. 2004; Perez-Losada et al. 2004; Drummond et al. 2006).
Discussion
We have evaluated and compared the performances of MultiDivTime and BEAST methods in timing evolutionary divergences when sequences have evolved with variable rates over time. We found that when the underlying assumptions of the method employed are met (e.g., ARs for MultiDivTime and highly variable yet uncorrelated rates for BEAST), the relaxed-clock methods produce increasingly better point estimates of time with larger numbers of genes. We have also shown that concatenated gene sets produce better time estimates, and smaller CrIs, than single genes. This is due to a reduction in sampling variance and the rate homogenization among lineages when rate variable sequences are concatenated.
We have also examined the relative usefulness of single and double calibrations when they are known with certainty. The means of the single-gene times as well as the failure rates of CrIs are similar for one and two calibration cases (fig. 2). The CrIs are narrower in the latter (by 24%; fig. 3, panel A), but they do not ameliorate the high CrI failure rates seen in some cases (Hedges and Kumar 2003; Near et al. 2005; Hug and Roger 2007). This lack of difference in results obtained using single and double calibrations may be attributed to the fact that we have used perfect calibration times, which is rarely the case in empirical data analysis (Benton et al. 2009). In a follow-up study, we plan to quantify the improvements afforded by the use of multiple calibrations when they are not known perfectly, and we will also examine whether it is preferable to use one (or a few) highly reliable calibration point(s) rather than many minimum calibration points.
We do, however, find that the phylogenetic depths and locations of the calibration points affect the time estimates significantly. The shallowest nodes (nodes j, k, and l) in the tree are grossly overestimated if the calibration point does not come from the group to which these nodes belong (taxa I–L). Shallow nodes belonging to different phylogenetic clusters tend to be separated by large branch lengths, as is the case in the model tree we used. This causes larger differences between the estimated and the true times due to the extent of extrapolation needed. In the same way, the use of shallow calibrations to estimate the deepest nodes leads to underestimation of older times. Therefore, time estimates far from the calibration node are expected to be unreliable, regardless of the accuracy of the calibration.
We then focused on those nodes that are closest to the calibrations used and evaluated whether the proximity of nodes and calibration points had an effect on the accuracy of the time estimates. We divided the calibration/estimated nodes into three groups: 1) the estimated node is the direct descendant of the calibration, 2) the estimated node is the direct ancestor of the calibration, and 3) the calibration and estimated nodes have a sister group relationship. We found no significant difference among these three types of calibration/estimated nodes. This suggests that the relative positions of calibration and estimated nodes do not affect the time estimations as long as the two nodes are closely related.
Overall, however, CrIs reported by relaxed-clock methods for multigene data sets are overly narrow (conservative). They fail to contain the true time in greater than 5% data sets. These failure rates become uncomfortably large when BEAST is used to analyze sequences that have evolved with ARs, and when MultiDivTime is used to analyze sequences that are a product of extensive, but uncorrelated, evolutionary rate changes over time. Therefore, the selection of the appropriate relaxed-clock method is important in generating correct time estimates and CrIs. As confirmed in this study, the estimated times are closer to the true times when the two methods produce similar time estimates regardless of the rate variation model followed by the sequences.
In the real data analysis, it is generally difficult to know the actual distribution underlying the changes in evolutionary rates among lineages. One approach is to evaluate Bayes factors for sequences evolving under different models (e.g., one that assumes autocorrelation and one that assumes uncorrelated rate changes). But this and other approaches are known to make contrasting predictions when applied to empirical data depending on the extent of taxonomic sampling, gene selection, and the taxonomic level considered (Drummond et al. 2006; Lepage et al. 2007; Brown et al. 2008; Ho 2009). Though BEAST does provide a means to estimate the model of evolutionary rate variation, it is not known to be very powerful (Drummond et al. 2006). Nonetheless, we examined our AR concatenated alignments, categorizing them as “autocorrelated” or “uncorrelated,” according to the 95% CrI of the covariance parameter (i.e., zero covariance indicates uncorrelation; a 95% CrI that includes zero does not allow to reject the hypothesis of uncorrelation). In all cases, BEAST did not detect significant autocorrelation (i.e., all covariance CrIs included zero), which confirms its powerlessness.
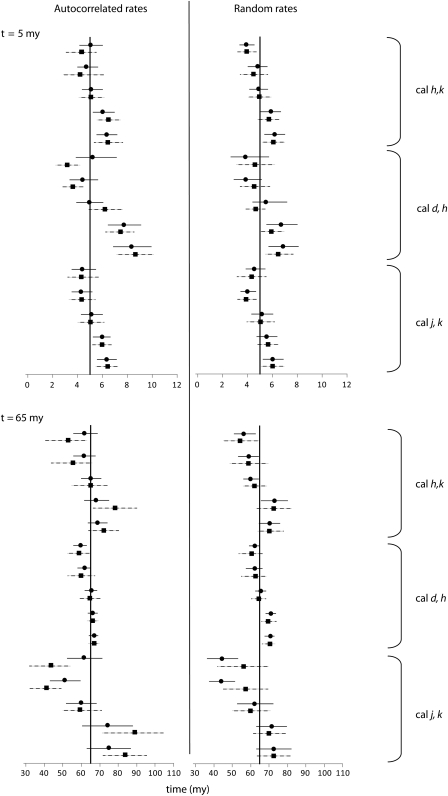
A simple strategy to get around this problem is revealed when one examines the CrIs reported by BEAST and MultiDivTime simultaneously for the same data set (fig. 8). It is clear that when the lineage rate model assumption is violated, relaxed-clock methods would produce biased CrIs. However, if AR and RR represent two extremes, then CrI from at least one of the two programs (MultiDivTime and BEAST) will be appropriate. It is therefore possible to reduce the CrI failure rates significantly by building a composite CrI derived from the two CrIs. In the composite credibility interval (cCrI), the lower bound is given by cCrIlower = minimum(BEAST-CrIlower, MultiDivTime-CrIlower) and the upper bound is given by cCrIupper = maximum(BEAST-CrIupper, MultiDivTime-CrIupper). By definition, cCrIs are wider than those from BEAST and the MultiDivTime alone. cCrIs are, on average, 12–37% wider than MultiDivTime and BEAST CrIs under AR and RR conditions. The largest increase in width is obtained compared with MultiDivTime CrIs with 37% (2–73%) wider intervals for AR simulations and 27% (2–53%) for RR simulations. Compared with BEAST CrIs, the increase of cCrIs width is 14% (0–35%) for AR and 12% (0–57%) for RR sequences.
FIG. 8.
A comparison of time estimates and CrIs produced by MultiDivTime (filled circles, with solid line) and BEAST (filled squares, with dotted line), for example, nodes. Each point (black symbol) and the associated 95% CrI are shown for five 10-gene concatenation data sets when using different sets of calibrations.
Application of the cCrIs strategy decreases the failure rate of CrIs for all data sets, with an overall success rate equal to 95% compared with the success rate of 88% when an investigator applies only one of the two methods with equal probability. The success rate for individual nodes also improves the range from 80% to 100% for both rate variation cases. The only exception is the autocorrelated data set in which both BEAST and MultiDivTime perform poorly for which only a slight improvement is noted with the composite CrIs (calibrations j and k) (table 1). Even in cases where cCrI failed to include the true time, this was, on average, no more than 10% away from the upper or the lower bound. These results are much better than those observed for cases where BEAST or MultiDivTime CrIs were used. Therefore, we recommend that cCrI be used to convey the uncertainty in time estimates, especially because the distribution of lineage rate is likely to be a mixture of correlated and uncorrelated rates. Furthermore, we recommend that as many genes as possible be used to make cCrIs narrower, which will improve the precision of the inferred time estimates.
Supplementary Material
Supplementary tables S1–S4 and figures S1–S12 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org).
Supplementary Material
Acknowledgments
We thank Joseph Felsenstein, Claudia Acquisti, and Antonio Marco for helpful comments or discussions and Naoko Takezaki and two anonymous reviewers for comments on a previous version of this manuscript. We are thankful to Kristi Garboushian for text editing support and Revak Raj for assistance in data preparation and analysis execution. Financial support for this work was provided in part by the National Institute of Health to S. K. (HG002096) and A. Escalante (GM080586), and the National Science Foundation to S.B.H. and S.K. (DBI-0850013).
References
- Ayala FJ. Molecular clock mirages. Bioessays. 1999;21:71–75. doi: 10.1002/(SICI)1521-1878(199901)21:1<71::AID-BIES9>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Benton MJ, Ayala FJ. Dating the tree of life. Science. 2003;300:1698–1700. doi: 10.1126/science.1077795. [DOI] [PubMed] [Google Scholar]
- Benton MJ, Donoghue PCJ, Asher RJ. Calibrating and constraining the molecular clock. In: Hedges SB, Kumar S, editors. The timetree of life. New York: Oxford University Press; 2009. [Google Scholar]
- Blair JE, Hedges SB. Molecular clocks do not support the Cambrian explosion. Mol Biol Evol. 2005;22:387–390. doi: 10.1093/molbev/msi039. [DOI] [PubMed] [Google Scholar]
- Brocks JJ, Pearson A. Building the biomarker tree of life. Rev Mineral Geochem. 2005;59:233–258. [Google Scholar]
- Bromham L, Penny D. The modern molecular clock. Nat Rev Genet. 2003;4:216–224. doi: 10.1038/nrg1020. [DOI] [PubMed] [Google Scholar]
- Brown JW, Rest JS, Garcia-Moreno J, Sorenson MD, Mindell DP. Strong mitochondrial DNA support for a cretaceous origin of modern avian lineages. BMC Biol. 2008;6:6. doi: 10.1186/1741-7007-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue PCJ, Benton MJ. Rocks and clocks: calibrating the tree of life using fossils and molecules. Trends Ecol Evol. 2007;22:424–431. doi: 10.1016/j.tree.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. Beast: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givnish TJ, Millam KC, Mast AR, Paterson TB, Theim TJ, Hipp AL, Henss JM, Smith JF, Wood KR, Sytsma KJ. Origin, adaptive radiation and diversification of the Hawaiian lobeliads (asterales: Campanulaceae) Proc R Soc B. 2009;276:407–416. doi: 10.1098/rspb.2008.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur D, Martin W. Reading the entrails of chickens: molecular timescales of evolution and the illusion of precision. Trends Genet. 2004;20:80–86. doi: 10.1016/j.tig.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano TA. Dating of the human ape splitting by a molecular clock of mitochondrial-DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Blair JE, Venturi ML, Shoe JL. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol. 2004;4:2. doi: 10.1186/1471-2148-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Kumar S. Genomic clocks and evolutionary timescales. Trends Genet. 2003;19:200–206. doi: 10.1016/S0168-9525(03)00053-2. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Kumar S. Precision of molecular time estimates. Trends Genet. 2004;20:242–247. doi: 10.1016/j.tig.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Kumar S. Discovering the timetree of life. In: Hedges SB, Kumar S, editors. The timetree of life. New York: Oxford University Press; 2009. [Google Scholar]
- Hedges SB, Parker PH, Sibley CG, Kumar S. Continental breakup and the ordinal diversification of birds and mammals. Nature. 1996;381:226–229. doi: 10.1038/381226a0. [DOI] [PubMed] [Google Scholar]
- Ho SYW. An examination of phylogenetic models of substitution rate variation among lineages. Biology Lett. 2009;5:421–424. doi: 10.1098/rsbl.2008.0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SYW, Larson G. Molecular clocks: when times are a-changin’. Trends Genet. 2006;22:79–83. doi: 10.1016/j.tig.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Ho SYW, Phillips MJ, Drummond AJ, Cooper A. Accuracy of rate estimation using relaxed-clock models with a critical focus on the early metazoan radiation. Mol Biol Evol. 2005;22:1355–1363. doi: 10.1093/molbev/msi125. [DOI] [PubMed] [Google Scholar]
- Hug LA, Roger AJ. The impact of fossils and taxon sampling on ancient molecular dating analyses. Mol Biol Evol. 2007;24:1889–1897. doi: 10.1093/molbev/msm115. [DOI] [PubMed] [Google Scholar]
- Kishino H, Thorne JL, Bruno WJ. Performance of a divergence time estimation method under a probabilistic model of rate evolution. Mol Biol Evol. 2001;18:352–361. doi: 10.1093/oxfordjournals.molbev.a003811. [DOI] [PubMed] [Google Scholar]
- Kodner RB, Summons RE, Pearson A, King N, Knoll AH. Sterols in a unicellular relative of the metazoans. Proc Natl Acad Sci U S A. 2008;105:9897–9902. doi: 10.1073/pnas.0803975105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. Molecular clocks: four decades of evolution. Nat Rev Genet. 2005;6:654–662. doi: 10.1038/nrg1659. [DOI] [PubMed] [Google Scholar]
- Kumar S, Filipski A, Swarna V, Walker A, Hedges SB. Placing confidence limits on the molecular age of the human-chimp divergence. Proc Natl Acad Sci U S A. 2005;102:18842–18847. doi: 10.1073/pnas.0509585102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Lepage T, Bryant D, Philippe H, Lartillot N. A general comparison of relaxed molecular clock models. Mol Biol Evol. 2007;24:2669–2680. doi: 10.1093/molbev/msm193. [DOI] [PubMed] [Google Scholar]
- Linder HP, Hardy CR, Rutschmann F. Taxon sampling effects in molecular clock dating: an example from the African restionaceae. Mol Phylogenet Evol. 2005;35:569–582. doi: 10.1016/j.ympev.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Morrison DA. How to summarize estimates of ancestral divergence times. Evol Bioinform. 2008;4:75–95. doi: 10.4137/ebo.s545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, Meylan PA, Shaffer HB. Assessing concordance of fossil calibration points in molecular clock studies: an example using turtles. Am Nat. 2005;165:137–146. doi: 10.1086/427734. [DOI] [PubMed] [Google Scholar]
- Nei M, Xu P, Glazko G. Estimation of divergence times from multiprotein sequences for a few mammalian species and several distantly related organisms. Proc Natl Acad Sci U S A. 2001;98:2497–2502. doi: 10.1073/pnas.051611498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Losada M, Hoeg JT, Crandall KA. Unraveling the evolutionary radiation of the thoracican barnacles using molecular and morphological evidence: a comparison of several divergence time estimation approaches. Syst Biol. 2004;53:244–264. doi: 10.1080/10635150490423458. [DOI] [PubMed] [Google Scholar]
- Peterson KJ, Cotton JA, Gehling JG, Pisani D. The Ediacaran emergence of bilaterians: congruence between the genetic and the geological fossil records. Philos T R Soc B. 2008;363:1435–1443. doi: 10.1098/rstb.2007.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poux C, Madsen O, Glos J, de Jong WW, Vences M. Molecular phylogeny and divergence times of Malagasy tenrecs: influence of data partitioning and taxon sampling on dating analyses. BMC Evol Biol. 2008;8:102. doi: 10.1186/1471-2148-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulquerio MJF, Nichols RA. Dates from the molecular clock: how wrong can we be? Trends Ecol Evol. 2007;22:180–184. doi: 10.1016/j.tree.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Grassly NC. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic frees. Comput Appl Biosci. 1997;13:235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- Reisz RR, Muller J. Molecular timescales and the fossil record: a paleontological perspective. Trends Genet. 2004;20:237–241. doi: 10.1016/j.tig.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Rosenberg MS, Kumar S. Heterogeneity of nucleotide frequencies among evolutionary lineages and phylogenetic inference. Mol Biol Evol. 2003;20:610–621. doi: 10.1093/molbev/msg067. [DOI] [PubMed] [Google Scholar]
- Rutschmann F, Eriksson T, Abu Salim K, Conti E. Assessing calibration uncertainty in molecular dating: the assignment of fossils to alternative calibration points. Syst Biol. 2007;56:591–608. doi: 10.1080/10635150701491156. [DOI] [PubMed] [Google Scholar]
- Sanderson M. A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol Biol Evol. 1997;14:1218–1231. [Google Scholar]
- Smith AB, Peterson KJ. Dating the time of origin of major clades: molecular clocks and the fossil record. Annu Rev Earth Pl Sc. 2002;30:65–88. [Google Scholar]
- Steiper ME, Young NM. Timing primate evolution: lessons from the discordance between molecular and paleontological estimates. Evol Anthropol. 2008;17:179–188. [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- Thorne JL, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- Wray GA, Levinton JS, Shapiro LH. Molecular evidence for deep precambrian divergences among metazoan phyla. Science. 1996;274:568–573. [Google Scholar]
- Yang ZH. Paml 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.