Abstract
Living organisms have evolved intricate systems to harvest trace elements from the environment, to control their intracellular levels, and to ensure adequate delivery to the various organs and cellular compartments. Copper is one of these trace elements. It is at the same time essential for life but also highly toxic, not least because it facilitates the generation of reactive oxygen species. In mammals, copper uptake in the intestine and copper delivery into other organs are mediated by the copper importer Ctr1. Drosophila has three Ctr1 homologs: Ctr1A, Ctr1B, and Ctr1C. Earlier work has shown that Ctr1A is an essential gene that is ubiquitously expressed throughout development, whereas Ctr1B is responsible for efficient copper uptake in the intestine. Here, we characterize the function of Ctr1C and show that it functions as a copper importer in the male germline, specifically in maturing spermatocytes and mature sperm. We further demonstrate that loss of Ctr1C in a Ctr1B mutant background results in progressive loss of male fertility that can be rescued by copper supplementation to the food. These findings hint at a link between copper and male fertility, which might also explain the high Ctr1 expression in mature mammalian spermatozoa. In both mammals and Drosophila, the X chromosome is known to be inactivated in the male germline. In accordance with such a scenario, we provide evidence that in Drosophila, the autosomal Ctr1C gene originated as a retrogene copy of the X-linked Ctr1A, thus maintaining copper delivery during male spermatogenesis.
Keywords: Copper, Drosophila Genetics, Metabolism, Metals, Transport Metals, Copper Import, Ctr1, Ctr1C, Fertility, X Chromosome Inactivation
Introduction
Copper is an essential nutrient for all eukaryotic organisms. Several vital processes depend on the ability of this trace metal to undergo changes in redox state. Copper serves as a catalytic cofactor in redox enzymes in respiration (cytochrome c oxidase), protection from oxidative stress (superoxide dismutase), melanization (tyrosinase and laccase), neuropeptide and peptide hormone processing (peptidylglycine α-amidating mono-oxygenase), and others. Although being essential for eukaryotic life, copper can also be highly toxic; it can disrupt the function of proteins by ectopically binding to them, and it catalyzes the formation of reactive oxygen species via the so-called Fenton reaction (1). Thus, eukaryotic organisms have evolved intricate systems to regulate copper homeostasis, including high affinity copper uptake systems, copper trafficking proteins (metallochaperones), metal-scavenging proteins (metallothioneins), copper exporters (P-type ATPases), and transcriptional regulators (2–4).
Given the importance of copper, organisms have evolved efficient copper uptake, as well as storage systems, that allow them to survive periods of copper scarcity. Copper can be taken up via lower affinity, less specific cation transporters, such as divalent metal transporter 1 (DMT1) (5, 6). There is, however, a dedicated high affinity transport system that is conserved from yeast to humans: the Ctr family of copper importers, small proteins that assemble into homotrimeric membrane complexes, forming a pore that allows an ATP-independent transport of copper (7). In yeast, three Ctr proteins are known. Two of these, yCtr1 and yCtr3, have been shown to localize to the plasma membrane and to import extracellular copper into the cytoplasm (8, 9), whereas the third family member, yCtr2, transports copper stored in the vacuole back into the cytoplasm (10). In mammals, including humans and mice, two Ctr proteins are present, Ctr1 and Ctr2. Ctr1 is an essential gene, localizing to the plasma membrane where it facilitates copper import into the cytoplasm (11–14). Recently, the mouse Ctr1 protein was also shown to be essential for intestinal copper uptake (15), its role in intestinal copper uptake, however, not being fully understood (16). Ctr2 was reported to localize to endosomes/lysosomes and to contribute to copper uptake by mobilization of intracellular copper stores in a manner similar to yCtr2 (17).
Recently, Drosophila has been introduced as a model system for the study of copper homeostasis. Three Ctr1-like genes are present in Drosophila: Ctr1A, Ctr1B, and Ctr1C (18). Ctr1A is constitutively and ubiquitously expressed, its loss resulting in developmental arrest and a general failure of copper-dependent processes (copper enzyme activity, neuropeptide maturation, heart function) (19). Ctr1B is responsible for copper uptake from the intestine. Ctr1B mutants are viable, but they have strongly reduced organismal copper levels, and they are sensitive to copper starvation. Ctr1B is transcriptionally activated upon copper starvation by the metal-responsive transcription factor MTF-1 (18, 20, 21).
In contrast to Ctr1A and Ctr1B, the third family member, Ctr1C, has not been further studied after an initial characterization in which it was shown to be able to complement copper import-deficient yeast and to be expressed in Drosophila third instar larvae and adult males (18). Here, we present a detailed analysis of the function of Ctr1C. We show that Ctr1C localizes to the plasma membrane when ectopically expressed and compare the effects of ectopic overexpression of Ctr1C with Ctr1A and Ctr1B. The results reveal that Ctr1C is a bona fide copper importer that is indistinguishable in its overexpression effects from those of Ctr1A and Ctr1B. We also show that Ctr1C can functionally replace Ctr1A in Ctr1A25 null mutants. Using a GFP2 knock-in mutant of Ctr1C and genomic Ctr1C-GFP transgenes, we demonstrate that the expression of Ctr1C is restricted to maturing spermatocyte cysts and mature sperm and that the protein is localized in the plasma membrane. We show that in a Ctr1B mutant background, i.e. in a situation of low intraorganismal copper, Ctr1C is required for male fertility, revealing a hitherto unrecognized aspect of copper biology. Finally, we explore the scenario that Ctr1C has originated as a retrogene during sex chromosome formation in the evolutionary precursor of the drosophilid lineage, providing a copper delivery function in the male germline where the X chromosome-linked Ctr1A cannot function due to X chromosome inactivation in the male germline.
EXPERIMENTAL PROCEDURES
Fly Culture
1 liter of standard fly food was composed of 55 g of corn, 10 g of wheat, 100 g of yeast, 75 g of glucose, 8 g of agar, and 15 ml of the antifungal agent nipagin (15% in ethanol). For experiments, food was supplemented with CuSO4 or bathocuproine disulfonate (BCS) disodium salt hydrate (Sigma-Aldrich number 14,662-5) to the indicated concentrations. BCS is a specific copper chelator used to deplete copper in the food. Flies were raised at 25 °C and 65% humidity.
Plasmids, Mutants, and Transgenic Fly Strains
To generate UAS-Ctr1C transgenes, a DNA segment corresponding to the Ctr1C-PA coding sequence (Flybase designation FBpp0085111, coordinates: 3R: complement(26789170..26789982)) was cloned into pUASTattB (22), pUAST, and pUASP vectors. To generate genomic Ctr1C transgenes, an 8-kb DNA segment corresponding to coordinates 3R: complement(26792528..26784388) was cloned into a pAttP vector (22). For the Ctr1C-GFP genomic rescue transgene, an enhanced GFP ORF was inserted in-frame at the C-terminal end of the Ctr1C ORF into the Ctr1C genomic transgene. Transgenic fly lines were generated using standard P element transgenesis and PhiC31-mediated transgenesis (22, 23). The Ctr1C6D mutant was generated by ends-in homologous recombination followed by allelic substitution (24, 25). The Ctr1C ORF is disrupted by precise integration of a DNA segment containing the enhanced GFP-ORF followed by an SV40 poly(A) signal sequence, deleting amino acids 1–13 of the Ctr1C ORF. Plasmid sequences are available upon request.
Determination of Survival Rates
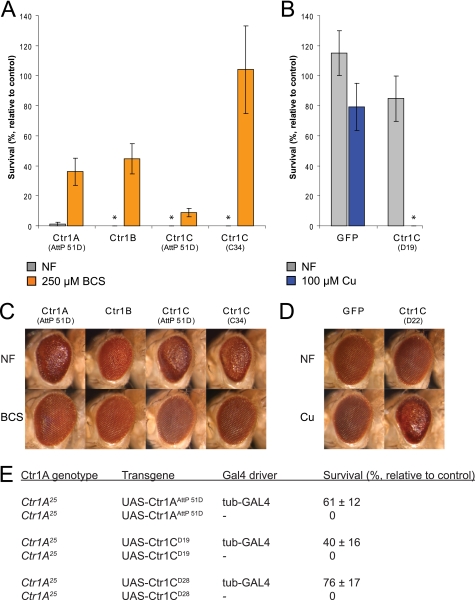
The survival rates in the actin-GAL4 overexpression assay (see Fig. 1, A and B) were calculated relative to the numbers of eclosing siblings (adult flies hatching from their pupal cases) that carried a balancer chromosome (CyO, y+) instead of the GAL4 driver, having otherwise identical genotypes. For the complementation assay (see Fig. 1E), a null mutant allele of Ctr1A (Ctr1A25) was used (19). The indicated transgenes were ubiquitously expressed, using a moderately strong tubulin-GAL4 driver in hemizygous Ctr1A25/Y males, and the number of rescued animals was scored. Survival rates were calculated relative to fully viable Ctr1A25/FM7; UAS-transgene/+; MKRS/+ sisters (these females carry the X chromosome balancer FM7 and the third chromosome balancer MKRS and show wild type survival).
FIGURE 1.
Ctr1C shows copper import activity comparable with Ctr1A and Ctr1B and can functionally replace mutant Ctr1A. A and B, strong ubiquitous expression of Ctr1C transgenes causes copper-dependent lethality. The indicated transgenes were expressed under the control of UAS enhancers by actin-GAL4. Flies were raised on food containing the indicated concentrations of copper or BCS. A, a Ctr1C transgene that was integrated into the same AttP landing site as the Ctr1A control (AttP 51D) showed slightly stronger lethality on NF and a less pronounced rescue of survival on BCS than Ctr1A, whereas a strongly expressed transgene on a P element (Ctr1C C34) showed a similarly high lethality as Ctr1A and Ctr1B, but a complete rescue on BCS. B, a weaker P element transgene (Ctr1C D19) was viable on NF but completely lethal on copper. The Ctr1A and Ctr1B controls were also shown in Ref. 27. Error bars indicate S.E. Data points with zero value are indicated with an asterisk. C and D, eye-specific expression of Ctr1C under control of GMR-GAL4 disrupts eye morphology in a copper-dependent fashion, comparable with Ctr1A and Ctr1B transgenes. C, BCS supplementation (250 μm) in the food completely rescued the severe rough eye phenotype that arises when strong Ctr1C transgenes (Ctr1C in AttP51D and the P element transgene Ctr1C C34) were expressed in flies raised on NF. D, the weaker P element transgene Ctr1C D22 only caused a rough eye phenotype on 100 μm copper. The Ctr1A and Ctr1B controls were also shown in Ref. 27. E, hemizygous lethal Ctr1A25 null mutant males can be rescued by ubiquitous expression of Ctr1C transgenes by the tub-GAL4 driver. Survival rates with S.E. are given relative to viable Ctr1A25/y w; UAS-transgene/+; MKRS/+ siblings.
Fertility Assays
Males were kept separate from females before mating (see Fig. 4, A and B, for 12 days; Fig. 4C, for 7 days). Single males were each crossed to two y w virgin females. After a mating period (see Fig. 4, A and B, 5 days; Fig. 4C, a first period of 3 days, a second period of 2 days), the parents were discarded. The offspring were left to develop and were counted after eclosion.
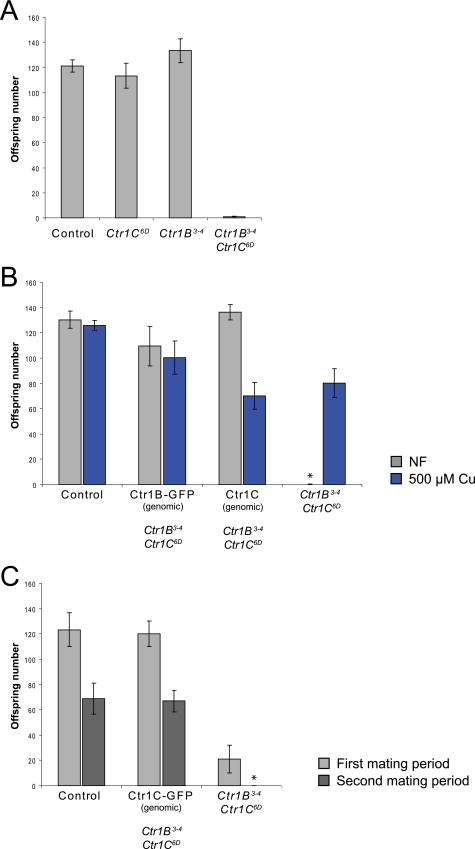
FIGURE 4.
Ctr1B Ctr1C double mutant males exhibit copper- and time-dependent loss of fertility. A, although Ctr1B3–4 and Ctr1C6D single mutants are normally fertile, Ctr1B3–4 Ctr1C6D double mutants show almost complete sterility. The graph shows average offspring numbers and S.E. from crosses of single males of the indicated genotypes to two y w virgins. Before mating, the males had been kept separate from females until an age of 12 days. B, the sterility of Ctr1B3–4 Ctr1C6D double mutant males can be rescued by the presence of a genomic Ctr1B-GFP, by a genomic Ctr1C WT transgene, or by keeping eclosed adult males on 500 μm copper food. Average offspring numbers and S.E. from single males of the indicated genotypes crossed to two y w virgins are given. Before mating, the males had been raised on NF, and after eclosion, kept on either NF or 500 μm copper food for 12 days. Data points with zero value are indicated with an asterisk. C, a genomic Ctr1C-GFP construct rescues the sterility phenotype of Ctr1B3–4 Ctr1C6D double mutant males, who themselves exhibit a rapid loss in fertility in an experiment using two consecutive mating rounds. Single males of the indicated genotypes at least 7 days old were mated to two y w virgins. The parents were together for 3 days for a first mating period and then used for a second mating period of 2 days. The distribution of offspring numbers for the different genotypes is shown in supplemental Fig. 1.
Flp-out Clones
UAS-Ctr1C(WT) UAS-GFP (see Fig. 2, A and B) and UAS-Ctr1C-FLAG UAS-GFP (see Fig. 2, C and D) clones were generated with the hs-flp actin-FRT-CD2-FRT-GAL4 flp-out technique (26). With this technique, the expression of UAS transgenes can be induced with a heat shock in cell clones. Ctr1C-overexpressing clones were marked by the coexpression of UAS-GFP. Because the cell clones only differ in the expression of actin-GAL4, UAS-Ctr1C, and UAS-GFP from their non-expressing neighbors, the antibody signal in non-expressing neighbors can serve as a direct internal control for antibody signal and specificity. To induce the clones, a 5-min heat shock at 37 °C was administered after a 24-h egg lay period. Salivary glands and fat bodies were harvested from wandering third instar larvae.
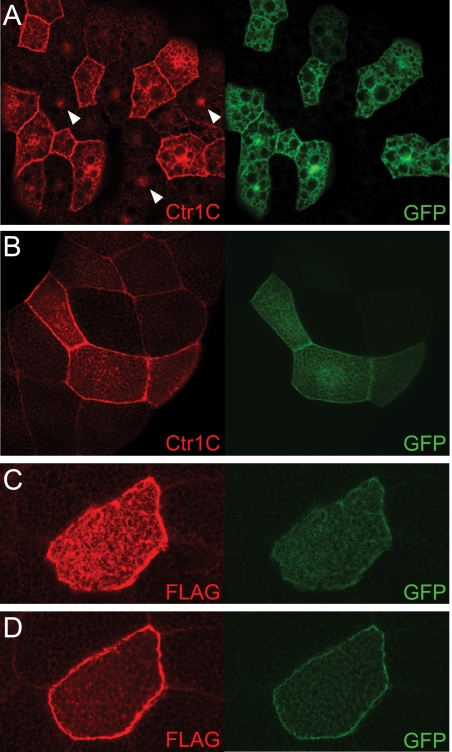
FIGURE 2.
Clonal expression of Ctr1C reveals predominant plasma membrane localization. A–D, Ctr1C WT transgenes (A and B) and C-terminally tagged Ctr1C-FLAG transgenes (C and D) were expressed in cell clones together with GFP, using the hs-flp actin-FRT-CD2-FRT-GAL4 flp-out technique (26). An antibody that recognizes Ctr1C clearly labels the plasma membrane but also structures in the cytoplasm (confocal sections of fat body cells in A and of salivary gland cells in B; lens magnification ×25). The antibody also detects a nuclear signal (indicated with arrows), which is not specific to Ctr1C because it is equally strong in Ctr1C6D mutants (data not shown). Ctr1C-FLAG predominantly localizes to the plasma membrane in salivary gland cells (C and D; lens magnification ×60). C, confocal section through the flat top membrane of a salivary gland cell (strong staining throughout the surface). D, section below the top of the same cell.
Tissue Preparation and Immunohistochemistry
Wandering third instar larvae (to harvest larval male gonads, fat bodies, and salivary glands) and adult males (to harvest adult testes and seminal vesicles) were anesthetized and dissected in ice-cold phosphate-buffered saline, pH 7.4. In cases where unfixed tissues were analyzed (see Figs. 3C and 5C), the tissues were directly mounted in 87% glycerol and microphotographed. Tissues that were subjected to DAPI staining (see Figs. 3B and 5A) were fixed in 4% paraformaldehyde for 10 min at room temperature and washed three times for 10 min with phosphate-buffered saline. In the middle wash step, DAPI was included at a concentration of 5 μg/ml. Tissues were mounted in 87% glycerol and microphotographed. Tissues subjected to antibody and DAPI stainings (see Figs. 2 and 5B) were fixed in 4% paraformaldehyde for 10 min at room temperature. The fixed tissues were washed twice for 10 min with washing solution (0.3% Triton X-100 in phosphate-buffered saline, pH 7.4). Blocking of the tissues was performed in 10% fetal calf serum (in washing solution) for 1 h at room temperature. After blocking, the tissue was washed once for 10 min and incubated with primary antibody (in washing solution) overnight at 4 °C. Subsequently, the tissue was washed twice for 10 min at room temperature. The tissues were incubated with secondary antibody (in washing solution) for 2–4 h at room temperature. The secondary antibody was washed three times for 10 min. In the middle washing step, DAPI was included at a concentration of 5 μg/ml. The dissected tissues were mounted in 87% glycerol. Antibodies and concentrations were: Fig. 2, A and B, rabbit anti-Ctr1C peptide (raised against the epitope GGRDQYNRPRRYREA), 1:500 and goat anti-rabbit Alexa Fluor 594 (Invitrogen, A11012), 1:1000; Fig. 2, C and D, mouse anti-FLAG (Sigma, F1804), 1:400 and goat anti-mouse Alexa Fluor 546 (Invitrogen, A11030), 1:1000; Fig. 5B, rabbit anti-GFP (MBL598), 1:500 and goat anti-rabbit Alexa Fluor 488 (Molecular Probes A-11008), 1:400. The anti-Ctr1C antibody only produced a signal when used against overexpressed Ctr1C protein, and it strongly cross-reacted with a nuclear protein.
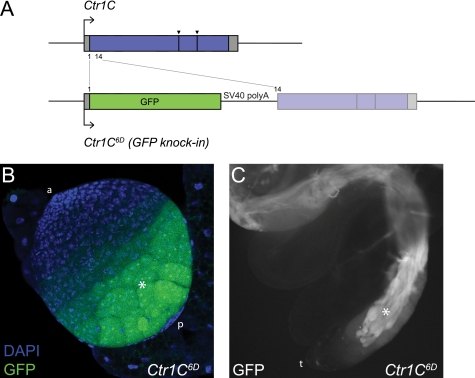
FIGURE 3.
Ctr1C is expressed in the male germline. A, structure of the Ctr1C6D GFP knock-in mutant. The Ctr1C ORF was disrupted by precise integration of a GFP transgene at the Ctr1C start (position 1). The remainder of the Ctr1C ORF is separated from the GFP ORF by an SV40 poly(A) signal sequence and is not in-frame with the GFP. Gray boxes indicate 5′- and 3′-untranslated regions, and blue and green boxes indicate ORFs. Large arrows indicate the transcription start site, and arrowheads indicate the position of splice sites in the Ctr1C ORF. B and C, GFP expression in Ctr1C6D mutant flies in the late germline cysts of the larval male gonads (a, anterior somatic cap; p, posterior somatic cap; asterisk, late germline cysts) (B) and in the elongating germline cysts of the adult testis (t, tip of the testis; asterisk, elongating germline cysts) (C). Pictures were taken with a lens magnification of ×25 (larval gonad) and ×10 (adult testis).
FIGURE 5.
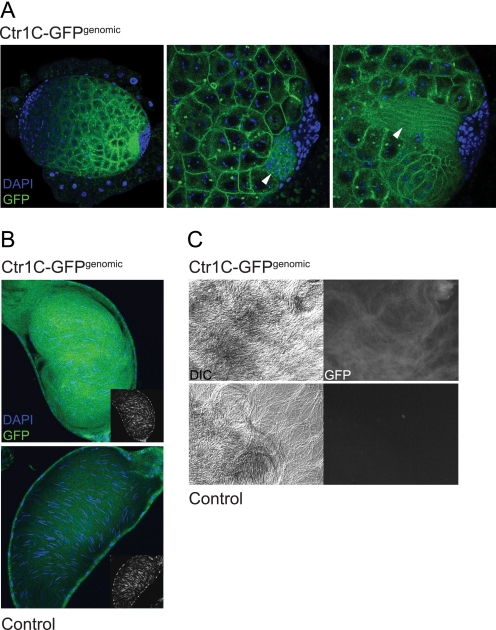
A genomic, C-terminally tagged Ctr1C-GFP transgene reveals Ctr1C localization in the plasma membranes of maturing male germline cysts and in mature sperm. A, localization of genomic Ctr1C-GFP in the male larval gonad. Ctr1C-GFP is present in the plasma membranes of the late spermatocyte cysts, around developing sperm heads (middle panel, arrow), and in the plasma membranes of elongating spermatocyte cysts (right panel, arrow). Pictures show confocal sections of a male gonad of a wandering third instar larva, with the left panel at ×25 and the middle and right panel at ×60. The three panels show different Z sections of the same gonad. The punctate staining visible in some cell bodies must be unspecific because it was also seen in the Ctr1C6D mutant male gonads (Fig. 3B). B, Ctr1C-GFP is present in mature sperm. Seminal vesicles of adult males carrying a genomic Ctr1C-GFP transgene show GFP-labeled mature sperm. Sperm head nuclei are labeled by DAPI. The small inlay pictures show the DAPI signal of the elongated sperm nuclei alone. C, presence of Ctr1C-GFP in sperm tails. Mature sperm were squeezed out of seminal vesicles and microphotographed. The left panels show Nomarski (differential interference contrast (DIC)) images, and the right panels show fluorescein isothiocyanate channel signals. Control panels in B and C show seminal vesicles and mature sperm from flies that are identical in genetic background but do not carry the genomic Ctr1C-GFP construct.
Microscopy
Eye pictures were recorded with a Leica MZ16 stereomicroscope and a Leica DFC280 camera; pictures of adult testes (see Fig. 3C) and mature sperm (see Fig. 5C) were recorded with a Zeiss Axioplan 2 microscope and an Axiocam MRm camera. Pictures of larval male gonads (see Figs. 3B and 5A) and of seminal vesicles (see Fig. 5B) were recorded with a Zeiss LSM 710 confocal microscope.
Quantitative Analysis of Transgene Expression
To quantitate the strength of the GFP signal in seminal vesicles, fluorescein isothiocyanate filter epifluorescence pictures from seminal vesicles were taken using a Zeiss Axioplan 2 microscope and a Zeiss Axiocam MRm camera. The exposure time was kept equal for all recordings. The average signal intensity for 50 × 50-pixel sections from pictures of individual seminal vesicles was determined with the histogram function of Adobe PhotoShop. To measure β-galactosidase activity (liquid lacZ assay), three female wandering third instar larvae per sample were lysed by successive freeze-thaw cycles in liquid nitrogen. 100 μl of Z-buffer (60 mm Na2HPO4, 40 mm NaH2PO4, 10 mm KCl, 1 mm MgSO4) was added to each sample, and the sample was vortexed for 20 s. 700 μl of Z-buffer (containing 50 mm β-mercaptoethanol) was added to each sample. Subsequently, 160 μl of 4 mg/ml ortho-nitrophenyl-β-galactoside substrate was added to start the reaction. The reaction was carried out at 30 °C for 5 min. At the end of the reaction, 400 μl of 1 m Na2CO3 was added, and the samples were centrifuged for 10 min at 13,000 rpm. The absorbance of the clear supernatant was measured at 420 nm.
Annotation of Ctr1 Genes in Insects
Ctr1-type genes in the different insect genomes were identified by standard BLAST searches (National Center for Biotechnology Information (NCBI) BLAST and Flybase BLAST) against the sequenced genomes and against protein databases. Sequences found in the BLAST searches were annotated using the Artemis software (Sanger Institute). Sequence alignments were generated with the ClustalW2 online software (available from the European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI)) with standard parameters.
Fly Genotypes Used in the Figures
See the supplemental data.
RESULTS
Ctr1C Functions as a Copper Importer
To characterize Ctr1C, we employed two overexpression assays that had been previously used to study Ctr1A and Ctr1B (20, 21, 27). In these assays, the copper transporters are expressed under the control of UAS enhancers either ubiquitously, with the GAL4 driver actin-GAL, or specifically in the eye with GMR-GAL4. Strong ubiquitous expression of Ctr1C transgenes was lethal when the flies were raised on normal food (NF). Such flies could be rescued by growing them on food supplemented with the copper chelator BCS, which was comparable with the effects on viability of Ctr1A and Ctr1B overexpression (Fig. 1A). The chelator rescue experiment suggests that the lethality is due to the copper toxicity imposed by overexpression of Ctr1C. A transgenic fly line expressing Ctr1C at lower levels was viable on NF but lethal if raised on food supplemented with copper (Fig. 1B), further demonstrating that the observed effect is copper-dependent. Similarly, eye-specific expression of strong Ctr1C transgenes led to severely disrupted eye morphology (the rigid, quasi-crystalline pattern of the ommatidia was lost and ommatidia were fused, leading to an uneven eye surface; in addition, overall eye shape was changed) on NF, comparable with the effect of Ctr1A or Ctr1B expression. This rough eye phenotype could be fully rescued by BCS supplementation (Fig. 1C). Consistent with the viability results, a weaker Ctr1C transgene did not alter eye morphology on NF but caused a strong rough eye phenotype on food containing elevated levels of copper (Fig. 1D). These data provide evidence that Ctr1C can function as a copper importer in a manner similar to Ctr1A and Ctr1B.
The notion that Ctr1C functions as a copper importer is further underscored by the observation that Ctr1C can functionally replace Ctr1A. As shown in Fig. 1E, ubiquitous expression of Ctr1C transgenes with a moderately strong GAL4 driver (tubulin-GAL4) restored the viability of the Ctr1A null mutant Ctr1A25 (19, 27).
Finally, in agreement with the proposed copper import function of Ctr1C described above and with previous data on the subcellular localization of Ctr1A and Ctr1B proteins (19, 21), we found that Ctr1C predominantly localizes to the plasma membranes when ectopically expressed in cell clones in larval fat bodies and salivary glands (Fig. 2). Of note, a Ctr1C-GFP genomic construct with endogenous spermatocyte cyst expression (see below) also showed clear plasma membrane localization (see Fig. 5A).
Taken together, these data demonstrate that Ctr1C functions as a bona fide copper importer.
Ctr1C Is Expressed in the Male Germline
To examine the in vivo role of Ctr1C, we used ends-in homologous recombination followed by allelic substitution (24, 25) to generate a Ctr1C mutant (Fig. 3A). In the resulting allele, Ctr1C6D, the Ctr1C open reading frame is disrupted by a precise integration of a GFP transgene, allowing expression of GFP in place of the original Ctr1C ORF. Residual expression of truncated Ctr1C is highly unlikely because the remainder of the Ctr1C ORF is separated from the GFP ORF by a DNA sequence containing an SV40 poly(A) signal sequence and several stop codons. Moreover, the two ORFs are not in-frame. Successful targeting of the Ctr1C gene was confirmed by PCR assays and sequencing (data not shown). Flies carrying the Ctr1C6D allele were homozygous viable and did not show sensitivity to either elevated or reduced copper levels in the food (data not shown), indicating that Ctr1C is not an essential gene.
Further analysis of the Ctr1C6D flies revealed that Ctr1C expression is essentially restricted to the male germline. In the male gonads of wandering third instar larvae, GFP could be observed in germline spermatocyte cysts, the signal gradually becoming stronger from early to late spermatocyte cysts (Fig. 3B). In adult testes, maturing elongating spermatocyte cysts are labeled by GFP (Fig. 3C). The finding that Ctr1C is expressed in the male germline was further confirmed by the expression pattern of genomic Ctr1C-GFP fusion transgenes (see Fig. 5 and below).
Ctr1C Is Required for Male Fertility
In light of the male germline-specific expression pattern of Ctr1C, we next examined whether the loss of Ctr1C would influence male fertility. Because Ctr1C6D mutants did not show reduced fertility when compared with WT controls, we combined the mutant allele with a Ctr1B null mutant (Ctr1B3–4). Ctr1B mutants exhibit strongly reduced organismal copper levels due to impaired intestinal copper uptake in the absence of Ctr1B (20). Although 12-day-old Ctr1B3–4 mutant males were normally fertile, Ctr1B3–4 Ctr1C6D double mutants displayed almost complete sterility, indicating that in the absence of Ctr1B, Ctr1C is required for male fertility (Fig. 4A). Importantly, this loss of fertility is dependent on copper status. If Ctr1B3–4 Ctr1C6D double mutant males were kept on food supplemented with copper after eclosion (emergence of the adult fly from its pupal case), i.e. as adult flies, for 12 days, fertility was largely restored (Fig. 4B). The fertility of Ctr1B3–4 Ctr1C6D double mutant males could also be restored by the presence of genomic Ctr1B-GFP, Ctr1C, and Ctr1C-GFP transgenes (Fig. 4, B and C), demonstrating that the observed mutant phenotype is a specific consequence of the concomitant loss of Ctr1B and Ctr1C.
In the experiments described above, fertility was assayed at an age of 12 days. At this age, Ctr1B3–4 Ctr1C6D double mutant males show almost complete sterility. Nevertheless, Ctr1B3–4 Ctr1C6D mutants can give rise to considerable offspring numbers at an earlier age. In the experiment shown in Fig. 4C and in supplemental Fig. S1, males were crossed at an adult age of 7 days. Single males were left together with female virgins for 3 days and then placed in a fresh food tube for another 2 days. For both mating periods, the offspring were counted. In the first mating period of 3 days, only a part of the Ctr1B3–4 Ctr1C6D double mutants were completely sterile. In the second period of 2 days immediately following the first, almost all double mutants were sterile, revealing a rapid loss of fertility. We examined the testes of these sterile males but could not find any obvious morphological differences to fertile control males (data not shown).
Ctr1C Is Present in Maturing Spermatocyte Cysts and in Mature Sperm
To gain further insights into the male germline function of Ctr1C, we studied the expression and subcellular localization of a genomic Ctr1C-GFP fusion transgene. The transgene consists of the Ctr1C genomic locus (∼3 kb upstream and downstream of the Ctr1C gene) with a GFP ORF C-terminally fused to the Ctr1C ORF. As shown in Fig. 5A, genomic Ctr1C-GFP recapitulates the male larval gonad expression seen with the Ctr1C6D GFP knock-in mutant in male larval gonads. Confocal sections showed that Ctr1C-GFP localizes to the plasma membranes of late spermatocyte cysts but is absent from early cysts and the brightly DAPI-labeled somatic cap of the gonads (Fig. 5A, middle and right panel), confirming the male germline expression pattern seen with the Ctr1C6D allele. In some larval gonads, elongating spermatocyte cysts are already present. Here, the Ctr1C-GFP signal could be detected around the developing sperm heads (Fig. 5A, middle panel) and in the elongating sperm tails (Fig. 5A, right panel). In the adult testes, elongating spermatocyte cysts are labeled (data not shown), giving a signal pattern similar to the one shown in Fig. 3C. Interestingly, the seminal vesicles (sperm storage organs situated immediately behind the testes, filled with mature sperm) of Ctr1C-GFP-expressing adult males showed a strong GFP signal, indicating that Ctr1C-GFP protein is present in mature sperm (Fig. 5B). In Ctr1C6D mutants, no signal of (soluble) GFP could be detected in the seminal vesicles (data not shown), which was to be expected because spermatozoa slough off most of their cytosol during maturation. The confocal sections in Fig. 5B show that the GFP expression is not restricted to the DAPI-labeled sperm heads but comprises a much larger domain, suggesting Ctr1C-GFP presence along the sperm tails. This finding was corroborated by squeezing out mature sperm from seminal vesicles to separate individual sperm tails (Fig. 5C). Taken together with the expression data from Ctr1C6D mutants (Fig. 3) and the finding that the genomic Ctr1C-GFP transgene is functional because it can rescue the sterility of Ctr1B3–4 Ctr1C6D double mutants (Fig. 4C), these data suggest that Ctr1C functions in maturing spermatocyte cysts and possibly in mature sperm.
X Chromosome Inactivation in the Male Germline Precludes the Expression of X-linked Ctr1C-GFP
Recently it has been shown that the X chromosome is inactivated in the Drosophila melanogaster male germline during spermatogenesis (28). In a process termed male meiotic sex chromosome inactivation (MSCI), genes residing on the X chromosome are silenced (29, 30). This finding provides a molecular basis for the earlier findings that there is a bias for retrogene formation with a gene parent on the X chromosome and that most retrogenes derived from a parent on the X chromosome have evolved testis-specific expression (31). Intriguingly, Ctr1C is encoded as a single open reading frame on an autosome, whereas Ctr1A, which is ubiquitously expressed (18, 19), is encoded on the X chromosome (supplemental Fig. S2, supplemental Table 1, and supplemental information) and thus presumably is inactive during spermatogenesis. It is therefore possible that Ctr1C originated as a retrogene in the drosophilid lineage (see below for a more detailed discussion). One of the predictions of such a hypothesis is that Ctr1C would not be expressed during spermatogenesis if transplanted onto the X chromosome. To test this hypothesis, we inserted genomic Ctr1C-GFP transgenes into AttP landing sites on the X (landing site 2A), on the second (landing site 51D), and on the third chromosome (landing site 86Fb) and compared the GFP signal intensities of mature sperm in the seminal vesicles of adult males (Fig. 6A). Indeed, the GFP signal of Ctr1C-GFP in the landing site on the X was only slightly above the background of autofluorescence and far below the signal intensity of the Ctr1C-GFP transgenes at the two autosomal sites. Importantly, the difference in signal intensity is not due to general silencing effects at the landing site on the X chromosome because ubiquitous expression of UAS-lacZ transgenes in females (to exclude dosage compensation effects) resulted in comparably high expression levels (Fig. 6B).
FIGURE 6.
A Ctr1C genomic transgene is marginally expressed if inserted in the X chromosome. Ctr1C-GFP genomic transgenes were inserted in AttP landing sites on the X chromosome and the second and third chromosome. The strength of the GFP signal in mature sperm in seminal vesicles was measured. A, although Ctr1C-GFP transgenes in AttP landing sites on the second and third chromosome exhibited equally strong GFP signals in seminal vesicles, the GFP signal for the transgene inserted into an AttP landing site on the X chromosome was barely elevated over background. FITC, fluorescein isothiocyanate. B, this effect is not due to generally lower expression of transgenes in the AttP landing site on the X chromosome because ubiquitous expression of UAS-lacZ transgenes by arm-GAL4 in females (to exclude dosage compensation effects) resulted in comparably high lacZ signals in a liquid lacZ assay. Red lines indicate background fluorescence levels of an “empty”-landing site control (A) and background lacZ signal levels from an average of the four controls (B). Error bars indicate S.E. in A and S.D. in B.
DISCUSSION
Ctr1C Is a Bona Fide Copper Importer
In this study, we present a detailed characterization of Ctr1C, the third member of the Drosophila Ctr1 copper importer family. Ctr1C was initially identified along with Ctr1A and Ctr1B (18) and, like these, was able to complement yeast mutants defective in copper import (18). However, ectopically expressed Ctr1C could not facilitate copper uptake in Drosophila S2 cells (18), raising the possibility that Ctr1C might not function as a copper transporter at the plasma membrane but rather in an intracellular compartment similar to yeast Ctr2. It is not clear why Ctr1C failed to mediate copper uptake in cell culture, but our in vivo data strongly suggest that Ctr1C functions as a copper transporter in a similar manner to Ctr1A and Ctr1B. Strikingly, ubiquitous expression of Ctr1C could rescue Ctr1A25 null mutants, demonstrating that Ctr1C can functionally replace Ctr1A. When strongly overexpressed, either ubiquitously or eye-specifically, Ctr1C led to copper toxicity phenotypes that were indistinguishable from those caused by Ctr1A or Ctr1B. The observed toxicity effects were dose-dependent, allowing us to estimate relative copper import efficiencies of differently expressing genomic P element insertions and genomic AttP landing sites. Of note, a Ctr1C transgene inserted into the genomic landing site AttP 51D showed similarly strong toxicity effects as a Ctr1A transgene in the same landing site, arguing for similar copper import capabilities of the two proteins. In addition to these findings, our data on the subcellular localization of Ctr1C (showing plasma membrane localization of a Ctr1C-GFP fusion protein with endogenous expression pattern and expression levels) support a role of Ctr1C in cellular copper uptake.
Evolution of the Ctr1 Homologs in the Insects
The occurrence of three Ctr1 homologs in the genetically streamlined model organism Drosophila is striking and has prompted us to examine the genome sequences of the other 11 sequenced drosophilid species as well as nine genomes from more distantly related insect species for the presence of Ctr1-type copper transporter genes (supplemental Fig. 2 and Table 1 and supplemental information). We could identify Ctr1A homologs in all sequenced genomes. Ctr1B was present in all drosophilids and also in some of the analyzed non-drosophilid insects. Ctr1B, the major intestinal copper importer, appears to be less well conserved than the ubiquitous Ctr1A but more so than the male germline-specific Ctr1C, which was only found among drosophilids. Ctr1B differs from both Ctr1A and Ctr1C by its shortened N terminus. Interestingly, the Ctr1B homologs show partial conservation of the exon-intron boundaries that are shared between human Ctr1 and insect Ctr1A genes (see below), making it plausible that the insect Ctr1B gene has arisen from a duplication of the genomic DNA segment containing Ctr1A.
Regarding Ctr1C, we wanted to investigate the possibility that Ctr1C originated as a retrogene and has acquired male germline function. An “out-of-X movement” of male germline-specific retrogenes is thought to be a general consequence of sex chromosome evolution, either due to sex chromosome inactivation in the male germline (28, 31) or due to evolutionary processes leading to a general demasculinization of the X chromosome (32); this phenomenon is also documented in mammals that independently evolved their sex chromosome system (33). The hypothesis of a retrogene origin of Ctr1C would thus be falsified if Ctr1C homologs were present in species not sharing the Drosophila X chromosome or if the cDNA-type single open reading frame configuration of D. melanogaster Ctr1C was not shared by other Ctr1C homologs. As noted above, we could find Ctr1A homologs in all examined insect species, but Ctr1C homologs were only present in the Drosophila genus, which shares an ancestral X chromosome (34–36). Ctr1A homologs have a tightly conserved exon-intron structure. Of note, the exon junctions in front of the first and the second transmembrane domains appear to be conserved even between human Ctr1 and the insect Ctr1As. In marked contrast, all Ctr1C genes possess a single long ORF. In some species, including D. melanogaster, a part of the variable intracellular loop sequence can be spliced out, but the splice sites of these apparent “neo-introns” are not conserved between the Ctr1C genes. Regarding chromosomal locations, all Ctr1A homologs among drosophilids display synteny; in all cases where scaffolds are mapped to chromosomes, Ctr1A is present on Muller element A (the ancestral X chromosome). The Ctr1C homologs also show synteny and map to Muller element E (an autosome). These data, taken together with our experimental finding that expression of a Ctr1C-GFP transgene in the male germline was at most marginal upon inserting it into the X chromosome, support the hypothesis of a retrogene origin of Ctr1C as a consequence of X inactivation in the male germline.
Copper, a New Player in Male Fertility?
Analysis of the endogenous Ctr1C expression pattern and loss of function analysis revealed that Ctr1C plays a specific role in spermatogenesis; it is expressed in the maturing spermatocyte cysts and present in mature sperm. Most importantly, in a Ctr1B mutant background when intraorganismal copper levels are strongly reduced, lack of Ctr1C leads to sterility. Fertility can be restored if the adult males are kept on food containing additional copper. These findings demonstrate that Ctr1C contributes to copper delivery in the male germline and that copper is required for the maintenance of normal fertility. The importance of the trace element zinc for male fertility has long been documented (reviewed in Ref. 37), and zinc was recently shown to be essential for spermatogenesis (38). Our findings are compatible with a specific role of copper in male fertility. At present, we can only speculate about the nature of such a role. Sperm motility depends on the function of the mitochondrial respiratory chain (39); Drosophila sperm contain a large so-called mitochondrial derivative in the head region (40) and might have a high copper demand for proper mitochondrial function. Intriguingly, high levels of Ctr1 are present in mature spermatozoa in the mouse (41), and Ctr2 levels are high in testis.3 It is therefore possible that the requirement for Ctr proteins in male fertility is conserved between insects and mammals.
Supplementary Material
Acknowledgments
We thank Drs. Johannes Bischof, Konrad Basler, and Peter Gallant for fly stocks and plasmids; Dr. Monica Steinmann-Zwicky and Margrith Cavegn for reagents; Corinna Oberle for help with experiments; Werner Boll for advice on microscopy; Till Strassen for help with stock keeping; and Drs. Haiqing Hua and George Hausmann for valuable comments on the manuscript.
This work was supported by grants from the Kanton Zürich and the Swiss National Science Foundation (to W. S.), and by National Institutes of Health Grant GM062555 (to D. J. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental information, Figs. S1 and S2, and Table 1.
D. J. Thiele, unpublished results.
- GFP
- green fluorescent protein
- ORF
- open reading frame
- BCS
- bathocuproine disulfonate
- DAPI
- 4′,6-diamidino-2-phenylindole
- NF
- normal food
- WT
- wild type.
REFERENCES
- 1.Halliwell B., Gutteridge J. M. (1990) Methods Enzymol. 186, 1–85 [DOI] [PubMed] [Google Scholar]
- 2.Balamurugan K., Schaffner W. (2006) Biochim. Biophys. Acta 1763, 737–746 [DOI] [PubMed] [Google Scholar]
- 3.Puig S., Thiele D. J. (2002) Curr. Opin. Chem. Biol. 6, 171–180 [DOI] [PubMed] [Google Scholar]
- 4.Kim B. E., Nevitt T., Thiele D. J. (2008) Nat. Chem. Biol. 4, 176–185 [DOI] [PubMed] [Google Scholar]
- 5.Southon A., Farlow A., Norgate M., Burke R., Camakaris J. (2008) J. Exp. Biol. 211, 709–716 [DOI] [PubMed] [Google Scholar]
- 6.Knöpfel M., Smith C., Solioz M. (2005) Biochem. Biophys. Res. Commun. 330, 645–652 [DOI] [PubMed] [Google Scholar]
- 7.De Feo C. J., Aller S. G., Siluvai G. S., Blackburn N. J., Unger V. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4237–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pena M. M., Puig S., Thiele D. J. (2000) J. Biol. Chem. 275, 33244–33251 [DOI] [PubMed] [Google Scholar]
- 9.Dancis A., Haile D., Yuan D. S., Klausner R. D. (1994) J. Biol. Chem. 269, 25660–25667 [PubMed] [Google Scholar]
- 10.Rees E. M., Lee J., Thiele D. J. (2004) J. Biol. Chem. 279, 54221–54229 [DOI] [PubMed] [Google Scholar]
- 11.Zhou B., Gitschier J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7481–7486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo Y. M., Zhou B., Cosco D., Gitschier J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6836–6841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J., Prohaska J. R., Thiele D. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6842–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J., Peña M. M., Nose Y., Thiele D. J. (2002) J. Biol. Chem. 277, 4380–4387 [DOI] [PubMed] [Google Scholar]
- 15.Nose Y., Kim B. E., Thiele D. J. (2006) Cell Metab. 4, 235–244 [DOI] [PubMed] [Google Scholar]
- 16.van den Berghe P. V., Klomp L. W. (2009) Nutr. Rev. 67, 658–672 [DOI] [PubMed] [Google Scholar]
- 17.van den Berghe P. V., Folmer D. E., Malingré H. E., van Beurden E., Klomp A. E., van de Sluis B., Merkx M., Berger R., Klomp L. W. (2007) Biochem. J. 407, 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou H., Cadigan K. M., Thiele D. J. (2003) J. Biol. Chem. 278, 48210–48218 [DOI] [PubMed] [Google Scholar]
- 19.Turski M. L., Thiele D. J. (2007) J. Biol. Chem. 282, 24017–24026 [DOI] [PubMed] [Google Scholar]
- 20.Selvaraj A., Balamurugan K., Yepiskoposyan H., Zhou H., Egli D., Georgiev O., Thiele D. J., Schaffner W. (2005) Genes Dev. 19, 891–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balamurugan K., Egli D., Hua H., Rajaram R., Seisenbacher G., Georgiev O., Schaffner W. (2007) EMBO J. 26, 1035–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bischof J., Maeda R. K., Hediger M., Karch F., Basler K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubin G. M., Spradling A. C. (1983) Nucleic Acids Res. 11, 6341–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rong Y. S., Titen S. W., Xie H. B., Golic M. M., Bastiani M., Bandyopadhyay P., Olivera B. M., Brodsky M., Rubin G. M., Golic K. G. (2002) Genes Dev. 16, 1568–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egli D., Selvaraj A., Yepiskoposyan H., Zhang B., Hafen E., Georgiev O., Schaffner W. (2003) EMBO J. 22, 100–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basler K., Struhl G. (1994) Nature 368, 208–214 [DOI] [PubMed] [Google Scholar]
- 27.Hua H., Georgiev O., Schaffner W., Steiger D. (2010) J. Biol. Inorg. Chem. 15, 107–113 [DOI] [PubMed] [Google Scholar]
- 28.Hense W., Baines J. F., Parsch J. (2007) PLoS Biol. 5, e273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner J. M. (2007) Development 134, 1823–1831 [DOI] [PubMed] [Google Scholar]
- 30.McKee B. D., Handel M. A. (1993) Chromosoma 102, 71–80 [DOI] [PubMed] [Google Scholar]
- 31.Betrán E., Thornton K., Long M. (2002) Genome Res. 12, 1854–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sturgill D., Zhang Y., Parisi M., Oliver B. (2007) Nature 450, 238–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potrzebowski L., Vinckenbosch N., Marques A. C., Chalmel F., Jégou B., Kaessmann H. (2008) PLoS Biol. 6, e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charlesworth B. (1996) Curr. Biol. 6, 149–162 [DOI] [PubMed] [Google Scholar]
- 35.Lucchesi J. C. (1978) Science 202, 711–716 [DOI] [PubMed] [Google Scholar]
- 36.Bachtrog D., Jensen J. D., Zhang Z. (2009) PLoS Biol. 7, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hidiroglou M., Knipfel J. E. (1984) J. Dairy Sci. 67, 1147–1156 [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi S., Miura C., Kikuchi K., Celino F. T., Agusa T., Tanabe S., Miura T. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10859–10864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruiz-Pesini E., Diez C., Lapeña A. C., Pérez-Martos A., Montoya J., Alvarez E., Arenas J., López-Pérez M. J. (1998) Clin. Chem. 44, 1616–1620 [PubMed] [Google Scholar]
- 40.Shoup J. R. (1967) J. Cell Biol. 32, 663–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuo Y. M., Gybina A. A., Pyatskowit J. W., Gitschier J., Prohaska J. R. (2006) J. Nutr. 136, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.