Abstract
The unidirectional translocation of messenger RNA (mRNA) through the aqueous channel of the nuclear pore complex (NPC) is mediated by interactions between soluble mRNA export factors and distinct binding sites on the NPC. At the cytoplasmic side of the NPC, the conserved mRNA export factors Gle1 and inositol hexakisphosphate (IP6) play an essential role in mRNA export by activating the ATPase activity of the DEAD-box protein Dbp5, promoting localized messenger ribonucleoprotein complex remodeling, and ensuring the directionality of the export process. In addition, Dbp5, Gle1, and IP6 are also required for proper translation termination. However, the specificity of the IP6-Gle1 interaction in vivo is unknown. Here, we characterize the biochemical interaction between Gle1 and IP6 and the relationship to Dbp5 binding and stimulation. We identify Gle1 residues required for IP6 binding and show that these residues are needed for IP6-dependent Dbp5 stimulation in vitro. Furthermore, we demonstrate that Gle1 is the primary target of IP6 for both mRNA export and translation termination in vivo. In Saccharomyces cerevisiae cells, the IP6-binding mutants recapitulate all of the mRNA export and translation termination defects found in mutants depleted of IP6. We conclude that Gle1 specifically binds IP6 and that this interaction is required for the full potentiation of Dbp5 ATPase activity during both mRNA export and translation termination.
Keywords: Nucleus/Nuclear Export, Nucleus/Nuclear Pore, Nucleus/Nuclear Transport, Nucleus/Nuclear Import, Inositol Phosphates, DEAD-box Protein, Inositol Kinase, Inositol Signaling
Introduction
Directional transport of mRNA from the nucleus to the cytoplasm is a highly orchestrated process that bridges nuclear mRNA processing events to protein synthesis in the cytoplasm and thus represents a central step in the regulation of gene expression (1–4). Properly capped, spliced, and polyadenylated mRNAs and their associated RNA-binding proteins constitute mature messenger ribonucleoprotein particles (mRNPs).4 Of importance for nuclear export, soluble mRNA export factors associate with mRNAs throughout this maturation process, some serving dual roles of mRNA processing and transport regulators (5–8). Export requires targeting to and translocation through NPCs, large hetero-oligomeric structures embedded in nuclear envelope pores. The conserved Saccharomyces cerevisiae Mex67/Mtr2 heterodimer (vertebrate TAP/p15 or NXF1/NXT1) is thought to be the major mRNA export receptor, directly binding the mRNP in the nucleus and facilitating NPC docking and translocation by interaction with NPC proteins (Nups) (1, 9, 10). Mex67-Mtr2 interacts preferentially with phenylalanine-glycine repeats found in Nup domains positioned on the nuclear NPC face and within the central NPC channel (11, 12).
In addition to critical interactions between Mex67-Mtr2 and phenylalanine-glycine repeat domains in the NPC, directional mRNP export is coincident with altered protein-protein and protein-RNA interactions in the mRNP complex (13). For example, Mex67 and the nuclear poly(A)+-binding protein Nab2 are removed from the mRNP during or after the terminal NPC export step (14, 15). Key factors that mediate this mRNP remodeling at the NPC include Gle1, Dbp5, and IP6. The conserved mRNA export factor Gle1 plays an essential role in mRNA export (16–19). Immunoelectron microscopy studies show that S. cerevisiae Gle1 is predominantly localized at the cytoplasmic side of the NPC, where it interacts with the cytoplasmic NPC protein Nup42 (16, 20, 21). Additionally, in S. cerevisiae, Gle1 has functional and physical interactions with several other mRNA export factors and NPC components including Dbp5, Nup159, Nup100, and Gfd1 (16, 17, 20, 22–24). Deficient Gle1 function results in nuclear mRNA accumulation in a variety of organisms (16, 17, 19, 25), underscoring the conserved role of Gle1 in mRNA export. Sequence analysis reveals a highly conserved C-terminal domain and a more divergent N-terminal domain (18). Interestingly, the conserved C-terminal domain of Gle1 is responsible for interaction with the conserved mRNA export factor Dbp5 (20, 22, 24, 26).
Dbp5, a conserved member of the DEAD-box helicase family of RNA-dependent ATPases (27, 28), mediates mRNP remodeling through the displacement of proteins from mRNAs (15). Dbp5 is possibly cotranscriptionally recruited to the mRNP (29, 30); however, its ATPase activity is activated specifically by Gle1 (23, 24). Importantly, although Dbp5 ATPase activity can be enhanced by Gle1 alone, it is maximally stimulated by the presence of both Gle1 and IP6. Thus, Gle1 and IP6 function as Dbp5 ATPase coactivating factors, promoting conversion to a Dbp5-ADP conformation (15). The ATP to ADP conformational change in Dbp5 is speculated to trigger Dbp5-dependent RNA-protein remodeling events and result in the removal of Nab2 and Mex67 from the exporting mRNP (14, 15, 31). Thus, the nuclear export of mRNAs from the nucleus to the cytoplasm is mediated by the temporal and spatial coordination of biochemical interactions between mRNPs, mRNA export factors, and Nups.
Despite great progress in defining components of the mRNA processing and export machinery, there remain large gaps in our knowledge regarding how mRNP translocation through the NPC and release are coupled. Moreover, the extent of functional connections between mRNA export and translation is unknown. Intriguingly, we and others have shown that Dbp5, Gle1, and IP6 have roles in translation independent from their roles in mRNA export (32, 33). Functionally, each of these factors is required for efficient translation termination. S. cerevisiae mutants for the genes encoding Gle1, Dbp5, and Ipk1 (the inositol 1,3,4,5,6-pentakisphosphate 2-kinase that generates IP6 (34)) show synthetic defects in combination with translation termination mutants. Moreover, Dbp5 and Gle1 physically interact with eRF1/Sup45 (32, 33). Gle1 is also involved in translation initiation by conserved physical interactions with eukaryotic translation initiation factor 3 (33). However, the role of Gle1 in translation initiation appears to be independent from Dbp5 and IP6, suggesting that Gle1 and IP6 do not always work together during gene expression (33).
IP6 and other inositol polyphosphates have key roles in gene expression through regulation of chromatin remodeling, transcription factor activation, mRNA editing, mRNA export, and translation (35). Functional in vivo links for IP6 production have come from using a S. cerevisiae ipk1-null (Δ) mutant that presumably lacks IP6 as well as its pyrophosphorylation products, with a coincident drastic elevation in levels of inositol phosphates upstream of IP6 (34, 36, 37). Thus, it is important to independently analyze the binding targets to clearly establish all potential roles for different inositol polyphosphates in a given cellular process. To date, gle1 mutants that lack IP6 binding have not been reported, and consensus binding motifs or properties of IP6-binding proteins have not been established. Here, we further elucidate the biochemical parameters for Gle1-IP6 binding and analyze this relationship in regard to Dbp5 activation. We find that Gle1 is the main target of IP6 in mRNA export and translation termination in S. cerevisiae. IP6 and the IP signaling pathway are thus positioned to regulate gene expression at multiple levels during the life of the mRNP.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Yeast Growth
The strains and plasmids used in this study are listed in Table 1. S. cerevisiae strains were grown in YPD (1% yeast extract, 2% peptone, 2% glucose) at 23 °C unless otherwise noted. Synthetic medium lacking the appropriate amino acids was supplemented with 2% glucose. Medium used for plasmid loss was supplemented with 1.0 mg/ml 5-fluoroorotic acid (U.S. Biological). Yeast cells were transformed using a lithium acetate method (38). To generate strains expressing different alleles of GLE1, we utilized a gle1Δ strain (16) with a plasmid harboring GLE1/URA3. This strain was transformed with plasmids containing wild type or mutant alleles of GLE1 and LEU2. The resulting colonies were selected in −Leu medium and streaked in 5-fluoroorotic acid plates to select for colonies that had lost the GLE1/URA3 plasmid. To generate a psi− strain of gle1-4, a PSI+ gle1-4 strain (16) was patched and grown up on YPD supplemented with 5 mm guanidine hydrochloride for three successive rounds and then restreaked to single colonies on YPD (39). Multiple isolates were tested for read-through activity. The rat8-2 (dbp5) strain was generated by six successive back-crosses of CSY550 (27) to a W303 wild type strain (40). The GLE1/LEU2/CEN (pSW399) and pMAL-TEV-Gle1 (pSW3242) plasmid vectors were used in oligonucleotide-based site-directed in vitro mutagenesis to generate yeast (pSW3343, pSW3344, and pSW3345) and bacterial (pSW3291, pSW3292, pSW3293, and pSW3295) plasmids that express mutant alleles of gle1.
TABLE 1.
Strains and plasmids in this study
| Genotype/description | Source | |
|---|---|---|
| Yeast strains used | ||
| SWY1831 | MATα ade2 ura3 his3 leu2 trp1 can1 psi−+ pSW410 | Ref. 16 |
| SWY3823 | MATα ade2 ura3 his3 leu2 trp1 can1 psi−+ pSW3343 | This study |
| SWY3824 | MATα ade2 ura3 his3 leu2 trp1 can1 psi−+ pSW3344 | This study |
| SWY3825 | MATα ade2 ura3 his3 leu2 trp1 can1 psi−+ pSW3345 | This study |
| SWY3826 | MATα ade2 ura3 his3 leu2 trp1 can1 psi−+ pSW399 | This study |
| W303 | MATa ade2-1 ura3-1 his3-11,15 leu2-3,112 trp1-1 can1-100 psi− | Ref. 40 |
| SWY4209 | MATα gle1-4 ade2 ura3 his3 leu2 trp1 psi− | This study |
| SWY4014 | MATα rat8-2 (dbp5) ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 psi− | This study |
| SWY1790 | MATα ipk1::KAN ade2 ura3 his3 leu2 trp1 psi− | Ref. 33 |
| Plasmids used | ||
| pSW399 | GLE1/CEN/LEU2 | Ref. 16 |
| pSW410 | GLE1/CEN/URA3 | Ref. 16 |
| pSW3343 | gle1-K377Q/CEN/LEU2 | This study |
| pSW3344 | gle1-K494Q/CEN/LEU2 | This study |
| pSW3345 | gle1-K377Q/K378Q/CEN/LEU2 | This study |
| pSW3409 | lacZ-linker-luciferase CEN/URA3 | Ref. 33 |
| pSW3410 | lacZ-TAG-luciferase CEN/URA3 | Ref. 33 |
| pSW3242 | pMAL-TEV-GLE1 | Ref. 15 |
| pSW3291 | pMAL-TEV-gle1-K377Q | This study |
| pSW3292 | pMAL-TEV-gle1-K494Q | This study |
| pSW3293 | pMAL-TEV-gle1-K377Q/K378Q | This study |
| pSW3295 | pMAL-TEV-gle1-R417Q | This study |
| pSW3319 | pGEX-DBP5 | Ref. 15 |
Protein Purification and IP6 Preparation
Recombinant, bacterially expressed, and untagged Dbp5 and Gle1 protein were purified as described previously (15). After purification, the proteins were dialyzed in Buffer B (20 mm Hepes-HCl, pH 7.5, 150 mm NaCl, 20% (w/v) glycerol). IP6 (Sigma) was prepared by resuspension in 50 mm Hepes-HCl, pH 7.5, and adjusting pH with 10 n NaOH to reach ∼pH 7.5. pH was tested by paper indicator (litmus). The IP6 solution was stored at 4 °C.
ATPase Assays
Dbp5 ATPase assays were conducted as described previously (41) with the stated modifications. The ATPase reaction was conducted in a total volume of 60 μl containing 10 mm Hepes-HCl, pH 7.5, 45 mm NaCl, 3 mm MgCl2, 1 mm dithiothreitol, 3 mm phosphoenolpyruvate, 0.21 mm NADH, 0.333 unit/μl SUPERasin (Ambion), 0.777 units of pyruvate kinase-lactate dehydrogenase (Sigma), and 1 mm ATP (Roche Applied Science). Varying amounts of protein, RNA (25-mer poly(A)), and IP6 were used, and the reactions were started by the addition of ATP/Mg2+. Measurements were taken every 20 s by detecting the A340 using a Synergy HT Multi-mode microplate reader (Biotek). The rate of A340 signal decline was then utilized to measure steady-state ATPase activity.
IP6 Binding Assays
Polyethylene glycol precipitation for IP6 binding assays were performed as described in Ref. 23 with minor modifications. In brief, 60-μl binding reactions were conducted with 10 nm [3H]IP6 (21.4 Ci/mmol; PerkinElmer Life Sciences) in Buffer A (16 mm Hepes, pH 7.5, 120 mm NaCl, 3 mm MgCl2, 16% (w/v) glycerol, and 1 mg/ml bovine serum albumin) supplemented with 1 unit/μl SUPERasin. Gle1, Dbp5, RNA (25-mer poly(A)+), with or without 1 mm nucleotides. The samples were mixed and incubated at room temperature for 10 min. The proteins were precipitated by adding 40 μl of 30% polyethylene glycol (Sigma), followed by thorough mixing. Precipitated proteins with bound [3H]IP6 were separated by centrifugation for 25 min at full speed at 4 °C in a Beckman bench top centrifuge. The supernatant was carefully aspirated, and the pellets were solubilized with 300 μl of 1% SDS overnight. Radioactivity was measured by scintillation counting (Beckman LS6500). Counting efficiency was determined empirically, and the specific activity conversion for [3H]IP6 stock was calculated as 19.5 cpm/fmol. Background correction and determination of equilibrium binding and dissociation constants were done using GraphPad Prism v.4 software. The data points were fitted to a single-binding site hyperbolic equation: Y = Bmax*X/(Kd + X).
Glutathione S-Transferase Pulldown Assays
For in vitro Dbp5-Gle1 binding assays, 500 nm recombinant glutathione S-transferase-Dbp5 and Gle1 were incubated in Buffer B with 3 mm MgCl2 in the presence or absence of 200 nm IP6 and glutathione resin at room temperature for 40 min. Supernatants and beads were transferred twice to fresh tubes to remove nonspecific Gle1 binding to the tubes, then washed four times with buffer, and resuspended in SDS sample buffer. The samples were diluted, run on SDS-PAGE, and immunoblotted using Dbp5 and Gle1 antibodies (15, 33). The majority of input Gle1 was bound to tubes and was not observed in either bound or unbound fractions (data not shown). We estimate that 25–35% of free Gle1 bound to Dbp5.
Microscopic Analysis of mRNA Export Defects and Gle1 Localization
The localization of poly(A)+ RNA was analyzed by growing strains in rich medium at 23 °C prior to shifting to 30 or 37 °C for 1 h. The cells were collected, fixed in 3.7% formaldehyde and 20% methanol, and washed before processing for indirect fluorescence in situ hybridizations using an digoxigenin-oligo(dT)30 probe as described previously (34). Indirect immunofluorescence was performed as described previously (42). The cells were incubated at 4 °C overnight with previously described Gle1 antibodies (33). Primary antibody was detected using fluorescein isothiocyanate-conjugated anti-guinea pig IgG, and the nuclei were visualized by DAPI staining. The cells were observed using a fluorescent microscope (model BX50; Olympus, Lake Success, NY) using an Uplan 100×/1.3 objective. The images were taken using a digital camera (Photometrics Cool Snap HQ; Roper Scientific) and processed using MetaVue software (Universal Imaging, West Chester, PA) and Adobe Photoshop 7.0. Analysis and quantification of mRNA export assays were done by identifying the percentages of cells with a visible concentration of nuclear oligo(dT) signal in DAPI-stained cells.
Immunoblot Analysis
Analysis of Gle1 protein levels in selected strains was performed using immunoblotting. The cells grown at 23 °C were shifted to higher temperatures for 1 h as indicated, collected, washed, and processed for crude cell lysis as described previously (43). The proteins were separated by electrophoresis in SDS-PAGE gels for immunoblot analysis with anti-Gle1 and anti-Pgk1 (mAb22c5; Molecular Probes).
Termination Read-through
Assays with tandem reporters were performed essentially as described in Refs. 33 and 44. Strains containing the reporter plasmids were grown in selective medium and then diluted overnight in YPD at 23 °C. The cultures were rapidly shifted by dilution with prewarmed medium and incubated at 37 °C for 30 min prior to harvest. Significance was determined by Student's t test.
Sequence Alignments
For the alignment in Fig. 2A and supplemental Fig. S2, sequences of GLE1 orthologues in fungal and metazoan species were obtained from the Fungal Genome Initiative, Broad Institute (Cambridge, MA), ZFIN (Eugene, Oregon), and NCBI (Bethesda, MD). Alignment was performed with a modified Clustal-W algorithm in AlignX (Invitrogen). For statistical analysis of the alignments, pair-wise BLAST similarity searches (NCBI) between scGle1 and its orthologues were performed (45). Expected (E) values for the highest scoring segment were obtained and converted to p values. The scored alignment covered 63–81% of the protein sequences for each pair and always included the C-terminal half of Gle1 and the sequences shown in Fig. 2A.
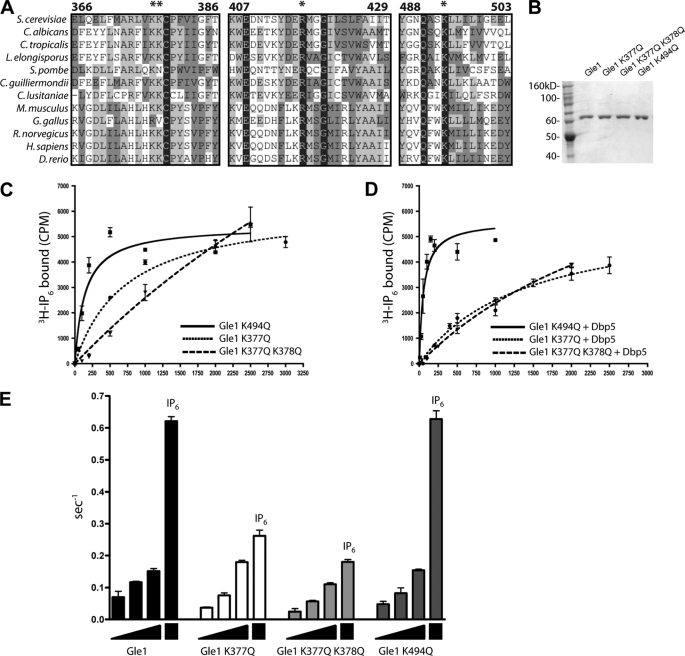
FIGURE 2.
Gle1 residues Lys377 and Lys378 are required for the IP6-mediated Dbp5 ATPase stimulation. A, sequence alignment of conserved regions in the C-terminal domain of Gle1 from selected fungal and metazoan species. Residues with greater homology identified by a modified Clustal-W alignment (see “Experimental Procedures”) are depicted in increasingly darker gray. The asterisks denote amino acids selected as putative IP6 binding residues. B, bacterially expressed, purified recombinant Gle1, gle1-K377Q, gle1-K377Q/K378Q, and gle1-K494Q were separated in a 7.5% SDS-polyacrylamide gel and stained with Coomassie Blue. C, equilibrium binding assays utilizing 10 nm [3H]IP6 and increasing amounts of Gle1 were used to calculate the Kd of Gle1 proteins for IP6. D, equilibrium binding assays as shown in C in the presence of 1 μm Dbp5. E, Dbp5 ATPase assays were conducted utilizing 100, 400, or 800 nm of Gle1, 1 mm ATP, 10 μm RNA, and 200 nm Dbp5. 100 nm IP6 was added to each sample containing 800 nm Gle1. The mean and standard error of the mean were calculated from three independent experiments in C–E.
RESULTS
Dbp5 Binding to Gle1 Enhances the Gle-IP6 Interaction
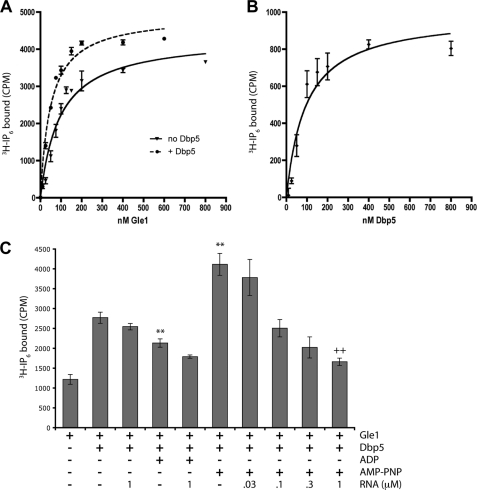
We and others have previously demonstrated that Gle1 binds IP6 in vitro and that IP6 production is important for Gle1 function (21, 23, 24, 34). Moreover, in vitro Dbp5 appeared to enhance Gle1 binding to IP6. To further define the biochemical interactions between these factors, we experimentally determined the apparent dissociation constant (Kd) between full-length Gle1 and IP6. Equilibrium binding assays with [3H]IP6 showed that Gle1 and IP6 interact with an apparent Kd of 94 nm (with a 95% confidence interval from 67 to 121 nm) (Fig. 1A and Table 2). Next we measured the impact of Dbp5 on the Gle1-IP6 interaction. In the presence of Dbp5, Gle1 bound IP6 with an apparent Kd of 50 nm and a 95% confidence interval from 39 to 60 nm (Fig. 1A). To gain insight into Gle1-Dbp5 association, we also analyzed the saturation of Gle1-IP6 binding enhancement by Dbp5. With a Gle1 concentration of 25 nm and an IP6 concentration of 10 nm, the half-maximal concentration of Dbp5 needed to impact Gle1-IP6 binding was 69 nm (Fig. 1B), with a 95% confidence interval from 51 to 137 nm. This result was consistent with a strong but transient Dbp5-Gle1 interaction. We did not detect an alteration in Gle1 binding to Dbp5 in vitro in the presence of IP6 (supplemental Fig. S1).
FIGURE 1.
Dbp5 stimulates binding of Gle1 and IP6. A, equilibrium binding assays utilizing 10 nm [3H]IP6 and Gle1 (solid line) or Gle1 + 1 μm Dbp5 (dashed line) were used to determine the dissociation constant (Kd) of Gle1 for IP6. 19.5 cpm = 1 fmol of IP6 bound. B, equilibrium binding assays as in A were used to measure the stimulation of Dbp5 for Gle1-IP6 binding using 25 nm Gle1 and 10 nm [3H]IP6. Gle1-IP6 binding in the absence of Dbp5 was subtracted out to achieve the saturation curve shown. C, equilibrium binding assays as in A were used to test the effect of known regulators of Dbp5 activity on Gle1-IP6 binding. 10 nm [3H]IP6, 25 nm Gle1, and 100 nm Dbp5 were used for these assays. **, p < 0.01 versus Gle1 + Dbp5; ++, p < 0.01 versus Gle1 + Dbp5 + AMP-PNP by Student's t test. The mean and standard error of the mean were calculated from three to eight independent experiments in A–C.
TABLE 2.
Dissociation constants for Gle1 binding to IP6
Equilibrium binding assays were performed with [3H]IP6 and Gle1 in the presence or absence of Dbp5. Dissociation constants (Kd) and 95% confidence intervals were determined using GraphPad Prism v.4 software.
| Protein | Kd of IP6 binding | 95% confidence interval |
|---|---|---|
| nm | nm | |
| Gle1 | 94 | 67–122 |
| gle1-K377Q | 724 | 245–1200 |
| gle1-K377Q/K378Q | 8250 | 0–16700 |
| gle1-K494Q | 141 | 65–217 |
| Gle1 + 1 μm Dbp5 | 50 | 39–60 |
| gle1-K377Q + 1 μm Dbp5 | 1380 | 811–1940 |
| gle1-K377Q/K378Q + 1 μm Dbp5 | 3370 | 1750–4990 |
| gle1-K494Q + 1 μm Dbp5 | 56 | 27–84 |
Because Dbp5 also binds both nucleotides and RNA, we determined whether the Dbp5-dependent stimulation of IP6 by Gle1 was affected by the presence of RNA or nucleotides (Fig. 1C). We tested the ability of Dbp5 to stimulate Gle1-IP6 binding in the presence of saturating levels of ADP or the nonhydrolyzable ATP analogue AMP-PNP, factors known to induce distinct structural changes in Dbp5 (15, 31, 46, 47). Interestingly, Dbp5-AMP-PNP further enhanced Gle1-IP6 binding, whereas Dbp5-ADP had a reduced ability to stimulate IP6 binding compared with Dbp5 in the absence of nucleotide (Fig. 1C). However, the addition of RNA to Dbp5-AMP-PNP resulted in a dramatic reduction of stimulation of Gle1-IP6 binding in a dose-dependent fashion. The addition of RNA to Dbp5-ADP or to Dbp5 in the absence of nucleotide had only small effects (Fig. 1C), likely because of the low RNA binding affinity of the apo and ADP-bound forms of Dbp5 (24). These results suggest that different conformations of Dbp5 can differentially affect Gle1-IP6 binding.
Critical Residues in the Gle1 C-terminal Domain Are Required for IP6 Binding
The high resolution crystal structures of the C-terminal domain for human ADAR2 (48) and the F-box protein subunit of the ubiquitin ligase complex SCF, Tir1 (49), reveal a molecule of IP6 embedded in the tertiary structure of the respective protein. In both proteins, a highly positively charged pocket lined with several lysine and arginine residues formed the IP6-binding site. However, comparison of the ADAR2 and Tir1 structures have not defined a simple IP6-binding consensus sequence or domain. Rather, the interacting K and R residues are dispersed throughout a domain of several hundred residues. However, in the case of ADAR2, the residues responsible for IP6 binding are highly conserved with the budding yeast tRNA editing enzyme Tad1 (48). Because nuclear mRNA accumulation is observed in both human cells with perturbed IP6 levels (50) and in Schizosaccharomyces pombe harboring a ipk1+ deletion (51), we hypothesized that the residues coordinating IP6 binding were also conserved among Gle1 homologues. Sequence alignments were conducted by comparing sequences of GLE1 orthologues from a variety of fungal and metazoan species with a modified Clustal-W algorithm (52). This identified four conserved positively charged residues that were present within stretches of medium to high conservation in the C-terminal domain of Gle1 (Fig. 2A and supplemental Fig. S2).
We targeted the sequence encoding these conserved residues for site-directed mutagenesis, replacing codons for lysine or arginine residues with glutamine-encoding codons to conserve the polarity and relative size of the side chains. To test for IP6 binding, recombinant proteins were expressed and purified from bacteria. Recombinant gle1-R417Q protein (harboring a Glu at position 417) was insoluble and was not analyzed further (data not shown). However, the gle1-K377Q, gle1-K377Q/K378Q, and gle1-K494Q proteins were soluble and could be purified to homogeneity (Fig. 2B). [3H]IP6 equilibrium binding assays were performed (Fig. 2C and Table 2). IP6 binding affinity was unaffected with the gle1-K494Q protein, and the addition of Dbp5 enhanced IP6 binding of gle1-K494Q to levels similar to that observed for wild type Gle1 (Fig. 2D). However, the gle1-K377Q and gle1-K377Q/K378Q proteins had nearly 8- and 90-fold reductions in Gle1-IP6 binding affinity, respectively (724 and 8250 nm). In addition, the gle1-K377Q and gle1-K377Q/K378Q proteins had a significant deficiency in Dbp5-stimulated IP6 binding. This defined Lys377 and Lys378 in Gle1 as critical residues for IP6 binding.
Conserved Gle1 Residues Lys377 and Lys378 Are Necessary for IP6-mediated Dbp5 Stimulation in Vitro
We and others have previously shown that Gle1 stimulates the ATPase activity of Dbp5 in an IP6-independent fashion; however, IP6 greatly potentiates this stimulation (23, 24). To test the role of Gle1 residues Lys494, Lys377, and Lys378 in the IP6-dependent stimulation of Dbp5 ATPase activity, we performed ATPase assays with increasing concentrations of altered gle1 proteins. Interestingly, wild type Gle1, gle1-K377Q, gle1-K377Q/K378Q, and gle1-K494Q exhibited roughly similar Dbp5 ATPase activation in the absence of IP6, although gle1-K377Q/K378Q appeared to be partially reduced (Fig. 2E). This indicated that the gle1 proteins were still functional for basic Dbp5 interaction and activation. We further tested for stimulation of Dbp5 ATPase activity by adding 100 nm IP6 to the samples containing the highest Gle1 concentrations (800 nm). In samples containing wild type Gle1 and gle1-K494Q, the addition of IP6 induced ATPase activity dramatically. However, little or no increase in ATPase activity was observed in samples containing gle1-K377Q or gle1-K377Q/K378Q protein after IP6 addition. These results show that it is possible to decouple IP6 binding from Dbp5 stimulation by Gle1. Taken together, these assays strongly argue that the conserved Lys377 and Lys378 residues in Gle1 are required for coordinating IP6 binding and potentiating Gle1 activation of Dbp5 ATPase activity in vitro.
Cells Harboring gle1-K377Q and gle1-K377Q/K378Q Mutants Have mRNA Export Defects
Our previous studies found that S. cerevisiae cells harboring the ipk1Δ mutation do not produce IP6 and exhibit an mRNA export defect at 37 °C (34). This phenotype is presumably due to a lack of cellular IP6 for Gle1 binding and thus diminished Dbp5 ATPase activation. The gle1 mutants with defects in IP6 binding allow a direct test for whether Gle1 is the major IP6 target in the mRNA export mechanism. If Gle1 is the main IP6 target, we speculated that S. cerevisiae cells expressing gle1-K377Q and gle1-K377Q/K378Q would have mRNA export defects when grown at 37 °C, similar to ipk1Δ cells. Further, these gle1 mutants should have a translation termination defect similar to the one shown in strains harboring the ipk1Δ mutation (33). On the other hand, cells expressing wild type GLE1 and gle1-K494Q should have wild type phenotypes. However, if IP6 targets proteins other than Gle1 in its regulation of export and translation, cells expressing gle1-K377Q and gle1-K377Q/K378Q might have weaker mRNA export and translation termination defects.
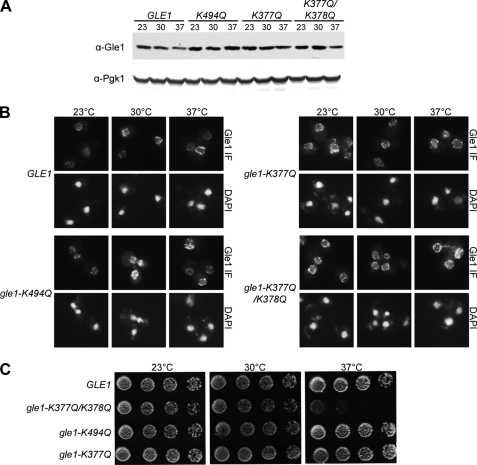
For in vivo functional studies, we generated S. cerevisiae strains using a plasmid shuffle strategy in which the chromosomal copy of GLE1 was deleted and either wild type or mutant alleles of GLE1 were expressed from a CEN plasmid (under control of the GLE1 promoter). Expression levels of Gle1 were tested by immunoblotting cell lysate from strains grown at 23 °C and shifted to 30 or 37 °C for 1 h (Fig. 3A). Expression of Pgk1 was used as a loading control. Gle1 levels did not drastically change in any condition tested; thus, the mutations introduced in GLE1 did not alter protein stability in vivo. We next analyzed the steady-state localization of Gle1 in the strains at 23, 30, and 37 °C. Immunofluorescence microscopy was conducted using an anti-Gle1 antibody, and all strains showed nuclear rim staining (Fig. 3B). Furthermore, all of the strains showed a distribution comparable with that of wild type Gle1 for localization at the nuclear rim and in the cytoplasm, suggesting that the overall interaction of Gle1 with NPC components was not affected.
FIGURE 3.
Protein stability, localization, and growth phenotype of strains expressing gle1 mutant alleles. A, indicated strains were grown at 23 °C and shifted to 30 and 37 °C for 1 h prior to immunoblot analysis utilizing anti-Gle1 polyclonal antibodies. Anti-Pgk1 was used as a loading control. B, Gle1 localization was observed by indirect immunofluorescence microscopy utilizing anti-Gle1 polyclonal antibodies, and the nuclei were visualized with DAPI. gle1Δ mutant strains carrying plasmids harboring GLE1, gle1-K377Q, gle1-K377Q/K378Q, or gle1-K494Q were grown at 23 °C and shifted to 30 and 37 °C for 1 h prior to processing. C, indicated strains were spotted in 5-fold serial dilutions and incubated in rich medium at 23, 30, and 37 °C.
To assess the viability of strains expressing the different GLE1 alleles, we spotted 5-fold serial dilutions of yeast S. cerevisiae cells harboring gle1 mutants and grew them on YPD plates for 3–4 days (Fig. 3C). No growth defects were observed in gle1-K377Q or gle1-K494Q strains; however, the gle1-K377Q/K378Q strain showed marked temperature sensitivity at 37 °C. We concluded that gle1-K377Q/K378Q is unable to fully stimulate Dbp5 in vivo, consistent with the partially diminished Dbp5 stimulation observed in vitro with the gle1-K377Q/K378Q protein (Fig. 2E). These results suggest that a threshold of Dbp5 activity might be required for viability, consistent with another recent report analyzing dbp5 mutants (26).
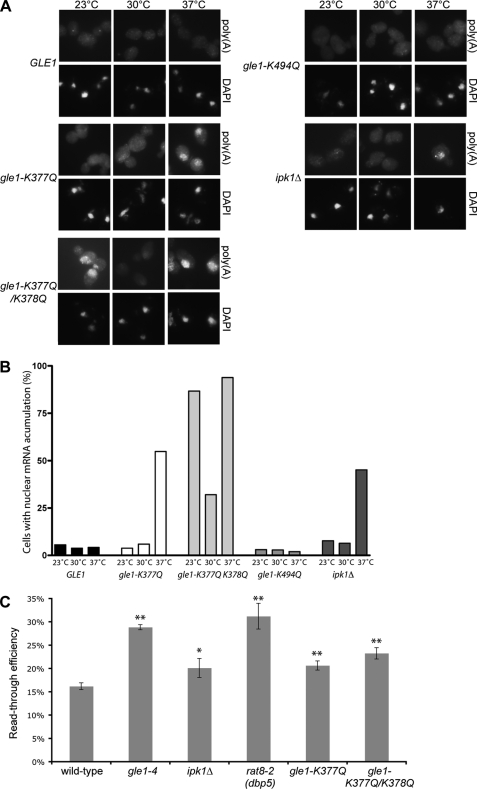
We next tested the mRNA export efficiency of cells harboring gle1 mutant alleles by in situ hybridization for poly(A)+ localization. In agreement with the lack of IP6 binding in vitro, nuclear mRNA accumulation was observed in both gle1-K377Q and gle1-K377Q/K378Q strains at 37 °C but not in GLE1 or gle1-K494Q strains (Fig. 4, A and B). Importantly, similar to ipk1Δ strains, all of the strains showed normal mRNA export at 30 °C. Thus, the IP6-binding mutants of gle1 phenocopied the mRNA export defects in ipk1Δ strains. This indicated that IP6 function in mRNA export is mediated primarily, if not solely, through Gle1. Notably, gle1-K377Q/K378Q also showed nuclear mRNA accumulation at 23 °C, a phenotype not detected in the ipk1Δ strain or in the other gle1 point mutant strains, suggesting that additional cold-sensitive defects may be present in the gle1-K377Q/K378Q mutant. Consistent with this interpretation, the gle1-K377Q/K378Q strain also had minor growth defects at 16 °C on synthetic medium (data not shown). Taken together, these observations confirm our biochemical analysis and corroborate the role of Gle1 residues Lys377 and Lys378 in IP6 binding and in stimulation of Dbp5 ATPase activity in vivo and in vitro.
FIGURE 4.
In vitro IP6 binding defects correlate with mRNA export and translation termination defects in vivo. A, an ipk1Δ strain and gle1Δ mutant strains carrying plasmids harboring either GLE1, gle1-K377Q, gle1-K377Q/K378Q, or gle1-K494Q were used for in situ hybridization with a digoxigenin-coupled oligo(dT) probe after growing at 23 °C and shifting to 30 or 37 °C for 1 h. Poly(A)+ RNA localization was visualized by indirect immunofluorescence microscopy with a fluorescein isothiocyanate-coupled anti-digoxigenin antibody, and the nuclei were visualized with DAPI staining as indicated. B, quantification of cells in A presenting nuclear mRNA accumulation. The bars represent the percentages of cells with mRNA export defects from a total of 50–130 cells/condition. C, wild type (W303), gle1-4, ipk1Δ, rat8-2 (dbp5), gle1-K377Q, and gle1-K377Q/K378Q strains transformed with TQ/U and TMV/U tandem reporter constructs were grown overnight at 23 °C and shifted to 37 °C for 30 min prior to harvest. Luciferase and β-galactosidase assays were performed, and the ratios of the activities were calculated. Read-through activity is expressed as the percentage of the TMV reporter compared with TQ control for each strain. The mean and standard error of the mean were calculated from three to eight independent experiments. *, p < 0.05; **, p < 0.01 by Student's t test.
The gle1-K377Q and gle1-K377Q/K378Q Mutants Have Translation Termination Defects
Similar to S. cerevisiae strains with mutations in GLE1 and DBP5, ipk1Δ cells show a defect in translation termination (32, 33). Additionally, coincident deletion of IPK1 aggravates the growth defects observed in budding yeast also harboring mutations in SUP45 (yeast eRF1), in striking resemblance to gle1 and dbp5 mutants. We speculated that IP6 specifically targets Gle1 during translation termination. To test this hypothesis, we utilized read-through termination assays as described previously (33, 44). A reporter containing a β-galactosidase/luciferase fusion separated by a linker sequence with an in-frame stop codon was transformed into the respective wild type and mutant strains. Comparing the ratio of β-galactosidase to luciferase activity with that from a second reporter lacking the stop codon gave a measure of the efficiency of translation termination. During our further analysis of the ipk1Δ strain, we found that several of the original parental strains used in Bolger et al. (33) were infected with Psi (Ψ), a prion form of the Sup35 termination factor (53). To address this issue, we undertook curing of the PSI+ strains by growth on medium containing guanidine hydrochloride (39) and retested the strains in termination assays. As shown in Fig. 4C, the defects in termination read-through were still detected, although the magnitude was reduced compared with our previous results (33). Notably, the defect in a gle1-4 strain was now more similar to that of the rat8-2 (dbp5) strain. Conversion from PSI+ to psi− did not affect synthetic growth defects in sup45-2 gle1-4 or sup45–2 ipk1Δ double mutants, nor did it qualitatively affect ade2-1 nonsense suppression (data not shown).
The gle1-K377Q and gle1-K377Q/K378Q strains were directly tested in the read-through assay (Fig. 4C). Interestingly, read-through in these mutant strains was significantly higher than that in wild type control strains, particularly in the gle1-K377Q/K378Q strain. Therefore, these strains have defects in termination. Of note, the read-through defects were not as high as that observed in gle1-4 and rat8-2 (dbp5) mutant strains. Instead, they were more similar to the level of defect observed in an ipk1Δ strain (Fig. 4C). Thus, as with mRNA export, the gle1 IP6-binding mutants recapitulated the defects observed in ipk1Δ cells for translation termination, suggesting that Gle1 is the primary IP6-binding target during translation.
DISCUSSION
Here we report that Gle1 is the critical target of IP6 for its roles in gene expression during mRNA export and translation termination. Importantly, we present biochemical and functional data that support a model in which Gle1 is engaged by IP6, and together Gle1-IP6 associates strongly with Dbp5 to stimulate Dbp5 ATPase activity both at the NPC and at sites of translation termination in the cytoplasm. We also find that the affinity of Gle1 for IP6 is very high (Kd of 94 nm) and that this association is enhanced in the presence of Dbp5 (Fig. 1A). Although in vitro studies have demonstrated that Gle1 is sufficient for IP6-mediated stimulation of Dbp5 activity (23, 24), whether IP6 has additional targets that regulate mRNA export or translation was unknown. Here, the mutant versions of Gle1 that have decreased IP6 interaction not only define key IP6 binding determinants but also allowed us to test for other such IP6 effectors in these mechanisms. These strains faithfully recapitulate the mRNA export and translation termination defect present in ipk1Δ strain (Fig. 4). This observation expands our in vitro studies and strongly suggests that Gle1, not another protein(s) in the export and translation pathways, is the main factor responsible for mediating IP6 roles in mRNA export and translation termination.
It has not been directly shown whether the IP6 binding by Gle1 is conserved in mammalian cells. Previous studies have revealed a conserved role for Gle1 and IP6 in mRNA export in S. cerevisiae, S. pombe, and human cells, placing both factors in the same pathway (16, 18, 19, 34, 51). We have now identified two positively charged residues in the C-terminal domain of Gle1 (Lys377 and Lys378) that are required for IP6 binding in S. cerevisiae and that are conserved in mammalian Gle1. Given the relatively high conservation of the C-terminal domain of Gle1, we suggest that mammalian Gle1 is also targeted by IP6 during mRNA export. However, the requirement of Gle1 for IP6 may be different across species.
We propose that the role of Gle1-IP6 is that of an ATPase-activating factor, stimulating Dbp5 activity and supporting a switch from Dbp5-ATP to Dbp5-ADP. But how is this activation achieved? Our data begin to address some possible mechanisms of action. First, the gle1 mutants that abrogate IP6 binding do not have an impact in IP6-independent Dbp5 stimulation in vitro (Fig. 2E), suggesting that the binding site in Gle1 for IP6 is, at least in part, distinct from the Dbp5 binding surface. Our data suggest that Dbp5 interacts with Gle1 very tightly (Fig. 1B), and we were not able to detect an IP6 alteration in the association of Gle1 and Dbp5 in vitro (supplemental Fig. S1). These results argue against a molecular glue model where IP6 binds a pocket formed between the two proteins. A second model is that Gle1-IP6 stabilizes a particular conformation of Dbp5. Our findings that Gle1-IP6 binding is differentially stimulated by the nucleotide and RNA binding state of Dbp5 (Fig. 1C) are consistent with this model. Although an indirect assessment, these binding data suggest that Dbp5-ATP and Dbp5-RNA-ATP represent distinct forms. The enhanced stimulation of Gle1-IP6 binding by Dbp5-AMP-PNP in the absence of RNA suggests that this is the preferential conformation for association. We speculate that dynamic associations could possibly sustain a rapid ATP hydrolysis cycle and guarantee the cycles required to sustain export rates through the NPC.
Other models for Gle1-IP6 interaction are also possible, such as allosteric changes or direct participation of IP6 in the Dbp5 ATPase reaction. Understanding the differences between in vitro and in vivo IP6 binding conditions will also be crucial for defining the full mechanism of Dbp5 activation. The nucleoporins Nup159 and Nup42 bind to Dbp5 and Gle1, respectively (16, 20, 22), providing a potential scaffold for the interaction of these proteins, and additional cellular factors that may regulate and participate in this interaction. Nonetheless, this study adds to previous work in supporting a pivotal role for the Gle1-IP6-driven ATPase stimulation of Dbp5 in control over mRNP remodeling events in mRNA export.
We also present further evidence of a direct role for Gle1 binding of IP6 in translation termination. As in mRNA export, we propose that this interaction potentiates the ATPase activity of Dbp5 during translation termination. A goal of our future studies will be to identify the targets of the potential Dbp5 remodeling activity during translation termination. Whether IP6 is an obligatory cofactor or a regulator of Gle1 is still unclear. Genetic manipulation of GLE1 has now allowed us to uncouple IP6 signaling from mRNA export and provides a platform to further investigate this possibility. IP6 might only associate with Gle1 to coordinate specific cellular functions. Our previous work suggests that Gle1 and IP6 do not always work together (33). Interestingly, Gle1 is required in translation initiation, but apparently Dbp5 and IP6 are not (33). A plausible explanation is that Gle1 targets a different factor that operates exclusively in translation initiation. Defining the mechanism by which Gle1 functions in translation initiation may help elucidate the linkage of IP6 regulation to Dbp5 during export and translation termination.
This study adds to the growing understanding of how phosphatidylinositol phosphates and inositol polyphosphates potentially coordinate the regulation of gene expression. Signal transduction pathways that utilize phosphatidylinositol phosphates and inositol polyphosphates as secondary messengers might not only regulate transcription factor activation but also orchestrate the different steps in gene expression. Future studies will be required to further delineate the full range of requirements for these small molecules in gene expression.
Supplementary Material
Acknowledgments
We thank Yingna Zhou for protein purification and other members of the Wente lab for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-GM51219 (to S. R. W.) and F32-GM082065 (to T. A. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- mRNP
- messenger ribonucleoprotein complex
- NPC
- nuclear pore complex
- Nup
- nuclear pore complex protein
- IP6
- inositol hexakisphosphate
- mRNA
- messenger RNA
- DAPI
- 4′,6′-diamino-2-phenylindole
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate.
REFERENCES
- 1.Köhler A., Hurt E. (2007) Nat. Rev. Mol. Cell Biol. 8, 761–773 [DOI] [PubMed] [Google Scholar]
- 2.Iglesias N., Stutz F. (2008) FEBS Lett. 582, 1987–1996 [DOI] [PubMed] [Google Scholar]
- 3.Kelly S. M., Corbett A. H. (2009) Traffic 10, 1199–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmody S. R., Wente S. R. (2009) J. Cell Sci. 122, 1933–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hieronymus H., Silver P. A. (2004) Genes Dev. 18, 2845–2860 [DOI] [PubMed] [Google Scholar]
- 6.Saguez C., Olesen J. R., Jensen T. H. (2005) Curr. Opin. Cell Biol. 17, 287–293 [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez M. S., Dargemont C., Stutz F. (2004) Biol. Cell 96, 639–655 [DOI] [PubMed] [Google Scholar]
- 8.McKee A. E., Silver P. A. (2007) Cell Res. 17, 581–590 [DOI] [PubMed] [Google Scholar]
- 9.Grüter P., Tabernero C., von Kobbe C., Schmitt C., Saavedra C., Bachi A., Wilm M., Felber B. K., Izaurralde E. (1998) Mol. Cell 1, 649–659 [DOI] [PubMed] [Google Scholar]
- 10.Segref A., Sharma K., Doye V., Hellwig A., Huber J., Lührmann R., Hurt E. (1997) EMBO J. 16, 3256–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katahira J., Strässer K., Podtelejnikov A., Mann M., Jung J. U., Hurt E. (1999) EMBO J. 18, 2593–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terry L. J., Wente S. R. (2007) J. Cell Biol. 178, 1121–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daneholt B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7012–7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lund M. K., Guthrie C. (2005) Mol. Cell 20, 645–651 [DOI] [PubMed] [Google Scholar]
- 15.Tran E. J., Zhou Y., Corbett A. H., Wente S. R. (2007) Mol. Cell 28, 850–859 [DOI] [PubMed] [Google Scholar]
- 16.Murphy R., Wente S. R. (1996) Nature 383, 357–360 [DOI] [PubMed] [Google Scholar]
- 17.Del Priore V., Snay C. A., Bahr A., Cole C. N. (1996) Mol. Biol. Cell 7, 1601–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watkins J. L., Murphy R., Emtage J. L., Wente S. R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6779–6784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moon D., Bae J. A., Cho H. J., Yoon J. H. (2008) J. Microbiol. 46, 422–428 [DOI] [PubMed] [Google Scholar]
- 20.Strahm Y., Fahrenkrog B., Zenklusen D., Rychner E., Kantor J., Rosbach M., Stutz F. (1999) EMBO J. 18, 5761–5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller A. L., Suntharalingam M., Johnson S. L., Audhya A., Emr S. D., Wente S. R. (2004) J. Biol. Chem. 279, 51022–51032 [DOI] [PubMed] [Google Scholar]
- 22.Hodge C. A., Colot H. V., Stafford P., Cole C. N. (1999) EMBO J. 18, 5778–5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alcázar-Román A. R., Tran E. J., Guo S., Wente S. R. (2006) Nat. Cell Biol. 8, 711–716 [DOI] [PubMed] [Google Scholar]
- 24.Weirich C. S., Erzberger J. P., Flick J. S., Berger J. M., Thorner J., Weis K. (2006) Nat. Cell Biol. 8, 668–676 [DOI] [PubMed] [Google Scholar]
- 25.Kendirgi F., Barry D. M., Griffis E. R., Powers M. A., Wente S. R. (2003) J. Cell Biol. 160, 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dossani Z. Y., Weirich C. S., Erzberger J. P., Berger J. M., Weis K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16251–16256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snay-Hodge C. A., Colot H. V., Goldstein A. L., Cole C. N. (1998) EMBO J. 17, 2663–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tseng S. S., Weaver P. L., Liu Y., Hitomi M., Tartakoff A. M., Chang T. H. (1998) EMBO J. 17, 2651–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Estruch F., Cole C. N. (2003) Mol. Biol. Cell 14, 1664–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J., Jin S. B., Björkroth B., Wieslander L., Daneholt B. (2002) EMBO J. 21, 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan J. S., Cheng Z., Zhang J., Noble C., Zhou Z., Song H., Yang D. (2009) J. Mol. Biol. 388, 1–10 [DOI] [PubMed] [Google Scholar]
- 32.Gross T., Siepmann A., Sturm D., Windgassen M., Scarcelli J. J., Seedorf M., Cole C. N., Krebber H. (2007) Science 315, 646–649 [DOI] [PubMed] [Google Scholar]
- 33.Bolger T. A., Folkmann A. W., Tran E. J., Wente S. R. (2008) Cell 134, 624–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.York J. D., Odom A. R., Murphy R., Ives E. B., Wente S. R. (1999) Science 285, 96–100 [DOI] [PubMed] [Google Scholar]
- 35.Alcázar-Román A. R., Wente S. R. (2008) Chromosoma 117, 1–13 [DOI] [PubMed] [Google Scholar]
- 36.Saiardi A., Erdjument-Bromage H., Snowman A. M., Tempst P., Snyder S. H. (1999) Curr. Biol. 9, 1323–1326 [DOI] [PubMed] [Google Scholar]
- 37.Mulugu S., Bai W., Fridy P. C., Bastidas R. J., Otto J. C., Dollins D. E., Haystead T. A., Ribeiro A. A., York J. D. (2007) Science 316, 106–109 [DOI] [PubMed] [Google Scholar]
- 38.Ito H., Fukuda Y., Murata K., Kimura A. (1983) J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chernoff Y. O., Uptain S. M., Lindquist S. L. (2002) Methods Enzymol. 351, 499–538 [DOI] [PubMed] [Google Scholar]
- 40.Du J., Nasir I., Benton B. K., Kladde M. P., Laurent B. C. (1998) Genetics 150, 987–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang T. G., Hackney D. D. (1994) J. Biol. Chem. 269, 16493–16501 [PubMed] [Google Scholar]
- 42.Wente S. R., Rout M. P., Blobel G. (1992) J. Cell Biol. 119, 705–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yaffe M. P., Schatz G. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 4819–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stahl G., Bidou L., Rousset J. P., Cassan M. (1995) Nucleic Acids Res. 23, 1557–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 46.von Moeller H., Basquin C., Conti E. (2009) Nat. Struct. Mol. Biol. 16, 247–254 [DOI] [PubMed] [Google Scholar]
- 47.Napetschnig J., Kassube S. A., Debler E. W., Wong R. W., Blobel G., Hoelz A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3089–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macbeth M. R., Schubert H. L., VanDemark A. P., Lingam A. T., Hill C. P., Bass B. L. (2005) Science 309, 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan X., Calderon-Villalobos L. I., Sharon M., Zheng C., Robinson C. V., Estelle M., Zheng N. (2007) Nature 446, 640–645 [DOI] [PubMed] [Google Scholar]
- 50.Feng Y., Wente S. R., Majerus P. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 875–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarmah B., Wente S. R. (2009) Eukaryot. Cell 8, 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tuite M. F., Cox B. S. (2007) Prion 1, 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.