Abstract
The lymphotoxin-β receptor (LTβR) activates the NF-κB2 transcription factors, p100 and RelB, by regulating the NF-κB-inducing kinase (NIK). Constitutive proteosomal degradation of NIK limits NF-κB activation in unstimulated cells by the ubiquitin:NIK E3 ligase comprised of subunits TNFR-associated factors (TRAF)3, TRAF2, and cellular inhibitor of apoptosis (cIAP). However, the mechanism releasing NIK from constitutive degradation remains unclear. We found that insertion of a charge-repulsion mutation in the receptor-binding crevice of TRAF3 ablated binding of both LTβR and NIK suggesting a common recognition site. A homologous mutation in TRAF2 inhibited cIAP interaction and blocked NIK degradation. Furthermore, the recruitment of TRAF3 and TRAF2 to the ligated LTβR competitively displaced NIK from TRAF3. Ligated LTβR complexed with TRAF3 and TRAF2 redirected the specificity of the ubiquitin ligase reaction to polyubiquitinate TRAF3 and TRAF2, leading to their proteosomal degradation. Stimulus-dependent degradation of TRAF3 required the RING domain of TRAF2, but not of TRAF3, implicating TRAF2 as a key E3 ligase in TRAF turnover. The combined action of competitive displacement of NIK and TRAF degradation halted NIK turnover, and promoted its association with IKKα and signal transmission. These results indicate the LTβR modifies the ubiquitin:NIK E3 ligase, and also acts as an allosteric regulator of the ubiquitin:TRAF E3 ligase.
Keywords: Cytokine, Immunology, NF-κB Transcription Factor, TRAF, Tumor Necrosis Factor (TNF), Ubiquitin Ligase
Introduction
The lymphotoxin-β receptor (LTβR),2 a member of the TNF receptor superfamily (1), is a key regulator of lymphoid organogenesis and homeostasis of the immune system(2, 3). Ligation of the LTβR activates a serine kinase cascade involving the NF-κB inducing kinase (NIK; MAPKKK14) and the inhibitor of κB kinase-α (IKKα; IKK1). Activation of kinase cascade culminates in the proteosome-dependent processing of p100 (NF-κB2), degrading the ankyrin inhibitory domain and releasing a 52-kDa fragment (p52) (4–6). The p52 fragment forms a heterodimer with RelB that controls transcription of numerous effector genes, such as tissue-organizing chemokines (CCL21, CXCL13) and integrins (intercellular adhesion molecule, ICAM1) that recruit lymphoid cells into the developing lymphoid organ.
NIK is the key molecule that controls the non-canonical pathway of NF-κB activation by several members of the TNF receptor superfamily through direct binding of the TRAF adaptors (7). In unstimulated cells, NIK is prevented from accumulating by constitutive proteosome degradation mediated by a cytosolic ubiquitin E3 ligase comprised of TRAF, a family of zinc RING finger proteins (8). The ubiquitin:NIK E3 ligase is a multisubunit complex comprised of TRAF3 and TRAF2 in association with the cellular inhibitors of apoptosis (cIAP)-1 and -2 (9, 10). In the complex, TRAF3 binds NIK, and TRAF2 engages cIAP. All three subunits contain RING and zinc finger motifs that are required for E3 ligase activity and NIK turnover. Although transcription of mRNA is constitutive in unstimulated cells, this highly efficient ubiquitin:NIK E3 ligase maintains NIK at vanishing low levels, below detection by the most sensitive assays. Thus, TRAF3 and TRAF2 function as inhibitors of NIK suppressing NF-κB activation.
Although it is well established that the trimeric ligands of the TNF superfamily initiate signaling by clustering of their cognate receptors, the translation of receptor ligation to the activation of intracellular signals is unknown. TRAF3 and TRAF2 directly associate with LTβR and other receptors rapidly after ligand binding, implicating their role in signaling (11). Structural studies revealed significant complexity of LTβR-TRAF interface. The surface crevices in TRAF3 and TRAF2 are homologous and located in the C-terminal TRAF domain. The TRAF crevice accommodates short peptides of limited sequence homology found in several TNF receptors and other unrelated regulatory molecules (8), demonstrating significant promiscuity in protein recognition. The location and function of the TRAF3-binding site for NIK is not defined, limiting our understanding of the relationship between receptor-TRAF recruitment and the activation of NIK.
Here, we demonstrate the TRAF3 binding site for NIK is located in the common receptor-binding crevice, thus the recruitment of TRAF3 to the ligated LTβR competes with NIK for TRAF3. Similarly, recruitment of TRAF2 to the LTβR displaced cIAP from TRAF2. Furthermore, TRAF2 and TRAF3 recruited to the LTβR cytosolic domain were polyubiquitinated and degraded. Polyubiquitination and degradation of TRAF3 and TRAF2 was dependent on the TRAF2 RING domain. NIK liberated from its association with TRAF3 engaged IKKα propagating the serine kinase cascade leading to the formation of the active NF-κB p52/RelB transcriptional complex. Together, these results indicate the LTβR serves as an allosteric regulator by competitively displacing the substrate NIK and redirecting the specificity of the ubiquitin:NIK E3 ligase to ubiquitinate the TRAF molecules.
EXPERIMENTAL PROCEDURES
Cell Culture, Plasmids, and Gene Transfer
Human embryonic kidney (HEK) 293T cells, mouse NIH3T3 cells, Ltβr−/− MEF, and Traf2−/−Traf5−/− MEF were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Human TRAF3, TRAF2, NIK, cIAP1, and LTβR cDNA were cloned into pcDNA3.1 or pEF (Invitrogen) expression vectors and transfected into HEK293T cells using the calcium phosphate method. For stable expression, various cDNA were cloned into pMSCV-IRES-GFP or pMSCV-IRES-CD2 retroviral vectors that were used to infect MEF or NIH3T3 cells. HA-tagged ubiquitin expression vector (pEF-HA-Ubiquitin) was kindly provided by Dr. Yun Cai Liu (LIAI). Short hairpin RNA (shRNA) for TRAF2 and TRAF3 were cloned into pMSCV-LMP retroviral vector (OPENBIOSYSTEMS).
Antibodies and Reagents
Rat anti-mouse LTβR monoclonal antibody (clone 4H8) (12) and soluble recombinant human LIGHT were used as LTβR agonists (13). Commercial sources of monoclonal antibodies included anti-HA.11 (Covance); TRAF2 (MBL); Pan cIAP (R&D); human TRAF2, mouse ICAM1 and human LTβR (BD Biosciences); rat CD2 (Biolegend); and ubiquitin (Santa Cruz Biotechnology); FLAG M2 and Actin (Sigma); anti-Myc 4A6 and human p100 (Millipore). The source of polyclonal antibodies included TRAF3, TRAF2, and NIK (Santa Cruz Biotechnology); NIK, p100 and IKKα (Cell Signaling Technology).
Cell Sorting and Flow Cytometry
Retrovirus-transduced cells were monitored and selected by cell sorting (FACS Aria, BD Biosciences) for expression of GFP or CD2. Flow cytometric analysis was performed by incubating cells with fluorochrome-conjugated ICAM1 antibody in buffer (1× phosphate-buffered saline, 3% bovine serum albumin, 5 mm EDTA, and 0.02% NaN3). The antibody-bound cells were analyzed with a FACS Calibur cytometer using Cell Quest (BD Biosciences) and FlowJo software (TreeStar).
Immunoprecipitation and Immunoblots
Cell lysates from MEF or NIH3T3 were prepared with TNE-N buffer (20 mm Tris-Cl, pH7.5, 150 mm NaCl, 5 mm EDTA, and 1% Nonidet P-40, supplemented with protease inhibitor mixture, 10 mm NaF, and 10 mm Na3VO4). For transfected 293T, cells were lysed with TNE-N lysis buffer plus 0.1% SDS. The lysates were precleared and immunoprecipitated with the indicated antibody, followed by incubation of protein G-Sepharose (GE Healthcare). The immunoprecipitates or cell lysates were subjected to immunoblotting and specific proteins were visualized with substrate from SuperSignal West Pico or Dura systems (Thermo Scientific). For detection of ubiquitinated NIK in transfected 293T, the cells were pretreated with 20 μm of MG132 (Calbiochem) for 4 h to block ubiquitin-proteosomal pathway before lysis. The cell lysates were prepared with TNE-N plus 0.1% SDS supplemented with protease inhibitor, 10 mm NaF, 10 mm Na3VO4, and 20 μm MG132. After the first immunoprecipitation with NIK antibody, the immunoprecipitates were mixed with TNE-N buffer containing 1% SDS and 0.5% b-ME and then boiled for 5 min to completely release NIK-bound protein(s). The supernatant was 10-fold diluted with TNE-N buffer, followed by the second immunoprecipitation with NIK antibody. Immunoblotting with anti-HA was performed to detect HA-tagged ubiquitin bound to NIK. For detection of ubiquitinated TRAF proteins in MEF, the cells were pretreated with 10 μm MG132 for 30 min, followed by the stimulation with soluble LIGHT (20 ng/ml). The cell lysates were prepared with TNE-N buffer supplemented with protease inhibitor, 10 mm NaF, 10 mm Na3VO4, and 20 μm MG132 and 10 mm NEM (Sigma). FLAG-tagged LTβR was immunoprecipitated with anti-FLAG M2-agarose, and the LTβR-depleted lysates were immunoprecipitated with specific antibody to either TRAF2 or TRAF3. After denaturation of the immunoprecipitates and a second immunoprecipitation, immunoblotting with anti-ubiquitin was performed to detect endogenous ubiquitin bound to TRAF3 or TRAF2.
Structure
Structural figures were prepared from the atomic coordinates and structural factors of the TRAF3-LTβR crystal structure (1RF3.pdb) as described previously (14). The program PyMOL was used to visualize TRAF3 and electrostatic surface potentials were calculated with the program APBS, together with the PDB2PQR server.
RESULTS
A Common Binding Site for LTβR and NIK in the TRAF3 Crevice
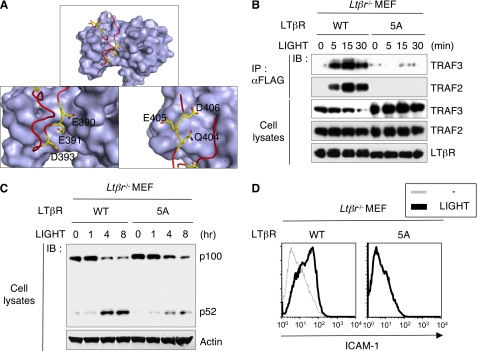
Binding of the ligands, LTαβ and LIGHT, or artificial activation with agonist antibody, rapidly recruits TRAF3 (11) and TRAF2 (15) to the LTβR. Crystallographic analysis previously demonstrated the TRAF3 binding region of the LTβR (385PYPIPEEGDPGPPGLSTPHQEDGK408) forms a reverse turn, with the change in direction occurring through residues 394PGPPG398 (Fig. 1A) (14). The residues 388IPEEGD393 in the LTβR provide several primary contacts with TRAF3, however, alanine substitution mutagenesis revealed an additional region located in the C-terminal LTβR strand required for signaling (Fig. S1A). We expressed these LTβR mutants in mouse embryonic fibroblasts (MEF) derived from Ltβr-deficient mice to assess signaling activity. Stable lines were selected by flow cytometry for receptor levels comparable to normal fibroblasts. Mutational inactivation of the TRAF binding region in the LTβR required multiple alanine substitution mutations in both the N-proximal region (Glu390, Glu391, and Asp393) and C-terminal region (404QED406; exemplified by the 5A mutant) to abolish recruitment of TRAF3, and also TRAF2 (Fig. 1B). Interestingly, ligation of wild-type LTβR, but not the 5A mutant, induced the specific depletion of endogenous TRAF3 (Fig. 1B). The position of the Ala substitutions in 404QED406 had less impact than the number of substitutions, which suggested a relatively high affinity interaction between LTβR and TRAF3 or TRAF2. In response to ligation with LIGHT, the LTβR-5A mutant was dramatically attenuated in its ability to activate IKKα-dependent processing of p100→p52 (Fig. 1C), which is required to form the transcriptionally active NF-κB RelB/p52 complex. Correspondingly, the 5A mutant failed to induce expression of the NF-κB regulated gene ICAM1 in fibroblasts (Fig. 1D). These observations suggested that TRAF3 can tolerate a high degree of structural variance in the LTβR-binding site, a concept supported by the ability of TRAF3 to accommodate multiple TNF receptors and regulatory molecules (8).
FIGURE 1.
The TRAF3-LTβR complex. A, TRAF3-LTβR structural diagram was generated from the cocrystal structure (PDB 1RF3). The image depicts the molecular surface of the TRAF domain of TRAF3 (gray) and the TRAF-binding domain of the LTβR peptide with colored α-C backbone (red) and charged residues (yellow). B and C, MEF from Ltβr−/− mice reconstituted with FLAG-tagged human LTβR WT or 5A mutants were treated with soluble LIGHT. The recruitment of TRAF molecules to LTβR was assessed by the coimmunoprecipitation with anti-FLAG antibody, and the processing of p100 (NF-κB2) was assessed by immunoblotting. D, flow cytometric analysis of ICAM-1 expression in the LTβR-reconstituted MEF treated with soluble LIGHT or medium for 3 days.
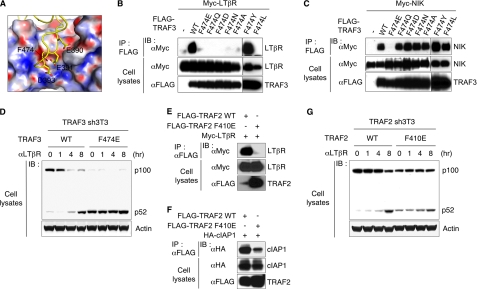
Mutagenesis studies indicated the TRAF domain of TRAF3 is required for interaction with the N-terminal region of NIK at position 78ISIIAQA84 (16). The shallow surface crevice in the TRAF domain of TRAF3 that binds LTβR contains three subregions of predominately polar, or hydrophobic, or mixed polar/charged residues (14). Mutations in mixed charge/polar (Tyr459) and hydrophobic (Phe512 and Phe521) subregions of TRAF3, but not the polar region, inhibited LTβR binding (14), yet none of these mutations impacted binding to NIK (supplemental Fig. S1B). However, substitution of F474E, centrally positioned on the floor of the TRAF3 crevice (Fig. 2A) ablated binding of both LTβR (Fig. 2B) and NIK (Fig. 2C). Substitution with Tyr retained binding of LTβR revealing the side chain phenyl group of Phe474 was important for activity. Other substitutions at Phe474, including Asp, did not alter NIK binding suggesting the extended acidic moiety of Glu474 may function as bulkier charge repulsion mutation compared with Asp474. The mutation at F474E did not appear to compromise the global integrity of TRAF3 as wild-type TRAF3 coimmunoprecipitated with the mutant, indicating the mutant assembled into its native trimeric conformation (supplemental Fig. S1C).
FIGURE 2.
A mutation in the TRAF3 crevice ablates binding of LTβR and NIK. A, surface charge distribution of the receptor-binding crevice in TRAF3. The LTβR peptide located within the TRAF domain is depicted as the electrostatic surface potential of TRAF3 (PDB2PQR server) where electronegative (red) and electropositive (blue) were scaled from −30 to +30 kT/e. The three acidic residues (Glu390, Asp391, and Glu393) in LTβR contact a stretch of neutral to positively charged surface of TRAF3. B and C, 293T cells were contransfected with LTβR (B) or NIK (C) along with various TRAF3 constructs. The binding ability of TRAF3 to LTβR or NIK was assessed by the coimmunoprecipitation with anti-FLAG antibody. D, NIH3T3 cells depleted of TRAF3 by shRNA (TRAF3 sh3T3) were reconstituted with wild-type (WT) or F474E mutant of TRAF3. TRAF3 sh3T3 cells were treated with LTβR agonistic antibody (aLTβR) for the indicated times and the processing of p100 (NF-κB2) was assessed by immunoblotting. E and F, F410E mutation in TRAF2 alters the binding to LTβR and cIAP1. Binding of TRAF2 to NIK or LTβR was determined in 293T cells contransfected with LTβR (E) or NIK (F) along with TRAF2 constructs. The binding ability of TRAF2 to LTβR or cIAP1 was assessed by coimmunoprecipitation with anti-FLAG antibody. G, TRAF2 sh3T3 cells were treated with αLTβR as in D.
To assess the functional impact of the F474E mutation, mouse 3T3 fibroblasts were depleted of endogenous TRAF3 using transcriptional interference with short hairpin RNA (sh3T3) and reconstituted with wild-type or mutant human TRAF3 (supplemental Fig. S2). TRAF3 sh3T3 cells expressing wild type human TRAF3 regained specific responsiveness to LTβR activation, whereas cells expressing the TRAF3-F474E mutant constitutively processed p100 → p52 (Fig. 2D). The constitutive p100-processing phenotype for the F474E mutant was similar to TRAF3−/− MEF (17) and TRAF3 shRNA cells (supplemental Fig. S2). This phenotype was expected because the binding of NIK to TRAF3 is essential for constitutive degradation of NIK (16). Importantly, these observations indicate that NIK engages the same crevice in TRAF3 bound by the LTβR.
cIAP Binds TRAF2 in the LTβR-binding Crevice
Sequence alignment among TRAF family members showed that Phe410 in TRAF2 is homologous to TRAF3-F474 and centrally positioned in the receptor-binding crevice of TRAF2 (18). As expected, the F410E mutation in TRAF2 ablated binding to LTβR (Fig. 2E). The interaction of cIAP with TRAF2 (19, 20) and the requirement of TRAF2-cIAP complex in the ubiquitination of NIK (9, 10, 21, 22) (supplemental Fig. S3) suggested that F410E mutation in TRAF2 may impact cIAP binding. Indeed, a coimmunoprecipitation experiment showed that the TRAF2 F410E mutation substantially reduced binding to cIAP1 (Fig. 2F). To assess the functional impact of F410E mutation, we generated TRAF2 sh3T3 cells, followed by reconstitution with wild type or the F410E mutant of TRAF2 (supplemental Fig. S2). As expected, the 3T3 cells reconstituted with TRAF2-F410E showed constitutive processing of p100 (Fig. 2G). These data clearly indicate that the TRAF2-cIAP1 interaction regulates signaling from NIK to p100, and remarkably, the F410E mutation present within the LTβR binding region altered cIAP binding to TRAF2.
LTβR Competitively Displaces NIK and cIAP from the TRAF Complex
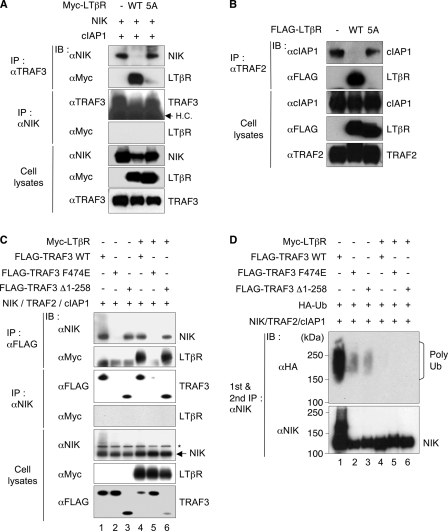
The loss of both LTβR and NIK binding to the TRAF3 F474E raised the possibility these molecules occupy the same surface crevice and compete for TRAF3. We reasoned that the activation of the LTβR should abrogate binding between TRAF3 and NIK, and similarly LTβR should compete with cIAP for binding TRAF2. To test this concept, 293T cells were transfected with wild-type or mutant LTβR-5A, and the interactions between NIK and TRAF3, or TRAF2 and cIAP were observed by coimmunoprecipitation and Western blot. Wild-type LTβR, but not the LTβR-5A mutant, blocked the coimmunoprecipitation of NIK with TRAF3, and conversely, LTβR coimmunoprecipitated with TRAF3 and at this time, NIK was not associated with LTβR (Fig. 3A). This result is consistent with the idea that LTβR displaced NIK from TRAF3 by competitively binding to the same surface on TRAF3. Thus, TRAF3 specifically exchanged NIK with the LTβR. Similarly, LTβR, but not the 5A mutant, blocked the association between TRAF2 and cIAP1 (Fig. 3B).
FIGURE 3.
Competitive displacement of NIK and cIAP1 from TRAF3 and TRAF2. A, 293T cells were cotransfected with the indicated plasmids, cultured for 2 days and prior to extraction treated with MG132 for 4 h to block the ubiquitin-proteosomal pathway. The dissociation of NIK from TRAF3-NIK complex in the presence of wild-type or mutant LTβR was assessed by coimmunoprecipitation with a specific antibody to TRAF3. B, dissociation of cIAP1 from the TRAF2-cIAP1 complex was assessed by coimmunoprecipitation with a specific antibody to TRAF2 (as in A). C, LTβR-induced competitive displacement does not require Zinc RING Finger domain of TRAF3. 293T cells were transfected with the indicated combination of expression plasmids including TRAF3Δ1–258 lacking the N terminus zinc RING finger domains. Anti-FLAG or anti-NIK were used to immunoprecipitate their respective antigens, followed by immunoblotting with anti-NIK, -Myc, or -FLAG, respectively (top three panels). The expression of NIK, LTβR, and TRAF3 was verified by immunoblotting of the whole cell extract (bottom three panels). D, 293T cells were transfected as in C with HA-tagged ubiquitin (Ub); after 2 days, the cells were treated with MG132 (20 μm) for 4 h, and anti-NIK was used to sequentially immunoprecipitate NIK from the cell extract, and ubiquitin detected by immunoblotting with anti-HA.
The N-terminal, RING, and zinc finger domains of TRAF3 are important for ubiquitin:NIK E3 ligase activity raising the question of whether NIK displacement required the N-terminal region of TRAF3. To address this question, we performed coimmunoprecipitation experiments to see whether a deletion mutant lacking RING and zinc finger domains of TRAF3 (TRAF3 Δ1–258)(23) impacted LTβR-mediated displacement of NIK (Fig. 3C). Wild-type TRAF3 (lane 1) and the TRAF3Δ1–258 mutant (lane 3), but not the F474E mutant (lane 2), coimmunoprecipitated with NIK in the absence of LTβR, confirming the TRAF domain is required to bind NIK. However, polyubiquitinated forms of NIK were clearly present when bound to wild-type TRAF3, but not either the TRAF3Δ1–258 or F474E mutants (Fig. 3D, lanes 1–3), indicating ubiquitination of NIK required binding the TRAF domain and the functionality of the RING zinc finger domains. Wild-type and Δ1–258 TRAF3, but not F474E, bound LTβR (Fig. 3C, lanes 4–6) and halted NIK ubiquitination (Fig. 3D, lanes 4–6). The Δ1–258 TRAF3 mutant revealed the essential role of the interaction of the NIK and TRAF domains, and separated competitive displacement of NIK from NIK ubiquitination.
LTβR Redirects Ubiquitination to TRAF3 and TRAF2
The relative abundance of TRAF3 compared with LTβR suggested a direct molar interaction alone was unlikely to liberate sufficient NIK to propagate signaling. We observed that TRAF3 and Δ1–258 mutant, but not the F474E mutant, were specifically depleted in cells cotransfected with LTβR (Fig. 3C, lower panel, lanes 4–6). The loss of TRAF3 in transfected cells suggested an enzymatic action may be involved in depleting TRAF3 from the cell, which in turn prevents reassociation with NIK.
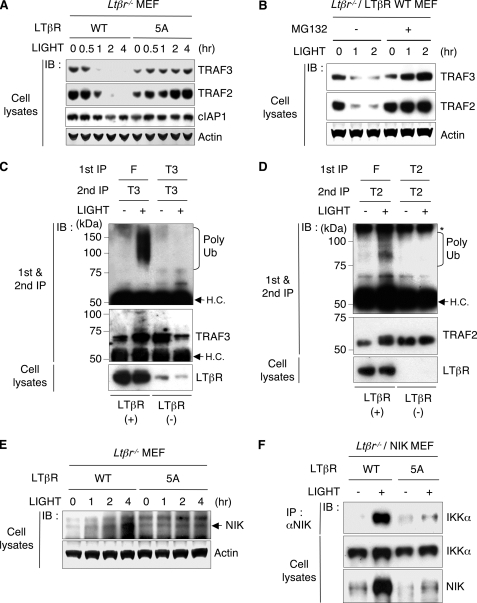
LIGHT treatment of Ltβr−/− MEF reconstituted with wild-type LTβR, but not the 5A mutant, rapidly induced the loss of TRAF3 and TRAF2 in the detergent soluble fraction (Fig. 4A). The proteosomal inhibitor, MG132 halted the LTβR-dependent loss of TRAF3 and TRAF2 (Fig. 4B). This result raised the issue of where the ubiquitination of the TRAF molecules occurred in the cell. To address this question, we analyzed the ubiquitination status of TRAF3 and TRAF2 that were either bound to the LTβR or free in the cytosol. Strikingly, polyubiquitinated TRAF3 and TRAF2 were observed only associated with the LTβR in LIGHT activated cells, but not in the receptor-depleted, cytosolic fraction (Fig. 4, C and D).
FIGURE 4.
LTβR-TRAF complex redirects the ubiquitination reaction. A, expression of TRAF3, TRAF2, and cIAP1 in LTβR-reconstituted MEF treated with soluble LIGHT was assessed by immunoblotting. B, LTβR-reconstituted MEF were pretreated with or without MG132 for 30 min, followed by stimulation with soluble LIGHT for the indicated times. C and D, LTβR-deficient MEF reconstituted with FLAG-tagged LTβR were pretreated with MG132 (10 μm) for 30 min, followed by stimulation with LIGHT (20 ng/ml) for 30 min. The LTβR was immunoprecipitated with FLAG M2 antibody-conjugated agarose (F). The LTβR-depleted lysate was then immunoprecipitated with anti-TRAF3 (T3) or anti-TRAF2 (T2). The immunoprecipitates were then denatured to dissociate the bound protein(s), and then re-immunoprecipitated with anti-TRAF3 or anti-TRAF2. The immunoprecipitates containing TRAF3 and TRAF2 from the LTβR (+) and LTβR (−) fractions were subjected to immunoblotting with anti-ubiquitin to detect polyubiquitination (poly Ub) of TRAF3 or TRAF2. HC, nonspecific heavy chain band. E, LTβR WT or 5A reconstituted MEF were treated with soluble LIGHT (20 ng/ml), and endogenous NIK expression in whole cell lysate was analyzed with anti-NIK. F, LTβR-reconstituted MEF stably expressing NIK were treated with soluble LIGHT for 6 h. The recruitment of IKKα to NIK was assessed by coimmunoprecipitation with anti-NIK and immunoblotting with anti-IKKα. Total IKKα and NIK expression was determined by immunoblotting of the whole cell lysate.
The displacement of NIK from TRAF3 and subsequent LTβR-dependent degradation of TRAF3 should allow NIK protein to accumulate and propagate downstream signaling. Ligation of wild-type LTβR with LIGHT in reconstituted Ltβr−/−MEF increased endogenous NIK expression in a time-dependent manner, but not in cells reconstituted with LTβR-5A mutant (Fig. 4E). To assess the activity of liberated NIK, we measured NIK-IKKα interaction by coimmunoprecipitation in the reconstituted Ltβr−/− MEF-expressing exogenous NIK (supplemental Fig. S4). When stimulated with LIGHT, NIK specifically accumulated and anti-NIK coimmunoprecipitated IKKα from cells expressing wild-type LTβR, but not the LTβR 5A mutant (Fig. 4F). These results indicate that the LTβR becomes a functional subunit of a TRAF3-TRAF2 complex that promotes the ubiquitination and degradation of TRAF2 and TRAF3.
TRAF2 Controls LTβR-dependent TRAF3 Degradation
The finding that receptor-bound TRAF3 and TRAF2 were substrates for polyubiquitination raised the possibility that TRAF3 or TRAF2 may serve as the E3 ligase when bound to the LTβR. We observed that the TRAF3Δ1–258 mutant, which lacks the RING and zinc finger domains, was polyubiquitinated (supplemental Fig. S5A) and degraded following LTβR activation (Fig. 3C, lane 6). Furthermore, TRAF2 degradation induced by LTβR proceeded normally in TRAF3 shRNA 3T3 cells (supplemental Fig. S5B). These findings strongly suggest that TRAF3 is unlikely to ubiquitinate and degrade itself or TRAF2 following LTβR activation.
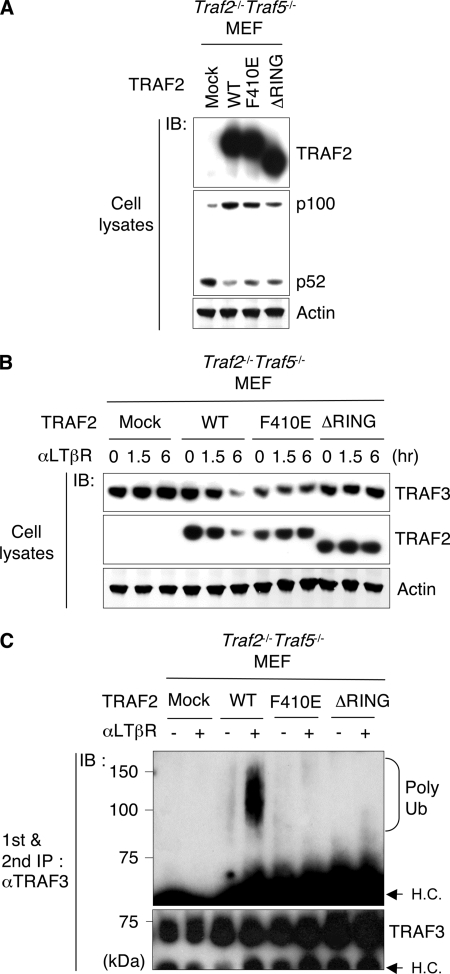
These results implicated TRAF2 as a likely candidate for the ubiquitin:TRAF3 E3 ligase involved in degrading TRAF3. To test this possibility, we expressed wild-type TRAF2, F410E, or RING domain deletion (ΔRING) mutants in MEF deficient in both Traf2 and Traf5. As expected, wild type TRAF2 halted p100 processing relative to the constitutive processing in mock transfected cells, whereas the F410E and ΔRING mutants showed limited ability block processing of p100 (Fig. 5A). As expected, LTβR stimulation induced the loss of TRAF3 in cells reconstituted with wild-type TRAF2, but not in cells expressing either the F410E or ΔRING mutants of TRAF2 (Fig. 5B). TRAF2-depleted 3T3 cells reconstituted with F410E mutant were also unable to degrade TRAF3 (supplemental Fig. S6). In response to LTβR stimulation the polyubiquitination of TRAF3 was halted in cells harboring either of the TRAF2 mutants (Fig. 5C). The F410E mutant indicates that TRAF2 recruitment to the LTβR is a necessary step for induction of TRAF3 degradation, and the ΔRING mutant indicated the RING finger domain of TRAF2 is crucial in promoting the ubiquitination and proteasomal degradation of TRAF3.
FIGURE 5.
TRAF2 controls LTβR-dependent TRAF3 degradation. A, cells were reconstituted with the indicated cDNA, and exogenous TRAF2 and NF-κB2 (p100 and p52) expression was assessed by immunoblotting. B, TRAF2-reconstituted Traf2−/−Traf5−/− MEF were stimulated with LTβR agonistic antibody (aLTβR) for the indicated time (hr), and TRAF3 and TRAF2 expression was assessed by immunoblotting. C, TRAF2-reconstituted cells were pretreated with MG132 for 30 min and incubated with αLTβR for 1.5 h. TRAF3-containing immunoprecipitates were denatured to dissociate the bound protein(s) and then re-immunoprecipitated with anti-TRAF3 antibody, followed by the immunoblotting to detect polyubiquitinylation (poly Ub) of TRAF3. HC, nonspecific heavy chain band.
We were unable to demonstrate any association of cIAP with the activated LTβR-TRAF2-TRAF3 complex (Fig. 3B and supplemental Fig. S5C). Furthermore, the demonstration that the polyubiquitination of TRAF3 required association with the LTβR suggested the possibility that cIAP might not be essential for LTβR-mediated degradation of TRAF3 and TRAF2.
DISCUSSION
Our results indicate LTβR regulates NIK by disrupting and modifying the ubiquitin:NIK E3 ligase. LTβR in complex with TRAF2 forms a distinct ubiquitin:TRAF3 E3 ligase that promotes TRAF3 ubiquitination and degradation. These two processes mediated by the LTβR are essential to liberate NIK from constitutive degradation. We suggest that the cytoplasmic domain of the LTβR serves as an allosteric3 regulator of the E3 ligases that control polyubiquitination of NIK and TRAF3. LTβR is an allosteric regulator because its binding site in the C-terminal TRAF domain is distinct from the zinc RING domain containing the E3 ligase catalytic site. This conclusion is based on the results demonstrating that the LTβR competitively displaced NIK from the ubiquitin:NIK E3 ligase, and the binding of LTβR to TRAF2 redirected the polyubiquitination reaction to TRAF3 and TRAF2. Hence, the LTβR-TRAF2 complex forms a ubiquitin:TRAF3 E3 ligase. Ubiquitination and degradation of TRAF3 depleted the entire cellular pool of TRAF2 and -3, preventing the reassociation of liberated NIK with TRAF3, and blocking the reassembly of the ubiquitin:NIK E3 ligase.
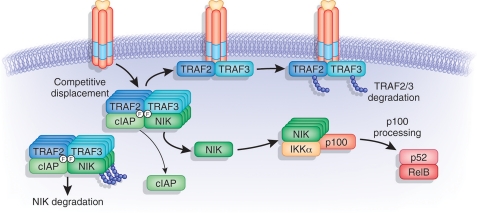
We suggest that TRAF recruitment to the cytosolic domain of the LTβR and the polyubiquitination of TRAF3 may function as an enzymatic cycle (Fig. 6) that degrades the relatively more abundant TRAF molecules. The rapid degradation of polyubiquitinated TRAF molecules may free LTβR to recruit additional molecules of TRAF3 and TRAF2 to the receptor for ubiquitination. Disruption of the ubiquitin:NIK E3 ligase by the activated LTβR allows NIK to accumulate and interact with the serine kinase, IKKα, propagating the extracellular signal to p100 and eventuating NF-κB activation. TRAF3 and TRAF2 degradation occurs in cells over 1–2-h post-receptor ligation, just preceding the accumulation of NIK (2–4 h) and processing of p100 and nuclear translocation of p52 RelB (4–8 h).
FIGURE 6.
Allosteric regulation of ubiquitin:NIK and ubiquitin:TRAF3 E3 ligase by the LTβR. NIK is maintained at low levels in non-stimulated cells by Ub-dependent degradation via Ubiquitin:NIK E3 ligase consisting of the TRAF3-TRAF2-cIAP complex. Ligation of LTβR recruits TRAF3 and TRAF2, with binding in the TRAF crevice, where NIK and cIAP also bind. LTβR competitively displaces NIK and cIAP, which halts the ubiquitination of NIK. Phe474 in TRAF3 and Phe410 in TRAF2 define the key binding sites for LTβR, NIK, and cIAP. The specificity of the ubiquitin ligase is redirected to TRAF3 and TRAF2 when bound to the LTβR, forming a ubiquitin:TRAF E3 ligase that catalyzes polyubiquitination of TRAF2 and TRAF3. Consequentially, TRAF3 and TRAF2 are rapidly degraded depleting the cellular pools of TRAF3 and TRAF2, and allowing ligated LTβR to bind more TRAF3. Liberated NIK binds IKKα promoting p100 processing, essential for the formation of the RelB/p52 transcription complex.
The competition for TRAF3 seems to favor ligated LTβR over NIK, which may reflect the higher avidity of the ligand-clustered receptors for the TRAF3 crevice. As evidence, we identified two separate regions in the TRAF3-interaction loop of LTβR that required multiple mutations to disrupt binding and signaling activity. We suggest that the inherent higher avidity of the multivalent cytosolic domains of LTβR displaces NIK from the TRAF3 surface crevice.
The receptor-binding crevice of TRAF3 can be considered as the substrate-binding site in the ubiquitin:NIK E3 ligase based on the mutants that affected binding of both NIK and LTβR. The stimulus-specific polyubiquitination of TRAF3 and TRAF2 can be viewed as evidence that the binding of the LTβR altered the substrate specificity of the E3 ligase from NIK to TRAF3. Furthermore, the results with the TRAF2 F410E mutant indicated that TRAF2 must bind the LTβR to promote both ubiquitination and degradation of TRAF3. Deletion of the RING finger domain of TRAF2 allowed LTβR binding, but inhibited TRAF3 ubiquitination and degradation, consistent with the idea that TRAF2 functions as the E3 ligase for TRAF3. That TRAF2 ubiquitination was also affected by deletion of the RING domain suggests TRAF2 may mediate auto-ubiquitination.
Interestingly, the TRAF3 Δ1–258 mutant, although capable of binding LTβR and NIK, was unable to participate in the constitutive ubiquitination and turnover of NIK, suggesting that the RING domain of TRAF3 is important in the ubiquitin:NIK E3 ligase complex involved in NIK degradation. However, TRAF3 ubiquitination and degradation did not require the TRAF3 RING domain indicating that TRAF3 does not mediate auto-ubiquitination and degradation. Taken together, our data support the concept that the activated LTβR disrupts and modifies the substrate specificity of the ubiquitin:NIK E3 ligase to a ubiquitin:TRAF3 E3 ligase, thus defining LTβR as an allosteric regulator of ubiquitin E3 ligases.
The role of cIAP in TRAF3 and TRAF2 ubiquitination and degradation may vary with different receptors. Although the LTβR displaced cIAP from TRAF2, and cIAP failed to coimmunoprecipitate with the activated LTβR-TRAF complex, we must consider other evidence before ascribing a direct competitive mechanism between LTβR and cIAP for TRAF2. Recent evidence indicates that cIAP-TRAF2 interaction requires a binding motif in the coiled coil (TRAF-N) domain (24), distant from the receptor-binding crevice in the C terminus of TRAF2. This result suggests an indirect mechanism may be operative in the displacement of cIAP from TRAF2 by the LTβR. cIAP are labile in the absence of TRAF2, yet their stability does not require direct interaction with TRAF2 (25). That mutations in distinct regions of TRAF2 affect cIAP binding may reflect interactions between cIAP, TRAF2 and TRAF3, and perhaps other components, in the ubiquitin:NIK E3 ligase complex. Experiments in progress seek an answer to this question.
In contrast to LTβR, CD40, another member of the TNFR superfamily, coimmunoprecipitated with cIAP, and induced TRAF3 degradation in a cIAP-dependent manner (9, 10). However, CD40 signaling domain also interacts with TRAF6, another E3 ligase, and has a second distinct binding site for TRAF2 (26), substantially different from the LTβR. Other TNF receptors, such as TNFR2 and FN14 induce TRAF2 degradation in a cIAP1-dependent fashion (27, 28). Interestingly, CD30 activation-induced degradation of the cIAP-TRAF2 complex required the RING domain of TRAF2, but not cIAP, yet cIAP antagonists induced cIAP degradation without TRAF2 (25, 29). The differences in modulating combinations of E3 ligases by the various TNFR may contribute to the unique aspects of transcription factor activation (e.g. duration and specificity) and ensuing cellular responses.
The competitive displacement and allosteric regulatory mechanisms described here may be applicable to other stimulus-dependent signaling systems that utilize TRAF molecules including most other TNFR and several innate defense sensors (30–32). For example, Cardiff (also known as IPS-1, MAVS, and VISA), a critical adaptor for the mitochondrial-localized cytoplasmic nucleic acid sensor systems required for the interferon response, engages the receptor-binding crevice of TRAF3 (33, 34). The activation of the NF-κB pathway is constrained by numerous extrinsic and autoregulatory loops (35) that could also influence the allosteric mechanism described here. Interestingly, viral proteins targeting TRAF molecules may usurp this allosteric mechanism to modulate host defense pathways (36–39). These results suggest distinct and common mechanisms may regulate TRAF-dependent signaling pathways, providing new opportunities to specifically modulate NF-κB-dependent and related inflammatory pathways in disease processes.
Supplementary Material
Acknowledgments
We thank Yun Cai Liu and Chris Benedict for comments, Klaus Pfeffer for the Ltβr−/− mice, Hiroyasu Nakano for the Traf2−/−Traf5−/− MEF, and Masato Kubo for the pMX-IRES-CD2 vector. The authors declare no competing financial interests.
This work was supported, in whole or in part, by Grants CA069381, R37AI033068, and AI048073 (to C. F. W.) from the US Public Health Service, National Institutes of Health. This work was also supported by the Uehara Memorial Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
Allostery is used here in the sense of “other site,” in reference to a modifier that binds at a site on the enzyme distinct from the catalytic site, as in the original definition by Monod, Changeux, and Jacob as detailed in A. Lehninger's Biochemistry text (1974) p. 253.
- LTβR
- lymphotoxin-β receptor
- cIAP
- cellular inhibitor of apoptosis
- ICAM1
- intercellular adhesion molecule
- LT
- lymphotoxin
- NIK
- NF-κB-inducing kinase
- TRAF
- TNF receptor-associated factors
- HA
- hemagglutinin.
REFERENCES
- 1.Bodmer J. L., Schneider P., Tschopp J. (2002) Trends Biochem. Sci. 27, 19–26 [DOI] [PubMed] [Google Scholar]
- 2.Vondenhoff M. F., Kraal G., Mebius R. E. (2007) Eur. J. Immunol. 37, Suppl. 1, S46–S52 [DOI] [PubMed] [Google Scholar]
- 3.Ware C. F. (2008) Immunol. Rev. 223, 186–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dejardin E., Droin N. M., Delhase M., Haas E., Cao Y., Makris C., Li Z. W., Karin M., Ware C. F., Green D. R. (2002) Immunity 17, 525–535 [DOI] [PubMed] [Google Scholar]
- 5.Müller J. R., Siebenlist U. (2003) J. Biol. Chem. 278, 12006–12012 [DOI] [PubMed] [Google Scholar]
- 6.Yilmaz Z. B., Weih D. S., Sivakumar V., Weih F. (2003) EMBO J. 22, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramakrishnan P., Wang W., Wallach D. (2004) Immunity 21, 477–489 [DOI] [PubMed] [Google Scholar]
- 8.Ely K. R., Kodandapani R., Wu S. (2007) Adv. Exp. Med. Biol. 597, 114–121 [DOI] [PubMed] [Google Scholar]
- 9.Zarnegar B. J., Wang Y., Mahoney D. J., Dempsey P. W., Cheung H. H., He J., Shiba T., Yang X., Yeh W. C., Mak T. W., Korneluk R. G., Cheng G. (2008) Nat. Immunol. 9, 1371–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vallabhapurapu S., Matsuzawa A., Zhang W., Tseng P. H., Keats J. J., Wang H., Vignali D. A., Bergsagel P. L., Karin M. (2008) Nat. Immunol. 9, 1364–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.VanArsdale T. L., VanArsdale S. L., Force W. R., Walter B. N., Mosialos G., Kieff E., Reed J. C., Ware C. F. (1997) Proc. Natl. Acad. Sci. 94, 2460–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Trez C., Ware C. F. (2008) Cytokine Growth Factor Rev. 19, 277–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rooney I. A., Butrovich K. D., Glass A. A., Borboroglu S., Benedict C. A., Whitbeck J. C., Cohen G. H., Eisenberg R. J., Ware C. F. (2000) J. Biol. Chem. 275, 14307–14315 [DOI] [PubMed] [Google Scholar]
- 14.Li C., Norris P. S., Ni C. Z., Havert M. L., Chiong E. M., Tran B. R., Cabezas E., Reed J. C., Satterthwait A. C., Ware C. F., Ely K. R. (2003) J. Biol. Chem. 278, 50523–50529 [DOI] [PubMed] [Google Scholar]
- 15.Kim Y. S., Nedospasov S. A., Liu Z. G. (2005) Mol. Cell. Biol. 25, 2130–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao G., Zhang M., Harhaj E. W., Sun S. C. (2004) J. Biol. Chem. 279, 26243–26250 [DOI] [PubMed] [Google Scholar]
- 17.He J. Q., Zarnegar B., Oganesyan G., Saha S. K., Yamazaki S., Doyle S. E., Dempsey P. W., Cheng G. (2006) J. Exp. Med. 203, 2413–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park Y. C., Burkitt V., Villa A. R., Tong L., Wu H. (1999) Nature 398, 533–538 [DOI] [PubMed] [Google Scholar]
- 19.Rothe M., Pan M. G., Henzel W. J., Ayres T. M., Goeddel D. V. (1995) Cell 83, 1243–1252 [DOI] [PubMed] [Google Scholar]
- 20.Uren A. G., Pakusch M., Hawkins C. J., Puls K. L., Vaux D. L. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4974–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varfolomeev E., Blankenship J. W., Wayson S. M., Fedorova A. V., Kayagaki N., Garg P., Zobel K., Dynek J. N., Elliott L. O., Wallweber H. J., Flygare J. A., Fairbrother W. J., Deshayes K., Dixit V. M., Vucic D. (2007) Cell 131, 669–681 [DOI] [PubMed] [Google Scholar]
- 22.Vince J. E., Wong W. W., Khan N., Feltham R., Chau D., Ahmed A. U., Benetatos C. A., Chunduru S. K., Condon S. M., McKinlay M., Brink R., Leverkus M., Tergaonkar V., Schneider P., Callus B. A., Koentgen F., Vaux D. L., Silke J. (2007) Cell 131, 682–693 [DOI] [PubMed] [Google Scholar]
- 23.Force W. R., Cheung T. C., Ware C. F. (1997) J. Biol. Chem. 272, 30835–30840 [DOI] [PubMed] [Google Scholar]
- 24.Vince J. E., Pantaki D., Feltham R., Mace P. D., Cordier S. M., Schmukle A. C., Davidson A. J., Callus B. A., Wong W. W., Gentle I. E., Carter H., Lee E. F., Walczak H., Day C. L., Vaux D. L., Silke J. (2009) J. Biol. Chem. 284, 35906–35915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Csomos R. A., Brady G. F., Duckett C. S. (2009) J. Biol. Chem. 284, 20531–20539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grammer A. C., Lipsky P. E. (2000) Adv. Immunol. 76, 61–178 [DOI] [PubMed] [Google Scholar]
- 27.Li X., Yang Y., Ashwell J. D. (2002) Nature 416, 345–347 [DOI] [PubMed] [Google Scholar]
- 28.Vince J. E., Chau D., Callus B., Wong W. W., Hawkins C. J., Schneider P., McKinlay M., Benetatos C. A., Condon S. M., Chunduru S. K., Yeoh G., Brink R., Vaux D. L., Silke J. (2008) J. Cell Biol. 182, 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Csomos R. A., Wright C. W., Galbán S., Oetjen K. A., Duckett C. S. (2009) Biochem. J. 420, 83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oganesyan G., Saha S. K., Guo B., He J. Q., Shahangian A., Zarnegar B., Perry A., Cheng G. (2006) Nature 439, 208–211 [DOI] [PubMed] [Google Scholar]
- 31.Häcker H., Karin M. (2006) Sci. STKE 2006, re13. [DOI] [PubMed] [Google Scholar]
- 32.Chung J. Y., Park Y. C., Ye H., Wu H. (2002) J. Cell Sci. 115, 679–688 [DOI] [PubMed] [Google Scholar]
- 33.Saha S. K., Pietras E. M., He J. Q., Kang J. R., Liu S. Y., Oganesyan G., Shahangian A., Zarnegar B., Shiba T. L., Wang Y., Cheng G. (2006) EMBO J. 25, 3257–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakhaei P., Mesplede T., Solis M., Sun Q., Zhao T., Yang L., Chuang T. H., Ware C. F., Lin R., Hiscott J. (2009) PLoS Pathog 5, e1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shih V. F., Kearns J. D., Basak S., Savinova O. V., Ghosh G., Hoffmann A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9619–9624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi S. H., Park K. J., Ahn B. Y., Jung G., Lai M. M., Hwang S. B. (2006) Mol. Cell. Biol. 26, 3048–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsumoto M., Hsieh T. Y., Zhu N., VanArsdale T., Hwang S. B., Jeng K. S., Gorbalenya A. E., Lo S. Y., Ou J. H., Ware C. F., Lai M. M. (1997) J. Virol. 71, 1301–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guasparri I., Wu H., Cesarman E. (2006) EMBO Rep. 7, 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu S., Xie P., Welsh K., Li C., Ni C. Z., Zhu X., Reed J. C., Satterthwait A. C., Bishop G. A., Ely K. R. (2005) J. Biol. Chem. 280, 33620–33626 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.