Abstract
OBJECTIVE
Cytochrome P450 17α-hydroxylases-C-17,20-lyase (CYP17) is a key enzyme involved with the androgen biosynthesis pathway and has recently been targeted for therapy in men with advanced prostate cancer (PCa). However, studies relating prostate cancer outcomes with CYP17 gene variants have conflicting results. In this study we analyzed Single Nucleotide Polymorphisms (SNPs) spanning the CYP17 gene for association with PCa survival.
METHODS
The cohort was comprised of Caucasian men, aged 40–64, diagnosed with PCa between 1993–1996 in King County, Washington who participated in a population-based case-control study. CYP17 SNPs were selected to capture variation across the gene and known regulatory regions. PCa-specific mortality (PCSM) was obtained by linking to the SEER cancer registry. Recurrence/progression of PCa was determined from patient survey data and medical records. Cox proportional hazards regression analysis was used to generate hazard ratios for patient outcomes.
RESULTS
Genotypes were available for 598 cases. With a median follow-up of 13.2 years, 44 PCa deaths were observed. Recurrence/progression events were observed in 30% of subjects. No genetic association with disease progression were identified. However, men with the variant A allele in rs10883783 had a 56% risk reduction in PCSM (HR 0.44, 95% CI 0.21–0.98).
CONCLUSION
These data suggest that genetic variation in the CYP17 gene in Caucasian men is associated with PCa survival.
Keywords: Prostate cancer, CYP17, Single nucleotide polymorphisms, Survival, Population based
Introduction
The role of androgens and androgen regulation in prostate cancer (PCa) development and progression is an area of strong interest. Cytochrome P450 17α-hydroxylase-C-17,20-lyase (CYP17) is a key enzyme in the androgen biosynthesis pathway.(1) Previous studies of CYP17 and prostate cancer risk focused on a single nucleotide polymorphism (SNP) in the 5′-untranslated (5′-UTR) promoter region (rs743572). The results were conflicting, with some studies finding lower risk in carriers of the wild-type allele(2–5), while others reported the variant allele was associated with reduced risk.(6–9) A meta-analysis involving 2,404 patients with PCa and 2,755 controls concluded that the rs743572 CYP17 polymorphism was unlikely to substantially alter the risk of prostate cancer occurence.(10)
Additional SNPs in the CYP17 gene have been identified, with subsequent studies highlighting specific variants purported to be associated with PCa risk and/or outcomes.(11–13) It is conceivable that men with genetic variants in CYP17 have altered enzymatic activity not only affecting their baseline hormone levels, but that such SNPs may also alter responsiveness to targeted therapies such as abiraterone, a CYP17 protein inhibitor. Recently, Hamada et al. reported an association between a CYP17 SNP and increased mortality in men with castrate-resistant PCa (CRPC).(14) Additionally, a phase I trial of abiraterone demonstrated anti-tumor activity in men with CRPC.(15) In this study, we have utilized a population-based cohort and a set of tagSNPs to test the relationship between CYP17 variation and PCa-specific survival and progression outcomes.
Materials and Methods
Study Population
The study population consisted of patients from a population-based, case-control study of PCa. Details of the study participants and data collection have been previously described.(16) Briefly, cases were residents of King County, Washington with histologically confirmed PCa identified from the Seattle-Puget Sound SEER cancer registry who were diagnosed between January 1, 1993, and December 31, 1996. Case selection was weighted such that men diagnosed before age 60 years (100%), African Americans (100%) and a random 75% sample of Caucasians aged 60–64 years at diagnosis were deemed eligible. A total of 917 eligible cases were identified and 753 (82%) participated. Controls were not included in these analyses which are focused on outcomes in cancer patients.
Genotyping
For men who consented (n = 630), DNA was isolated from peripheral blood samples using standard methods, aliquoted, and stored at −80°C. SNPs in CYP17 were selected using publicly available data from the Genome Variation Server (http://gvs.gs.washington.edu/GVS/). Haplotype tagging SNPs (tagSNPs) with a minor allele frequency > 5.0% were selected to maximize coverage of genetic variation (r2>0.80) in a region encompassing the transcript of interest (+ 5 kb upstream and downstream). The Applied Biosystems (ABI) SNPlex™ Genotyping System was used to determine SNP genotypes. Proprietary GeneMapper® software was used for calling alleles (www.appliedbiosystems.com). The SNPlex™ assay is an allele-specific hybridization that brings two oligonucleotides close enough to each other to allow ligation. Discrimination of the specific SNP allele is carried out with the ABI 3730xl DNA Analyzer and is based on the presence of a unique sequence assigned to the original allele-specific oligonucleotide. Quality control included genotyping of blind duplicate samples (n = 75), which revealed > 96% agreement of genotyping calls across the SNPs assayed. CYP17 SNP data were available from 598 cases. A total of five tagSNPs were selected from three haplotype blocks spanning the coding region and adjacent regulatory regions. Two sets of SNPs chosen from the same blocks were in perfect linkage disequilibrium, so only one SNP from each block was included in the analyses. Redundant SNPs were selected in the event of genotyping failure. Of the three SNPs, two (rs10883783 and rs17115100) are located in intron segments of the CYP17 gene and one (rs743572) is located in the 5′-UTR.
Data Collection
Subjects completed in-person interviews which collected information about demographic and lifestyle factors, medical and family history, and PCa screening within the five year period prior to diagnosis (PSA and DRE). Clinical information on PCa cases was obtained from the SEER cancer registry, including Gleason score and tumor stage. The SEER summary stage was used for analyses and is defined as localized (confined to the prostate, stages A and B), regional (regional spread outside the prostatic capsule, and/or regional lymph node involvement, stage C), and distant (metastatic, stage D). For men who did not undergo radical prostatectomy (RP), staging was based on clinical information, whereas pathological stage was used for cases undergoing radical prostatectomy. The majority of cases (79%) staged as regional (extracapsular spread and/or node positivity) underwent RP, suggesting that these men initially underwent surgery for clinically localized disease, but were upstaged upon pathology review to have regional disease.
A follow-up survey was sent to all living cases from the parent study in January 2004 who had previously consented to future contact. Physician diagnoses of PCa recurrence/progression, use of secondary therapies, PSA results, diagnostic procedures (biopsy, bone scan, MRI or CT scan) performed, along with dates and results, were collected. Five hundred-twenty of the 631 men contacted completed the survey. Thirty-one (5%) men refused to participate, 70 (11%) did not return the questionnaire, and 10 (2%) had died. Men who did not complete the survey were typically younger at diagnosis (< 50 years), of African-American race, and less well-educated compared to responders (p-values < 0.05). However, there were no significant differences between tumor stage, Gleason score or primary treatment.
Disease recurrence/progression events were recorded by multiple criteria, including the development of metastatic disease in men with locoregional disease at diagnosis. Recurrence times were imputed for 13 patients who died from prostate cancer but for whom time to recurrence was unknown. We used a multiple imputation approach (Harrell(17)) based on data from patients who died of prostate cancer and had a valid recurrence time using a linear regression model that included attempted curative treatment (yes or no) and Gleason score (<7 or ≥ 7). In two cases where the imputed time to recurrence was longer than the time to prostate cancer death, a uniformly distributed error from 0 to 1 year was subtracted from the time to prostate cancer death and used as the imputed time to recurrence. PSA progression was defined by a PSA value of ≥ 0.2 in men who had radical prostatectomy (RP) as primary therapy, or by nadir PSA + 2 ng/mL (Phoenix criteria)(18) for men who received radiation (XRT) as primary therapy. The date of last follow-up for recurrence/progression events was December 31, 2005. PCa-specific mortality was determined using data from the SEER registry, which links quarterly with the Washington State computerized mortality database. Underlying cause of death was then verified by a review of death certificates, which confirmed 99% agreement for PCa-specific deaths. The date of last follow-up for survival was December 1, 2008.
Statistical Analysis
The study was restricted to Caucasians because allelic frequencies differed between Caucasians and African-Americans for all CYP17 SNPs (data not shown) and there were insufficient numbers of African Americans to analyze separately. Alleles were all found to be in Hardy-Weinberg Equilibrium (HWE) as measured by the Fisher’s exact test. Cox proportional hazards models were created to estimate the hazard ratios (HR) and 95% confidence intervals (95% CI) separately for recurrence/progression events and PCa-specific mortality. A simple model adjusting only for age and a second multivariate model adjusting for age, PSA screening history, Gleason score, stage, diagnostic PSA, BMI and primary treatment were created. Potential interaction between genotype and treatment group was evaluated with the likelihood ratio test. The proportional hazards assumption for Cox regression was evaluated with Schoenfeld goodness-of-fit testing and was met for all analyses (global test p-value was 0.34, 0.48, and 0.69 for all three SNPs respectively in the mortality models). All statistical analyses were conducted using STATA software, Version 8 (Stata, Inc., College Station, TX).
Results
A total of 598 (95%) Caucasian men had sufficient data for analysis. After accounting for dropouts, genotyping data were obtained for 93%, 96% and 93% of patients for rs743572, rs10883783 and rs17115100, respectively. The median follow-up for survival for the cohort was 13.2 years (range 1.26 – 15.9). Of the 468 cases for which recurrence/progression data were available, 142 (30%) events occurred during a median follow-up of 8.8 years. Table I lists the clinicopathologic factors of the patients. The majority (72%) had a PSA <10, Gleason 6 or less disease (62%) and organ-confined tumors (74%). Gleason score was missing for one patient and PSA was missing for 58 (9.7%) patients. The most common treatment was radical prostatectomy (70%). As of December 2008, 113 (19%) deaths had occurred, with 39% of these deaths attributable to metastatic PCa. For six deceased patients, cause of death could not be determined.
Table 1.
Distribution of Clinicopathologic Characteristics in 598 Caucasian Prostate Cancer Patients with CYP17 SNP Genotypes *
| Alive | Other/Unknown Cause of Death |
Prostate Cancer Specific Death |
|
|---|---|---|---|
| N (%) | N (%) | N (%) | |
| Total patients | 485 | 69 | 44 |
| Age | |||
| 40–49 | 26 (5.4) | 1 (1.5) | 9 (20.5) |
| 50–54 | 115 (23.7) | 6 (8.7) | 5 (11.4) |
| 55–59 | 164 (33.8) | 23 (33.3) | 14 (31.8) |
| 60–64 | 180 (37.1) | 39 (56.5) | 16 (36.4) |
| Family History of PCa | |||
| Negative | 385 (79.4) | 58 (84.1) | 40 (90.9) |
| Positive | 100 (20.6) | 11 (15.9) | 4 (9.1) |
| PCa Screening ± | |||
| None | 24 (5.0) | 3 (4.4) | 12 (27.3) |
| DRE only | 92 (19.0) | 22 (31.9) | 7 (15.9) |
| PSA + DRE | 369 (76.1) | 44 (63.8) | 25 (56.8) |
| BMI | |||
| < 25.0 | 177 (36.5) | 19 (27.5) | 16 (36.4) |
| 25.0 – 29.9 | 235 (48.5) | 37 (53.6) | 20 (45.5) |
| >= 30.0 | 73 (15.1) | 13 (18.8) | 8 (18.2) |
| PSA at Diagnosis | |||
| 0.0 – 3.9 | 67 (15.3) | 11 (18.0) | 1 (2.4) |
| 4.0 – 9.9 | 265 (60.5) | 33 (54.1) | 10 (24.4) |
| >= 10 | 106 (24.2) | 17 (27.9) | 30 (73.1) |
| Gleason Score | |||
| 2 – 6 | 323 (66.6) | 42 (60.9) | 7 (16.3) |
| 3 + 4 | 121 (25.0) | 15 (21.7) | 12 (27.9) |
| 4 + 3 | 15 (3.1) | 4 (5.8) | 6 (14.0) |
| 8 – 10 | 26 (5.4) | 8 (11.6) | 18 (41.9) |
| Tumor Stage | |||
| Local | 382 (78.8) | 48 (69.6) | 10 (22.7) |
| Regional | 99 (20.4) | 20 (29.0) | 21 (47.7) |
| Distant | 4 (0.8) | 1 (1.5) | 13 (29.6) |
| Treatment | |||
| Radical Prostatectomy | 366 (75.5) | 40 (58.0) | 15 (34.1) |
| Radiation +/− Hormones | 80 (16.5) | 20 (29.0) | 12 (27.3) |
| Androgen Deprivation | 8 (1.7) | 4 (5.8) | 17 (38.6) |
| Other | 4 (0.8) | 0 (0.0) | 0 (0.0) |
| Watchful Waiting | 27 (5.6) | 5 (7.3) | 0 (0.0) |
Data may not sum to 598 due to missing values
Screening within the 5-year period prior to diagnosis date
Recurrence/Progression data available for 468 patients
The allelic frequencies of the three CYP17 SNPs are given in Table 2. For SNPs rs743572 and rs10883783, PCSM was more common in those homozygous for the wild-type allele compared to patients with any copies of the less common alleles. The results of the Cox proportional hazards analyses for PCa-specific mortality are shown in Table 2. In the multivariate analysis, carriers of any copies of the variant allele for SNP rs10883783 had a 56% risk reduction in PCSM (HR 0.44, 95% CI 0.20 – 0.95). The disease-specific Kaplan-Meier curve for rs10883783 is shown in Figure 1. For the three main treatment groups (RP, XRT and androgen deprivation therapy (ADT)) there was no significant interaction between treatment and genotype on survival (p = 0.06). Although numbers of patients in each treatment group are limited, stratification by treatment revealed similar HRs for the rs10883783 in the RP (HR 0.28, 95% CI 0.07 – 1.03), XRT (HR 0.23, 95% CI 0.05 – 1.13) and ADT (HR 0.59, 95%CI 0.18 – 2.02) groups. In Table 3, the results of the Cox proportional hazards analyses for recurrence/progression are shown. None of the SNPs were significantly associated with the risk of recurrence/progression.
Table 2.
Risk of Prostate Cancer-Specific Mortality by CYP17 Genotypes in Caucasian Patients
| Allelic Frequencies | Risk of Prostate Cancer-Specific Mortality |
||||
|---|---|---|---|---|---|
| PCa-Specific Death | Age Adjusted | Adjusted* | |||
| SNP | No | Yes | HR (95% CI) | HR (95% CI) | |
| rs743572 | |||||
| AA | 202 (90.6) | 21 (9.4) | 1.00 (referent) | 1.00 (referent) | |
| AG + GG | 313 (94.6) | 18 (5.4) | 0.58 (0.31 – 1.09) | 0.68 (0.33 – 1.41) | |
| rs10883783 | |||||
| TT | 273 (91.0) | 27 (9.0) | 1.00 (referent) | 1.00 (referent) | |
| AT + AA | 261 (95.3) | 13 (4.7) | 0.53 (0.27 – 1.02) | 0.44 (0.20 – 0.95) | |
| rs17115100 | |||||
| GG | 424 (93.8) | 30 (6.6) | 1.00 (referent) | 1.00 (referent) | |
| GT + TT | 95 (92.2) | 8 (7.8) | 1.16 (0.53 – 2.53) | 1.44 (0.56 – 3.69) | |
Adjusted for age, Gleason score, PSA, stage, primary treatment, BMI
Figure 1.
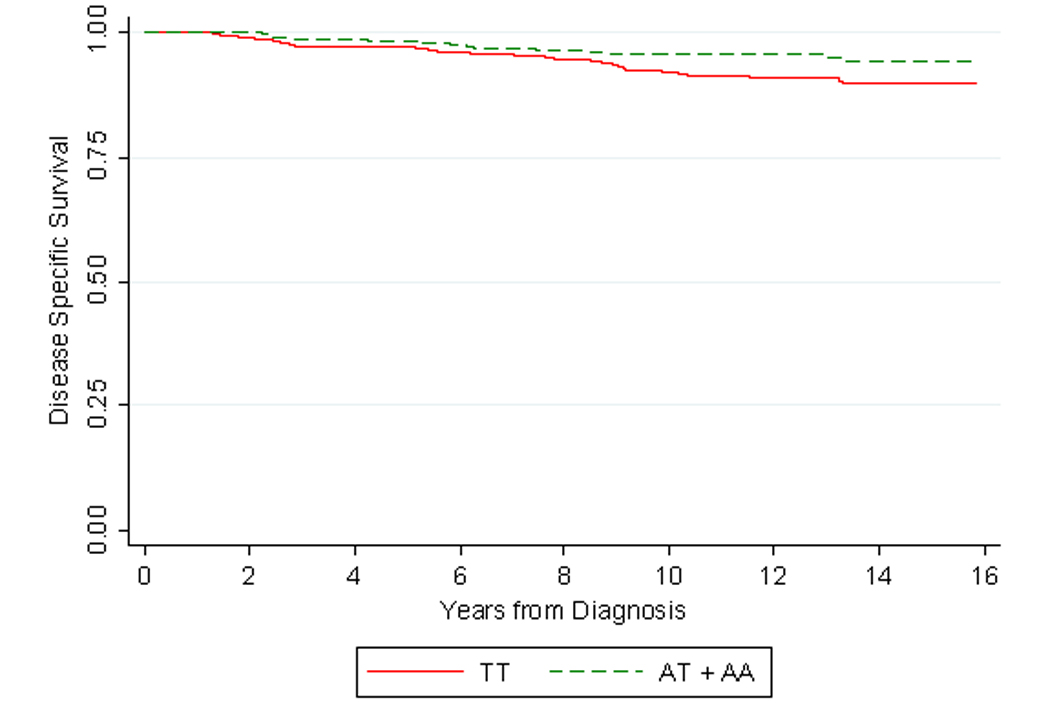
Kaplan-Meier estimates of prostate-cancer specific survival by SNP rs10883783 stratified by homozygous carriers of the wild type allele (TT) versus heterozygous and/or homozygous carries of the variant allele (AT + AA).
Table 3.
Risk of Prostate Cancer Recurrence/Progression by CYP17 Genotypes in Caucasian Patients
| Allelic Frequencies | Risk of Prostate Cancer Recurrence/Progression |
|||
|---|---|---|---|---|
| Recurrence/Progression | Age Adjusted | Adjusted* | ||
| SNP | No | Yes | HR (95% CI) | HR (95% CI) |
| rs743572 | ||||
| AA | 128 (72.7) | 48 (27.3) | 1.00 (referent) | 1.00 (referent) |
| AG + GG | 175 (67.8) | 83 (32.2) | 1.03 (0.71 – 1.50) | 0.86 (0.58 – 1.27) |
| rs10883783 | ||||
| TT | 173 (72.1) | 67 (27.9) | 1.00 (referent) | 1.00 (referent) |
| AT + AA | 140 (66.7) | 70 (33.3) | 1.11 (0.79 – 1.56) | 0.84 (0.59 – 1.21) |
| rs17115100 | ||||
| GG | 245 (69.0) | 110 (31.0) | 1.00 (referent) | 1.00 (referent) |
| GT + TT | 59 (72.0) | 23 (28.0) | 0.85 (0.54 – 1.34) | 0.67 (0.40 – 1.14) |
Adjusted for age, Gleason score, PSA, stage, primary treatment, BMI
Discussion
In this population-based cohort of patients with PCa followed for a median of 13.2 years, we observed differences in PCSM based on variant alleles within the CYP17 gene, even after adjusting for other potential confounding and pathological factors. If confirmed, these findings may have implications in the future management of PCa patients given the role of CYP17 in androgen biosynthesis.
Androgen signaling and regulation has a critical role in prostate cancer, and CYP17 is a microsomal enzyme that catalyses two steps in the biosynthesis of androgens.(1) The relationship between CYP17 and PCa-specific mortality has received recent attention because of drug treatments designed to target the CYP17 enzyme in an effort to decrease local tumor production of androgens despite castrate serum testosterone levels. Ketoconazole is often used in treatment of advanced PCa and has non-specific CYP17 inhibitory properties. Abiraterone acetate has recently been introduced as a selective CYP17 inhibitor and shown to have antitumor activity in a Phase I study of 21 men with castrate resistant PCa.(15) If genetic variations in CYP17 lead to increased PCa-specific survival through alterations in enzyme activity, the impact this may have on the action of CYP17 inhibitors becomes important.
The mechanism by which the specific SNPs studied here affect protein expression or function has not been defined. Two of the SNPs are located in introns and no function has been ascribed to them at this time. However, because of the extent of linkage disequilibrium in the region, the SNPs under study may not directly affect the gene or its product, but rather be indirectly correlated with a causal variant elsewhere in this or an adjacent gene. Intronic SNPs may have functional consequences as well as SNPs in close proximity to intron-exon boundaries that may alter splicing.(19,20) However, the most significant SNPs in this study (rs10883783), at approximately 110 base-pairs from the intron-exon boundary, is further than traditionally described for altered splicing to occur. The SNP, rs743572, occurs in the 5′-UTR and previous work by Carey et al., suggests that this change leads to an additional binding site in the promoter region with resultant effects on androgen levels.(21) However, subsequent work does not support that conclusion by failing to find alterations in hormone levels or promoter binding.(22–24)
Studies of CYP17 mRNA expression and prostate cancer have been performed. A recent study of 60 patients undergoing RP for PCa evaluated CYP17 expression in both neoplastic and non-neoplastic prostate tissue.(25) CYP17 mRNA was detected in all tumors and was found to be upregulated in tumor compared to normal tissues. In addition, CYP17 mRNA expression was increased in higher grade (>7) and higher stage tumors when compared to those of lower stage and lower grade (p<0.005). Further, 86% of the patients with low CYP17 expression (n = 35) remained disease free at two-years, whereas 96% of those with high levels of expression (n = 25) experienced biochemical relapse (p < 0.005).
Other lines of evidence support a role for CYP17 in advanced prostate cancer. Montgomery and colleagues have shown that CYP17 expression is 17-fold higher in CRPC versus untreated primary tumors. (26) Similarly, it has been shown that CRPC tumors have high tissue androgen levels, despite castrate serum levels.(26–28) These findings support the notion that there is local, intracrine production of testosterone, and CYP17 is a crucial enzyme in the testosterone biosynthesis pathway. If variant CYP17 alleles impair this process, there may be a decrease in local testosterone production, which in turn may reduce cellular proliferation and thereby prolong patient survival.
Recently, two studies have evaluated SNPs within the CYP17 gene in relation to mortality in men with PCa. Hamada studied 222 Caucasian men with CRPC and focused on a SNP in the 5′-UTR (rs743572),(14) which some have argued results in altered androgen levels.(21) They did not identify any differences in disease characteristics by alleles. With a median follow-up of 7.7 years, 187 total deaths occurred. Assuming a dominant model, men carrying the variant C allele had a 26% reduction in risk of overall mortality in an unadjusted model (p = 0.04). The median survival from diagnosis for patients homozygous for the wild-type genotype was 6.7 years compared to 8.9 years for those with one or two copies of the variant C allele. These results suggest that this CYP17 polymorphism is associated with overall survival in men with PCa, but effects are limited to those with CRPC; and they did not explore PCSM which is a more relevant endpoint. A second study of a Swedish population-based cohort of 2,761 men with PCa and 300 PCa deaths analyzed seven CYP17 SNPs, including two of the SNPs in our study (rs10883783 and rs17115100).(11) None were found to be significantly associated with PCSM in this study. Our findings of a reduced risk for PCSM associated with rs10883783 (HR 0.44, 95% CI 0.20 – 0.95) may be due to differences in study population or chance. In the Swedish study, 70% of deaths were due to PCa, whereas in our cohort, only 40% of deaths were attributed to PCa. The PCSM rate in this recent Swedish CYP17 study is 10 – 15% higher than an earlier Swedish population-based study(29) suggesting that patients analyzed by CYP17 genotypes represent a cohort with more aggressive disease.
We did not find any differences in the risk of recurrence/progression by genotype. This may partially be explained by the varying differences in this outcome by treatment groups and respective definitions of recurrence/progression. Given that this is an observational cohort with recurrence/progression events ascertained by self-report and availability of medical records, there may also be some misclassification that would bias results toward the null. Alternatively, biochemical recurrence may not be a good surrogate for PCa survival as many patients who have PSA-based evidence for recurrence/progression do not die of their disease.(30) Moreover, reported progression rates may vary by up to 35% depending on how biochemical recurrence is defined.(31)
If variants within the CYP17 gene affect enzymatic activity at baseline, some subsets of patients may be variably responsive to drugs that target the CYP17 enzyme. Thus, it may be important to understand the genetic background of prospective patients with regard to CYP17 before embarking on targeted therapy. Indeed, some drug therapies are already based on molecular status, such as Her2-neu status in breast cancer. Genetic information regarding CYP17 status may also dictate choice of therapy as well as provide important information on prognosis and predicted disease response.
There are limitations to this study. Misclassification of underlying cause of death can occur in the SEER registry. However, all deaths in this cohort were confirmed with death certificates and a review of underlying cause showed 99% agreement. In addition, the findings of our study could be due to chance alone given the relatively few events due to the generally good prognosis of PCa patients, although the results are biologically plausible given the relationship between CYP17, the androgen pathway and PCa. However, the SNPs studied have unknown function and even if associated with survival, may not be causal but rather reflect linkage disequilibrium with the causal SNP such that all of the genetic variability in the studied SNP is inherited with the causal SNP. Due to concerns about population stratification, our analyses were limited to Caucasian men and these findings may only apply to this group. Finally, the men in this cohort were selected from one geographic area (King County, Washington).
Conclusion
Specific polymorphisms in the CYP17 gene are associated with improved PCa-specific survival. With increased attention given to CYP17 inhibitors in the treatment of advanced PCa, CYP17 genetic status may yield important prognostic information, particularly in lending insight on responsiveness to targeted therapies. It will be important in subsequent studies to further characterize this gene and gain an understanding of the true causative variant(s) and how they function. Attempts to replicate these provocative findings should be undertaken in similar, larger datasets to confirm our findings.
Acknowledgments
NIH Grants: R01 CA 56678; R01 CA 092579; P50 CA097186, T32 CA009168-30; with additional support from the Fred Hutchinson Cancer Research Center, the Intramural Program of the National Human Genome Research Institute, and the many men who generously participated in this study.
References
- 1.Picado-Leonard J, Miller WL. Cloning and sequence of the human gene for P450c17 (steroid 17 alpha-hydroxylase/17,20 lyase): similarity with the gene for P450c21. DNA. 1987;6(5):439–448. doi: 10.1089/dna.1987.6.439. [DOI] [PubMed] [Google Scholar]
- 2.Yamada Y, Watanabe M, Murata M, Yamanaka M, Kubota Y, Ito H, Katoh T, Kawamura J, Yatani R, Shiraishi T. Impact of genetic polymorphisms of 17-hydroxylase cytochrome P-450 (CYP17) and steroid 5alpha-reductase type II (SRD5A2) genes on prostate-cancer risk among the Japanese population. Int J Cancer. 2001;92(5):683–686. doi: 10.1002/1097-0215(20010601)92:5<683::aid-ijc1255>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 3.Kittles RA, Panguluri RK, Chen W, Massac A, Ahaghotu C, Jackson A, Ukoli F, Adams-Campbell L, Isaacs W, Dunston GM. Cyp17 promoter variant associated with prostate cancer aggressiveness in African Americans. Cancer Epidemiol Biomarkers Prev. 2001;10(9):943–947. [PubMed] [Google Scholar]
- 4.Lunn RM, Bell DA, Mohler JL, Taylor JA. Prostate cancer risk and polymorphism in 17 hydroxylase (CYP17) and steroid reductase (SRD5A2) Carcinogenesis. 1999;20(9):1727–1731. doi: 10.1093/carcin/20.9.1727. [DOI] [PubMed] [Google Scholar]
- 5.Gsur A, Bernhofer G, Hinteregger S, Haidinger G, Schatzl G, Madersbacher S, Marberger M, Vutuc C, Micksche M. A polymorphism in the CYP17 gene is associated with prostate cancer risk. Int J Cancer. 2000;87(3):434–437. doi: 10.1002/1097-0215(20000801)87:3<434::aid-ijc19>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 6.Stanford JL, Noonan EA, Iwasaki L, Kolb S, Chadwick RB, Feng Z, Ostrander EA. A polymorphism in the CYP17 gene and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2002;11(3):243–247. [PubMed] [Google Scholar]
- 7.Wadelius M, Andersson AO, Johansson JE, Wadelius C, Rane E. Prostate cancer associated with CYP17 genotype. Pharmacogenetics. 1999;9(5):635–639. [PubMed] [Google Scholar]
- 8.Habuchi T, Liqing Z, Suzuki T, Sasaki R, Tsuchiya N, Tachiki H, Shimoda N, Satoh S, Sato K, Kakehi Y, Kamoto T, Ogawa O, Kato T. Increased risk of prostate cancer and benign prostatic hyperplasia associated with a CYP17 gene polymorphism with a gene dosage effect. Cancer Res. 2000;60(20):5710–5713. [PubMed] [Google Scholar]
- 9.Antognelli C, Mearini L, Talesa VN, Giannantoni A, Mearini E. Association of CYP17, GSTP1, and PON1 polymorphisms with the risk of prostate cancer. Prostate. 2005;63(3):240–251. doi: 10.1002/pros.20184. [DOI] [PubMed] [Google Scholar]
- 10.Ntais C, Polycarpou A, Ioannidis JP. Association of the CYP17 gene polymorphism with the risk of prostate cancer: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2003;12(2):120–126. [PubMed] [Google Scholar]
- 11.Lindstrom S, Adami HO, Balter KA, Xu J, Zheng SL, Stattin P, Gronberg H, Wiklund F. Inherited variation in hormone-regulating genes and prostate cancer survival. Clin Cancer Res. 2007;13(17):5156–5161. doi: 10.1158/1078-0432.CCR-07-0669. [DOI] [PubMed] [Google Scholar]
- 12.Setiawan VW, Schumacher FR, Haiman CA, Stram DO, Albanes D, Altshuler D, Berglund G, Buring J, Calle EE, Clavel-Chapelon F, Cox DG, Gaziano JM, Hankinson SE, Hayes RB, Henderson BE, Hirschhorn J, Hoover R, Hunter DJ, Kaaks R, Kolonel LN, Kraft P, Ma J, Le Marchand L, Linseisen J, Lund E, Navarro C, Overvad K, Palli D, Peeters PH, Pike MC, Riboli E, Stampfer MJ, Thun MJ, Travis R, Trichopoulos D, Yeager M, Ziegler RG, Spencer Feigelson H, Chanock SJ. CYP17 genetic variation and risk of breast and prostate cancer from the National Cancer Institute Breast and Prostate Cancer Cohort Consortium (BPC3) Cancer Epidemiol Biomarkers Prev. 2007;16(11):2237–2246. doi: 10.1158/1055-9965.EPI-07-0589. [DOI] [PubMed] [Google Scholar]
- 13.Douglas JA, Zuhlke KA, Beebe-Dimmer J, Levin AM, Gruber SB, Wood DP, Cooney KA. Identifying susceptibility genes for prostate cancer--a family-based association study of polymorphisms in CYP17, CYP19, CYP11A1, and LH-beta. Cancer Epidemiol Biomarkers Prev. 2005;14(8):2035–2039. doi: 10.1158/1055-9965.EPI-05-0170. [DOI] [PubMed] [Google Scholar]
- 14.Hamada A, Danesi R, Price DK, Sissung T, Chau C, Venzon D, Sparreboom A, Dahut WL, Figg WD. Association of a CYP17 polymorphism with overall survival in Caucasian patients with androgen-independent prostate cancer. Urology. 2007;70(2):217–220. doi: 10.1016/j.urology.2007.06.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Attard G, Reid AH, Yap TA, Raynaud F, Dowsett M, Settatree S, Barrett M, Parker C, Martins V, Folkerd E, Clark J, Cooper CS, Kaye SB, Dearnaley D, Lee G, de Bono JS. Phase I clinical trial of a selective inhibitor of CYP17, abiraterone acetate, confirms that castration-resistant prostate cancer commonly remains hormone driven. J Clin Oncol. 2008;26(28):4563–4571. doi: 10.1200/JCO.2007.15.9749. [DOI] [PubMed] [Google Scholar]
- 16.Stanford JL, Wicklund KG, McKnight B, Daling JR, Brawer MK. Vasectomy and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 1999;8(10):881–886. [PubMed] [Google Scholar]
- 17.Harrell F. Regression Modeling Strategies. New York: Springer; 2001. [Google Scholar]
- 18.Roach M, 3rd, Hanks G, Thames H, Jr, Schellhammer P, Shipley WU, Sokol GH, Sandler H. Defining biochemical failure following radiotherapy with or without hormonal therapy in men with clinically localized prostate cancer: recommendations of the RTOG-ASTRO Phoenix Consensus Conference. Int J Radiat Oncol Biol Phys. 2006;65(4):965–974. doi: 10.1016/j.ijrobp.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 19.Sawa H, Ohshima TA, Ukita H, Murakami H, Chiba Y, Kamada H, Hara M, Saito I. Alternatively spliced forms of cyclin D1 modulate entry into the cell cycle in an inverse manner. Oncogene. 1998;16(13):1701–1712. doi: 10.1038/sj.onc.1201691. [DOI] [PubMed] [Google Scholar]
- 20.Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WD, Heighway J. Alternate splicing produces a novel cyclin D1 transcript. Oncogene. 1995;11(5):1005–1011. [PubMed] [Google Scholar]
- 21.Carey AH, Waterworth D, Patel K, White D, Little J, Novelli P, Franks S, Williamson R. Polycystic ovaries and premature male pattern baldness are associated with one allele of the steroid metabolism gene CYP17. Hum Mol Genet. 1994;3(10):1873–1876. doi: 10.1093/hmg/3.10.1873. [DOI] [PubMed] [Google Scholar]
- 22.Kakinuma H, Tsuchiya N, Habuchi T, Ohyama C, Matsuura S, Wang L, Nakamura A, Kato T. Serum sex steroid hormone levels and polymorphisms of CYP17 and SRD5A2: implication for prostate cancer risk. Prostate Cancer Prostatic Dis. 2004;7(4):333–337. doi: 10.1038/sj.pcan.4500753. [DOI] [PubMed] [Google Scholar]
- 23.Nedelcheva Kristensen V, Haraldsen EK, Anderson KB, Lonning PE, Erikstein B, Karesen R, Gabrielsen OS, Borresen-Dale AL. CYP17 and breast cancer risk: the polymorphism in the 5' flanking area of the gene does not influence binding to Sp-1. Cancer Res. 1999;59(12):2825–2828. [PubMed] [Google Scholar]
- 24.Severi G, Hayes VM, Tesoriero AA, Southey MC, Hoang HN, Padilla EJ, Morris HA, English DR, Sutherland RL, Boyle P, Hopper JL, Giles GG. The rs743572 common variant in the promoter of CYP17A1 is not associated with prostate cancer risk or circulating hormonal levels. BJU Int. 2008;101(4):492–496. doi: 10.1111/j.1464-410X.2007.07272.x. [DOI] [PubMed] [Google Scholar]
- 25.Stigliano A, Gandini O, Cerquetti L, Gazzaniga P, Misiti S, Monti S, Gradilone A, Falasca P, Poggi M, Brunetti E, Agliano AM, Toscano V. Increased metastatic lymph node 64 and CYP17 expression are associated with high stage prostate cancer. J Endocrinol. 2007;194(1):55–61. doi: 10.1677/JOE-07-0131. [DOI] [PubMed] [Google Scholar]
- 26.Montgomery RB, Mostaghel EA, Vessella R, Hess DL, Kalhorn TF, Higano CS, True LD, Nelson PS. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res. 2008;68(11):4447–4454. doi: 10.1158/0008-5472.CAN-08-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mostaghel EA, Page ST, Lin DW, Fazli L, Coleman IM, True LD, Knudsen B, Hess DL, Nelson CC, Matsumoto AM, Bremner WJ, Gleave ME, Nelson PS. Intraprostatic androgens and androgen-regulated gene expression persist after testosterone suppression: therapeutic implications for castration-resistant prostate cancer. Cancer Res. 2007;67(10):5033–5041. doi: 10.1158/0008-5472.CAN-06-3332. [DOI] [PubMed] [Google Scholar]
- 28.Locke JA, Guns ES, Lubik AA, Adomat HH, Hendy SC, Wood CA, Ettinger SL, Gleave ME, Nelson CC. Androgen levels increase by intratumoral de novo steroidogenesis during progression of castration-resistant prostate cancer. Cancer Res. 2008;68(15):6407–6415. doi: 10.1158/0008-5472.CAN-07-5997. [DOI] [PubMed] [Google Scholar]
- 29.Gronberg H, Damber L, Jonson H, Damber JE. Prostate cancer mortality in northern Sweden, with special reference to tumor grade and patient age. Urology. 1997;49(3):374–378. doi: 10.1016/S0090-4295(96)00508-0. [DOI] [PubMed] [Google Scholar]
- 30.Collette L. Prostate-specific antigen (PSA) as a surrogate end point for survival in prostate cancer clinical trials. Eur Urol. 2008;53(1):6–9. doi: 10.1016/j.eururo.2007.08.041. [DOI] [PubMed] [Google Scholar]
- 31.Stephenson AJ, Kattan MW, Eastham JA, Dotan ZA, Bianco FJ, Jr, Lilja H, Scardino PT. Defining biochemical recurrence of prostate cancer after radical prostatectomy: a proposal for a standardized definition. J Clin Oncol. 2006;24:3973–3978. doi: 10.1200/JCO.2005.04.0756. [DOI] [PubMed] [Google Scholar]


