Abstract
Microscopy has always been an obligate tool in the field of developmental biology, a goal of which is to elucidate the essential cellular and molecular interactions that coordinate the specification of different cell types and the establishment of body plans. The 2008 Nobel Prize in chemistry was awarded ‘for the discovery and development of the green fluorescent protein, GFP′ in recognition that the discovery of genetically encoded fluorescent proteins (FPs) has spearheaded a revolution in applications for imaging of live cells. With the development of more-sophisticated imaging technology and availability of FPs with different spectral characteristics, dynamic processes can now be live-imaged at high resolution in situ in embryos. Here, we review some recent advances in this rapidly evolving field as applied to live-imaging capabilities in the mouse, the most genetically tractable mammalian model organism for embryologists.
Seeing is believing
The phenomenon of color has always fascinated researchers and academics of all disciplines. The discovery of fluorescent proteins (FPs) and the cloning of the first FP, wild-type green fluorescent protein (wtGFP), from the jellyfish Aequorea victoria [1] in the early 1990 s particularly excited life-scientists. Since then, FPs have proven a useful tool and made a tremendous impact on molecular biology. The original laboratory FP has served as a reagent for the production of many chemical modifications, producing, among other desirable features, spectral variants [2–4]. Other organisms, especially the Anthozoans (corals), have been exploited in the search for new FPs, with the desired properties focusing most of all on improved brightness but also on their excitation and emission spectra, monomerization, folding dynamics and reduced photobleaching [4–6]. GFP and its variants have remained the workhorse of life-science investigations because, unlike other FPs, they have been shown to be non-toxic in vivo. GFP variants have been used to make ubiquitously and constitutively expressing transgenic plants or animals, such as worms, fruit flies, zebrafish, frogs and mice [7–14]. Since then, FPs have been applied in a variety of experimental settings, for example in promoter function studies [15,16], as fusion-protein tags to elucidate basic cellular functions [4,17] and in an organismal context for investigating tumor–host interactions [18,19].
Combined with the imaging technology that is now available, FPs have become indispensable tools for exploring cellular function in real-time and at high resolution. It is widely recognized that genetic approaches are central in deciphering the roles of specific genes and gene networks operating during mouse embryonic development. However, a dynamic understanding of the early morphogenetic events that create the three-dimensional (3D) organization of the animal are lacking owing to the static nature of established protocols for dissecting spatially and temporally regulated events. Live imaging using FP reporters combined with the power of mouse genetics represents the essential next step forward towards unraveling the mechanisms regulating embryonic development.
Although the most recent advancements in microscopy are only found in specialized laboratories or core environments, suitable equipment for capturing the wealth of data revealed by FP technologies can be found in most academic campuses. Thus, this review is centered on providing a survey of widely available probes and readily implementable technologies (see Box 1) for investigators interested in establishing live-imaging experiments in their own laboratories.
Box 1. The tool box of a live cell imager.
The confocal microscope has become a mainstay of contemporary biological science. Confocal microscopes function to exclude light that originates from outside of the plane of focus. By changing the level of the focal plane, several optical sections can be gathered and digitally reconstructed into a 3D representation of the sample. Confocal images can be generated by several methods, including laser-scanning point, multipoint or slit-scanning modalities. Point scanning, the most commonly available modality, passes a laser beam across a sample, exposing all points within the narrow cylinder of the laser light perpendicular to the focal plane. Fluorophores contained within this cylinder emit light that is subsequently focused through a pinhole, eliminating photons originating from above and below the plane of focus. This process is repeated as the laser is scanned across the specimen. The process is relatively slow owing to the amount of time required to draw the laser across the entire sample.
Increases in the speed of acquisition have been accomplished by the development of slit-scanning and multipoint confocal microscopy. In the former, a line of laser light rather than a point is drawn across the sample. Light is passed through a slit that serves as a conventional pinhole and is then detected by a unique CCD (charged-coupled device) camera. By collecting a 512 pixel line as a batch, slit-scanning microscopy reduces acquisition time. Selective plane illumination microscopy (SPIM), uses 2D illumination with orthogonal camera-based detection, thereby providing greater depth penetration [92]. This allows high-speed imaging at cellular resolution of samples up to a few mm in size. Multipoint microscopy is typified by the Nipkow-type spinning disc systems. This technology passes a laser through a spinning disc that contains several thousand pinholes. Emitted light is returned through the same holes and collected on a CCD camera, effectively producing a ‘real-time’ confocal image. This technology has been further developed with digital scanned light sheet microscopy (DSLM) [93].
Multiphoton microscopes, like confocal instruments, collect data only from a given plane within the sample. Unlike confocal microscopes, multiphoton technology induces fluorescence only at finite points within the 3D space of the sample. Although this technology uses higher-powered lasers than conventional single photon microscopy, the limited point of excitation reduces problems associated with photobleaching and phototoxicity. Excitation is produced only when two coincident photons of wavelengths twice that required for single photon excitation strike a fluorophore at nearly the same moment. Because two-photon emission must inherently occur at a discrete point, pinholes are not required.
A palette of colors for tagging and tracking
The use of FPs as tags has accelerated in the past decade. FP reporters have been used to study gene expression, protein localization and cell behaviors. Different applications have been developed to investigate protein–protein interactions, track and quantify proteins, track cells and perform lineage-tracing experiments. These methods often use photomodulation of FPs (e.g. photobleaching) or photomodulatable FPs whose emission spectra can be varied in response to variable incident light wavelengths. Fusions of FPs to other proteins have proven useful in different contexts of cellular function. Nuclear labels, such as fusions of FPs and human histone H2B protein (e.g. H2B–GFP) that are bound to active chromatin in the nucleus, allow investigators to study cell division and apoptosis and are essential for cell tracking [20,21] (Figure 1). FP fusions that label proteins located at the plasma membrane, such as lipid-modified fusions including a myristoylation (myr) sequence and glycosylphosphatidylinositol (GPI) anchor fusions, enable the gathering of information on cell morphology and cell migration [22,23]. Furthermore, cytoskeletal dynamics and the movements of motile cilia can be visualized by fusions to the microtubule-binding protein tau [24]. Indeed, among the unrealized goals of live imaging technology is the idea that nearly all components and processes of a cell could be labeled and visualized simultaneously to generate a complex ‘four-dimensional’ (4D) understanding of cellular dynamics in a complex cell population or within the context of an organism. In practice, this dream has been grounded by the limited availability of genetically encoded FP reporters exhibiting robust reporter expression, the technologies for excitation and detection of multiple fluorophores, and the limited ability to deliver these constructs to living tissues through transient or stable transgenesis. To this end, multiplexing spectrally distinct subcellularly localized FP probes provides an unprecedented resolution in live-imaging studies in embryonic stem (ES) cells (Figure 2) and in embryos (Figure 3) [25,26].
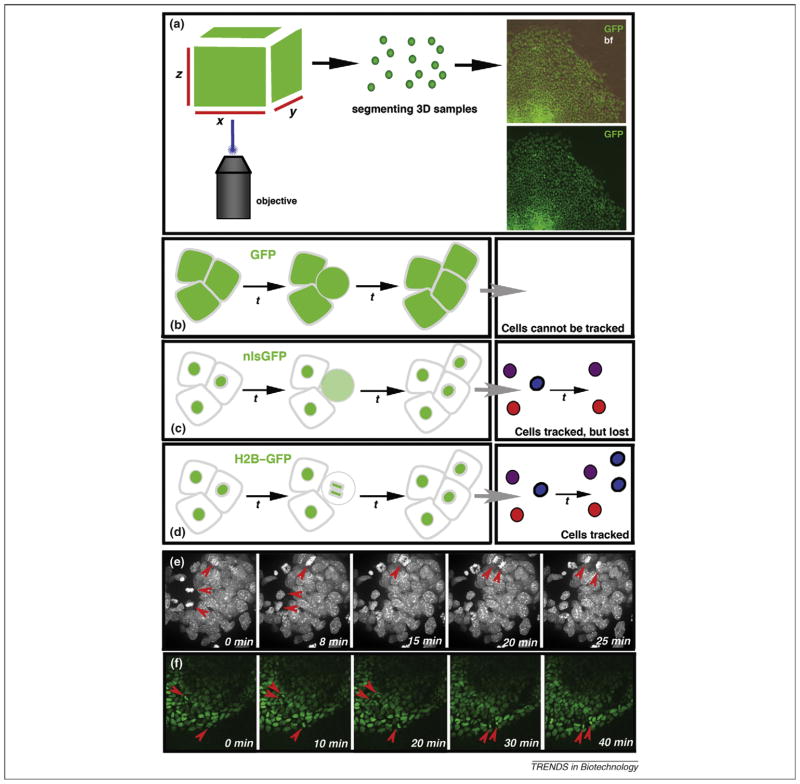
Figure 1.
Histone fusions localize fluorescent proteins (FPs) to chromatin and make them ideal for cell tracking. (a) Arrangement for imaging FPs expressed in the mouse embryo. 2D raw data (2D x-y images taken at different z stacks) of cells of interest in the embryo is rendered into 3D data using software packages. Images on the right depict 3D rendered images of GFP-expressing cells. The top panel is the green channel overlayed on a bright field image illustrating the contour of the ES cell colony, and the bottom panel shows the green channel only. (b–f) Cell tracking using fluorescent fusion proteins. Nuclear localization sequences (NLSs) are commonly used for the nuclear localization of gene-based reporters; however, when a cell divides (the round cell in the schematic drawing is destined to divide by mitosis) and the nuclear envelope breaks down, the reporter protein becomes distributed throughout the cell, causing a reduction in the fluorescence intensity. (b) No individual cells can be identified with cytoplasmically expressed GFP. Cells cannot be tracked. (c) Individual cells can be identified (schematized in different colors), counted and tracked using GFP fusion to NLS sequences (nlsGFP), especially when used in combination with software applications that can perform particle tracking functions. Because nuclei are single entities, they are ideal for assigning computational ‘particle’ status. Such a ‘particle’ can be tagged and followed computationally through a time-lapse series so that a set of tracks can be generated, with each track representing positional information (over time) for any given cell. However, computational methods designed to track cells usually lose track of an NLS-reporter-expressing cell that has divided, because it essentially disappears. This is a common problem, and such computational methods usually cannot distinguish between cells that divide and cells that die because, in both cases, the tracks terminate within the experimental time frame. (d) Histone fusions (H2B–GFP) remain bound to chromatin during cell division. Therefore, the computer can keep track of the cells expressing an H2B–GFP fusion reporter during cell division. In addition, they also provide information on the plane of cell division and the designation of daughter cells. (e,f) Each panel represents a time-point from a rendered z-stack of an x-y-z-t (4D) experiment imaging cell dynamics in cell populations expressing an H2B–GFP fusion reporter. Expression of an H2B-GFP fusion reporter and 3D time-lapse imaging provides information of single cell position and orientation of cell divisions within a population of cells. (e) Bright field of a time-lapse sequence of an ES cell colony. (f) Green channel of a time-lapse sequence of a gastrulating mouse embryo.
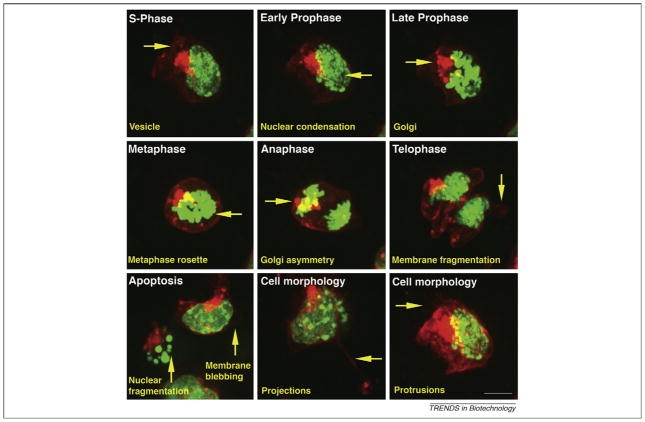
Figure 2.
Dual-tagged ES cells with H2B–GFP as a nuclear marker and myr–RFP as a label for the plasma membrane. ES cells expressing H2B–GFP and myr–RFP provide information about cell divisions (each phase of mitosis can be distinguished), apoptosis (nuclear fragmentation can be visualized) and cell morphology (cell protrusions and projections, membrane fragmentation), as well as the formation and breakdown of the Golgi apparatus. Scale bar represents 20 μm.
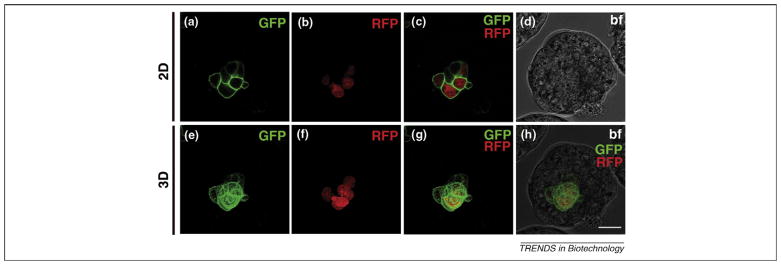
Figure 3.
Chimeric blastocysts containing cells expressing H2B–Cherry as a nuclear marker and GPI–GFP as a label for the plasma membrane. (a–d) 2D images of chimeric mouse blastocyst. (a) Green channel showing cells of blastocysts expressing GPI–GFP labeling the plasma membrane. (b) Red channel showing expression of H2B–Cherry in the nuclei of cells. (c) Merge of green and red channel. (d) Bright field. (e–h) 3D-rendered images of chimeric mouse blastocyst. (e) Green channel. (f) Red channel. (g) Merge of green and red channel. (h) Merge of bright field, green and red channel. Scale bar represents 20 μm.
Limiting the photon exposure of samples in live-imaging applications helps to maintain fidelity, so minimizing the number of fluorophores helps to meet this goal. This can be achieved by using the same fluorophore to label and acquire differential information in distinct cell types. Therefore dual spectrally and subcellularly distinct labels afford a high-resolution binary color-code for live imaging [26]. Theoretically, using just two spectrally distinct (e.g. GFP versus red FP [RFP]) FPs and two distinct spatial locations (e.g. nuclear versus plasma membrane), up to four different cell populations can be dual-tagged (Figure 4), with eight different possible populations by combining single and dual tagging.
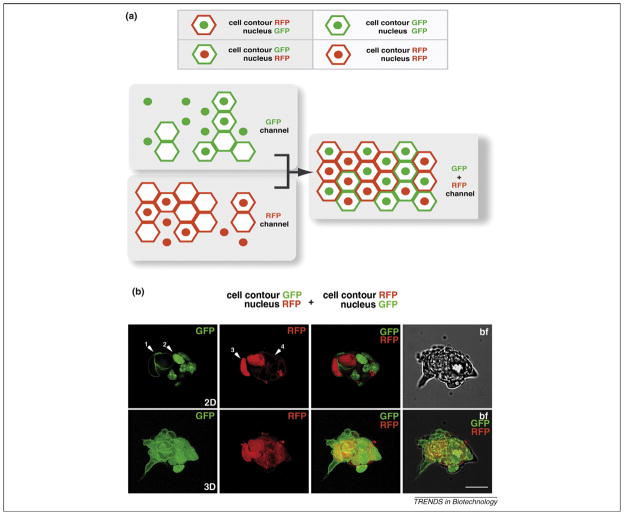
Figure 4.
Binary color coding for distinguishing different cell populations. (a) Schematic representation of using a binary color code to distinguish different cell populations. Using only two colors, it is possible to distinguish up to eight different cell populations. (b) 2D (top row) and 3D (bottom row) representations of green, red (top and bottom row) and bright-field (top row) channels and a merge of bright field, green and red channels (bottom row). The panels show an ES cell colony expressing a plasma membrane GFP (GPI–GFP) and plasma membrane RFP (myr–RFP) and nuclear GFP (H2B-GFP) and nuclear RFP (H2B-Cherry), respectively. White arrowheads: 1, GPI–GFP; 2, H2B–GFP; 3, H2B–Cherry; 4, myr–RFP. Scale bar represents 20 μm.
Next, we review of some of the commonly used FPs and their applicability in live cell imaging in the mouse embryo.
The green fluorescent protein (GFP) and its variants
The cloning of wtGFP as the first genetically encoded FP kicked off a revolution in the use of fluorescent markers in molecular biology. wtGFP was originally isolated from the bioluminescent jellyfish A. victoria [1]. wtGFP emits green light when ultraviolet (UV) or blue light is absorbed (Table 1). Stability over a broad range of pH and temperatures, as well as cofactor independent folding and maturation, made wtGFP attractive for molecular biology and biochemical experiments [27,28]. Improved versions of wtGFP (e.g. enhanced GFP [EGFP] and emerald GFP [EmGFP]) with brighter fluorescence and greater photostabilities were developed through random site-directed mutagenesis of wtGFP or other green FPs, such as Azami-Green (AG) [29–31]. However, the most popular variant of GFP to date is EGFP due to its simple excitation and emission spectra, as well as its simplicity for use in labeling experiments.
Table 1.
Absorbance and emission spectra of common, currently available fluorescent proteins
| Fluorescent protein | Absorbance (nm) | Emission (nm) | Oligomeric state | Commercially available | Refs |
|---|---|---|---|---|---|
| EBFP2 | 384 | 448 | Monomer | Invitrogena | [84] |
| ECFP | 433 | 475 | Monomer | Invitrogena | [29] |
| Cerulean | 433 | 475 | Monomer | n/aa | [37] |
| TagCFP | 458 | 480 | Monomer | Evrogen | [85] |
| TurboGFP | 482 | 502 | Dimer | Evrogen | [86] |
| TagGFP | 482 | 505 | Monomer | Evrogen | [85] |
| mEmerald | 487 | 509 | Monomer | Invitrogen | [87] |
| mEGFP | 488 | 507 | Monomer | Invitrogena | [30] |
| mAzami-Green (mAG) | 492 | 505 | Monomer | MBL Intl | [31] |
| mHoneydew | 504 | 562 | Monomer | n/aa | [43] |
| TagYFP | 508 | 524 | Monomer | Evrogen | [85] |
| EYFP | 512 | 528 | Monomer | Invitrogena | [38] |
| mVenus | 512 | 528 | Monomer | No | [38] |
| mCitrine | 516 | 529 | Monomer | n/aa | [88] |
| TurboYFP | 525 | 538 | Dimer | Evrogen | [86] |
| mBanana | 540 | 553 | Monomer | Clontech | [43] |
| mKO | 548 | 559 | Monomer | MBL Intl | [89] |
| mOrange | 548 | 562 | Monomer | Clontech | [43] |
| TurboRFP | 553 | 574 | Dimer | Evrogen | [90] |
| tdTomato | 554 | 581 | Dimer | n/a | [43] |
| TagRFP | 555 | 584 | Monomer | Evrogen | [90] |
| DsRed | 558 | 583 | Tetramer | Evrogena | [43] |
| mTangerine | 568 | 585 | Monomer | n/aa | [43] |
| mStrawberry | 574 | 596 | Monomer | Clontech | [43] |
| Katushka (TurboFP602) | 574 | 602 | Dimer | Evrogen | [48] |
| mRFP1 | 584 | 607 | Monomer | n/a | [40] |
| mCherry | 587 | 610 | Monomer | Clontech | [43] |
| mKate (TagFP635) | 588 | 635 | Monomer | Evrogen | [48] |
| mPlum | 595 | 655 | Monomer | Clontech | [43] |
Websites: Clontech, http://www.clontech.com; Evrogen, http://www.evrogen.com; Invitrogen, http://www.invitrogen.com; MBL International, http://mblintl.com.
Available from the non-profit plasmid repository ‘Addgene’ (http://www.addgene.org).
The need for FPs with different spectral characteristics, especially absorbance and emission spectra in the blue and red spectra, led to site-directed mutagenesis of GFP. These efforts have resulted in variants such as blue FP (BFP) [32,33], cyan FP (CFP) and yellow FP (YFP), respectively (Table 1). BFP, despite the disadvantage of being dim and easily photobleachable, was one of the first FPs used in multicolor imaging [33,34] and fluorescence resonance energy transfer (FRET) experiments [35,36] because it is demonstrably spectrally distinct from EGFP. The CFPs –CFP, ECFP and their successor Cerulean – are characterized by absorbance and emission spectra that are intermediate to those of EGFP and BFP [32]. Due to its relatively bright imaging characteristics and its significantly better signal-to-noise ratio in FRET experiments, Cerulean is now the CFP of choice in most laboratories [37] (Table 1). YFPs have been useful tools for studying and monitoring protein–protein interactions and signal transduction. However, first-generation YFPs were very pH sensitive and suffered from decreased photostablility. To overcome these disadvantages, new variants were generated through mutagenesis. The most prominent, Venus, is less acid sensitive and brighter than YFP and is ideal for labeling proteins in secretory organelles due to the acidic pH in these vesicles [38]. Prominent variants of the above-mentioned FPs are listed in Table 1.
Orange and red fluorescent proteins
Due to their long-wavelength emission, and thus reduced cell toxicity, FPs with emission peaks in the red and far-red spectrum have been of special interest for live-cell or whole-animal imaging.
The first successful RFP, DsRed, was isolated from the sea anemone Discosoma sp. [39]. DsRed, like many of the discovered yellow to red wild-type FPs, is autotetrameric. Such tetramers have often been shown to be toxic to the cell and are not suitable for use in fusion proteins owing to steric hindrance. The use of directed evolution to obtain monomeric variants of DsRed led to the first red monomer, mRFP1[40]. Widespread expression of this variant has been successful in the mouse, where, when expressed without fusions, it has been confirmed to be non-teratogenic and non-toxic during postnatal life [41]. A stable variant sacrificing the monomeric nature of mRFP, tandem dimer (td)RFP, has also been applied successfully in generating knock-in Cre reporter mice that are useful for lineage tracing [42]. However, widespread expression of mRFP1 in fusions in mouse, including relatively small fusions such as myr–mRFP1 and H2B–mRFP1, are teratogenic [26], highlighting the need for alternative RFPs.
Other currently available orange and red FPs, called the ‘fruit series’, were reported after a screen of directed mutagenesis of mRFP. These variants exhibit excitation–emission maxima between 537 and 610 nm. Out of this series, tdTomato, which exhibits great photostability, and mStrawberrry and mCherry are considered to be the most applicable orange and red FPs for live imaging [43]. In particular, owing to its fast maturation and high photostability, mCherry is currently the most suitable RFP for time-lapse imaging. mCherry has also been shown to work well in fusion proteins [44,45]. Strains of mice with widespread expression of mCherry labeling the plasma membrane and the nucleus have been generated [44]. However, similar to mRFP1, constitutive widespread expression of H2B–mCherry is also teratogenic [26]. Other currently available variants of red FPs, either generated by mutagenesis or isolated from other organisms, are listed in Table 1.
Far-red fluorescent proteins
Bright FPs with excitation–emission spectra in the near-infrared region of the spectrum are of great interest for live-imaging approaches. They provide greater tissue penetration and reduced autofluorescence. Currently, there are only a few FPs available that cover the far-red region of the spectrum. The first monomeric far-red FP, mPlum, was genetically engineered through iterative somatic hypermutation [46] of a blue chromoprotein of the sea anemone Actinia equina. Although its brightness is only 10% that of GFP, it can be advantageous when spectral separation is crucial [47]. The recently engineered dimer Katushka is the brightest far-red protein available so far. It is highly pH- and photo-stable, as is its monomeric version mKate. Their brightness and photostability make them ideal for cell labeling or protein tagging [48].
Spatiotemporal control of cell labeling with photomodulatable fluorescent proteins
Photomodulatable proteins provide the possibility to selectively label and trace cells and proteins in a spatio-temporal manner, yielding either a change of fluorescence or increase of intensity of the fluorescent signal after the photoconversion or photoactivation, respectively [49]. Pulse-chase labeling and tracking cells in live embryos has been achieved using invasive techniques such as dye injections or tissue grafts or genetically using binary site-specific recombinase systems [50]. There are two major categories of photomodulatable proteins – photoactivatable and photoconvertible proteins – of which the most common are described below (Table 2). Photoactivatable proteins change from a non-fluorescent to a fluorescent state upon irradiation with short-wavelength light. By contrast, photoconvertible proteins convert from one fluorescent state to another and change color. Below, the most prominent photomodulatable proteins to date are introduced.
Table 2.
Properties of currently available photomodulatable fluorescent proteins
| Photoactivatable fluorescent protein | Change of absorbance (nm) | Change of emission (nm) | Oligomeric state | Organism of origin | Commercially available | Refs |
|---|---|---|---|---|---|---|
| PAGFP | 400 to 504 | Increase at 517 | Monomer | Variant of GFP | a | [51] |
| PSCFP2 | 400 to 490 | 470 to 511 | Monomer | Variant of GFP | Evrogen | [61] |
| KFP | Increase at 590 | Increase at 600 | Tetramer | Anemonia sulcata | Evrogen | [56,57] |
| Kaede | 508 to 572 | 518 to 580 | Tetramer | Trachyphyllia geoffroyi | MBL Intl | [62] |
| mEosFP | 505 to 569 | 516 to 581 | Monomer | Lobophyllia hemprichii | MBL Intl | [91] |
| Dronpa | Increase at 503 | Increase at 518 | Monomer | Pectinidae spec. | MBL Intl | [58] |
| Dendra2 | 490 to 553 | 507 to 573 | Monomer | Dendronephthya spec. | n/a | [61] |
| KikGR | 507 to 583 | 517 to 593 | Tetramer | Favia favus | MBL Intl | [67] |
| PAmCherry | 404 to 564 | Increase at 595 | Monomer | Variant of mCherry | n/a | [55] |
Available from the non-profit plasmid repository ‘Addgene’ (http://www.addgene.org).
Irreversible photoactivatable proteins
Photoactivatable GFP (PAGFP) is a variant of GFP resulting from a single-residue substitution [51]. The mutation results in a non-fluorescent neutral fluorophore that, upon exposure to short-wavelength light, is irreversibly converted into an anionic form, resulting in a 100-fold increase in green fluorescence. PAGFP has been successfully applied in different organisms. In chick, cell migratory behavior in the hindbrain has been studied by labeling single cells or small groups of cells with PAGFP [52]. In Drosophila, an α-tubulin–PAGFP fusion has been used to study migration of mesodermal cells [53]. One of the few reports of successful PAGFP utilization in developing mammals is its use to examine in vivo protein dynamics during murine postnatal neocortex development [54]. The recent development of proteins such as PAmCherry1 paves the way for two-color photoactivation [55].
Reversible photoactivatable FPs: kindling FP and Dronpa
The tetrameric kindling FP (KFP) was engineered from the natural chromoprotein asulCP. AsulCP converts to a red fluorescent state after exposure to green light. This fluorescent state is unstable and converts back to a non-fluorescent state in the dark, whereas KFP is capable of both reversible and irreversible photoactivation depending on the intensity of the activating light [56,57]. Another reversibly photoswitchable FP, Dronpa, can change reversibly from a green to non-fluorescent form upon exposure to blue light [58]. Neither protein has, as yet, been reported effective in mice. However, Dronpa is used frequently and successfully in frog and zebrafish models [59].
Photoconvertible proteins
To date, several photoconvertible proteins have been isolated or engineered. The most prominent are photoswitchable CFP (PSCFP) and its successor PSCFP2, Kaede, EosFP (named after the goddess of dawn in Greek mythology), Kikume Green-Red (KikGR), Dendra and Dendra2. PSCFP and PSCFP2 are monomers and variants of GFP. These proteins fluoresce in the cyan spectra before photoconversion, but after exposure to UV light, they convert irreversibly to green with an accompanying increase in the green-to-cyan fluorescent ratio [60,61]. Kaede emits green fluorescent light and converts to a red fluorescent state upon irradiation with UV or violet light. This results in a 2000-fold increase of the red–green fluorescent ratio [62,63]. Kaede has been used in live-imaging studies of zebrafish embryogenesis to trace neurons, look at cell movements and label somas to visualize the growth of neurites [64].
mEosFP (a monomeric variant of EosFP), the tetramer KikGR (engineered from the natural protein KikG), Dendra and Dendra2 convert similarly to Kaede. EosFP has been successfully applied in Xenopus to label different germ layers at various developmental stages to follow the morphogenetic movements and formation of embryonic organs [65]. KikGR is the only photoconvertible protein so far that has been used in ES cells and mouse (Figure 5). A transgenic mouse strain that expresses KikGR in a widespread fashion was used to demonstrate that the specification of the embryonic–abembryonic axis in the mouse is independent of the early cell lineage [66]. KikGR is more advantageous for cell labeling and optical marking than Kaede because its photoconversion is more efficient, with both of its fluorescent states being brighter [67]. A direct comparison of the four photomodulatable proteins KikGR, PAGFP, PSCFP2 and Kaede in live imaging of neural crest cell migration in the avian embryo highlighted distinct advantages of each protein for certain developmental applications. Due to their high photoefficiency (i.e. the amount of light required to achieve photoconversion), KikGR and Kaede have been reported as more suitable for monitoring cell migratory behavior, whereas PSCFP2 and PAGFP are more photostable, which allows their use in long-term studies, such as cell-lineage analysis in chick embryos [68].
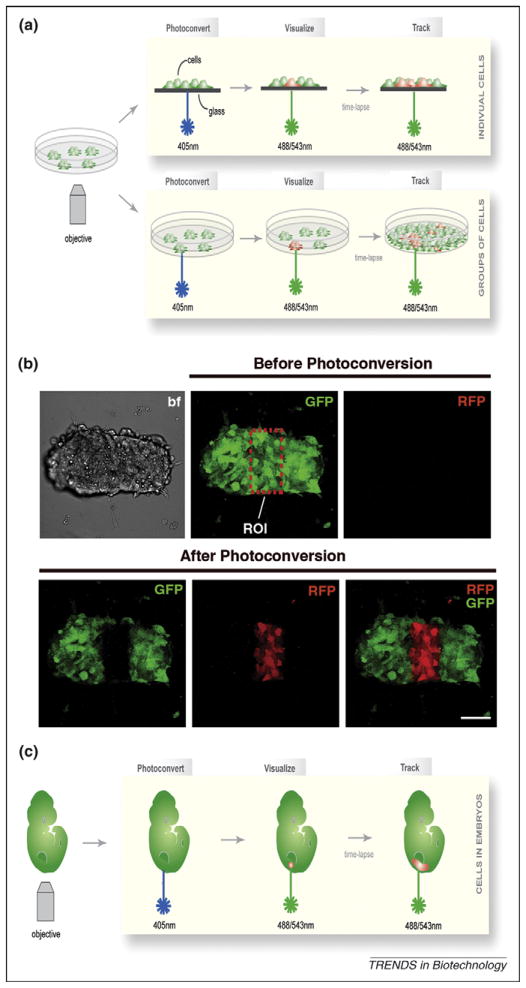
Figure 5.
Photoconversion of ES cells expressing the photomodulatable fluorescent protein KikGR. (a) Schematic representation of the photoconversion of a single cell or a group of cells constitutively expressing KikGR. Before photoconversion, all cells fluoresce green. After exposure to short-wavelength light (405 nm), single cells or a group of cells in the chosen region of interest (ROI) are efficiently photoconverted and emit red fluorescence. (b) 3D images of the photoconversion of a group of cells in an ES cell colony (ROI). Images show the green and red channel before photoconversion (all cells fluoresce green, no red fluorescence is detectable), as well as green, red and a merge of both channels after photoconversion, showing efficient photoconversion of cells in the ROI. (c) Schematic representation of photoconversion in a mouse embryo constitutively expressing KikGR. Photoconverted cells can be tracked over time. A similar approach was used to demonstrate that the specification of the embryonic–abembryonic axis in the mouse is independent of the early cell lineage [66]. Scale bar represents 50 μm.
Dendra and its successor Dendra2 are fairly new engineered PAFPs. Owing to the high photostability of their red fluorescentstates, they should have good potential as alternatives for long-term protein tracking in living cells [61,69].
The variety of PAFP derivatives now available forms a set of powerful tools for tracking and labeling cell populations, single cells and subcellular organelles in vivo in a nonintrusive manner and for following them over time. The PAFPs differ in certain qualities, for example in photoefficiency and photostability, and many are still tetramers. Thus, in their present form they might not be suitable for protein fusions due to steric hindrance that could disrupt protein localization or function. To label subcellular organelles, a monomeric PAFP might be a better choice.
Alternatives to conventional genetically encoded fluorescent proteins
Quantum dots (QDs) are inorganic crystals that can be made to emit light from blue to the infrared part of the spectrum (450–900 nm). Multiple colors can be excited simultaneously with blue-violet light. Their high photostability can be advantageous for tracking proteins for minutes to hours without much loss of fluorescence [70]. In Xenopus, it has been shown that QDs encapsulated in phospholipid micelles can effectively be used to label cells in live embryos with little toxicity and photobleaching compared with that generated by other fluorophores. This makes them worth considering as an alternative for long-term imaging [71]. QDs have been shown not to affect the course of development during early mouse embryogenesis [72]. Applying in utero electroporation and ultrasound-guided in vivo delivery techniques, QDs have been used to label neural stem and progenitor cells, presenting an alternative to FPs for cell-fate mapping or studies of cell migratory behavior during development [72].
Another alternative to FPs are the small molecule probes used in the biarsenical–tetracysteine system. These could circumvent the problem of steric hindrance that afflicts large molecules such as GFP. These probes comprise a small tetracysteine motif fused to a protein of interest and can be covalently bound through a pair of cysteine residues to a membrane-permeable biarsenic dye applied to a living cell. The resulting protein–dye complex emits fluorescence. Administration of an antidote, ethane-dithiol, excludes any interaction of the dye with endogenous cysteine residues. This ensures that the dye remains non-fluorescent until it has bound its specific target. Biar-senical dyes with different spectral properties have been synthesized [73]. Of these, a resorufin-based red probe (ReAsH) and a green probe (FlAsH) have been applied in studies of protein trafficking in the cell. However, the application of these small-molecule probes in mouse might be hampered by the fact that their brightness is not comparable to that of GFP [74].
Applications exploiting widespread FP reporters to gain cell-specific information
Various FP strategies for cell fate mapping and live imaging of embryonic development in mouse have emerged so far and have shown their potential. The following studies are some noteworthy examples that illustrate how FPs, in combination with mouse genetics, can lead to a deeper understanding of mammalian biology.
In a recent report, mice coined Brainbow, in which spectrally distinct FPs were expressed in the neurons of a mouse brain using two different recombinase-mediated strategies, stochastically expressed multiple FPs from a single transgene. The differential expression of multiple copies of FP-encoding constructs in the mouse was shown to label individual neurons in as many as 90 different ‘colors’, and hence it was possible to trace the origin and destination of those neurons [75]. The Brainbow constructs are available from the non-profit plasmid repository Addgene (http://www.addgene.org). The Brainbow strategy can in principle be used to address a variety of morphogenetic questions if placed under different gene-specific promoters for lineage-restricted expression.
As discussed earlier, subcellularly localized FPs are particularly attractive tools for examining cell biology in heterogeneous 3D tissue environments and, in combination with live imaging, promise high-resolution data in four dimensions (i.e. 3D time-lapse) in an organismal context. In a striking application, Sakaue-Sawano and colleagues [76] used genetically encoded fluorescent reporters that mark cell-cycle transitions to visualize the spatio-temporal dynamics of cell-cycle progression. By generating fluorescent probes fused to the oscillating cell-cycle proteins Cdt1 and Geminin (substrates of the two protein complexes APCCDH1 and SCFSkp2, which are E3 ubiquitin ligases that regulate each other through feedback inhibition), cell lines and transgenic mice were generated that constitutively express these probes in cell nuclei. Cdt1, which accumulates in the G1 phase of the cell cycle, was fused to an RFP (monomeric Kusabira Orange 2 [mKO2]), whereas Geminin, which is upregulated in the S, G2 and M phases of the cell cycle, was fused to a GFP (mAG). Hence, cell nuclei in the G1 phase of the cell cycle are labeled in red, whereas nuclei of the S/M and G2 phases of the cell cycle become labeled in green. In combination with time-lapse imaging, these fluorescent probes reveal interplay between cell-cycle dynamics and differentiation, morphogenesis and cell death in tissues [76]. However, although this is a big step towards understanding the in vivo dynamics of cell-cycle progression, probes that distinguish among the S (using cyclin B1, commercially available as a fusion), G2 and M phases would be desirable to develop a more complete picture of cell-cycle dynamics and are undoubtedly currently under development in several laboratories.
Fluorescent probes have also become indispensable tools for mapping the fate of cells. In another visually stunning application that yielded unexpected results, spectrally distinct FP reporters were used to analyze the contributions of clonal progenitors to yolk sac blood islands, the initial sites of development of hematopoietic and endothelial cells [77]. Suprisingly, it was shown that these cell lineages do not arise from a single clonal precursor, as had been proposed previously, but from multiple progenitors [77].
Applications exploiting gene-specific promoters for lineage-restricted FP expression
Expression of FPs under the control of cis-regulatory elements provides additional spatiotemporal information. An H2B–EGFP fusion reporter, which labels nuclei and thus allows the identification of single cells, has been knocked into the mouse PDGFRα locus, which encodes the α-type platelet-derived growth factor receptor. It recapitulated expression of endogenous PDGFRα and has been used as a reporter for the primitive endoderm lineage and its derivatives [78]. Such studies have uncovered novel cell behaviors and have led to new models for lineage development. Moreover, together with live imaging, lineage-specific reporters can help to visualize morphogenetic processes taking place in situ in wild-type embryos and contrast these with those in mutants. For example, an EGFP knock-in into the mouse Noto locus (which encodes a developmental transcription regulator) was used to unravel the morphogenetic processes underlying midline (node and notochord) formation in embryos. Live imaging of the Noto–GFP reporter revealed three previously unrecognized regions of the midline, each formed by distinct morphogenetic cell behaviors [79]. Two studies have used Venus-based FP reporters to look at somite formation using 3D time-lapse imaging. In one, Venus was placed under the control of the promoter from Lunatic fringe (encoding a developmental glycosyltransferase) and used to study the clock that regulates somite morphogenesis, revealing that its oscillations are independent of β-catenin protein levels [80]. In the second study, Venus was placed under the control of the promoter for Mesp2 (encoding a developmental transcription factor) and used to help determine the role of Mesp2 in somite border formation [81].
Imag(in)ing the future
Although the promise of spectrally diverse fluorophores and reporter strains is exciting for the study of mouse embryology, the establishment of mouse strains that can be used for live imaging has been a rate-limiting step in many applications. Impediments to the development of live imaging include: (i) the timeline for generating transgenic or gene targeted mouse strains; (ii) the higher levels of readily detectable fluorescence that are required for live imaging, which are not afforded by all promoters; (iii) the toxicity of many FP fusion proteins when expressed at readily detectable levels; and (iv) the availability of reagents for directing lineage-specific FP expression. However, the strains that have been successfully used for live-imaging experiments hint at the tremendous utility of using live-imaging approaches for understanding basic biological processes in the context of a living mammal. To this end, the versatility of spectrally diverse fluorophores has already given ground-breaking insight into processes of mouse development and will be pivotal in answering fundamental questions through an in-depth understanding of mouse embryonic development.
An alternative way of using fluorescent probes for fate mapping exploits photomodulatable FPs, which offer the advantage of labeling single or cell groups of interest in a tissue; in combination with single or multiphoton confocal microscopy, this approach should prove very powerful for cell fate mapping, as well as for studying cell migration events in the chick embryo [52,68]. It is likely that photomodulatable FPs will also soon see widespread use in live imaging approaches in mice [82,83] (Figure 5).
The process of embryonic development involves rapid changes in cell behavior reflected through changes in cell shape, organization, migration, proliferation and death. Classic studies have relied on the analysis of dead embryos with dynamics inferred from sequentially staged samples. Advances in live imaging are facilitating the visualization of development at single-cell resolution in living embryos. As reflected by the studies discussed, FPs have immense potential for a myriad of live-imaging applications. When applied in the mouse, they synergize the fields of cell and organismal biology. One might predict that future studies exploiting an ever-increasing technicolor array of transgenic and gene-targeted reporters will form the cornerstone of a deeper understanding of mammalian embryology.
Acknowledgments
We apologize to the many authors whose work we have omitted owing to space constraints. Work in our laboratory is supported by the National Institutes of Health (RO1-HD052115). S.N. is supported by a postdoctoral fellowship from the American Heart Association.
References
- 1.Prasher DC, et al. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 2.Chudakov DM, et al. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 2005;23:605–613. doi: 10.1016/j.tibtech.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Shaner NC, et al. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 4.Shaner NC, et al. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 5.Verkhusha VV, Lukyanov KA. The molecular properties and applications of Anthozoa fluorescent proteins and chromoproteins. Nat Biotechnol. 2004;22:289–296. doi: 10.1038/nbt943. [DOI] [PubMed] [Google Scholar]
- 6.Muller-Taubenberger A, Anderson KI. Recent advances using green and red fluorescent protein variants. Appl Microbiol Biotechnol. 2007;77:1–12. doi: 10.1007/s00253-007-1131-5. [DOI] [PubMed] [Google Scholar]
- 7.Chalfie M, et al. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 8.Hadjantonakis AK, et al. Generating green fluorescent mice by germline transmission of green fluorescent ES cells. Mech Dev. 1998;76:79–90. doi: 10.1016/s0925-4773(98)00093-8. [DOI] [PubMed] [Google Scholar]
- 9.Ju B, et al. Faithful expression of green fluorescent protein (GFP) in transgenic zebrafish embryos under control of zebrafish gene promoters. Dev Genet. 1999;25:158–167. doi: 10.1002/(SICI)1520-6408(1999)25:2<158::AID-DVG10>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 10.Kinoshita M, et al. Transgenic medaka enables easy oocytes detection in live fish. Mol Reprod Dev. 2009;76:202–207. doi: 10.1002/mrd.20942. [DOI] [PubMed] [Google Scholar]
- 11.Marsh-Armstrong N, et al. Germ-line transmission of transgenes in Xenopus laevis. Proc Natl Acad Sci U S A. 1999;96:14389–14393. doi: 10.1073/pnas.96.25.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okabe M, et al. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 13.Stewart CN., Jr The utility of green fluorescent protein in transgenic plants. Plant Cell Rep. 2001;20:376–382. doi: 10.1007/s002990100346. [DOI] [PubMed] [Google Scholar]
- 14.Yeh E, et al. Green fluorescent protein as a vital marker and reporter of gene expression in Drosophila. Proc Natl Acad Sci U S A. 1995;92:7036–7040. doi: 10.1073/pnas.92.15.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srinivas S, et al. Expression of green fluorescent protein in the ureteric bud of transgenic mice: a new tool for the analysis of ureteric bud morphogenesis. Dev Genet. 1999;24:241–251. doi: 10.1002/(SICI)1520-6408(1999)24:3/4<241::AID-DVG7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 16.Yoshimizu T, et al. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev Growth Differ. 1999;41:675–684. doi: 10.1046/j.1440-169x.1999.00474.x. [DOI] [PubMed] [Google Scholar]
- 17.Patterson GH, et al. Transport through the Golgi apparatus by rapid partitioning within a two-phase membrane system. Cell. 2008;133:1055–1067. doi: 10.1016/j.cell.2008.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perentes JY, et al. In vivo imaging of extracellular matrix remodeling by tumor-associated fibroblasts. Nat Methods. 2009;6:143–145. doi: 10.1038/nmeth.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang M, et al. Whole-body subcellular multicolor imaging of tumor–host interaction and drug response in real time. Cancer Res. 2007;67:5195–5200. doi: 10.1158/0008-5472.CAN-06-4590. [DOI] [PubMed] [Google Scholar]
- 20.Kanda T, et al. Histone–GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 21.Hadjantonakis AK, Papaioannou VE. Dynamic in vivo imaging and cell tracking using a histone fluorescent protein fusion in mice. BMC Biotechnol. 2004;4:33. doi: 10.1186/1472-6750-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhee JM, et al. In vivo imaging and differential localization of lipid-modified GFP-variant fusions in embryonic stem cells and mice. Genesis. 2006;44:202–218. doi: 10.1002/dvg.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muzumdar MD, et al. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 24.Hadjantonakis AK, et al. Tbx6 regulates left/right patterning in mouse embryos through effects on nodal cilia and perinodal signaling. PLoS One. 2008;3:e2511. doi: 10.1371/journal.pone.0002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trichas G, et al. Use of the viral 2A peptide for bicistronic expression in transgenic mice. BMC Biol. 2008;6:40. doi: 10.1186/1741-7007-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nowotschin SE, et al. Dual transgene strategy for live visualization of chromatin and plasma membrane dynamics in murine embryonic stem cells and embryonic tissues. Genesis. doi: 10.1002/dvg.20500. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bokman SH, Ward WW. Renaturation of Aequorea green-fluorescent protein. Biochem Biophys Res Commun. 1981;101:1372–1380. doi: 10.1016/0006-291x(81)91599-0. [DOI] [PubMed] [Google Scholar]
- 28.Ward WW, Bokman SH. Reversible denaturation of Aequorea green-fluorescent protein: physical separation and characterization of the renatured protein. Biochemistry. 1982;21:4535–4540. doi: 10.1021/bi00262a003. [DOI] [PubMed] [Google Scholar]
- 29.Cubitt AB, et al. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 30.Heim R, et al. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 31.Karasawa S, et al. A green-emitting fluorescent protein from Galaxeidae coral and its monomeric version for use in fluorescent labeling. J Biol Chem. 2003;278:34167–34171. doi: 10.1074/jbc.M304063200. [DOI] [PubMed] [Google Scholar]
- 32.Heim R, et al. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci U S A. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 34.Rizzuto R, et al. Double labelling of subcellular structures with organelle-targeted GFP mutants in vivo. Curr Biol. 1996;6:183–188. doi: 10.1016/s0960-9822(02)00451-7. [DOI] [PubMed] [Google Scholar]
- 35.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 36.Lippincott-Schwartz J, et al. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- 37.Rizzo MA, et al. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 38.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 39.Matz MV, et al. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 40.Campbell RE, et al. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long JZ, et al. Genetic and spectrally distinct in vivo imaging: embryonic stem cells and mice with widespread expression of a monomeric red fluorescent protein. BMC Biotechnol. 2005;5:20. doi: 10.1186/1472-6750-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luche H, et al. Faithful activation of an extra-bright red fluorescent protein in ‘knock-in’ Cre-reporter mice ideally suited for lineage tracing studies. Eur J Immunol. 2007;37:43–53. doi: 10.1002/eji.200636745. [DOI] [PubMed] [Google Scholar]
- 43.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 44.Egli D, et al. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature. 2007;447:679–685. doi: 10.1038/nature05879. [DOI] [PubMed] [Google Scholar]
- 45.Provost E, et al. Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis. 2007;45:625–629. doi: 10.1002/dvg.20338. [DOI] [PubMed] [Google Scholar]
- 46.Wang L, et al. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc Natl Acad Sci U S A. 2004;101:16745–16749. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shkrob MA, et al. Far-red fluorescent proteins evolved from a blue chromoprotein from Actinia equina. Biochem J. 2005;392:649–654. doi: 10.1042/BJ20051314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shcherbo D, et al. Bright far-red fluorescent protein for whole-body imaging. Nat Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 49.Lukyanov KA, et al. Innovation: photoactivatable fluorescent proteins. Nat Rev Mol Cell Biol. 2005;6:885–891. doi: 10.1038/nrm1741. [DOI] [PubMed] [Google Scholar]
- 50.Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- 51.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 52.Stark DA, Kulesa PM. Photoactivatable green fluorescent protein as a single-cell marker in living embryos. Dev Dyn. 2005;233:983–992. doi: 10.1002/dvdy.20385. [DOI] [PubMed] [Google Scholar]
- 53.Murray MJ, Saint R. Photoactivatable GFP resolves Drosophila mesoderm migration behaviour. Development. 2007;134:3975–3983. doi: 10.1242/dev.005389. [DOI] [PubMed] [Google Scholar]
- 54.Gray NW, et al. Rapid redistribution of synaptic PSD-95 in the neocortex in vivo. PLoS Biol. 2006;4:e370. doi: 10.1371/journal.pbio.0040370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subach FV, et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods. 2009;6:153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chudakov DM, et al. Kindling fluorescent proteins for precise in vivo photolabeling. Nat Biotechnol. 2003;21:191–194. doi: 10.1038/nbt778. [DOI] [PubMed] [Google Scholar]
- 57.Chudakov DM, et al. Chromophore environment provides clue to ‘kindling fluorescent protein’ riddle. J Biol Chem. 2003;278:7215–7219. doi: 10.1074/jbc.M211988200. [DOI] [PubMed] [Google Scholar]
- 58.Ando R, et al. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 59.Marriott G, et al. Optical lock-in detection imaging microscopy for contrast-enhanced imaging in living cells. Proc Natl Acad Sci U S A. 2008;105:17789–17794. doi: 10.1073/pnas.0808882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chudakov DM, et al. Photoswitchable cyan fluorescent protein for protein tracking. Nat Biotechnol. 2004;22:1435–1439. doi: 10.1038/nbt1025. [DOI] [PubMed] [Google Scholar]
- 61.Chudakov DM, et al. Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat Protocols. 2007;2:2024–2032. doi: 10.1038/nprot.2007.291. [DOI] [PubMed] [Google Scholar]
- 62.Ando R, et al. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mizuno H, et al. Photo-induced peptide cleavage in the green-tored conversion of a fluorescent protein. Mol Cell. 2003;12:1051–1058. doi: 10.1016/s1097-2765(03)00393-9. [DOI] [PubMed] [Google Scholar]
- 64.Hatta K, et al. Cell tracking using a photoconvertible fluorescent protein. Nat Protocols. 2006;1:960–967. doi: 10.1038/nprot.2006.96. [DOI] [PubMed] [Google Scholar]
- 65.Wacker SA, et al. A green to red photoconvertible protein as an analyzing tool for early vertebrate development. Dev Dyn. 2007;236:473–480. doi: 10.1002/dvdy.20955. [DOI] [PubMed] [Google Scholar]
- 66.Kurotaki Y, et al. Blastocyst axis is specified independently of early cell lineage but aligns with the ZP shape. Science. 2007;316:719–723. doi: 10.1126/science.1138591. [DOI] [PubMed] [Google Scholar]
- 67.Tsutsui H, et al. Semi-rational engineering of a coral fluorescent protein into an efficient highlighter. EMBO Rep. 2005;6:233–238. doi: 10.1038/sj.embor.7400361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stark DA, Kulesa PM. An in vivo comparison of photoactivatable fluorescent proteins in an avian embryo model. Dev Dyn. 2007;236:1583–1594. doi: 10.1002/dvdy.21174. [DOI] [PubMed] [Google Scholar]
- 69.Gurskaya NG, et al. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 70.Bruchez MP. Turning all the lights on: quantum dots in cellular assays. Curr Opin Chem Biol. 2005;9:533–537. doi: 10.1016/j.cbpa.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 71.Dubertret B, et al. In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science. 2002;298:1759–1762. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- 72.Slotkin JR, et al. In vivo quantum dot labeling of mammalian stem and progenitor cells. Dev Dyn. 2007;236:3393–3401. doi: 10.1002/dvdy.21235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adams SR, et al. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J Am Chem Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 74.Zhang J, et al. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 75.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 76.Sakaue-Sawano A, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 77.Ueno H, Weissman IL. Clonal analysis of mouse development reveals a polyclonal origin for yolk sac blood islands. Dev Cell. 2006;11:519–533. doi: 10.1016/j.devcel.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 78.Plusa B, et al. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development. 2008;135:3081–3091. doi: 10.1242/dev.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamanaka Y, et al. Live imaging and genetic analysis of mouse notochord formation reveals regional morphogenetic mechanisms. Dev Cell. 2007;13:884–896. doi: 10.1016/j.devcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 80.Aulehla A, et al. A β-catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nat Cell Biol. 2008;10:186–193. doi: 10.1038/ncb1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morimoto M, et al. The Mesp2 transcription factor establishes segmental borders by suppressing Notch activity. Nature. 2005;435:354–359. doi: 10.1038/nature03591. [DOI] [PubMed] [Google Scholar]
- 82.Tomura M, et al. Monitoring cellular movement in vivo with photoconvertible fluorescence protein ‘Kaede’ transgenic mice. Proc Natl Acad Sci U S A. 2008;105:10871–10876. doi: 10.1073/pnas.0802278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shigematsu Y, et al. Novel embryonic stem cells expressing tdKaede protein photoconvertible from green to red fluorescence. Int J Mol Med. 2007;20:439–444. [PubMed] [Google Scholar]
- 84.Ai HW, et al. Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry. 2007;46:5904–5910. doi: 10.1021/bi700199g. [DOI] [PubMed] [Google Scholar]
- 85.Xia NS, et al. Bioluminescence of Aequorea macrodactyla, a common jellyfish species in the East China Sea. Mar Biotechnol (NY) 2002;4:155–162. doi: 10.1007/s10126-001-0081-7. [DOI] [PubMed] [Google Scholar]
- 86.Shagin DA, et al. GFP-like proteins as ubiquitous metazoan superfamily: evolution of functional features and structural complexity. Mol Biol Evol. 2004;21:841–850. doi: 10.1093/molbev/msh079. [DOI] [PubMed] [Google Scholar]
- 87.Cubitt AB, et al. Understanding structure-function relationships in the Aequorea victoria green fluorescent protein. Methods Cell Biol. 1999;58:19–30. doi: 10.1016/s0091-679x(08)61946-9. [DOI] [PubMed] [Google Scholar]
- 88.Griesbeck O, et al. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J Biol Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- 89.Karasawa S, et al. Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem J. 2004;381:307–312. doi: 10.1042/BJ20040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Merzlyak EM, et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 91.Wiedenmann J, et al. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci U S A. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huisken J, et al. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 93.Keller PJ, et al. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]