Abstract
Cellular responses to carcinogens are typically studied in transformed cell lines, which do not reflect the physiological status of normal tissues. To address this question, we have characterized the transcriptional program and cellular responses of human lung WI-38 fibroblasts upon exposure to the ultimate carcinogen benzo[a]pyrene diol epoxide (BPDE). In contrast to observations in cell lines, we find that BPDE treatment induces a strong inflammatory response in these normal fibroblasts. Whole-genome microarrays show induction of numerous inflammatory factors, including genes that encode interleukins (ILs), growth factors and enzymes related to prostaglandin synthesis and signaling. Real-time reverse transcription–polymerase chain reaction and enzyme-linked immunosorbent assay (ELISA) revealed a time- and dose-dependent-induced expression and production of cyclooxygenase 2, prostglandin E2 and IL1B, IL6 and IL8. In parallel, cell cycle progression and DNA repair processes were repressed, but DNA damage signaling was increased via p53-Ser15 phosphorylation and induced expression levels of GADD45A, CDKN1A, BTG2 and SESN1. Network analysis suggested that activator protein 1 transcription factors may link the cell cycle response and DNA damage signaling with the inflammatory stress–response in these cells. We confirmed this hypothesis by showing that p53-dependent signaling through c-jun N-terminal kinase (JNK) led to increased cJun-Ser63 phosphorylation and that inhibition of JNK-mediated cJun activation using p53- or JNK-specific inhibitors significantly reduced IL gene expression and subsequent production of IL8. This is the first demonstration that a strong inflammatory response is triggered in normal fibroblasts by BPDE and that this occurs through coordinated regulation with other cellular processes.
Introduction
Cigarette smoking is the primary cause of lung cancer and the lung inflammatory diseases emphysema and chronic bronchitis (1,2). Polycyclic aromatic hydrocarbons (PAHs) are among the many chemical components found in cigarette smoke that induce tumor formation and inflammation in experimental animals, supporting the notion that they play a significant role as etiological agents in human disease (3). In fact, the International Agency for Research on Cancer reports that ∼15% of the carcinogenic compounds found in cigarette smoke are PAHs (1); hence, the PAHs most probably contribute to the etiology of cancers associated with the well-known correlation between tobacco smoke and lung cancer (4).
PAHs are readily metabolized to water-soluble compounds in a process that produces carcinogenic diol epoxide (DE) intermediates that react with nucleic acids, producing covalently modified bases in the genome (5). In the case of high levels of PAH–DNA lesions, p53-associated responses block the cell cycle, providing an opportunity for the DNA repair machinery to restore genome integrity or direct the cell to undergo apoptosis (6–9). PAHs also activate production of inflammatory mediators through an aryl hydrocarbon receptor-dependent pathway, either directly or by downstream activation of phosphorylation-mediated signaling through mitogen-activated protein kinases that further stimulate stress-responsive transcription factors (10–12). Frequent exposure to agents that damage DNA can lead to chronic inflammation that has been linked to the progression of several pathological conditions, including cardiovascular and pulmonary diseases and cancer (13,14).
Gene profiling studies on PAH DE in different cell types, including normal epithelial cells and carcinoma cell lines, have been reported to show alterations in the expression of genes involved in the cell cycle, apoptosis, DNA repair and p53 pathways (15–22). However, the genome-wide response of lung fibroblasts to PAHs has not been studied. Lung fibroblasts confer structural support to the lung's connective tissue and play a role in stimulating and amplifying inflammatory signals, including those that arise as responses to cigarette smoke (23). To examine this, we exposed normal human WI-38 lung fibroblasts to three different concentrations of benzo[a]pyrene diol epoxide (BPDE). Using whole-genome oligonucleotide microarrays to measure changes in gene expression, we found strong induction of genes involved in inflammatory response and parallel alterations in processes related to cell cycle regulation and DNA repair. To identify the essential signaling events that mediate these responses, we performed directed assays of key members of multiple pathways to quantify messenger RNA (mRNA) and protein levels and to assess their activation status. These experiments revealed that the mechanism underlying the response of normal lung fibroblasts to BPDE involves the coordinated regulation of multiple signaling pathways.
Material and methods
Chemicals and reagents
Racemic anti-BPDE [(±)-7r,8t-dihydroxy-9t,10t-epoxy-7,8,9,10-tetrahydro benzo[a]pyrene] was obtained from the National Cancer Institute Chemical Carcinogen Reference Standard Repository (Kansas City, MO). Stock solutions of BPDE were prepared in dimethyl sulfoxide (DMSO) and stored at −20°C. Protease Inhibitor Cocktail Set I, pifithrin-α (PFT-α) and c-jun N-terminal kinase (JNK) inhibitor VIII (JNKi) were from Calbiochem (Gibbstown, NJ). The antibodies against human p53 (DO-1) were from Santa Cruz Biotechnology (Santa Cruz, CA). The antibodies against phospho-p53 (Ser15), phospho-p44/42 (Thr202/Tyr204), phospho-p38 (Thr180/Tyr182), phospho-stress-activated protein kinase/JNK (Thr183/Tyr185), stress-activated protein kinase/JNK (56G8), phospho-c-Jun (Ser63) and c-Jun (60A8) were from Cell Signaling Technology (Danvers, MA). The antibodies against prostaglandin H2 and β-tubulin were from Cayman Chemicals (Ann Arbor, MI) and NeoMarkers (Fremont, CA), respectively. Additional chemicals and reagents were from Sigma–Aldrich (St Louis, MO) and Fisher Scientific (Fair Lawn, NJ).
Cell culture
Normal human diploid WI-38 lung fibroblasts, CCL-75™, were obtained from American Type Culture Collection (Manassas, VA) and grown in Eagle’s minimum essential medium supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and 1% penicillin/streptomycin (Invitrogen, Carlsbad CA). WI-38 cells between passages 7 to 12 were used in all experiments to ensure proper cell characteristics as determined by American Type Culture Collection. Prior to BPDE exposure, 106 WI-38 cells were plated onto 10 cm dishes and incubated at 37°C for 48 h. The cells were treated with BPDE as described previously, with some modifications (24). The cells were incubated with BPDE (added in 10 μl DMSO) for 20 min; 0.1% DMSO alone was added to cells to act as a control. The exposure medium was then replaced with fresh medium, and at selected time points, the medium was removed, and cells were washed with phosphate-buffered saline and harvested.
Cell viability
WI-38 cells were exposed to BPDE as described above. Cell viability was determined using a MTS assay in 96-well plates according to manufacturer's instruction (CellTiter 96 AQueous One Solution Cell Proliferation Assay; Promega, Madison, WI). Senescence status was determined by staining for senescence-associated β-galactosidase as described in ref. 25.
Western blot analysis
Cells were lysed in ice-cold RIPA buffer (50 mM Tris–HCl, 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate and 0.5% Na-deoxycholate) with 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, 1 mM NaF and 10 μl/ml of Protease Inhibitor Cocktail Set I. The lysate was boiled for 5 min in Laemmli buffer, and proteins were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis followed by immunoblotting. Proteins were visualized using ECL Plus™ reagents (GE Healthcare, Piscataway, NJ).
ELISA
Cells seeded in six-well plates (2 × 105 cells per well) were exposed to BPDE as described above. Extracellular concentrations were measured using a human IL8 (RayBiotech, Norcross, GA) or a human prostglandin E2 (PGE2) ELISA kit (Cayman Chemicals, Ann Arbor, MI). Absolute amounts were calculated using recombinant human IL8 and PGE2 as standards and normalized to number of cells.
RNA purification and real-time reverse transcription–polymerase chain reaction
Total cellular RNA was prepared using the RNeasy Mini Kit (QIAGEN, Valencia, CA) and further treated with TURBO DNA-free™ (Ambion, Austin, TX). Complementary DNA (cDNA) was generated using 1 μg of total RNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) according to protocol. Subsequently, quantification of gene expression was performed in duplicates using iQ SYBR Green Supermix (Bio-Rad) with detection on an iCycler MyiQ (Bio-Rad). The reaction cycles used were 95°C for 10 min and then 40 cycles at 95°C for 15 s and 60°C for 1 min followed by melt curve analysis. The primer sequences for ILs 1B, 6, 8, cyclooxygenase 2 (COX2) and GAPDH were taken from (26). Gene expression was further studied using a 96-well DNA damage signaling RT2Profiler™ PCR array together with the associated ReactionReady™ First Strand cDNA synthesis kit and real-time reverse transcription (RT2) Real-Time™ SYBR Green PCR master mix (all from SAbiosciences, Frederick, MD). Relative gene expression quantification was based on the comparative threshold cycle method (2−ΔΔCt) (27) with normalization of the raw data to the included housekeeping gene.
Microarray hybridization
Experimental procedures were performed according to the GeneChip® Expression Analysis Technical Manual using the One-Cycle Target Labeling and Control Reagents kit (Affymetrix, Santa Clara, CA). Total RNA (5 μg) was reverse transcribed into cDNA, which subsequently was used for synthesis of biotin-labeled complementary RNA. The Human Genome U133 Plus 2.0 microarray was then hybridized with 15 μg fragmented complementary RNA, followed by washing and staining in a fluidics station (all from Affymetrix) according to standard protocols. Hybridization of total RNA from control and BPDE-treated cells was performed in triplicate.
Analysis of microarray data
The microarrays were scanned using a GeneChip® Scanner 3000 (Affymetrix) and subsequent image analysis was performed using Affymetrix GCOS software. The resulting .cel files were normalized with the robust multichip average method (28,29). Probe sets were filtered (30) and subsequently analyzed for changes in expression using significance analysis of microarrays (SAM) (31). The raw data discussed in this publication have been deposited into National Center for Biotechnology Information Gene Expression Omnibus (Series GSE19510). Gene Ontology (GO) overrepresentation was analyzed by using GenMAPP/MAPPFinder version 2.1 (32). All genes identified as significantly affected by SAM were included in the analysis using a ≥2-fold change as analysis criteria. GO terms with a Z-score > 2 were assumed to be significantly affected by the treatment.
Network analysis
A human protein–protein interaction network was obtained from the Human Protein Reference Database, release 7. Proteins whose gene expression significantly changed according to microarray analysis and were annotated with significantly enriched GO Biological Process terms (specified in supplementary Table S5, available at Carcinogenesis Online) were selected from the network. This set was supplemented by genes/proteins identified as being involved in the response to BPDE in the current study (supplementary Table S6 is available at Carcinogenesis Online), resulting in 153 proteins. This core network was supplemented with direct interactors (local 1-hop neighbors) from Human Protein Reference Database, resulting in 1339 proteins with 7778 interactions. In Figure 6, the network was reduced to only include significantly changed genes together with the supplemented nodes. To develop more specific response networks, three separate cell cycle, DNA repair and stress–response subnetworks with 121, 89 and 147 proteins, respectively, were created (found in supplementary Figures S4–S6, available at Carcinogenesis Online).
Fig. 6.
Composite network presentation of the response to BPDE in normal fibroblasts. A PPI network was created with genes whose expression changed significantly and were annotated with functions most affected by BPDE exposure, namely those related to the cell cycle, DNA repair and stress–response as described in Materials and Methods. Nodes (genes) colored green were downregulated by one or more doses of BPDE; red were upregulated and yellow displayed complex regulation. Lighter colors did not fulfill the 2-fold change criteria but were identified as significant by SAM. Gray diamond-shaped nodes were identified as being involved in signaling by western blot analysis.
Results
Normal human lung fibroblasts exposed to BPDE show significant changes in their global gene expression profile
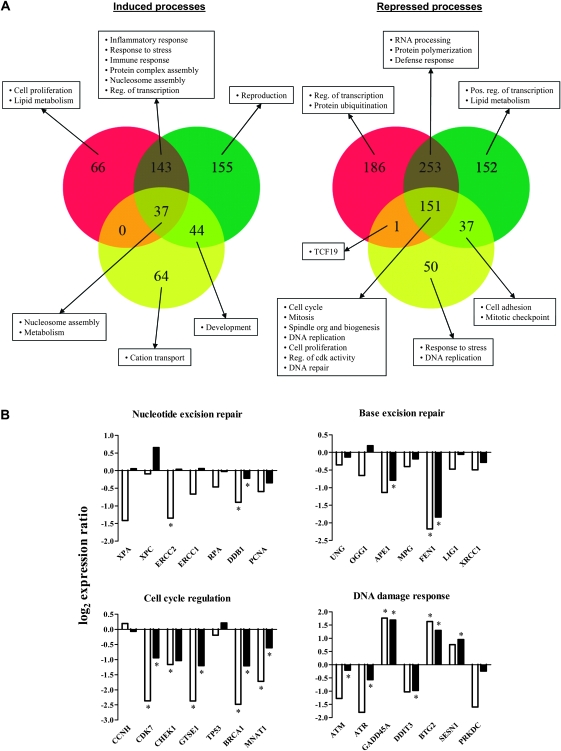
To test the effects of a PAH DE on lung fibroblasts, we used oligonucleotide microarrays representing >38 000 well-characterized human genes to measure gene expression in WI-38 cells exposed to 0.1, 0.5 or 1 μM BPDE for 24 h. We note that exposure to 1 μM BPDE for 24 or 48 h led to a 20% decrease in viability, whereas lower doses of BPDE (0.1 and 0.01 μM) were minimally cytotoxic with a <10% decrease in viability (supplementary Figure S1 is available at Carcinogenesis Online). Detection call filtering followed by SAM identified 1332 genes whose expression was significantly changed following exposure to at least one concentration of BPDE when compared with control cells (supplementary Table S1 is available at Carcinogenesis Online). A significant change in gene expression was defined as ≥2-fold change in expression with <10% false discovery rate (FDR). Exposure to 0.1, 0.5 or 1 μM BPDE resulted in 384, 972 and 837 differentially expressed genes, respectively (Figure 1A). Of these, 37 genes were upregulated and 151 genes were downregulated in response to all doses of BPDE. Hierarchical clustering revealed a significant similarity in responses induced by the two higher doses when compared with the lower dose of 0.1 μM BPDE (supplementary Figure S2, is available at Carcinogenesis Online).
Fig. 1.
Global changes in gene expression after BPDE treatment. (A) Venn diagram illustration of the number of genes identified as either significantly induced (left) or repressed (right) (≥2-fold and <10% false discovery rate) in WI-38 cells following exposure to BPDE. The yellow, green and red circles correspond to 0.1, 0.5 or 1.0 μM BPDE exposure, respectively. The overlapping regions contain genes that are modulated at more than one dose. Text boxes show examples of common and specific processes that are significantly affected by the different BPDE concentrations as determined by GO analysis (Z-score > 2). (B) Comparison of gene expression changes related to DNA damage signaling and repair obtained using microarray analysis (black bars) and RT2–PCR (white bars) 24 h after treatment with 1 μM BPDE. *P < 0.05 (RT2–PCR) or FDR < 10% (microarray) compared with vehicle control by Student's t-test or SAM, respectively.
BPDE induces changes in gene expression related to cell cycle regulation, DNA repair and cell–cell signaling
To elucidate the fundamental cellular processes affected in WI-38 cells following exposure to BPDE, we examined functional annotations of genes with significant changes in expression using GO Biological Process terms (written in italic). Downregulated genes were mainly involved in mitosis, cell cycle checkpoints, spindle organization and biogenesis, DNA replication and DNA repair, among others. Upregulated genes were involved in nucleosome assembly, cell proliferation and extracellular processes such as cell–cell signaling and stress, inflammatory and immune responses. Both negative and positive regulators of proliferation were among the induced genes involved in cell proliferation; induced expression of cytokines and growth factors were mainly responsible for the effect on cell–cell signaling and the stress–responses and the effect on nucleosome assembly was principally a function of upregulation of histone genes. A complete accounting of significantly affected GO terms and associated genes are provided in supplementary Figure S3 and Table S2 (available at Carcinogenesis Online).
To identify dose-dependent effects of BPDE on gene expression, we analyzed GO functional annotations for gene clusters in Figure 1A. Nucleosome assembly and metabolism were positively affected by all three BPDE concentrations, but stress, inflammatory and immune response processes were only upregulated following exposure to 0.5 or 1 μM BPDE. All three BPDE doses led to downregulation of processes related to the cell cycle and intracellular organization (i.e. cytoskeleton and chromosome organization). These results indicate that the threshold dose of BPDE required to produce an inflammatory response is higher than that required to shut down cell cycle related processes in lung fibroblasts.
DNA repair genes show time- and dose-dependent downregulation in response to BPDE
While microarray results showed that genes involved in DNA damage repair were generally downregulated, WI-38 cells are capable of repairing DNA adducts formed by BPDE in a biphasic manner, with ∼50% of the adducts removed within 6 h following exposure (K.Dreij, N.E.Geacintov and D.A.Scicchitano, unpublished results). This is in agreement with earlier studies looking at repair of DNA adducts derived from BPDE in cellular systems (24,33). Two genes directly involved in DNA repair were induced by BPDE: RRM2B (encoding ribonucleotide reductase RIR2B) and POLG2 (encoding the mitochondrial DNA polymerase accessory subunit DPOG2). In addition, exposure to 0.5 or 1 μM BPDE significantly induced genes whose products are involved in suppressing cell proliferation and the cell cycle in response to DNA damage, including BTG2, SESN1, CDKN1A and GADD45A.
To confirm and extend our microarray results, we used a real-time reverse transcription (RT2)–polymerase chain reaction (PCR) array covering 84 genes involved in DNA damage signaling and repair to study the effects of BPDE on their expression in more detail. Since the two higher doses induced similar overall expression responses based on microarray data, we extended the range of exposure to 0.01, 0.1 and 1 μM BPDE for 6, 24 and 48 h. The results revealed both time- and dose-dependent downregulation of expression, with 22 genes significantly downregulated (≥2-fold and P < 0.05; supplementary Table S3 is available at Carcinogenesis Online). Consistent with microarray results, only BTG2 and GADD45A were upregulated in response to 1 μM BPDE; these showed increased expression by 6 h and sustained levels of induction up to 48 h posttreatment. (Lower doses did not affect their expression levels significantly.) Although the RT2–PCR showed a more pronounced effect the overall results were in good agreement with the microarray analysis (Figure 1B).
Factors involved in prostaglandin and IL signaling exhibit time- and dose-dependent induction of mRNA and protein levels
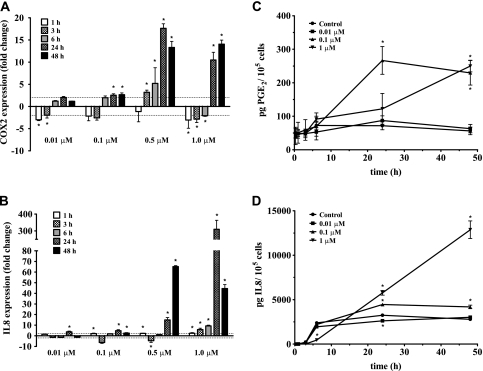
BPDE's most striking effect on gene expression in WI-38 cells was the upregulation of genes involved in cell–cell signaling, the immune response and the inflammatory response. Among the induced genes were ILs 6, 8 and 11; matrix metallopeptidases 1, 3 and 10; growth factors BMP2, FGF9 and GDF15 and genes involved in prostaglandin synthesis and its regulation, COX2, PTGES and EDN1. Due to the physiological importance of IL8 and the COX2-driven prostaglandins (PGE2) as regulators of inflammatory signaling, we performed a more in-depth analysis of the time–response and dose–response of IL8 and COX2 using both RT2–PCR and ELISA.
In RT2–PCR assays, COX2 mRNA was repressed during the first 6 h after BPDE exposure, followed by induced expression at 24 and 48 h; however, only the two highest BPDE doses led to extensive upregulation, with a maximal 15-fold increase in mRNA at 24 and 48 h (Figure 2A). This result was consistent with the microarray results, which likewise showed a significant increase in COX2 mRNA only at the two higher doses (3.9-fold and 12.8-fold, respectively). PGE2 levels measured by ELISA also increased over time, consistent with microarray and RT2–PCR results. The highest concentration of extracellular PGE2 was detected from cells exposed to 0.1 μM BPDE at 24 h (3.7-fold increase, P < 0.05; Figure 2C). Exposure to the highest dose of BPDE (1 μM) led to a slower response that peaked after 48 h (4.4-fold increase, P < 0.05). This elevation in extracellular PGE2 was not due to an increase in the amount of enzyme responsible for its synthesis since western blot analysis using antibodies against prostaglandin G/H synthase 2 (prostaglandin H2, encoded by COX2) showed no increase in protein levels (data not shown).
Fig. 2.
Time–response and dose–response of mRNA and extracellular levels of COX2/PGE2 and IL8 in WI-38 cells. (A and B) Fold changes in expression of COX2 and IL8 determined by RT2–PCR, respectively. The dashed line represents a 2-fold change. Results are represented as mean ± SD; experiments were performed at least twice in duplicates. *P < 0.05 and ≥2-fold change compared with vehicle control by Student's t-test. (C and D) Extracellular levels of PGE2 and IL8 determined by ELISA, respectively. Results are represented as mean ± SD, n = 3. *P < 0.05 compared with vehicle control by Student's t-test.
In the case of IL8, RT2–PCR assays showed a soaring 309-fold elevation in mRNA levels at 24 h followed by a 44.5-fold elevation at 48 h after exposure to 1 μM BPDE when compared with DMSO controls (all P < 0.05; Figure 2B). Microarray results displayed a parallel but less dramatic pattern, with significantly increased expression at 24 h of 11.2-fold and 41.9-fold in response to 0.5 and 1 μM BPDE, respectively. Extracellular IL8 protein levels were also significantly elevated in response to BPDE treatment as measured by ELISA (Figure 2D). Exposure to 1 μM BPDE produced the largest effect on IL8 excretion, displaying maximum levels of ∼5-fold over control (P < 0.05) after 48 h.
A secretory phenotype consisting of increased signaling of inflammatory mediators has been associated with senescence in response to genotoxic stress in normal human fibroblasts (34). To test whether the increased signaling of inflammatory mediators in this study was coupled with senescence, exposed cells were stained for senescence-associated β-galactosidase (25). The highest dose of BPDE (1 μM) induced senescence in a very low number of cells (≤4%), indicating that the observed increase in signaling of inflammatory mediators was not related to senescence (supplementary Table S4 is available at Carcinogenesis Online).
BPDE induces p53-dependent signaling through JNK with subsequent activation of cJun
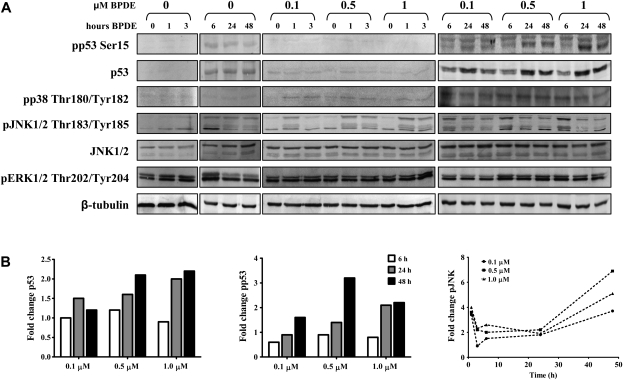
To identify the regulatory pathways responsible for gene expression changes in WI-38 cells following BPDE exposure, we measured levels and activation status of p53 and mitogen-activated protein kinase proteins, including phosphorylation of p53 at Ser15, which is mediated by ataxia telangiectasia mutated and ataxia telangiectasia and Rad3 related in response to DNA damage and promotes activation of p53-dependent pathways (35,36). As expected, we observed an increase in both p53 protein levels and phosphorylation of Ser15 after 24 and 48 h exposure to 0.5 and 1 μM BPDE, but not at 6 h (Figure 3A and B). The lower dose of 0.1 μM BPDE resulted in increased levels of p53 protein, but no detectable Ser15 phosphorylation, after 24 h. JNK protein levels remained constant after exposing WI-38 cells to BPDE, but increased JNK Thr183/Tyr185 phosphorylation was detected beginning 1 h after treatment (Figure 3A) and was observed up to 24 and 48 h after exposure to 0.5 and 1 μM BPDE (Figure 3B). Phosphorylation status of ERK and p38 was basically unaffected (Figure 3A).
Fig. 3.
Western blot analysis of signaling through p53 and mitogen-activated protein kinases in response to BPDE treatment. (A) WI-38 cells were exposed to up to 1 μM BPDE for up to 48 h (as indicated) and total levels and phosphorylation status of the indicated proteins were measured using specific antibodies. β-Tubulin was used as loading control. (B) Densitometric analysis of the p53, phosphorylated p53 and phosphorylated JNK bands shown in (A), respectively. Bands were normalized to the loading control and compared with corresponding vehicle control.
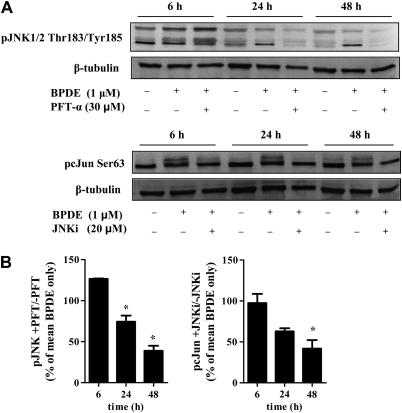
We further investigated whether the signaling through JNK was p53-dependent and would therefore result in downstream activation of the activator protein 1 (AP-1) transcription factor cJun. Indeed, pretreatment of WI-38 cells with PFT-α and JNKi—which inhibit p53 and JNK (37–40)—strongly reduced signaling through JNK and cJun (Ser63), respectively (Figure 4A). Based on densitometric analysis, the inhibitors did not affect the extent of JNK and cJun phosphorylation at 6 h. However, signaling was significantly reduced at the later time points, displaying >2-fold reduction of phosphorylation for both JNK and cJun at 48 h (39 ± 6% and 42 ± 10%, respectively, P < 0.05; Figure 4B).
Fig. 4.
Signaling through JNK was p53 dependent and resulted in downstream activation of cJun. (A) Western blot analysis of inhibition of JNK and cJun phosphorylation by PFT-α and JNKi, respectively, using specific antibodies. β-Tubulin was used as loading control. (B) Densitometric analysis of the response to BPDE in the presence of PFT-α or JNKi from three independent experiments, mean ± SD. *P < 0.05 compared with BPDE only treatment by Student's t-test.
Increased IL expression and signaling are mediated through a p53 and JNK signaling pathway
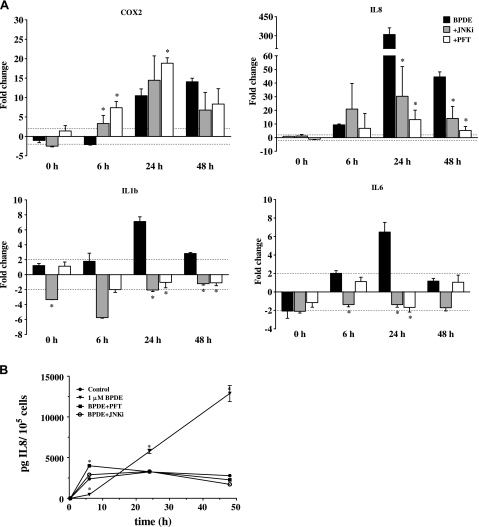
We next studied the effect of interrupted p53 and JNK signaling on COX2 and ILs 1B, 6 and 8 signaling using RT2–PCR and ELISA. BPDE induction of COX2 mRNA remained the same or increased when cells were pretreated with JNKi and PFT-α (Figure 5A). In contrast, inhibition of p53 and JNK significantly reduced the induction of IL mRNA levels in response to BPDE treatment. While IL8 expression still was induced in WI-38 cells treated with inhibitor plus BPDE, mRNA levels were significantly lower compared with cells exposed to BPDE alone at 24 and 48 h (P < 0.05; Figure 5A). Blocking the signaling through p53 and JNK significantly reduced mRNA levels of IL1B at 24 and 48 h (P < 0.05), similarly the expression of IL6 was significantly reduced by JNKi at 6 h and by both inhibitors at 24 h (P < 0.05).
Fig. 5.
Effect of p53 and JNK inhibition on the expression and signaling of inflammatory mediators. (A) Effects of PFT-α and JNKi on COX2, IL8, IL1B and IL6 expression in response to BPDE measured by RT2–PCR. WI-38 cells were pretreated with 20 μM JNKi (gray bars) or 30 μM PFT-α (white bars) for 2 h followed by exposure to 1 μM BPDE. The dashed line represents a 2-fold change. Results are represented as mean ± SD; experiments were performed at least twice in duplicates. *P < 0.05 compared with BPDE only treatment by Student's t-test. (B) Effect of PFT-α and JNKi on extracellular levels of IL8 in response to BPDE measured by ELISA. Results are represented as mean ± SD, n = 3. *P < 0.05 compared with vehicle control by Student's t-test.
Pretreatment with inhibitor also resulted in a significant reduction of extracellular IL protein levels, further confirming their negative effect on IL signaling: in the presence of either PFT-α or JNKi, extracellular IL8 was reduced to the same level as controls at 24 and 48 h (Figure 5B). These results clearly show that signaling through p53 and JNK plays a major role in the induction of IL expression and signaling in response to BPDE treatment of normal lung fibroblasts.
Discussion
Fibroblasts synthesize and maintain stromal support for most renewable epithelial tissue, including the lungs, where they modulate the inflammatory response to many inhaled agents such as cigarette smoke that contains a wide array of PAHs (23). Moreover, lung fibroblasts metabolize PAHs to DEs, leading to the formation of PAH–DNA adducts (7). The studies reported here using genome-wide expression analysis show that BPDE, the ultimate carcinogenic metabolite of the PAH benzo[a]pyrene (BP), induces a dose-dependent inflammatory response in normal human lung fibroblasts that operates parallel to downregulation of DNA repair and cell cycle progression. Furthermore, BPDE induces the activation of JNK, which in turn increases the expression of several ILs. Abrogation of the JNK pathway using p53- and JNK-specific inhibitors dramatically reduces gene expression and subsequent production of ILs 1B, 6 and 8. This is the first time such a strong inflammatory response has been observed in response to a PAH DE in human lung fibroblasts.
While cellular inflammatory responses to cigarette smoke and PAHs such as BP have been reported, there are important differences in how inflammation is mediated upon exposure to a DE metabolite like BPDE. First, inflammatory responses to BP and other PAHs are mediated through aryl hydrocarbon receptor-dependent signaling (10–12). BPDE interacts with the aryl hydrocarbon receptor to less extent and is furthermore a poor inducer of downstream metabolic events because of its short intracellular half-life compared with BP (41). Furthermore, PAH metabolism via cytochrome P450 (CYP450) or aldo–keto reductase generates reactive oxygen species (ROS), which are known to induce inflammatory responses (42–44); however, BPDE does not contribute to ROS production (44,45). The analysis reported here supports the limited contribution of ROS to the BPDE inflammatory response. Transcriptional changes in processes related to ROS or oxidative stress remain unaffected by BPDE treatment, whereas the parent PAH BP induces the expression of genes related to the response to ROS and oxidative stress (46). Consequently, BPDE should induce inflammatory responses by pathways that operate independently from those induced by ROS.
Increases in ILs are implicated in the development of several diseases related to inflammation, including the progression of lung cancer (reviewed in ref. 47). This may be important since IL8 expression in normal bronchial and amnion epithelial cells increases in response to BPDE (20,22). However, the intracellular signaling pathways regulating the induction of IL expression in response to BPDE have not been reported. The results presented here show that increased IL signaling mediates the inflammatory response in human lung fibroblasts following exposure to BPDE. Furthermore, the response is regulated through a pathway that includes p53 and JNK, leading to transactivation of cJun.
Our data also show that concomitant induction of COX2 in these cells leads to increased prostaglandin production independently of regulation through p53 and JNK. These results are consistent with the findings that BPDE-induced COX2 expression is mediated through nuclear factor-kappaB signaling in mouse epidermal cells and rat astrocytes (48,49). High expression levels of COX2 and the formation of prostaglandins are implicated in cancer development. Hence, it is not necessarily surprising that COX2 inhibitors are associated with lowering the risk of certain cancers, including colorectal cancer (50).
To extend our analysis and visualize the results for WI-38 fibroblast exposure to BPDE, we used publicly available data to construct an in silico protein–protein interaction (PPI) network for annotated genes whose expression was most affected by BPDE. These included primarily genes related to the cell cycle, DNA repair and stress–response (Figure 6; supplementary Figures S4–S6 is available at Carcinogenesis Online).
For the cell cycle network, genes involved in replication and mitosis were downregulated and genes that inhibit G1/S and G2/M transitions were upregulated in our experiments. These aggregate changes result in an overall slowing or arresting of cell cycle progression, which is consistent with previous reports (16,19,21,22). Cellular responses to DNA damage are frequently mediated through pausing or stalling the cell cycle, allowing time for repair or permitting apoptosis or senescence to occur when damage is extensive. Nonetheless, in WI-38 lung fibroblasts exposed to BPDE, most genes involved in DNA repair are generally downregulated or relatively unaffected (Figure 6), which is consistent with results in other BPDE-exposed cells (17–19). For nucleotide excision repair, which is the main DNA repair pathway that operates on PAH-damaged DNA, the primary regulatory mechanism may well involve rapid posttranslational modifications and PPIs as opposed to altered transcription (51).
Several stress–response mediators found in the PPI network (Figure 6) physically interact with factors related to the cell cycle and DNA repair, including GADD45B and the AP-1 family members ATF3 and FOS. Activation of AP-1 occurs through increased expression of FOS and the phosphorylation of cJun (52). The Fos–Jun complex binds DNA with high affinity and induces the expression of many of the inflammatory mediators in this network, including the ILs, thus connecting the cell cycle and DNA repair with the stress–response through transcriptional regulation (not represented in the PPI network) (53).
The data reported here emphasize that caution should be used in extrapolating results from transformed cells to normal cells following exposure to DNA damaging agents. The strong inflammatory response observed in normal lung fibroblasts after BPDE exposure agrees with findings from genome-wide studies of other normal human cell types (20,22). However, these results are in contrast with transformed or carcinoma cell lines which exhibit a much less pronounced upregulation of inflammatory mediators (18,19,21). Similarly, changes in histone gene expression in response to BPDE from this and other studies show induction in normal cells and repression in carcinoma cell lines (19–22). These differences are not unique to BPDE since gene expression responses related to CYP450 metabolism, cell cycle checkpoints and apoptosis signaling also differ between normal and corresponding transformed cells following ultraviolet B irradiation and exposure to cigarette smoke condensate (54,55).
Cell senescence is one of the many mechanisms that an organism employs for tumor suppression in response to genotoxic exposure (56). Moreover, senescence leads to increased secretion of inflammatory mediators (34). In fact, the addition of antioxidants diminishes senescence in ultraviolet B and bisulfan-treated cells (57,58), which provides persuasive evidence that ROS are involved in triggering senescence. WI-38 lung fibroblasts display a similar secretory phenotype in response to BPDE exposure, but there is no associated significant increase in senescence due to the fact that BPDE does not participate in the production of ROS. This supports the findings that oxidative stress and the formation of ROS are important contributing factors in activating senescence signaling in response to DNA damage.
In conclusion, the studies reported here show that PAH DEs contribute to inflammatory responses in fibroblasts, independent of senescence and any metabolic byproducts such as ROS that are made during biotransformation of the parent PAH to the ultimate carcinogenic DE. Therefore, our current study shows that PAH DEs cannot only induce carcinogenesis through the formation of DNA damage but also by activating intracellular and intercellular signaling, inducing tumor-promoting inflammation. These results have important implications concerning the stromal–epithelial interactions in lung cancer development as a result of PAH exposure.
Supplementary material
Supplementary Figures S1–S6 and Tables S1–S6 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (ES010581) to D.A.S.
Supplementary Material
Acknowledgments
We thank the members of the Scicchitano laboratory John Burns and Laura Cartularo for critical reading and evaluation of the manuscript. The (±)-7r,8t-dihydroxy-9t,10t-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene was obtained from the National Cancer Institute Chemical Carcinogen Reference Standard Repository.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AP-1
activator protein 1
- BP
benzo[a]pyrene
- BPDE
benzo[a]pyrene diol epoxide
- COX2
cyclooxygenase 2
- JNK
c-jun N-terminal kinase
- cDNA
complementary DNA
- DMSO
dimethyl sulfoxide
- DE
diol epoxide
- ELISA
enzyme-linked immunosorbent assay
- IL
interleukin
- GO
Gene Ontology
- mRNA
messenger RNA
- PAH
polycyclic aromatic hydrocarbon
- PCR
polymerase chain reaction
- PFT-α
pifithrin-α
- PGE2
prostglandin E2
- PPI
protein–protein interaction
- ROS
reactive oxygen species
- RT2
real-time reverse transcription
References
- 1.IARC. Tobacco Smoke and Involuntary Smoking. Lyon, France: IARC; 2004. [PMC free article] [PubMed] [Google Scholar]
- 2.Fletcher C, et al. The natural history of chronic airflow obstruction. Br. Med. J. 1977;1:1645–1648. doi: 10.1136/bmj.1.6077.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harvey RG. Polycyclic Aromatic Hydrocarbons. Cambridge: Cambridge University Press; 1991. Chemistry and carcinogenicity. [Google Scholar]
- 4.Hecht SS. Tobacco smoke carcinogens and lung cancer. J. Natl Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 5.Sims P, et al. Metabolic activation of benzo(a)pyrene proceeds by a diol-epoxide. Nature. 1974;252:326–328. doi: 10.1038/252326a0. [DOI] [PubMed] [Google Scholar]
- 6.Luch A, et al. The level of DNA modification by (+)-syn-(11S,12R,13S,14R)- and (-)-anti-(11R,12S,13S,14R)-dihydrodiol epoxides of dibenzo[a,l]pyrene determined the effect on the proteins p53 and p21WAF1 in the human mammary carcinoma cell line MCF-7. Carcinogenesis. 1999;20:859–865. doi: 10.1093/carcin/20.5.859. [DOI] [PubMed] [Google Scholar]
- 7.Binkova B, et al. The effect of dibenzo[a,1]pyrene and benzo[a]pyrene on human diploid lung fibroblasts: the induction of DNA adducts, expression of p53 and p21(WAF1) proteins and cell cycle distribution. Mutat. Res. 2000;471:57–70. doi: 10.1016/s1383-5718(00)00111-x. [DOI] [PubMed] [Google Scholar]
- 8.Lloyd DR, et al. p53 controls global nucleotide excision repair of low levels of structurally diverse benzo(g)chrysene-DNA adducts in human fibroblasts. Cancer Res. 2002;62:5288–5294. [PubMed] [Google Scholar]
- 9.Xiao H, et al. p53 regulates cellular responses to environmental carcinogen benzo[a]pyrene-7,8-diol-9,10-epoxide in human lung cancer cells. Cell Cycle. 2007;6:1753–1761. doi: 10.4161/cc.6.14.4430. [DOI] [PubMed] [Google Scholar]
- 10.N'Diaye M, et al. Aryl hydrocarbon receptor- and calcium-dependent induction of the chemokine CCL1 by the environmental contaminant benzo[a]pyrene. J. Biol. Chem. 2006;281:19906–19915. doi: 10.1074/jbc.M601192200. [DOI] [PubMed] [Google Scholar]
- 11.Oesterling E, et al. Benzo[a]pyrene induces intercellular adhesion molecule-1 through a caveolae and aryl hydrocarbon receptor mediated pathway. Toxicol. Appl. Pharmacol. 2008;232:309–316. doi: 10.1016/j.taap.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Podechard N, et al. Interleukin-8 induction by the environmental contaminant benzo(a)pyrene is aryl hydrocarbon receptor-dependent and leads to lung inflammation. Toxicol. Lett. 2008;177:130–137. doi: 10.1016/j.toxlet.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Aggarwal BB. Nuclear factor-kappaB: the enemy within. Cancer Cell. 2004;6:203–208. doi: 10.1016/j.ccr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Coussens LM, et al. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Z, et al. Identification of genes responsive to BPDE treatment in HeLa cells using cDNA expression assays. Environ. Mol. Mutagen. 2000;36:201–205. doi: 10.1002/1098-2280(2000)36:3<201::aid-em3>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 16.Wang A, et al. Response of human mammary epithelial cells to DNA damage induced by BPDE: involvement of novel regulatory pathways. Carcinogenesis. 2003;24:225–234. doi: 10.1093/carcin/24.2.225. [DOI] [PubMed] [Google Scholar]
- 17.Akerman GS, et al. Gene expression profiles and genetic damage in benzo(a)pyrene diol epoxide-exposed TK6 cells. Mutat. Res. 2004;549:43–64. doi: 10.1016/j.mrfmmm.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 18.Luo W, et al. Phenotypic anchoring of global gene expression profiles induced by N-hydroxy-4-acetylaminobiphenyl and benzo[a]pyrene diol epoxide reveals correlations between expression profiles and mechanism of toxicity. Chem. Res. Toxicol. 2005;18:619–629. doi: 10.1021/tx049828f. [DOI] [PubMed] [Google Scholar]
- 19.Hockley SL, et al. AHR- and DNA-damage-mediated gene expression responses induced by benzo(a)pyrene in human cell lines. Chem. Res. Toxicol. 2007;20:1797–1810. doi: 10.1021/tx700252n. [DOI] [PubMed] [Google Scholar]
- 20.Belitskaya-Levy I, et al. Gene profiling of normal human bronchial epithelial cells in response to asbestos and benzo(a)pyrene diol epoxide (BPDE) J. Environ. Pathol. Toxicol. Oncol. 2007;26:281–294. doi: 10.1615/jenvironpatholtoxicoloncol.v26.i4.50. [DOI] [PubMed] [Google Scholar]
- 21.Hockley SL, et al. Identification through microarray gene expression analysis of cellular responses to benzo(a)pyrene and its diol-epoxide that are dependent or independent of p53. Carcinogenesis. 2008;29:202–210. doi: 10.1093/carcin/bgm227. [DOI] [PubMed] [Google Scholar]
- 22.Lu X, et al. Early whole-genome transcriptional response induced by benzo[a]pyrene diol epoxide in a normal human cell line. Genomics. 2009;93:332–342. doi: 10.1016/j.ygeno.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 23.Martey CA, et al. Cigarette smoke induces cyclooxygenase-2 and microsomal prostaglandin E2 synthase in human lung fibroblasts: implications for lung inflammation and cancer. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004;287:L981–L91. doi: 10.1152/ajplung.00239.2003. [DOI] [PubMed] [Google Scholar]
- 24.Dreij K, et al. Differential removal of DNA adducts derived from anti-diol epoxides of dibenzo[a,l]pyrene and benzo[a]pyrene in human cells. Chem. Res. Toxicol. 2005;18:655–664. doi: 10.1021/tx0497090. [DOI] [PubMed] [Google Scholar]
- 25.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dussault AA, et al. Rapid and simple comparison of messenger RNA levels using real-time PCR. Biol. Proced. Online. 2006;8:1–10. doi: 10.1251/bpo114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, et al. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Bolstad BM, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 29.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 30.McClintick JN, et al. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinformatics. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tusher VG, et al. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doniger SW, et al. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagerqvist A, et al. Both replication bypass fidelity and repair efficiency influence the yield of mutations per target dose in intact mammalian cells induced by benzo[a]pyrene-diol-epoxide and dibenzo[a,l]pyrene-diol-epoxide. DNA Repair (Amst.) 2008;7:1202–12. doi: 10.1016/j.dnarep.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 34.Coppe JP, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banin S, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 36.Tibbetts RS, et al. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komarov PG, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 38.Vivanco I, et al. Identification of the JNK signaling pathway as a functional target of the tumor suppressor PTEN. Cancer Cell. 2007;11:555–569. doi: 10.1016/j.ccr.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 39.Szczepankiewicz BG, et al. Aminopyridine-based c-Jun N-terminal kinase inhibitors with cellular activity and minimal cross-kinase activity. J. Med. Chem. 2006;49:3563–3580. doi: 10.1021/jm060199b. [DOI] [PubMed] [Google Scholar]
- 40.Walton MI, et al. An evaluation of the ability of pifithrin-alpha and -beta to inhibit p53 function in two wild-type p53 human tumor cell lines. Mol. Cancer Ther. 2005;4:1369–1377. doi: 10.1158/1535-7163.MCT-04-0341. [DOI] [PubMed] [Google Scholar]
- 41.Bigelow SW, et al. The Ah regulatory gene product. Survey of nineteen polycyclic aromatic compounds' and fifteen benzo[a]pyrene metabolites' capacity to bind to the cytosolic receptor. Toxicol. Lett. 1982;10:109–118. doi: 10.1016/0378-4274(82)90276-4. [DOI] [PubMed] [Google Scholar]
- 42.Dalton TP, et al. Induction of cellular oxidative stress by aryl hydrocarbon receptor activation. Chem. Biol. Interact. 2002;141:77–95. doi: 10.1016/s0009-2797(02)00067-4. [DOI] [PubMed] [Google Scholar]
- 43.Kepley CL, et al. Environmental polycyclic aromatic hydrocarbons, benzo(a) pyrene (BaP) and BaP-quinones, enhance IgE-mediated histamine release and IL-4 production in human basophils. Clin. Immunol. 2003;107:10–19. doi: 10.1016/s1521-6616(03)00004-4. [DOI] [PubMed] [Google Scholar]
- 44.Park JH, et al. Evidence for the aldo-keto reductase pathway of polycyclic aromatic trans-dihydrodiol activation in human lung A549 cells. Proc. Natl Acad. Sci. USA. 2008;105:6846–6851. doi: 10.1073/pnas.0802776105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao D, et al. Benzo[a]pyrene and its metabolites combined with ultraviolet A synergistically induce 8-hydroxy-2′-deoxyguanosine via reactive oxygen species. Free Radic. Biol. Med. 2005;39:1177–1183. doi: 10.1016/j.freeradbiomed.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 46.Castorena-Torres F, et al. Changes in gene expression induced by polycyclic aromatic hydrocarbons in the human cell lines HepG2 and A549. Toxicol. In Vitro. 2008;22:411–421. doi: 10.1016/j.tiv.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 47.Raman D, et al. Role of chemokines in tumor growth. Cancer Lett. 2007;256:137–165. doi: 10.1016/j.canlet.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weng MW, et al. Benzo[a]pyrene diol epoxide up-regulates COX-2 expression through NF-kappaB in rat astrocytes. Toxicol. Lett. 2004;151:345–355. doi: 10.1016/j.toxlet.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 49.Ouyang W, et al. Benzo[a]pyrene diol-epoxide (B[a]PDE) upregulates COX-2 expression through MAPKs/AP-1 and IKKbeta/NF-kappaB in mouse epidermal Cl41 cells. Mol. Carcinog. 2007;46:32–41. doi: 10.1002/mc.20260. [DOI] [PubMed] [Google Scholar]
- 50.Thun MJ, et al. Nonsteroidal anti-inflammatory drugs as anticancer agents: mechanistic, pharmacologic, and clinical issues. J. Natl Cancer Inst. 2002;94:252–266. doi: 10.1093/jnci/94.4.252. [DOI] [PubMed] [Google Scholar]
- 51.Nouspikel T. DNA repair in mammalian cells: nucleotide excision repair: variations on versatility. Cell. Mol. Life Sci. 2009;66:994–1009. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Angel P, et al. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 53.Roebuck KA, et al. Stimulus-specific regulation of chemokine expression involves differential activation of the redox-responsive transcription factors AP-1 and NF-kappaB. J. Leukoc. Biol. 1999;65:291–298. doi: 10.1002/jlb.65.3.291. [DOI] [PubMed] [Google Scholar]
- 54.Dazard JE, et al. Genome-wide comparison of human keratinocyte and squamous cell carcinoma responses to UVB irradiation: implications for skin and epithelial cancer. Oncogene. 2003;22:2993–3006. doi: 10.1038/sj.onc.1206537. [DOI] [PubMed] [Google Scholar]
- 55.Nagaraj NS, et al. Cigarette smoke condensate induces cytochromes P450 and aldo-keto reductases in oral cancer cells. Toxicol. Lett. 2006;165:182–194. doi: 10.1016/j.toxlet.2006.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 57.Ho JN, et al. Protective effects of aucubin isolated from Eucommia ulmoides against UVB-induced oxidative stress in human skin fibroblasts. Biol. Pharm. Bull. 2005;28:1244–1248. doi: 10.1248/bpb.28.1244. [DOI] [PubMed] [Google Scholar]
- 58.Probin V, et al. Busulfan-induced senescence is dependent on ROS production upstream of the MAPK pathway. Free Radic. Biol. Med. 2007;42:1858–1865. doi: 10.1016/j.freeradbiomed.2007.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.