Abstract
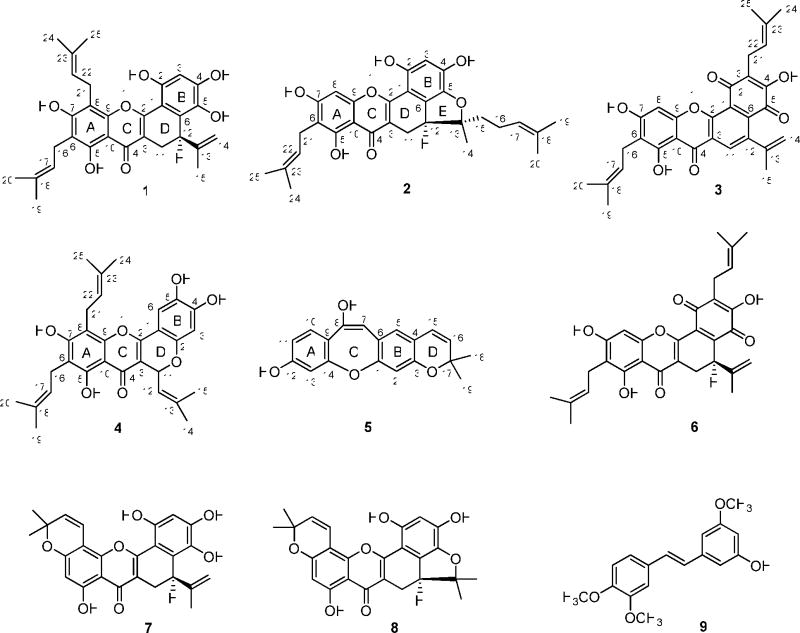
Four new prenylated flavonoids (1–4), a new stilbenoid (5), and nine known compounds were isolated from the twigs of Artocarpus rigida, collected in Indonesia. The structures of the new compounds were determined by analysis of their spectroscopic data, and the absolute configuration at C-12 of 1 and 2 and the known compounds artonin O (6), artobiloxanthone (7), and cycloartobiloxanthone (8), was determined by analysis of their CD and NMR spectroscopic data. Several of the compounds were cytotoxic towards HT-29 human colon cancer cells, with the most potent being compound 2 and the known compounds 6 and 8. Of the substances obtained, compounds 1 and 7 were the most active in the NF-κB p50 and p65 assay, respectively.
The genus Artocarpus (Moraceae), consisting of around 50 species, is a rich source of prenylated flavonoids1–7 and stilbenoids.8–12 Several of the compound types found in Artocarpus have been found to exhibit cytotoxicity against cancer cells,3,5–7,8–11 including artonin E (ED50 value of 0.06 μg/mL against P388 leukemia cells)5 and 3-hydroxy-4,3′,5′-trimethoxy-trans-stilbene (9), which showed cytotoxicity with ED50 values of 0.01 μg/mL and 0.1 μg/mL in the 9KB and 9PS in vitro test system, respectively.8
Artocarpus rigida Blume (Moraceae) is an evergreen tree distributed in tropical regions and Asia, and is a rare plant in Indonesia.12 Several prenylated flavonoids and stilbenoids have been reported from this plant previously.13–19 As a continuation of a research program on the discovery of potential new natural product anticancer agents from diverse organisms,20 it was found that a crude methanol extract of the twigs of A. rigida, collected in Indonesia, exhibited cytotoxicity toward HT-29 human colon cancer cells. Using column chromatography, four new prenylated flavonoids (1–4), a new stilbenoid (5), and several known compounds were isolated from this plant. Herein, we report the isolation and structure elucidation of compounds 1–5 and the biological evaluation of all metabolites obtained in this investigation.
Results and Discussion
A chloroform-soluble extract of the methanol extract of the dried ground twigs of A. rigida was separated by column chromatography over silica gel and yielded nine pooled fractions. Combined fractions three and four were further purified by silica gel column chromatography to afford the new compounds artorigidin B (2) and artorigidin C (3), along with the known compounds artonin O (6),14 3-hydroxy-4,3′,5′-trimethoxy-trans-stilbene (9),21 artonin K,2 and betulinic acid.22 Combined fractions five and six also were further purified by silica gel column chromatography to afford the new compounds artorigidin A (1), cyclorigidol (4), and artoristilbene (5), along with the known compounds artobiloxanthone (7),2, 23 cycloartobiloxanthone (8),24 artonin N,14 artonin G,14 and ursolic acid.25 The known compounds were identified by comparison of their spectroscopic data with literature values.
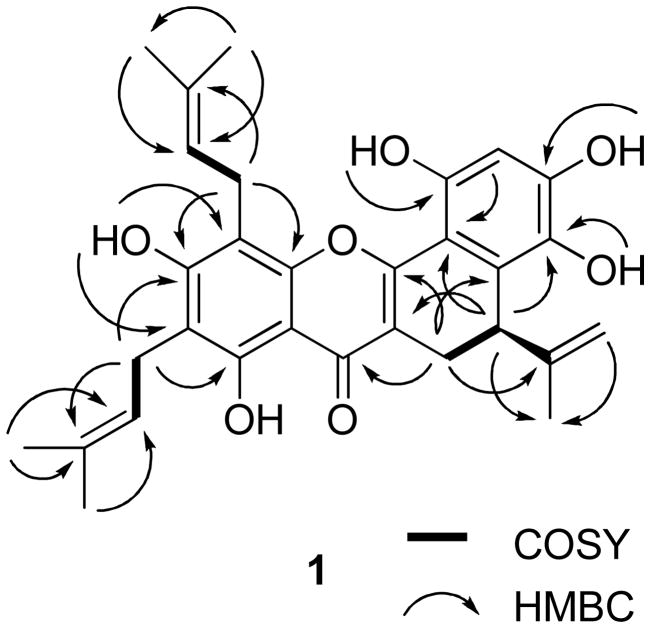
Compound 1 was isolated as an amorphous yellow powder, and showed a purple color under UV light at 365 nm. The HRESIMS exhibited a sodiated molecular ion peak at m/z 527.2051 (calcd 527.2046), consistent with a molecular formula of C30H32O7. The UV (λmax 230, 266, 278, 368 nm) and IR (νmax 3364, 1646, 1615, 1557, 1473 cm−1) spectra showed the typical absorption characteristics of a prenylated flavonoid,2,13,14,26 similar to analogous data for artonin B.26 The 1H and 13C NMR spectra gave evidence of 1 being a 3,5,6,7,8,2′,4′,5′,6′-nonasubstituted prenylflavonoid.26 The 1H NMR spectrum showed a signal due to a hydrogen-bonded phenolic OH group (δ 13.25) and other signals for other groups of a prenylflavonoid (Table 1). The 13C NMR and DEPT spectra contained signals consistent with the presence of a flavonoid unit and three prenyl groups (Table 2). Moreover, in the 1H NMR spectrum, two singlets at δ 4.47 and 4.75, due to two terminal olefinic protons, a singlet at δ 1.72 for a methyl group, together with an ABX system manifesting as a double doublet at δ 2.64 (J = 7.2 and 16.5 Hz, H-11), a doublet at δ 3.38 (J = 16.2, H-11), and another doublet at δ 3.85 (J = 7.2 Hz, H-12). These indicated a partially cyclized isoprenyl group, namely, a D-ring formed by linking C-3 and C-6′.1,2,14 This was confirmed by the HMBC correlations between H-11 and C-2, C-4, and C-6′ (Figure 1). In addition, the HMBC correlations between two prenyl methylene protons (H2-16) and C-5 and C- 7 and between another two prenyl methylene protons (H2-21) and C-7 and C-9 (Figure 1) supported the placement of these two prenyl groups at C-6 and C-8, respectively, which was confirmed by NOESY correlations between H2-16 and H3-20 and the C-7 hydroxy proton, and between H2-21 and H3-25 and the C-7 hydroxy proton (Figure 2). Comparison of the 1H and 13C NMR data of 1 with those of artonin B26 indicated the same structures for the A, B, C, and D ring systems, but with a different substituent at C-6 for these compounds. The signals at δ 116.4 (CH) and 128.9 (CH) due to a 2,2-dimethylpyran ring in artonin B26 were replaced by those at δ 120.7 (CH) and 136.1 (C) assignable to a 3,3-dimethylallyl group at C-6 in 1. Finally, the NOESY correlations between H-17 and H3-19 and H-22 and H3-24, together with analysis of the HMQC and HMBC spectra, enabled the assignment of the individual methyl groups in the prenyl substuents of 1 (Tables 1 and 2).
Table 1.
1H NMR Spectroscopic Data of Compounds 1–5
| position | 1a | 2a | 3b | 4a | 5a |
|---|---|---|---|---|---|
| 2 | 6.84 d (1.0) | ||||
| 5 | 13.25 s (OH) | 13.35 s (OH) | 12.97 s (OH) | 13.10 (OH) | 6.79 d (1.0) |
| 6 | |||||
| 7 | 6.36 s (OH) | 10.18 s (OH) | 6.60 (OH) | 6.86 dd (0.6, 1.0) | |
| 8 | 6.37 s | 6.70 s | |||
| 10 | 7.38 d (8.4) | ||||
| 2′ | 7.54 s (OH) | 6.65 s (OH) | |||
| 3′ | 6.50 s | 6.34 s | 6.50 s | ||
| 4′ | 4.97 s (OH) | 6.18 s (OH) | 9.25 s (OH) | ||
| 5′ | 6.47 s (OH) | ||||
| 6′ | 7.26 s | ||||
| 11 | 2.64 dd (7.2, 16.5) 3.38 d (16.5) |
2.51 t (15.6) 3.26 dd (7.2, 15.6) |
8.20 s | 6.23 d (9.0) | 6.76 dd (2.1, 8.4) |
| 12 | 3.85 d (7.2) | 3.46 m | 5.46 d (9.0) | ||
| 13 | 6.95 d (2.1) | ||||
| 14a | 4.47 s | 1.32 s | 4.90 brs | 1.68 s | |
| 14b | 4.75 s | 5.17 brs | |||
| 15 | 1.72 s | 1.94 m | 2.09 s | 1.95 s | 6.64 dd (0.6, 9.9) |
| 16 | 3.46 m | 2.14 m | 3.39 d (7.2) | 3.44 d (7.2) | 5.63 d (9.9) |
| 17 | 5.09 t (7.0) | 5.12 t (7.2) | 5.28 t (7.2) | 5.27 d (7.2) | |
| 18 | 1.44 s | ||||
| 19 | 1.76 s | 1.83 s | 1.65 s | 1.76 s | 1.44 s |
| 20 | 1.83 s | 1.77 s | 1.79 s | 1.83 s | |
| 21 | 3.46 m | 3.46 d (7.2) | 3.30 d (7.2) | 3.56 d (7.2) | |
| 22 | 5.23 t (7.0) | 5.28 t (7.2) | 5.25 t (7.2) | 5.27 d (7.2) | |
| 24 | 1.76 s | 1.69 s | 1.65 s | 1.76 s | |
| 25 | 1.80 s | 1.62 s | 1.79 s | 1.87 s |
Data were measured in CDCl3 at 300 MHz.
Data were measured in acetone-d6 at 300 MHz. Chemical shifts (δ) are expressed in ppm, with s = singlet, brs = broad singlet, d = doublet, t = triplet, m = multiplet, dd = double doublet. J values are presented in Hz and are omitted if the signals were overlapped as multiplets.
Table 2.
13C NMR Spectroscopic Data of Compounds 1–5
| position | 1a | 2a | 3b | 4a | 5a |
|---|---|---|---|---|---|
| 1 | 151.5 C | ||||
| 2 | 159.8 C | 159.4 C | 153.8 C | 155.3 C | 101.6 CH |
| 3 | 110.2 C | 112.2 C | 113.7 C | 109.2 C | 153.7 C |
| 4 | 180.4 C | 180.5 C | 179.7 C | 178.6 C | 109.7 C |
| 5 | 157.1 C | 157.2 C | 161.4 C | 156.9 C | 103.9 CH |
| 6 | 110.4 C | 110.2 C | 112.3 C | 110.0 C | 131.1 C |
| 7 | 159.1 C | 161.1 C | 165.1 C | 158.6 C | 105.7 CH |
| 8 | 104.5 C | 93.7 CH | 95.2 CH | 107.4 C | 154.3 C |
| 9 | 152.0 C | 154.1 C | 157.0 C | 151.0 C | 122.9 C |
| 10 | 105.4 C | 104.3 C | 103.9 C | 105.0 C | 121.2 CH |
| 1′ | 105.1 C | 104.3 C | 124.4 C | 105.3 C | |
| 2′ | 151.2 C | 149.6 C | 181.9 C | 152.0 C | |
| 3′ | 103.1 CH | 104.3 CH | 125.9 C | 104.2 CH | |
| 4′ | 149.7 C | 145.0 C | 153.8 C | 150.0 C | |
| 5′ | 134.7 C | 138.0 C | 181.9 C | 139.5 C | |
| 6′ | 127.7 C | 132.5 C | 132.0 C | 108.8 CH | |
| 11 | 21.9 CH2 | 20.7 CH2 | 132.3 CH | 69.1 CH | 112.1 CH |
| 12 | 38.1 CH | 44.7CH | 140.7 C | 120.9 CH | 155.7 C |
| 13 | 144.9 C | 97.5 C | 147.8 C | 138.2 C | 98.2 CH |
| 14 | 112.6 CH2 | 20.7 CH3 | 113.7 CH2 | 25.5 CH3 | 154.8 C |
| 15 | 20.9 CH3 | 41.5 CH2 | 23.8 CH3 | 18.4 CH3 | 116.2 CH |
| 16 | 21.8 CH2 | 23.0 CH2 | 22.0 CH2 | 21.3 CH2 | 129.6 CH |
| 17 | 120.7 CH | 120.9 CH | 122.9 CH | 121.4 CH | 76.2 C |
| 18 | 136.1 C | 137.9 C | 132.3 C | 134.3 C | 27.9 CH3 |
| 19 | 25.7 CH3 | 25.8 CH3 | 25.9 CH3 | 25.6 CH3 | 27.9 CH3 |
| 20 | 18.2 CH3 | 18.0 CH3 | 18.0 CH3 | 17.7 CH3 | |
| 21 | 21.7 CH2 | 21.6 CH2 | 23.1 CH2 | 21.7 CH2 | |
| 22 | 121.4 CH | 123.3 CH | 121.4 CH | 121.6 CH | |
| 23 | 136.3 C | 136.6 C | 133.4 C | 133.5 C | |
| 24 | 25.9 CH3 | 25.7 CH3 | 25.9 CH3 | 25.6 CH3 | |
| 25 | 18.0 CH3 | 17.8 CH3 | 17.9 CH3 | 17.6 CH3 |
Data were measured in CDCl3 at 75.5 MHz.
Data were measured in acetone-d6 at 75.5 MHz. Chemical shifts (δ) are presented in ppm.
Figure 1.
COSY and key HMBC correlations of 1.
Figure 2.
Selected NOESY correlations of 1.
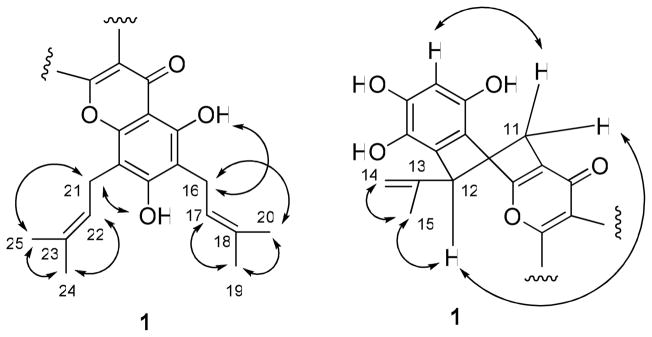
The absolute configuration at C-12 of 1 was determined by comparison of its CD spectrum with that of a model compound, (+)-(5R)-tert-butyl-1,3-cyclohexadiene, for which a specific rotation value ([α]25 D) of +187 and a positive Cotton effect at 270 nm in its CD spectrum have been reported.27 Compound 1 contains one stereogenic center, the same as that of (+)-(5R)-tert-butyl- 1,3-cyclohexadiene, and showed [α]20 D +80 and a positive Cotton effect at 270 nm. Therefore, the absolute configuration at C-12 was determined as R. In the 1H NMR spectrum, the signals at δ 2.64 and δ 3.38 were ascribed to the pseudo-axial H-11a and pseudo-equatorial H-11e,2 which were supported by the NOESY correlation between H-11a and H-3′ (Figure 2) and H-11e and H-12, and the allyl group at C-12 in 1 was assigned in an equatorial orientation (Figure 2). Consequently, the structure of 1 was determined as (5R)-5,6-dihydro-1,3,4,8,10-pentahydroxy-5-isopropenyl-9,11-bis(3-methyl-2-butenyl)-7H-benzo[c]xanthen-7-one, and this substance has been accorded the trivial name, artorigidin A.28
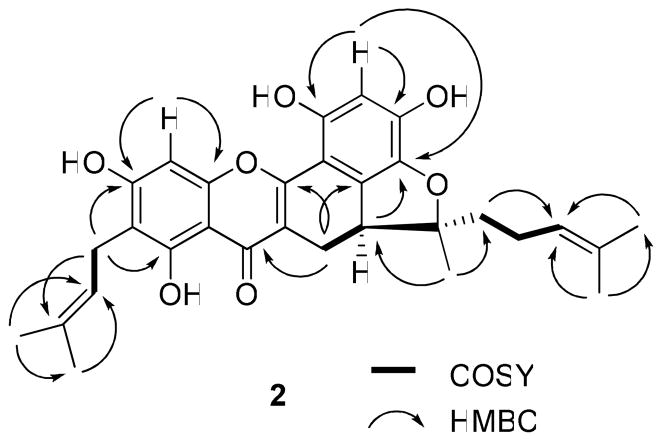
Compound 2 was isolated as an amorphous yellow powder, and its molecular formula of C30H32O7 was determined by HRESIMS analysis. Similarities of its UV, IR, MS, and NMR spectra with those of 1 indicated that 2 is also a tetraprenylflavonoid. The 1H NMR spectrum showed signals due to a substituted flavonoid unit, two tetra-3,3-dimethylallyl groups, and a furan ring fused with an ABX system (Table 1). Also observed in the 13C NMR and DEPT spectra, were signals due to five methyl, four methylene, five methine, and 16 quaternary carbons, including an oxygenated quaternary carbon (δ 97.5) (Table 2). On comparing the 13C NMR spectrum of 2 with that of 1, the signals at δ 144.9 (C, C-13) and δ 112.6 (CH2, C-14), due to a terminal double carbon bond in 1, were replaced by signals at δ 97.5 (C, C-13, an oxygenated quaternary carbon) and δ 41.5 (CH2, C-15, a methylene), in 2. On considering an oxidative cyclization between a hydroxy group at C-5′ and a prenyl group at C-13,26 a 2,2-dimethyldihydrofuran unit linking C-5′, C-6′, and C-13 could be proposed. Additionally, in the 13C NMR spectrum, the signal at δ 93.7 (CH) was assigned unambiguously to the non-substituted C-8, based on both the assignments for 2 and artonin M14 and the HMBC correlations between H-8 and C-7 and C-9 (Figure 3). A prenyl group was apparent at C-6, according to the HMBC correlations between H2-21 and C-5, C-7, and C-23 (Figure 3). Another prenyl group was assumed to be affixed to C-15 because of the observation of HMBC correlations between H2-15 and C-14 and C-17, and between H-17 and C-19 and 20 (Figure 3). Comparison of the 1H and 13C NMR spectra of 2 with those of artonin M14 showed that both compounds have similar NMR characteristics, except for the replacement of the signals at δ 116.0 (CH) and 129.2 (CH), due to a 2,2-dimethylpyran ring in artonin M,14 with the signals at δ 123.3 (CH, C-22) and 136.6 (C, C-23) being assigned to a 3,3-dimethylallyl group at C-6 in 2. This facilitated an assignment of the planar structure of artorigidin B (2).
Figure 3.
COSY and key HMBC correlations of 2.
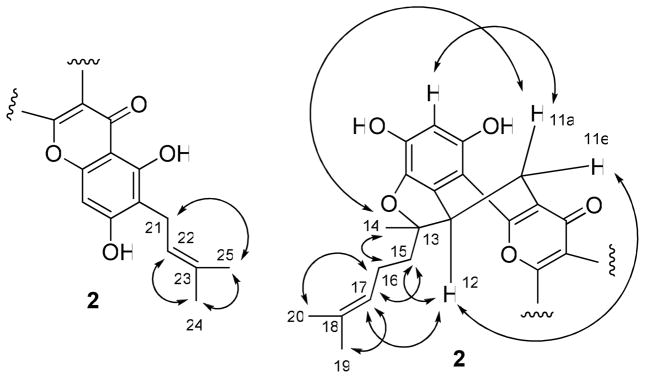
The NOESY correlations between H2-21 and H3-25, and H-22 and H3-24 (Figure 4) indicated a cis arrangement between H-22 and H3-24. The signals at δ 2.51 and 3.26 were assigned to a pseudo-axial H-11a and a pseudo-equatorial H-11e, respectively, the same as in 1. A NOESY correlation between H-11e and H-12 suggested their cis relationship. A positive Cotton effect at 262 nm in the CD spectrum of 2 indicated the S configuration of C-12.27 Other NOESY correlations between H-11a and H-14 and H-3′ were supportive of an equatorial methyl linked at C-13. Therefore, the absolute configuration at C-13 was determined as R, which was substantiated by the NOESY correlations between H-15 and H-12 and H-17, H-14 and H-16, H- 16 and H-20, and H-17 and H-19. Accordingly, compound 2 was determined28 as (5S)-5,6- dihydro-1,3,8,10-tetrahydroxy-9-(3-methyl-2-butenyl)-5-[(1R)-1-methyl-1-(3-methyl-2- butenyl)]-7H-benzofuro[3,3a,4:ab]xanthen-7-one.
Figure 4.
Selected NOESY correlations of 2.
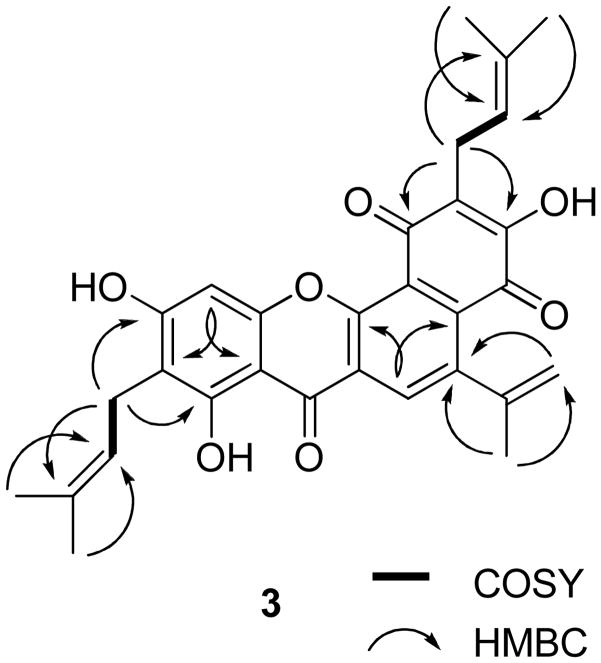
Compound 3 was isolated as an amorphous red powder. Both the IR spectrum, with absorptions typical of hydroxy (νmax 3303 cm−1) and conjugated carbonyl (νmax 1646 cm−1) groups, and the UV spectrum, with absorptions at 271 and 362 nm, indicated a quinonoid prenyflavonoid derivative.2,14,29 The molecular formula of C30H28O7 was determined by HRESIMS (two mass units less than 6), consistent with a C-15 flavonoid unit and three prenyl groups, which was supported by the 1H and 13C NMR spectra (Tables 1 and 2). HMBC correlations between H-11 and C-2 and C-6′, H2-16 and C-5, C-7, and C-18, and H2-21 and C-2′, C-4′, and C-23 (Figure 5) indicated the linkage of three prenyl groups at C-3, C-6, and C-3′ of 3, respectively. The signals at δH 6.70 (s) and δ 8.20 (s) were assigned to H-8 and H-11, respectively, because of the HMBC correlations between H-8 and C-9 and C-10 and H-11 and C-2 and C-6′ (Figure 5). In the 13C NMR spectrum, three signals due to carbonyl groups were observed at δ 179.7, 181.9, and 181.9 (Table 2). One of these could be assigned to the carbonyl carbon of a chromone unit (C-4), with the others attributable to ring B.14,29 Comparison of the 13C NMR spectrum of 3 with that of artonin O (6)14 indicated the structures of the A-, B-, and C- ring systems of these compounds to be the same, with the D-ring being different. Also, C-11 and C-12 were found to be unsaturated in 3, which was implied by the replacement of 13C NMR signals at δ 21.4 (CH2, C-11) and 35.3 (CH, C-12) in artonin O (6) by signals at δ 132.3 (CH, C- 11) and 140.7 (C, C-12) in 3. Finally, the correlations between H-11 and C-2 and C-6′, H-14 and C-12, H-15 and C-12 and C-14, H-19 and H-20 and C-17, H-24 and H-25 and C-22 supported the structure, 4,8,10-trihydroxy-5-isopropenyl-2,9-bis(3-methyl-2-butenyl)-7-benzo[c]xanthen- 2,4,7-trione for 3 (artorigidin C). 28
Figure 5.
COSY and key HMBC correlations of 3.
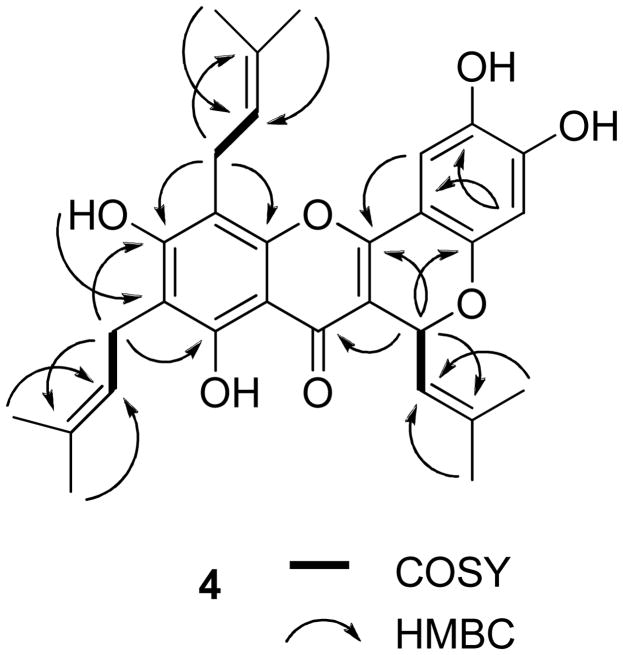
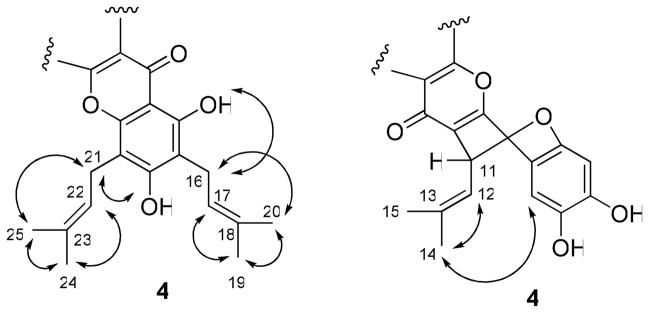
Compound 4 was obtained as an amorphous yellow powder. Its similar IR, UV, and NMR spectra to those of related compounds26,30–32 indicated this compound to be a prenylated flavonoid. The HRESIMS showed a sodiated molecular ion peak at m/z 527.2073, consistent with a molecular formula of C30H32O7. The 13C NMR spectrum revealed the presence of 30 carbons, including six methyl groups and a carbonyl group (δ 178.6), as expected for a triprenylated flavonoid. The 1H NMR spectrum exhibited six singlets at δ 1.68, 1.76, 1.76, 1.83, 1.87, and 1.95 for six vinyl methyl groups and two doublets at δ 6.23 (J = 9.0 Hz, H-11) and 5.46 (J = 9.0 Hz, H-12), consistent with a substituted 2H-benzopyran ring system, that would arise from oxidative cyclization between the prenyl group at C-3 and the C-2′ hydroxy group.31,32 This was confirmed by long-range cross-peaks of H-11 (δ 6.23) with C-2, C-4, C-2′, and C-13. Moreover, two aromatic singlets were observed, with that at δ 6.50 assigned to H-3′ since it correlated with C-1′ and C-5′, and the second at δ 7.26 assigned to H-6′ because of correlations observed with C-2, C-2′ and C-4′. Comparison of the 1H and 13C NMR spectra of 4 with those of 1 suggested that both compounds have the same substitution pattern at the A-ring. The presence of a prenyl group connected to C-6, and another at C-8, was supported by the HMBC correlations between H2-16 and C-5, C-7, and C-18, and H2-21 and C-7, C-9, and C-23 in the HMBC spectrum of 4 (Figure 6).
Figure 6.
COSY and key HMBC correlations of 4.
A set of NOESY correlations, as described for 1, was used to confirm the assignments of two prenyl groups at C-6 and C-8 in compound 4. The signal at δ 6.23 was assigned to an equatorial H-11, and the isobutylene group at C-11 was assigned to an axial form, confirmed by the observation of the NOESY correlations between H-12 and H-14, H-14 and H-6′, which supported a relative configuration of 4, as shown in Figure 7. The absolute configuration at C-11 was not determined because of the apparent unavailability of relevant reference CD or specific optical rotation data in the literature. Accordingly, the structure of 4 was proposed as 2,3,8,10- tetrahydroxy-6-(2-methylpropenyl)-9,11-bis(3-methyl-2-butenyl)-6H[1]benzopyrano[4,3-b]benzopyran-7-one, and the compound has been named cyclorigidol.33
Figure 7.
Selected NOESY correlations of 4.
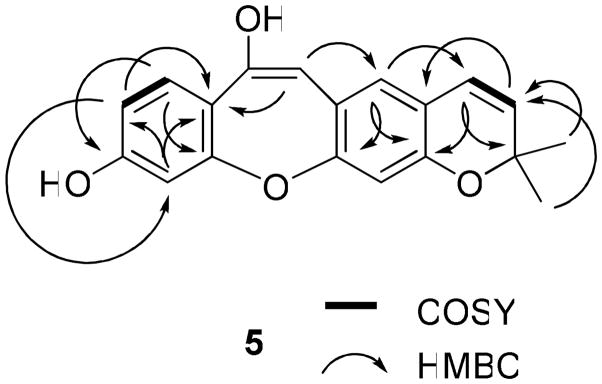
Compound 5 was isolated as a colorless powder. The UV (λmax 234, 340, and 360 nm) and IR (νmax 3364, 1615, 1456 cm−1) spectra showed absorption characteristics of a stilbenoid derivative.15–19 The molecular formula of C19H16O4 was determined by HRESIMS, and was consistent with a 14-carbon stilbene nucleus bearing a prenyl group substituent. The 1H and 13C NMR signals of 5 were assigned by analysis of the HMQC and HMBC spectra. The 1H NMR spectrum (Table 1) showed signals due to a 2,2-dimethylpyran ring (the D ring, connected to the B ring at C-3 and C-4), including two gem-dimethyl groups at δ 1.44 (s) and two typical cis-olefinic protons at δ 6.64 (dd, J = 0.6 and 9.9 Hz, H-15) and δ 5.63 (d, J = 9.9 Hz, H-16).14,26 A COSY correlation between H-15 and H-16 and HMBC correlations between H-15 and C-3, C-5 and C-17, H-16 and C-4 and C-17, and H3-18 or H3-19 and C-16 and C-17 (Figure 8) were observed. The 1H NMR spectrum also displayed resonances at δ 7.38 (1H, d, J = 8.4 Hz, H-10), 6.76 (1H, dd, J = 2.1 and 8.4 Hz, H-11), and 6.95 (1H, d, J = 2.1 Hz, H-13), due to the ABX spin system of a 1,3,4-trisubstituted benzene ring (the A ring, substituted by a hydroxy group at C-12 and connected to the C ring at C-9 and C-14). This was supported by the COSY correlation between H-10 and H-11 and the HMBC correlations between H-10 and C-12 and C-14, H-11 and C-9 and C-13, and H-13 and C-9 and C-11 (Figure 8). Additionally, the absence of any signal for a pair of cis or trans vinyl protons8–11 or any other ortho aromatic protons in the 1H NMR spectrum indicated one substituent to be linked to C-8, and the B ring to be connected with the C ring at C-1 and C-6 and with the D ring at C-3 and C-4. Comparison of the NMR and MS data of 5 with those of artocarpol E16 indicated that these two compounds contain the same structure of a stilbene core. However, artocarpol E has a prenyl group at C-4 and a geranyl group at C-7, which were either absent (geranyl group) or changed to a 2,2-dimethylpyran ring in 5.16 These inferences were confirmed by the HMBC correlations between H-5 and C-1, C-3 and C-15, and between H-7 and C-5 and C-9, as well as the appearance of five signals due to oxygen-substituted olefinic or aromatic carbons in the 13C NMR spectrum (Table 2) of 5, and its molecular weight of 308. Therefore, the structure of 5, artoristilbene,34 was determined as 2,2-dimethyl-2H-benzo{6,7-oxepino[3,2-g]}chromene-7,10-diol.
Figure 8.
COSY and key HMBC correlations of 5.
Previously, prenylflavonoids and stilbenoids have been isolated from A. rigida.13–19 Lin and associates investigated the root bark of this species collected in Taiwan, and reported several stilbenoids and prenylated flavonoids.15–19 Nomura’s group studied the bark of A. rigida, as obtained from Java in Indonesia, and reported several prenylated flavonoids,13,14 but no stilbenoids. In the present study, both prenylated flavonoids and stilbenoids were isolated from the twigs of A. rigida, collected in Java. It was found that prenylflavonoids are the main components, with stilbenoids being trace components. Therefore, it may be proposed that A. rigida contains different components in its various anatomical parts when obtained from different locations. The absolute configuration at C-12 of the prenylflavonoids has not been determined by previous investigators. The CD and NOESY spectra were used for the determination of the C-12 absolute configuration of five representatives of this class of compounds (1, 2, 6–8) for the first time in this study.
All compounds isolated from A. rigida twigs were evaluated for their cytotoxicity against the HT-29 human colon cancer cell line, using paclitaxel as the positive control (Table 3). Compounds 2, 6 and 8 were found to be cytotoxic with ED50 values of 3.4, 3.2, and 3.7 μM, respectively. The compounds isolated from A. rigida twigs were also evaluated for their ability to inhibit NF-κB activity in both the NF-κB p50 and NF-κB p65 assays, using rocaglamide as the positive control (Table 3). Compounds 1 and 7 exhibited NF-κB inhibitory activity in the NF-κB p50 assay, with IC50 values of 2.0 μM (1) and 3.7 μM (7), respectively, and compound 7 exhibited NF-κB inhibitory activity in the NF-κB p65 assay, with an IC50 value of 2.3 μM.
Table 3.
Cytotoxicity for HT-29 Cells and NF-κB Inhibition of Compounds 1–9a
| compound | cytotoxicityb | NF-κB p50 inhibitionc | NF-κB p65 inhibitionc |
|---|---|---|---|
| 1 | 9.3 | 2.0 | >10 |
| 2 | 3.4 | NTd | NTd |
| 3 | >10 | NTd | NTd |
| 4 | >10 | >10 | >10 |
| 5 | 10.7 | >10 | >10 |
| 6 | 3.2 | >10 | >10 |
| 7 | >10 | 3.7 | 2.3 |
| 8 | 3.7 | >10 | >10 |
| 9 | 8.7 | >10 | >10 |
All other compounds isolated in this study were inactive in these assay systems.
Data are represented as ED50 values (μM), and paclitaxel was used as positive control (ED50, 0.10 nM).
Data are represented as IC50 values (μM), and rocaglamide was used as positive control (IC50 = 4.0 μM in the NF-κB p50 assay and IC50 = 0.075 μM in the NF-κB p65 assay).
Not tested.
A number of prenylated oxygen heterocyclic constituents with cytotoxic effects against cancer cell lines have been isolated previously from Artocarpus species.3, 5–7 When the cytotoxicity data against the HT-29 colon cancer cell line, as obtained in the present investigation for the compounds isolated from A. rigida (Table 3), are compared with the previous literature, a number of preliminary observations about the structural requirements for activity may be made. In the current study, the known compound, cycloartobiloxanthone (8) was found to be cytotoxic (ED50 3.7 μM), but the closely related dihydrobenzoxanthone, artonin K, was inactive (ED50 >10 μM), indicating the potentiating effects on resultant biological potency of the two prenyl groups at C-2 and C-15 present in the structure of compound 8. Compounds 6 (artonin O) and 3 (artorigidin C) have the exact same structure apart from the presence of an olefinic group between C-11 and C-12 in 3, and exhibited ED50 values of 3.2 and >10 μM. Therefore, within this series, the occurrence of a C-12-affixed β-oriented substituent seems to be necessary. Compounds 7 (ED50 >10 μM) and 8 are also closely related structurally, with the C-12 isoprenyl group of 7 cyclized with the C-5′ hydroxy group to form a dihydrofuran ring in 8. Since only compound 8 was active, then this additional cyclization enhances the resultant cytotoxicity. Although not isolated in the present study, artonin E, with a C-3 prenyl group and a tricyclic overall composition, appears to be the most potent cytotoxic prenylated oxygen heterocyclic compound to have been isolated from the genus Artocarpus, being approximately two orders of magnitude more potent than artonin O (6) and cycloartobiloxanthone (8), when evaluated against the P-388 murine lymphocytic leukemia cell line.5
Experimental Section
General Experimental Procedures
Optical rotations were measured with a Perkin-Elmer model 343 polarimeter. UV spectra were recorded on an UV-Hitachi U2910 spectrophotometer. CD measurements were performed using a JASCO J-810 spectropolarimeter. IR spectra were recorded on a Nicolet 6700 FT-IR spectrometer. 1H, 13C, DEPT, HMQC, HMBC, NOESY, and COSY NMR spectra were recorded at room temperature on a Bruker Avance DRX-300 MHz NMR spectrometer, with TMS as internal standard. ESIMS and HRESIMS were recorded on a LCT-TOF mass spectrometer. Column chromatography was conducted using silica gel (70–230 mesh, Merck, Darmstadt, Germany). Analytical and preparative thin-layer chromatography (TLC) was performed on precoated silica gel 60 F254 plates (Sorbent Technologies, Atlanta, GA; 0.20 mm layer thickness). Sephadex LH-20 was purchased from Amersham Biosciences, Uppsala, Sweden. For visualization of TLC plates, sulfuric acid reagent was used. Fluorescence was tested using a Spectroline (model ENF-260C) UV light source.
Plant Material
The twigs of Artocarpus rigida were collected at Gunung Kancana, Cianjur, West Java, Indonesia in August, 2003, and identified by S. R. A voucher specimen (A05753) has been deposited at the Field Museum of Natural History, Chicago, IL, under the accession number SR-CJR 82 (2285411).
Extraction and Isolation
The milled, air-dried twigs of A. rigida (800 g) were extracted with MeOH (3 L × 5) at room temperature, and the solvent evaporated in vacuo. The dried MeOH extract (77.6 g, 9.8%) was resuspended in 10% H2O in MeOH (600 mL) and partitioned with n-hexane (500 mL and 3 × 300 mL) to yield a hexane-soluble residue (D001, 9.9 g, 1.3%). The aqueous-MeOH layer was then partitioned with CHCl3 (500, 400, 300, 200 mL) to afford a chloroform-soluble extract (D002, 19.8 g, 2.5%), which was followed by washing with a 1% aqueous solution of NaCl, to partially remove tannins. The chloroform-soluble extract exhibited cytotoxicity towards the HT29 cell line (ED50 < 20 μg/mL). Both the n-hexane-soluble and aqueous-soluble extracts were inactive in the bioassay system used.
The chloroform-soluble extract (19.0 g) was subjected to silica gel column chromatography (9.0 × 45 cm) and eluted with a gradient of n-hexane–acetone (100:1, 80:1, 60:1, 30:1, 20:1, 10:1, 5:1, 3:1, 1:1; 500 mL each). Fractions were pooled by TLC analysis to give nine combined fractions (D002F01-D002F09). Of these, D002F03 and D002F04 (ED50 < 20 μg/mL) were combined and further chromatographed over a silica gel column (2.5 × 20 cm), eluted with a gradient of n-hexane–acetone (20:1, 15:1, 10:1, 8:1, 5:1, 3:1, 1:1, 300 mL each), to yield seven combined fractions (D002F0301–D002F0307). Fraction D002F0301 was chromatographed over silica gel using n-hexane-acetone (10:1) as solvent, and then purified by separation over a Sephadex LH-20 column, eluted with CHCl3-MeOH (1:1), affording 6 (20 mg). Fraction D002F0302 was separated by silica gel chromatography, eluted with n-hexane–acetone (10:1), and then purified by passage over a Sephadex LH-20 column, eluted with a mixture of CHCl3- MeOH (1:1), to afford 2 (2.0 mg). Fraction D002F0303 was chromatographed over silica gel, eluted by n-hexane–acetone (8:1), and then purified using a Sephadex LH-20 column, eluted with a mixture of CHCl3-MeOH (1:1), to afford 3 (2.0 mg). Fraction D002F0304 was chromatographed over silica gel, eluted by n-hexane–acetone (3:1), and then purified by separation over a Sephadex LH-20 column, using CHCl3-MeOH (1:1) for elution, affording 9 (2.5 mg). Fraction D002F0305 was chromatographed over silica gel, eluted by n-hexane–acetone (3:1), and then purified over a Sephadex LH-20 column, eluted by CHCl3-MeOH (1:1), furnishing betulinic acid (25 mg). Fraction D002F0306 was chromatographed over silica gel, eluted by n-hexane–acetone (3:1), yielding artonin K (5.0 mg). D002F05 and D002F06 (ED50 < 20 μg/mL) were combined and further chromatographed over a silica gel column, eluted with a gradient of n-hexane–acetone (12:1, 10:1, 8:1, 6:1, 5:1, 4:1, 3:1, 2:1, and 1:1, 300 mL each), to yield nine combined fractions (D002F0501-D002F0509). Fraction D002F0501 was chromatographed over silica gel using n-hexane–acetone (10:1) as solvent, and then purified by passage over a Sephadex LH-20 column, eluted with CHCl3-MeOH (1:1), affording artonin N (3.5 mg). Fraction D002F0502 was purified by silica gel chromatography, using n-hexane– acetone (10:1) for elution, and then purified using Sephadex LH-20, eluted with a mixture of CHCl3-MeOH (1:1), to afford artonin G (5.0 mg). Fraction D002F0503 was chromatographed over silica gel, eluted by n-hexane–acetone (8:1), and then purified by passage over Sephadex LH-20, eluted with a mixture of CHCl3-MeOH (1:1), to afford cycloartobiloxanthone (8, 6 mg). Fraction D002F0504 was chromatographed over silica gel, eluted by n-hexane–acetone (3:1), and then purified with a column containing Sephadex LH-20, using CHCl3-MeOH (1:1) for elution, affording 7 (21 mg). Fraction D002F0505 was chromatographed over silica gel, with nhexane –acetone (3:1) as solvent, and then purified over a Sephadex LH-20 column, eluted by CHCl3-MeOH (1:1), producing 1 (25 mg). Fractions D002F0506 and D002F0507 were chromatographed over silica gel, eluted by n-hexane–acetone (3:1), and then purified over Sephadex LH-20, using CHCl3-MeOH (1:1) as solvent, affording 4 (3.5 mg). Fraction D002F0508 was chromatographed over silica gel, eluted by n-hexane–acetone (3:1), and then purified over a Sephadex LH-20 column, using CHCl3-MeOH (1:1) as solvent, affording 5 (2 mg). Fraction D002F0509 was chromatographed over silica gel, eluted by n-hexane–acetone (3:1), and then purified over a column containing Sephadex LH-20, using CHCl3-MeOH (1:1) as solvent, affording ursolic acid (10 mg).
Artorigidin A (1)
Amorphous yellow powder (n-hexane), showing a purple color under UV light at 365 nm; [α]20 D +80 (c 0.2, CH2Cl2); UV (CH2Cl2) λmax (log ε) 230 (4.18), 266 (4.18), 278 (4.15), 368 (4.08) nm; CD (MeOH, nm) λmax (Δε) 228 (+1.62), 270 (+1.04), 295 (−0.55), 312.5 (−1.07); IR (dried film) νmax 3364, 1646, 1615, 1557, 1473, 1282, 1096 cm−1; 1H and 13C NMR data, see Tables 1 and 2; positive ESIMS m/z 527.2 [M + Na]+; positive HRESIMS m/z 527.2051, calcd for C30H32O7Na, 527.2046.
Artorigidin B (2)
Amorphous yellow powder (n-hexane), showing a purple color under UV light at 365 nm; [α]20 D +10 (c 0.1, CH2Cl2); UV (CH2Cl2) λmax (log ε) 233 (4.33), 268 (4.42), 326 (4.18), 374 (4.30) nm; CD (MeOH, nm) λmax (Δε) 223.5 (+0.87), 243 (−0.47), 262 (+0.30), 294 (−0.68), 314 (−0.38); IR (dried film) νmax 3354, 1645, 1606, 1471, 1272, 1170 cm−1; 1H and 13C NMR data, see Tables 1 and 2; positive ESIMS m/z 505.2 [M + H]+; positive HRESIMS m/z 505.2222, calcd for C30H32O7H, 505.2226.
Artorigidin C (3)
Amorphous red powder (n-hexane), showing a purple color under UV light at 365 nm; UV (CH2Cl2) λmax (log ε) 271 (4.52), 362 (3.78) nm; IR (dried film) νmax 3303, 1646, 1463, 1268, 1078 cm−1; 1H and 13C NMR data, see Tables 1 and 2; positive ESIMS m/z 523.1 [M + Na]+; positive HRESIMS m/z 523.1725, calcd for C30H28O7Na, 523.1733.
Cyclorigidol (4)
Amorphous yellow powder (n-hexane), showing a purple color under UV light at 365 nm; [α]20 D +20 (c 0.2, CH2Cl2); UV (CH2Cl2) λmax (log ε) 228 (4.28), 282 (4.35), 386 (4.21) nm; CD (MeOH, nm) λmax (Δε) 222 (+1.13), 235 (−0.54), 253.5 (−0.30), 269 (−0.11), 294.5 (−0.39), 310 (−0.33); IR (dried film) νmax 3335, 1647, 1556, 1472, 1290, 1213 cm−1; 1H and 13C NMR data, see Tables 1 and 2; positive ESIMS m/z 527.2 [M + Na]+; positive HRESIMS m/z 527.2073, calcd for C30H32O7Na, 527.2046.
Artoristilbene (5)
Amorphous white powder (n-hexane), showing a blue color under UV light at 365 nm; UV (CH2Cl2) λmax (log ε) 234 (3.57), 340 (3.99), 360 (3.87) nm; IR (dried film) νmax 3364, 1615, 1558, 1456, 1262, 1065 cm−1; 1H and 13C NMR data, see Tables 1 and 2; positive ESIMS m/z 331.1 [M + Na]+; positive HRESIMS m/z 331.0968, calcd for C19H16O4Na, 331.0946.
Cytotoxicity Assay
Cytotoxicity of the samples was screened against HT-29 human colon cancer cells by a previously reported procedure.35 The cells were cultured under standard conditions and trypsinized. Then, the harvested cells were added to 96-well plates and treated by either the test compounds dissolved in DMSO or by DMSO (the negative control). The plates were incubated at 37 °C in 5% CO2 for three days, and then the cells were fixed, then incubated at room temperature for 30 min, washed with water once, dried at room temperature overnight, and then dyed using sulforhodamine B. After the dyed cells were lysed in tris-base buffer, the plates were read at 515 nm with an ELISA reader. Paclitaxel was used as a positive control, and cytotoxicity was calculated based on the values read from the samples and the negative control.
Enzyme-Based ELISA NF-κB Assays
The NF-κB assays were carried out as in a previous procedure with minor modification.36 Using an EZ-Detect Transcription Factor Assay System ELISA kit (Pierce Biotechnology, Rockford, IL), a nuclear extract of HeLa cells (ATCC, American Type Culture Collection) was treated by positive control and the list samples. The specific binding ability of activated p50 or p65 subunits of NF-κB to the biotinylated-consensus sequence was measured by detecting the chemiluminescent signal in a Fluostar Optima plate reader (BMG Labtech Inc., Durham, NC). Rocaglamide was used as a positive control in both the NF-kB p50 and p65 assays.
Supplementary Material
Acknowledgments
This investigation was supported by grants U01 CA52956 and P01 CA125066, funded by the National Cancer Institute, NIH, Bethesda, MD. We wish to acknowledge the late Mr. Agus Ruskandi of the Herbarium Bogoriense for assistance with the plant collection. We thank Jack Fowble, College of Pharmacy, The Ohio State University, for access to the NMR spectrometer. We also thank Dr. Kari Green-Church, Mass Spectrometry and Proteomics Laboratory, Campus Chemical Instrument Center, The Ohio State University, for the MS measurements.
Footnotes
Supporting Information Available: Mass spectrometric data and 1H and 13C NMR spectra of compounds 1–5. Physical and spectroscopic data of known prenylflavonoids and stilbene from A. rigida. 1H and 13C NMR data of known prenylflavonoids and stilbene from A. rigida. Structures of literature compounds in comparison to 2, 3, 6, and 8. This information is available free-of-charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Aida M, Shinomiya K, Matsuzawa K, Hano Y, Nomura T. Heterocycles. 1994;39:847–858. [Google Scholar]
- 2.Aida M, Yamaguchi N, Hano Y, Nomura T. Heterocycles. 1997;45:163–175. [Google Scholar]
- 3.Hakim EH, Fahriyati A, Kau MS, Achmad SA, Makmur L, Ghisalberti EL, Nomura T. J Nat Prod. 1999;62:613–615. doi: 10.1021/np980279l. [DOI] [PubMed] [Google Scholar]
- 4.Makmur L, Syamsurizal, Tukiran, Achmad SA, Aimi N, Hakim EH, Kitajima M, Takayama H. J Nat Prod. 2000;63:243–244. doi: 10.1021/np990220u. [DOI] [PubMed] [Google Scholar]
- 5.Suhartati T, Achmad SA, Aimi N, Hakim EH, Kitajima M, Takayama H, Takeya K. Fitoterapia. 2001;72:912–918. doi: 10.1016/s0367-326x(01)00343-4. [DOI] [PubMed] [Google Scholar]
- 6.Wang YH, Hou AJ, Chen L, Chen D-F, Sun HD, Zhao QS, Bastow KF, Nakanish Y, Wang XH, Lee KH. J Nat Prod. 2004;67:757–761. doi: 10.1021/np030467y. [DOI] [PubMed] [Google Scholar]
- 7.Ko HH, Lu YH, Yang SZ, Won SJ, Lin CN. J Nat Prod. 2005;68:1692–1695. doi: 10.1021/np050287j. [DOI] [PubMed] [Google Scholar]
- 8.Gill MT, Bajaj R, Chang CJ, Nichols DE, McLaughlin JL. J Nat Prod. 1987;50:36–40. doi: 10.1021/np50049a006. [DOI] [PubMed] [Google Scholar]
- 9.Likhitwitayawuid K, Sritularak B. J Nat Prod. 2001;64:1457–1459. doi: 10.1021/np0101806. [DOI] [PubMed] [Google Scholar]
- 10.Su BN, Cuendet M, Hawthorne ME, Kardono LBS, Riswan S, Fong HHS, Mehta RG, Pezzuto JM, Kinghorn AD. J Nat Prod. 2002;65:163–169. doi: 10.1021/np010451c. [DOI] [PubMed] [Google Scholar]
- 11.Roberti M, Pizzirani D, Simoni D, Rondanin R, Baruchello R, Bonora C, Buscemi F, Grimaudo S, Tolomeo M. J Med Chem. 2003;46:3546–3554. doi: 10.1021/jm030785u. [DOI] [PubMed] [Google Scholar]
- 12.Suhartati T, Yandri, Hadi S. Eur J Sci Res. 2008;23:330–337. [Google Scholar]
- 13.Hano Y, Inami R, Nomura T. Heterocycles. 1990;31:2173–2179. [Google Scholar]
- 14.Hano Y, Inami R, Nomura T. Heterocycles. 1993;35:1341–1350. [Google Scholar]
- 15.Chung MI, Ko HH, Yen MH, Lin CN, Yang SZ, Tsao LT, Wang JP. Helv Chim Acta. 2000;83:1200–1204. [Google Scholar]
- 16.Ko HH, Lin CN, Yang SZ. Helv Chim Acta. 2000;83:3000–3005. [Google Scholar]
- 17.Ko HH, Yang SZ, Lin CN. Tetrahedron Lett. 2001;42:5269–5270. [Google Scholar]
- 18.Lu YH, Lin CN, Ko HH, Yang SZ, Tsao LT, Wang JP. Helv Chim Acta. 2002;85:1626–1632. [Google Scholar]
- 19.Lu YH, Lin CN, Ko HH, Yang SZ, Tsao LT, Wang JP. Helv Chim Acta. 2003;86:2566–2572. [Google Scholar]
- 20.Kinghorn AD, Carcache-Blanco EJ, Chai HB, Orjala J, Farnsworth NR, Soejarto DD, Oberlies NH, Wani MC, Kroll DJ, Pearce CJ, Swanson SM, Kramer RA, Rose WC, Fairchild CR, Vite GD, Emanuel S, Jarjoura D, Cope FO. Pure Appl Chem. 2009;81:1051–1063. doi: 10.1351/PAC-CON-08-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mannila E, Talvitie A, Kolehmainen E. Phytochemistry. 1994;33:813–816. [Google Scholar]
- 22.Fujioka T, Kashiwada Y, Kilkuskie RE, Cosentino LM, Ballas LM, Jiang JB, Janzen WP, Chen IS, Lee KH. J Nat Prod. 1994;57:243–247. doi: 10.1021/np50104a008. [DOI] [PubMed] [Google Scholar]
- 23.Fujimoto Y, Zhang XX, Kirisawa M, Uzawa J, Sumatra M. Chem Pharm Bull. 1990;38:1787–1789. [Google Scholar]
- 24.Aida M, Shinomiya K, Hano Y, Nomura T. Heterocycles. 1993;36:575–583. [Google Scholar]
- 25.Ma CM, Cai SQ, Cui JR, Wang RQ, Tu PF, Hattori M, Daneshtalab M. Eur J Med Chem. 2005;40:582–589. doi: 10.1016/j.ejmech.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Hano Y, Aida M, Shiina M, Nomura T, Kawai T, Ohe H, Kagei K. Heterocycles. 1989;29:1447–1453. [Google Scholar]
- 27.Lightner DA, Bouman TD, Gawronski JK, Gawronska K, Chappuis JL, Crist BV, Hansen AE. J Am Chem Soc. 1981;103:5314–5327. [Google Scholar]
- 28.The systematic names for 1–3 are based on a numbering scheme published in reference 7.
- 29.Shieh WL, Lin CN. Phytochemistry. 1992;31:364–367. [Google Scholar]
- 30.Ferrari F, Messana I, Mesquita de Araujo MC. Planta Med. 1989;55:70–72. doi: 10.1055/s-2006-961830. [DOI] [PubMed] [Google Scholar]
- 31.Chen CC, Huang YL, Ou J-C, Lin CF, Pan TM. J Nat Prod. 1993;56:1594–1597. [Google Scholar]
- 32.Achmad SA, Hakim EH, Juliawaty LD, Makmur L, Suyatno, Aimi N, Ghisalberti EL. J Nat Prod. 1996;59:878–879. [Google Scholar]
- 33.The systematic name for 4 is based on a numbering scheme published in reference 6.
- 34.The systematic name for 5 is based on a numbering scheme published in reference 15.
- 35.Seo EK, Kim NC, Mi QW, Chai HB, Wall ME, Wani MC, Navarro HA, Burgess JP, Graham JG, Cabieses F, Tan GT, Farnsworth NR, Pezzuto JM, Kinghorn AD. J Nat Prod. 2001;64:1483–1485. doi: 10.1021/np0103158. [DOI] [PubMed] [Google Scholar]
- 36.Deng Y, Balunas MJ, Kim J-A, Lantvit DD, Chin YW, Chai H-B, Sugiarso S, Kardono LBS, Fong HHS, Pezzuto JM, Swanson SM, Carcache de Blanco EJ, Kinghorn AD. J Nat Prod. 2009;72:1165–1169. doi: 10.1021/np9001724. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.