Abstract
The glutamate transporter excitatory amino acid carrier 1 (EAAC1) catalyzes the co-transport of three Na+ ions, one H+ ion, and one glutamate molecule into the cell, in exchange for one K+ ion. Na+ binding to the glutamate-free form of the transporter generates a high affinity binding site for glutamate and is thus required for transport. Moreover, sodium binding to the transporters induces a basal anion conductance, which is further activated by glutamate. Here, we used the [Na+] dependence of this conductance as a read-out of Na+ binding to the substrate-free transporter to study the impact of a highly conserved amino acid residue, Thr101, in transmembrane domain 3. The apparent affinity of substrate-free EAAC1 for Na+ was dramatically decreased by the T101A but not by the T101S mutation. Interestingly, in further contrast to EAAC1WT, in the T101A mutant this [Na+] dependence was biphasic. This behavior can be explained by assuming that the binding of two Na+ ions prior to glutamate binding is required to generate a high affinity substrate binding site. In contrast to the dramatic effect of the T101A mutation on Na+ binding, other properties of the transporter, such as its ability to transport glutamate, were impaired but not eliminated. Our results are consistent with the existence of a cation binding site deeply buried in the membrane and involving interactions with the side chain oxygens of Thr101 and Asp367. A theoretical valence screening approach confirms that the predicted site of cation interaction has the potential to be a novel, so far undetected sodium binding site.
Keywords: Glutamate, Neurotransmitter Transport, Pre-steady-state Kinetics, Site-directed Mutagenesis, Sodium Transport
Introduction
Glutamate transporters belong to SLC1 (solute carrier family 1), which consists of glutamate transporters and neutral amino acid transporters (1). One of the major functions of glutamate transporters is to remove the excitatory neurotransmitter glutamate from the synaptic cleft, after it is released from presynaptic neurons, to prevent its concentration from reaching neurotoxic levels (2). Mammalian glutamate transporters consist of five members, excitatory amino acid transporter 1 (EAAT1 or GLAST),3 EAAT2 (GLT-1), EAAT3 (EAAC1 or excitatory amino acid carrier 1), EAAT4, and EAAT5 (3–7). According to the stoichiometry of these transporters, which co-transport three Na+ ions and one proton with one glutamate into the cell and counter-transport one K+ out of the cell (8, 9), transport of glutamate is electrogenic (5, 10, 11), namely, two positive charges are moved into the cell during each transport cycle. In addition to this stoichiometric, or coupled current, glutamate transporters also catalyze a sodium-dependent anion conductance, which is further activated by the substrates of the transporter (4). This anion conductance is specific for hydrophobic anions and is uncoupled from substrate transport itself, a process unaffected by the absence of chloride from both sides of the membrane (12). Glutamate transport is a sequential process where the co-transport of glutamate with three Na+ and a proton is followed by the counter-transport of K+ to reset the empty transporter in the outward-facing conformation (13–15) (see Fig. 1A). It is thought that the anion conductance is associated with specific states in the transport cycle, in particular those that are populated during the Na+/glutamate half-cycle (16–19). Bulky glutamate analogues, such as d,l-threo-β-benzoylaspartate (TBOA), can bind to the transporter but are too large to be transported and therefore “lock” the transporter in an outward-facing conformation. These analogues are not only competitive inhibitors of the two types of substrate-induced current but also inhibit the basal sodium-dependent anion conductance (20, 21).
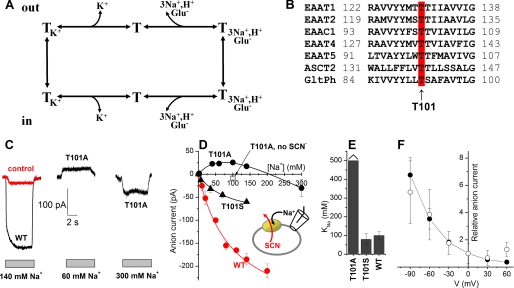
FIGURE 1.
Mutation of Thr101 to alanine interferes with the ability of Na+ to induce anion current. A, illustration of the transport cycle. B, multiple sequence alignment of EAATs and GltPh shows that Thr101 (red, arrow) is conserved within the glutamate transporter family. C, typical leak anion currents induced by Na+ application to EAAC1WT in the absence of external substrate (left panel, black), nontransfected control cells (left panel, red), and EAAC1T101A (middle and right panels). The gray bars indicate the length of Na+ application. The extracellular solution contained MES as the anion and Na+/NMG+. The intracellular solution contained 140 mm SCN− and 10 mm glutamate (see inset in C). D, [Na+] dependence of specific leak anion currents (red circles, WT; black circles, T101A; black triangles, T101S, control responses were subtracted; open square, control response in the absence of SCN−). The lines represent fits to the Michaelis-Menten equation (WT, T101S) or a sum of two Michaelis-Menten equations (T101A). E, KNa+ values determined from the fits. F, voltage dependence of the 60 mm Na+-induced apparent outward current (T101A, open circles) indicates that it is mediated by inhibition of a tonic inward anion conductance. For comparison, inhibition of the tonic leak anion conductance of EAAC1WT by TBOA as a function of the voltage is shown as the solid circles.
The identification of cation binding sites is a key requirement for our understanding of the coupling of substrate transport to Na+ transport in Na+-driven, secondary active transporters. Recently, a crystal structure of GltPh (aspartate transporter from Pyrococcus horikoshii) in complex with two Tl+ ions was published (22). Functional studies indicate that, like in the mammalian counterparts, transport of one aspartate molecule by GltPh is driven by co-transport of at least two Na+ ions (23). Although substrate transport could not be measured in the presence of Tl+, it was proposed that the two Tl+ ions bind to sites (termed Tl1 and Tl2 sites) that also bind Na+ (22). This proposal was supported by the fact that mutation of one of the amino acids coordinating Tl1, Asp405, to asparagine altered the experimentally observed Na+:Asp coupling ratio of 2:1 to a coupling ratio of 1.5:1 (22). This change in coupling ratio caused by the D405N mutation, which is equivalent to the EAAC1 D454N mutation (24), was interpreted as partial coupling of the Na+ bound to the Tl1 site to aspartate binding (22). Binding of Na+ to the Tl1 and Tl2 sites is also supported by computational studies (25, 26) and a valence scanning analysis (27). Moreover, a recent study on EAAC1 indicates that the conserved aspartate from the Tl1 site participates in an overlapping sodium and potassium binding site (28).
Functional studies have identified a highly conserved aspartate residue in TMD7, Asp367 (29), which upon mutagenesis to asparagine led to a >16-fold reduction in sodium affinity (24). In contrast to this dramatic effect of the D367N mutation, the D454N exchange did not reduce the Na+ affinity of EAAC1 (24), whereas both mutations potently reduced the affinity of EAAC1 for Tl+ (30). Based on these data, it was hypothesized that an additional Na+ binding site exists that is buried deeply in the membrane, which involves the side chain of Asp367 (24). Analyses of the crystal structure of GltPh and homology models of EAAC1 built from this structure reveal that another conserved amino acid side chain, Thr101 of EAAC1, protrudes into a hydrophilic cavity also occupied by the Asp367 side chain. Therefore, it is possible that the Thr101 hydroxy oxygen also contributes to coordinating a cation.
In this work, we have tested this hypothesis by studying the effects of mutations to residue Thr101 in EAAC1. Thr101 is highly conserved among all SLC1 family members (see Fig. 1B). The apparent affinity of the glutamate-free/bound form of the transporter for Na+ ions was determined, showing a drastic increase in the Km of the empty transporter for Na+ upon mutating Thr101 to alanine. Furthermore, the data showed that two Na+ ions bind to the glutamate-free, empty form of the T101A transporter. These results suggest the existence of an additional cation binding site not observed in the GltPh crystal structure, which is in agreement with calculations from a valence mapping approach.
MATERIALS AND METHODS
Molecular Biology and Transient Expression
Wild type EAAC1 cloned from rat retina was subcloned into pBK-CMV (Stratagene) as described previously (20) and was used for site-directed mutagenesis according to the QuikChange protocol (Stratagene, La Jolla, CA) as described by the supplier. The primers for mutation experiments were obtained from the DNA core lab of the Department of Biochemistry at the University of Miami School of Medicine. The complete coding sequences of mutated EAAC1 clones were subsequently sequenced. Wild type and mutant EAAC1 constructs were used for transient transfection of subconfluent human embryonic kidney cell (HEK293T/17, ATCC number CRL 11268) cultures using FuGENE 6 transfection reagent (Roche Applied Science) according to the supplied protocol. Electrophysiological recordings were performed between days 1 and 3 post-transfection.
For uptake experiments, the C-terminal histidine-tagged version of rabbit EAAC1 (WT) in the vector pBluescript was used as a parent vector for site-directed mutagenesis (31, 32). This was followed by subcloning of the mutation into the WT construct residing in the oocyte expression vector pOG1 (49), using the unique restriction enzymes MluI and PflMI. The subcloned DNA fragment was sequenced between these unique restriction sites.
Uptake Assays
HeLa cells were cultured (33), infected with the recombinant vaccinia/T7 virus vTF7–3 (34), and transfected with the plasmid DNA harboring the WT or Thr101 mutants or with the plasmid vector alone (33). Transport of l-[3H]aspartate was done as described (32). l-[3H]Aspartate was used rather than l-[3H]glutamate, because of the low background values obtained in HeLa cells transfected with the vector alone. Briefly, HeLa cells were plated on 24-well plates and washed with transport medium containing 150 mm NaCl, and 5 mm potassium phosphate, pH 7.4. Each well was then incubated with 200 μl of transport medium supplemented with 0.4 μCi of l-[3H]aspartate and incubated for 3 min (because transport as a function of time is linear up to 3 min), followed by washing, solubilization of the cells with SDS, and scintillation counting. To determine the rate dependence of transport on the sodium ion concentration, NaCl was replaced by choline chloride to a total concentration of 150 mm, during both washing and transport.
Electrophysiology
Glutamate-induced EAAC1 currents were recorded with an Adams & List EPC7 amplifier under voltage clamp conditions in the whole cell current recording configuration (20). The typical resistance of the recording electrode was 2–3 MΩ; the series resistance was 5–8 MΩ. Because the glutamate-induced currents were small (typically <500 pA), series resistance (RS) compensation had a negligible effect on the magnitude of the observed currents (<4% error). Therefore, RS was not compensated. If not otherwise noted, the extracellular solution contained 140 mm NaCl or 140 mm NaMES, 2 mm CaCl2, 2 mm MgCl2, and 30 mm HEPES (pH 7.4/NaOH). Two different pipette solutions were used depending on whether mainly the noncoupled anion current (with thiocyanate) or the coupled transport current (with MES−) was investigated (18). These solutions contained 140 mm KSCN or KMES, 2 mm MgCl2, 10 mm EGTA, and 10 mm HEPES (pH 7.4/KOH). Thiocyanate was used because it enhances glutamate transporter-associated currents and allows the detection of the EAAC1 anion conducting mode (35).
For the electrophysiological investigation of the Na+/glutamate homoexchange mode, the pipette solution contained 140 mm NaMES or NaSCN, 2 mm MgCl2, 10 mm EGTA, 10 mm glutamate, and 10 mm HEPES (pH 7.4/NaOH). In this transport mode, a constant intracellular concentration of 140 mm Na+ is used, and concentrations of glutamate are used on the intra- and extracellular sides that saturate their respective binding site. The apparent affinity of EAAC1 for cytoplasmic glutamate is 280 μm (19). Therefore, we used 10 mm cytoplasmic glutamate to ensure saturation of the intracellular binding site. In contrast, the apparent affinity for extracellular glutamate is ∼6 μm (20), and 100–200 μm extracellular glutamate is sufficient for saturation of this binding site. Initially, glutamate was omitted from the extracellular solution. Under these conditions, the high intracellular Na+ and glutamate concentrations will drive the majority of glutamate binding sites to face the extracellular side. Subsequently, application of extracellular glutamate resulted in a redistribution of these binding sites to reach a new steady state. According to the nature of the exchange mode, it is not associated with steady-state transport current. However, transient transport currents were observed when glutamate was applied to the extracellular side of the membrane, because of the electrogenic redistribution of Na+ and glutamate binding sites within the membrane (18). When permeating anions, such as SCN−, are present, establishment of homoexchange conditions leads to the permanent activation of an anion current (18, 36). This permanent anion current was used here as a tool to study the behavior of mutant transporters in the homoexchange mode. If not stated otherwise, the pH of the solutions was 7.4.
The currents were low pass filtered at 1–10 kHz (Krohn-Hite 3200) and digitized with a digitizer board (Axon, Digidata 1200) at a sampling rate of 10–50 kHz, which was controlled by software (Axon PClamp). All of the experiments were performed at room temperature.
Laser Pulse Photolysis and Rapid Solution Exchange
Rapid solution exchange was performed as described previously (20). Briefly, substrates were applied to the EAAC1-expressing cell by means of a quartz tube (opening diameter, 350 μm) positioned at a distance of ∼0.5 mm to the cell. The linear flow rate of the solutions emerging from the opening of the tube was ∼5–10 cm/s, resulting in typical rise times of the whole cell current of 30–50 ms (10–90%). Laser pulse photolysis experiments were performed according to previous studies (37, 38). 4-Methoxy-7-nitroindolinyl-caged glutamate (39) (Tocris) in concentrations of 1–4 mm or free glutamate were applied to the cells, and photolysis of the caged glutamate was initiated with a light flash (351 nm, diode-pumped Nd:YAG-laser, frequency-tripled; Continuum). The light was coupled into a quartz fiber (diameter, 365 μm) that was positioned at a distance of 300 μm from the cell. The laser energy was adjusted with neutral density filters (Andover Corp.). With maximum light intensities of 500–600 mJ/cm2, saturating glutamate concentrations could be released, which was tested by comparison of the steady-state current with that generated by rapid perfusion of the same cell with a glutamate concentration that saturated the transporter under study. In rare cases HEK293 cells respond to the laser flash with small transient current responses (<10 pA) in the absence of caged compound. Such cells were discarded.
Valence Mapping
To test for potential cation binding sites, we performed valence mapping according to the procedure developed by Page and Di Cera (40). We used the Vale program for screening the GltPh crystal structure for coordinates with appropriate valences (41). Valences between 0.8 and 1.2 were then mapped onto the GltPh structure file (Protein Data Bank entry 1XFH) (42). After candidates were found, the theoretical valence for the cation binding site was calculated using the following equation.
Here, Ri is the distance between the cation and a neighboring oxygen atom, and R0 is the value of Ri for νCation+ = 1. R0 and N are empirical values for cation-oxygen pairs, which depend on the identity of the bound cation. A suitable monovalent cation binding site is expected to have a valence of close to 1 with a R0 of 1.622 Å and n = 4.29 (Na+-O pair) or with R0 = 2.276 Å and n = 9.1 (K+-O pair) (40). According to this procedure, the valence of the proposed cation+ binding site was calculated as 1.17 for K+ and 0.59 for Na+.
Data Analysis
Nonlinear regression fits of experimental data were performed with Origin (OriginLab, Northampton, MA) or Clampfit (pClamp8 software; Axon Instruments, Foster City, CA). Pre-steady-state currents were fitted with sums of two or three exponential terms. Dose-response relationships of currents were fitted with a Michaelis-Menten-like equation, yielding Km and Imax. The errors in kinetic parameters are given as standard deviations and were determined from at least four independent experiments from at least three different cells.
RESULTS
The Apparent Affinity of the Transporter for Na+ Ion(s) Is Decreased by the T101A Mutation
The apparent affinity of the glutamate-free form of EAAC1T101A was detected by using the [Na+]-induced anion current as an assay, as detailed in Ref. 24. Anion currents induced by application of different [Na+] to EAAC1T101A-expressing HEK293 cells were measured with 140 mm NaSCN and 10 mm glutamate in the pipette solution (Vm = 0 mV) to ensure that the cation binding sites are exposed to the extracellular side (in the absence of external glutamate). Typical current recordings are shown in Fig. 1C. As in EAAC1WT, the Na+-induced anion current in EAAC1T101A was strongly dependent on the sodium concentration, as shown in Fig. 1C. However, whereas the anion current induced by Na+ was always inwardly directed in EAAC1WT (because of SCN− outflow), apparent outward current was observed in EAAC1T101A at [Na+] below 200 mm, which reverted to the expected inward current at higher [Na+] (Fig. 1, C and D) in a biphasic nature. Outward current was never observed in the absence of the permeant anion SCN− (Fig. 1D, open square) and in control cells (Fig. 1C, left panel, red trace), suggesting that this current component is anion current specifically carried by the mutant transporter.
As described previously (24), the [Na+] dependence of the EAAC1WT anion current was well described with a simple Michaelis-Menten-type equation. In contrast, the EAAC1T101A data required a sum of two Michaelis-Menten terms for a satisfactory fit of the biphasic curve, suggesting the existence of two separate Na+ binding processes. The apparent dissociation constant of the low affinity Na+ association process could not be determined precisely, because of our inability to apply [Na+] higher than 300 mm, but it can be estimated as KNa > 500 mm (Fig. 1, D and E). The KNa for the higher affinity step was estimated to 110 mm from a fit of a sum of two Michaelis-Menten equations to the data. For wild type EAAC1, the KNa was ∼90 mm (Fig. 1, D and E), indicating that one Na+ binds to EAAC1T101A with similar affinity as to the wild type transporter, whereas binding of a second Na+ ion is severely impaired in its affinity.
In contrast to T101A, the more conservative T101S mutation had little effect on the [Na+] dependence of the leak anion current (KNa = 105 ± 10 mm), although the amplitude of the leak current was reduced compared with EAAC1WT (Fig. 1, D and E). No apparent outward current was observed in EAAC1T101S. The KNa data are summarized in Fig. 1E.
The apparent outward current induced by [Na+] lower than 200 mm in EAAC1T101A (Fig. 1, C and D) is most likely due to the inhibition of a tonic outflow of SCN− in the empty transporter by binding of one Na+ ion. Consistent with this hypothesis, the voltage dependence of 100 mm [Na+]-induced current in EAAC1T101A showed an enhanced effect at more negative membrane potential but the absence of reversal at positive potentials (Fig. 1F). At −90 mV, the apparent outward current was 57 ± 24 pA, whereas at +30 mV the current was only 7 ± 5 pA. This result was expected if this current is caused by the inhibition of outflow of SCN−, as shown by the similar I-V relationship of inhibition of the Na+-induced anion current (Fig. 1F) by TBOA, which is a nontransportable inhibitor (43). TBOA and other competitive blockers are known to block the anion conductance of glutamate transporters (16, 20).
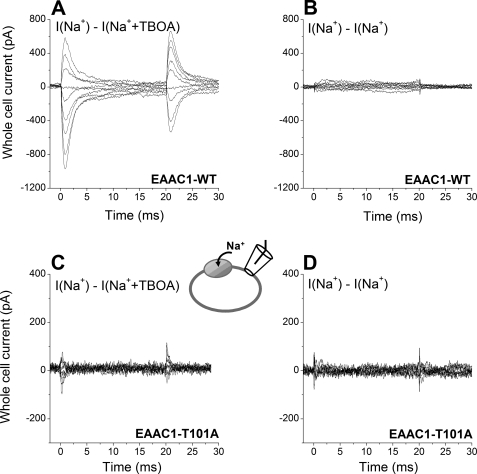
It is known that the binding of sodium ions or conformational changes associated with it can induce charge movement in EAAC1 (11, 18). This charge movement can be blocked in the presence of TBOA. At 140 mm Na+, the Na+ binding site should be approximately half-saturated at 0 mV (44). Thus, voltage-induced charge movements should be large at this [Na+]. Consistent with this hypothesis, large TBOA-sensitive charge movements were observed in response to voltage jumps applied to HEK293 cells expressing EAAC1 wild type transporters (Fig. 2A). To exclude a change in membrane capacitance during the voltage jump protocol, the voltage jump-induced transient currents measured before and after TBOA application were subtracted from each other, showing no difference (Fig. 2, B and D). Transient currents were also absent in control, nontransfected cells. In contrast to EAAC1WT, little TBOA-sensitive charge movement was detected in EAAC1T101A-expressing cells (Fig. 2C; little charge movement can be induced even by a driving force as high as −150 mV). This result is consistent with a low saturation of at least one Na+ binding site, because the Na+ concentration (140 mm) is far below the apparent KNa for Na+ binding. This indicates that the affinity of the glutamate-free form of EAAC1T101A for Na+ is very low, consistent with the result from the [Na+] jump experiment described above.
FIGURE 2.
Na+-dependent transient currents in response to voltage jumps are abolished by the T101A mutation. A, EAAC1-specific transient currents measured in EAAC1WT in response to voltage jumps at time 0 from 0 mV to voltages ranging from +60 to −100 mV in 20-mV increments. I(Na+) − I(Na+ + TBOA) represents the current in 140 mm extracellular Na+, from which the current in the presence of 140 mm Na+ + 0.1 mm TBOA (a saturating concentration) was subtracted. TBOA blocks only the EAAC1-specific component of the current response. The extracellular solution contained 140 mm NaMES. The intracellular solution contained 140 mm NaMES and 10 mm glutamate. B, to test for changes in membrane capacitance during the voltage jump protocol, currents were measured before and after TBOA application. Upon subtraction from each other (I(Na+) − I(Na+), these currents show no difference. C and D, similar experiments as in A and B for EAAC1T101A.
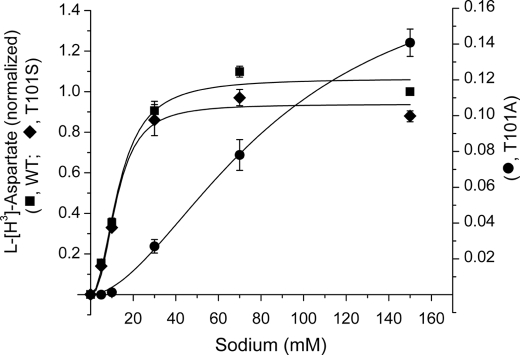
We next tested the effect of [Na+] on substrate uptake, as shown in Fig. 3. In agreement with the effect of the T101A mutation on Na+ affinity predicted by the above experiments, EAAC1T101A required significantly higher [Na+] to activate l-aspartate uptake than the wild type transporter (Fig. 3), whereas the T101S mutation had little effect on [Na+] dependence of uptake. The positive cooperativity observed, with nH values of 2.42 ± 0.55, 1.78 ± 0.15, and 2.13 ± 0.42 for T101A, T101S, and WT, respectively (n = 3), is consistent with the requirement of three sodium ions/substrate molecule transported (8, 9).
FIGURE 3.
Sodium concentration dependence of l-[3H]aspartate transport. The rates of transport of EAAC1WT (squares), EAAC1T101S (diamonds), and EAAC1T101A (circles), expressed in HeLa cells, on the sodium concentration were measured as described under “Experimental Procedures” after subtraction of the values for HeLa cells transfected with the vector alone. All of the values, including those of the T101A mutant (right ordinate; note the different scale caused by the lower uptake of the mutant), are normalized to those of EAAC1WT at 150 mm NaCl and are the averages ± S.E. of three different experiments each done in quadruplicate.
Affinities of the Glutamate-bound Transporter for Glutamate and Na+
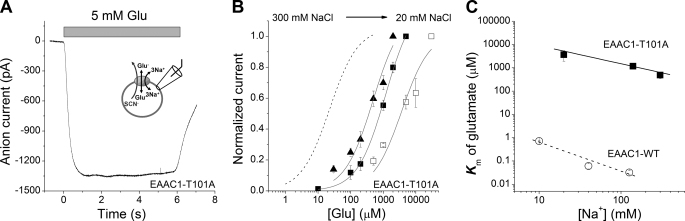
In addition to the two Na+ ions binding to the empty transporter, it was proposed that a third Na+ ion associates with the glutamate-bound form of EAAC1 (18). The apparent affinity of EAAC1 for this third sodium can be determined when saturating glutamate concentration is used. These conditions lead to a shift in the Na+ binding equilibrium of the empty transporter to the fully Na+-bound form, thus eliminating the effects of these initial Na+ binding steps on the [Na+] dependence of the current. To determine which glutamate concentration is saturating, we first measured the apparent dissociation constant (Km) of the transporter for glutamate as a function of [Na+], as shown in Fig. 4. Whole cell anion currents were recorded in the exchange mode with 140 mm NaSCN and 10 mm glutamate in the pipette solution, Vm = 0 mV. Fig. 4A shows a typical current induced by application of 5 mm glutamate to a EAAC1T101A-expressing HEK293T cell. The current-[glutamate] relationships of EAAC1T101A at 20, 140, and 300 mm [Na+] are shown in Fig. 4B. At 140 mm [Na+], the Km of EAAC1T101A for glutamate was 1.2 ± 0.1 mm (n = 3). This value is ∼160-fold higher than that of wild type EAAC1. When [Na+] was raised, the Km of EAAC1T101A for glutamate decreased (Fig. 4, B and C), as expected if Na+ binding to the empty transporter is required for the generation of a high affinity glutamate binding site. The Km-[Na+] relationship was linear on a log scale, as shown in Fig. 4C. This is consistent with the result obtained previously with EAAC1WT and EAAC1D367N. According to this Km-[Na+] relationship, 5.3 m [Na+] would be needed in order for the Km of EAAC1T101A for the substrate to become similar to that of wild type transporter at 140 mm [Na+] (Km = 7.2 μm), consistent with the idea that a lack of saturation of the initial Na+ binding site(s) reduces the apparent affinity for the substrate.
FIGURE 4.
Increased occupancy of the Na+ binding site(s) increases the glutamate affinity of EAAC1. A, typical anion current in response to 5 mm glutamate applied to EAAC1T101A in the exchange mode (see inset). B, normalized glutamate dose-response curves as a function of extracellular [Na+] (300 mm (▴), 140 mm (■), and 20 mm (□)) in exchange mode. The dotted line represents the wild type curve at 140 mm Na+. The currents were normalized to the response at 30 mm glutamate (20 mm Na+), 5 mm glutamate (140 mm Na+), and 2 mm glutamate (300 mm Na+). The extracellular solution contained MES as the anion and the indicated Na+ concentration supplemented with NMG+ to 300 mm. The composition of the internal solution was 140 mm NaSCN and 10 mm glutamate, and the transmembrane potential was 0 mV. C, [Na+] dependence of the Km of EAAC1WT (open circles) and EAAC1T101A (closed squares) for glutamate.
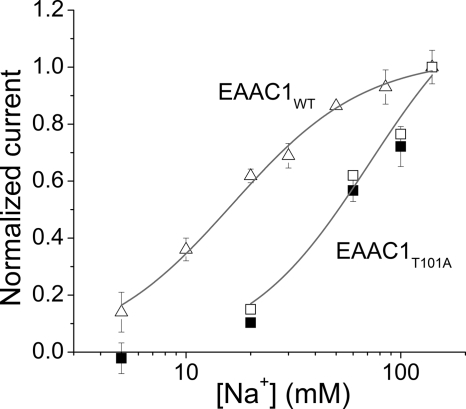
As shown in Fig. 5(closed squares), a saturating glutamate concentration induced anion currents in EAAC1T101A that were highly [Na+]-dependent. The data could be represented by the Hill equation with a Hill co-efficient of n = 1.7 and an apparent dissociation constant of KNa = 61 ± 12 mm. For EAAC1WT, the dissociation constant of Na+ to glutamate-bound form transporter was 18.0 ± 2.0 mm (Fig. 5, triangles), with a Hill co-efficient of 1.4. Therefore, the KNa of EAAC1T101A was approximately three times higher than that of wild type EAAC1, indicating that the effect of the mutation on Na+ binding to the empty transporter is more dramatic than that on Na+ binding to glutamate-bound EAAC1.
FIGURE 5.
Determination of the affinity of the glutamate-bound transporter for Na+. Anion currents were determined as a function of [Na+] at a close to saturating concentration of 30 mm glutamate (EAAC1WT, triangles; EAAC1T101A, closed squares; no currents were induced when the glutamate was substituted by 30 mm MES−). The open squares represent Imax values calculated by extrapolation according to the Km values for glutamate at each [Na+] shown in Fig. 4C. The solid lines represent fits to the Hill equation (fit parameters in main text). All of the experiments were performed in the exchange mode and at 0 mV transmembrane potential. The currents were normalized to the response induced by 140 mm Na+.
The T101A Mutation Slows but Does Not Eliminate Steady-state Glutamate Transport
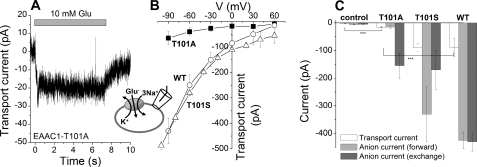
As shown in Fig. 6 (A and C), application of 10 mm glutamate to EAAC1T101A-expressing HEK293T cells (the pipette solution contained KMES, Vhold = 0 mV) generated a small, but significant transport current (−15 ± 2 pA, which is 3.5-fold larger than the current in nontransfected cells). Because there was no permeable anion present, only the coupled transport component of the current should be observed under these conditions. Increasing the electrical driving force enhanced the transport current (Fig. 6B, closed squares). This behavior was very similar to that of wild type EAAC1 (Fig. 6B, open circles), although transport currents in EAAC1WT were on average 6-fold larger than those observed in EAAC1T101A. This result indicates that steady-state substrate transport is inhibited but not eliminated by the T101A mutation.
FIGURE 6.
The T101A mutation inhibits steady-state glutamate transport. A, typical transport current induced by 10 mm glutamate application to EAAC1T101A (Vm = 0 mV; the extracellular solution contained MES− as the anion and 140 mm Na+, and the intracellular solution contained 140 mm KMES). B, transport current-voltage relationships for EAAC1T101A (solid squares), EAAC1T101S (open triangles), and EAAC1WT (open circles). C, average transport currents (white bars) and anion currents in the forward transport mode (light gray bars, 140 mm KSCN internal) and exchange mode (dark gray bars, 140 mm NaSCN, 10 mm glutamate internal) at 0 mV transmembrane potential. The control cells were nontransfected. The glutamate concentration was 10 mm.
To determine which step of the transport cycle is affected by the mutation, we measured steady-state anion current in response to glutamate application. It was previously shown that transporters with mutations impairing the K+-dependent relocation but not Na+-dependent glutamate exchange lack steady-state anion current in the presence of intracellular K+ but catalyze anion flux in the presence of intracellular Na+ and glutamate (exchange conditions) (45). EAAC1T101A shows a similar phenotype (Fig. 6C), consistent with the possibility that K+-dependent relocation is slowed but not abolished in the mutant transporter.
An alternative explanation for the low transport activity would be that the expression level of EAAC1T101A in the cell membrane is much lower than that of wild type EAAC1. To test this possibility, the expression levels of wild type and T101A mutant transporters were compared by isolating the membrane fraction using cell surface biotinylation labeling, followed by Western blotting. The result shows that the expression levels of the wild type and mutant transporters are similar (see supplemental Fig. S1). Thus, reduced cell surface expression is not responsible for the low transport activity in EAAC1T101A. In contrast to T101A, the T101S mutation had little effect on transport current, voltage dependence of transport, and anion current in the exchange and forward transport modes (Fig. 6, B and C).
The T101A Mutation Slows Glutamate Translocation
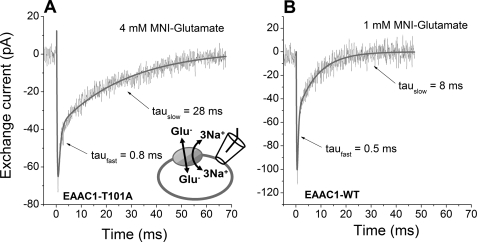
To determine the rate of glutamate translocation, we analyzed pre-steady-state currents in response to glutamate concentration jumps. As shown in Fig. 7A, in exchange transport mode (140 mm NaMES and 10 mm glutamate in pipette solution, without permeable anion SCN−) glutamate released from 4 mm 4-methoxy-7-nitroindolinyl-glutamate by laser pulse photolysis induced a transient inward current. After reaching a peak value of approximately −70 pA, the current decayed to a steady state of 0 pA, because there is no net glutamate transport in the exchange mode. This transient current was similar to that of EAAC1WT (Fig. 7B) in that it decayed with two exponential components, reflecting separate steps in the glutamate translocation process. However, in comparison with wild type EAAC1, the decay steps for EAAC1T101A were slower. In particular the rate-limiting, slowly decaying phase was associated with an average time constant of 26 ± 4 ms, which was approximately three times larger than that of wild type EAAC1 (8.7 ± 1.7 ms). These results show a significant although not dramatic reduction of the exchange rate by the T101A mutation.
FIGURE 7.
EAAC1T101A is active in exchange mode, but with slowed exchange kinetics. A, typical transport current elicited by a glutamate concentration jump at time t = 0, generated by laser flash photolysis of 4 mm 4-methoxy-7-nitroindolinyl-caged glutamate. The intracellular solution contained 140 mm NaMES and 10 mm glutamate, and the extracellular solution contained 140 mm NaMES (Vm = 0 mV). The solid line is a fit with a sum of three exponential terms (time constants for the decaying phases stated; the third phase represents the rising phase, which is most likely related to glutamate binding). B, similar experiment with EAAC1WT, 1 mm 4-methoxy-7-nitroindolinyl-caged glutamate under conditions that match the fractional occupation of the glutamate binding site to that of EAAC1T101A in A.
DISCUSSION
We studied the effects of mutation of a conserved amino acid residue, Thr101, which we hypothesize to contribute its side chain oxygen atom to the coordination of a cation bound to glutamate transporters. The results obtained in this work support this hypothesis for the following reason: the T101A mutation had a dramatic effect on the Na+ apparent affinity of the empty transporter, determined in the absence of glutamate, reducing this apparent affinity ∼10-fold. This result is in agreement with a previous report on mutation of the conserved Asp367 residue (24), whose side chain is close in space to the Thr101 side chain, as predicted by the GltPh crystal structure. The D367N mutation also led to a large reduction of the Na+ apparent affinity of EAAC1 (24).
In addition, the T101A mutation reduced Na+-dependent charge movements in response to voltage jumps, as expected if the Na+ binding site is little occupied at 140 mm extracellular Na+. Like in EAAC1D367N, the effect of the mutation is more pronounced for Na+ binding to the empty transporter than to the glutamate-bound EAAC1. In the latter case, the apparent affinity was only reduced by a factor of ∼3 (Fig. 5). This result is consistent with the idea that the cation binding site equivalent to the Tl2 site of GltPh, which requires bound substrate to be occupied, is located in proximity to the substrate binding site, but several Angstroms removed from the location of the Thr101 mutation. Thus, the effect on Na+ association with the Tl2 site appears to be indirect, as proposed for EAAC1D367N (24).
The Binding of Two Na+ Ions to the Glutamate-free Form of EAAC1T101A Is Required for Anion Conductance Activation
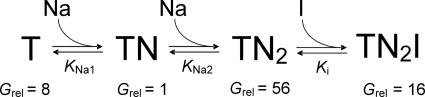
Anion current induced by applying different [Na+] to EAAC1T101A showed biphasic dose dependence (Fig. 1D), a behavior that was not observed in wild type EAAC1 or other mutant transporters studied so far. This biphasic behavior cannot be explained by assuming that only one Na+ binds to the glutamate-free form of the transporter. However, the data can be very well explained by assuming that two Na+ ions bind to the transporter in the absence of glutamate, in agreement with a recent proposal based on fluorescence labeling experiments (46). A kinetic model depicting this binding sequence is shown in Fig. 8, and simulations of anion current according to this model are shown in supplemental Fig. S2, suggesting that binding of two Na+ ions to the glutamate-free transporter is required to form a high affinity substrate binding site. Therefore it is likely that under forward transport conditions the sequence of binding events is as follows: two Na+ ions associate sequentially with the transporter, followed by binding of the transported substrate (Fig. 8). Once the Na2-transporter-Glu complex is formed, a third binding site for Na+ becomes available that must be occupied to allow translocation. This third binding site is possibly analogous to the Tl2 site of GltPh, which is only occupied in the presence of transported substrates but which is not occupied in the presence of nontransportable inhibitors, such as TBOA (22). Why is the biphasic behavior of the [Na+] dependence of the anion current not observed in EAAC1WT? The most likely reason is that Na+ ions associate with EAAC1WT with much higher affinity than with the T101A mutant transporter, as shown in Fig. 8. In fact, simulations of anion current amplitudes as a function of [Na+] demonstrate that the experimental results can be reproduced by assuming a 7–14-fold reduction of Na+ affinity by the T101A mutation (supplemental Fig. S2). However, it should be noted that anion conductance levels are also altered by the T101A mutation (supplemental Fig. S2) and that it cannot be ruled out that the T101A mutation alters the sequence of sodium binding.
FIGURE 8.
Simplified model for the sequence of Na+ and inhibitor binding to EAAC1T101A. Grel denotes the conductance values for the SCN− conductance of each state relative to the TN state, which was arbitrarily set to 1.
Our results obtained with EAAC1T101A give further insights into the mechanism of the anion conductance of the glutamate transporter, which is known to be uncoupled from glutamate transport (35). It is also known that the anion conductance requires the binding of Na+ to the transporter (18) and that it can already be activated in the absence of glutamate (leak anion conductance) (16, 17). According to our proposed sequential binding model deduced from the T101A data (Fig. 8), the anion conductance is modulated as the transporter occupies the several states included in the model, in agreement with previous suggestions (36, 47). The state with only one Na+ ion bound has the lowest anion conductance, whereas full activation of the Na+-dependent anion conductance requires binding of two Na+ ions. According to the data shown in Fig. 1, the Na+-free state must also have residual anion conductance, because binding of one Na+ ion inhibits tonic anion flux-mediated inward current. Thus, it appears that Na+ binding is not absolutely required for the activation of anion conductance, in contrast to a previous proposal (18), but that Na+ binding strongly enhances the anion conductance intrinsic to the transporter.
The T101A Mutation Does Not Eliminate Substrate Transport
Transport current recordings, as well as analysis of aspartate uptake, demonstrate that EAAC1T101A is able to transport glutamate across the cell membrane (Figs. 3 and 6A), although the transport rate is significantly lower than that of the wild type transporter. Analysis of pre-steady-state and steady-state currents suggests that the reduction of the transport rate is caused by both a 3-fold slowing of the Na+-dependent glutamate translocation reaction and inhibition of the K+-dependent relocation process (Fig. 6). For technical reasons we could not determine the affinity of the mutant transporter for potassium. It remains therefore an open question whether Thr101 participates in an overlapping sodium and potassium binding site as has been suggested for the putative Na1 binding site (28). The impact of the T101A mutation is more subtle than that of the D367N mutation, which completely eliminates glutamate transport and dramatically slows translocation (24). Why do the T101A and D367N mutations impair the binding of Na+ to the glutamate-free form of the transporter but have differential effects on glutamate uptake? We propose that negative charge on the side chain of Asp367 and also on other acidic amino acid residues located in the transmembrane domain is absolutely necessary for the full function of EAAC1. Thus, charge-altering mutations, such as D367N, dramatically alter transporter properties. In contrast, the T101A mutation is not expected to result in a substantially different charge distribution in the transmembrane domains, suggesting that the lack of electrostatic effects results in a more subtle phenotype.
Does the Side Chain of Thr101 Contribute to a Cation Binding Site?
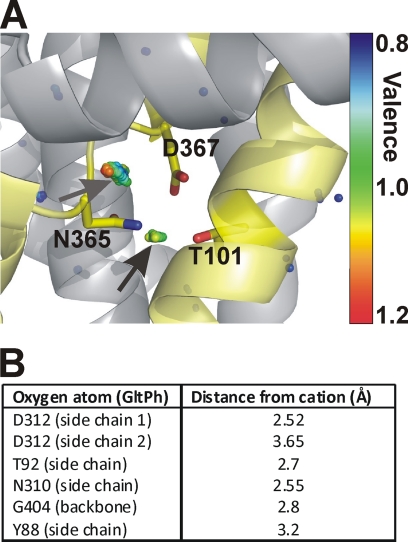
Although indirect effects of the T101A mutation on Na+ binding cannot per se be excluded, mounting evidence points to the existence of a cation binding site constituted by the side chain oxygens of Asp367 and Thr101 and possibly other ligands (main chain or side chain oxygens) that have not yet been identified. In the crystal structure of GltPh, the analogous Asp312 and Thr92 side chains both point toward a hydrophilic pocket. To test whether this pocket has the potential to harbor a cation binding site, we performed a valence screening analysis (40) of the GltPh structure, using the empirical parameters described in Equation 1 for Na+ and K+. Interestingly, we found a point of suitable but not optimal valence for K+ (1.17) in a position that is surrounded by the side chains of Thr92, Asp312, and Asn310 (Fig. 9A), as indicated by the arrow and the colored dots overlaid on the GltPh structure. The Gly404 main chain carbonyl oxygen, as well as the Tyr88 side chain oxygen, also contribute to electrostatic compensation of the positive charge of the cation. The distances of the ligand atoms to the center of this potential cation binding site are listed in Fig. 9B. These distances are in the range expected for an average K+-O pair of 2.8 Å (41). The valence for Na+ at the same position was 0.59. This analysis suggests that this could be a potential cation binding site. However, the site does not have an optimal valence for either Na+ or K+, consistent with the lack of effect of K+ on GltPh-dependent aspartate transport (23), as well as the lack of previous identification of this site through crystallography. Why does the valence screening procedure yield valences that deviate from unity for sodium? A number of possibilities have to be considered in isolation or in combination to explain this effect: 1) GltPh binds only two Na+ ions, whereas EAAC1 binds three. If this is the case, subtle differences in geometry between the GltPh and EAAC1 structures at this site, leading to a closer spacing of the ligands to the center of the site, may result in a more optimal Na+ valence for EAAC1. 2) The cation binding site has a low affinity for cations, consistent with the experimental results showing an apparent Km of 100 mm. 3) A Na+ ligand in form of an additional water molecule is missing in the crystal structure, because of the limited resolution. This ligand would contribute additional electrostatic compensation, increasing the valence for sodium. The last possibility is supported by recent results from molecular dynamics simulations (48).
FIGURE 9.
Valence mapping of the GltPh structure predicts a potential cation binding site with the side chains of Asp367 and Thr101 as ligands. A, valence map for K+ (colored dots) in overlay with the GltPh structure. The dots illustrate points in three-dimensional space that were mapped to a valence of 0.8 (blue) to 1.2 (red) (the valence color code is shown on the right side of the figure). An optimal valence of 1 is color-coded green. Thus, green dots indicate a suitable binding site for K+, as indicated by the arrows. Residues Thr92, Asn310, and Asp312 of GltPh (Thr101, Asn365, and Asp367 in EAAC1) are highlighted as sticks. B, distances of the coordinating oxygen atom to the center of the cation used for valence calculation.
A third cation binding site has also been proposed by Holley and Kavanaugh (27). This site is in the proximity of the substrate binding site. Our mutagenesis data do not allow insight into this other potential cation binding site. While this work was in preparation, a molecular dynamics study suggested a third sodium binding site in the exact same location as the one proposed here (48).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants 2R01NS049335-06A1 (to C. G.) and NS 16708 (to B. I. K.). This work was also supported by Binational Science Foundation Grant 2007051 (to C. G. and B. I. K.) and American Heart Association Grant 0525485B (to Z. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- EAAT
- excitatory amino acid transporter
- EAAC
- excitatory amino acid carrier
- TBOA
- d,l-threo-β-benzoylaspartate
- WT
- wild type
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Kanai Y., Hediger M. A. (2003) Eur. J. Pharmacol. 479, 237–247 [DOI] [PubMed] [Google Scholar]
- 2.Bergles D. E., Diamond J. S., Jahr C. E. (1999) Curr. Opin. Neurobiol. 9, 293–298 [DOI] [PubMed] [Google Scholar]
- 3.Arriza J. L., Eliasof S., Kavanaugh M. P., Amara S. G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4155–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fairman W. A., Vandenberg R. J., Arriza J. L., Kavanaugh M. P., Amara S. G. (1995) Nature 375, 599–603 [DOI] [PubMed] [Google Scholar]
- 5.Kanai Y., Hediger M. A. (1992) Nature 360, 467–471 [DOI] [PubMed] [Google Scholar]
- 6.Pines G., Danbolt N. C., Bjørås M., Zhang Y., Bendahan A., Eide L., Koepsell H., Storm-Mathisen J., Seeberg E., Kanner B. I. (1992) Nature 360, 464–467 [DOI] [PubMed] [Google Scholar]
- 7.Storck T., Schulte S., Hofmann K., Stoffel W. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10955–10959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy L. M., Warr O., Attwell D. (1998) J. Neurosci. 18, 9620–9628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zerangue N., Kavanaugh M. P. (1996) Nature 383, 634–637 [DOI] [PubMed] [Google Scholar]
- 10.Kanner B. I., Sharon I. (1978) Biochemistry 17, 3949–3953 [DOI] [PubMed] [Google Scholar]
- 11.Wadiche J. I., Arriza J. L., Amara S. G., Kavanaugh M. P. (1995) Neuron 14, 1019–1027 [DOI] [PubMed] [Google Scholar]
- 12.Kanner B. I., Sharon I. (1978) FEBS Lett. 94, 245–248 [DOI] [PubMed] [Google Scholar]
- 13.Kanner B. I., Bendahan A. (1982) Biochemistry 21, 6327–6330 [DOI] [PubMed] [Google Scholar]
- 14.Kavanaugh M. P., Bendahan A., Zerangue N., Zhang Y., Kanner B. I. (1997) J. Biol. Chem. 272, 1703–1708 [DOI] [PubMed] [Google Scholar]
- 15.Pines G., Kanner B. I. (1990) Biochemistry 29, 11209–11214 [DOI] [PubMed] [Google Scholar]
- 16.Otis T. S., Jahr C. E. (1998) J. Neurosci. 18, 7099–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wadiche J. I., Kavanaugh M. P. (1998) J. Neurosci. 18, 7650–7661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watzke N., Bamberg E., Grewer C. (2001) J. Gen. Physiol. 117, 547–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watzke N., Grewer C. (2001) FEBS Lett. 503, 121–125 [DOI] [PubMed] [Google Scholar]
- 20.Grewer C., Watzke N., Wiessner M., Rauen T. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9706–9711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otis T. S., Kavanaugh M. P. (2000) J. Neurosci. 20, 2749–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boudker O., Ryan R. M., Yernool D., Shimamoto K., Gouaux E. (2007) Nature 445, 387–393 [DOI] [PubMed] [Google Scholar]
- 23.Ryan R. M., Compton E. L., Mindell J. A. (2009) J. Biol. Chem. 284, 17540–17548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao Z., Zhang Z., Grewer C. (2006) J. Biol. Chem. 281, 10263–10272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu Y., Shrivastava I. H., Amara S. G., Bahar I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2589–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Z., Tajkhorshid E. (2008) Biophys. J. 95, 2292–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holley D. C., Kavanaugh M. P. (2009) Philos. Trans. R. Soc. Lond. B. Biol. Sci. 364, 155–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teichman S., Qu S., Kanner B. I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14297–14302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosental N., Bendahan A., Kanner B. I. (2006) J. Biol. Chem. 281, 27905–27915 [DOI] [PubMed] [Google Scholar]
- 30.Tao Z., Gameiro A., Grewer C. (2008) Biochemistry 47, 12923–12930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunkel T. A., Roberts J. D., Zakour R. A. (1987) Methods Enzymol. 154, 367–382 [DOI] [PubMed] [Google Scholar]
- 32.Pines G., Zhang Y., Kanner B. I. (1995) J. Biol. Chem. 270, 17093–17097 [DOI] [PubMed] [Google Scholar]
- 33.Keynan S., Suh Y. J., Kanner B. I., Rudnick G. (1992) Biochemistry 31, 1974–1979 [DOI] [PubMed] [Google Scholar]
- 34.Fuerst T. R., Niles E. G., Studier F. W., Moss B. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 8122–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wadiche J. I., Amara S. G., Kavanaugh M. P. (1995) Neuron 15, 721–728 [DOI] [PubMed] [Google Scholar]
- 36.Bergles D. E., Tzingounis A. V., Jahr C. E. (2002) J. Neurosci. 22, 10153–10162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grewer C., Madani Mobarekeh S. A., Watzke N., Rauen T., Schaper K. (2001) Biochemistry 40, 232–240 [DOI] [PubMed] [Google Scholar]
- 38.Mim C., Balani P., Rauen T., Grewer C. (2005) J. Gen. Physiol. 126, 571–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Canepari M., Nelson L., Papageorgiou G., Corrie J. E., Ogden D. (2001) J. Neurosci. Methods 112, 29–42 [DOI] [PubMed] [Google Scholar]
- 40.Page M. J., Di Cera E. (2006) Physiol. Rev. 86, 1049–1092 [DOI] [PubMed] [Google Scholar]
- 41.Nayal M., Di Cera E. (1996) J. Mol. Biol. 256, 228–234 [DOI] [PubMed] [Google Scholar]
- 42.Yernool D., Boudker O., Jin Y., Gouaux E. (2004) Nature 431, 811–818 [DOI] [PubMed] [Google Scholar]
- 43.Shimamoto K., Lebrun B., Yasuda-Kamatani Y., Sakaitani M., Shigeri Y., Yumoto N., Nakajima T. (1998) Mol. Pharmacol. 53, 195–201 [DOI] [PubMed] [Google Scholar]
- 44.Mim C., Tao Z., Grewer C. (2007) Biochemistry 46, 9007–9018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grewer C., Watzke N., Rauen T., Bicho A. (2003) J. Biol. Chem. 278, 2585–2592 [DOI] [PubMed] [Google Scholar]
- 46.Koch H. P., Hubbard J. M., Larsson H. P. (2007) J. Biol. Chem. 282, 24547–24553 [DOI] [PubMed] [Google Scholar]
- 47.Tao Z., Grewer C. (2007) J. Gen. Physiol. 129, 331–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang Z., Tajkhorshid E. (2010) in 54th Annual Meeting of the Biophysical Society, February 20–24, 2010, San Francisco, Biophysical Society, Rockville, MD [Google Scholar]
- 49.Borre L., Kanner B. I. (2004) J. Biol. Chem. 279, 2513–2519 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.