Abstract
Peroxisome proliferator-activated receptor γ co-activator-1α (PGC-1α) plays a central role in the regulation of cellular energy metabolism and metabolic adaptation to environmental and nutritional stimuli. We recently described a novel, biologically active splice variant of PGC-1α (NT-PGC-1α, amino acids 1–270) that retains the ability to interact with and transactivate nuclear hormone receptors through its N-terminal transactivation domain. Whereas PGC-1α is an unstable nuclear protein sensitive to ubiquitin-mediated targeting to the proteasome, NT-PGC-1α is relatively stable and predominantly cytoplasmic, suggesting that its ability to interact with and activate nuclear receptors and transcription factors is dependent upon regulated access to the nucleus. We provide evidence that NT-PGC-1α interacts with the nuclear exportin, CRM1, through a specific leucine-rich domain (nuclear export sequence) that regulates its export to the cytoplasm. The nuclear export of NT-PGC-1α is inhibited by protein kinase A-dependent phosphorylation of Ser-194, Ser-241, and Thr-256 on NT-PGC-1α, which effectively increases its nuclear concentration. Using site-directed mutagenesis to prevent or mimic phosphorylation at these sites, we show that the transcriptional activity of NT-PGC-1α is regulated in part through regulation of its subcellular localization. These findings suggest that the function of NT-PGC-1α as a transcriptional co-activator is regulated by protein kinase A-dependent inhibition of CRM1-mediated export from the nucleus.
Keywords: Methods/Microscopic Imaging, Protein/Post-translational Modification, Regulation/Covalent Modification, Signal Transduction/Cyclic Nucleotides/Cyclic AMP, Signal Transduction/Protein Kinases, Transcription/Coactivators, CRM1
Introduction
Many biological programs are regulated at the transcriptional level by interaction of transcription factors with transcriptional co-activators. For example, the peroxisome proliferator-activated receptor γ co-activator-1 (PGC-1)2 family is integral to the coordinated activation of transcription complexes (1, 2) that regulate global responses such as mitochondrial biogenesis (3–7) and tissue-specific responses such as adaptive thermogenesis (3), fatty acid oxidation (8, 9), hepatic gluconeogenesis (10–14), fiber type switching in muscle (15), reactive oxygen species metabolism (16), and clock gene expression (17).
PGC-1α contains a powerful autonomous transactivation domain at its N terminus (aa 1–200) that binds two other co-activators with acetyl transferase activity, SRC-1 and CBP/p300 (18). The leucine-rich motifs located downstream of the transactivation domain allow the co-activator to interact with ligand-activated nuclear receptors, as well as various other transcription factors such as NRF1, Host Cell factor, MEF2C, and FOXO1 (4, 8, 19–22). The central region of PGC-1α (aa 200–400) is required for interaction with p160 myb-binding protein (p160MBP), which acts as a repressor of PGC-1α (23), whereas the C-terminal end of PGC-1α (aa 400–797) contains RNA recognition motifs and arginine/serine-rich domains characteristic of splicing factors (24).
PGC-1α is regulated by signaling inputs from cAMP, calcium, nitro-oxide, and cGMP that induce its expression and/or increase its activity through post-translational modifications (10, 25–27). PGC-1α protein is relatively short-lived (∼2.3 h) due to rapid targeting to ubiquitin/proteasome-mediated proteolysis in the nucleus (28–30). Various post-translational inputs are known to affect proteasomal targeting and stability, most notably p38 MAPK, which phosphorylates PGC-1α on Thr-262, Ser-265, and Thr-298 and enhances its stability (23, 28, 31). In addition, PGC-1α expression and/or stability are also modulated by AMP-dependent phosphorylation at residues Thr-177 and Ser-538 (32), Akt2/protein kinase B-dependent phosphorylation at Ser-570 (33), and glycogen synthase kinase 3β-dependent phosphorylation at Thr-295 (30). In addition to phosphorylation, protein arginine methyltransferase 1-mediated methylation of PGC-1α at residues Arg-665, Arg-667, and Arg-669 activates transcriptional activity (34), whereas acetylation by GCN5 decreases PGC-1α activity (35).
We recently identified a novel, biologically active 270-aa isoform of PGC-1α (NT-PGC-1α) produced by alternative splicing that introduces a premature stop codon between exons 6 and 7 (36). NT-PGC-1α retains the N-terminal transactivation and nuclear receptor interaction domains of PGC-1α and is functionally active (36). In contrast to PGC-1α, NT-PGC-1α is relatively stable and predominantly sequestered in the cytoplasm (36). cAMP triggers nuclear accumulation of NT-PGC-1α and enhances the recruitment of NT-PGC-1α to the promoters of consensus PGC-1α target genes such as UCP1 and CPT1β (36). These findings suggest that nuclear translocation of NT-PGC-1α is a critical regulatory step in its response to cAMP-dependent signaling input, whereas cAMP signaling increases PGC-1α activity in part by stabilizing the protein in the nucleus. Sequestering transcription factors outside the nucleus is a common mode of regulation. For example, NF-κB, FOXO1, c-Fos, and FKHRL1 are retained in the cytosol, and their activity is limited by binding partners that mask the nuclear localization signals or promote nuclear export. Thus, as seen with NT-PGC-1α, signaling input regulates their activity by regulating their nuclear translocation (37–41).
In this report, we investigated how post-translational input from PKA regulates the subcellular localization and function of NT-PGC-1α. We found that NT-PGC-1α interacts with CRM1 through the N-terminal CRM1-binding motif and is actively exported to the cytoplasm. PKA-dependent phosphorylation of NT-PGC-1α on Ser-194, Ser-241, and Thr-256 prevents nuclear export and allows a greater proportion of cellular NT-PGC-1α to remain in the nucleus. Using site-directed mutagenesis to prevent PKA-dependent phosphorylation of these sites, we find a close relationship between the ability of PKA to increase nuclear content of NT-PGC-1α and its ability to enhance transcriptional activation of the UCP1 and CIDEA genes. These findings support PKA-dependent inhibition of CRM1 mediated-NT-PGC-1α nuclear export as the primary mechanism through which PKA regulates the transcriptional activity of NT-PGC-1α. In addition, we show that nuclear NT-PGC-1α regulates genes that are both distinct from (e.g. Cox8b) and common to those that are regulated by PGC-1α. Thus, this PKA-dependent activation of NT-PGC-1α would be important to induction of the complete set of genes that are transcriptionally regulated by PGC-1α isoforms.
EXPERIMENTAL PROCEDURES
Cell Cultures and Adipocyte Differentiation
CHO-K1 cell lines were maintained in culture in F-12K supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Invitrogen) and transfected using FuGENE 6 (Roche Applied Science). An immortalized PGC-1α-deficient brown preadipocyte cell line (42) was a gift from Bruce Spiegelman (Harvard Medical School) and was maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% glutamine, and 1% penicillin/streptomycin. Preadipocytes were grown to confluence in culture medium supplemented with 20 nm insulin and 1 nm T3 (differentiation medium). Differentiation of adipocytes was induced (day 1) by incubating the cells in differentiation medium supplemented with 0.5 mm isobutylmethylxanthine, 0.5 μm dexamethasone, and 0.125 mm indomethacin for 48 h. Thereafter, the cells were maintained in differentiation medium until day 7.
Plasmids
Expression plasmids for pcDNA3.1-NT-PGC-1α-HA were described previously (36). Site-directed mutagenesis of pcDNA3.1-NT-PGC-1α-HA with the QuikChange mutagenesis system (Stratagene, La Jolla, CA) was performed to generate leucine to alanine mutations (NES1, NES2, and NES1/2) in the consensus CRM1 nuclear export sites, and serine/threonine to alanine or aspartate mutations in the PKA phosphorylation sites (Ser-194, Ser-241, and Thr-256). pcDNA3.1-myc-CRM1 and pQE32-HIS6-RanQ69L were kindly provided by S. Meloche (Université de Montréal). To construct retroviral plasmids of pBABE-NT-PGC-1α-HA and pBABE-NT-PGC-1α-S194A/S241A/T256A-HA, BamHI and XhoI fragments of wild-type and mutant NT-PGC-1α were subcloned into a BamHI- and SalI-digested pBABE-neo vector. Gal4-DBD fusions of wild-type and mutant NT-PGC-1α and PGC-1α were generated by subcloning into a pCMV-DB vector. (PPRE)3-TK-luc, pSV sport-RXRα, and pSV sport-PPARγ1 were from Addgene Inc.
Retroviral and Adenoviral Infection
To produce retrovirus, GP-293 cells grown to 80% confluence were co-transfected with pVSV-G and pBABE-neo, pBABE-NT-PGC-1α-HA, or pBABE-NT-PGC-1α-S194A/S241A/T256A-HA by co-precipitation with calcium phosphate (Promega, Madison, WI). Following transfection, the cells were incubated at 32 °C to increase viral titer. Virus-containing medium was collected, filtered through the 0.45-μm low protein binding filter, and used to infect target cells. Immortalized PGC-1α-deficient brown preadipocyte cells (30–40% confluence) were infected with the viral supernatant supplemented with 8 μg/ml Polybrene by a centrifugation method (1800 rpm × 45 min). The medium was aspirated after 2 h and replaced with fresh viral supernatant, and the procedure was repeated. After 8 h of infection, the cells were replaced with fresh Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% glutamine, and 1% penicillin/streptomycin. Selection was initiated with 800 μg/ml G418 (Invitrogen) 48 h after infection.
Adenoviral infection of immortalized PGC-1α-deficient brown preadipocyte or differentiated brown adipocyte cells with NT-PGC-1α-FLAG was conducted using a poly-l-lysine (Mr 30,000–70,000)-assisted transduction procedure (43). Adenoviral supernatants were mixed in Opti-MEM containing poly-l-lysine (0.5 μg/ml) and incubated for 1.5 h at room temperature. Cells were washed with PBS, and the adenoviral mixture was added and the mixture was incubated for 2 h at 37 °C followed by addition of Dulbecco's modified Eagle's medium and incubation overnight. The culture medium was replaced with a fresh medium. Each experiment was performed at 48 h after viral infection.
Immunocytochemistry
Cells on coverslips were washed with 1× PBS, fixed with 4% paraformaldehyde for 10 min, and permeabilized with 0.5% Triton X-100. The fixed cells were first incubated with the blocking buffer (1× PBS, 5% normal goat serum, and 1% bovine serum albumin) and then with a 1:4000 diluted rabbit anti-HA antibody (Abcam) or a 1:1000 diluted mouse anti-FLAG M2 antibody (Sigma) in the same buffer for 2 h. After incubation with primary antibodies, cells were washed three times with 1× PBS and incubated with an Alexa Fluor 488-conjugated anti-rabbit or mouse immunoglobulin secondary antibody for 1 h (Invitrogen). Cells were washed three times with 1× PBS and mounted with Vectashield (Vector Labs). Nuclei were counterstained with 4,6-diamidino-2-phenylindole. Subcellular localization of NT-PGC-1α was examined with a Plan-Neofluar 40×/0.85 numerical aperture objective on a Zeiss LSM510 confocal microscope. The relative intensity of the fluorescence in the nucleus and cytoplasm was quantified using the National Institutes of Health ImageJ program as described before (36, 44, 45). Briefly, the fluorescence signals of NT-PGC-1α in the nucleus and entire cell were measured using the NIH ImageJ in 150–200 (CHO-K1) or 20–60 (brown preadipocytes/adipocytes) randomly selected positively transduced cells, and the nuclear and whole cell areas were recorded. The total nuclear fluorescence intensity was normalized for nuclear area. The total cytoplasmic fluorescence intensity (total cellular fluorescence minus nuclear fluorescence) was normalized for cytoplasmic area (whole cell area minus nuclear area). The relative nuclear/cytoplasmic ratio of fluorescence intensity was determined by dividing the nuclear fluorescence intensity per unit area by the cytoplasmic fluorescence intensity per unit area.
Western Blot and Immunoprecipitation
Cells were harvested, washed with ice-cold PBS, and lysed in 50 mm Tris-HCl, pH 7.5, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride supplemented with a protease inhibitor mixture (Roche Applied Science). Antibodies used were as follows: monoclonal anti-PGC-1α (36), anti-Myc (9E10), and anti-CRM1(H-300) (Santa Cruz Biotechnology), anti-HA (Convance), and anti-tubulin (Abcam). Immunoprecipitations were performed in 20 mm HEPES, pH 7.0, 150 mm NaCl, 0.2% Nonidet P-40 supplemented with protease and phosphatase inhibitors. Lysates were precleared with protein G-agarose beads and immunoprecipitated with antibody-coated beads for 3 h at 4 °C. After washings, immunoprecipitates were subjected to Western blot analysis.
Recombinant GST Fusion Protein Purification and GST Pulldown Assay
Overnight BL21 cultures were diluted and grown in LB+AMP at 30 °C to an A600 of 0.5. Protein expression was induced with isopropyl 1-thio-β-d-galactopyranoside for 3 h at 30 °C. Cells were harvested, washed, resuspended in 1× PBS buffer supplemented with 2.5 mm phenylmethylsulfonyl fluoride, 5 mm dithiothreitol, and a protease inhibitor mixture, lysed by sonication, and treated with 1% Triton X-100. The lysates were then incubated with glutathione beads for 2 h at 4 °C. GST, GST-PGC-1α (1–200 aa), and GST-NT-PGC-1α fusion proteins immobilized on the beads were incubated with the CHO-K1 cell lysates expressing Myc-CRM1 or brown preadipocyte cell lysates for 3 h at 4 °C. After washings, the bound proteins were subjected to Western blot analysis.
[32P]Orthophosphate Metabolic Labeling
CHO-K1 cells transiently expressing pcDNA3.1, NT-PGC-1α-HA, or NT-PGC-1α-S194A/S241A/T256A-HA were washed three times with pre-warmed phosphate-free Dulbecco's modified Eagle's medium containing 1% glutamine, 0.1% bovine serum albumin, and 1% (10 mm) Hepes, pH 7.4, and then incubated in the same phosphate-free medium for 1 h. During the incubation, H89 was added in some experiments. The medium was then replaced with fresh phosphate-free medium containing [32P]orthophosphate (0.25 mCi/ml, PerkinElmer Life Sciences) with the addition of Bt2cAMP (Sigma) alone or Bt2cAMP and H89 (Calbiochem) and incubated for 3 h. Cells were washed three times with ice-cold 0.15 m NaCl solution and lysed in cold lysis buffer (1% Nonidet P-40, 0.15 m NaCl, 20 mm Hepes, pH 7.4) with protease (Roche Applied Science) and phosphatase (10 mm NaF, 0.1 mm Na3VO4) inhibitors. Lysates were precleared with protein G-agarose beads and immunoprecipitated with anti-HA antibody-coated beads for 3 h at 4 °C. The immunoprecipitates were washed eight times with the same buffer and subjected to autoradiography and Western blot analyses.
In Vitro Phosphorylation of NT-PGC-1α by PKA
NT-PGC-1α-HA and its PKA mutants were in vitro transcribed/translated using TnT T7 Coupled Reticulocyte Lysate System (Promega) and immunoprecipitated in immunoprecipitation buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.2% Nonidet P-40, 5 mm MgCl2, and protease inhibitors) with anti-HA antibodies. The immunoprecipitates were washed four times with the same buffer and washed once with the kinase reaction buffer (25 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 2 mm dithiothreitol, 5 mm β-glycerophosphate, 0.1 mm Na3VO4). Kinase reactions were initiated by addition of 100 μm ATP, 10 μCi (0.74 MBq) of [γ-32P]ATP (PerkinElmer Life Sciences), and 1 μg of affinity-purified bovine PKA in a final volume of 40 μl. After 30-min incubation at 30 °C, the immunoprecipitates were washed and subjected to autoradiography and Western blot analyses.
Luciferase Reporter Assay
CHO-K1 cells were transiently transfected with (PPRE)3-TK-luc, pSV sport-RXRα, pSV sport-PPAPγ1, pRL-SV40, and pcDNA3.1/NT-PGC-1α-HA/NT-PGC-1α-S194A/S241A/T256A-HA/NT-PGC-1α-S194D/S241D/T256D-HA using Fugene6 (Roche). 24 h after transfection, cells were treated with vehicle, BRL49653/9-cis-RA or Bt2cAMP. 48h later, cells were harvested for assay of luciferase activity using a Promega Dual Luciferase assay kit (Promega). Firefly luciferase activity was normalized using Renilla luciferase and protein expression. Data represent mean ± S.D. of at least three independent experiments.
Real-time PCR Analysis
Total RNA was isolated from differentiated brown adipocyte cell lines using RNeasy kits (Qiagen). For real-time PCR analysis, RNA samples were reverse transcribed using oligo(dT) primers and M-MLV reverse transcriptase (Promega), and 20 ng of cDNA was used in quantitative PCR reactions in the presence of a fluorescent dye (Cybergreen, Takara) on a Smart Cycler (Cepheid). Relative abundance of mRNA was normalized to cyclophilin mRNA. The following primers were used: UCP1 (36), cyclophilin (36), CIDEA (42), CytC (42), Cox8b (46), and ERRα (46).
RESULTS
CRM1 Promotes Cytoplasmic Translocation of NT-PGC-1α
Proteins of <50 kDa are able to pass through nuclear pores and equilibrate across the nuclear membrane by passive diffusion (47, 48). Although NT-PGC-1α lacks the four consensus nuclear localization sequences identified in PGC-1α (20, 22), its predicted size (∼30 kDa) should not preclude its diffusion through the nuclear pore complex (36). However, our initial hypothesis that the protein would equilibrate across the nuclear membrane was not supported by our finding that NT-PGC-1α was primarily localized to the cytosol (36). Two potential explanations for this unexpected finding are that 1) NT-PGC-1α is retained in the cytoplasm by interaction with a protein localized to the cytosol or 2) NT-PGC-1α is excluded from the nucleus by a specific transport process that translocates the protein to the cytosol. Support for the second possibility became evident upon inspection of NT-PGC-1α and finding two consensus CRM1-binding leucine-rich nuclear export sequences (NES) near the N terminus.
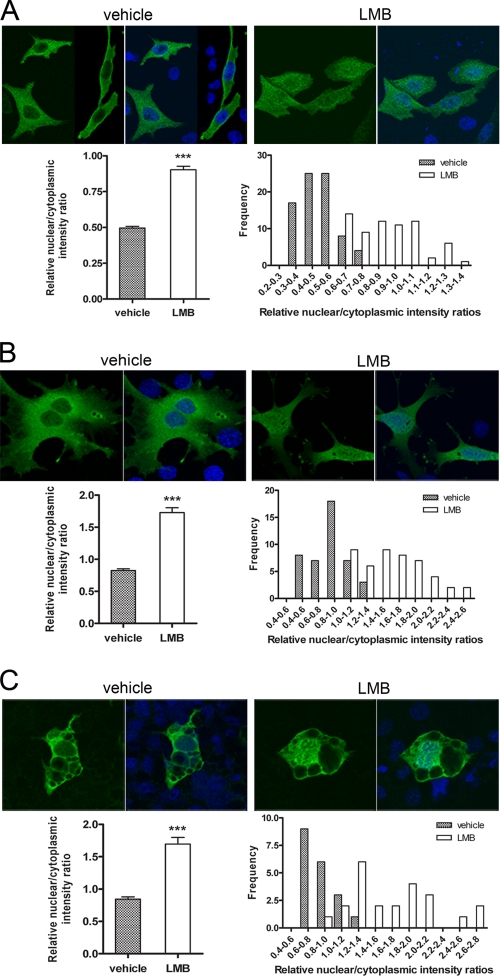
To investigate whether NT-PGC-1α is exported from the nucleus by CRM1, we examined the ability of leptomycin B (LMB), a specific inhibitor of CRM1-dependent nuclear export (49), to affect subcellular distribution of the protein. In CHO-K1 cells expressing NT-PGC-1α-HA, LMB produced a redistribution of NT-PGC-1α within the cell, reducing the cytosolic concentration of the protein while increasing its concentration in the nucleus (Fig. 1A, top panels). This finding supports the hypothesis that CRM1 concentrates NT-PGC-1α in the cytosol by efficiently exporting it from the nucleus. Using confocal microscopy to measure the concentration of NT-PGC-1α in the respective compartments, we found that the nuclear/cytoplasmic ratio of NT-PGC-1α was increased ∼1.8-fold by LMB (Fig. 1A, left bottom panel). By comparing the distribution of nuclear/cytoplasmic intensity ratios in LMB-treated cells, we found a significant increase in the population of cells with higher ratios compared with vehicle-treated cells (Fig. 1A, right bottom panel). We observed a similar response to LMB in brown adipocytes immortalized from PGC-1α null mice and transiently transduced with NT-PGC-1α-HA (Fig. 1, B and C). In both preadipocytes (Fig. 1B) and differentiated adipocytes (Fig. 1C), LMB increased the nuclear/cytoplasmic ratios of NT-PGC-1α by 2.1- and 2.0-fold, respectively. These data are uniformly consistent with the hypothesis that CRM-1 reduces the nuclear content of NT-PGC-1α by effectively exporting it to the cytoplasm.
FIGURE 1.
Cytoplasmic localization of NT-PGC-1α is sensitive to LMB. A, translocation of NT-PGC-1α-HA from nucleus to the cytoplasm is inhibited following treatment with LMB in CHO-K1 cells. CHO-K1 cells were transiently transfected with NT-PGC-1α-HA and treated with vehicle alone or LMB for 1 h. Quantitative determination of the relative nuclear to cytoplasmic intensity ratios was performed as described under “Experimental Procedures.” The bar diagram of the relative ratios of nuclear to cytoplasmic fluorescence intensity of NT-PGC-1α is shown with the mean value and standard error. Significant deference by Student's t test is shown; ***, p < 0.0001 versus control. The nuclear to cytoplasmic ratio values were presented as frequency distribution. B, PGC-1α-null brown preadipocyte cells were adenovirally transduced with Ad-NT-PGC-1α-FLAG and treated with vehicle alone or LMB for 1 h. The bar diagram of the relative ratios of nuclear to cytoplasmic fluorescence intensity of NT-PGC-1α is shown with the mean ± S.D. Significant deference by Student's t test is shown; ***, p < 0.0001 versus control. The nuclear to cytoplasmic ratio values are presented as frequency distribution. C, fully differentiated PGC-1α-deficient brown adipocyte cells were transduced with Ad-NT-PGC-1α-FLAG and treated with vehicle alone or LMB for 1 h. The bar diagram of the relative ratios of nuclear to cytoplasmic fluorescence intensity of NT-PGC-1α is shown with the mean ± S.D. Significant deference by Student's t test is shown; ***, p < 0.0001 versus control. The nuclear to cytoplasmic ratio values are presented as frequency distribution.
To investigate whether the previously reported nuclear localization of PGC-1α (3) is affected by CRM1, CHO-K1 cells expressing PGC-1α-HA were treated with LMB. As expected, PGC-1α-HA was localized to the nucleus in vehicle-treated cells, and LMB had no effect on the nuclear/cytoplasmic ratio of PGC-1α (supplemental Fig. S1). These findings indicate that the nuclear localization signals of PGC-1α override the effect of the NES on full-length protein.
NT-PGC-1α Physically Interacts with CRM1
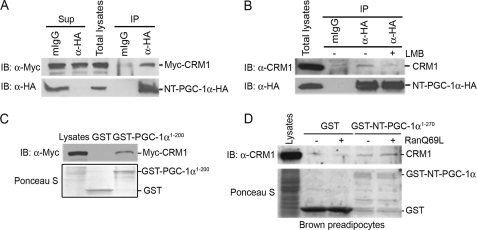
To test for a protein-protein interaction between CRM1 and NT-PGC-1α, we co-transfected CHO-K1 cells with epitope-tagged NT-PGC-1α and CRM1, followed by immunoprecipitation. We found that Myc-tagged CRM1 was co-immunoprecipitated with HA-tagged NT-PGC-1α, but not with nonspecific IgG (Fig. 2A). Given that LMB inhibits CRM1 binding to the NES site (49), we examined its effect in this system and found that LMB blocked the interaction between CRM1 and NT-PGC-1α (Fig. 2A). To further confirm the interaction of NT-PGC-1α with CRM1 in vitro, GST pulldown assays were performed using a GST-PGC-1α (aa 1–200) fusion that contains both putative NES sequences. Using lysates from CHO-K1 cells transfected with Myc-CRM1, we found that GST-PGC-1α (aa 1–200) effectively pulled down Myc-CRM1 (Fig. 2C). Previous studies have shown that the interaction between CRM1- and NES-containing proteins is regulated by Ran and that a GTPase-deficient form of Ran (RanQ69L) prolongs the formation of CRM1·Ran-GTP·NES protein complexes (50). As predicted, we found that CRM1 binding to immobilized GST-NT-PGC-1α was enhanced by the presence of RanQ69L (Fig. 2D). Moreover, no CRM1 binding was detected to GST alone in the absence or presence of RanQ69L (Fig. 2D).
FIGURE 2.
Interaction of NT-PGC-1α with CRM1. A, coimmunoprecipitation of NT-PGC-1α-HA and Myc-CRM1 in transfected CHO-K1 cells. Immunoprecipitates with anti-HA antibody were separated by SDS-PAGE and immunoblotted with anti-Myc antibody (top). The same blot was reprobed with anti-HA antibody to confirm the presence of NT-PGC-1α-HA (bottom). B, coimmunoprecipitation of NT-PGC-1α-HA with endogenous CRM1 in CHO-K1 cells that express NT-PGC-1α-HA. Immunoprecipitates with anti-HA antibody were separated by SDS-PAGE and immunoblotted with anti-CRM1 antibody (top). The same blot was reprobed with anti-HA antibody to confirm the presence of NT-PGC-1α-HA (bottom). C, in vitro interaction of GST-PGC-1α (1–200) with Myc-CRM1. GST or GST-PGC-1α (aa 1–200) fusion proteins immobilized on the beads were incubated with the CHO-K1 cell lysates that express Myc-CRM1. GST pulldown assays were separated by SDS-PAGE and immunoblotted with anti-Myc antibody (top). Expression of GST and GST-PGC-1α (aa 1–200) is shown in Ponceau S staining (bottom). D, GST-NT-PGC-1α interacts with CRM1 in a RanGTP-regulated manner. GST or GST-NT-PGC-1α fusion proteins immobilized on the beads were incubated with brown preadipocyte cell lysates without or with addition of affinity-purified RanQ69L. GST pulldown assays were separated by SDS-PAGE and immunoblotted with anti-CRM1 antibody (top). Expression of GST and GST-NT-PGC-1α is shown in Ponceau S staining (bottom).
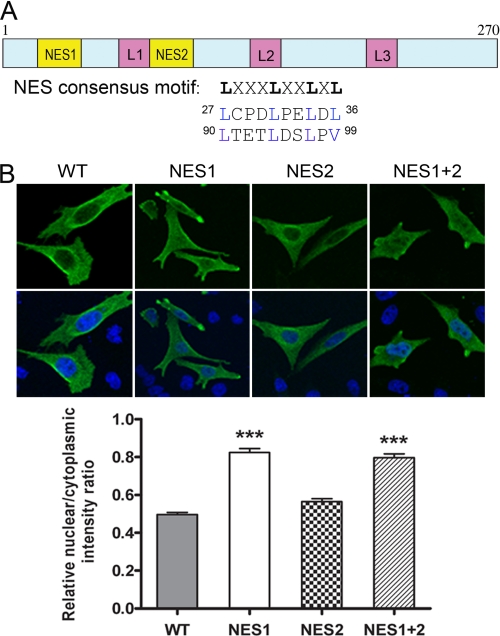
NT-PGC-1α Contains a Functional NES
In general, the NES recognized by CRM1 is composed of leucine- and other hydrophobic residue-rich motifs in the configuration LXXXLXXLXL) (51). NT-PGC-1α contains two similarly configured domains in its N terminus: residues 27–36 (NES1), LCPDLPELDL and residues 90–99 (NES2), LTETLDSLPV (Fig. 3A). To test whether these consensus NES sequences are involved in the nuclear export of NT-PGC-1α, we mutated the leucines/valine to alanines singly and in combination, and analyzed the subcellular localization of each mutated form of NT-PGC-1α. Mutations within the NES1 site significantly increased the nuclear/cytoplasmic ratio (1.7-fold), consistent with inhibition of nuclear export, whereas mutations within the putative NES2 site had no effect on the nuclear/cytoplasmic ratio (Fig. 3B). Moreover, mutation of both sites (NES1 and NES2) produced no additional increase in the nuclear/cytoplasmic ratio (Fig. 3B). These results identify the first NES motif (NES1) as the functional NES which mediates CRM1-dependent nuclear export of NT-PGC-1α.
FIGURE 3.
Identification of NES within NT-PGC-1α. A, schematic representation of the structure of NT-PGC-1α. Two putative NES consensus sequences (Lx(1–3)[livfm]x(2–3)lx[lIV]) are indicated. B, effects of single or double NES mutations (Leu/Val to Ala) on nuclear export of NT-PGC-1α, CHO-K1 cells were transiently transfected with NT-PGC-1α-HA, NT-PGC-1α-NES1-HA. NT-PGC-1α-NES2-HA, or NT-PGC-1α-NES1 plus 2-HA. The bar diagram of the relative ratios of nuclear to cytoplasmic fluorescence intensity is shown with the mean ± S.E. Significant deference by one-way analysis of variance is shown; ***, p < 0.0001 versus control.
Nuclear Export of NT-PGC-1α Is Inhibited by PKA-dependent Phosphorylation of NT-PGC-1α
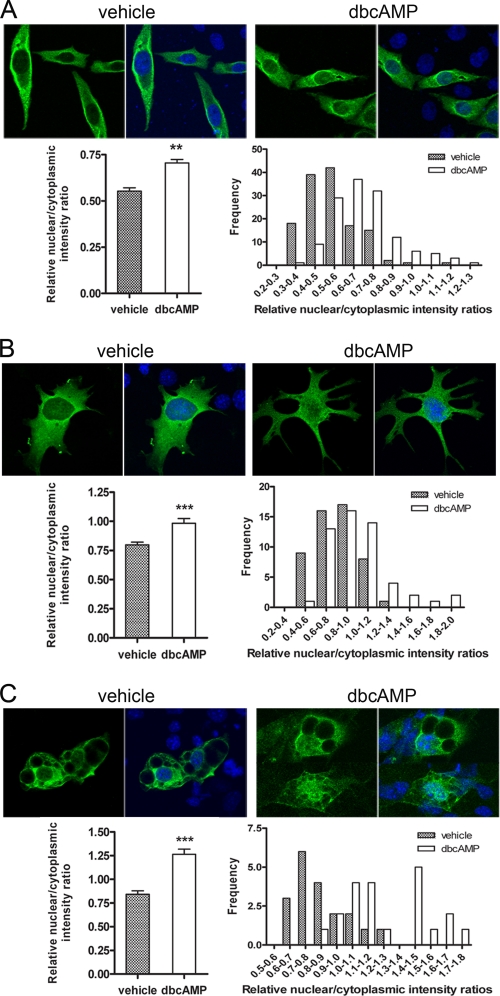
In our initial description of NT-PGC-1α, activation of PKA-regulated expression and subcellular localization of the protein (36). In addition, we established that a significant portion of the PKA-mediated increase in nuclear NT-PGC-1α was independent of downstream activation of p38 MAPK (36). To explore the mechanism in greater detail, we examined the acute response to Bt2cAMP (1 h) and found a rapid increase in nuclear content of NT-PGC-1α-HA in CHO-K1 cells (Fig. 4A, top panels). The relative nuclear/cytoplasmic intensity ratio of NT-PGC-1α was increased ∼1.4-fold by Bt2cAMP (Fig. 4A, left bottom panel), with an increase in the frequency of cells with higher nuclear/cytoplasmic intensity ratios (Fig. 4A, right bottom panel). Bt2cAMP also produced a rapid accumulation of NT-PGC-1α-HA in nuclei of brown adipocytes derived from PGC-1α-null mice, with a ∼1.3-fold increase in preadipocytes (Fig. 4B) and a ∼1.5-fold increase in differentiated adipocytes (Fig. 4C, left bottom panels). As with CHO cells, distribution analysis of adipocytes treated with vehicle or Bt2cAMP showed that higher nuclear/cytoplasmic intensity ratios were more frequently associated with cells treated with the cAMP analogue (Fig. 4, B and C, right bottom panels). These data establish that PKA produces a rapid change in subcellular distribution of NT-PGC-1α, presumably through a phosphorylation mechanism that inhibits nuclear export.
FIGURE 4.
Nuclear accumulation of NT-PGC-1α is increased by cAMP analogue. A, nuclear accumulation of NT-PGC-1α occurs following treatment with Bt2cAMP. CHO-K1 cells were transiently transfected with NT-PGC-1α-HA and treated with vehicle alone or Bt2cAMP for 1 h. The bar diagram of the relative ratios of nuclear to cytoplasmic fluorescence intensity of NT-PGC-1α is shown with the mean ± S.D. Significant deference by Student's t test is shown; **, p = 0.0044 versus control. The nuclear to cytoplasmic ratio values are presented as frequency distribution. B, nuclear accumulation of NT-PGC-1α following treatment with vehicle alone or Bt2cAMP in PGC-1α-null brown preadipocyte cells transduced with Ad-NT-PGC-1α-FLAG. The bar diagram of the relative ratios of nuclear to cytoplasmic fluorescence intensity of NT-PGC-1α is shown with the mean ± S.D. Significant deference by Student's t test is shown; **, p = 0.001 versus control. The nuclear to cytoplasmic ratio values are presented as frequency distribution. C, nuclear accumulation of NT-PGC-1α following treatment with vehicle alone or Bt2cAMP in PGC-1α-deficient fully differentiated brown adipocyte cells transduced with Ad-NT-PGC-1α-FLAG. The bar diagram of the relative ratios of nuclear to cytoplasmic fluorescence intensity of NT-PGC-1α is shown with the mean ± S.D. Significant deference by Student's t test is shown; ***, p < 0.0001 versus control. The nuclear to cytoplasmic ratio values were presented as frequency distribution.
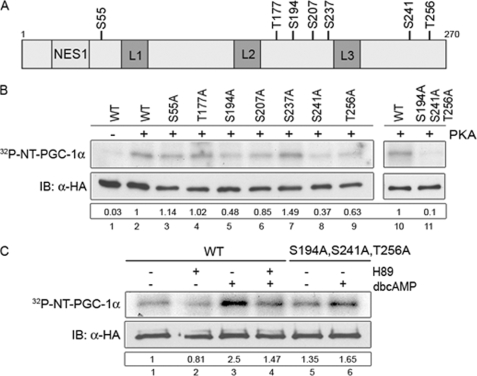
To test whether PKA produced this response through a direct effect on NT-PGC-1α, we examined its sequence for consensus PKA phosphorylation sites ((R/K)XX(S/T)) and found 7 potential sites at residues Ser-55, Thr-177, Ser-194, Ser-207, Ser-237, Ser-241, and Thr-256 (Fig. 5A). To assess PKA input at these sites and assess their relevance to subcellular localization, alanine substitution mutants were generated singly or in combination and examined for PKA-dependent phosphorylation. Using in vitro transcribed/translated NT-PGC-1α and an in vitro kinase assay, we found that wild-type (WT) NT-PGC-1α was efficiently phosphorylated by the purified catalytic subunit of PKA (Fig. 5B, lanes 2 and 10), whereas phosphorylation of NT-PGC-1α was significantly reduced by single S194A, Ser-241A, and T256A substitutions (Fig. 5B, lanes 5, 8, and 9). Also, phosphorylation was almost completely eliminated in the triple S194A/S241A/T256A mutant (Fig. 5B, lane 11). Therefore, the primary sites within NT-PGC-1α that are phosphorylated in vitro by PKA are Ser-194, Ser-241, and Thr-256.
FIGURE 5.
PKA-dependent phosphorylation of NT-PGC-1α. A, schematic diagram shows seven putative PKA consensus phosphorylation sites ([R,K]XX[S,T]) in NT-PGC-1α. B, in vitro PKA-mediated phosphorylation of NT-PGC-1α. NT-PGC-1α-HA and its serine/threonine to alanine mutants were in vitro transcribed/translated using a TnT T7 Coupled Reticulocyte Lysate System, immunoprecipitated, and subjected for the in vitro kinase assay with the purified bovine catalytic subunit of PKA. Autoradiography (top) shows phosphorylated NT-PGC-1α by PKA. Western blot (bottom) shows expression of NT-PGC-1α-HA and its serine/threonine to alanine mutants. Numbers in the box represent the relative phosphorylation that is normalized by NT-PGC-1α protein. C, in situ PKA-dependent phosphorylation of NT-PGC-1α. Autoradiography (top) shows phosphorylated NT-PGC-1α in 32P-labeled CHO-K1 cells ectopically expressing NT-PGC-1α-HA (untreated and H89/Bt2cAMP-treated) and NT-PGC-1α-S194A/S241A/T256A-HA (untreated and Bt2cAMP-treated). Western blot (bottom) shows expression of NT-PGC-1α-HA and NT-PGC-1α-S194A/S241A/T256A-HA. Numbers in the box represent the relative phosphorylation that is normalized by NT-PGC-1α protein.
Next, to determine whether NT-PGC-1α is phosphorylated by PKA in situ, CHO-K1 cells expressing NT-PGC-1α were treated with Bt2cAMP and/or the PKA inhibitor, H89 in the presence of [32P]orthophosphate. Basal phosphorylation was evident in cells treated with vehicle, and the addition of Bt2cAMP produced a significant increase in phosphorylation of NT-PGC-1α (Fig. 5C, compare lanes 1 to 3). H89 produced a slight decrease in basal phosphorylation (Fig. 5C, lanes 1 and 2), and blocked the major proportion of Bt2cAMP-induced phosphorylation of NT-PGC-1α (Fig. 5C, lane 4). NT-PGC-1α with alanine substitutions at residues 194, 241, and 256 was used to test for residual PKA-dependent phosphorylation at other sites. In cells expressing the triple alanine mutant, significant basal phosphorylation was evident in vehicle-treated cells, similar to levels seen in cells expressing WT NT-PGC-1α and treated with H89 (Fig. 5C, compare lanes 5 to lanes 1 and 2). Bt2cAMP produced a small increase in phosphorylation of the triple alanine mutant (Fig. 5C, lanes 5 and 6), perhaps due to PKA-dependent activation of p38 MAPK, which is known to phosphorylate residues Thr-262 and Ser-265 (27, 28). Collectively, these findings establish that Ser-194, Ser-241, and Thr-256 are the primary sites in NT-PGC-1α that are phosphorylated by PKA.
PKA-dependent Phosphorylation of NT-PGC-1α Reduces Nuclear Export of NT-PGC-1α and Increases Its Transcriptional Activity
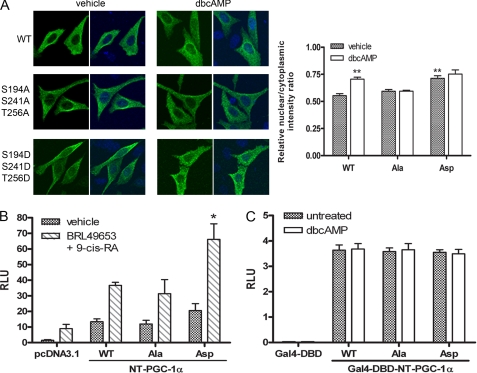
To assess whether PKA-dependent phosphorylation of NT-PGC-1α Ser-194, Ser-241, and Thr-256 is responsible for increasing its nuclear content, wild-type NT-PGC-1α, phosphorylation-resistant NT-PGC-1α-S194A/S241A/T256A, and phosphomimetic NT-PGC-1α-S194D/S241D/T256D were expressed in CHO-K1 cells and their subcellular localization was analyzed in the absence and presence of Bt2cAMP. In vehicle-treated cells, NT-PGC-1α and NT-PGC-1α-S194A/S241A/T256A predominantly reside in the cytoplasm with nuclear/cytoplasmic intensity ratios of 0.54 and 0.59, respectively (Fig. 6A). NT-PGC-1α-S194A/S241A/T256A remained primarily cytoplasmic even in the presence of Bt2cAMP (Fig. 6A). In contrast, the nuclear content of NT-PGC-1α-S194D/S241D/T256D in vehicle-treated cells was significantly higher (nuclear/cytoplasmic intensity ratio of 0.72) than vehicle-treated WT and NT-PGC-1α-S194A/S241A/T256A and was comparable to the nuclear content of the WT construct in Bt2cAMP-treated cells (Fig. 6A). Moreover, Bt2cAMP had no additional effect on nuclear content of NT-PGC-1α-S194D/S241D/T256D (Fig. 6A). Together, these results are uniformly consistent with the idea that phosphorylation of NT-PGC-1α on Ser-194, Ser-241, and Thr-256 by PKA regulates the relative nuclear content of NT-PGC-1α by regulating CRM1-dependent nuclear export of the protein.
FIGURE 6.
PKA-dependent regulation of subcellular localization and transcriptional activity of NT-PGC-1α. A, localization of NT-PGC-1α-HA, NT-PGC-1α-S194A/S241A/T256A-HA, and NT-PGC-1α-S194D/S241D/T256D-HA in CHO-K1 cells. The bar diagram of the relative ratios of nuclear to cytoplasmic fluorescence intensity is shown with the mean ± S.D. Significant deference by Student's t test; **, p < 0.01. B, transcriptional co-activation assay using a luciferase reporter assay. pcDNA3.1, NT-PGC-1α-HA, NT-PGC-1α-S194A/S241A/T256A-HA, or NT-PGC-1α-S194D/S241D/T256D-HA were co-transfected in CHO-K1 cells with a PPRE luciferase reporter (PPRE X3-TK-luc), RXRα, PPAPγ1, and a Renilla luciferase reporter. 24 h after transfection cells were treated with vehicle or with BRL49653 and 9-cis-retinoic acid. Luciferase activity was determined 48 h after transfection and the relative luciferase units were calculated as described under “Experimental Procedures.” Data represent mean ± S.D. of at least three independent experiments. Significant deference by Student's t test: *, p < 0.05. C, transcriptional activation assay. Gal4-DBD, Gal4-NT-PGC-1α, Gal4-NT-PGC-1α-S194A/S241A/T256A, and Gal4-NT-PGC-1α-S194D/S241D/T256D were co-transfected in CHO-K1 cells with a luciferase reporter containing Gal4 DNA binding sites and a Renilla luciferase reporter. Luciferase activity was determined as above. Data represent mean ± S.D. of at least five independent experiments.
We have shown that recruitment of NT-PGC-1α to multiprotein complexes involving nuclear receptors (e.g. PPARγ and PPARα) and co-activators (CBP and SRC1) enhances transactivation of some PGC-1α-target genes (36). To examine whether the co-activator function of NT-PGC-1α is regulated by subcellular localization, the ability of NT-PGC-1α, NT-PGC-1α-S194A/S241A/T256A, and NT-PGC-1α-S194D/S241D/T256D to enhance the activation of PPARγ and RXRα was examined by transient co-transfection assays in CHO-K1 cells. A (PPRE)3-TK-luc reporter construct containing a PPAR-response element was used (8). Using transcriptional activity of (PPRE)3-TK-luc observed with co-expression of PPARγ and RXRα as basal, addition of ligands for PPARγ and RXRα (BRL49653 and 9-cis-retinoic acid) increased activity of the reporter by 6-fold (Fig. 6B). Co-expression of WT NT-PGC-1α with PPARγ and RXRα increased reporter activity by 9-fold, and addition of the ligand mixture produced a further 2.7-fold transactivation of the reporter (Fig. 6B). The alanine substitution mutant of NT-PGC-1α (S194A/S241A/T256A) increased PPARγ/RXRα-mediated transcription of the reporter to the same extent as WT NT-PGC-1α in both vehicle (8-fold) and ligand-treated cells (2.6-fold) (Fig. 6B). In contrast, transactivation of the reporter by the phosphomimetic mutant of NT-PGC-1α-S194D/S241D/T256D was higher enhanced ∼2-fold in both vehicle (14-fold) and ligand-treated cells (3.2-fold) (Fig. 6B), suggesting a close correlation between nuclear localization and co-activation function of NT-PGC-1α. Addition of Bt2cAMP produced an additional 3-fold increase in activity of the reporter that was independent of NT-PGC-1α, and likely reflected direct post-translational activation of the nuclear receptor complex by PKA. This characteristic makes it difficult to resolve the direct effects of PKA on the PPARγ/RXRα complex from the effects of PKA on localization of NT-PGC-1α in this model.
To address the question of whether the transactivation property of NT-PGC-1α is also elevated by phosphorylation, we tested the ability of Gal4-NT-PGC-1α, Gal4-NT-PGC-1α-S194A/S241A/T256A, and Gal4-NT-PGC-1α-S194D/S241D/T256D to transactivate a reporter gene containing Gal4 DNA binding sites. As expected, all the fusion proteins with Gal4-DBD were localized in the nucleus (supplemental Fig. S2). Expression of Gal4-NT-PGC-1α, Gal4-NT-PGC-1α-S194A/S241A/T256A, and Gal4-NT-PGC-1α-S194D/S241D/T256D increased reporter gene expression to the same extent (∼120-fold), and no additional activation of the reporter was observed with Bt2cAMP (Fig. 6C).
Transcriptional Activation of UCP1 and CIDEA by NT-PGC-1α Is Mediated in Part by PKA Phosphorylation of NT-PGC-1α
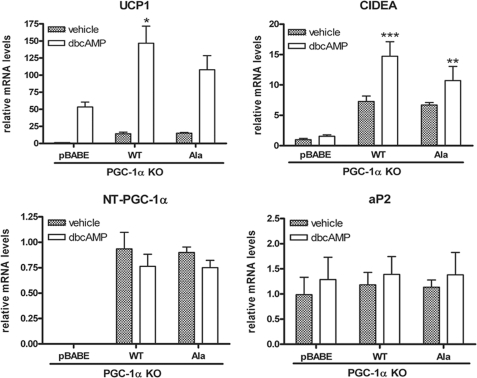
It is well established that increased sympathetic nervous system activity after cold exposure engages the transcriptional program of thermogenesis in brown adipose tissue (BAT) through PKA-dependent signaling mechanisms. For example, transactivation of UCP1 by PKA involves increased p38 MAPK-dependent phosphorylation of ATF-2 and PGC-1α (27). Additionally, we have shown that PKA provides direct signaling input to NT-PGC-1α, and that ectopic expression of NT-PGC-1α in brown adipocytes increases induction of UCP1 expression by cAMP (36). To further assess the relationship between PKA-dependent regulation of subcellular localization and transcriptional function of NT-PGC-1α, we examined the ability of PKA to induce transcription of UCP1 in brown adipocytes expressing the WT or triple alanine mutant of NT-PGC-1α. PGC-1α-deficient brown preadipocytes were retrovirally transduced with pBABE, NT-PGC-1α, and NT-PGC-1α-S194A/S241A/T256A and differentiated for 7 days. As shown in Fig. 7, basal UCP1 mRNA expression was low in brown adipocytes transduced with empty vector but was increased 45-fold after addition of Bt2cAMP. As noted previously, the induction of UCP1 mRNA by Bt2cAMP under these conditions reflects the PGC-1-independent component of the response (27, 42). In contrast, overexpression of NT-PGC-1α and NT-PGC-1α-S194A/S241A/T256A produced a similar 12-fold increase in basal expression of UCP1 compared with empty vector (Fig. 7). Bt2cAMP produced an additional 10-fold increase in UCP1 mRNA over basal levels in cells expressing NT-PGC-1α, whereas the induction of UCP1 by Bt2cAMP in cells expressing NT-PGC-1α-S194A/S241A/T256A was ∼30% lower (Fig. 7). Basal expression of CIDEA, another PGC-1α target gene (52), was increased 7-fold by NT-PGC-1α or NT-PGC-1α-S194A/S241A/T256A in brown adipocytes lacking PGC-1α (Fig. 7). However, as noted with UCP1, the induction of CIDEA mRNA by Bt2cAMP was less robust in cells expressing NT-PGC-1α-S294A/S241A/T256A (1.6-fold) compared with NT-PGC-1α (2-fold) (Fig. 7). Expression of aP2, a marker of adipocyte differentiation, was comparably expressed among the cell lines, and mRNA levels of NT-PGC-1α and NT-PGC-1α-S194A/S241A/T256A were also comparable between the lines (Fig. 7). Considered together, these results show that a significant proportion of the cAMP-dependent induction of UCP1 and CIDEA expression by NT-PGC-1α is directly dependent on phosphorylation of NT-PGC-1α by PKA. The findings further suggest that PKA-dependent regulation of nuclear content of NT-PGC-1α through these sites contributes to cAMP-dependent regulation of UCP1 and CIDEA expression in brown adipocytes.
FIGURE 7.
NT-PGC-1α-dependent induction of UCP1 and CIDEA is reduced by mutation of sites phosphorylated by PKA. UCP1, CIDEA, aP2, and NT-PGC-1α mRNA levels were assessed by real-time PCR analysis in differentiated PGC-1α KO brown adipocyte cells where pBABE, NT-PGC-1α-HA, or NT-PGC-1α-S194A/S241A/T256A-HA was retrovirally expressed. On the seventh day of differentiation, cells were treated with or without Bt2cAMP for 4 h. Relative abundance of mRNA was normalized to cyclophilin mRNA. Data represent mean ± S.D. of at least three independent experiments. Significant difference of UCP1 and CIDEA gene expression was determined by Student's t test; *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Preferential Regulation of Target Genes by NT-PGC-1α and PGC-1α
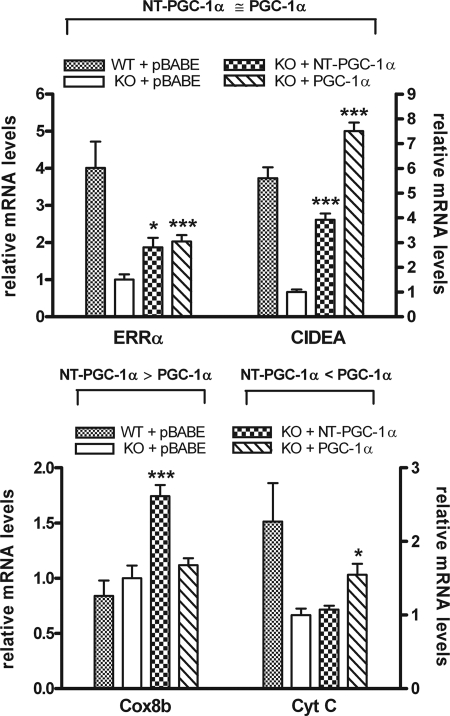
In response to cold exposure, it is expected that PGC-1α and NT-PGC-1α, which are fully activated by transcriptional induction and post-translational modification, play a cooperative role in the regulation of adaptive thermogenesis by promoting mitochondrial biogenesis, respiration, and the uncoupling of oxidative phosphorylation. To investigate whether specific genes are preferentially affected by PGC-1α or NT-PGC-1α, we examined expression of a number of genes involved in these processes in fully differentiated wild-type brown adipocytes and PGC-1α-deficient brown adipocytes expressing empty vector, NT-PGC-1α, or PGC-1α. Many genes, including ERRα and CIDEA (Fig. 8, top panel), and Cox7al and PPARα (data not shown), were comparably induced by NT-PGC-1α and PGC-1α. However, Cox8b mRNA was induced 1.8-fold by NT-PGC-1α while PGC-1α was without effect (Fig. 8, bottom panel). In contrast, CytC mRNA expression was increased 1.6-fold by PGC-1α while NT-PGC-1α had no effect (Fig. 8, bottom panel). Considered together, the results show examples of genes that are both comparably and differentially regulated by NT-PGC-1α and PGC-1α, and argue that both protein isoforms contribute to the full transcriptional response mediated by their induction in BAT after cold exposure.
FIGURE 8.
Genes that are comparably (top panel) or differentially (bottom panel) regulated by NT-PGC-1α and PGC-1α. ERRα, CIDEA, Cox8b, and CytC mRNA levels were assessed by real-time PCR in differentiated brown adipocytes from either wild-type or PGC-1α null mice. On day 7 of differentiation, wild-type cells expressing empty vector (pBABE) or PGC-1α null cells expressing empty vector, NT-PGC-1α, or PGC-1α were treated with Bt2cAMP for 4 h. Relative abundance of mRNA was normalized to cyclophilin mRNA. Data represent mean ± S.D. of at least three independent experiments. Significant deference by Student's t test is shown; *, p < 0.05 and ***, p < 0.001.
DISCUSSION
Our initial description of NT-PGC-1α reported that it is a novel, biologically active splice variant of PGC-1α that is co-expressed with PGC-1α in all tissues so far examined (36). NT-PGC-1α mRNA and protein are expressed at levels comparable to PGC-1α, and their expression is regulated by the same physiological signals that regulate the full-length isoform of the protein. For example, expression of NT-PGC-1α and PGC-1α is highly induced in brown adipose tissue of mice by cold exposure. p38 MAPK is known to be the central downstream mediator of β-adrenergic receptor-dependent induction of PGC-1α gene expression and protein stability (27). In our original description of NT-PGC-1α, we found that PKA provides signaling inputs that are independent of p38 MAPK and that regulate the function of NT-PGC-1α (36). The absence of nuclear localization sequences results in a cytoplasmic distribution for NT-PGC-1α where it may be far less effectively targeted to the proteasome for degradation. Activation of PKA increases the nuclear content of NT-PGC-1α in part through a p38 MAPK-independent mechanism. This suggests that the transcriptional activity of the respective PGC-1α isoforms may be differentially regulated, based on how signaling input from PKA is integrated, with p38 MAPK diminishing ubiquitin/proteasome-mediated degradation of PGC-1α and PKA decreasing translocation of NT-PGC-1α to the cytoplasm.
In this report, we demonstrate that NT-PGC-1α contains a leucine-rich CRM1-binding site (LCPDLPELDL) at the N terminus and is actively transported out of the nucleus by binding to CRM1. The functional significance of this interaction is only evident in NT-PGC-1α, which lacks the strong nuclear localization signals of PGC-1α. In fact, fusion of NT-PGC-1α to a Gal4 DNA binding domain that contains a strong nuclear localization signal makes NT-PGC-1α insensitive to the CRM1-mediated nuclear export. Why is NT-PGC-1α excluded from the nucleus? Previous transactivation studies using a Gal4-DBD fusion strategy found that PGC-1α120, PGC-1α170, and PGC-1α284 were much more active than full-length PGC-1α (8, 18), suggesting that NT-PGC-1α (aa 1–270) might be more potent than PGC-1α in the nucleus. Thus, one possibility is that export of NT-PGC-1α from the nucleus to the cytosol by CRM1 may be a mechanism to negatively regulate transcription by limiting assembly of active transcriptional complexes. This inhibitory mode of regulation is common among a number of transcription factors, including NF-κB, FOXO1, c-Fos, and FKHRL1. These transcription factors are sequestered from the nucleus by binding partners that mask nuclear localization signals or promote nuclear export, and their nuclear translocation is controlled by multiple signaling inputs (37–41).
An additional feature of NT-PGC-1α is that its CRM1-binding site (NES, 27LCPDLPELDL36) overlaps its transactivation domain (30DLPELDLSELD40) (53). This raises the interesting possibility that CRM1 may negatively regulate NT-PGC-1α by both exporting it from the nucleus and masking the transactivation domain.
Our initial description of NT-PGC-1α reported that PKA activation by a cell-permeant cAMP analogue induced nuclear accumulation of NT-PGC-1α and enhanced its recruitment to the promoters of two consensus PGC-1α target genes (36). We have extended those findings in the present report by showing that NT-PGC-1α is a substrate for PKA and is directly phosphorylated by the purified catalytic subunit of PKA in vitro. The serines at residues 194 and 241, and the threonine at position 256, are phosphorylated in vitro, and the same sites are phosphorylated in situ in response to PKA activation by Bt2cAMP. Although it is evident that NT-PGC-1α is phosphorylated by other kinases in the cell, the overall PKA-dependent phosphorylation of NT-PGC-1α is essentially eliminated by mutation of the identified residues to alanine in both in vitro and in situ assays.
The significance of phosphorylation at these sites derives from the effect of this post translational input on interaction of NT-PGC-1α with CRM1. Our findings show that NT-PGC-1α interacts with CRM1, which efficiently exports the protein to the cytosol, so we interpret this to mean that CRM1 exports NT-PGC-1α at a rate that exceeds the rate at which the protein diffuses into the nucleus. This conclusion is supported by our finding that NT-PGC-1α equilibrates across the nuclear membrane when its interaction with CRM1 is blocked with LMB. Blocking PKA-dependent phosphorylation of NT-PGC-1α at residues 194, 241, and 256 prevents its nuclear accumulation in response to PKA. Moreover, mutation of these sites to aspartates to mimic phosphorylation increases nuclear content of NT-PGC-1α and eliminates the ability of PKA to increase nuclear content. These data are uniformly consistent with the conclusion that PKA-dependent phosphorylation of NT-PGC-1α increases its nuclear content by reducing the ability of CRM1 to export it from the nucleus.
There are several precedents showing that phosphorylation negatively regulates CRM1-mediated nuclear export (54, 55). At G2/M transition, phosphorylation of the cyclin B cytoplasmic retention sequence that is required for CRM1-mediated nuclear export reduces nuclear export by preventing its interaction with CRM1 (54). Although the NES at the N terminus is separated from the PKA phosphorylation sites by 150–200 aa, phosphorylation might affect the accessibility of the NES to CRM1 by inducing a conformational change in NT-PGC-1α or favoring its interaction with other nuclear proteins. Further experiments will be required to clarify the mechanism by which PKA phosphorylation of NT-PGC-1α negatively regulates CRM1-mediated nuclear export.
Two approaches were used to assess the biological significance of regulated nuclear localization of NT-PGC-1α. In the first we used a (PPRE)3-TK-luc reporter to examine the relationship between nuclear content of NT-PGC-1α and its ability to transactivate PPARγ/RXRα-dependent transcriptional activation of the reporter. By modulating the nuclear content of NT-PGC-1α in the absence of PKA input, we found that the enhanced nuclear content of the phosphomimetic form of NT-PGC-1α-S194D/S241D/T256D led to greater transactivation of the reporter (Fig. 6B). In the second approach, we used PGC-1α null adipocytes and retroviral expression of either WT NT-PGC-1α or an alanine mutant of NT-PGC-1α-S194A/S241A/T256A, which limits the effect of PKA to increase its nuclear content. We found that a significant component (∼30%) of the Bt2cAMP-dependent induction of UCP1 and CIDEA was dependent upon regulated access of NT-PGC-1α to the nucleus. A limitation of these studies is that the conditions don't faithfully reproduce what occurs with endogenous protein when cAMP is elevated in BAT. For example, our initial report showed that in vivo and in situ PKA activation produces a significant induction of both NT-PGC-1α and full-length PGC-1α (36). Thus, the combination of increased expression of both isoforms in conjunction with decreased nuclear export of NT-PGC-1α makes it difficult to assess the relative contribution of diminished export to changes in transcriptional activity under these conditions.
The data presented here make a compelling case that the transcriptional activity of NT-PGC-1α is regulated in part through PKA-dependent regulation of its subcellular localization. Under basal conditions, CRM1-dependent nuclear export of NT-PGC-1α concentrates the protein in the cytosol, whereas PKA activation increases the nuclear content of NT-PGC-1α. We show that phosphorylation of NT-PGC-1α on three residues by PKA reduces the ability of CRM1 to export NT-PGC-1α from the nucleus. More importantly, we show that this spatio-temporal regulation of NT-PGC-1α by PKA is coupled to regulation of its transcriptional potential. Our findings indicate that many genes are regulated comparably by NT-PGC-1α and PGC-1α, but we also provide examples that are preferentially regulated by each isoform. Global expression analysis will be required to expand the list of genes in each category, but because of the different half lives of PGC-1α and NT-PGC-1α, it seems likely that differences in target gene regulation may extend to duration of transcriptional activation by the respective protein isoforms. It is particularly interesting that the co-activation function of a novel isoform of PGC-1α is regulated differently from full-length PGC-1α. Two different modes of regulation might be a way to coordinate the timing and full transcriptional potential of both PGC-1α isoforms in response to physiological signals. For example, in BAT after cold exposure, cAMP increases the transcriptional induction of both NT-PGC-1α and PGC-1α, and mediates the post-translational activation of both proteins. Given that approximately half of the cold-induced PGC-1α total mRNA encodes for NT-PGC-1α, we predict that the post-translational input by PKA, which regulates subcellular localization of NT-PGC-1α, is important not only for achieving full PGC-1α activity in animals exposed to cold, but also for induction of the complete set of genes that are transcriptionally regulated by PGC-1α isoforms.
Supplementary Material
Acknowledgments
We thank Dr. Bruce Spiegelman for providing cell lines from PGC-1α null mice. pcDNA3.1-myc-CRM1 and pQE32-HIS6-RanQ69L were kindly provided by S. Meloche (Université de Montréal). We thank Anik Boudreau, Jeho Shin, Aaron Adamson, and David Burk for their technical contributions and advice. We thank Anne Gooch for excellent administrative support.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 DK 074772 and P20-RR021945 (to T. W. G.), NIH Clinical Nutrition Research Unit Center Grant 1P30 DK072476, and NIH Grant DK064951 (to A. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- PGC-1
- peroxisome proliferator-activated receptor γ co-activator-1
- PPAR
- peroxisome proliferator-activated receptor
- PPRE
- PPAR response element
- Bt2cAMP
- dibutyryl cAMP
- aa
- amino acid(s)
- MAPK
- mitogen-activated protein kinase
- PKA
- protein kinase A
- CHO
- Chinese hamster ovary
- PBS
- phosphate-buffered saline
- GST
- glutathione S-transferase
- HA
- hemagglutinin
- NES
- nuclear export sequence
- LMB
- leptomycin B
- RXR
- retinoid X receptor
- BAT
- brown adipose tissue.
REFERENCES
- 1.Finck B. N., Kelly D. P. (2006) J. Clin. Invest. 116, 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Handschin C., Spiegelman B. M. (2006) Endocr. Rev. 27, 728–735 [DOI] [PubMed] [Google Scholar]
- 3.Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. (1998) Cell 92, 829–839 [DOI] [PubMed] [Google Scholar]
- 4.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., Spiegelman B. M. (1999) Cell 98, 115–124 [DOI] [PubMed] [Google Scholar]
- 5.Lehman J. J., Barger P. M., Kovacs A., Saffitz J. E., Medeiros D. M., Kelly D. P. (2000) J. Clin. Invest. 106, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C. (2003) Nat. Genet. 34, 267–273 [DOI] [PubMed] [Google Scholar]
- 7.Schreiber S. N., Emter R., Hock M. B., Knutti D., Cardenas J., Podvinec M., Oakeley E. J., Kralli A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6472–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vega R. B., Huss J. M., Kelly D. P. (2000) Mol. Cell Biol. 20, 1868–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y. X., Lee C. H., Tiep S., Yu R. T., Ham J., Kang H., Evans R. M. (2003) Cell 113, 159–170 [DOI] [PubMed] [Google Scholar]
- 10.Herzig S., Long F., Jhala U. S., Hedrick S., Quinn R., Bauer A., Rudolph D., Schutz G., Yoon C., Puigserver P., Spiegelman B., Montminy M. (2001) Nature 413, 179–183 [DOI] [PubMed] [Google Scholar]
- 11.Yoon J. C., Puigserver P., Chen G., Donovan J., Wu Z., Rhee J., Adelmant G., Stafford J., Kahn C. R., Granner D. K., Newgard C. B., Spiegelman B. M. (2001) Nature 413, 131–138 [DOI] [PubMed] [Google Scholar]
- 12.Kressler D., Schreiber S. N., Knutti D., Kralli A. (2002) J. Biol. Chem. 277, 13918–13925 [DOI] [PubMed] [Google Scholar]
- 13.Puigserver P., Spiegelman B. M. (2003) Endo. Rev. 24, 78–90 [DOI] [PubMed] [Google Scholar]
- 14.Rhee J., Inoue Y., Yoon J. C., Puigserver P., Fan M., Gonzalez F. J., Spiegelman B. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4012–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin J., Wu H., Tarr P. T., Zhang C. Y., Wu Z., Boss O., Michael L. F., Puigserver P., Isotani E., Olson E. N., Lowell B. B., Bassel-Duby R., Spiegelman B. M. (2002) Nature 418, 797–801 [DOI] [PubMed] [Google Scholar]
- 16.St-Pierre J., Drori S., Uldry M., Silvaggi J. M., Rhee J., Jäger S., Handschin C., Zheng K., Lin J., Yang W., Simon D. K., Bachoo R., Spiegelman B. M. (2006) Cell 127, 397–408 [DOI] [PubMed] [Google Scholar]
- 17.Liu C., Li S., Liu T., Borjigin J., Lin J. D. (2007) Nature 447, 477–481 [DOI] [PubMed] [Google Scholar]
- 18.Puigserver P., Adelmant C., Wu Z., Fan M., Xu J., O'Malley B., Spiegelman B. M. (1999) Science 286, 1368–1371 [DOI] [PubMed] [Google Scholar]
- 19.Delerive P., Wu Y., Burris T. P., Chin W. W., Suen C. S. (2002) J. Biol. Chem. 277, 3913–3917 [DOI] [PubMed] [Google Scholar]
- 20.Michael L. F., Wu Z., Cheatham R. B., Puigserver P., Adelmant G., Lehman J. J., Kelly D. P., Spiegelman B. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3820–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J., Puigserver P., Donovan J., Tarr P., Spiegelman B. M. (2002) J. Biol. Chem. 277, 1645–1648 [DOI] [PubMed] [Google Scholar]
- 22.Puigserver P., Rhee J., Donovan J., Walkey C. J., Yoon J. C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D., Spiegelman B. M. (2003) Nature 423, 550–555 [DOI] [PubMed] [Google Scholar]
- 23.Fan M., Rhee J., St-Pierre J., Handschin C., Puigserver P., Lin J., Jaeger S., Erdjument-Bromage H., Tempst P., Spiegelman B. M. (2004) Genes Dev. 18, 278–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monsalve M., Wu Z., Adelmant G., Puigserver P., Fan M., Spiegelman B. M. (2000) Mol. Cell 6, 307–316 [DOI] [PubMed] [Google Scholar]
- 25.Handschin C., Rhee J., Lin J., Tarr P. T., Spiegelman B. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7111–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nisoli E., Clementi E., Paolucci C., Cozzi V., Tonello C., Sciorati C., Bracale R., Valerio A., Francolini M., Moncada S., Carruba M. O. (2003) Science 299, 896–899 [DOI] [PubMed] [Google Scholar]
- 27.Cao W., Daniel K. W., Robidoux J., Puigserver P., Medvedev A. V., Bai X., Floering L. M., Spiegelman B. M., Collins S. (2004) Mol. Cell Biol. 24, 3057–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puigserver P., Rhee J., Lin J., Wu Z., Yoon J. C., Zhang C. Y., Krauss S., Mootha V. K., Lowell B. B., Spiegelman B. M. (2001) Mol. Cell 8, 971–982 [DOI] [PubMed] [Google Scholar]
- 29.Sano M., Tokudome S., Shimizu N., Yoshikawa N., Ogawa C., Shirakawa K., Endo J., Katayama T., Yuasa S., Ieda M., Makino S., Hattori F., Tanaka H., Fukuda K. (2007) J. Biol. Chem. 282, 25970–25980 [DOI] [PubMed] [Google Scholar]
- 30.Olson B. L., Hock M. B., Ekholm-Reed S., Wohlschlegel J. A., Dev K. K., Kralli A., Reed S. I. (2008) Genes Dev. 22, 252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knutti D., Kressler D., Kralli A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9713–9718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jäger S., Handschin C., St-Pierre J., Spiegelman B. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12017–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X., Monks B., Ge Q., Birnbaum M. J. (2007) Nature 447, 1012–1016 [DOI] [PubMed] [Google Scholar]
- 34.Teyssier C., Ma H., Emter R., Kralli A., Stallcup M. R. (2005) Genes Dev. 19, 1466–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y., Huypens P., Adamson A. W., Chang J. S., Henagan T. M., Boudreau A., Lenard N. R., Burk D., Klein J., Perwitz N., Shin J., Fasshauer M., Kralli A., Gettys T. W. (2009) J. Biol. Chem. 284, 32813–32826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biggs W. H., 3rd, Meisenhelder J., Hunter T., Cavenee W. K., Arden K. C. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7421–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunet A., Kanai F., Stehn J., Xu J., Sarbassova D., Frangioni J. V., Dalal S. N., DeCaprio J. A., Greenberg M. E., Yaffe M. B. (2002) J. Cell Biol. 156, 817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayden M. S., Ghosh S. (2004) Genes Dev. 18, 2195–2224 [DOI] [PubMed] [Google Scholar]
- 40.Sasaki T., Kojima H., Kishimoto R., Ikeda A., Kunimoto H., Nakajima K. (2006) Mol. Cell 24, 63–75 [DOI] [PubMed] [Google Scholar]
- 41.Roux P., Blanchard J. M., Fernandez A., Lamb N., Jeanteur P., Piechaczyk M. (1990) Cell 63, 341–351 [DOI] [PubMed] [Google Scholar]
- 42.Uldry M., Yang W., St-Pierre J., Lin J., Seale P., Spiegelman B. M. (2006) Cell Metab. 3, 333–341 [DOI] [PubMed] [Google Scholar]
- 43.Béréziat V., Moritz S., Klonjkowski B., Decaudain A., Auclair M., Eloit M., Capeau J., Vigouroux C. (2005) Biochimie 87, 951–958 [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y., Xiong Y. (2001) Science 292, 1910–1915 [DOI] [PubMed] [Google Scholar]
- 45.Qu L., Huang S., Baltzis D., Rivas-Estilla A. M., Pluquet O., Hatzoglou M., Koumenis C., Taya Y., Yoshimura A., Koromilas A. E. (2004) Genes Dev. 18, 261–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villena J. A., Hock M. B., Chang W. Y., Barcas J. E., Giguère V., Kralli A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panté N., Aebi U. (1996) Crit. Rev. Biochem. Mol. Biol. 31, 153–199 [DOI] [PubMed] [Google Scholar]
- 48.Cyert M. S. (2001) J. Biol. Chem. 276, 20805–20808 [DOI] [PubMed] [Google Scholar]
- 49.Kudo N., Matsumori N., Taoka H., Fujiwara D., Schreiner E. P., Wolff B., Yoshida M., Horinouchi S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9112–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sachdev S., Bagchi S., Zhang D. D., Mings A. C., Hannink M. (2000) Mol. Cell Biol. 20, 1571–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stade K., Ford C. S., Guthrie C., Weis K. (1997) Cell 90, 1041–1050 [DOI] [PubMed] [Google Scholar]
- 52.Hallberg M., Morganstein D. L., Kiskinis E., Shah K., Kralli A., Dilworth S. M., White R., Parker M. G., Christian M. (2008) Mol. Cell Biol. 28, 6785–6795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sadana P., Park E. A. (2007) Biochem. J. 403, 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J., Bardes E. S., Moore J. D., Brennan J., Powers M. A., Kornbluth S. (1998) Genes Dev. 12, 2131–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kilstrup-Nielsen C., Alessio M., Zappavigna V. (2003) EMBO J. 22, 89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.