Abstract
The seven-transmembrane receptor PalH and its coupled, positive-acting arrestin-like protein PalF are key components of a molecular sensor that in Aspergillus nidulans and other ascomycete fungi mediates activation of an intracellular signaling cascade by alkaline ambient pH. PalF is ubiquitinated in an alkaline pH- and PalH-dependent manner. We show here that PalF assists the plasma membrane localization of PalH and that PalF overexpression slightly hypersensitizes the pathway to alkaline pH but does not bypass the need for the ambient pH signal receptor in signaling. In contrast, covalent attachment of Ub to PalF activates the signaling pathway under acidic pH conditions in which the pathway is normally inactive, demonstrating a positive role for ubiquitination. We further show that PalF acts upstream of, or in concert with, the Bro1 domain-containing pH signaling protein PalC, which is normally recruited to cortical structures likely to represent active pH signaling foci under neutral/alkaline pH conditions. In agreement with its pathway-activating consequences, expression of PalF-Ub also promotes PalC cortical recruitment under acidic conditions. Notably, our data establish that expression of PalF-Ub, at approximately physiological levels, in a null palH background leads to a considerable degree of signaling even in the complete absence of the receptor. Thus PalF ubiquitination is a key, perhaps the sole, molecular trigger required for transmitting the alkaline pH signal to the downstream elements of the pathway.
Keywords: Fungi, 7-Helix Receptor, Plasma Membrane, Signal Transduction, Ubiquitination, Arrestin, ESCRT
Introduction
The fungal ambient pH signaling pathway has been intensively studied in Aspergillus nidulans and Saccharomyces cerevisiae (1, 2). In A. nidulans, activation of the pathway by alkaline ambient pH leads to the proteolytic activation of the zinc-finger transcription factor PacC (3, 4) (see Fig. 1A), which mediates transcriptional responses to environmental pH.
FIGURE 1.
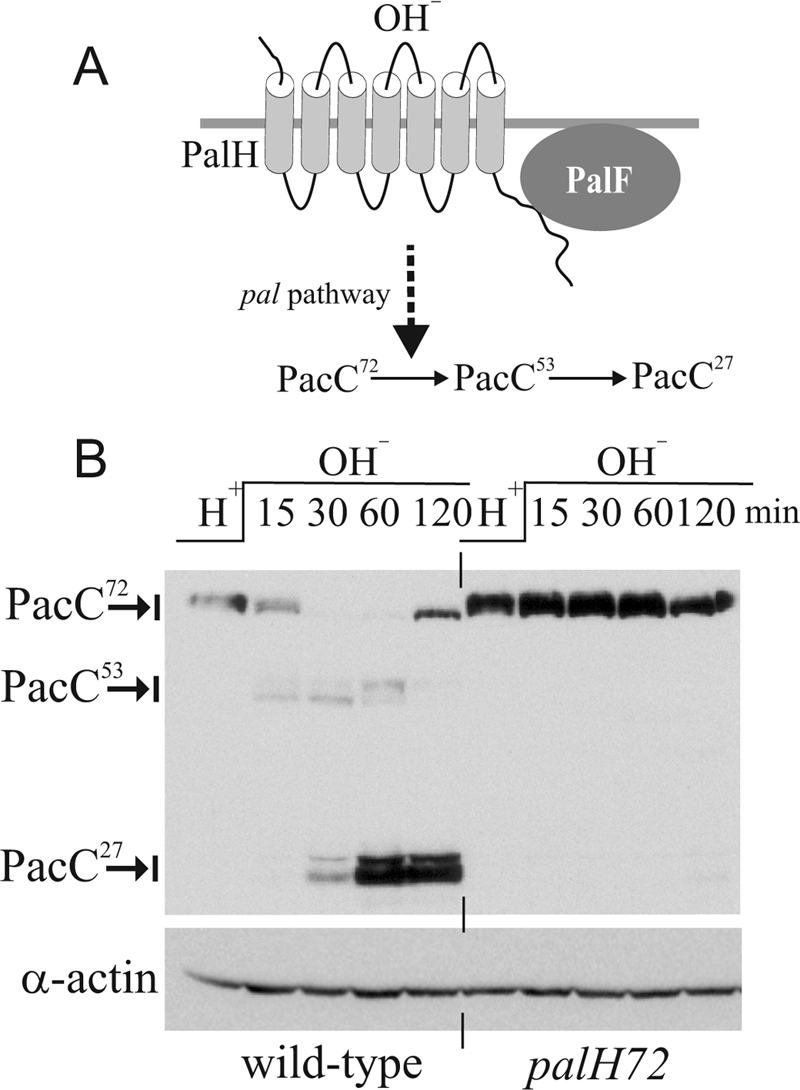
PalH dependence of PacC processing. A, sketch of the A. nidulans pH signaling pathway. Upon alkaline pH stimulation, the pal pathway (PalH, PalF, PalI, PalC, PalA, and PalB) mediates conversion of PacC72 (i.e. 72 kDa) into the committed 53-kDa intermediate PacC53, which is further processed to 27-kDa PacC27. B, Western analysis of Myc3-PacC processing in wild-type and palH− strains. Cells were cultured at pH 4.2 (H+) and shifted to pH 8.2 (OH−) for the indicated times. Actin levels were used as loading control.
pH signaling involves a plasma membrane complex, including the 7-TMD2 receptor (7-TMDR) and almost certain pH sensor PalH, the helper protein PalI, and the positive-acting arrestin-like protein PalF (5). Three additional pH-signaling proteins associate with ESCRT-III, one of four complexes involved in multivesicular body biogenesis (6). These are two Vps32 (a key component of ESCRT-III) interactors, PalA and PalC, and the Vps24 interactor and calpain 7 orthologue PalB (7–11). PalA binding to PacC through two YPX(L/I) motifs in the latter is essential for proteolytic activation, suggesting that PalA recruits PacC to the signaling protease, almost certainly PalB (7, 8). Elucidating how the plasma membrane complex communicates with ESCRT-III-associated proteins is crucial to understanding this important example of ESCRT-III playing a positive-acting role in signal transduction (12).
Metazoan arrestins show strong conservation of functionally important residues/motifs (13). Although distantly related, A. nidulans arrestin-like PalF (Rim8 in S. cerevisiae) interacts strongly with its 7-TMDR PalH, contains arrestin N-terminal (PFAM02752) and C-terminal (PFAM00339) domains and has convincing amino acid sequence similarity to mammalian arrestins (5), indicating that PalF is genuinely related to arrestins.
S. cerevisiae has, in addition to Rim8, a family of nine arrestin-related traffic adaptors (ARTs) (14–16). ARTs contain, included within an arrestin fold that was predicted by in silico modeling, a partially conserved 29-amino acid “arrestin motif” (16). However, the finding that this motif is present in different places within the arrestin fold predicted in ARTs (16) in conjunction with their lower sequence similarity to arrestins (some do not contain detectable N-terminal and/or C-terminal arrestin domains) suggests that ARTs are more distantly related to bona fide arrestins than PalF/Rim8. Moreover, the physiological role of ARTs is to mediate the vacuolar targeting of transporters by serving as adaptors for their ubiquitination by the Rsp5 ubiquitin (Ub) ligase (14–16). Thus, ARTs do not appear to bind 7-TMDRs.
Also resembling mammalian β-arrestins, PalF is itself ubiquitinated in a signal (alkaline ambient pH) and receptor (PalH)-dependent manner (5), suggesting that the 7-TMDR mediates PalF accessibility to a ubiquitin ligase. Work with S. cerevisiae has shown that the Rsp5 Ub ligase ubiquitinates Rim8 and that a mono-ubiquitinated Lys acts in concert with a C-terminal SXP motif to mediate Rim8 binding to ESCRT-I Vps23, such that this ESCRT-I protein preferentially binds Rim8-Ub (17). These data, and the finding that the A. nidulans Vps32 interactor PalC localizes to cortical structures at alkaline pH (10), support a model in which ubiquitinated arrestin-like proteins hijack ESCRT complexes for pH signaling purposes at the plasma membrane. We used the robust physiological response to pH changes of A. nidulans to demonstrate that covalent attachment of mono-Ub to PalF results in receptor-independent pH signaling, establishing that arrestin-like ubiquitination plays a key positive role in pH signaling.
EXPERIMENTAL PROCEDURES
Plasmids and Strains
palF500 encoding PalF-HA3 and its derivatives encoding PalF-HA3-Ub and PalF-HA3-UbK48R were constructed by gene replacement, using strain MAD1732 (pyrG89) (supplemental Table 1) as recipient for transformation. DNA fragments encoding the corresponding fusion proteins followed by the A. fumigatus pyrG and the 3′-untranslated region of palF were assembled by fusion PCR, cloned (plasmids p1808, p1809, and p1809′, respectively), and sequenced to verify the absence of mutations. Linear DNA fragments were used for transformation of strain MAD1732 (pacC900 pyrG89; pacC900 encodes Myc3-PacC) (4). Gene-replaced pyrimidine-independent transformants were identified by Southern blotting. Plasmids driving expression of PalF-HA3 (p1914), PalF-HA3-Ub (p1861), and PalF-HA3-UbK48R (p1915) under the control of the gpdAmini promoter were derivatives of pgpdA003 (18), which targets integration to the pyroA locus. Single copy transformants of MAD1730 (pacC900 pyroA4) were confirmed by Southern. Progeny of crosses with a palH72 strain were analyzed by Western blots, diagnostic PCR (HA3 and pacC900), and DNA sequencing (genotyping of palH72). Plasmids driving PalF-FLAG (p1818) or PalF-HA (p1946) under the control of the alcA promoter were targeted to pyroA as described before (19). These plasmids were originally transformed into MAD1288 (pyroA4 argB2::[argB*-alcAp::palH-gfp]) to obtain strains co-expressing PalH-GFP and PalF-HA or PalF-FLAG at similar levels from single copy transgenes, respectively, integrated at the argB (PalH-GFP) and pyroA (PalF-HA or PalF-FLAG) loci, respectively. Strains carrying palC-GFP and gpdAmini::PalF-HA3 or gpdAmini::PalF-HA3-UbK48R were obtained by crossing. A complete list of strains constructed for this work is presented in supplemental Table 1.
Cultures, Protein Isolation, and Western Blots
Overnight cultures for acidic (final pH 5.1–5.4), neutral (final pH ∼ 6.5), and mildly alkaline (pH 7.1–7.4) pH conditions, and cultures for pH shift experiments were made as described previously (3, 4). In pH shift experiments, the pH of the cultures before shifting mycelia to alkaline conditions (pH ∼ 8.4) was ∼4.3. Soluble protein isolation and Myc3-PacC and actin Western analyses were also as described (4). PalF-containing extracts were prepared using a modified version of the protocol used for soluble protein extracts: mycelia were collected by filtration, frozen, and lyophilized. Lyophilized cells (2-ml screw-cap tubes) were broken using a Fast Prep cell disruptor FP120 and a 0.5 mm ceramic bead, with a 10-s pulse at a setting of 4. Glass beads (0.7 ml, 0.45 mm) were added per sample, and the volume of the tube was completed with buffer A50AR containing 25 mm HEPES, pH 7.5, 50 mm KCl, 5 mm MgCl2, 0.1 mm EDTA, 10% (v/v) glycerol, 0.5 mm dithiothreitol, 1 mm Pefabloc, 1 μm pepstatin, 0.6 μm leupeptin, 5 mm N-ethylmaleimide, 2 μm MG132, 0.1% (v/v) Triton X-100, and the Roche Applied Science EDTA-free protease inhibitor mixture. The resulting suspensions were further homogenized with three 30-s pulses at a setting of 6.5. Cell debris was removed after centrifugation at 14,000 × g for 30 min at 4 °C, and the protein concentration in the supernatants was estimated by Bradford. Proteins were resolved in 7.5% SDS-polyacrylamide gels, which were transferred to nitrocellulose. PalF-HA3 blots were reacted with rat anti-HA (clone 3F10, Roche Applied Science) antibody (1:1000) and developed with Southern Biotech peroxidase-coupled goat anti-rat IgM+IgG secondary antiserum (1:4000) and Amersham Biosciences ECL. If needed, chemiluminescence was quantified using a TDI 06-LAS3000Q chemiluminescence image analyser with Fuji Multi Gauge software.
Growth Tests
Diagnostic growth tests for pH regulation were carried out as described before (20, 21). Plates were incubated at 37 °C.
Microscopy
Recruitment of PalC-GFP to cortical structures was monitored as described (10). Briefly, germlings were cultured overnight in watch minimal medium (18) containing glucose 0.1% (w/v) as carbon source and adjusted to acidic pH with 25 mm NaH2PO4 (pH 5.2–5.3) before being shifted to the same medium adjusted to acidic (5.2), neutral (7–7.2, with 12.5 mm NaH2PO4 plus 12.5 mm Na2HPO4) or alkaline (8.2–8.3, with 25 mm Na2HPO4) pH, where they were incubated for 5–30 min before taking photographs, using a Nikon Eclipse epifluorescence microscope equipped with a 100 × 1.40 numerical aperture plan-apochromat objective and a Semrock Brightline GFP-3035B filter. Images were recorded with an Orca-ER camera (Hamamatsu) driven by Metamorph (Invitrogen) before being analyzed. PalC-GFP punctate structures were counted in at least 25 germlings per strain and pH condition and normalized to the germling length.
For experiments on PalH-GFP localization, cells were cultured overnight at 25 °C in appropriately supplemented acid watch minimal medium containing 0.01% yeast extract with 0.05% (w/v) glucose and 10 mm NaNO3 as carbon and nitrogen sources, respectively. Cells were then transferred to the same medium containing 1.2% (v/v) ethanol, which resulted in the full induction of the alcAp driving expression of PalH-GFP and PalF-HA or PalF-FLAG. Cells were photographed after a 3-h induction period.
Statistical Analysis
Data were analyzed using GraphPad Prism 5.00 for Windows (GraphPad Software, San Diego, CA). Statistical significance of the differences observed when more than two groups were compared was estimated using the one-way analysis of the variance with a Bonferroni multiple comparisons test.
RESULTS
In S. cerevisiae, pH signaling involves two PalH paralogues, Dfg16p and Rim21 (22). We reported previously that the A. nidulans sole ambient pH signaling 7-TMD receptor PalH is required for PacC processing (23). Using more sensitive pH shift experiments we now show (Fig. 1B) that processing is abolished in null palH72 cells, establishing that signaling is crucially dependent on PalH.
PalF Facilitates PalH Localization to the Plasma Membrane
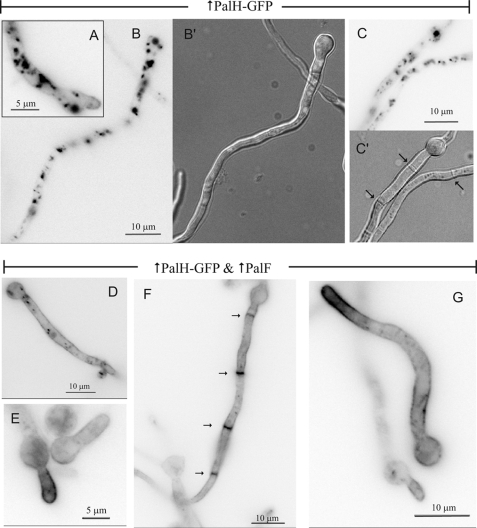
Endogenously tagged PalH-GFP is undetectable by epifluorescence (data not shown). When PalH-GFP is transiently overexpressed, the steady-state level at the plasma membrane is low, and the receptor predominates in endosomes and vacuoles unless the helper plasma membrane protein PalI is co-overexpressed (19) (Fig. 2, A–C). Because β-arrestins (β-ARs) bind plasma membrane phosphoinositides and such binding possibly contributes to their plasma membrane recruitment (24), we hypothesized that PalF might also stabilize PalH at this locale. This was indeed the case: in strains co-overexpressing PalH-GFP and PalF, PalH-GFP, although still reaching endosomes, predominates in the plasma membrane, strongly labeling septae (Fig. 2, D–G). Because we used pH 5 conditions, under which little signaling takes place, these experiments strongly suggest that PalF and PalH interact in the absence of receptor stimulation.
FIGURE 2.
PalH plasma membrane localization is assisted by PalF. GFP epifluorescence microscopy images (inverted contrast) of cells overexpressing PalH-GFP alone (A–C) or in combination with PalF-FLAG or PalF-HA3 (D–G). B′ and C′ are Nomarski images. A and B, PalH-GFP predominates in internal endosomes/vacuoles and localizes to the plasma membrane in growing tips (arrow). C, several septae (arrowed in C′) that show no PalH labeling. D and E, germlings showing that co-overexpression of PalF-FLAG promotes PalH-GFP plasma membrane localization. F, a long PalF-HA3 cell showing PalH-GFP plasma membrane labeling, which is particularly prominent in septae (arrowed). G, example of PalH-GFP germling co-overexpressing PalF-HA3.
C-terminal Ub Attachment to PalF Leads to Protein Instability
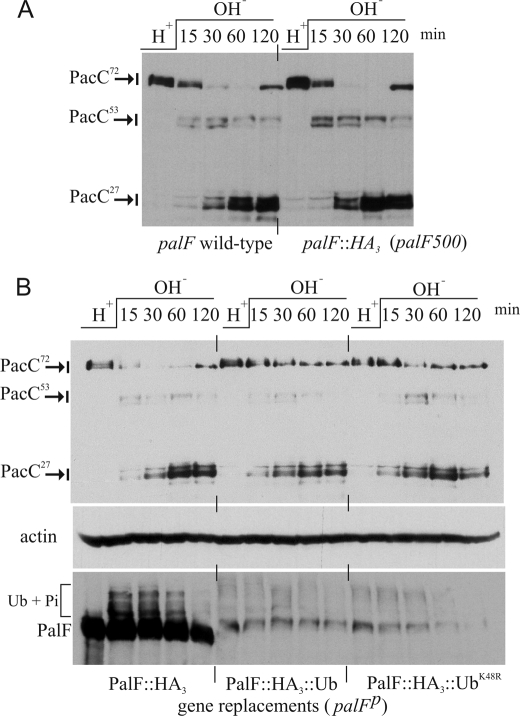
To investigate the role of ubiquitination in PalF function, we constructed, by gene replacement, two HA-tagged palF alleles respectively encoding wild-type (wt) PalF (palF500) and a protein fusion consisting of wt PalF with a single Ub moiety attached to its C terminus (palF-Ub). Gene-replaced strains also expressed Myc3-PacC, allowing us to monitor the two-step processing of the transcription factor as readout of alkaline pH signaling (4). palF500 strains are phenotypically wild-type in their PacC processing pattern (Fig. 3A).
FIGURE 3.
Effect of HA3 tagging and of covalent Ub attachment on PalF levels and PacC processing. A, C-terminal HA tagging of palF by gene replacement does not impair pH signaling, as determined by PacC processing analysis in pH shift experiments. B, upper panel, PacC processing is slightly delayed in palF-Ub and palF-UbK48R gene replaced strains relative to the palF500 control without any Ub tag. Lower panel, covalent attachment of Ub or UbK48R leads to a marked reduction in PalF levels in the corresponding gene replaced alleles.
PalF is detected as a single band at acidic pH (Fig. 3B, lower panel). Shifting cells to alkaline pH led to detection of lower mobility phosphorylated and oligo-ubiquitinated species, as described (5). C-terminal Ub attachment to PalF led to decreased mobility, but additionally resulted in a marked reduction in PalF levels (Fig. 3B). This reduction was not unexpected, because a similar finding had been reported for a mammalian β-AR2-Ub chimera (25). Very likely, additional Ub moieties are conjugated to Lys residues in β-AR2- and PalF-attached Ub, thus targeting the proteins to the proteasome.
Although all seven Lys residues within Ub itself can serve as acceptors for another Ub (26), Lys-48-linked polyubiquitin chains with four or more units appear to be the most common proteasome targeting signal (27). Because the predominant site for additional Ub conjugation within yeast Ub is Lys-48 (26) and there are only sporadic reports of the same polyubiquitin chain containing more than one linkage (28), we hypothesized that Lys-48 → Arg substitution within PalF-attached Ub might detectably impair polyubiquitination/proteasome targeting of the protein. However, Fig. 3B shows that this substitution did not increase PalF levels (Fig. 3B), suggesting that other Lys residues in PalF-UbK48R are poly-ubiquitinated. Thus, in view of the similar steady-state levels of PalF-Ub and PalF-UbK48R, the corresponding gene-replaced strains were expected to be phenotypically indistinguishable. Indeed, in agreement with this and with the low levels of PalF seen with both strains, PacC processing was slightly and similarly delayed by palF-Ub and palF-UbK48R (Fig. 3B, upper panel).
Forced Expression of PalF-Ub or UbK48R Results in Constitutive Pathway Activation
We next made constructs in which expression of the wt and the two Ub versions of PalF was driven by the moderately strong promoter gpdAmini (18), with the aim of compensating for decreased stability with increased synthesis of the Ub fusion proteins. These constructs were targeted in a single copy to the pyroA locus as reported previously. In palF500 gene-replaced cells, wt PalF levels decreased as the culture pH was increased (Fig. 4). This reduction in PalF levels possibly reflects that alkaline ambient pH triggers a negative transcriptional feedback mechanism that operates through the palF promoter.3 In contrast, because the gpdAmini promoter is pH-independent, such transcriptional feedback was expected to be bypassed with the constructs. Fig. 4 illustrates the degree of overexpression attained for wild-type PalF with the gpdAmini driver compared with the gene replacement and shows that PalF levels driven by the constitutive promoter are virtually indistinguishable at the three tested pH values. We conclude that gpdAmini drives pH-independent PalF overexpression.
FIGURE 4.
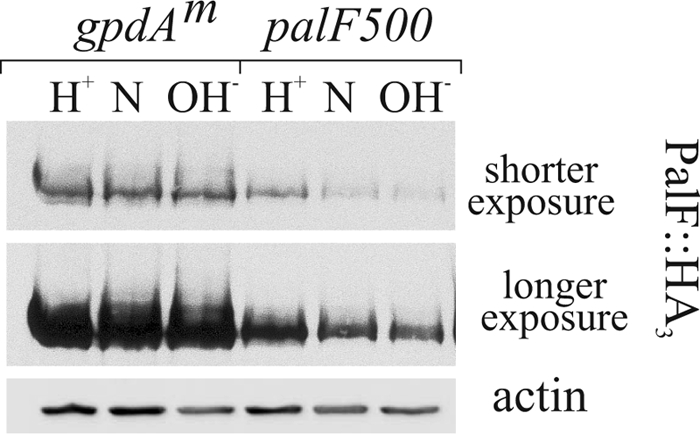
pH independent overexpression of PalF using the gpdAmini driver. Western analysis of cells expressing PalF-HA3 from the gene replacement allele palF500 or under the control of the gpdAmini promoter. H+, N, and OH−: acidic, neutral, and alkaline growth conditions, respectively. Two different exposures of the same PalF-HA3 blot are shown as indicated. Actin was used as loading control.
Western blots of strains expressing PalF-ubiquitin fusion proteins driven by gpdAmini showed that attachment of Ub or UbK48R still led to a marked reduction in PalF levels relative to the wild-type, confirming that the chimeras are unstable (Fig. 5A, lanes 3–5 and 7–9, α-HA). However, using the gpdAmini driver, PalF-Ub and PalF-UbK48R levels approached the physiological levels of PalF (Fig. 3A, α-HA blots, lanes 2, 4, 5, 6, 8, and 9). Thus, we will hereafter use the term “forced expression” to underline the fact that, in contrast to the wild-type, the PalF-Ub fusion proteins are not actually overexpressed in terms of their steady-state levels.
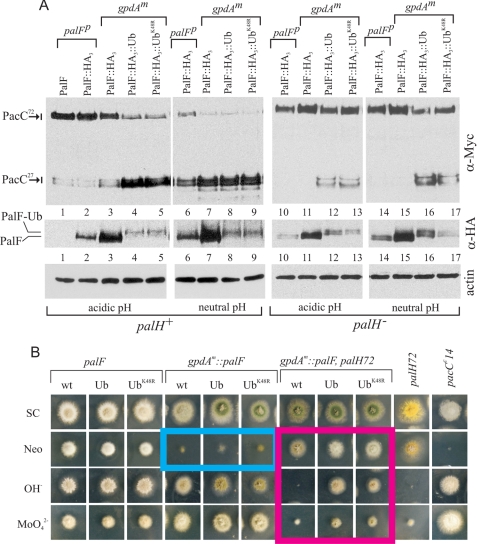
FIGURE 5.
Constitutive activation of the pathway by forced expression of PalF-Ub. A, Western blot analyses of PacC forms (α-Myc), PalF-HA3 (α-HA), and actin (as loading control) in wild-type and null palH72 cells, which were cultured at acidic or neutral conditions, under which considerable pH signaling takes place. Note that palH72 cannot grow under alkaline conditions. B, diagnostic growth tests of pH regulation. SC, synthetic complete. Neo, SC plus 1 mg/ml neomycin sulfate; OH−, medium adjusted to ∼pH 8 with 0.2 m Na2HPO4. MoO24−, SC containing 12.5 mm sodium molybdate. pacCc14 is an alkalinity-mimicking pacC mutation leading to constitutive activation of the pathway.
We next examined the effects of Ub attachment to PalF on PacC processing. At pH 5.2, leading to low pH signaling activity, unprocessed PacC72 largely predominates in wt (untagged) and palF500 controls, with only a small fraction of PacC seen as processed PacC27 (Fig. 5A, lanes 1 and 2). Overexpression of wt PalF led to a weak increase in PacC27 (Fig. 5A, lane 3). In sharp contrast, in strains expressing PalF-Ub or PalF-UbK48R it was the PacC27 form that largely predominated, indicating that these proteins lead to signaling at acidic pH (Fig. 5A, lanes 4–5), despite the fact that the level of these proteins were, approximately, physiological. At pH 6.8 substantial pH signaling takes place (3), but overexpression of wt PalF clearly leads to more efficient conversion of PacC72 to PacC27 (Fig. 5A, compare lanes 6 and 7), similar to that of the Ub constructs. Thus PalF levels seem limiting for pH signaling, and, importantly, PalF-Ub proteins lead to pH-independent processing even though they are not actually overexpressed.
Diagnostic growth tests (Fig. 5B) agreed with biochemical data. Molybdate and neomycin are toxic for A. nidulans. Strains carrying pacCc mutations leading to constitutive activity of the pathway at any ambient pH are more tolerant of molybdate and more sensitive to neomycin than the wild-type (21) (Fig. 5B; pacCc14 (last column) should be compared with the wt (first column)). In contrast, strains carrying pal− mutations preventing signaling are unable to grow at alkaline pH and are more sensitive to molybdate and more resistant to neomycin than the wild type (21) (Fig. 5B, palH72 (penultimate column) should be compared with the wt (first column)). Thus, these toxicity tests are diagnostic of the mutational activation or impairment of the pathway (29). The PalF-overexpressing and the two PalF-Ub (with or without K48R) strains were slightly more tolerant of molybdate than the palF500 wt control and, unlike the latter, did not grow on neomycin (Fig. 5B, blue box), indicating that the three transgenes activate the pathway to a detectable and similar extent. As diagnostic growth tests are carried out at pH 6.5, these data agree with PacC processing Western blots corresponding to neutral pH (∼6.8) conditions, which showed that the three transgenes promoted PacC processing to a similar extent (Fig. 5A, compare lanes 7–9 with lane 6).
We next constructed Myc3-PacC strains carrying PalF, PalF-Ub, and PalF-UbK48R transgenes in a null palH72 mutant background. palH72 is lethal at alkaline pH, but palH null mutant cells can grow at neutral pH, enabling PacC processing analysis under pH signaling conditions. PacC processing analysis at pH ∼ 6.8 demonstrated that, in cells overexpressing wt PalF, PacC72 processing is abolished when PalH is absent (Fig. 5A, lanes 7 and 15), demonstrating that PalF overexpression does not bypass the ambient pH receptor to any significant extent. In contrast, considerable PacC72 processing was detected in palH72 strains upon forced expression of PalF-Ub or PalF-UbK48R at either acidic (lanes 12–13) or neutral (lanes 16–17) pH, demonstrating that the PalF chimeras bypass the ambient pH 7-TMDR to a considerable extent, despite the fact that their steady-state levels are approximately physiological (Fig. 5A, lanes 10, 12, and 13; lanes 14, 16, and 17; and α-HA blots). In agreement, palH72 strains overexpressing PalF resembled single palH72 mutants in that they were slightly more resistant to neomycin than the wt (Fig. 5B, compare with first column) and could not grow on molybdate or alkaline pH plates, indicating that these strains are unable to activate the pathway. In contrast, PalF-Ub and PalF-UbK48R constructs permitted growth at alkaline pH and increased molybdate tolerance of palH72 strains (Fig. 5B, red box), demonstrating that the transgenes activate the pathway to a detectable extent even in the absence of PalH. We conclude that PalF-Ub and PalF-UbK48R lead to “true” constitutive activation of pH signaling, conferring a considerable degree of 7-TMDR independence; however, we also note that PalH contributes detectably to the PalF-Ub-mediated constitutive activation of signaling at acidic pH (Fig. 5A, lanes 4, 5, 12, and 13), indicating that the presence of the receptor further promotes signaling by PalF-Ub mutants. Finally the finding that the promoting effect on signaling resulting from PalF (without Ub) overexpression is strictly PalH-dependent further suggests that PalF is limiting and that its overexpression hypersensitizes the pathway to the pH signal.
pH shift experiments in which cells were pre-cultured at pH ∼ 4.2 and subsequently transferred to pH ∼ 8.2 further supported the above conclusions: Compared with the control expressing physiological levels of PalF (displayed in Fig. 3, A and B), wt PalF overexpression accelerated PacC72 processing in response to alkaline pH in a strictly PalH-dependent manner and alkaline ambient pH boosted the processing-promoting effect of forced PalF-Ub expression in the presence but not in the absence of the PalH receptor (supplemental Fig. 1).
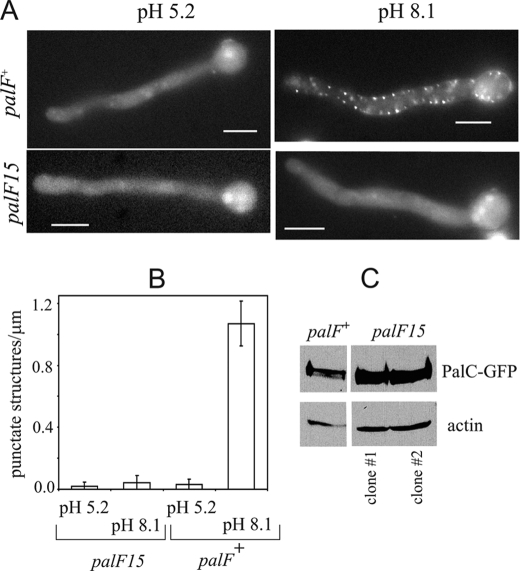
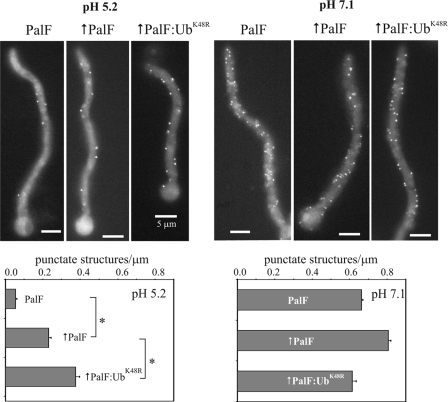
Recruitment of PalC to Cortical Structures under Acidic Conditions by Forced Expression of PalF-Ub
pH signaling can be monitored at the subcellular level by determining the alkaline ambient pH- and PalH-dependent recruitment of PalC-GFP to cortical structures by epifluorescence (10) (Fig. 6, wild-type control). We showed that this recruitment is prevented by the nonsense mutation palF15, truncating PalF within the N-terminal arrestin domain, without affecting PalC-GFP steady-state levels (Fig. 6); this demonstrated that PalF acts upstream of or at the level of PalC in this process. Therefore we predicted that expression of PalF-Ub resulting in alkalinity mimicry would lead to the cortical recruitment of PalC-GFP under acidic conditions. Fig. 7 shows that this is indeed the case. In wt cells expressing physiological levels of PalF the number of PalC cortical structures seen under moderately acidic (pH 5.2) conditions was ∼10% of that under neutral (pH 7.1) conditions. wt PalF overexpression (↑PalF in Fig. 7) significantly increased the number of cortical structures under acidic conditions (and also appeared to increase this number slightly under neutral conditions), in agreement with the conclusion, derived from Western analyses of PacC72 processing, that PalF overexpression slightly hypersensitizes the pathway. Notably, the increase in PalC-GFP structures under acidic conditions was even higher upon forced expression of PalF-UbK48R (Fig. 7, ↑PalF-UbK48R; this difference was statistically significant, bottom left panel). Indeed PalF-UbK48R cells showed, under acidic (low signaling) conditions a density of cortical structures amounting to 57% of those counted in the wild-type under neutral (signaling-inducing) conditions (Fig. 7, lower bar diagrams; compare PalF with ↑PalF-UbK48R), despite the fact that the steady state of PalF- UbK48R is similar to the physiological levels of PalF (see above). In contrast, the number of cortical PalC structures in PalF-UbK48R cells was not significantly different from the wt under neutral pH conditions.
FIGURE 6.
Recruitment of PalC-GFP to cortical structures is dependent on palF. A, germlings were cultured under acidic conditions and shifted to alkaline conditions before being photographed. B, PalC-GFP cortical punctate structures were counted in n = 23 germlings from palF+ and palF15 strains and normalized to the length of the germ tubes. Error bars indicate standard error. The difference between palF15 and palF+ cells observed at alkaline pH was statistically significant (p < 10−10; Student's test). C, Western blot analyses of PalC-GFP in the palF+ and in two independent palC-gfp palF15 clones isolated from the progeny of the cross. Actin was used as loading control.
FIGURE 7.
PalF-UbK48R promotes PalC-GFP recruitment to cortical structures under acidic conditions. Micrographs are representative GFP fluorescence images. Forced expression is indicated by up-pointing arrows. Bar diagrams plot the number of these structures in a sample of 25–30 germlings, normalized to the germ tube length. Error bars indicate standard error. *, denotes that differences between the indicated samples are statistically significant (p < 0.001).
DISCUSSION
Unlike other fungal arrestins mediating the endocytic down-regulation of plasma membrane transporters (14, 16), PalF plays a positive role in ambient pH signaling. Because PalF is ubiquitinated in a strictly alkaline ambient pH- and 7-TMDR PalH-dependent manner (5), and because covalent attachment of Ub to PalF results in constitutive pathway activation, we conclude that ubiquitination of PalF promotes downstream ambient pH signaling events.
Mammalian β-ARs are ubiquitinated in an agonist-dependent manner (30). β-AR ubiquitination stabilizes 7-TMD·β-AR complexes, regulates their intracellular trafficking, and, importantly, plays a major role in stabilizing downstream MAPK signaling complexes (25, 31). Our work, in conjunction with a recent study on S. cerevisiae Rim8 (17), strongly indicates that PalF ubiquitination promotes the assembly of cortical pH signaling complexes. Rim8 binds the ESCRT-I subunit Vps23 through interaction between an SXP motif in the former with the UEV domain in the latter. An ubiquitinated Lys in Rim8 contributes to this binding. Two components of the pal pathway, PalC and PalA, interact with Vps32. We show here that the alkaline pH-mediated recruitment of PalC to cortical structures (10) is dependent on PalF and that PalC recruitment is strongly promoted by PalF-Ub under acidic conditions. Together with the finding that Rim8 overexpression drives S. cerevisiae Vps23 to cortical sites (17), our results strongly support a model in which activation of A. nidulans PalH by alkaline pH leads to PalF ubiquitination, subsequent recruitment of AnVps23 to cortical sites, and sequential assembly of ESCRT-I, -II, and -III at these sites. Vps32-containing ESCRT-III polymers would facilitate the localization of PalC to these cortical structures. In view of the demonstrated structural similarity between arrestins and Vps26, a component of the retromer complex that regulates traffic from endosomes to the Golgi (32), it is tempting to speculate that PalF might be involved in the endocytic trafficking of cortical pH signaling structures. However, we would like to underscore that whether cortical pH-signaling complexes undergo endocytosis to mediate downstream signaling steps remains to be determined.
Unlike that of PalF, Rim8 ubiquitination is ambient pH- and 7-TMDR-independent. (Recall that S. cerevisiae has two ambient pH 7-TMDR, Rim21 and Dfg16; Rim8 ubiquitination is unaffected in the double deletion mutant (17).) This suggests that in S. cerevisiae the pH-signaling, arrestin-containing module is activated by inputs other than alkaline pH. One likely possibility for such alternative input is cell wall stress. It is well established that alkaline ambient pH leads to cell wall damage activating the Slt2 MAPK cell integrity pathway through the membrane sensor Wsc1. However, it has also been reported that calcofluor induces cell wall stress responses mediated by the PalH homologue Rim21 (33, 34). Thus, the actual signals and receptors regulating the interaction between Vps23 and Rim8 in S. cerevisiae remain to be clarified.
Our data show that PalF commands, through its ubiquitination, the cascade of intracellular events that mediate fungal adaptation to environmental pH. Our conclusion that expression of PalF-Ub leads to constitutive activation of pH signaling even in the absence of its cognate 7-TMDR should be considered in the context of the important positive-acting role that mammalian arrestins play as scaffolds of MAPK signaling complexes. It has been established that β-AR2 promotes activation of the c-Jun N-terminal MAP kinase (JNK3) in the absence of receptor stimulation (35). Indeed Song et al. (36) demonstrated that mutant β-AR2 defective in receptor binding is capable of promoting JNK3 phosphorylation, thus showing that the free arrestin is able to act as scaffold for the JNK3 activation cascade.
Supplementary Material
Acknowledgments
Thanks are due to Joan Tilburn and Herb Arst for critically reading the manuscript and to Elena Reoyo for technical assistance.
This work was supported by Grant BIO2009-7281 from Dirección General de Investigación (Spain) and by Comunidad de Madrid Grant SAL/0246/2006 (to M. A. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Table 1.
T. Múnera-Huertas, H. N. Arst, and J. Tilburn, personal communication.
- 7-TMD
- seven-transmembrane domain
- 7-TMDR
- 7-TMD receptor
- β-AR
- β-arrestin
- ESCRT
- endosomal sorting complex required for transport
- HA3
- three tandem copies of the human influenza virus hemagglutinin epitope
- JUN3
- c-Jun N-terminal kinase 3
- MAPK
- mitogen-activated protein kinase
- Myc3
- three tandem copies of the c-Myc epitope
- Ub
- ubiquitin
- UbK48R
- ubiquitin carrying a Lys-48 → Arg substitution
- wt
- wild type
- ART
- arrestin-related traffic adaptor.
REFERENCES
- 1.Peñalva M. A., Arst H. N., Jr. (2004) Annu. Rev. Microbiol. 58, 425–451 [DOI] [PubMed] [Google Scholar]
- 2.Peñalva M. A., Tilburn J., Bignell E., Arst H. N., Jr. (2008) Trends Microbiol. 16, 291–300 [DOI] [PubMed] [Google Scholar]
- 3.Orejas M., Espeso E. A., Tilburn J., Sarkar S., Arst H. N., Jr., Peñalva M. A. (1995) Genes. Dev. 9, 1622–1632 [DOI] [PubMed] [Google Scholar]
- 4.Hervás-Aguilar A., Rodríguez J. M., Tilburn J., Arst H. N., Jr., Peñalva M. A. (2007) J. Biol. Chem. 282, 34735–34747 [DOI] [PubMed] [Google Scholar]
- 5.Herranz S., Rodríguez J. M., Bussink H. J., Sánchez-Ferrero J. C., Arst H. N., Jr., Peñalva M. A., Vincent O. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12141–12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams R. L., Urbé S. (2007) Nat. Rev. Mol. Cell. Biol. 8, 355–368 [DOI] [PubMed] [Google Scholar]
- 7.Vincent O., Rainbow L., Tilburn J., Arst H. N., Jr., Peñalva M. A. (2003) Mol. Cell. Biol. 23, 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu W., Mitchell A. P. (2001) J. Bacteriol. 183, 6917–6923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boysen J. H., Mitchell A. P. (2006) Mol. Biol. Cell 17, 1344–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galindo A., Hervás-Aguilar A., Rodríguez-Galán O., Vincent O., Arst H. N., Jr., Tilburn J., Peñalva M. A. (2007) Traffic 8, 1346–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodríguez-Galán O., Galindo A., Hervás-Aguilar A., Arst H. N., Jr., Peñalva M. A. (2009) J. Biol. Chem. 284, 4404–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu W., Smith F. J., Jr., Subaran R., Mitchell A. P. (2004) Mol. Biol. Cell 15, 5528–5537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurevich E. V., Gurevich V. V. (2006) Genome Biol. 7, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikko E., Sullivan J. A., Pelham H. R. (2008) EMBO. Rep. 9, 1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikko E., Pelham H. R. (2009) Traffic 10, 1856–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin C. H., MacGurn J. A., Chu T., Stefan C. J., Emr S. D. (2008) Cell 135, 714–725 [DOI] [PubMed] [Google Scholar]
- 17.Herrador A., Herranz S., Lara D., Vincent O. (2010) Mol. Cell. Biol. 30, 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pantazopoulou A., Peñalva M. A. (2009) Mol. Biol. Cell 20, 4335–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calcagno-Pizarelli A. M., Negrete-Urtasun S., Denison S. H., Rudnicka J. D., Bussink H. J., Múnera-Huertas T., Stanton L., Hervás-Aguilar A., Espeso E. A., Tilburn J., Arst H. N., Jr., Peñalva M. A. (2007) Eukaryot. Cell 6, 2365–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peñas M. M., Hervás-Aguilar A., Múnera-Huertas T., Reoyo E., Peñalva M. A., Arst H. N., Jr., Tilburn J. (2007) Eukaryot. Cell 6, 960–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tilburn J., Sarkar S., Widdick D. A., Espeso E. A., Orejas M., Mungroo J., Peñalva M. A., Arst H. N., Jr. (1995) EMBO. J. 14, 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barwell K. J., Boysen J. H., Xu W., Mitchell A. P. (2005) Eukaryot. Cell 4, 890–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Negrete-Urtasun S., Reiter W., Diez E., Denison S. H., Tilburn J., Espeso E. A., Peñalva M. A., Arst H. N., Jr. (1999) Mol. Microbiol. 33, 994–1003 [DOI] [PubMed] [Google Scholar]
- 24.Gaidarov I., Krupnick J. G., Falck J. R., Benovic J. L., Keen J. H. (1999) EMBO. J. 18, 871–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shenoy S. K., Lefkowitz R. J. (2003) J. Biol. Chem. 278, 14498–14506 [DOI] [PubMed] [Google Scholar]
- 26.Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 27.Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) EMBO. J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pickart C. M., Fushman D. (2004) Curr. Opin. Chem. Biol. 8, 610–616 [DOI] [PubMed] [Google Scholar]
- 29.Peñalva M. A., Arst H. N., Jr. (2002) Microbiol. Mol. Biol. Rev. 66, 426–446, table of contensts [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lefkowitz R. J., Rajagopal K., Whalen E. J. (2006) Mol. Cell 24, 643–652 [DOI] [PubMed] [Google Scholar]
- 31.Shenoy S. K., Barak L. S., Xiao K., Ahn S., Berthouze M., Shukla A. K., Luttrell L. M., Lefkowitz R. J. (2007) J. Biol. Chem. 282, 29549–29562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi H., Rojas R., Bonifacino J. S., Hurley J. H. (2006) Nat. Struct. Mol. Biol. 13, 540–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serrano R., Martín H., Casamayor A., Ariño J. (2006) J. Biol. Chem. 281, 39785–39795 [DOI] [PubMed] [Google Scholar]
- 34.Castrejon F., Gomez A., Sanz M., Duran A., Roncero C. (2006) Eukaryot. Cell 5, 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller W. E., McDonald P. H., Cai S. F., Field M. E., Davis R. J., Lefkowitz R. J. (2001) J. Biol. Chem. 276, 27770–27777 [DOI] [PubMed] [Google Scholar]
- 36.Song X., Coffa S., Fu H., Gurevich V. V. (2009) J. Biol. Chem. 284, 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.