Abstract
The DNA polymerase encoded by bacteriophage T7 has low processivity. Escherichia coli thioredoxin binds to a segment of 76 residues in the thumb subdomain of the polymerase and increases the processivity. The binding of thioredoxin leads to the formation of two basic loops, loops A and B, located within the thioredoxin-binding domain (TBD). Both loops interact with the acidic C terminus of the T7 helicase. A relatively weak electrostatic mode involves the C-terminal tail of the helicase and the TBD, whereas a high affinity interaction that does not involve the C-terminal tail occurs when the polymerase is in a polymerization mode. T7 gene 2.5 single-stranded DNA-binding protein (gp2.5) also has an acidic C-terminal tail. gp2.5 also has two modes of interaction with the polymerase, but both involve the C-terminal tail of gp2.5. An electrostatic interaction requires the basic residues in loops A and B, and gp2.5 binds to both loops with similar affinity as measured by surface plasmon resonance. When the polymerase is in a polymerization mode, the C terminus of gene 2.5 protein interacts with the polymerase in regions outside the TBD. gp2.5 increases the processivity of the polymerase-helicase complex during leading strand synthesis. When loop B of the TBD is altered, abortive DNA products are observed during leading strand synthesis. Loop B appears to play an important role in communication with the helicase and gp2.5, whereas loop A plays a stabilizing role in these interactions.
Keywords: DNA, DNA Polymerase, DNA-Protein Interaction, DNA Replication, Surface Plasmon Resonance (SPR)
Introduction
Protein-protein interactions are essential for coordination of the multiple reactions that occur at a replication fork. The economy of proteins involved in bacteriophage T7 DNA replication has made it an attractive model for the study of these interactions. T7 DNA polymerase (gp5), an 80-kDa product of gene 5 of the phage, forms a tight 1:1 complex with the host protein, thioredoxin (trx)3 (1, 2). This interaction stimulates gp5 activity by increasing the processivity of nucleotide polymerization (3–5). gp5/trx physically interacts with the hexameric gene 4 protein (gp4) that possesses both helicase activity, required for unwinding of duplex DNA, and primase activity, required for the initiation of Okazaki fragment synthesis on the lagging strand (1, 6). The interaction of helicase and primase is essential to coordinate DNA synthesis and unwinding on the leading strand and primer handoff to the lagging strand DNA polymerase. The gene 2.5 protein (gp2.5) is a single-stranded DNA (ssDNA)-binding protein that interacts with both gp4 and gp5/trx and stimulates their DNA unwinding and polymerization activities, respectively (7, 8). We have recently shown that gp2.5 plays a role in the loading of T7 gp5/trx and helicase at a nick in duplex DNA (9).
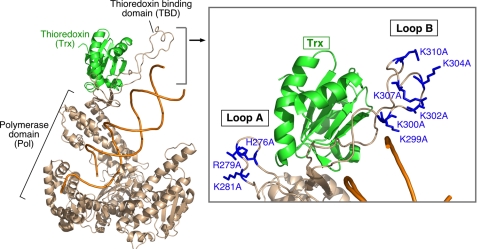
The communication between gp5, trx, gp4, and gp2.5 is dynamic and complex (4, 5, 10, 11). Although a member of the DNA polymerase I family, gp5 is unique for a 76-amino acid segment located between helices H and H1 in the thumb subdomain (see Fig. 1). Thioredoxin binds with high affinity to this segment designated the thioredoxin-binding domain (TBD) (12, 13). The binding of trx to the TBD structures this region such that its basic residues point toward the DNA binding crevice (see Fig. 1) (12).
FIGURE 1.
Crystal structure of gp5/trx in the presence of primer-template. gp5 is displayed in brown, trx is in green, and the primer-template is in orange. trx binds to the unique 76-residue segment (TBD) at the tip of the thumb (brown). The inset shows an enlargement of the TBD and indicates the position of the two basic loops within the TBD. Loop A is formed by residues 275–285. Residues 299–314 form loop B. The basic residues in loops A and B of the TBD that were replaced with alanine are indicated and shown in blue.
Both gp4 and gp2.5 have acidic C termini that are involved in interactions with gp5 (15, 16). Deletion mutations in the TBD revealed that the acidic tails of both gp4 and gp2.5 interact with the TBD of gp5. The C-terminal 21 residues of gp4 and gp2.5 have no sequence homology and are most likely unstructured; gp2.5 only crystallized when the C terminus was deleted (17), and the C-terminal tail of gp4 did not diffract in the crystals of gp4 (18). The interaction between the TBD of gp5 and the C-terminal tail of gp4 has a moderate affinity (KD = 90 nm) and is electrostatic in nature (11). A more stable interaction between gp5/trx and gp4 is observed when gp5/trx engages the primer-template strand during the nucleotide polymerization (half-life more than 10 min). Interestingly, this stable interaction does not involve the C-terminal tail of gp4 (4, 5). Collectively the two binding modes provide a processivity of greater than 17,000 nucleotides for the gp5/trx-helicase complex during leading strand DNA synthesis (3, 5, 14).
The interaction between the TBD of gp5 and the C-terminal tail of gp2.5 has an affinity (KD = 130 nm) similar to that between the TBD and the C-terminal tail of gp4, and it is also electrostatic in nature (11). However, in contrast to gp4, the C terminus of gp2.5 remains essential for the interaction with gp5/trx when gp5/trx binds to a primer-template strand (11). Furthermore, the affinity between gp5/trx and gp2.5 when gp5/trx is bound to DNA is not significantly increased as is the case for the interaction of gp5/trx with gp4. Interestingly, recent genetic studies identified another site of interaction of gp2.5 with gp5/trx. Two mutations, both of which reside in gene 5, suppress a dominant lethal point mutation in the C-terminal tail of gp2.5. These mutations affect residues in gp5 that reside near the tip of the thumb of gp5, in close proximity to its duplex DNA binding crevice (9, 19). The altered gp2.5 has a lower affinity for gp5/trx, and this affinity is partially restored by the suppressor changes in gp5 (9). The role of this interaction is crucial in enabling the polymerase to displace sufficient DNA for loading of the helicase (9).
The interactions of the primer-template, trx, gp4, and gp2.5 with portions of the TBD of gp5 are quite remarkable. Clearly this unique insert in the thumb subdomain plays a pivotal role in the assembly of the replisome, and it is a misnomer to consider it only a trx-binding domain, the first function of this segment to be identified. The crystal structure of gp5/trx bound to a primer-template reveals two small, solvent-exposed basic loops (loops A and B) in the TBD (see Fig. 1) (5, 12). Recent studies examined the effect of the basic residues in loop A and B on the interaction with gp4 (5). In these studies, basic charges in loop A were eliminated by substituting Ala for His-276, Lys-278, and Lys-281, forming a gp5 variant designated gp5-loopA. Similar removal of charges in loop B, where Ala was substituted for Lys-302, Lys-304, Arg-307, and Arg-310, yielded gp5-loopB (see Fig. 1). gp4 helicase binds to both gp5/trx variants but with 2–3-fold less affinity (5). However, when the charges are eliminated in both loops A and B (gp5-loopAB), gp4 fails to bind (5). Elimination of the charges in loops A and B of the TBD does not significantly affect the affinity of trx for the TBD (20).
The collective roles of the interaction of loop A and B with the acidic C-terminal tail of gp4 during DNA synthesis has been examined in leading strand synthesis. During leading strand synthesis, the polymerase dissociates from the primer-template strands every 5 kb on average (5, 14). Interestingly, the polymerase remains bound at the replication fork via the electrostatic interaction between loops A and B and the C-terminal tail of gp4, allowing gp5/trx to quickly rebind the primer-template strand and to continue DNA synthesis (4, 5). The communication between loops A and B with gp4 increases the processivity of leading strand synthesis from 5,000 to 17,000 bases on average per single copy of gp4 and gp5/trx (5, 13, 14).
It seems likely that the interaction of the acidic C-terminal tail of gp2.5 with the TBD of gp5 also involves the basic loops A and B in the TBD. However, such a role for loops A and B in an interaction with gp2.5 has not been shown. Furthermore, the role of the overlapping interactions between TBD and the C-terminal tails of gp4 and gp2.5 during DNA synthesis is not understood. In the present study we show that in the absence of DNA, gp2.5 interacts with loops A and B of the TBD in a manner similar to that seen between gp4 and gp5/trx. We find that gp2.5, like gp4, adopts a second mode of binding to gp5/trx when the latter is in a polymerization mode bound to a primer-template. In contrast to gp4, gp2.5 mediates both of these interactions with gp5/trx by its C-terminal tail. We also show that the communication between the C-terminal tails of gp4 and gp2.5 with loops A and B is required for appropriate assembly of the replication fork.
EXPERIMENTAL PROCEDURES
Preparation of Plasmids and Overproduction and Purification of Recombinant Proteins
Site-directed point mutations in gene 5 were constructed using polymerase chain reaction with plasmid pGP5 harboring gene 5. The mutagenesis, using a “Megaprimer” method, requires two separate polymerase chain reactions using PfuTurbo DNA polymerase (2, 20). Several constructs were made by changing the basic residues on loop A: His-276, Lys-278, and Arg-281, and those of loop B: Lys-299, Lys-300, Lys-302, Lys-304, Lys-307, Lys-310, to alanine. The construct with H276A, K278A, and R281A is referred to as gp5-loopA. The constructs made by various combinations of changes in the six residues in loop B changed to alanine are: K299A,K300A, referred to as gp5-loopB1; K302A,K304A, referred to as gp5-loopB2; K299A,K300A,K302A,K304A, referred to as gp5-loopB1+2; K302A,K304A,K307A, referred to as gp5-loopB3; K302A,K304A,K307A,K310A, referred to as gp5-loopB; and K304A,K307A,K310A, referred to as gp5-loopB4. Two additional constructs were made where charges were removed from both loops A and B, H276A, K278A, R281A, K304A, K307A, and K310A, referred to as gp5-loopAB4; and H276A, K278A, R281A, K302A, K304A, K307A, and K310A, referred to as gp5-loopAB. The identity of the constructs was confirmed by sequencing. gp5 variants were overproduced in Escherichia coli strain A307(DE3) that does not express trxA and then purified as previously described (2). trx was overproduced in E. coli strain A307(DE3) that does not express trxA and then purified using the procedures described previously (2, 12). gp2.5 was purified from E. coli BL21(DE3)pLysS cells overexpressing gene 2.5 as previously described (21). gp4 was overproduced and purified as described (15).
DNA Polymerase Assay
DNA polymerase activity was measured using M13 ssDNA annealed to a 24-nucleotide oligonucleotide primer as described previously (2, 20). The reaction contained 50 mm Tris-HCl (pH 7.5); 10 mm MgCl2; 5 mm dithiothreitol; 50 mm NaCl; 20 nm M13 mGP1–2 ssDNA annealed to a 24-nucleotide oligonucleotide with 500 μm each of dATP, dCTP, dGTP, and [3H]dTTP (2 cpm/pmol); 50 μg/ml bovine serum albumin; and 5 nm of gp5/trx in a total volume of 10 μl. The reaction mixtures were incubated at 37 °C for the indicated times and stopped by the addition of 5 μl of 0.25 m EDTA (pH 7.5). The incorporation of [3H]dTMP was measured on DE81 filter disks as described (20). To examine the role of gp2.5 proteins in this reaction, varying amounts of gp2.5 were added to the reaction.
Leading Strand DNA Synthesis Assay
Leading strand DNA synthesis catalyzed by gp5/trx and gene 4 helicase was measured using circular M13 containing a preformed replication fork (5) (see inset to Fig. 5). The replication fork was constructed by annealing M13 mGP1–2 ssDNA to an oligonucleotide (5′-TAATTCGTAATCATCATGGTCATAGCTGTTTCCT-3′).
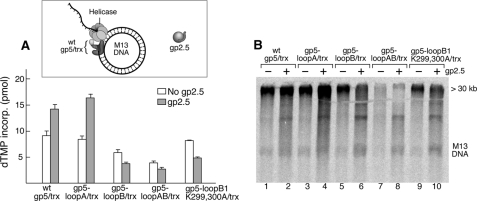
FIGURE 5.
Effect of gp2.5 on leading strand synthesis catalyzed by gp4 and gp5/trx. A, leading strand synthesis catalyzed by wild-type (wt) gp5/trx and the variants with alterations in loops A and B was measured using circular M13 double-stranded DNA bearing a preformed replication fork as shown in the inset. The reactions (10 μl) contained 20 nm DNA; 0.5 mm dATP, dGTP, dCTP, and [α-32P]dATP; 5 nm gp4 (hexamer); 5 nm of the indicated gp5; 500 nm trx; and 4 μm of gp2.5. After incubation at 37 °C for 10 min, the amount of [α-32P]dAMP incorporated into DNA was measured and presented as a bar graph. B, gel analysis of products of leading strand DNA synthesis. The radioactive reaction products from A were denatured and analyzed on a 0.6% alkaline-agarose gel by autoradiography.
The oligoribonucleotide was then extended by T7 DNA polymerase to obtain double-stranded DNA. Strand displacement DNA synthesis was carried out in a reaction mixture (10 μl) containing 20 nm circular M13 containing the M13 DNA with a preformed fork; 50 mm Tris-HCl (pH 7.5); 10 mm MgCl2; 5 mm dithiothreitol; 50 mm potassium glutamate; 500 μm each dCTP, dGTP, and dTTP; and 0.05 μCi of [α-32P]dATP, 5 nm gp4 (hexamer), and 2.5–20 nm of gp5/trx. gp5/trx and gp4 were incubated on ice for 15 min, and the reactions were initiated by transferring to 37 °C. After 10 min, the reaction was stopped by the addition of EDTA to a final concentration of 125 mm. The assays to study the effect of gp2.5 on leading strand synthesis assay contain varying amounts of the protein in the reaction. DNA synthesis was monitored by the amount of [α-32P]dAMP incorporated into DNA (20). To visualize the products of DNA synthesis, the DNA products were denatured and analyzed by electrophoresis in a 0.6% alkaline-agarose gel.
Physical Interaction of Proteins
Protein interactions were measured using surface plasmon resonance (SPR). SPR was performed using a Biacore 3000 instrument. Wild-type gp2.5 and gp4 were immobilized (150 and 3000 response units (RU), N-(3-dimethylaminopropyl-N′-ethylcarbodiimide/N-hydroxysuccinimide respectively) on a CM-5 (carboxymethyl-5) chip using chemistry. Immobilization was performed in 10 mm sodium acetate (pH 5.0) at a flow rate of 10 μl/min. Binding studies were performed in 20 mm Hepes (pH 7.5), 10 mm MgCl2, 250 mm potassium glutamate, and 5 mm dithiothreitol at a flow rate of 40 μl/min (5, 11). The chip surface was regenerated using 1 m NaCl at a flow rate of 100 μl/min. As a control, a flow cell was activated and blocked in the absence of protein to account for changes in the bulk refractive index. Apparent binding constants were calculated under steady-state conditions, and the data were fitted using BIAEVAL 3.0.2 software (Biacore).
To examine the binding of gp5/trx to gp4 and gp2.5 in the presence of primer/template, biotinylated DNA was coupled to a streptavidin-coated chip as previously described (see Fig. 5A) (5, 20). A template strand was used with a biotin group attached to the 5′ end and an annealed primer (11). The template DNA was coupled at a concentration of 0.25 μm in HBS-EP buffer (10 mm Hepes, pH 7.4, 150 mm NaCl, and 0.005% (v/v) Tween 20) at a flow rate of 10 μl/min. Binding studies of gp5/trx were performed at a concentration of 0.25 μm in 20 mm Hepes (pH 7.4), 5 mm MgCl2, 2.5 mm dithiothreitol, 200 mm potassium glutamate, and 1% (w/v) glycerol at a flow rate of 10 μl/min. gp4 was injected over the chip in the above buffer containing 0.1 mm ddGTP and 2 mm dTTP. gp2.5 was also injected under the same conditions. A flow cell blocked with biotin was used as a control to measure the nonspecific interaction and bulk refractive index of the sample buffer containing gp4. The chip surface was stripped of bound proteins by sequential injections of 150 μl of 1 m NaCl at a flow rate of 100 μl/min.
RESULTS
gp5 forms a tight complex with trx (KD of 5 nm) via the TBD and structures the TBD so that the basic residues now face the DNA binding cleft through which the primer-template passes (2, 12). Inspection of the crystal structure of gp5/trx revealed that some of these basic residues do not interact with trx or DNA (12). These residues are located in two loops (Fig. 1) comprised of portions of the TBD; loop A (residues 275–285) contains four basic residues, and loop B (residues 299–314) contains six basic residues (9). Using in vitro mutagenesis, the residues not critical for binding trx and DNA were identified and replaced with alanine to generate three altered gp5s (9): gp5-loopA/trx, where three residues in loop A (His-276, Lys-278, and Arg-281) were replaced by alanine; gp5-loopB/trx, where four of the basic residues in loop B (Lys-302, Lys-304, Arg-307, and Arg-310) were substituted by alanine; and gp5-loopAB/trx, where all of these seven residues in loop A and B were replaced with alanine (Fig. 1) (9). In the current study we have constructed six additional gp5 variants, gp5-loopB1 (Lys-299 and Lys-300 to Ala), gp5-loopB2 (Lys-302 and Lys-304 to Ala), gp5-loopB1+2 (Lys-299, Lys-300, Lys-302, and Lys-304 to Ala), gp5-loopB3 (Lys-302, Lys-304, and Arg-307 to Ala), gp5-loopB4 (Lys-304, Arg-307, and Arg-310 to Ala), and gp5-loopAB4 (His-276, Lys-278, Arg-281, Lys-304, Arg-307, and Arg-310 to Ala).
Affinity of gp2.5 for the Basic Loops in the TBD of gp5
Both gp2.5 and gp4 bind to gp5/trx (11). Earlier studies examined the roles of loops A and B in the interaction of gp5/trx with gp4 (15, 16). gp4 has two modes of interaction with gp5/trx: the electrostatic mode that does not require DNA with a moderate affinity (KD of 90 nm) and a much tighter mode (half-life of at least 10 min) when gp5 is in a polymerizing mode on a primer-template (15, 16). During the electrostatic mode, in the absence of primer-template, both loops A and B of gp5 individually contribute to the binding of gp4. When the basic charges in either loop are eliminated, the interaction is reduced by ∼50% relative to wild-type gp5/trx. Absence of any stable complex is observed only when the charges on both loops A and B (gp5-loopAB/trx) are removed.
SPR has also been used to show that gp2.5 physically interacts with gp5/trx with a KD of 130 nm (11). Furthermore, gp2.5 lacking the C-terminal acidic tail does not bind to gp5/trx, and wild-type gp2.5 does not bind to gp5/trx in which a portion of the TBD has been deleted. In contrast to gp4 there is no high affinity complex of gp5/trx and gp2.5 lacking its C-terminal tail observed when gp5/trx is bound to a primer-template in a polymerizing mode (11). However, the specific regions in the TBD that contribute to the binding of gp2.5 were not identified. Furthermore, the lack of binding of the truncated TBD used in these early studies to trx and to DNA negated any studies on the effect of the interaction of gp5/trx with a primer-template on the interaction with gp2.5. By eliminating only the basic charges in loops A and B, the binding of gp5 to trx and to DNA is not affected (20), and thus an examination of the binding of gp2.5 to gp5/trx in the presence of a primer-template is now feasible. Consequently we set the goal to use the gp5 proteins with alterations in loops A and B to first determine whether loops A and B are the sites of interaction with gp2.5 in the absence of the primer-template strand. The second goal was to examine the effect of binding of gp5/trx to the primer-template strand on its interaction with gp2.5.
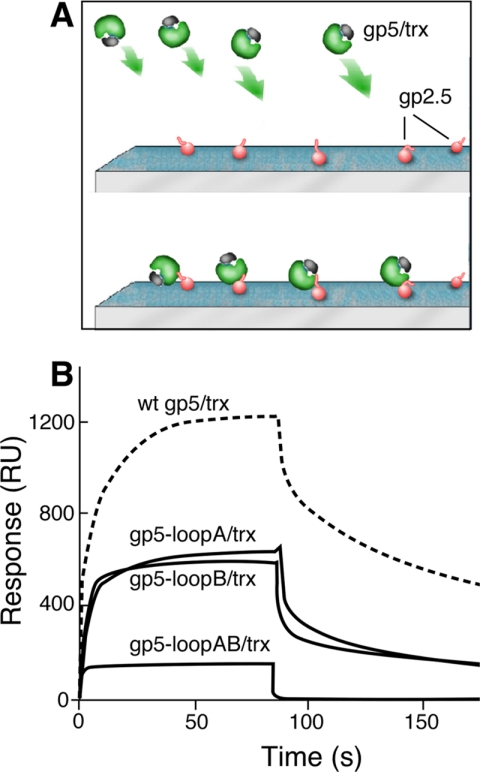
To examine the interaction between gp5/trx variants and gp2.5, we used SPR where gp2.5 was amine-coupled to a CM-5 chip and gp5/trx was flowed over the bound gp2.5 (Fig. 2). gp5-loopA/trx and gp5-loopB/trx bind ∼50% as well as does the wild-type gp5/trx. gp5-loopAB/trx in which the basic charges in both loops are eliminated does not form a stable complex with gp2.5. The decreased binding of these gp5/trx variants to gp2.5 is not due to a defect in the binding of gp5 to trx; wild-type gp5 binds to trx with a KD of 198 nm, gp5-loopA binds trx with a KD of 247 nm, gp5-loopB binds trx with a KD of 134 nm, and gp5-loopAB binds trx with a KD of 154 nm measured by observing the ability of trx to stimulate DNA synthesis catalyzed by gp5 (5, 20). These results demonstrate that the basic residues of loops A and B of the TBD are indeed the site of interaction of gp5 with the C-terminal tail of gp2.5, as is the case with gp4. Furthermore, their cooperative effect is also identical to that found for the interactions of gp5/trx with gp4 (Fig. 2) (5).
FIGURE 2.
Binding of gp2.5 to gp5/trx variants. The binding of gp5/trx to gp2.5 immobilized on a chip is measured by SPR. A, schematic representation of gp2.5 immobilized via its amino groups the sensor chip and gp5/trx flowing over immobilized gp2.5. B, binding studies were carried out as described under “Experimental Procedures.” 500 RU of gp2.5 are coupled to the chip, and the concentration of the gp5/trx variants in the flow buffer was 0.2 μm. A control flow cell lacking gp2.5 is used to subtract the RU resulting from the nonspecific interaction and bulk refractive index. The gp5 variants used in this study are wild-type (wt) gp5/trx, gp5-loopA/trx, gp5-loopB/trx, and gp5-loopAB/trx.
gp2.5 Uses Two Binding Modes in the Interaction with gp5/trx
The basic residues in loops A and B of the TBD bind gp4 and gp2.5 via the respective C-terminal tail when gp5/trx is not bound to DNA. In the interaction of gp5/trx with gp4, the binding of gp5/trx to primer-template in a polymerization mode switches the binding from the electrostatic interaction mode to a higher affinity mode that does not involve the C-terminal tail (4, 5). To determine whether loops A and B of the TBD are involved in the binding of gp2.5 to gp5/trx bound to DNA, we examined the binding of gp5-loopAB/trx to gp2.5 when the polymerase is in complex with a primer-template. A stable complex of gp5/trx or gp5-loopAB/trx and a primer-template was formed as described for obtaining stable complexes of gp5/trx-DNA for crystallization (5, 11, 12). gp5/trx forms a stable complex with a primer-template in which the primer strand is terminated by a 2′,3′-dideoxynucleotide (ddGMP in this experiment), provided that the next dNTP specified in the template (dTTP in this experiment) is present (5). Fig. 3A depicts gp5/trx bound to this construct. A total of 1000 RU of either gp5 or gp5-loopAB/trx was assembled onto 150 RU of the primer/template bound to the chip. The stable complex shown in Fig. 3 is dependent on the next incoming nucleotide, dTTP (data not shown). gp2.5 binds to both the preassembled gp5/trx and gp5-loopAB/trx on a primer-template as seen by the slow dissociation after the end of the injection of gp2.5 (Fig. 3B). The binding of gp2.5 to gp5-loopAB/trx in the presence of DNA is strikingly different from the lack of binding observed with gp5-loopAB in the absence of DNA (Fig. 2). These results demonstrate that gp5/trx also utilizes two different binding modes to interact with gp2.5 depending on whether it is bound to DNA or not as is the case for its interaction with gp4.
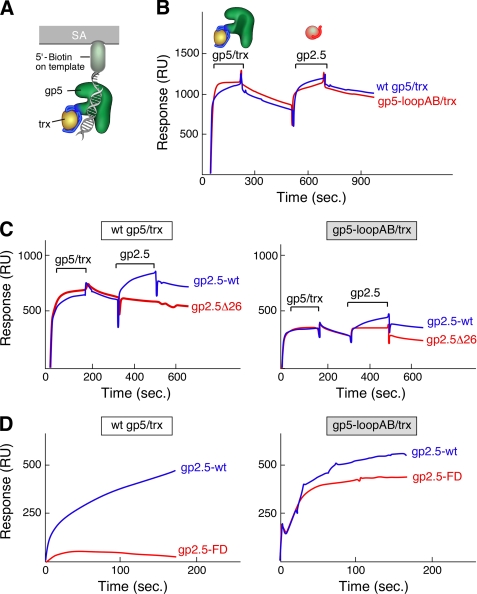
FIGURE 3.
Binding of gp2.5 to gp5/trx and gp5-loopAB/trx in presence of primer-template. A, schematic of the immobilized primer-template. The primer-template with biotin attached to the 5′ end of the template-strand is immobilized on the SA sensor chip. gp5/trx or gp5-loopAB/trx was flowed over the chip, followed by gp2.5 or gp2.5Δ26 (lacking the C-terminal tail). Binding studies were carried out as described under “Experimental Procedures.” B, 125 RU of the primer/template was coupled to a SA-coated chip. gp5/trx or gp5-loopAB/trx was injected at a concentration of 0.2 μm in a flow buffer containing 1 mm 2′,3′-dideoxy-GTP and 10 μm dTTP. The 125 RU resulting from the coupling of the primer-template was subtracted from the base line. gp2.5 or gp2.5Δ26 was then injected at a concentration of 4 μm in a flow buffer containing 1 mm 2′,3′-dideoxy-GTP and 10 μm dTTP. C, gp5/trx or gp5-loopAB/trx was bound to the immobilized primer-template as described in B. gp2.5 or gp2.5Δ26 were injected at a concentration of 4 μm in a flow buffer containing 1 mm 2′,3′-dideoxy-GTP and 10 μm dTTP. D, gp5/trx or gp5-loopAB/trx was bound to the immobilized primer-template as described in B. gp2.5 or gp2.5-FD was injected at a concentration of 4 μm in a flow buffer containing 1 mm 2′,3′-dideoxy-GTP and 10 μm dTTP. wt, wild type.
gp2.5Δ26 lacking the C-terminal tail does not form a stable complex with either wild-type gp5/trx or gp5-loopAB/trx bound to DNA in a polymerizing mode (Fig. 3C). Thus the acidic C-terminal tail is required for the interaction with gp5/trx both in the presence and in the absence of DNA. These results demonstrate that gp2.5 interaction is different from gp4 in that it always utilizes the C-terminal tail to interact with gp5/trx regardless of whether gp5/trx is bound to the primer-template strand or not.
We recently showed that gp2.5 was essential for the initiation of lagging strand DNA synthesis at a nick in duplex DNA (9). gp5/trx and gp4 helicase together mediate leading strand DNA synthesis with high processivity. However, gp5/trx can only idle at a nick, and gp4 requires a 5′-single-stranded tail of at least 36 nucleotides to assemble as a functional hexamer. In the presence of gp2.5, gp5/trx can catalyze sufficient strand displacement synthesis to generate the 5′-tail for the binding of gp4. An altered gp2.5 in which the C-terminal phenylalanine is switched with the adjacent aspartate (gp2.5-FD) cannot support T7 phage growth (19). gp2.5-FD, lacking this essential C-terminal phenylalanine, does not enable gp5/trx and gp4 to initiate leading strand DNA synthesis at a nick. The binding of gp2.5-FD to gp5/trx is greatly reduced relative to that observed with wild-type gp2.5 and gp2.5-FD. Suppressor mutations in gene 5 give rise to altered gp5s whose affinity for gp2.5-FD is increased and that can partially interact with gp2.5-FD to allow for the initiation of leading strand DNA synthesis (9, 22). Interestingly, the suppressor mutations do not reside within the TBD, suggesting that other sites on gp5/trx may be involved in interactions with gp2.5. Therefore we were intrigued to examine the interaction of gp5/trx and gp5-loopAB/trx bound to a primer/template with gp2.5-FD.
In these studies we injected gp2.5-FD over gp5/trx bound to primer-template (Fig. 3D). The wild-type gp2.5 is able to form a stable complex with the polymerase; however, gp2.5-FD fails to do so. Thus gp2.5-FD cannot bind with any higher affinity to gp5/trx bound to DNA than to gp5/trx in the absence of DNA (9). This result is not surprising considering the similar behavior we observed with gp2.5Δ26 lacking the C-terminal tail (Fig. 3, B and C). However, when gp2.5-FD was flowed over gp5-loopAB/trx bound to primer-template, a stable complex that is similar to that found with wild-type gp2.5 is observed, demonstrating that the loss of basic residues in loops A and B mimic the suppressor mutations in gp5 that partially restored the interaction of gp5-FD with gp5/trx. These results are surprising because one would have predicted that gp5-loopAB would not have an effect on the interaction with gp2.5-FD because the gp5 suppressor mutants are in a region other than the TBD. Nonetheless, the result of this experiment provides additional evidence of a second binding site on gp5 outside the TBD.
Stimulation of the Polymerase Activity of gp5/trx by gp2.5
When gp5/trx catalyzes the synthesis of DNA on ssDNA templates, it encounters impediments caused by the secondary structure in the DNA. gp2.5, upon binding to ssDNA, removes the secondary structures in the DNA, thus enhancing the processivity of the polymerase. It is not clear whether a physical interaction between gp5 and gp2.5 is important for this stimulation. In previous studies, the elimination of the C-terminal tail of gp2.5 resulted in inhibition of gp5/trx polymerization activity (9, 23). However, gp2.5 lacking the C-terminal tail not only is defective in its interaction with gp5/trx but also binds considerably more tightly to ssDNA (19). Consequently, the inhibition could arise via either mechanism or both mechanisms. Therefore, we have examined the ability of wild-type gp2.5 to stimulate polymerase activity of gp5-loopAB/trx in which the basic residues in loops A and B of the TBD were replaced with alanine (Fig. 4). By removing the basic residues of loops A and B, the interaction of gp2.5 with gp5/trx is eliminated without affecting the binding of gp2.5 to ssDNA.
FIGURE 4.
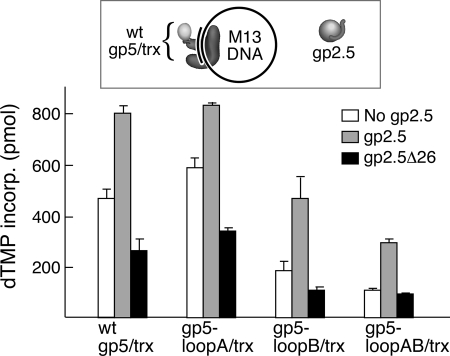
Effect of gp2.5 on DNA synthesis catalyzed by gp5/trx on ssDNA templates. The ability of wild-type (wt) gp5/trx and gp5/trx with alterations in loops A and B of the TBD to stimulate polymerase activity on ssDNA templates was examined as described under “Experimental Procedures.” The reactions (10 μl) contained M13 ssDNA annealed to 24-mer oligonucleotide (see inset) at a concentration of 10 nm; 0.5 mm each of dATP, [3H] dTTP, dGTP, and dCTP; and 1 and 0 nm of the indicated gp5/trx in the presence and absence of 4 μm gp2.5. After incubation at 37 °C for 3 min, the amount of DNA synthesis was determined by the amount of [3H]dTMP incorporated into DNA.
The activity of wild-type gp5/trx, gp5-loopA/trx, gp5-loopB/trx, and gp5-loopAB/trx on primed M13 ssDNA in the absence and presence of gp2.5 is shown in Fig. 4. Although gp5-loopA/trx supports polymerization equally as well as wild-type gp5/trx, gp5-loopB/trx and gp5-loopAB/trx show 2-fold decreases in synthesis (5). gp2.5 stimulates the polymerase activity of all of the proteins ∼2-fold. We conclude that the stimulatory affect of gp2.5 does not require a physical interaction of the C-terminal tail with gp5/trx and is most likely solely because of the removal of secondary structures. Replacing wild-type gp2.5 with gp2.5Δ26 lacking the C-terminal tail leads to inhibition of the polymerase activity for each of the polymerases.
Competition between gp2.5 and gp4 during Leading Strand DNA Synthesis
The C termini of gp4 and gp2.5 are quite similar in that both are abundant in acidic residues and contain a C-terminal phenylalanine. Therefore it is not surprising that both proteins interact with loops A and B of the TBD of gp5 (4, 5, 9, 16, 20, 24). Indeed, chimeric proteins in which the C-terminal tails of gp2.5 and gp4 have been exchanged support the growth of T7 phage lacking the corresponding wild-type protein (19, 20). To investigate the potential role for such overlapping binding sites at the replication fork, we investigated the effect of gp2.5 on leading strand DNA synthesis mediated by gp4 and wild-type gp5/trx and the variants of gp5/trx (Fig. 5A). In these experiments gp2.5 was added at a 1000-fold higher concentration than gp4 because it also binds to the ssDNA formed during leading strand synthesis in the absence of lagging strand synthesis. gp2.5 stimulates leading strand DNA synthesis catalyzed by wild-type gp5/trx ∼40% and that by gp5-loopA a little more than 50%. Interestingly, however, gp2.5 is inhibitory to gp5-loopB/trx and to gp5-loop-AB/trx.
Analysis of the products of the reaction by electrophoresis through 0.6% agarose gels reveals that those synthesized by gp5-loopB/trx or gp5-loopAB/trx are of considerably shorter length than those synthesized in the presence of wild-type gp5/trx (Fig. 5B). In the latter case they exceed 30 kb (as observed by the high molecular mass band), whereas with gp5-loopB/trx they fail to form the discrete high molecular length product and form a smear of heterogeneous lengths below 30 kb. The effect on leading strand synthesis is more severe with gp5-loopAB/trx polymerase where very little synthesis is observed. The abortive effect of gp2.5 on gp5-loopAB/trx most likely arises from the absence of basic charges on loop B and an additive effect from the removal of charges on loop A.
Lysines 299 and 300 of Loop B Are Communicating with gp4 and gp2.5
The above results suggest that one or more of the lysines in loop B of the TBD are involved in an interaction with gp4 and gp2.5 during leading strand DNA synthesis. The residues on loop B that may not be involved in interaction with DNA or trx are lysines at positions 299, 300, 302, 304, 307, and 310. To gain a more clear understanding of the role of these residues in leading strand synthesis, a series of gp5 mutants were created using an alanine scanning method whereby the charges were removed in different combinations, and the effects of these changes on leading strand synthesis were observed.
All of the gp5 variants carrying different group mutations of residues in loop B had a negative effect similar to that of gp5-loopB/trx on leading strand synthesis in the presence of gp2.5. This inhibitory effect became progressively greater as the number of negative charges were increased. However, among the combinations of amino acids examined, the effect of gp2.5 on lowering the processivity of the polymerase in leading strand synthesis is pronounced in a mutant in which only Lys-299 and Lys-300 in loop B were substituted with alanine (gp5-loopB1 K299A,K300A/trx). As shown in Fig. 5, gp5-loopB1 K299A,K300A/trx is almost indistinguishable from gp5-loopB/trx both with regard to the decrease in leading strand synthesis in the presence of gp2.5 and in the abortive DNA synthesis observed on gel analysis (Fig. 5B, lanes 9 and 10). In control experiments we found that DNA synthesis catalyzed by gp5-loopB1 K299A,K300A/trx was identical to that of wild-type gp5/trx and that gp5-loopB1 K299A,K300A had the same affinity for trx (data not shown).
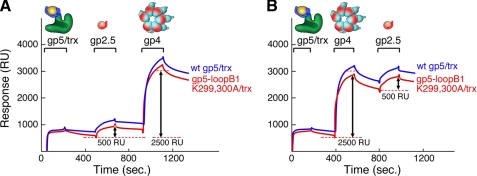
gp4 and gp2.5 Bind to Two Independent Sites on gp5/trx in Presence of Primer-Template
The above results combined with previous reports (5, 11) demonstrate that when gp5/trx binds to DNA, it switches its binding mode with gp4 and gp2.5 to a non-TBD-dependent interaction mode. To address whether gp4 and gp2.5 are competing for the same binding site when gp5/trx is bound to DNA, we utilized the ability of SPR to detect binding in real time and investigated how gp4 and gp2.5 interact if they are sequentially introduced to the gp5/trx-primer-template complex. In these experiments, the primer-template described in Fig. 3A was immobilized on a SA chip, and wild-type gp5 or gp5-loopB1 K299A,K300A/trx, which mimics gp5-loopB/trx in its inhibitory effect on leading strand DNA synthesis, was flowed over the chip. Both polymerases exhibit the same strong affinity for the primer-template in the presence of the next incoming nucleotide as described in Fig. 3A; there is little dissociation after the injection is completed. gp2.5 and gp4 were then consecutively flowed over the stably bound gp5/trx. Both proteins form stable complexes with gp5/trx and gp5-loopB1 K299A,K300A/trx, regardless of the order in which they were injected (Fig. 6). Identical results were observed with gp5-loopAB/trx (data not shown). These results suggest that gp4 and gp2.5 bind to separate sites beyond the TBD in the presence of primer-template. Consequently the inhibition of leading strand synthesis observed upon the addition of gp2.5 is not due to a competition of gp4 and gp2.5 for the same site on gp5/trx once gp5/trx is bound to DNA.
FIGURE 6.
Binding of gp2.5 and gp4 to gp5/trx and gp5-loopB1 K299A,K300A/trx bound to a primer-template. The primer-template with biotin attached to the 5′ end of the template-strand is immobilized on the SA sensor chip. gp5/trx or gp5-loopB1 K299A,K300A/trx was flowed over the chip, followed by gp2.5 or gp4. Binding studies were carried out as described under “Experimental Procedures.” A, 125 RU of the primer/template was coupled to a SA-coated chip that was subtracted from the base line. gp5/trx or gp5-loopB1 K299A,K300A/trx was injected at a concentration of 0.2 μm in a flow buffer containing 1 mm 2′,3′-dideoxy-GTP and 10 μm dTTP. gp2.5 was then injected at a concentration of 4 μm in a flow buffer containing 1 mm 2′.3′-dideoxy-GTP and 10 μm dTTP. gp4 was injected at a concentration of 0.7 μm (monomer) in the same flow buffer. B, gp5/trx or gp5-loopB1 K299A,K300A/trx was bound to the immobilized primer-template as described for A. gp4 was injected at a concentration of 0.7 μm (monomer) in a flow buffer containing 1 mm 2′,3′-dideoxy-GTP and 10 μm dTTP. gp2.5 was then injected at a concentration of 4 μm in the same flow buffer. The start and end of the injections of gp5/trx, gp2.5, and gp4 are indicated.
gp2.5 Assists gp5/trx in Loading onto DNA during DNA Synthesis
The observation that gp5/trx bound to DNA can interact noncompetitively with gp2.5 and gp4 simultaneously indicates that the latter two proteins interact with independent sites outside the TBD when gp5/trx is bound to DNA. Therefore the stimulation of leading strand synthesis by gp2.5 must result from its binding to this non-TBD site. This interaction may allow for a conformational change of gp5/trx that leads to better utilization of the primer for initiation of DNA synthesis, or it could result in a conformational change in gp5/trx that enhances its interaction with gp4 helicase during strand displacement synthesis.
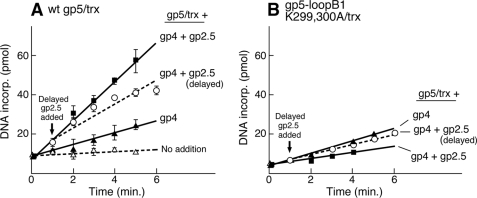
To examine the mechanism of stimulation by gp2.5, we investigated the effect of gp2.5 on the rate of leading strand synthesis when gp2.5 is added either at the start of the reaction or to an ongoing leading strand synthesis (Fig. 7). When gp2.5 is present prior to the start of the reaction, it enhances the rate of DNA synthesis 3-fold relative to that observed in its absence (Fig. 7A). When gp2.5 is added 1 min after the start of the reaction, there is only a 2-fold stimulation (Fig. 7A). When gp5/trx is replaced by gp5-loopB1 K299A,K300A/trx under identical conditions, we observe a different effect. When gp2.5 is present at the start of the reaction, it inhibits leading strand synthesis catalyzed by gp5-loopB1 K299A,K300A/trx (Fig. 7B). However, this effect is not seen when gp2.5 is added 1 min after the start of the reaction (Fig. 7B). These results suggest that the presence of gp2.5 at the initial stage of the reaction assists in the loading of gp5/trx onto the fork. This loading most likely requires binding of the polymerase via loop B to the C-terminal tail of gp4.
FIGURE 7.
Effect of gp2.5 on leading strand DNA synthesis. The rate of leading strand synthesis catalyzed by gp5/trx (A) and gp5-loopB1 K299A,K300A/trx (B) was measured using circular M13 double-stranded DNA bearing a preformed replication fork as shown in Fig. 5. The reactions (10 μl) contained 20 nm DNA; 0.5 mm dATP, dGTP, dCTP, and [3H]dTTP; 5 nm gp4 (hexamer); 5 nm of the indicated gp5; and 500 nm trx. The reaction mixtures were incubated at 37 °C for the indicated times and stopped by the addition of 5 μl of 0.25 m EDTA (pH 7.5). The incorporation of [3H]dTMP was measured on DE81 filter disks as described (2). The amount of [3H]dTMP incorporated into DNA was measured and presented as a bar graph. To study the effect of gp2.5 in the reaction, 4 μm of protein was added to the reaction prior to the start of the reaction (▾) and 1 min after the start of the reaction (♦). wt, wild type.
DISCUSSION
Both gp2.5 and gp4 physically interact with the TBD of gp5/trx (4, 5, 8, 10, 11). Previous studies have shown that in the case of gp4, the electrostatic interaction involves the acidic C-terminal tail of gp4 and the two basic loops in the TBD (5). Because gp2.5 also has an acidic C-terminal tail, it seemed likely that it too uses this motif to interact with loops A and B in the TBD. However, until the present study little was known about the site of interaction of gp2.5 with gp5/trx.
gp5/trx can interact with gp4 not only through this electrostatic mode but also through a much more stable mode that is observed when gp5/trx is bound to a primer-template during polymerization (4, 5). This latter mode does not involve the C-terminal tail of gp4 or the two basic loops of the TBD and can support nearly 5 kb of leading strand DNA synthesis in a single DNA binding event before dissociation from the polymerase. The electrostatic interaction of gp5/trx with gp4 allows for the transient capture of the dissociating polymerase and its rapid return to the primer-template to continue polymerization of nucleotides. The electrostatic interaction also provides sites for the assembly of accessory gp5/trx on one or more of the six subunits of the helicase to assure an available polymerase in the event that the polymerizing gp5/trx dissociates into solution. Thus the electrostatic mode increases the processivity of leading strand DNA synthesis at least 4-fold with products greater than 17 kb (3, 5, 14).
The TBD serves as the docking site for trx, which binds with high affinity (5 nm) to this 76-amino acid insert in the thumb subdomain of gp5. The binding of trx models the TBD such that basic residues on the surface contact the duplex region of the primer-template and lock it into the DNA binding crevice of gp5. Thus the TBD not only serves as a dynamic site for the electrostatic interaction with gp4 and gp2.5, but it also contacts DNA to increase processivity. trx is also involved in the electrostatic interaction of gp4 and gp5/trx in that the affinity of gp4 for loops A and B in the TBD is increased when trx is bound to gp5, most likely reflecting a conformational change in the TBD that alters the position of the residues on loops A and B (11). However, trx itself most likely also electrostatically interacts with gp4. Reversal of the charge of lysine at position 36 to glutamate obliterates the interaction of gp4 with gp5-loopB and leads to a reduction in the efficiency of leading strand synthesis (20).
An earlier study revealed that one interaction of gp2.5 with gp5/trx involved the acidic C-terminal tail of gp2.5 and the TBD of gp5/trx (11). However, the role of loops A and B and the effect of binding of the TBD to DNA on the interaction have not been studied previously. The present study was designed to identify the specific sites of interaction of gp2.5 with gp5/trx and to examine the binding of both gp2.5 and gp4 to gp5/trx and the consequences of these interactions during progression of the replisome in the presence of these three proteins.
As mentioned above, gp5/trx uses the TBD to interact with the acidic C-terminal tail of gp2.5 (11, 19). Deletion of the TBD of gp5 or of the C-terminal tail of gp2.5 abolishes the interaction between the two proteins. This electrostatic interaction displays a moderate affinity (KD = 130 nm). The deletion of the TBD eliminates many residues besides those constituting loops A and B. Thus the interaction with each loop was not revealed in the earlier study. By employing altered forms of gp5 where the basics residues in the TBD are eliminated by substitution of alanine for lysine, we show that gp5/trx uses loops A and B of the TBD to interact with the acidic C-terminal tail of gp2.5. The contribution of loops A and B to the interaction with gp5/trx is additive. Furthermore, omitting the charges on loop A and B has the same effect on the interaction of gp5 with gp2.5 as the deletion of the entire TBD. These results indicate that both loops A and B are responsible for the binding to the C-terminal tail of gp2.5.
The two loops are the same binding site for the C-terminal tail of gp4 described previously (5), and they account for the majority of the interaction of TBD with the C-terminal tail of gp4. The ability of gp5-loopA/trx, gp5-loopB/trx, and gp5-loopAB/trx to bind to DNA equally as well as the wild-type gp5/trx enabled us to investigate the interaction of gp2.5 on the binding of gp5/trx to DNA in a polymerizing mode. Such an examination would not be possible with the altered gp5 lacking the entire TBD because the lack of the TBD dramatically reduces the binding of trx that, in turn, decreases the affinity for DNA. We constructed a stable complex of gp5/trx and gp5-loopAB/trx with a primer-template in the presence of the incoming nucleotide and then examined its interaction with gp2.5 using SPR. gp2.5 forms equally stable complexes with wild-type gp5/trx and gp5-loopAB/trx. Therefore gp2.5, like gp4, also uses two binding modes in its interaction with gp5/trx: one in the absence of DNA and the other when the TBD is engaging a primer-template. However, unlike gp4, the C-terminal tail of gp2.5 always mediates this interaction. gp2.5 lacking the C-terminal tail fails to interact with gp5/trx in the presence of a primer-template. Thus, although gp2.5 adopts two binding modes in the interaction with gp5/trx, its C-terminal tail is essential for both modes, a clear distinction from gp4.
To gain insight into the role of having two overlapping binding sites of the C-terminal tails of gp2.5 and gp4, we investigated the sequential binding of gp2.5 and gp4 with the complex of gp5/trx and DNA by SPR. In addition we also examined leading strand DNA synthesis, catalyzed by gp5/trx and gene 4 helicase, in the presence of wild-type gp5 and those altered in loop A and B. gp2.5 stimulates leading strand synthesis mediated by wild-type gp5/trx and gp5-loopA/trx. Interestingly, in the case of either gp5-loopB/trx or gp5-loopAB/trx, this effect is reversed; gp2.5 inhibits leading strand synthesis. gp5-loopB/trx and gp5-loopAB/trx form abortive short DNA products in the presence of gp2.5 relative to the high molecular mass products in the absence of gp2.5. DNA synthesis catalyzed by gp5-loopA/trx is, however, stimulated by gp2.5, demonstrating the direct involvement of only loop B in the interaction of gp5/trx with gp4 and gp2.5 during leading strand synthesis. Using several point mutations in loop B, we show that this inhibitory action of gp2.5 on leading strand synthesis can be localized to two residues in loop B, Lys-299 and Lys-300. When gp5/trx is bound to DNA, it can bind sequentially to the same extent to both gp2.5 and gp4. These results confirm that the two proteins indeed have separate binding sites when gp5/trx is bound to the DNA. Furthermore, the results indicate that the inhibition of leading strand synthesis when loop B is altered is not due to competitive displacement of proteins from the polymerase. Examination of this inhibitory action of gp2.5 during leading strand synthesis by adding gp2.5 to gp5-loopB1 K299A,K300A to ongoing leading strand synthesis suggests that the inhibition is caused by the initial step of assembling the proteins at the replication fork.
gp2.5 binds to ssDNA (KD = 3 μm) and removes hairpins, secondary structures formed on ssDNA (19). These secondary structures impede the progress of the polymerase. The molecular mechanism by which gp2.5 stimulates the polymerization activity of gp5/trx is not clear. To determine whether the physical interaction between gp2.5 and gp5/trx is required for this stimulation, the effect of different mutations in the C-terminal tail of gp2.5 that result in a loss of interaction with gp5/trx were tested for their effect on the polymerization activity of gp5/trx.4 The extent of stimulation of the polymerase activity in the presence of gp2.5-FD, lacking the essential C-terminal phenylalanine, was almost the same as that observed with wild-type gp2.5, but gp2.5Δ26, lacking the entire C-terminal tail, inhibits DNA synthesis. However, gp2.5-FD has a slightly increased affinity for ssDNA, and gp2.5Δ26 has a much stronger affinity to ssDNA compared with wild-type gp2.5, making it difficult to evaluate the results. Our results show that gp2.5 can stimulate the polymerization activities of gp5-loopA/trx, gp5-loopB/trx, and gp5-loopAB/trx to the same extent, despite the fact that an altered protein such as gp5-loopAB/trx shows a defect in its interaction with gp2.5. This result suggests that the stimulation of polymerase activity by gp2.5 does not involve any physical interaction with the polymerase. The C-terminal tail of gp2.5 mimics DNA binding (25). In the absence of DNA it occupies the DNA binding crevice of gp2.5, and in the presence of DNA it is free for interaction with other proteins. A model was proposed where the C-terminal tail competes and displaces the ssDNA from the DNA binding crevice of gp2.5. Elimination of the physical interaction between gp2.5 and gp5/trx still results in stimulation of DNA synthesis. This finding suggests that the mechanism by which the polymerase interacts with the C-terminal tail of gp2.5 during DNA synthesis involves repulsive interactions between acidic and/or hydrophobic residues on gp5/trx and the C-terminal tail of gp2.5 to compete with the ssDNA and displace it.
The phenylalanine residue at the C terminus of gp2.5 is involved in direct interaction with the polymerase irrespective of the presence of DNA (19, 24). Relocation of the phenylalanine from the terminal position to the penultimate position of gp2.5 creates a dominant lethal mutant, gp2.5-FD, that does not enable gp5/trx and gp4 helicase to catalyze strand displacement synthesis from nicked DNA. A suppressor mutation screen identified mutations in gene 5 that resulted in amino acid alterations in the thumb region near the DNA binding crevice (9, 22). In the real time interaction studies, gp2.5-FD fails to interact with wild-type gp5 even in presence of DNA. Removal of the charges on the TBD can restore binding of gp2.5-FD to gp5-loopAB in a similar way as seen for the suppressor mutants of gp5 in the thumb region. This result strongly suggests a second site of interaction outside the TBD. The region of the C-terminal tail of gp2.5 binding to TBD may be constrained such that the phenylalanine can only bind to gp5 when it is positioned at the terminal position. In this model removal of the charges from the TBD removes this restraint so that the C-terminal tail can now bind gp5 by accommodating the phenylalanine residue at the penultimate position. Further studies using a variant of the polymerase encompassing the residues of the TBD domain and the suppressor mutations possibly could help understand the second site binding of the gp2.5 with gp5/trx in the presence of gp4.
Acknowledgments
We thank all members of the laboratory for helpful discussions, Boriana Marintcheva and Masateru Takahashi for gp2.5 protein, and Steven Moskowitz (Advanced Medical Graphics, Boston, MA) for help with figure preparation.
This work was supported, in whole or in part, by National Institutes of Health Grant GM 54937.
S. Ghosh and C. C. Richardson, unpublished data.
- trx
- thioredoxin
- TBD
- thioredoxin-binding domain
- ssDNA
- single-stranded DNA
- SPR
- surface plasmon resonance
- RU
- response units
- SA
- streptavidin.
REFERENCES
- 1.Richardson C. C. (1983) Cell 33, 315–317 [DOI] [PubMed] [Google Scholar]
- 2.Tabor S., Huber H. E., Richardson C. C. (1987) J. Biol. Chem. 262, 16212–16223 [PubMed] [Google Scholar]
- 3.Wuite G. J., Smith S. B., Young M., Keller D., Bustamante C. (2000) Nature 404, 103–106 [DOI] [PubMed] [Google Scholar]
- 4.Hamdan S. M., Richardson C. C. (2009) Annu. Rev. Biochem. 78, 205–243 [DOI] [PubMed] [Google Scholar]
- 5.Hamdan S. M., Johnson D. E., Tanner N. A., Lee J. B., Qimron U., Tabor S., van Oijen A. M., Richardson C. C. (2007) Mol. Cell 27, 539–549 [DOI] [PubMed] [Google Scholar]
- 6.Frick D. N., Baradaran K., Richardson C. C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7957–7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim Y. T., Tabor S., Bortner C., Griffith J. D., Richardson C. C. (1992) J. Biol. Chem. 267, 15022–15031 [PubMed] [Google Scholar]
- 8.Nakai H., Richardson C. C. (1988) J. Biol. Chem. 263, 9831–9839 [PubMed] [Google Scholar]
- 9.Ghosh S., Marintcheva B., Takahashi M., Richardson C. C. (2009) J. Biol. Chem. 284, 30339–30349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamdan S. M., Loparo J. J., Takahashi M., Richardson C. C., van Oijen A. M. (2009) Nature 457, 336–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamdan S. M., Marintcheva B., Cook T., Lee S. J., Tabor S., Richardson C. C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5096–50101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doublié S., Tabor S., Long A. M., Richardson C. C., Ellenberger T. (1998) Nature 391, 251–258 [DOI] [PubMed] [Google Scholar]
- 13.Yang X., Richardson C. C. (1996) J. Biol. Chem. 271, 24207–24212 [DOI] [PubMed] [Google Scholar]
- 14.Lee J. B., Hite R. K., Hamdan S. M., Xie X. S., Richardson C. C., van Oijen A. M. (2006) Nature 439, 621–624 [DOI] [PubMed] [Google Scholar]
- 15.Notarnicola S. M., Mulcahy H. L., Lee J., Richardson C. C. (1997) J. Biol. Chem. 272, 18425–18433 [DOI] [PubMed] [Google Scholar]
- 16.Kim Y. T., Tabor S., Churchich J. E., Richardson C. C. (1992) J. Biol. Chem. 267, 15032–15040 [PubMed] [Google Scholar]
- 17.Hollis T., Stattel J. M., Walther D. S., Richardson C. C., Ellenberger T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9557–9562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sawaya M. R., Guo S., Tabor S., Richardson C. C., Ellenberger T. (1999) Cell 99, 167–177 [DOI] [PubMed] [Google Scholar]
- 19.Marintcheva B., Hamdan S. M., Lee S. J., Richardson C. C. (2006) J. Biol. Chem. 281, 25831–25840 [DOI] [PubMed] [Google Scholar]
- 20.Ghosh S., Hamdan S. M., Cook T. E., Richardson C. C. (2008) J. Biol. Chem. 283, 32077–32084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bedford E., Tabor S., Richardson C. C. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 479–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marintcheva B., Qimron U., Yu Y., Tabor S., Richardson C. C. (2009) Mol. Microbiol. 72, 869–880 [DOI] [PubMed] [Google Scholar]
- 23.Kim Y. T., Richardson C. C. (1994) J. Biol. Chem. 269, 5270–5278 [PubMed] [Google Scholar]
- 24.Lee S. J., Marintcheva B., Hamdan S. M., Richardson C. C. (2006) J. Biol. Chem. 281, 25841–25849 [DOI] [PubMed] [Google Scholar]
- 25.Marintcheva B., Marintchev A., Wagner G., Richardson C. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]