Abstract
Ten new diterpenes, 1–10, having a dolabellane skeleton were isolated from a Colombian gorgonian coral of the genus Eunicea. Their structures, as well as those of known compounds 11–18, were determined based on spectroscopic analysis and, in some instances, by chemical conversion and X-ray crystallographic analysis. The absolute structure of 7 was established by chemical conversion from 11, a co-occurring dolabellane congener of known absolute structure. Most of these diterpenoids showed antimalarial activity against the protozoan parasite Plasmodium falciparum.
Gorgonian octocorals belonging to the genus Eunicea, which can be found in great abundance in waters of the Caribbean Sea, have been recognized as rich sources for marine natural products, most of which possess unique structural features and remarkable biological activities.1,2 A variety of chemically complex diterpenoids, such as cembrane lactones, dolabellanes, dolastanes, cubitanes, dilophols, and fuscols have been isolated since the early 1960's from several species of Eunicea, including E. succinea, E. mammosa, E. laciniata, E. palmeri, E. calyculata, E. asperula, E. fusca, and E. tourneforti.1–10 As part of a study on the chemical constituents of Caribbean gorgonians of this genus, we reported in 2004 the isolation and structure characterization of five new representatives of the cembrane class of diterpenes from an unknown species of Eunicea collected near the Colombian Southwestern Caribbean Sea.11 During our continuing studies on the chemical constituents of this gorgonian species, 10 new dolabellane-type diterpenoids (1–10) were isolated, as well as eight known dolabellane and dolastane congeners (11–18). This paper describes the isolation and structure determination of these compounds.12 The structures of 1–18 were established by detailed spectroscopic analysis, including extensive examination of 2D NMR (1H–1H COSY, HMQC, HMBC, and NOESY) correlations, chemical conversion and, in the case of metabolites 1, 2, 8 and 12, by X-ray crystallographic analysis. Identification of known compounds was based on comparison of their physical properties (mp, [α]D, MS and NMR) with those reported in the literature, by direct comparison with authentic samples, and in the case of 12, by X-ray crystallography. It is to be noted that, except for 7, only the relative configuration of the new compounds has been determined. However, in light of the established absolute structures of the co-occurring 11 and 16, the configurations shown are considered more probable than the enantiomeric ones.13,14 The antiprotozoan activity of 1–17 against the Plasmodium falciparum W2 strain (chloroquine resistant) was also studied. The results displayed that these compounds are generally toxic toward P. falciparum, with 1 and 9 being the most potent.
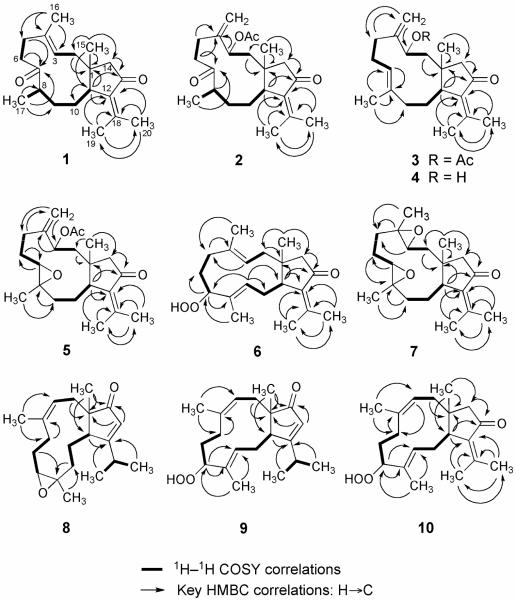
The HREIMS (m/z 302.2252) of 1 established the molecular formula C20H30O2, appropriate for six degrees of unsaturation, and the IR spectrum revealed the presence of carbonyl (1700 cm−1) and olefin (1621 cm−1) groups. The 13C NMR (Table 3) and DEPT spectroscopic data showed signals of five methyls, six sp3 methylenes, two sp3 methines, one sp2 methine, one sp3 and five sp2 quaternary carbons (including two ketone carbonyls). The NMR signals (Tables 1 and 3) observed at δC 206.6 (C, C-13), 147.9 (C, C-18), 137.6 (C, C-12), 24.3 (CH3, C-19), and 21.5 (CH3, C-20), and δH 2.20 (s, 3H, H3-20) and 1.88 (s, 3H, H3-19) revealed the presence of the α-isopropylidenyl ketone functionality.3 Furthermore, the chemical shift for the C-13 carbonyl carbon in the 13C NMR spectrum of 1 (206.6 ppm), the IR absorption at 1700 cm−1, and the UV absorption data were virtually identical with those reported for known compounds 11–14, thus allowing the further assignment of this chromophore as α-isopropylidenylcyclopentanone. Signals appearing at δC 135.7 (C, C-4), 125.1 (CH, C-3), δH 5.27 (dd, 1H, J = 3.2, 11.8 Hz, H-3) and 1.63 (s, 3H, H3-16) revealed the presence of one trisubstituted carbon-carbon double bond. One α-methylene-α'-methyl substituted ketone was also identified from the NMR signals at δC 215.2 (C, C-7), 47.2 (CH, C-8), 40.3 (CH2, C-6), 18.0 (CH3, C-17); δH 2.89 (dt, 1H, J = 2.7, 14.0 Hz, H-6β), 2.66 (m, 1H, H-8), 2.38 (dq, 1H, J = 2.1, 14.0 Hz, H-6α) and 1.05 (d, 3H, J = 7.1 Hz, H3-17). In the 1H–1H COSY spectrum, it was possible to identify three different structural units, which were assembled with the assistance of an HMBC experiment (Figure 1). Key HMBC correlations of H3-15 to C-1, C-2, C-11, and C-14; H3-16 to C-5, H2-6 to C-7; H3-17 to C-7, C-8, and C-9; and H-11 to C-1, C-10, C-12, C-13, C-14, and C-18 permitted connection of the carbon skeleton. Furthermore, the geminal methyl groups positioned at C-18 were confirmed from the simultaneous HMBC correlations of H3-19 and H3-20 (δH 1.88 and 2.20, respectively) to the sp2 quaternary carbons C-18 (δC 147.9) and C-12 (δC 137.6). On the basis of the above analysis, the planar structure of 1 was established unambiguously.
Table 3.
13C NMR (75 MHz) Assignments for Compounds 1–10 in CDCl3 [δc, mult]a
| C | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 40.5, C | 38.1, C | 38.0, C | 40.4, C | 38.3, C | 39.3, C | 37.9, C | 50.8, C | 53.5, C | 39.0, C |
| 2 | 40.3, CH2 | 45.3, CH2 | 43.8, CH2 | 41.0, CH2 | 43.5, CH2 | 42.8, CH2 | 40.3, CH2 | 36.4, CH2 | 33.8, CH2 | 37.4, CH2 |
| 3 | 125.1, CH | 70.9, CH | 74.4, CH | 74.8, CH | 75.0, CH | 125.1, CH | 63.0, CH | 122.4, CH | 122.9, CH | 121.6, CH |
| 4 | 135.7, C | 145.8, C | 148.7, C | 153.9, C | 148.3, C | 133.8, C | 60.3, C | 136.7, C | 134.8, C | 135.0, C |
| 5 | 35.6, CH2 | 30.7, CH2 | 35.1, CH2 | 34.5, CH2 | 33.6, CH2 | 38.5, CH2 | 37.3, CH2 | 28.9, CH2 | 27.1, CH2 | 27.4, CH2 |
| 6 | 40.3, CH2 | 37.2, CH2 | 28.5, CH2 | 34.4, CH2 | 29.3, CH2 | 28.0, CH2 | 23.4, CH2 | 26.1, CH2 | 30.6, CH2 | 27.6, CH2 |
| 7 | 215.2, C | 211.7, C | 127.7, CH | 123.5, CH | 64.4, CH | 80.6, CH | 63.8, CH | 67.8, CH | 80.6, CH | 80.6, CH |
| 8 | 47.2, CH | 45.8, CH | 134.6, C | 136.7, C | 60.7, C | 137.5, C | 60.6, C | 60.6, C | 138.8, C | 138.2, C |
| 9 | 30.9, CH2 | 31.9, CH2 | 37.9, CH2 | 37.9, CH2 | 36.3, CH2 | 129.3, CH | 36.7, CH2 | 35.9, CH2 | 124.0, CH | 125.4, CH |
| 10 | 28.6, CH2 | 27.0, CH2 | 27.5, CH2 | 30.2, CH2 | 28.0, CH2 | 30.0, CH2 | 27.9, CH2 | 23.6, CH2 | 23.6, CH2 | 30.4, CH2 |
| 11 | 44.4, CH | 43.2, CH | 42.4, CH | 44.0, CH | 42.1, CH | 47.9, CH | 43.0, CH | 45.5, CH | 46.7, CH | 43.2, CH |
| 12 | 137.6, C | 136.6, C | 137.9, C | 137.6, C | 137.4, C | 137.0, C | 137.1, C | 190.4, C | 187.2, C | 134.5, C |
| 13 | 206.6, C | 205.9, C | 206.9, C | 206.3, C | 206.2, C | 205.5, C | 205.4, C | 125.6, CH | 123.6, CH | 205.5, C |
| 14 | 55.3, CH2 | 54.6, CH2 | 55.7, CH2 | 56.0, CH2 | 54.8, CH2 | 57.5, CH2 | 54.8, CH2 | 213.4, C | 212.4, C | 52.8, CH2 |
| 15 | 21.3, CH3 | 23.0, CH3 | 23.0, CH3 | 22.1, CH3 | 23.2, CH3 | 23.1, CH3 | 23.6, CH3 | 21.1, CH3 | 24.2, CH3 | 25.0, CH3 |
| 16 | 15.6, CH3 | 118.7, CH2 | 115.0, CH2 | 110.7, CH2 | 117.4, CH2 | 15.5, CH3 | 15.6, CH3 | 23.9, CH3 | 22.9, CH3 | 23.3, CH3 |
| 17 | 18.0, CH3 | 14.5, CH3 | 16.8, CH3 | 15.9, CH3 | 17.3, CH3 | 11.2, CH3 | 17.5, CH3 | 17.5, CH3 | 10.1, CH3 | 10.6, CH3 |
| 18 | 147.9, C | 150.4, C | 148.1, C | 147.8, C | 148.6, C | 145.9, C | 149.5, C | 29.0, CH | 29.4, CH | 148.4, C |
| 19 | 24.3, CH3 | 25.0, CH3 | 24.7, CH3 | 24.1, CH3 | 25.1, CH3 | 23.8, CH3 | 25.2, CH3 | 22.5, CH3 | 21.7, CH3 | 24.7, CH3 |
| 20 | 21.5, CH3 | 21.3, CH3 | 21.3, CH3 | 21.6, CH3 | 21.3, CH3 | 21.4, CH3 | 21.7, CH3 | 21.2, CH3 | 20.7, CH3 | 22.5, CH3 |
| 21 | 170.7, C | 170.6, C | 170.5, C | |||||||
| 22 | 21.4, CH3 | 21.4, CH3 | 21.4, CH3 |
Chemical shifts values are in ppm relative to TMS. Spectra were recorded at 25 °C. Atom multiplicities were obtained from DEPT NMR experiments. Assignments were aided by 1H–1H COSY, HMQC, HMBC, and NOESY experiments.
Table 1.
1H NMR (300 MHz) Assignments for Compounds 1–5 in CDCl3 [δH, mult (J in Hz)]a
| H | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 2α | 2.06, m, | 2.06, m | 2.17, m | 2.19, m | 2.20, m |
| 2β | 1.72, dd (13.1, 4.2) | 1.35, m | 1.28, m | 1.85, m | 1.30, m |
| 3 | 5.27, dd (11.8, 3.2) | 5.25, m | 5.39, dd (10.6, 2.2) | 4.01, dd (5.1, 3.9) | 5.48, dd (11.4, 1.5) |
| 5α | 2.07, m | 2.35, m | 2.49, m | 2.13, m | 2.63, m |
| 5β | 2.72, m | 3.04, m | 2.15, m | 2.03, m | 2.22, m |
| 6α | 2.38, dq (14.0, 2.1) | 2.40, m | 2.38, m | 2.28, m | 2.10, m |
| 6β | 2.89, dt (14.0, 2.7) | 3.28, m | 2.01, m | 1.94, m | 1.82, m |
| 7 | 5.18, m | 5.34, br s | 3.00, d (8.1) | ||
| 8 | 2.66, m | 2.67, m | |||
| 9α | 2.24, m | 2.28, m | 2.28, m | 2.18, m | 2.08, m |
| 9β | 1.38, m | 1.76, m | 2.12, m | 2.18, m | 1.40, m |
| 10α | 1.54, m | 1.58, m | 1.68, m | 1.72, m | 1.75, m |
| 10β | 1.26, m | 1.32, m | 1.35, m | 1.56, m | 1.41, m |
| 11 | 2.30, m | 2.29, m | 3.28, br d (9.4) | 2.48, m | 3.23, br d (12.2) |
| 14α | 2.28, d (17.6) | 2.37, d (18.0) | 2.36, d (18.2) | 2.25, br s | 2.45, br d (18.2) |
| 14β | 2.11, d (17.6) | 1.92, d (18.0) | 2.00, d (18.2) | 2.25, br s | 2.00, br d (18.2) |
| 15 | 1.06, s | 1.42, s | 1.30, s | 1.16, s | 1.34, s |
| 16α | 1.63, s | 5.32, br s | 5.18, br s | 5.27, br s | 5.34, br s |
| 16β | 5.18, br s | 5.14, br s | 5.02, br s | 5.28, br s | |
| 17 | 1.05, d (7.1) | 1.08, d (6.8) | 1.62, s | 1.54, s | 1.31, s |
| 19 | 1.88, s | 1.69, s | 1.76, s | 1.83, s | 1.82, s |
| 20 | 2.20, br s | 2.17, s | 2.21, s | 2.17, s | 2.24, s |
| 22 | 1.99, s | 2.00, s | 2.01, s |
Chemical shift values are in ppm relative to TMS. Spectra were recorded at 25 °C. Proton assignments were aided by 1H—1H COSY, HMQC, HMBC, and NOESY experiments.
Figure 1.
1H–1H COSY and HMBC correlations for compounds 1–10.
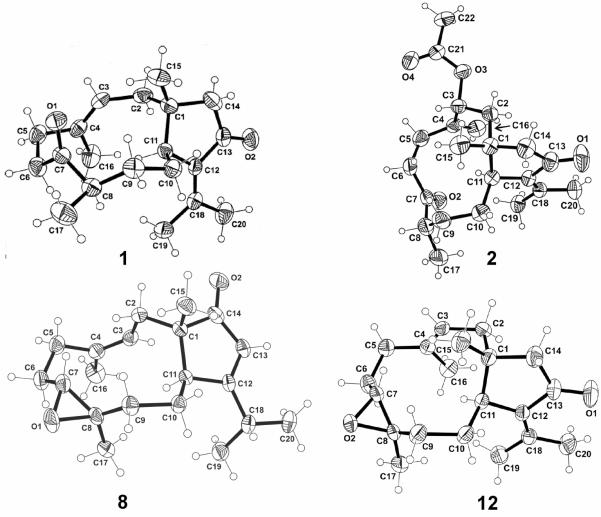
The relative configuration of three asymmetric centers (C-1, C-8, and C-11), as well as the configuration of the trisubstituted double bond in 1, were determined by X-ray crystallographic analysis on a single crystal of 1. The results of the X-ray analysis is shown in Figure 2, disclosing the 1R*, 8R* and 11S* relative configurations for the asymmetric centers as well as the E configuration for the double bond. Compound 1 was thus elucidated to be 7,13 diketo-1R*,8R*,11S*-dolabell-3E,12(18)-diene, a derivative most likely stemming from the acid promoted epoxide rearrangement of 12, the main compound isolated during this investigation.3
Figure 2.
Molecular structures of compounds 1, 2, 8 and 12, which define only the relative configuration, with atom labeling scheme. The carbon and oxygen atoms are drawn as 30% thermal ellipsoids.
The molecular formula of C22H32O4 for 2 was demonstrated by HREIMS and the 13C NMR spectrum. All 22 carbons appeared in the 13C NMR spectrum, and DEPT experiments indicated the presence of five methyls (including one acetate methyl), six sp3 methylenes, one sp2 methylene, three sp3 methines (including one oxymethine), one sp3 and six sp2 quaternary carbons (including one ester and two ketone carbonyls). The 1H and 13C NMR spectra (Tables 1 and 3) showed the presence of an α-isopropylidenecyclopentanone system [δC 205.9 (C, C-13), 150.4 (C, C18), 136.6 (C, C-12), 54.6 (CH2, C-14), 25.0 (CH3, C-19), and 21.3 (CH3, C-20); δH 2.37 (d, 1H, J = 18.0, H-14α), 1.92 (d, 1H, J = 18.0, H-14β), 2.17 (s, 3H, H3-20) and 1.69 (s, 3H, H3-19)] which was demonstrated to be the same as that in 1 by the UV [λmax (log ε) 203 (3.4), 255 (3.2) nm] and IR (1698, 1622 cm−1) spectra. 1H and 13C NMR spectra also showed signals due to a terminal olefin [δC 145.8 (C, C-4), 118.7 (CH2, C-16); δH 5.32 (br s, 1H, H-16α) and 5.18 (br s, 1H, H-16β)] and due to an α-methylene-α'-methyl substituted ketone [δC 211.7 (C, C-7), 45.8 (CH, C-8), 37.2 (CH2, C-6), 14.5 (CH3, C-17); δH 3.28 and 2.40 (each, m, 1H, H2-6), 2.67 (m, 1H, H-8), and 1.08 (d, 3H, J = 6.8 Hz, H3-17)]. The presence of a secondary acetate group was demonstrated by the IR absorption (1735 cm−1), the low-field 1H signal at δ 5.25 (m, 1H) due to the oxymethine proton H-3 in addition to the 1H and 13C NMR data [δH 1.99 (s, 3H, H3-22); δC 170.7 (C, C-21), 70.9 (CH, C-3), 21.4 (CH3, C-22)]. These spectroscopic findings coupled with 1H–1H correlations observed in the 1H–1H COSY spectrum gave several partial structures, as shown in Figure 1. After assignments of the direct 13C–1H correlations were made based on the HMQC analysis, the HMBC spectrum of 2 was measured and analyzed to obtain the gross structure of 2 (Figure 1). The relative configuration of the four asymmetric centers (C-1, C-3, C-8, and C-11) was determined by X-ray crystallographic analysis on a single crystal of 2. The result of the X-ray analysis is shown in Figure 2, demonstrating 1R*,3R*,8S*, and 11S* relative configurations for the asymmetric centers.
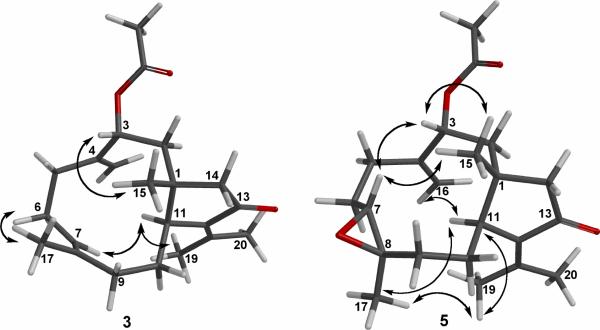
Compound 3 analyzed for C22H32O3 as revealed by the HREIMS (m/z 344.2346) and 13C NMR data, a molecular formula possessing one oxygen atom less in comparison to that of compound 2. Of the seven degrees of unsaturation implied by the molecular formula, five were accounted for by multiple bonds; three carbon-carbon double bonds [δC 148.7 (C, C-4), 148.1 (C, C-18), 137.9 (C, C-12), 134.6 (C, C-8), 127.7 (CH, C-7), 115.0 (CH2, C-16)], a keto group [δC 206.9 (C, C-13)], and an acetate function [δC 170.6 (C, C-21), 21.4 (CH3, C-22)], indicating 3 to be bicyclic. Comparison of all spectroscopic data of 3 with those of 2, and the results of a 2D 1H–1H COSY experiment, revealed that the two molecules were identical, except for the fact that the keto group at C-7 in 2 was replaced by a carbon-carbon double bond in 3. On the basis of the overall 2D NMR data it was apparent that the new olefin functionality in compound 3 had to reside between carbons C-7 and C-8. From the NOESY spectrum, it was found that H3-15 (δ 1.30, s, 3H) showed an NOE interaction with H-3 (δ 5.39, dd, 1H, J = 2.2, 10.6 Hz), but not with H-11 (δ 3.28, br d, 1H, J = 9.4 Hz), revealing the β-orientation of H-3. Furthermore, we concluded that the acetate function had to be α, based on a 10.6 Hz coupling between H-2α (δ 2.17) and H-3, as well as a 2.2 Hz coupling between H-2β (δ 1.28) and H-3. The E geometry of the trisubstituted double bond was also assigned from the NOE correlation of H3-17 (δ 1.62, s, 3H) with H-6α (δ 2.38), but not with olefinic proton H-7 (δ 5.18, m, 1H), and also the upper field chemical shift of C-17 (δ 16.8). On the basis of the above findings and detailed examination of other NOE correlations (Figure 3), we concluded that the relative structure of compound 3 is 3R*-acetoxy-13-keto-1R*,11S*-dolabell-4(16),7E,12(18)-triene.
Figure 3.
Key NOESY correlations for 3 and 5.
Compound 4 was shown by HREIMS to possess the molecular formula C20H30O2 (m/z 302.2242). The IR spectrum of 4 revealed the presence of hydroxy (3434 cm−1), ketone carbonyl (1701 cm−1) and olefin (1620 cm−1) groups. Comparison of the 1H and 13C NMR data (Tables 1 and 3) of compounds 3 and 4 showed that the structure of 4 should be very close to that of 3, with the exception of signals assigned to C-3, where an acetate methyl (δ 2.00) and acetoxymethine (δ 5.39, dd, 1H, J = 2.2, 10.6 Hz) in 3 are replaced by a hydroxymethine (δ 4.01, dd, 1H, J = 3.9, 5.1 Hz) in 4. The overall planar structure of 4 was fully established by analyzing the 1H–1H COSY and HMBC correlations (Figure 1). The relative configuration of 4 was confirmed to be 1R*, 3R* and 11S* from the following NOESY correlations: both H-3 (δ 4.01) and H3-15 (δ 1.16) with H-7 (δ 5.34), and H3-15 with H-3. The Δ7,8 double bond was assigned with the E configuration based on the 13C NMR chemical shift of C-17 (δC 15.9), which is in the same range as that of compound 3. These results, together with other detailed 2D NMR correlations of 4 (Figure 1), unambiguously established the structure of this secondary metabolite as shown in formula 4. The structural relationship of compounds 3 and 4 was decisively demonstrated when 4, upon acetylation at room temperature, yielded acetate 3 quantitatively. Thus, 4 is the 3-deacetyl derivative of 3.
Compound 5 possessed the same molecular formula (C22H32O4) as that of compound 2, as revealed from HREIMS, suggesting 5 to be an isomer of 2. Furthermore, it was found that the NMR spectroscopic data of 5 were very similar to those of 2. The four oxygen atoms in 5 were readily assigned to an acetate, a trisubstituted epoxide, and an α,β-unsaturated ketone constellation by evaluation of spectral information. 13C NMR bands (Table 3) at δ 64.4 (CH, C-7) and 60.7 (C, C-8) in conjunction with 1H NMR resonances at δ 3.00 (d, 1H, J = 8.1 Hz, H-7) and δ 1.31 (s, 3H, H3-17), illustrated 5 to possess a methyl-substituted epoxide group. By 2D NMR spectroscopic data, including 1H–1H COSY, HMQC and HMBC, compound 5 was shown to possess many of the same structural features as those of 2. Comparison of the 13C NMR data for the two compounds revealed that the major differences between them lay in the vicinity of the C-7,8 junction (see Table 3). On the basis of 1H*#x2013;1H couplings observed in the 1H NMR spectrum and overall HMBC spectral data of 5 (Figure 1) it was apparent that the trisubstituted epoxide group had to reside at the C -7,8 position. The HMBC spectrum of 5 showed heteronuclear couplings between C-8 (δC 60.7, C) and H2-9 (δH 2.08 and 1.40), as well as from C-7 (δC 64.4, CH) to H2-6 (δH 2.10 and 1.82), which allowed the epoxide group to be unambiguously positioned across C-7 and C-8. The relative configuration of 5 at C-1, C-3, and C-11 was identical to that of 2 on the basis of comparable NOE interactions (Figure 3) and inter-proton coupling patterns. For C-7 and C-8 it was concluded that the epoxide function had to possess the 7R*, 8R* relative configuration, based on a 8.1 Hz coupling between H-7 (δ 3.00) and H-6α (δ 2.10), as well as strong NOE effects observed between H3-15 (δ 1.34), H-3 (δ 5.48) and H-7 (δ 3.00), and between H-11 (δ 3.23) and H3-17 (δ 1.31). Thus, compound 5, a likely precursor to 2 upon acid-promoted epoxide rearrangement, is 3R* acetoxy-7R*,8R*-epoxy-13-keto-1R*,11S*-dolabell-4(16),12(18)-diene.
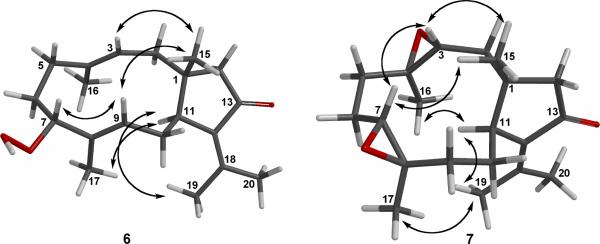
Mass spectrometry and NMR data revealed compound 6 to have a molecular formula of C20H30O3. The presence of seven sp2 hybridized carbon atoms in the molecule, as deduced from the 13C and DEPT NMR spectra, being for three carbon-carbon double bonds and one carbon-oxygen bond as the only multiple bonds, indicated compound 6 to be bicyclic. The IR spectrum showed the presence of hydroxy and α,β unsaturated ketone functionalities (vmax 3397, 1704 cm−1). The 1H and 13C NMR spectra (Tables 2 and 3) resembled those of known dolabellatrienone (11).3 However, a secondary hydroperoxy [δH 4.33 (dd, 1H, J = 2.6, 10.7, H-7); δC 80.6 (CH, C-7)] adjacent to a methyl bearing E trisubstituted double bond [δH 5.41 (t, 1H, J = 7.8 Hz, H-9), 1.68 (s, 3H, H3-17); δC 137.5 (C, C-8), 129.3 (CH, C-9), 11.2 (CH3, C-17)] in 6 replaced the E trisubstituted Δ7,8 double bond in 11. The presence of a secondary hydroperoxy group in 6 was confirmed by the EIMS data which showed pronounced fragment ions at m/z 301 and 284 through elimination of HOM· and H2O2, respectively, from the molecular ion of 6 at m/z 318. 1H–1H COSY cross-peaks between H-7 (δ 4.13) and H2-6 (δ 1.92 and 1.59) and between H-9 (δ 5.41) and H2-10 (δ 2.40 and 1.78), as well as HMBC correlations between H-7 and C-6, C-8, C-9, C-17; H-9 and C-7, C-8, C-10, C-11 and C-17 (Figure 1), positioned the secondary hydroperoxy group at C-7 and the nearby trisubstituted double bond between C-8 and C-9. In the NOESY spectrum (Figure 4), NOEs between H-7 and H-9; between H-7 and H3-15, between H-9 and H3-15, and between H-11 and H3-17, which indicated that the 11-membered ring in 6 adopts a crown conformation, allowed the configuration of the hydroperoxy and Δ8,9 moieties to be assigned as 7R* and E, respectively. Correspondingly, the triene 6 was fully characterized as 7R*-hydroperoxy-13 keto-1R*,11S*-dolabell-3E,8E,12(18)-triene. The co-ocurrence of 6 and 11 within the same Eunicea sp. extract suggests that 6 might be formed from 11 during the isolation process upon air-oxidation on the double bond at C-7 of 11.
Table 2.
1H NMR (300 MHz) Assignments for Compounds 6–10 in CDCl3 [δH, mult (J in Hz)]a
| H | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|
| 2α | 2.20, m | 1.77, m | 2.51, m | 2.47, br d (14.5) | 2.42, dd (11.8, 11.6) |
| 2β | 1.97, m | 1.37, m | 2.34, m | 2.18, dd (14.6, 12.3) | 1.85, m |
| 3 | 5.09, dd (11.6, 4.5) | 3.06, dd (11.2, 2.9) | 4.68, br d (11.7) | 4.66, br d (12.2) | 5.08, br d (11.3) |
| 5α | 2.33, m | 2.27, m | 1.89, m | 1.78, m | 1.79, m |
| 5β | 2.22, m | 1.42, m | 2.53, m | 2.30, m | 2.27, m |
| 6α | 1.92, m | 1.76, m | 1.48, m | 1.83, m | 1.83, m |
| 6β | 1.59, m | 1.47, m | 2.03, m | 1.83, m | 1.83, m |
| 7 | 4.13, dd (10.7, 2.6) | 2.96, br d (7.7) | 2.93, br d (10.3) | 4.02, dd (10.0, 5.2) | 4.03, dd (10.0, 5.1) |
| 9α | 5.41, t (7.8) | 2.12, m | 2.15, m | 5.68, t (7.4) | 5.66, t (7.4) |
| 9β | 1.43, m | 1.42, m | |||
| 10α | 2.40, m | 1.78, m | 1.94, m | 2.62, m | 2.80, m |
| 10β | 1.78, m | 1.57, m | 1.53, m | 1.95, m | 1.82, m |
| 11 | 2.77, br d (11.8) | 2.81, br d (11.4) | 2.98, br d (11.0) | 2.98, dt (10.2, 2.6) | 3.01, br d (11.1) |
| 13 | 5.97, br s | 5.83, br s | |||
| 14α | 2.37, d (16.3) | 2.44, d (18.4) | 2.53, d (15.7) | ||
| 14β | 2.10, d (16.3) | 2.13, d (18.4) | 1.81, d (15.7) | ||
| 15 | 1.08, s | 1.33, s | 1.04, s | 1.06, s | 1.01, s |
| 16 | 1.62, s | 1.21, s | 1.64, s | 1.64, s | 1.75, s |
| 17 | 1.68, s | 1.43, s | 1.30, s | 1.58, s | 1.62, s |
| 18 | 2.62, m | 2.62, m | |||
| 19 | 1.96, s | 1.94, s | 1.13, d (6.8) | 1.18, d (6.3) | 1.98, br s |
| 20 | 2.14, s | 2.24, s | 1.20, d (6.8) | 1.20, d (6.7) | 2.20, br s |
Chemical shift values are in ppm relative to TMS. Spectra were recorded at 25 °C. Proton assignments were aided by 1H—1H COSY, HMQC, HMBC, and NOESY experiments.
Figure 4.
Key NOESY correlations for 6 and 7.
Compound 7 was isolated as a colorless oil. HREIMS, 13C NMR, and DEPT spectra established the molecular formula of 7 as C20H30O3. The IR and UV absorptions clearly defined the analogous α-isopropylidenylcyclopentanone group as in 1−6. The NMR data of compound 7 were strongly reminiscent of those of known epoxide 12, apart from the replacement of the NMR bands of the trisubstituted olefin [δH 5.43 (dd, 1H, J = 4.7, 11.6 Hz, H-3), 1.57 (br s, 3H, H3-16); δC 124.7 (CH, C-3, 135.9 (C, C-4), 15.7 (CH3, C-16)]3 by those of an additional epoxide [δH 3.06 (dd, 1H, J = 2.9, 11.2 Hz, H-3), 1.21 (s, 3H, H3-16); δC 63.0 (CH, C-3), 60.3 (C, C-4), 15.6 (CH3, C-16)]. Based upon this very favorable comparison, dolabellanone 7 was formulated as the C-3,4 epoxy derivative of 12. The proposed structural type and substitution pattern of 7 was confirmed by means of 2D NMR correlated spectroscopy including HMQC, HMBC, and 1H−1H COSY (Figure 1). The relative structure of diepoxide 7 was elucidated by the analysis of NOE correlations, as shown in Figure 4. It was found that epoxymethine protons H-3 (δ 3.06) and H-7 (δ 2.96) each showed NOE interactions with H3-15 (δ 1.33); therefore, assuming a β-orientation of H3-15, H-3 and H-7 should also be positioned on the β-face. NOE correlations observed between H-11 (δ 2.81) and H3-16 (δ 1.21), and H-11 and H3-17 (δ 1.43), reflected the α-orientations of H3-16 and H3-17. Furthermore, H-3 exhibited interaction with H-7 and H3-19 (δ 1.94) exhibited NOE interaction with H3-17, all of which pointed toward identical R* relative configuration at C-3, C-4, C-7 and C-8. To confirm this contention, a solution of known dolabellatrienone (11) in CH2Cl2 was treated with m-CPBA at 25° C to find a 20% conversion of 11 to 7, whose NMR data and the [α]D value were identical to those of 7 obtained by the above-mentioned isolation. Because the absolute configuration of 11 was previously established by synthesis,13 the absolute configuration of 7 was determined by correlation. Therefore, 7 was found to be 3R,4R,7R,8R-diepoxy-13-keto 1R,11S-dolabell-12(18)-ene.
The molecular formula of compound 8 was established as C20H30O2 from its HREIMS data. The 1H and 13C NMR spectra suggested the same dolabellane diterpenoid skeleton of 1–7 with signals for an epoxy functionality, five methyl groups, two trisubstituted double bonds, and one carbonyl group [δ 213.4 (C, C-14)]. While the 1H NMR spectrum of 8 (Table 2) displayed an unusually shielded broad doublet at δ 4.68 (1H, J = 11.7 Hz) for H-3 (cf. δ 5.27 for H-3 of 1), it contained an additional one-proton vinyl singlet at δ 5.97 and an additional one-proton multiplet at δ 2.62. The usual methyl signals near δ 2.20 and 1.90, ascribable to the vinyl methyl groups of the α-isopropylidenylcyclopentanone moiety of 1–7 (H3-19 and H3-20) were conspicuously absent in the 1H NMR spectrum of 8 and showed in their place two doublets at δ 1.13 and 1.20 (each, 3H, J = 6.8 Hz). Coupling was observed in the 1H–1H COSY spectrum between these methyl signals and the multiplet at δ 2.62 indicating the presence of an isopropyl group at C-12 of the dolabellane skeleton. The 13C NMR spectrum (Table 3) showed, in addition to the carbonyl absorption at δ 213.4, sp2 carbon signals at δ 190.4 (C, C-12), 136.7 (C, C-4), 125.6 (CH, C-13), and 122.4 (CH, C-3). All of these features were suggestive of a structure isomeric with that of known compound 15. Further analysis of the 2D NMR (1H–1H COSY, HMQC, and HMBC) correlations showed compound 8 to possess the same planar structure as that of 15 (Figure 1), thus the differences between them must be stereochemical. The relative configuration, 1R*, 7R*, 8R*, 11S*, as well as the Z configuration of the Δ3,4 trisubstituted double bond in 8 were determined by X-ray crystallographic analysis on a single crystal of 8 (Figure 2). Compound 8 was thus elucidated to have a structure corresponding to the Δ3,4 Z-isomer of 15.
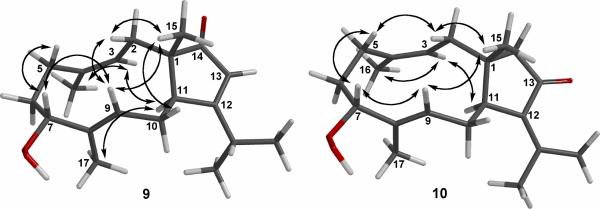
The molecular formula of C20H30O3 for 9 was determined by HRESIMS [found m/z 319.2259 (M+1)+; calcd for C20H31O3 3319.2273]. All 20 carbons appeared in the 13C NMR spectrum, and DEPT experiments indicated the presence of five methyls, four sp3 methylenes, three sp3 methines, three sp2 methines, one sp3 quaternary carbon, and four sp2 quaternary carbons (Table 3). The presence of a β-isopropyl-α,β-cyclopentenone system, the same as that in compound 8, was demonstrated by UV, IR, 13C NMR (Table 3), and 1H NMR (Table 2) data. The 1H and 13C NMR spectra showed signals due to two methyl-bearing trisubstituted olefins [δC 134.8 (C, C-4), 122.9 (CH, C-3), 22.9 (CH3, C-16); δH 4.66 (br d, 1H, J = 12.2 Hz, H-3), 1.64 (s, 3H, H3-16)] and [δC 138.8 (C, C-8), 124.0 (CH, C-9), 10.1 (CH3, C-17); δH 5.68 (t, 1H, J = 7.4 Hz, H-9), 1.58 (s, 3H, H3-17)], and due to a secondary hydroperoxide group [δC 80.6 (CH, C-7); δH 4.02 (dd, 1H, J = 5.2, 10.0 Hz, H-7)]. These spectroscopic findings coupled with proton-proton and long-range 13C–1H correlations observed in the 1H–1H COSY and HMBC spectra gave a series of partial structures, as shown in Figure 1, to give a gross structure for 9. In the NOESY experiment (Figure 5), the secondary hydroperoxide at C-7 was assigned as α on the basis of NOEs between H-7 and H-9, H 5β; between H-11 and H-3, H3-17; between H3-15 and H-9, H-10β; and between H-2β and H-9, H3-15. The geometries of the trisubstituted Δ3,4 and Δ8,9 double bonds were certified to be Z and E, respectively, on the basis of NOE experiments and the 13C chemical shift values of the corresponding vinyl methyls. Compound 9 is thus 7R*-hydroperoxy-14-keto-1S*,11S*-dolabell-3Z,8E,12Z-triene.
Figure 5.
Key NOESY correlations for 9 and 10.
Compound 10 was shown by HREIMS to possess the same molecular formula (C20H30O3) as those of 6 and 9. Furthermore, it was found that the NMR spectroscopic data of 10 were very similar to those of 6 suggesting that these compounds possess the same planar structure. Comparison of the 1H and 13C NMR data (Tables 2 and 3) of compounds 6 and 10 showed that the structure of 10 should be very close to that of 6, with the exception of signals assigned to the methyl bearing E trisubstituted olefin Δ3,4 in 6, which were replaced by resonances for a Z trisubstituted olefin [δH 5.08 (br d, 1H, J = 11.3 Hz, H-3), 1.75 (s, 3H, H3-16); δC 135.0 (C, C-4), 121.6 (CH, C-3), 23.3 (CH3, C-16)] in 10. The planar structure of 10 was fully established by analyzing the 1H–1H COSY and HMBC correlations (Figure 1), and its relative configuration was confirmed to be 1R*, 7R*, and 11S* from the following NOESY correlations: both H-5β (δ 2.27) and H-9 (δ 5.66) with H-7 (δ 4.03), both H-11 (δ 3.01) and H-14α (δ 2.53) with H-3 (δ 5.08), and H3-16 (δ 1.75) with H-3. These results, together with other detailed NOE correlations of 10 (Figure 5), unambiguously established that compound 10 is 7R*-hydroperoxy-13-keto-1R*,11S*-dolabell-3Z,8E,12(18)-triene.
The spread of drug resistance in Plasmodium falciparum has made it essential to look into new effective chemotherapeutic agents that possess antimalarial activity with favorable pharmacokinetic properties.15 Thus, Eunicea sp. dolabellanes 1–16 as well as dolastane 17 were tested for their inhibitory activity toward the growth of P. falciparum W2 (chloroquine-resistant). Interestingly, most of the compounds were active with IC50 values ranging from 0.01–0.05 μM. The two most active metabolites were 1 and 9 (IC50 = 0.01 μM), and the least active compound was epoxide 8 (IC50 ≥ 0.20 μM).16 Some of the present isolates (1, 3, 6, 8, 9, 11, 12, 16, 17 and 18) were also tested for their inhibitory activity toward the growth of Mycobacterium tuberculosis, but were deemed inactive (% of inhibition at 6.25 μg/mL ≤ 50%). Furthermore, dolabellanes 1, 7, 11, and 12 were evaluated in a 3-cell line panel consisting of the MCF-7 breast cancer, NCI-H460 non small cell lung cancer, and SF-268 (CNS). However, results from the one dose primary anticancer assay showed a lack of significant cytotoxicity when compared to the untreated control cells. Likewise, at a concentration of 30 mM, compounds 1, 6, 9, 11, and 16 were not active against the hepatitis B virus (HBV).
Experimental Section
General Experimental Procedures
Optical rotations were recorded with a Rudolph Autopol IV polarimeter and melting point determinations with an electrothermal IA 900 digital apparatus. The UV spectra were recorded with a Shimadzu UV-2401PC spectrometer, and the IR analyses were performed with a Nicolet Magna IR 750FT-IR spectrometer. 1D and 2D NMR data were recorded on a Bruker DRX-300 spectrometer. 1H-NMR and 13C-NMR chemical shifts were recorded at 25 °C with respect to TMS. Mass spectrometric measurements were generated at the Mass Spectrometry Laboratory of the University of Illinois at Urbana–Champaign. Column chromatography was performed using Si gel (35–75 mesh), and TLC analysis was carried out using glass pre-coated Si gel plates and the spots were visualized using a UV lamp at λ = 254 nm or by exposure to I2 vapor. HPLC was performed using either an Ultrasphere polar–bonded Cyano semi-preparative column (5 μ, 10 mm × 25 cm), an Ultrasphere normal–phase Si gel semi-preparative column (5 μ, 10 mm × 25 cm) or an Ultrasphere ODS reversed-phase Si gel semi-preparative column (5 μ, 10 mm × 25 cm). All HPLC separations were monitored simultaneously with a refractive index detector and a UV detector set at λ = 220 nm using a flow rate = 2 mL/min with isocratic elution of the mobile phase. All solvents were distilled from glass prior to use. The percentage yield of each compound is based on the weight of the hexane extract.
Animal Material
A detailed taxonomical description of the biological specimens used in this investigation has been disclosed previously.11 A photograph of the gorgonian coral voucher specimen is available as Supporting Information.
Collection, Extraction and Isolation
Medium to large colonies (0.5–1.3 m) of the gorgonian coral Eunicea sp. (undescribed species; Order: Gorgonacea; Family: Gorgoniidae, Phylum: Cnidaria) were collected by SCUBA at depths of 23–26 m from the coral reefs off Old Providence Island (March, 2002), Colombia located off the Nicaraguan shelf in the Southwestern Caribbean Sea. A voucher specimen (No. Eunicea sp. 2) is on deposit at the Chemistry Department of the University of Puerto Rico. The gorgonian specimens were partially air-dried, freeze-dried, and then kept frozen prior to extraction. The dried animal (1.7 kg) was blended with a mixture of 1:1 MeOH/CH2Cl2 (5 × 4 L) and, after filtration, the combined extracts were concentrated in vacuo to afford a gummy green residue (74 g). The extract was suspended in H2O (1 L) and extracted successively with hexane (4 × 2 L), CH2Cl2 (4 × 2 L), and EtOAc (3 × 2 L). The hexane extract (46 g) was chromatographed on a large Si gel column by stepwise elution with 100% hexane, hexane/EtOAc mixtures (100–0%), and then 100% MeOH. Fractions were pooled based on their TLC and NMR profile to yield 15 primary fractions, denoted I–XV. Purification of fraction III (1.5 g) by Si gel CC with 100:1 hexane/EtOAc led to the isolation of known compounds dolabellatrienone (11, 7.6 mg, 0.02% yield), epoxide 12 (800 mg, 1.8% yield), and palominol (16, 5.3 mg, 0.01% yield).3–5 A solution of fraction VII (420 mg) in toluene was filtered under vacuum and purified by size-exclusion chromatography on a Bio-Beads SX-3 (90 g) column with toluene as eluant. The combined portions (NMR and TLC guided) were concentrated to yield four subfractions, designated as A (80 mg), B (139 mg), C (155 mg), and D (40 mg). Subfraction C was purified successively by Si gel (18 g) CC with 80:1 hexane-acetone and reversed-phase HPLC (Ultrasphere ODS column with 65:35 MeOH/H2O) to afford pure compound 1 (33.0 mg, 0.07% yield) as a white crystalline solid. Plate crystals of dolabellanone 1 suitable for X-ray crystallographic analysis were obtained upon recrystallization from a 90:10 MeOH/H2O mixture. Purification of fraction IX (0.6 g) by size-exclusion chromatography on a Bio-Beads SX-3 column (toluene) led to four subfractions. The penultimate subfraction was eluted through a Si gel column (100:1 hexane-acetone) and then the mixture obtained was separated by successive HPLC analyses (Ultrasphere Si gel column with 95:5 hexane/2-isopropanol followed by a Cyano column with 97:3 hexane/2-isopropanol) affording pure compounds 3 (5 mg, 0.01% yield) and 10 (4 mg, 0.009 yield%). Fraction X (0.6 g) was chromatographed directly over Si gel (30 g) and separated by stepwise elution with hexane/acetone (2–3%) to yield three fractions (A, B and C). Fraction B (0.1 g) was purified successively by Si gel CC with 125:1 hexane/2-isopropanol and then by HPLC (Cyano column with 95:5 hexane/2-propanol) to afford pure compound 6 (15 mg, 0.03% yield) along with the known compounds 12 (10 mg, 0.02% yield), 17 (1 mg, 0.002% yield) and 18 (1 mg, 0.002% yield).3 As crystals of suitable quality were obtained for previously reported compound 12, a single crystal X-ray analysis was undertaken (Figure 2).17 Fraction XI (1.2 g) was purified by size exclusion chromatography (Bio-Beads SX-3, toluene) to yield five fractions, denoted A–E. Fraction D (0.5 g) was fractionated successively by normal-phase HPLC (Si gel column with 95:5 hexane/2-propanol), polar-bonded HPLC (Cyano column with 95:5 hexane/2-propanol), and reversed-phase HPLC (Ultrasphere ODS column with 75:25 MeOH/H2O). The main fraction obtained (100 mg) was loaded onto a Si gel column and separated by stepwise elution with hexane/acetone (2–100%) to afford pure dolabellanones 8 (14 mg, 0.03 yield%) and 9 (28 mg, 0.06% yield). Needle crystals of dolabellanone 8 suitable for X-ray crystallographic analysis were subsequently obtained upon recrystallization from a 80:20 MeOH/CHCl3 mixture. Size exclusion CC (Bio-Beads SX-3 with toluene) of fraction XII (614 mg) followed by successive reversed-phase HPLC (Ultrasphere ODS column with 75:25 MeOH/H2O) and Si gel CC (with 50:1 hexane/acetone) yielded known dolabellane 15 (3.4 mg, 0.007% yield).8 Fraction XIII (900 mg) was purified upon elution through a Bio-Beads SX-3 column with toluene. The penultimate subfraction (564 mg) was subsequently purified by Si gel CC and normal-phase HPLC (Cyano column with 97:3 hexane/2-propanol) to yield pure compound 2 (8 mg, 0.02% yield) and known dolabellane 14 (3.6 mg, 0.008% yield).8 Recrystallization of dolabellanone 2 from 90:10 MeOH/CHCl3 afforded white cubic crystals. After fraction XIV (1.5 g) was fractionated by size-exclusion chromatography on a Bio-Beads SX-3 column (toluene), the penultimate fraction eluted (0.5 g) was purified by Si gel CC and separated by stepwise elution with hexane-acetone mixtures (5–100%). The second subfraction isolated was purified by HPLC (Cyano column with 97:3 hexane/2-propanol) to yield pure dolabellanone 5 (13 mg, 0.03% yield). The third subfraction was purified by HPLC (Cyano column with 95:5 hexane/2-propanol) to afford dolabellanone 7 (15 mg, 0.03% yield) along with known compound 13 (10 mg, 0.02% yield),8 and the last subfraction was purified by Si gel CC with 20:1 hexane/acetone followed by HPLC (Cyano column with 93:7 hexane/2-propanol) yielding dolabellanone 4 (8 mg, 0.02% yield).
Dolabellanone 1
white crystalline solid; mp 128.5–128.9 °C; [α]25D +41.2 (c 1.0, CHCl3); UV (MeOH) λmax (log ε) 203 (3.1), 254 (3.7) nm; IR (film) νmax 2975, 2957, 2929, 2873, 1700, 1621, 1459, 1406, 1369, 1273, 1182 cm−1; 1H NMR (300 MHz, CDCl3) and 13C NMR (75 MHz, CDCl3) (see Tables 1 and 3, respectively); EIMS m/z [M]+ 302 (100), 287 (7), 269 (3), 231 (7), 219 (14), 163 (53), 151 (42), 137 (36), 121 (23), 109 (24), 93 (33); HREIMS m/z [M]+ 302.2252 (calcd for C20H30O2, 302.2245).
Dolabellanone 2
white crystalline solid; mp 169–170 °C; [α]25D +26.3 (c 1.3, CHCl3); UV (MeOH) λmax (log ε) 203 (3.4), 255 (3.2) nm; IR (film) νmax 2981, 2969, 2930, 1735, 1698, 1622, 1441, 1412, 1371, 1242, 1224, 1186, 1014, 958, 912 cm−1; 1H NMR (300 MHz, CDCl3) and 13C NMR (75 MHz, CDCl3) (see Tables 1 and 3, respectively); EIMS m/z [M]+ 360 (21), 318 (29), 300 (95), 285 (14), 189 (23), 163 (58), 151 (100), 136 (50), 121 (34), 107 (49); HREIMS m/z [M]+ 360.2306 (calcd for C22H32O4, 360.2301).
Dolabellanone 3
colorless oil, [α]25D +5.7 (c 1.3, CHCl3); UV (MeOH) λmax (log ε) 203 (3.5), 247 (3.2) nm; IR (film) νmax 3077, 2963, 2931, 2869, 1737, 1702, 1624, 1447, 1370, 1280, 1243, 1180, 1016, 966, 906 cm−1; 1H NMR (300 MHz, CDCl3) and 13C NMR (75 MHz, CDCl3) (see Tables 1 and 3, respectively); EIMS m/z [M]+ 344 (9), 302 (18), 284 (87), 269 (21), 241 (26), 201 (38), 187 (30), 150 (96), 135 (74), 121 (82), 93 (100), 81 (76), 69 (55); HREIMS m/z [M]+ 344.2346 (calcd for C22H32O3, 344.2351).
Dolabellanone 4
colorless oil, [α]25D +11.8 (c 0.83, CHCl3); UV (MeOH) λmax (log ε) 202 (3.9), 235 (3.7) nm; IR (film) νmax 3434, 2933, 2873, 1701, 1620, 1453, 1373, 1242, 1188, 1037, 903 cm−1; 1H NMR (300 MHz, CDCl3) and 13C NMR (75 MHz, CDCl3) (see Tables 1 and 3, respectively); EIMS m/z [M]+ 302 (16), 284 (6), 189 (19), 175 (25), 161 (40), 150 (54), 135 (47), 121 (54), 107 (61), 93 (87), 81 (70), 61 (100); HREIMS m/z [M]+ 302.2242 (calcd for C20H30O2, 302.2246).
Dolabellanone 5
colorless oil, [α]25D −7.4 (c 1.3, CHCl3); UV (MeOH) λmax (log ε) 204 (3.9), 235 (3.4) nm; IR (film) νmax 3079, 2971, 2933, 2864, 1735, 1706, 1626, 1454, 1372, 1282, 1247, 1190, 1018, 978, 931, 914, 756 cm−1; 1H NMR (300 MHz, CDCl3) and 13C NMR (75 MHz, CDCl3) (see Tables 1 and 3, respectively); EIMS m/z [M]+ 360 (2), 318 (1), 300 (3), 257 (1), 215 (2), 189 (4), 163 (5), 149 (11), 135 (7), 107 (11), 91 (11), 61 (100); HREIMS m/z [M]+ 360.2298 (calcd for C22H32O4, 360.2301).
Dolabellanone 6
colorless oil, [α]25D −29.5 (c 1.0, CHCl3); UV (MeOH) λmax (log ε) 206 (3.3), 234 (3.7), 235 (3.6) nm; IR (film) νmax 3397, 2969, 2935, 2874, 1704, 1619, 1457, 1376, 1040 cm−1; 1H NMR (300 MHz, CDCl3) and 13C NMR (75 MHz, CDCl3) (see Tables 2 and 3, respectively); EIMS m/z [M]+ 318 (21), 302 (36), 301 (22), 284 (64), 189 (24), 149 (43), 136 (100), 121 (26), 109 (30), 83 (36); HRESIMS m/z [M–1]− 317.2102 (calcd for C20H29O3, 317.2117).
Dolabellanone 7
colorless oil, [α]25D −26 (c 0.9, CHCl3); UV (MeOH) λmax (log ε) 203 (3.9), 254 (3.8) nm; IR (film) νmax 2968, 2932, 2859, 1701, 1621, 1454, 1385, 1272, 1256, 1188, 1086, 1057, 945, 888 cm−1; 1H NMR (300 MHz, CDCl3) and 13C NMR (75 MHz, CDCl3) (see Tables 2 and 3, respectively); EIMS m/z [M]+ 318 (54), 163 (37), 149 (79), 135 (73), 121 (59), 109 (79), 93 (72), 81 (70), 69 (80), 55 (100); HREIMS m/z [M]+ 318.2197 (calcd for C20H30O3, 318.2195). The physical data of dolabellanone 7 were found to be identical with those of a diepoxide which was chemically derived from dolabellatrienone (11) isolated from the Caribbean gorgonian Eunicea calyculata and reported with incomplete relative configuration about the epoxides.3
Dolabellanone 8
white crystalline solid, mp 102.2–102.7 °C, [α]20D + 93.0 (c 1.0, CHCl3); UV (MeOH) λmax (log ε) 229 (3.7) nm; IR (film) νmax 3077, 2986, 2957, 2919, 2862, 1686, 1601, 1447, 1376, 1266, 1217, 1190, 1162, 1095, 1083, 1024, 868 cm−1; 1H NMR (300 MHz, CDCl) and 13C NMR (75 MHz, CDCl3) (see Tables 2 and 3, respectively); EIMS m/z [M]+ 302 (19), 287 (8), 269 (5), 241 (8), 217 (9), 203 (20), 189 (15), 161 (33), 150 (100), 135 (54), 121 (41), 107 (41), 95 (45), 81 (51), 55 (69); HREIMS m/z [M]+ 302.2252 (calcd for C20H30O2, 302.2246).
Dolabellanone 9
colorless oil, [α]25D +43.1 (c 1.0, CHCl3); UV (MeOH) λmax (log ε) 203 (4.2), 229 (4.1) nm; IR (film) νmax 3419, 3075, 2969, 2932, 2875, 1697, 1600, 1458, 1381, 1341, 1264, 1214, 1190, 1110, 1090, 1040, 1014, 864, 825, 810, 758 cm−1; 1H NMR (300 MHz, CDCl3) and 13C NMR (75 MHz, CDCl3) (see Tables 2 and 3, respectively); EIMS m/z [M]+ 318 (1), 302 (2), 301(1), 284 (1), 269 (2), 241 (2), 203 (2), 175 (3), 161 (5), 150 (37), 135 (11), 121 (6), 85 (72), 83 (100); HRESIMS m/z [M+1]+ 319.2259 (calcd for C20H31O3, 319.2273).
Dolabellanone 10
colorless oil, [α]25D −15.0 (c 1.0, CHCl3); UV (MeOH) λmax (log ε) 202 (3.3), 238 (3.1) nm; IR (film) νmax 3426, 2965, 2935, 2874, 1712, 1621, 1452, 1377, 1242, 1038, 756 cm−1; 1H NMR (300 MHz, CDCl3) and 13C NMR (75 MHz, CDCl3) (see Tables 2 and 3, respectively); EIMS m/z [M]+ 318 (4), 302 (18), 301 (5), 284 (12), 269 (7), 189 (22), 150 (84), 136 (45), 121 (35), 107 (46), 95 (39), 83 (100), 69 (44), 55 (63); HREIMS m/z [M–H2O2]+ 284.2131 (calcd for C20H28O, 284.2140).
Acetylation of Dolabellanone 4
Compound 4 (3.0 mg, 0.01 mmol) was dissolved in a mixture of dry pyridine (500 μL) and acetic anhydride (500 μL) and stirred at rt for 12 h. The reaction mixture was concentrated in vacuo and the oily residue obtained was purified by CC over Si gel (1 g) using 5% EtOAc in hexane to yield a homogeneous oil (3.3 mg, quantitative yield) whose NMR (1H and 13C) data, TLC retention time, and [α]D value were identical to those of dolabellanone 3.
Epoxidation of Dolabellatrienone (11)
We followed the general procedure reported by Look and Fenical.3 A mixture of dolabellatrienone (11, 0.33 g, 1.2 mmol), anhydrous Na2HPO4 (0.17 g, 1.2 mmol) and m-chloroperbenzoic acid (0.20 g, 1.2 mmol) in dry CH2Cl2 (54 mL) was stirred at 25 °C for 1 h. The reaction mixture was diluted and washed with 10% Na2SO3 (2 × 25 mL), 5% NaHCO3 (2 × 25 mL), and brine (2 × 25 mL). The organic layer was dried over MgSO4, filtered and concentrated. The product mixture (305 mg) was chromatographed on a Si gel (15 g) column by elution with benzene–EtOAc (95:5) to afford pure diepoxide 7 (73.7 mg, 20%), C 7R,8R monoepoxide 12 (71.2 mg, 21%), and claenone (19, 145 mg, 42%);18,19 partial data for claenone (19) [3R,4R-epoxy-13-keto-1R,11S-dolabell-7E,12(18)-diene]: [α]25D −64 (c 0.2, CHCl3); 13C NMR (75 MHz, CDCl3) δ 205.9 (C, C-13), 148.5 (C, C-18), 137.0 (C, C-12), 132.8 (C, C-8), 128.2 (CH, C-7), 63.6 (CH, C-3), 61.2 (C, C-4), 55.4 (CH2, C-14), 42.2 (CH, C-11), 40.7 (C, C-1), 38.4 (CH2, C-9), 37.4 (CH2, C-2), 37.1 (CH2, C-5), 27.2 (CH2, C-6), 24.4 (CH3, C-19), 24.3 (CH2, C-10), 23.2 (CH3, C-15), 21.2 (CH3, C-20), 16.5 (CH3, C-17), 15.4 (CH3, C-16). The [α]D, IR, UV, HREIMS, and 1H NMR (cDCl3) data for 19 have been previously described.3,18
Single Crystal X-Ray Analysis of Dolabellanones 1, 2, 8 and 12
The X-ray data were collected at 298° K with a Bruker SMART 1 K CCD diffractometer equipped with a graphite monochromator and Mo-Kα radiation (λ = 0.71073 Å) using the SMART software. Final values of the cell parameters were obtained from least-squares refinement. The frames were processed using the SAINT software to give the hkl file corrected for Lorentz and polarization effects. No absorption correction was applied. The structures were solved by direct methods with the SHELX-90 program and refined by least-squares methods on F2, SHELXTL-93, incorporated in SHELXTL, Version 5.1.20 The initial E-maps yielded all non-hydrogen atom positions. All non-hydrogen atoms were refined anisotropically, and the H atoms were positioned geometrically and treated as riding, with C-H distances in the range 0.93–0.98 Å and with Uiso(H) = 1.2 or 1.5 Ueq(C). The crystallographic data for dolabellanones 1, 2, 8, and 12 reported in this article (Table 4) have been deposited at the Cambridge Crystallographic Data Centre, under the reference numbers CCDC 709008–709011. Copies of the data can be obtained, free of charge, on application to the Director, CCDC, 12 Union Road, Cambridge CB2 1EZ, UK (fax: +44 1223 336033 or deposit@ccdc.cam.ac.uk). ORTEP representations of 1, 2, 8, and 12 (Figure 2) show the relative configuration for each of these molecules.
Table 4.
Single Crystal X-Ray Diffraction Data for Compounds 1, 2, 8, and 12a
| Parameters | Compound |
|||
|---|---|---|---|---|
| 1 | 2 | 8 | 12 | |
| Empirical formula | C20H30O2 | C22H32O4 | C20H30O2 | C20H30O2 |
| Formula weight | 302.44 | 360.48 | 302.44 | 302.44 |
| Temperature (K) | 299 (2) | 298 (2) | 299 (2) | 298 (2) |
| Wavelength (Å) | 0.71073 | 0.71073 | 0.71073 | 0.71073 |
| Crystal system | Monoclinic | Orthorhombic | Orthorhombic | Monoclinic |
| Space group | P21 (No.4) | P212121 (No.l9) | P21212 (No.l8) | P21 (No.4) |
| Unit cell dimensions | ||||
| a (Å) | 9.777 (2) | 8.994 (2) | 11.149 (2) | 8.267 (2) |
| b (Å) | 10.719 (2) | 12.140 (2) | 21.716 (3) | 8.670 (2) |
| c (Å) | 17.521 (3) | 18.745 (3) | 7.590 (2) | 12.790 (3) |
| α (dge) | 90 | 90 | 90 | 90 |
| β (dge) | 90.174 (3) | 90.174 (3) | 90 | 90.451 (4) |
| γ (dge) | 90 | 90 | 90 | 90 |
| Volume (Å3) | 1836.2 (5) | 2046.8 (6) | 1837.5 (4) | 904.3 (3) |
| Z | 4 | 4 | 4 | 2 |
| Density (calc.) (Mg m−3) | 1.094 | 1.170 | 1.093 | 1.111 |
| Absorption coefficient (mm−1) | 0.068 | 0.079 | 0.068 | 0.069 |
| F (000) | 664 | 784 | 664 | 332 |
| Crystal size (mm) | 0.15 × 0.14 × 0.14 | 0.46 × 0.09 × 0.05 | 0.34 × 0.29 × 0.24 | 0.19 × 0.16 × 0.08 |
| θ range for data collection (dge) | 1.16 to 23.25 | 2.00 to 25.52 | 1.88 to 27.99 | 1.61 to 23.33 |
| Index ranges | −10 ≤ h ≤ 10, −11 ≤ k ≤ 9, −19 ≤ 1 ≤ 18 | −10 ≤ h ≤ 10, −12 ≤ k ≤ 14, −22 ≤ 1 ≤ 21 | −14 ≤ h ≤ 14, −26 ≤ k ≤ 28, −10 ≤ 1 ≤ 8 | −7 ≤ h ≤ 9, −9 ≤ k ≤ 9, −14 ≤ 1 ≤ 14 |
| Reflections collected | 8096 | 11514 | 12536 | 4050 |
| Independent reflections | 4306 [R(int)=0.0235] | 3795 [R(int)=0.0688] | 4358 [R(int)=0.0217] | 2282 [R(int)=0.0379] |
| Independent reflections [I > 2 σ (I)] | 3121 [R(int)=0.0372] | 2439 [R(int)=0.0630] | 3596 [R(int)=0.0206] | 1913 [R(int)=0.0397] |
| Max. and min. transmission | 0.0233, 0.0075 | |||
| Refinement method | Full-matrixleast-squares on F2 | Full-matrixleast-squares on F2 | Full-matrixleast squares on F2 | Full-matrixleast-squares on F2 |
| Data/restraints/parameters | 4306 / 1 / 407 | 3795 / 0 / 240 | 4358 / 0 / 205 | 2282 / 1 / 204 |
| Goodness-of-fit on F2 | 0.976 | 1.263 | 1.036 | 1.096 |
| Final R indices [I > 2 σ(I)] | Rl = 0.0418, wR2 = 0.0942 | Rl = 0.0657, wR2 = 0.1278 | Rl = 0.0416, wR2 = 0.0973 | Rl = 0.0479, wR2 = 0.1168 |
| R indices (all data) | Rl = 0.0675, wR2 = 0.1049 | Rl = 0.1405, wR2 = 0.1435 | Rl = 0.0541, wR2 = 0.1035 | Rl = 0.0618 wR2 = 0.1356 |
| Largest diff. peak and hole(e Å−3) | 0.175 and −0.119 | 0.139 and −0.114 | 0.175 and −0.119 | 0.148 and −0.131 |
Atomic coordinates for all X-ray structures have been deposited with the Cambridge Crystallographic Data Centre and can be obtained on request from Dr. Olga Kennard, University Chemical Laboratory, Lensfield Road, Cambridge CB2 1EZ, UK.
Biological Screening Assays
For a general description of the approach used by the NIAID's Antimicrobial Acquisition and Coordinating Facility (AACF) for determining antiviral activity and toxicity for hepatitis B virus, visit: http://niaid-aacf.org/protocols/HBV.htm. Anticancer activity screening by the Developmental Therapeutics Program (DTP) of the National Cancer Institute is conducted following this general protocol: most of the compounds screened have no antiproliferative activity (up to 85%). In order to avoid screening inactive compounds across all the cell lines, a prescreen is done using three highly sensitive cell lines (breast MCF-7, lung NCI-H640, CNS SF-268). Antiproliferative activity must be seen in these cell lines in order to continue to the 60 cell line panel. The 60 different human tumor lines are incubated with five different doses of compound and a sulforhodamine blue (SRB) assay is performed after 48 hours to determine cytotoxicity. From the five point curve, the following concentrations are extrapolated: GI50 (inhibits growth by 50%), TGI (totally inhibits growth), LC50 (kills 50% of cells). For the specific screening methods from the DTP website, visit: http://www.dtp.nci.nih.gov/branches/btb/ivclsp.html. Compounds shown to have anticancer activity in cell lines within the NCI 60 panel may then move on to animal trials and if successful, may eventually move on to be tested in clinical trials. Additional experimental details for our primary in vitro antimicrobial assays against Mycobacterium tuberculosis and Plasmodium falciparum have been previously described.21,22
Supplementary Material
Acknowledgement
This work was supported by a grant from the National Institutes of Health-SCORE Program (Grant S06GM08102) awarded to A. D. Rodríguez. We thank J. A. Sánchez for assistance during the collection and identification of the gorgonian coral, E. González for the chemical conversion of 11 to 7, and K. Nieves for measuring the specific rotation of 8. Mass spectrometry determinations were provided by the Mass Spectrometry Laboratory of the University of Illinois at Urbana–Champaign. We gratefully acknowledge the assistance of E. Ortega Barria and J. González from the Instituto de Investigaciones Avanzadas y Servicios de Alta Tecnología in Panama for the antimalarial bioassays. The National Cancer Institute (NCI), the National Institute of Allergy and Infectious Diseases (NIAID), and the TAACF provided in vitro cytotoxicity, antiviral, and antituberculosis activity data, respectively.
Footnotes
Supporting Information Available: 1H and 13C NMR spectra in CDCl3, including representative 2D NMR spectra, for compounds 1–10, as well as a photograph of the air dried voucher specimen Eunicea sp. 2. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- (1).Rodríguez AD. Tetrahedron. 1995;51:4571–4618. doi: 10.1016/0040-4020(95)00216-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Berrue F, Kerr RG. Nat. Prod. Rep. 2009;26:681–710. doi: 10.1039/b821918b. [DOI] [PubMed] [Google Scholar]
- (3).Look SA, Fenical W. J. Org. Chem. 1982;47:4129–4134. [Google Scholar]
- (4).Cáceres J, Rivera ME, Rodríguez AD. Tetrahedron. 1990;46:341–348. [Google Scholar]
- (5).Shin J, Fenical W. J. Org. Chem. 1991;56:3392–3398. [Google Scholar]
- (6).Rodríguez AD, Acosta AL, Dhasmana H. J. Nat. Prod. 1993;56:1843–1849. doi: 10.1021/np50100a031. [DOI] [PubMed] [Google Scholar]
- (7).Rodríguez AD, González E, González C. J. Nat. Prod. 1995;58:226–232. doi: 10.1021/np50116a010. [DOI] [PubMed] [Google Scholar]
- (8).Govindan M, Govindan GN, Kingston DGI. J. Nat. Prod. 1995;58:1174–1184. doi: 10.1021/np50122a004. [DOI] [PubMed] [Google Scholar]
- (9).Bashyal B, Desai P, Rao KV, Hamann MT, Avery BA, Reed JK, Avery MA. J. Chem. Res. 2006:165–167. [Google Scholar]
- (10).Xiang W, Chang LC. Planta Med. 2006;72:735–739. doi: 10.1055/s-2006-931584. [DOI] [PubMed] [Google Scholar]
- (11).Wei X, Rodríguez AD, Baran P, Raptis RG, Sánchez JA, Ortega-Barria E, González J. Tetrahedron. 2004;60:11813–11819. [Google Scholar]
- (12).Rodríguez AD, González E, Ramírez C. Tetrahedron. 1998;54:11683–11729. [Google Scholar]
- (13).Corey EJ, Kania RS. J. Am. Chem. Soc. 1996;118:1229–1230. [Google Scholar]
- (14).Snyder SA, Corey EJ. J. Am. Chem. Soc. 2006;128:740–742. doi: 10.1021/ja0576379. [DOI] [PubMed] [Google Scholar]
- (15).(a) Enserink M. Science. 2008;322:1776. doi: 10.1126/science.322.5909.1776. [DOI] [PubMed] [Google Scholar]; (b) Noedl H, Socheat D, Satimai W. N. Engl. J. Med. 2009;361:540–541. doi: 10.1056/NEJMc0900231. [DOI] [PubMed] [Google Scholar]
- (16).The antimalarial activity of the compounds tested was as follows: 1 (IC50 0.01 μM); 2 (IC50 0.05 μM); 3 (IC50 0.02 μM); 4 (IC50 0.04 μM); 5 (IC50 0.04 μM); 6 (IC50 0.05 μM); 7 (IC50 0.04 μM); 8 (IC50 ≥ 0.20 μM); 9 (IC50 0.01 μM); 10 (IC50 0.04 μM); 11 (IC50 0.05 μM); 12 (IC50 0.02 μM); 13 (IC50 0.03 μM); 14 (IC50 0.03 μM); 15 (IC50 0.04 μM); 16 (IC50 0.03 μM); 17 (IC50 0.06 μM).
- (17).Although the exact same X-ray structure of 12 has been already described in the literature by Xiang and Chang (see ref. 10), they wrongfully referred it in the text as the enantiomer 1S,7S,8S,11R.
- (18).Mori K, Iguchi K, Yamada N, Yamada Y, Inouye Y. Chem. Pharm. Bull. 1988;36:2840–2852. doi: 10.1248/cpb.36.2840. [DOI] [PubMed] [Google Scholar]
- (19).Miyaoka H, Isaji Y, Kajiwara Y, Kunimune I, Yamada Y. Tetrahedron Lett. 1998;39:6503–6506. [Google Scholar]
- (20).(a) Data Collection: SMART NT Software Reference Manual, version 5.0. Bruker AXS, Inc.; Madison, WI: 1998. [Google Scholar]; (b) Data Reduction: SAINT NT Software Reference Manual, verion 4.0. Bruker AXS, Inc.; Madison, WI: 1996. [Google Scholar]; (c) Sheldrick GM. SHELXTL NT, version 5.1. Bruker AXS, Inc.; Madison, WI: 1999. [Google Scholar]
- (21).Collins LA, Franzblau SG. Antimicrob. Agents Chemother. 1997;41:1004–1009. doi: 10.1128/aac.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Corbett Y, Herrera L, González J, Cubilla L, Capson T, Colley PD, Kursar TA, Romero LI, Ortega-Barria E. J. Trop. Med. Hyg. 2004;70:119–124. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.