Abstract
Rift Valley fever virus (RVFV) causes large outbreaks of acute febrile and often fatal illness among humans and domesticated animals in sub-saharan Africa and the Arabian peninsula. RVFV is a member of the family Bunyaviridae, genus Phlebovirus. Like all members of this large virus family, it contains a three-segmented genome of negative/ambisense strand RNA, packaged into viral nucleocapsid protein, and enveloped by a lipid bilayer containing two viral glycoproteins. During the past years, there was an increased interest in RVFV epidemiology, molecular biology, and virulence mechanisms. Here, we will try to provide an overview over the basic features of this significant pathogen, and review the latest developments in this highly active research field.
Keywords: Rift Valley fever, reverse genetics, non-structural proteins, interferon antagonism.
INTRODUCTION
Rift Valley fever virus (RVFV) is a serious emerging pathogen affecting humans and livestock in sub-Saharan Africa, Egypt, Yemen, and Saudi Arabia. Since the first description of an outbreak in Kenya 1931 [1], recurrent epidemics have killed hundreds of thousands of animals, more than a thousand humans, and caused significant economic losses [2, 3]. To animals, RVFV is mainly transmitted by a range of mosquito species (Aedes, Anopheles, Culex, Eretmapoites, Mansonia), but has also been shown to be transmitted by other vectors e.g. sandflies [4, 5]. Infected animals develop necrotic hepatitis, hemorrhage and abortion, with death rates up to 100% among newborn animals [3]. Humans are often infected by close contact with sick animals [6, 7] and the disease is associated with symptoms ranging from uncomplicated acute febrile illness to retinitis, hepatitis, renal failure, meningoencephalitis, severe hemorrhagic disease, and death [3]. Although the mortality rate for humans was reported to be approximately 2%, in recent outbreaks it went up to 45% [8, 9]. The severity of RVFV zoonosis, its capability to cause major epidemics among livestock and humans, and the lack of efficient prophylactic and therapeutic measures make infection with this pathogen a serious public health concern not only in endemic, developing countries, but also in many non-endemic industrial countries. Authorities have therefore listed RVF as a notifiable disease and classified RVFV as a BSL-3 or BSL-3+/4 agent respectively, in Europe and in the US, and a potential biological weapon [10].
During the past years, there was an increased interest in RVFV epidemiology, molecular biology, and virulence mechanisms. Here, we will try to provide an overview over the basic features of this significant pathogen, and review the latest developments in this highly active research field.
THE FAMILY BUNYAVIRIDAE
RVFV belongs to the genus Phlebovirus, family Bunyaviridae [11]. Bunyaviruses are enveloped and have a tri-segmented single-stranded RNA genome of negative or ambisense polarity. Transcription and replication take place in the cytoplasm. The genome segments of bunyaviruses encode four structural proteins: the viral polymerase (L) on the large (L) segment, two glycoproteins (Gn and Gc) on the medium (M) segment, and the viral nucleocapsid protein (N) on the smallest (S) segment [12]. RVFV additionally expresses two nonstructural proteins encoded on the M segment, a 78kDa protein and a 14kDa protein that we propose to name NSm1 and NSm2, as suggested initially by Gerrard and Nichol [13], and one on the S segment (termed NSs). These nonstructural proteins are dispensable for viral multiplication in cell culture, but play important roles for pathogenesis in vivo [14-17].
Virion Structure
Early ultrastructural studies by electron microscopy and negative staining [18] described RVFV particles measuring 90-110 nm in diameter. The envelope is composed of a lipid bilayer containing the Gn and Gc glycoproteins forming surface subunits, 5-8 nm in length, regularly arranged on its surface, similar to those reported for the related Uukuniemi phlebovirus by von Bonsdorff and Pettersson [19]. The viral ribonucleoproteins (RNPs) composed of the three genomic segments associated with numerous copies of the nucleoprotein N and the RNA dependent RNA polymerase L are packaged into the virion. More recent studies by cryo-electron microscopy [20-22] have changed the former view that phleboviruses are pleiomorphic. Instead, these studies indicated that the virion is likely to have an icosahedral symmetry: the structure of RVFV is surprisingly ordered and the surface covered by a shell of 122 glycoprotein capsomers arranged in an icosahedral lattice with T=12. A similar structure was reported for Uukuniemi virus [23]. For RVFV, three-dimensional reconstructions at 2.1 or 2.3 nm resolution revealed that the capsomers resemble hollow cylinders situated at five- and six-coordinated positions. Strikingly, inside the envelope a layer of RNPs is visible which bends the inner leaflet membrane towards the glycoproteins, of which even the transmembrane segments are discernible [21]. This suggests a strong interaction between the cytoplasmic tail of the glycoproteins (most likely Gn) and the RNPs which is known to enable packaging of the genome into the particles [24, 25].
The Viral Genome
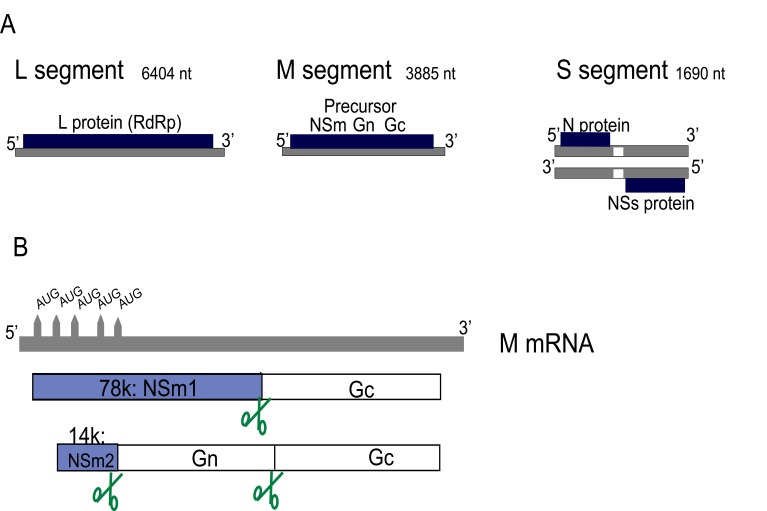
The three genomic RNA segments of bunyaviruses contain untranslated regions (UTRs) at the 3’ and 5’ ends which are segment-specific and serve as promoters for transcription and replication by the viral polymerase. The termini of the UTRs are conserved and form panhandle structures [26-28] which give the RNPs a circular appearance when observed by electron microscopy [29-31]. For RVFV, the L and M segments are of negative polarity while the S segment utilizes an ambisense strategy to code for both N and NSs [32]. The coding capacity of the genome is depicted in Fig. (1A). The general view that only the viral, negative-sense genome is incorporated into the mature particle must be revised as a small but significant fraction of the antigenomes i.e. replicative intermediates have been detected in purified RVFV particles [33], confirming past studies on Uukuniemi virus [34].
Fig. (1).
Genome and expression strategies of RVFV. (A) Schematic representation of the three genomic segments L (large), M (medium) and S (small) and the proteins encoded on them. (B) The mRNA transcribed from the M segment contains five in-frame start codons which gove rise to the 78kDa and the 14 kDa proteins, as well as some minor products [13]. Scissor indicate host cell protease sites for cleavage of the polyproteins.
Viral Replication Cycle and the Role of Viral Gene Products
Transcription and Replication
The general features of RVFV transcription and replication are similar to those of other negative stranded RNA viruses [35]. During the replication cycle, each segment is transcribed into a mRNA and is replicated through a process which involves the synthesis of the exact copy of the genome, called complementary RNA (cRNA) or antigenome. For Phleboviruses, and RVFV in particular, the cRNA representing the copy of the S ambisense segment serves as template for the synthesis of the NSs mRNA. Since some cRNA is also present in the input virus [33], the virulence factor NSs is expressed immediately after the virus has entered the host cell. Bunyaviral mRNA synthesis in general is initiated through a cap-snatching mechanism by which host mRNAs are cleaved through an endonuclease activity of the L protein [36] to obtain 10 to 18 nucleotide-long primers for transcription [36-40]. Synthesis of cRNA and vRNA does not require an oligonucleotide primer and is directly initiated with 5’ nucleoside triphosphates. cRNA represents a complete copy of the vRNA, whereas mRNAs terminate before the 5’ end of the template. The mechanism of the switch between transcription and replication remains ill-characterized. Moreover, although RNA-dependent polymerase consensus motifs are present in the RVFV L protein [41], the domains responsible for the different activities have not been defined precisely. However, minigenome systems have been helpful to analyze some steps in RNA synthesis i.e. transcription, replication, transcription termination, and packaging. Minigenomes are similar to viral genome segments but the viral ORF is replaced by a reporter gene. Minigenomes are usually expressed either from T7- or from Pol I-based promoter plasmids, or transcribed in vitro and transfected as RNA. Early on, studies with minigenomes have established that transcription and replication of the bunyaviral RNA requires the N and L proteins [42, 43]. Together with the vRNA, L and N are needed to reconstitute RNPs since, as for all negative stranded genomes, naked vRNA cannot be transcribed. Analysis of the RVFV RNPs showed that the N protein is able to establish intermolecular interactions through several amino-acids located in the N terminal region of the protein which are conserved among phlebovirus N sequences [44]. The ability of N to form oligomers may be a conserved feature in bunyaviruses [45-48]. The L protein, probably in association with N, is able to perform both transcription and replication, excluding the possibility that the L protein had to be modified by a third viral factor to function as a replicase. For RVFV, the viral protein NSs was reported to promote viral RNA replication and transcription in a minigenome system [49]. However, these experiments were performed in human 293T cells expressing T7 polymerase. RNAs produced by the T7 polymerase contain 5’ triphosphate ends which are strong inducers of the antiviral type I interferon (IFN) response [50, 51, 51a]. 293T cells, like most other cells, are capable to respond to 5’triphosphate RNAs, whereas the BHK-derived cells used by most researchers to express T7 polymerase are unresponsive due to a defect in the RIG-I pattern recognition pathway [52]. Moreover, the stimulatory effect of NSs was not confirmed in BHK cells (MB, unpublished data) but rather inhibited minireplicon activity in a manner similar to the NSs of the related viruses Bunyamwera [53] and La Crosse [54]. In line with this, when infectious RVFV particles were produced by T7-based systems in BHK-derived cells, RVFV NSs was shown to be either inhibitory [55], or irrelevant for generating recombinant viruses [52, 56]. It is thus conceivable that the observed stimulatory effect of RVFV NSs is indirect, namely through the well-established ability to suppress the antiviral IFN response (see below).
When compared to each other, the L, M and S segment-based minigenomes do not express identical levels of reporter gene. This indicates differential promoter activities associated with the UTRs [57]. Although the mutagenesis analysis was not as extensive as the one carried out for Uukuniemi virus [58], it appeared that some of the conserved UTR nucleotides play an important role in promoter activity and the regulation of gene expression [59].
Bunyavirus mRNAs (except for Sin Nombre hantavirus [60]) are not polyadenylated. They have a shortened 3’ end relative to the full-length cRNA, suggesting that a specific termination signal is recognized during transcription. Preliminary data had indicated that a conserved motif is present in the intergenic region of the S segment of the phleboviruses Toscana (TOSV), Sandfly fever Sicilian (SFSV), and RVFV [32, 38]. Albarino et al. [61] mapped precisely the 3’ ends of the L, M, N and NSs mRNAs of TOSV, SFSV and RVFV, and identified a sequence motif 3’-C1-3GUCG/A-5’ which is conserved on the M and S segments of phleboviruses. The phleboviral L segment mRNA, by contrast, was found to have a full-length 3’ end. Ikegami et al. [62] came to similar conclusions for the termination of the M and S segment mRNAs of RVFV, but claimed that the L mRNA lacks the last 16-41 nucleotides when compared to the full-length cRNA copy, and that the termination signal corresponds to two 13-nucleotide-long complementary sequences present in the 5’ non-coding region of the L genomic segment.
Glycoproteins and Particle Formation
The RVFV envelope glycoproteins mediate particle entry into cells through receptors which, like for most bunyaviruses, remain to be identified. RVFV entry is predicted to employ a class II fusion mechanism that is activated by low pH following endocytosis of the virion [63]. The RVFV M segment encodes Gn (encoded by amino-terminal sequences) and Gc (encoded by carboxy-terminal sequences) as well as two nonstructural proteins NSm1 (78kDa) and NSm2 (14kDa). The NSm1 and NSm2 proteins are produced by alternative use of the first and second of the 5 in-frame AUG codons, respectively, which are present at the 5’ end of the M mRNA i.e. upstream of the Gn sequence (Fig. 1B). The role of these proteins will be discussed further below.
Like all bunyaviruses, RVFV particles usually bud into the lumen of the Golgi apparatus [35]. Budding at the plasma membrane was however observed in rat hepatocytes [64]. Both RVFV Gn and Gc localize to the Golgi apparatus when expressed together from polyprotein-expressing plasmids [65]. When expressed alone, Gn could be transported to the Golgi apparatus due to a specific Golgi localization motif, whereas Gc localized to the endoplasmic reticulum (ER) due to the presence of a specific ER retention signal [66]. This suggests that, similar to other bunyaviruses [67, 68], Gc moves to the Golgi apparatus via its physical association with Gn.
The minigenome systems expressing a reporter gene were also useful to investigate viral packaging activity. In that case, the glycoproteins were co-expressed with the transcription machinery to allow formation of viral like particles (VLPs) released into the medium [69]. When observed by electron microscopy and negative staining, these particles resembled RVFV particles in size and morphology. They were able to infect naïve cells and to undergo the first step of the replication cycle, i. e. primary transcription. If both L and N were provided in trans, replication of minigenomes also occured [69]. Using these VLPs, it was shown that the interferon-induced antiviral protein MxA inhibits both primary and secondary transcription of RVFV. Interestingly, similar RVFV VLPs were produced in insect cells infected with a dual baculovirus vector expressing Gn/Gc and N [70]. Another vector expressing N and Gc only resulted in particles which were more pleiomorphic than full-fledged VLPs and RVFV particles, suggesting that both Gn and Gc contribute to the assembly process and likely interact with N.
Reverse Genetics
Reverse genetics systems allow to freely manipulate the viral genome and to dissect individual steps in the viral multiplication cycle. The minireplicon and VLP systems mentioned above were proven useful to study primary transcription, cell-autonomous antiviral responses, genome replication, and RNP packaging [49, 57, 69]. Fully infectious, recombinant virus particles can be recovered from cloned cDNA plasmids if the minireplicon is replaced by constructs for the full-length viral RNA segments. This technique was first established for Bunyamwera virus, the prototype of the Bunyaviridae [71, 72], followed by the related La Crosse virus [73]. These rescue systems were based on T7-transgenic cells to express the constructs for the viral genome segments and the helper proteins N and L. T7 systems were also used to recover recombinant RVFV [15, 55], but promoters for the cellular pol I could also be used to express the genome segments [52, 56]. This indicates that RVFV, a cytoplasmic virus, does not contain critical sequences e.g. cryptic splice sites, which could interfere with nuclear transcription of the genome. In fact, T7- and Pol I/Pol II-driven systems were shown to be of similar efficiency [52]. It should be noted that the Pol I system requires co-transfection of the N and L helper plasmids, whereas in the T7 system helper plasmids are either not necessary or even inhibitory. However, in T7-driven systems the T7 polymerase has to be provided in trans, usually by stably expressing cell lines. As outlined above, the cell lines used for T7 expression [55, 74] are deficient in the RIG-I pathway [52] which would normally respond to the 5’ triphosphorylated T7 transcripts with the production of the antiviral IFNs.
Depending on the laboratory which developed the rescue system, the genetic backbone of the recombinant virus was based either on the attenuated strain MP12 [55, 56], or the virulent strains ZH501 [15] or ZH548 [52, 56].
Role of the Non-Structural Proteins
The establishment of reverse genetics systems has opened the door for studies of RVFV molecular biology and pathogenesis which were unthinkable a few years ago.
The Role of NSs in Virulence
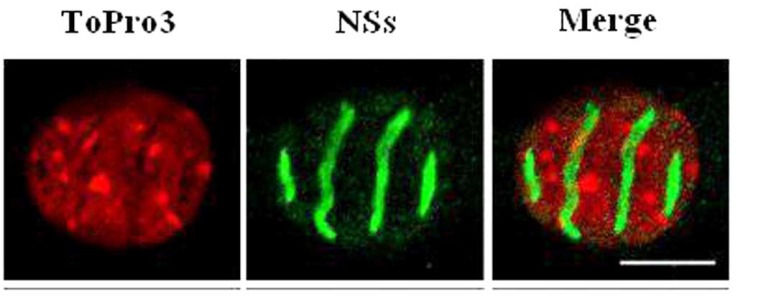
NSs of RVFV forms a ribbon-like filament in the nucleus [12] (Fig. 2). This feature is unexpected for a virus replicating in the cytoplasm. Moreover, it is particular to RVFV and not shared with NSs proteins of other bunyaviruses. NSs was characterized as a major virulence factor counteracting the antiviral IFN system [16, 75, 76]. It has become clear meanwhile that NSs is a multifunctional protein inhibiting cellular transcription in general and suppressing two different arms of the IFN response, namely (i) IFN induction by a specific, early, mechanism (ii) the antiviral protein PKR.
Fig. (2).
RVFV NSs forms a filamentous structure in the nucleus. Confocal microscopy picture showing a section of a L929 cell nucleus infected by ZH548 and stained with the DNA intercalating dye ToPro3 (red), with anti-NSs antibodies (green) and merge. The cellular DNA is predominantly excluded from the filament (from Mansuroglu et al., J. Virol. in press).
With respect to specific suppression of IFN induction, the cellular protein SAP30 was identified as interacting with NSs. SAP30 belongs to the Sin3A/NCoR/HDAC repressor complexes intervening in gene transcription regulation [77]. Moreover, it is known that SAP30 interacts directly with YY1 [78], a transcription factor involved in the regulation of expression of numerous genes, including IFN-β [79]. Through a series of co-immunoprecipitation, confocal microscopy and chromatin immunoprecipitations, it was demonstrated that NSs, SAP30, YY1 and Sin3A-associated corepressors are recruited on the IFN-β promoter inhibiting CBP recruitment, histone acetylation and transcriptional activation [77]. Using reverse genetics, a recombinant ZH548 in which NSs was deleted of the SAP30 interaction domain was produced. In contrast with the wt virus, this mutant, ZH548-NSs∆210-230, induced IFN-β expression and was unable to kill mice. Although this mutant protein was still located in the nucleus, it was not able to form filaments. Further analyses on the wt NSs filament indicated that, even though cellular DNA is mostly excluded from the filament, heterochromatin clusters of pericentromeric gamma satellite sequences appeared intimately associated with NSs [79a]. This results in a high incidence of nuclear anomalies, translating chromosome cohesion and segregation defects which could contribute to RVFV pathology.
NSs also suppresses cellular gene expression by a more general mechanism which relies on the interaction with the host cell protein p44. p44 is a subunit of the TFIIH basal transcription factor and becomes sequestered into the NSs filamentous structure characteristic of RVFV infection [80]. As a consequence, TFIIH cannot assemble and its concentration drops rapidly, explaining the drastically reduced transcriptional activity of cells infected with RVFV. Interestingly, NSs thus functions as a general inhibitor of Pol II as well as Pol I, since TFIIH is involved in transcription by both these host polymerases. Inhibition of TFIIH and hence general transcription is a relatively late event occurring after 8 h of infection, whereas the specific inhibition of IFN-β transcription (mediated by SAP30 recruitment) is in place as early as 3-4 h after infection.
Very recently, a novel function of NSs was described, the targeting of the antiviral, IFN-induced protein kinase PKR [81, 82]. PKR is a serine-threonine kinase activated by viral RNAs and mediating a stop of translation [83]. NSs-dependent degradation is specific for PKR and occurs through the proteasome. Hence, cells infected with RVFV are devoid of PKR, whereas PKR was clearly detected and activated by infection with NSs-deleted virus strains e.g. Clone 13. This anti-PKR activity is important for viral replication and pathogenesis, since mice expressing PKR clear infection by Clone 13, whereas knockout mice lacking PKR succumb to Clone 13 and develop high virus titers in the liver [81]. The fact that NSs itself provokes a PKR-knockout-like situation is likely to contribute to the pathogenicity of wt RVFV. Interestingly, it was also observed that this activity is specific to RVFV NSs and not shared with other Phleboviruses like Sandfly fever Sicilian virus or the orthobunyavirus La Crosse [81].
Altogether, it appears that NSs has multiple functions to counteract the IFN system, either at the transcriptional level or at the translational level by degrading PKR.
The Role of the NSm Protein as a Virulence Factor and a Suppressor of Virus-Induced Apoptosis
The biological function of the NSm1 and NSm2 proteins was addressed by producing RVFV mutants with either a deletion in the pre-Gn region [15, 84] or point mutations in the start codons of the two respective reading frames [17]. In all cases, virus growth in cell culture was similar to the parental viruses. However, when the pre-Gn deletion was introduced into the MP12 strain, plaques were larger than for the parental MP12 strain and a more extensive apoptosis due to elevated caspase activity was observed [84]. Curiously, a similar deletion introduced into the virulent strain ZH501 did not show any differences in plaque size, and an increase in apoptosis was not reported [15]. Moreover, if the AUG-inactivating point mutations in the pre-Gn region, which abrogated the NSm1 and NSm2 expression, were introduced into the MP12 background, they also led to a plaque phenotype indistinguishable from the parental virus [17]. The differences between the pre-Gn deletion and the AUG point mutant of MP12 can be explained by residual expression of a truncated version of the NSm1 protein, the 73-kDa protein [84]. Why this is not happening for ZH501 remains to be investigated. In any case, all available data are congruent in indicating that the NSm1 and NSm2 proteins of RVFV are dispensable for virus growth in cell culture.
NEW INSIGHTS FOR THE DEVELOPMENT OF VACCINES
A natural virus, Clone 13, lacks more than 70% of the coding sequence for the IFN antagonist NSs and is avirulent for mice, sheep and cattle, but highly immunogenic [85]. The anti-IFN function of NSs is conserved among the bunyaviruses and has been described for another phlebovirus, Punta Toro virus [86] as well as for the orthobunyaviruses Bunyamwera [87, 88] and La Crosse [89]. NSs is thus a prime target for the development of attenuated virus strains. In addition, deletion of the NSm2 gene confers an additional level of safety. NSm2 was shown to severely hamper immune responses to a DNA vaccine construct encoding the M segment [90]. The mechanism behind this interesting observation is not clear so far. In contrast to the full-length construct which conferred only poor immunogenicity, NSm2-deleted M segment cDNA was very immunogenic and gave 100 % survival rate upon challenge [90]. With respect to live viruses, RVFV mutants lacking NSm2 are somewhat attenuated compared to wt RVFV but can still cause either acute lethal hepatic necrosis or delayed lethal neurologic disease [14]. NSs-NSm2 double deletion mutants induced robust anti-RVF virus antibody responses and 100% protection from a lethal challenge [91]. Replacement of NSs by a marker gene e.g. GFP, could be helpful to screen antivirals [91]. Based on the absence of NSs-specific antibodies, an ELISA was developed to distinguish infected from vaccinated animals [92].
An even more radical approach is represented by transcriptionally active RVFV VLPs which were produced by reverse genetics [69]. The VLPs resemble authentic RVFV particles and are able to express a reporter gene, but lack all the genes necessary for virus spread or pathogenesis. Three injections of VLPs induced neutralizing antibodies and were shown to be highly protective against a lethal challenge [93].
Thus, the recently gained knowledge on RVFV molecular biology already proved useful for the understanding and development of modern vaccine candidates [94, 95], and future findings will certainly help to further improve our repertoire of strategies to combat this important pathogen.
ACKNOWLEDGEMENTS
Work in our laboratories is supported by grants We2616/5-1 and We2616/5-2 from the Deutsche Forschungsgemeinschaft to F.W., and by grants ANR-08-MIE-022 from Agence Nationale de la Recherche (ANR) and 7U01AI066327 from the National Institute of Health to MB.
REFERENCES
- 1.Daubney R, Hudson JR, Gamham PC. Enzootic hepatitis of Rift Valley fever: an undescribed virus disease of sheep, cattle and man from East Africa. J Pathol Bacteriol. 1931;34:545–9. [Google Scholar]
- 2.Balkhy HH, Memish ZA. Rift Valley fever: an uninvited zoonosis in the Arabian peninsula. Int J Antimicrob Agents. 2003;21:153–7. doi: 10.1016/s0924-8579(02)00295-9. [DOI] [PubMed] [Google Scholar]
- 3.Bird BH, Ksiazek TG, Nichol ST, Maclachlan NJ. Rift Valley fever virus. J Am Vet Med Assoc. 2009;234:883–93. doi: 10.2460/javma.234.7.883. [DOI] [PubMed] [Google Scholar]
- 4.Fontenille D, Traore-Lamizana M, Diallo M, Thonnon J, Digoutte JP, Zeller HG. New vectors of Rift Valley fever in West Africa. Emerg Infect Dis. 1998;4:289–93. doi: 10.3201/eid0402.980218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moutailler S, Krida G, Schaffner F, Vazeille M, Failloux AB. Potential vectors of Rift Valley fever virus in the Mediterranean region. Vector Borne Zoonotic Dis Larchmont, NY. 2008;8:749–53. doi: 10.1089/vbz.2008.0009. [DOI] [PubMed] [Google Scholar]
- 6.Morrill JC, McClain DJ. Epidemiology and pathogenesis of Rift Valley fever and other phleboviruses. In: Elliott RM, editor. The bunyaviridae. Vol. 281. New York: Plenum Press; 1996. p. 93. [Google Scholar]
- 7.Wilson ML, Chapman LE, Hall DB, et al. Rift Valley fever in rural northern Senegal: human risk factors and potential vectors. Am J Trop Med Hyg. 1994;50:663–75. doi: 10.4269/ajtmh.1994.50.663. [DOI] [PubMed] [Google Scholar]
- 8.LaBeaud AD, Muchiri EM, Ndzovu M, et al. Interepidemic Rift Valley fever virus seropositivity, northeastern Kenya. Emerg Infect Dis. 2008;14:1240–6. doi: 10.3201/eid1408.080082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO. Rift Valley fever outbreak--Kenya, November 2006-January 2007. MMWR Morb Mortal Wkly Rep. 2007;56:73–6. [PubMed] [Google Scholar]
- 10.Borio L, Inglesby T, Peters CJ, et al. Hemorrhagic fever viruses as biological weapons: medical and public health management. JAMA. 2002;287:2391–405. doi: 10.1001/jama.287.18.2391. [DOI] [PubMed] [Google Scholar]
- 11.Elliott RM. Emerging viruses: the Bunyaviridae. Mol Med. 1997;3:572–7. [PMC free article] [PubMed] [Google Scholar]
- 12.Struthers JK, Swanepoel R, Shepherd SP. Protein synthesis in Rift Valley fever virus-infected cells. Virology. 1984;134:118–24. doi: 10.1016/0042-6822(84)90277-0. [DOI] [PubMed] [Google Scholar]
- 13.Gerrard SR, Nichol ST. Synthesis, proteolytic processing and complex formation of N-terminally nested precursor proteins of the Rift Valley fever virus glycoproteins. Virology. 2007;357:124–33. doi: 10.1016/j.virol.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bird BH, Albarino CG, Nichol ST. Rift Valley fever virus lacking NSm proteins retains high virulence in vivo and may provide a model of human delayed onset neurologic disease. Virology. 2007;362:10–5. doi: 10.1016/j.virol.2007.01.046. [DOI] [PubMed] [Google Scholar]
- 15.Gerrard SR, Bird BH, Albarino CG, Nichol ST. The NSm proteins of Rift Valley fever virus are dispensable for maturation, replication and infection. Virology. 2007;359:459–65. doi: 10.1016/j.virol.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vialat P, Billecocq A, Kohl A, Bouloy M. The S segment of rift valley fever phlebovirus (Bunyaviridae) carries determinants for attenuation and virulence in mice. J Virol. 2000;74:1538–43. doi: 10.1128/jvi.74.3.1538-1543.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Won S, Ikegami T, Peters CJ, Makino S. NSm and 78-kilodalton proteins of Rift Valley fever virus are nonessential for viral replication in cell culture. J Virol. 2006;80:8274–8. doi: 10.1128/JVI.00476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellis DS, Simpson DI, Stamford S, Abdel Wahab KS. Rift Valley fever virus: some ultrastructural observations on material from the outbreak in Egypt 1977. J Gen Virol. 1979;42:329–37. doi: 10.1099/0022-1317-42-2-329. [DOI] [PubMed] [Google Scholar]
- 19.von Bonsdorff CH, Pettersson R. Surface structure of Uukuniemi virus. J Virol. 1975;16:1296–307. doi: 10.1128/jvi.16.5.1296-1307.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freiberg AN, Sherman MB, Morais MC, Holbrook MR, Watowich SJ. Three-dimensional organization of Rift Valley fever virus revealed by cryoelectron tomography. J Virol. 2008;82:10341–8. doi: 10.1128/JVI.01191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huiskonen JT, Overby AK, Weber F, Grunewald K. Electron cryo-microscopy and single-particle averaging of Rift Valley fever virus: evidence for GN-GC glycoprotein heterodimers. J Virol. 2009;83:3762–9. doi: 10.1128/JVI.02483-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherman MB, Freiberg AN, Holbrook MR, Watowich SJ. Single-particle cryo-electron microscopy of Rift Valley fever virus. Virology. 2009;387:11–5. doi: 10.1016/j.virol.2009.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Overby AK, Pettersson RF, Grunewald K, Huiskonen JT. Insights into bunyavirus architecture from electron cryotomography of Uukuniemi virus. Proc Natl Acad Sci USA. 2008;105:2375–9. doi: 10.1073/pnas.0708738105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overby AK, Pettersson RF, Neve EP. The glycoprotein cytoplasmic tail of Uukuniemi virus (Bunyaviridae) interacts with ribonucleoproteins and is critical for genome packaging. J Virol. 81:3198–205. doi: 10.1128/JVI.02655-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Overby AK, Popov VL, Pettersson RF, Neve EP. The cytoplasmic tails of Uukuniemi Virus (Bunyaviridae) G(N) and G(C) glycoproteins are important for intracellular targeting and the budding of virus-like particles. J Virol. 81:11381–91. doi: 10.1128/JVI.00767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pardigon N, Vialat P, Girard M, Bouloy M. Panhandles and hairpin structures at the termini of germiston virus RNAs (Bunyavirus) Virology. 1982;122:191–7. doi: 10.1016/0042-6822(82)90388-9. [DOI] [PubMed] [Google Scholar]
- 27.Patterson JL, Kolakofsky D, Holloway BP, Obijeski JF. Isolation of the ends of La Crosse virus small RNA as a double-stranded structure. J Virol. 1983;45:882–4. doi: 10.1128/jvi.45.2.882-884.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolakofsky D, Hacker D. Bunyavirus RNA synthesis: genome transcription and replication. Curr Top Microbiol Immunol. 1991;169:143–59. doi: 10.1007/978-3-642-76018-1_5. [DOI] [PubMed] [Google Scholar]
- 29.Pettersson RF, von Bonsdorff CH. Ribonucleoproteins of Uukuniemi virus are circular. J Virol. 1975;15:386–92. doi: 10.1128/jvi.15.2.386-392.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obijeski JF, Bishop DH, Palmer EL, Murphy FA. Segmented genome and nucleocapsid of La Crosse virus. J Virol. 1976;20:664–75. doi: 10.1128/jvi.20.3.664-675.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samso A, Bouloy M, Hannoun C. Circular ribonucleoproteins in the virus Lumbo (Bunyavirus) Comptes rendus hebdomadaires des seances de l'Academie des sciences. 1975;280:779–82. [PubMed] [Google Scholar]
- 32.Giorgi C, Accardi L, Nicoletti L, et al. Sequences and coding strategies of the S RNAs of Toscana and Rift Valley fever viruses compared to those of Punta Toro, Sicilian Sandfly fever, and Uukuniemi viruses. Virology. 1991;180:738–53. doi: 10.1016/0042-6822(91)90087-r. [DOI] [PubMed] [Google Scholar]
- 33.Ikegami T, Won S, Peters CJ, Makino S. Rift Valley fever virus NSs mRNA is transcribed from an incoming anti-viral-sense S RNA segment. J Virol. 2005;79:12106–11. doi: 10.1128/JVI.79.18.12106-12111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simons JF, Hellman U, Pettersson RF. Uukuniemi virus S RNA segment: ambisense coding strategy, packaging of complementary strands into virions, and homology to members of the genus Phlebovirus. J Virol. 1990;64:247–55. doi: 10.1128/jvi.64.1.247-255.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elliott RM. The Bunyaviridae. New York: Plenum Press; 1996. [Google Scholar]
- 36.Patterson JL, Holloway B, Kolakofsky D. La Crosse virions contain a primer-stimulated RNA polymerase and a methylated cap-dependent endonuclease. J Virol. 1984;52:215–22. doi: 10.1128/jvi.52.1.215-222.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouloy M, Pardigon N, Vialat P, Gerbaud S, Girard M. Characterization of the 5' and 3' ends of viral messenger RNAs isolated from BHK21 cells infected with Germiston virus (Bunyavirus) Virology. 1990;175:50–8. doi: 10.1016/0042-6822(90)90185-t. [DOI] [PubMed] [Google Scholar]
- 38.Gro MC, Di Bonito P, Accardi L, Giorgi C. Analysis of 3' and 5' ends of N and NSs messenger RNAs of Toscana Phlebovirus. Virology. 1992;191:435–8. doi: 10.1016/0042-6822(92)90205-4. [DOI] [PubMed] [Google Scholar]
- 39.Jin H, Elliott RM. Characterization of Bunyamwera virus S RNA that is transcribed and replicated by the L protein expressed from recombinant vaccinia virus. J Virol. 1993;67:1396–404. doi: 10.1128/jvi.67.3.1396-1404.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simons JF, Pettersson RF. Host-derived 5' ends and overlapping complementary 3' ends of the two mRNAs transcribed from the ambisense S segment of Uukuniemi virus. J Virol. 1991;65:4741–8. doi: 10.1128/jvi.65.9.4741-4748.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muller R, Poch O, Delarue M, Bishop DH, Bouloy M. Rift Valley fever virus L segment: correction of the sequence and possible functional role of newly identified regions conserved in RNA-dependent polymerases. J Gen Virol. 1994;75(Pt 6):1345–52. doi: 10.1099/0022-1317-75-6-1345. [DOI] [PubMed] [Google Scholar]
- 42.Jin H, Elliott RM. Expression of functional Bunyamwera virus L protein by recombinant vaccinia viruses. J Virol. 1991;65:4182–9. doi: 10.1128/jvi.65.8.4182-4189.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez N, Muller R, Prehaud C, Bouloy M. The L protein of Rift Valley fever virus can rescue viral ribonucleoproteins and transcribe synthetic genome-like RNA molecules. J Virol. 1995;69:3972–9. doi: 10.1128/jvi.69.7.3972-3979.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le May N, Gauliard N, Billecocq A, Bouloy M. The N terminus of Rift Valley fever virus nucleoprotein is essential for dimerization. J Virol. 2005;79:11974–80. doi: 10.1128/JVI.79.18.11974-11980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alfadhli A, Love Z, Arvidson B, Seeds J, Willey J, Barklis E. Hantavirus nucleocapsid protein oligomerization. J Virol. 2001;75:2019–23. doi: 10.1128/JVI.75.4.2019-2023.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaukinen P, Koistinen V, Vapalahti O, Vaheri A, Plyusnin A. Interaction between molecules of hantavirus nucleocapsid protein. J Gen Virol. 2001;82(Pt 8):1845–53. doi: 10.1099/0022-1317-82-8-1845. [DOI] [PubMed] [Google Scholar]
- 47.Leonard VH, Kohl A, Osborne JC, McLees A, Elliott RM. Homotypic interaction of Bunyamwera virus nucleocapsid protein. J Virol. 2005;79:13166–72. doi: 10.1128/JVI.79.20.13166-13172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mir MA, Panganiban AT. Trimeric hantavirus nucleocapsid protein binds specifically to the viral RNA panhandle. J Virol. 2004;78:8281–8. doi: 10.1128/JVI.78.15.8281-8288.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ikegami T, Peters CJ, Makino S. Rift valley fever virus nonstructural protein NSs promotes viral RNA replication and transcription in a minigenome system. J Virol. 2005;79:5606–15. doi: 10.1128/JVI.79.9.5606-5615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hornung V, Ellegast J, Kim S, et al. 5'-Triphosphate RNA Is the Ligand for RIG-I. Science. 2006;314:994–7. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 51.Pichlmair A, Schulz O, Tan CP, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 51a.Plumet S, Herschke F, Bourhis JM, Valentin H, Longhi S, Gerlier D. Cytosolic 5'-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS Onel. 2007;2(3):e279. doi: 10.1371/journal.pone.0000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Habjan M, Penski N, Spiegel M, Weber F. T7 RNA polymerase-dependent and -independent systems for cDNA-based rescue of Rift Valley fever virus. J Gen Virol. 2008;89:2157–66. doi: 10.1099/vir.0.2008/002097-0. [DOI] [PubMed] [Google Scholar]
- 53.Weber F, Dunn EF, Bridgen A, Elliott RM. The Bunyamwera virus nonstructural protein NSs inhibits viral RNA synthesis in a minireplicon system. Virology. 2001;281:67–74. doi: 10.1006/viro.2000.0774. [DOI] [PubMed] [Google Scholar]
- 54.Blakqori G, Kochs G, Haller O, Weber F. Functional L polymerase of La Crosse virus allows in vivo reconstitution of recombinant nucleocapsids. J Gen Virol. 2003;84(Pt 5):1207–14. doi: 10.1099/vir.0.18876-0. [DOI] [PubMed] [Google Scholar]
- 55.Ikegami T, Won S, Peters CJ, Makino S. Rescue of infectious rift valley fever virus entirely from cDNA, analysis of virus lacking the NSs gene, and expression of a foreign gene. J Virol. 2006;80:2933–40. doi: 10.1128/JVI.80.6.2933-2940.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Billecocq A, Gauliard N, Le May N, Elliott RM, Flick R, Bouloy M. RNA polymerase I-mediated expression of viral RNA for the rescue of infectious virulent and avirulent Rift Valley fever viruses. Virology. 2008;378:377–84. doi: 10.1016/j.virol.2008.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gauliard N, Billecocq A, Flick R, Bouloy M. Rift Valley fever virus noncoding regions of L, M and S segments regulate RNA synthesis. Virology. 2006;351:170–9. doi: 10.1016/j.virol.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 58.Flick R, Elgh F, Pettersson RF. Mutational analysis of the Uukuniemi virus (Bunyaviridae family) promoter reveals two elements of functional importance. J Virol. 2002;76:10849–60. doi: 10.1128/JVI.76.21.10849-10860.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prehaud C, Lopez N, Blok MJ, Obry V, Bouloy M. Analysis of the 3' terminal sequence recognized by the Rift Valley fever virus transcription complex in its ambisense S segment. Virology. 1997;227:189–97. doi: 10.1006/viro.1996.8324. [DOI] [PubMed] [Google Scholar]
- 60.Hutchinson KL, Peters CJ, Nichol ST. Sin Nombre virus mRNA synthesis. Virology. 1996;224:139–49. doi: 10.1006/viro.1996.0515. [DOI] [PubMed] [Google Scholar]
- 61.Albarino CG, Bird BH, Nichol ST. A shared transcription termination signal on negative and ambisense RNA genome segments of rift valley Fever, sandfly Fever sicilian, and toscana viruses. J Virol. 2007;81:5246–56. doi: 10.1128/JVI.02778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ikegami T, Won S, Peters CJ, Makino S. Characterization of rift valley fever virus transcriptional terminations. J Virol. 2007;81:8421–38. doi: 10.1128/JVI.02641-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Filone CM, Heise M, Doms RW, Bertolotti-Ciarlet A. Development and characterization of a Rift Valley fever virus cell-cell fusion assay using alphavirus replicon vectors. Virology. 2006;356:155–64. doi: 10.1016/j.virol.2006.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anderson GW Jr, Smith JF. Immunoelectron microscopy of Rift Valley fever viral morphogenesis in primary rat hepatocytes. Virology. 1987;161:91–100. doi: 10.1016/0042-6822(87)90174-7. [DOI] [PubMed] [Google Scholar]
- 65.Wasmoen TL, Kakach LT, Collett MS. Rift Valley fever virus M segment: cellular localization of M segment-encoded proteins. Virology. 1988;166:275–80. doi: 10.1016/0042-6822(88)90174-2. [DOI] [PubMed] [Google Scholar]
- 66.Gerrard SR, Nichol ST. Characterization of the Golgi retention motif of Rift Valley fever virus G(N) glycoprotein. J Virol. 2002;76:12200–10. doi: 10.1128/JVI.76.23.12200-12210.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi X, Elliott RM. Golgi localization of Hantaan virus glycoproteins requires coexpression of G1 and G2. Virology. 2002;300:31–8. doi: 10.1006/viro.2002.1414. [DOI] [PubMed] [Google Scholar]
- 68.Shi X, Lappin DF, Elliott RM. Mapping the Golgi targeting and retention signal of Bunyamwera virus glycoproteins. J Virol. 2004;78:10793–802. doi: 10.1128/JVI.78.19.10793-10802.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Habjan M, Penski N, Wagner V, et al. Efficient production of Rift Valley fever virus-like particles: The antiviral protein MxA can inhibit primary transcription of bunyaviruses. Virology. 2009;385:400–8. doi: 10.1016/j.virol.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 70.Liu L, Celma CC, Roy P. Rift Valley fever virus structural proteins: expression, characterization and assembly of recombinant proteins. Virol J. 2008;5:82. doi: 10.1186/1743-422X-5-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bridgen A, Elliott RM. Rescue of a segmented negative-strand RNA virus entirely from cloned complementary DNAs. Proc Natl Acad Sci USA. 1996;93:15400–4. doi: 10.1073/pnas.93.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lowen AC, Noonan C, McLees A, Elliott RM. Efficient bunyavirus rescue from cloned cDNA. Virology. 2004;330:493–500. doi: 10.1016/j.virol.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 73.Blakqori G, Weber F. Efficient cDNA-based rescue of La Crosse bunyaviruses expressing or lacking the nonstructural protein NSs. J Virol. 2005;79:10420–8. doi: 10.1128/JVI.79.16.10420-10428.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73:251–9. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Billecocq A, Spiegel M, Vialat P, et al. NSs protein of Rift Valley Fever Virus blocks interferon production by inhibiting host gene transcription. J Virol. 2004;78:9798–806. doi: 10.1128/JVI.78.18.9798-9806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bouloy M, Janzen C, Vialat P, et al. Genetic evidence for an interferon-antagonistic function of rift valley fever virus nonstructural protein NSs. J Virol. 2001;75:1371–7. doi: 10.1128/JVI.75.3.1371-1377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Le May N, Mansuroglu Z, Leger P, et al. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog. 2008;4:e13. doi: 10.1371/journal.ppat.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang NE, Lin CH, Lin YS, Yu WC. Modulation of YY1 activity by SAP30. Biochem Biophys Res Commun. 2003;306:267–75. doi: 10.1016/s0006-291x(03)00966-5. [DOI] [PubMed] [Google Scholar]
- 79.Weill L, Shestakova E, Bonnefoy E. Transcription factor YY1 binds to the murine beta interferon promoter and regulates its transcriptional capacity with a dual activator/repressor role. J Virol. 2003;77:2903–14. doi: 10.1128/JVI.77.5.2903-2914.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79a.Mansuroglu Z, Josse T, Gilleron J, Billecocq A, Leger P, Bouloy M, Bonnefoy E. Nonstructural NSs protein of rift valley fever virus interacts with pericentromeric DNA sequences of the host cell, inducing chromosome cohesion and segregation defects. J Virol. 2010;84:928–39. doi: 10.1128/JVI.01165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Le May N, Dubaele S, De Santis LP, Billecocq A, Bouloy M, Egly JM. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell. 2004;116:541–50. doi: 10.1016/s0092-8674(04)00132-1. [DOI] [PubMed] [Google Scholar]
- 81.Habjan M, Pichlmair A, Elliott RM, et al. NSs protein of rift valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J Virol. 2009;83:4365–75. doi: 10.1128/JVI.02148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ikegami T, Narayanan K, Won S, Kamitani W, Peters CJ, Makino S. Rift Valley fever virus NSs protein promotes post-transcriptional downregulation of protein kinase PKR and inhibits eIF2alpha phosphorylation. PLoS Pathog. 2009;5:e1000287. doi: 10.1371/journal.ppat.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–68. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Won S, Ikegami T, Peters CJ, Makino S. NSm protein of Rift Valley fever virus suppresses virus-induced apoptosis. J Virol. 2007;81:13335–45. doi: 10.1128/JVI.01238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Muller R, Saluzzo JF, Lopez N, et al. Characterization of clone 13, a naturally attenuated avirulent isolate of Rift Valley fever virus, which is altered in the small segment. Am J Trop Med Hyg. 1995;53:405–11. doi: 10.4269/ajtmh.1995.53.405. [DOI] [PubMed] [Google Scholar]
- 86.Perrone LA, Narayanan K, Worthy M, Peters CJ. The S segment of Punta Toro virus (Bunyaviridae, Phlebovirus) is a major determinant of lethality in the Syrian hamster and codes for a type I interferon antagonist. J Virol. 2007;81:884–92. doi: 10.1128/JVI.01074-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bridgen A, Weber F, Fazakerley JK, Elliott RM. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc Natl Acad Sci USA. 2001;98:664–9. doi: 10.1073/pnas.98.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weber F, Bridgen A, Fazakerley JK, et al. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J Virol. 2002;76:7949–55. doi: 10.1128/JVI.76.16.7949-7955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Blakqori G, Delhaye S, Habjan M, et al. La Crosse Bunyavirus nonstructural protein NSs serves to suppress the Type I interferon system of mammalian hosts. J Virol. 2007;81:4991–9. doi: 10.1128/JVI.01933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Spik K, Shurtleff A, McElroy AK, Guttieri MC, Hooper JW, SchmalJohn C. Immunogenicity of combination DNA vaccines for Rift Valley fever virus, tick-borne encephalitis virus, Hantaan virus, and Crimean Congo hemorrhagic fever virus. Vaccine. 2006;24:4657–66. doi: 10.1016/j.vaccine.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 91.Bird BH, Albarino CG, Hartman AL, Erickson BR, Ksiazek TG, Nichol ST. Rift valley fever virus lacking the NSs and NSm genes is highly attenuated, confers protective immunity from virulent virus challenge, and allows for differential identification of infected and vaccinated animals. J Virol. 2008;82:2681–91. doi: 10.1128/JVI.02501-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McElroy AK, Albarino CG, Nichol ST. Development of a RVFV ELISA that can distinguish infected from vaccinated animals. Virol J. 2009;6:125. doi: 10.1186/1743-422X-6-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Naslund J, Lagerqvist N, Habjan M, et al. Vaccination with virus-like particles protects mice from lethal infection of Rift Valley Fever Virus. Virology. 2009;385:409–15. doi: 10.1016/j.virol.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 94.Bouloy M, Flick R. Reverse genetics technology for Rift Valley fever virus: current and future applications for the development of therapeutics and vaccines. Antiviral Res. 2009;84:101–18. doi: 10.1016/j.antiviral.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ikegami T, Makino S. Rift valley fever vaccines. Vaccine. 2009;27(Suppl 4):D69–72. doi: 10.1016/j.vaccine.2009.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]