Introduction
As students of mitosis, we seek to identify the DNA and protein components required for chromosome segregation and to design experiments in order to test our latest theory on how these components fit together. The identification of what is approaching a complete parts list has provided major advances in the past few years. The problem on the horizon is to understand how the parts fit together to segregate the complete genetic complement to daughter cells with the accuracy required to form complex organisms. Historically, science is guided by our observations, thus much of our intuition stems from how we interact with our environment. We make analogies based on our experiences and use these to help us begin to think about processes inside a cell. The problem lies in that life inside a cell is nothing like our life.
There are several physical concepts to consider as we delve into the life of a cell. One is the sense of scale. As we think small (i.e., micrometers), consider that structures do not scale in linear proportion upon miniaturization. As described in a classic lecture by Richard Feynman (There’s plenty of room at the bottom, 1959 in Engineering and Science, Caltech, 1960), if you build a billion structures at 1/4,000 the scale of the original, the sum of the new structure occupies less than 2% of the original volume. This reflects the fact that volume scales to the power of 3 (the volume of a sphere being 4/3πr3). Secondly, molecules stick together. Experimental scientists spend a great deal of time trying to identify binding partners (e.g., yeast two-hybrid, co-immunoprecipitation), to identify functional complexes and deciphering networks. What might be overlooked is that van der Waals forces (0.01–1 kBT) are significant on the force scales inside a cell (see below). For instance, it is important to use the right nut and bolt combination (angle, diameter, pitch) in construction. However, were you to unscrew a nut from a bolt in the cell, you would not have to worry about losing it. The problem will be that you will not be able to get the nut off the bolt. The point is that viscous forces dominate; it is thick and crowded in the cell. Finally, cellular structures are in constant flux. Try to imagine a racecar where the pit crew is following the car around, changing tires, refueling, and hammering bent fenders at 200 mph. This is what it is like in the cell. Our challenge is to design experiments to test mechanisms that move billions of nucleotides on timescales of a few minutes in a very small world.
What is it really like to segregate a chromosome? If there is no weight, what does it mean that the chromosome is long? How does this influence our thinking about the mechanism of mitosis? How much force is required to segregate a chromosome? Why does not a chromosome fragment as it gets dragged across the cell?
In this review, we consider mitosis in a world very foreign to us. We need to know the composition of the materials, the scale of force required, and how these parameters shape questions for future studies. The review is focused on chromosome segregation in budding yeast where we know the exact number of kinetochore microtubule attachments per chromosome (one) and the number of spindle microtubules and can emphasize the quantitative properties of the system. It is likely that the principle derived from this simple system will lead the way for understanding complex kinetochores.
Mechanical properties of the major spindle components: DNA and microtubules
The mitotic spindle is comprised of tubulin protein dimers that polymerize to form the dynamic microtubule polymer. The other major polymer in the spindle, often overlooked as a structural component, are the chromosomes, comprised of an equal mass of DNA and proteins (including histone and nonhistone protein). To understand how the spindle works as a molecular segregation apparatus, we must understand the physical properties of its composite elements. The language of polymer mechanics includes stress, strain, persistence length, and Young’s modulus (see “Appendix”).
DNA has a Young’s modulus on the order of 0.3–1 GPa, similar to hard plastic. Unlike microtubules, DNA is extremely flexible, with a persistence length of 50 nm. As fragments less than the persistence length are on average straight, we consider chromosomal DNA as a freely jointed chain of many short stiff elements (see “Appendix”). For the bacterial chromosome (e.g., Escherichia coli), there are 27,000 segments, 50 nm in length (4.5×106 base pairs (bp)/150 bp one persistence length). These rods adopt a random walk with a radius of gyration defined by R2=nb2 (n = number of segments, b = Kuhn length, 2×persistence length). The radius of gyration for the bacterial chromosome, were it to adopt an ideal random chain in vitro, is 13 μm. This is ten times the length of a typical bacterial cell. Additional proteins must further compact or organize the genome to fit inside a 1–2-μm microbe. From the perspective of polymer dynamics, it is readily apparent that proximity of any two regions is not a function of contour length; rather, it is the number of different states the polymer samples. The random walk is like a bowl of boiled spaghetti. Contacts between regions of different strands are equally likely as contacts within a strand.
The eukaryotic chromosome is more difficult to preserve (or reconstitute) outside of the cell and hence more difficult to obtain accurate measurements of its physical characteristics. The estimates for the Young’s modulus of the chromosome range between 40 and 400 Pa (see “Appendix”; Marshall et al. 2001; Nicklas 1983), orders of magnitude lower than DNA. The persistence length is 150–200 nm for the 30-nm chromatin fiber (compacted ~40-fold relative to B-form DNA, 40×3.4 bp/nm=136 bp/nm). Thus, the eukaryotic chromosome is a very soft, compliant material.
Force generation of microtubules, DNA, and the spindle
Is it simply mechanochemical forces, like adenosine triphosphate (ATP) hydrolysis of motor proteins, that provide the force for chromosome segregation or do the physical properties of the two predominant components, microtubules and chromatin, contribute to the generation of force? Rather than thinking about change in momentum, we should consider that when our polymer or material is compressed or stretched (Δ,L) there is force exerted. The relationship between force (F) and the material property (Young’s modulus, E) F=EA ΔL/L shows us that the Young’s modulus of a material can be used to calculate the force exerted under a specific strain. Interestingly, this formula is essentially Hooke’s law F=(EA/L)ΔL=κx, where the spring constant κ, is the restoring force exerted by a material following deformation. This restoring force and the force that drives most biological processes arise from changes in free energy (ΔG).
For microtubules, the gain in free energy (ΔG) connected with the addition of a single GTP–tubulin dimer to a growing microtubule is of the order of 5–7 kBT. A force of 40 pN can be produced if a microtubule grows by a distance (d)=8 nm for every 13 dimers added (Fmax= NΔG/d; Dickinson et al. 2004). Recent estimates of the depolymerization force are also in the range of 30–65 pN (Grishchuk et al. 2005). These are relatively large forces, indicative of the fact that microtubules are stiff, and relatively incompressible. It is estimated that the maximum force exerted by microtubules on a chromosome is 47 pN (Nicklas 1983).
DNA, in contrast, is not thought of as a force-producing polymer. However, DNA can both store and release entropy and in this way can perform mechanical work. As DNA (and chromosomes) are highly flexible, they will adopt a random walk (defined mathematically by the radius of gyration, see above) thereby maximizing entropy. Upon exertion of force via protein machines (i.e., polymerases) or growing and shortening microtubules in the mitotic spindle, the path of chromosomal DNA is straightened and the number of entropic states that DNA can adopt is reduced. The tendency to return to a state of maximum entropy is known as an entropic spring. The spring constant κ for naked DNA or chromatin is proportional to kBT/persistence length. For a 10-kb DNA strand, in the low to moderate force regime, κ is on the order of femtoNewton per micrometer (see “Appendix”; for reference, a slinky has a spring constant of ~1 μN/μm, six orders of magnitude >DNA). Note that, as persistence length increases, the spring constant decreases. DNA is therefore a long, weak spring. The spring constant is linear over a very limited range of extension. As the molecule approaches its full B-form length, it becomes taut; the spring constant increases exponentially with much more force required to extend DNA to its B-form and beyond. Even in the linear regime, however, this weak spring may contribute to chromosome segregation. The organization of DNA into nucleosomes and the higher order of compaction and organization further complicate force estimates inherent in chromosomes.
The spindle is a molecular machine that does the work of segregating chromosomes. How powerful is this machine and how does it compare to other nanomachines? The performance of a machine over a range of sizes can be compared by power output or volume (Nicklas 1984, 1988) (Table 1). This measure includes force and velocity and, by including volume, reveals how concentrated the force generators are. The bacterial flagellum has a power output per volume of 108 erg s−1/cm3. Muscle is 106 erg s−1/cm3 and a eukaryotic flagellum is 105 erg s−1/cm3. The grasshopper spindle is 6 erg s−1/cm3. The power output of the spindle is five orders of magnitude less than muscle and eight orders of magnitude weaker than the bacteria flagellum. Why is the spindle so weak? What must we consider in understanding the biological role of such a relatively powerless machine? It is apparent that the spindle sacrifices speed for accuracy.
Table 1.
Specific power output (erg s−1/cm3)
| Source | Power output |
|---|---|
| Gas turbine enginea | 8×107 |
| Race car enginea | 2×107 |
| Passenger car enginea | 3×106 |
| Flagelluma | 3×105 |
| Striated musclea | 2×106 |
| Cytokinetic furrowa | 3×102 |
| Spindle motor (grasshopper)/chromosomea | 6 |
| Spindle motor (yeast)/16 chromosomesb | 7 |
Calculation for power output for yeast spindle: force×velocity/volume, F (force=ma=g cm/s−2 )×velocity=g cm2/s−3. This converts to dyne cm/s= erg/s. Force in spindle = 10 pN (Fisher and Bloom, unpublished). Velocity of chromosome to pole movement 0.3 μm/min = 0.0053 μm/s = 5.3 × 10−7 cm/s (Pearson et al. 2001); 10 pN= 10−6 dyne. Thus, 10−6 dyne × 5.3 × 10−7 cm/s = 5.3 × 10−13 dyne cm/s = erg/s. Volume of spindle 250 nm in diameter×1,500 nm in height (O’Toole et al. 1999); (125 nm2 × 3.14 × 1, 500 nm) = 7.4 × 10−14 cm3. Dyne cms−1/cm3 = 5.3 × 10−13 dyne cm/s/7.4 × 10−14 cm3 = 7 erg s/cm3
The classic experiments demonstrating the elastic properties of chromosome were micromanipulation experiments of R.B. Nicklas on grasshopper chromosomes (Nicklas 1963, 1965). The maximum amount of force the spindle can generate was determined by the amount of external force required to stop chromosome movement. This number is 700 pN (Jannink et al. 1996; Nicklas 1983). The 15 microtubules per chromosome tells us that each microtubule can exert 47 pN of force (Jannink et al. 1996; Nicklas 1983). How much force is actually required to segregate a chromosome? To calculate this, one needs to estimate the drag force, fdrag=viscosity×shape×velocity (Stokes’ law). The force for chromosome segregation of a large Melanoplus chromosome is on the order of 0.2–0.5 pN (Nicklas 1965), much smaller than Fmax (Jannink et al. 1996; Nicklas 1983). The large forces that are capable of being generated might be relevant in metaphase, when the major effort is to measure tension between sister chromatids. Stretching the tension sensors for the spindle checkpoint, which includes strain in protein bonds, may require larger forces than that needed for segregation in anaphase. Twenty-five ATP molecules are enough to supply the energy to move a large Melanoplus bivalent chromosome 20 μm (Nicklas 1965). The force to move chromosomes may be smaller than the force to stretch or strain covalent bonds in the protein sensors.
Tensional integrity: the balance between tension and compression
Tension and compression are the key forces involved in building a stable structure. Tension elements resist pulling, while compression elements resist pushing. Mitotic spindles require strong tension elements in order to resist the pulling forces used in orienting the spindle with respect to cortical cues as well as compression to resist collapse from forces directed inward bringing sister kinetochores and/or spindle poles together. As spindles vary in size throughout phylogeny, an important consideration in functional comparisons will be to understand how compression and tension elements scale with size. Structures that experience tensile loads face different scaling rules than that of those facing compressive loads. A taller column is more prone to breakage than a shorter one, all other dimensions being equal, while the force to break a rope is independent of the length of the rope.
Microtubules are stiff, hard polymers and are excellent in resisting compression (Gittes et al. 1993). What is the tension structure in the spindle? Tension is the reaction force applied by the structure when it is pulled. One source of tension element in the spindle is the kinetochore. The kinetochore is the DNA–protein complex that is built at the centromere and is comprised of over 60 proteins, many of which change conformation as a function of attachment and pulling by microtubules, and generate an opposing tension force in their attempt to return to their unstretched state. A second source of tension structure may be chromatin DNA in the pericentric region. DNA is highly compacted in chromatin, is highly extensible, and, as discussed above, is a spring that will return to a state of maximal entropy.
One typically thinks about DNA in the context of mitosis as the site for kinetochore assembly. However, chromatin is very compliant (large ΔL) and likely to be the tension-bearing polymer in the mitotic spindle. As discussed above, naked DNA behaves as an entropic spring. A tension element by definition has a restoring force when a structure is pulled. While individual strands of DNA may be quite weak springs, the spring constant of parallel arrays of springs is the sum of the individual spring constants (parallel Ksum = K1 + K2 + … + Kn; series Ksum = K1 × K2 × … × Kn/K1 + K2 + … + Kn). Let us now consider that, for each microtubule binding site within a kinetochore, there is a DNA strand under tension. For a mammalian kinetochore with approximately 30 microtubule binding sites, our tension spring constant is now the sum of 30 DNA strands. This spring is surely strong enough to be a restoring force. Sister kinetochores “spring back” from a distance of separation of 2 μm under tension to ~1-μm rest length (Waters et al. 1996). This oscillation between sister kinetochores illustrates the intrinsic restorative force at the pericentric region.
DNA as a structural component of the spindle
The contribution of DNA to spindle structure and function has been restricted to the role of centromere DNA in specifying deposition of kinetochore protein binding. Does pericentric chromatin buffer genes on chromosome arms from the pulling forces at the kinetochore or centromere? Does pericentric chromatin provide a mechanical linkage (e.g., shock absorber) that helps prevent chromosome breakage in mitosis? Is pericentric chromatin organized into parallel arrays of springs that contribute to the mechanical properties of chromatin? If we knew the path of DNA at the kinetochore–microtubule interface, we might be able to distinguish between these and other possibilities. Recent studies have indicated that DNA may be mechanically linked to mitotic pulling forces (Baumann et al. 2007). It has been proposed that pericentric chromatin is organized into an intramolecular loop, bringing sister chromatids together in the vicinity of the centromere in the budding yeast Saccharomyces cerevisiae (Bloom et al. 2006).
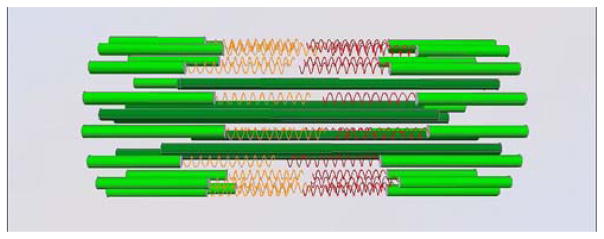
From a structural perspective, the simple budding yeast spindle with its complete genomic sequence including the centromeres is an excellent model from which to calculate the contribution of microtubules and structural DNA to spindle function. There are 16 kinetochore microtubules, on average 0.35 μm in length in the half spindle, and four interpolar microtubules ~1 μm in length from each spindle pole. This yields ~20 μm of microtubule polymer in the spindle. At 8 nm per tubulin dimer, 13 protofilaments per microtubule, 110 kDa per dimer=3.25×106 kDa of tubulin polymerized in the spindle. If we assume that pericentric chromatin adopts a loop configuration as proposed by Bloom et al. (2006), we calculate for 16 chromosomes, 20 kb per sister chromatid×2 sister chromatids=640 kb DNA. This is roughly 4.5% of the genome (6.4×105/1.4× 107=3.2% of the genome) or 640 kb×660 Da/bp=0.4× 106 kDa, assuming an equal weight of histone yields a chromatin mass of 0.8×106 kDa. This structural consideration predicts that pericentric chromatin constitutes roughly one fifth the mass of the segregation apparatus (0.8× 106 kDa/0.8+3.25×106 kDa), a significant fraction relative to spindle microtubules (see Fig. 1).
Fig. 1.
Proposed view of DNA strands as tension elements in the budding yeast mitotic spindle. Dark green—interpolar microtubules, light green—kinetochore microtubules, orange and red—DNA strands representing intramolecular loops of DNA
The concept that the minimal sequences required for chromosome segregation is not simply the centromere as the binding site for kinetochore protein but comprises structural pericentric chromatin may help explain a longstanding paradox in centromere evolution. Centromeres in budding yeast are small (125 bp) compared to other fungi (30–40 kb, Schizosaccharomyces pombe) and mammalian cells (~5 Mb). In contrast, the number of microtubules or chromosome is one in budding yeast, two to three in fission yeast and 25–30 in mammalian cells. Why such a large disparity in centromere DNA content and not in microtubule number? This range of DNA sequences specifying kinetochore formation has led to the classification of point vs. regional centromeres (Pluta et al. 1995). If one considers that the centromere is comprised of the site for kinetochore protein binding as well as pericentric flanking DNA, we see that the ratio of pericentric DNA or microtubule may indeed scale throughout phylogeny (see Table 2). The centromere region of chromosomes may be much more conserved than what an initial inspection reveals.
Table 2.
The centromere DNA–microtubule paradox
| Centromere DNA | Microtubule/chromosome | Centromere DNA/microtubule (kb) | Structural pericentric DNA (kb) | Centromere region/microtubule (kb) |
|---|---|---|---|---|
| Budding yeast 0.125 kb | 1 | 0.125 | 20 | 20 |
| Fission yeast 30–40 kb | 2–3 | 15–20 | 30–40 | 15–20 |
| Mammalian cell 5,000 kb | 25–30 | 165–200 | 1,000 | 33–40 |
The size of the centromere DNA from budding yeast to mammalian cells spans almost six orders of magnitude, from budding yeast (Fitzgerald-Hayes et al. 1982) and fission yeast (Baum et al. 1994) to mammalian cells (Willard 1990). The size of structural pericentric chromatin in budding yeast is based upon the increased concentration of cohesin around centromere (Blat and Kleckner 1999) and the organization of pericentric chromatin as proposed by Bloom et al. (2006). The number of microtubules per kinetochore ranges from one in budding yeast (O’Toole et al. 1999) to two to three in fission yeast (Ding et al. 1993) to 25–30 in newt spermatocytes (Hays and Salmon 1990). The centromere region per microtubule is the amount of structural pericentric chromatin per average microtubule number.
Linking tension and compression elements: the kinetochore
The kinetochore is the multiprotein complex that provides the physical linkage between chromosomes and microtubules. The kinetochore is built at the centromere of eukaryotic chromosomes and links the chromosomes to dynamic microtubule plus-ends. While there is a great deal of information regarding the mechanism of plus-end attachment and the role of subcomplexes within the kinetochore, there is very little known about the structural linkage to DNA beyond the basic centromere-specific nucleosome. A recent study to determine the number of kinetochore proteins helps us understand the molecular architecture of the yeast kinetochore (Joglekar et al. 2006). Microtubule-proximal components might be arranged cylindrically around the microtubule plus-end (Fig. 2), in a fashion that allows access of tubulin to the growing microtubule plus-end. Extending toward the DNA is more speculative; however, the number of protein components decreases. A cylinder that extends from the microtubule to the centromere DNA is the simplest geometry consistent with the protein counts. Interestingly, if the DNA adopts an intramolecular loop, the diameter of two nucleosomal structures will be 23 nm. Considering that the microtubule is 25 nm, one might imagine the kinetochore sleeve as a cylindrical array that connects these two polymers. CBF3 kinks the centromere DNA at the site of Cse4 deposition (Pietrasanta et al. 1999) and may bring the centromere-specific histone H3 variant (Cse4) quite close to the microtubule plus-end (Fig. 2).
Fig. 2.
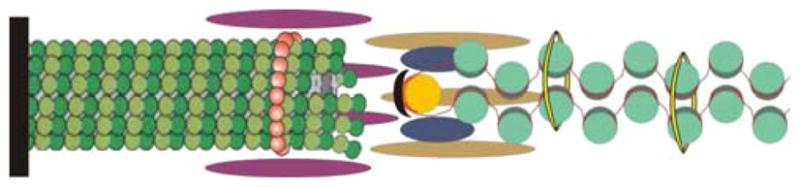
A schematic representation of the interface between kinetochore microtubule, kinetochore, and centromere DNA. The microtubule (green polymer, left) is nucleated from the spindle pole body (black rod). Chromatin DNA is depicted as a red line wrapped around histone proteins (turquoise). At the centromere (125 bp in budding yeast), there is a unique set of histone proteins Cse4 (Meluh et al. 1998) and scm3 (Camahort et al. 2007; Mizuguchi et al. 2007; Stoler et al. 2007) depicted in yellow. If the DNA adopts an intramolecular loop, the dimension of a loop of nucleosomal chromatin DNA is approximately 2 × 11.5 nm = 23 nm. The diameter of the microtubule is 25 nm. The kinetochore is depicted as an array of elongated helices based upon the hydrodynamic properties of the isolated proteins (McAinsh et al. 2003) and determination of molecular protein counts (Joglekar et al. 2006)
The role of entropy and/or elasticity in chromosome segregation
Once kinetochores have moved to the spindle poles, there is still 95% of the genome to be segregated. How does this work? Chromosomes could be very sticky, and upon segregating a piece of the chromosome, i.e., the centromere, the remaining arms are passively distributed. Alternatively, just a small bit of the chromosome is actively segregated, and the remainder of the DNA works its way to the pole in a random walk. When two polymers are brought together in a confined space in close proximity, they lose some of their entropy (Jun and Mulder 2006). Jun and Mulder have proposed that entropy-driven global movement of the bacterial chromosome contributes to the mechanism of chromosome segregation in bacterial cells. The random walk, as defined by the radius of gyration (see above) does not include volume taken up by the chain. The real volume of the chain is only a very small fraction of the volume encompassed by the radius of gyration, equal to (4/3)πRg3. The molecular volume for DNA can be determined from (π)(0.001 μm)2(length). For the E. coli genome (4 × 106 bp = 1, 320 μm), the molecular volume = 0.004 μm3. The volume occupied by Rg (13 μm) = (4/3)πRg3~ 10,000 um3. Only 0.004/10,000 or 0.00004% of the random coil volume is actually occupied. The point is there is ample room for interaction of chains. This interaction takes the form of intramolecular expansion (α, Rfloury=αRg) as well as intermolecular repulsion Rfloury=N3/55. There is a free energy cost for chains overlapping, and when confined this repulsion between two chains can be very high. A reasonable mechanism for segregating two polymers in a confined space can be made from free energy and polymer behavior (Jun and Mulder 2006). Is this why the nuclear envelop does not break down in fungi? Perhaps there is a need to maintain spatial confinement in situations where forces are small and one relies on entropic mechanisms.
Alternatively, chromatin is elastic due to nucleosome release or assembly. Release of a single nucleosome results in a 65-nm extension (from nucleosomal to B-form DNA). Loss of 20 nucleosomes increases sister centromere separation by 650 nm, a value in range of centromere DNA dynamics in live cells (Pearson et al. 2001). Chromatin remodeling complexes found at the centromere (Sharp et al. 2002) could function in nucleosome reassembly upon loss (or decrease) of tension. The force required to release wrapped DNA from the nucleosome is approximately 20 pN (Brower-Toland et al. 2002; Cui and Bustamante 2000), well within the force generated by single microtubule binding. Recent studies in cell extracts indicate that forces as low as 3.5 pN may be able to release nucleosomes (Yan et al. 2006), indicating that much smaller forces may potentiate nucleosome release or reassembly events in vivo. Secondly, the elastic properties of chromatin may result from supercoiling. Chromatin is negatively supercoiled in vivo. It has recently been shown that chromatin becomes overwound when stretched (Gore et al. 2006). The superhelicity of small circular plasmids (1.4–2.3 kb) with wild-type and mutant centromeres has been examined (Bloom et al. 1984). All similar-sized circular molecules had the same number of topoisomers, leading to the conclusion that the centromere was likely to be wound around a histone core. However, the distribution of topoisomers from plasmids with functional centromeres was altered. The presence of centromere may optimize positioning and/or stability of nearby nucleosomes. Consequently, more negative supercoils (nucleosomes) become stabilized in chromatin regions close to a centromere element. Alternatively, the linking number shift reflects a geometric property of the centromere DNA–protein complex or the organization of pericentric chromatin.
Conclusion
Chromosomes are soft, elastic materials in comparison to stiff, rigid microtubules. Based upon the mechanical properties of DNA, we propose that the segregation apparatus is a composite structure of compliant DNA elements and rigid microtubules. A cylindrical kinetochore sleeve encircles both the microtubule plus-end and the apex of an intramolecular loop of pericentric chromatin. As proposed by Zinkowski and Brinkely (Zinkowski et al. 1991), the mammalian kinetochore is based on a repeat subunit structure. The single microtubule binding site in budding yeast may be the conserved repeat subunit, with the cluster of 16 yeast kinetochores comparable to one mammalian kinetochore. The number of microtubules in a typical mammalian kinetochore is 25–30 (twice the number of yeast kinetochores per genome). The power output for kinetochore spindle fibers of a single Melanoplus chromosome was estimated to be 6 erg s−1/cm3 (Nicklas 1984, 1988), comparable to that estimated for kinetochore spindle fibers of the 16 yeast chromosomes, 7 erg s−1/cm3 (Table 1). Sister centromeres are separated by similar distances when under tension (~2 μm, newt-lung cell (Waters et al. 1996), vs. ~0.8 μm, budding yeast (Pearson et al. 2001)) despite extreme disparity in spindle size. Finally, there is evolutionary precedence for the repeat subunit model for kinetochore function. Indian muntjac kinetochores (2n=6) are thought to represent a centromere fusion evolved from the smaller Chinese muntjac progenitor (2n=46; He and Brinkley 1996). This view of the segregation apparatus as a repeat, composite structure of DNA and microtubules should encourage students of mitosis to consider the role of DNA in mitosis beyond the code.
Acknowledgments
I would like to thank Dr. Elaine Yeh, Dr. Jay Fisher, Rachael Bloom, Julian Haase, and Ben Harrison for discussion and critical comments on the manuscript and Julian Haase for artwork.
Appendix
Explanatory box: life at the nanoscale
Life inside a cell is dominated by viscosity and thermal motion. Weight and inertia, properties familiar in our world as the force of gravity is acting upon us at all times, are hardly relevant. The most important expression for the role of viscous forces at the nanoscale comes from Osborne Reynolds (1883). Reynolds number (Fi/Fv) is the expression of inertial force (Fi) to viscous force (Fv). Inertial force, the classical Newtonian physics law of F=ma, is very small at the cellular level as mass is infinitesimal. At low Reynolds number, viscosity trumps inertia. When a bacterial flagellum ceases to turn, the bacterium coasts less than an angstrom (the width of an H-bond; Purcell 1977). Elastic, viscous, and thermal forces dominate at this scale. The force equation for understanding how cellular materials respond to being pulled or pushed is the elastic modulus of the material (E=Young’s modulus) times the area, force= EA. To solve this equation, we must know the physical properties and the size of the materials in question.
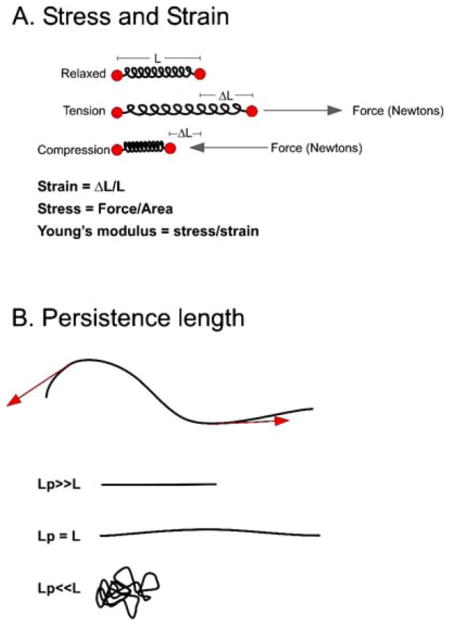
The Young’s modulus is the relation of stress to strain. Stress is the distribution of force per unit area (F/A; inset A, below) and is a measure of how a material reacts to external load. Strain is the geometric expression of deformation (ΔL/L) of the material. Microtubules and DNA have a Young’s modulus of approximately 1–2 GPa, much like hard plastics that have a Young’s modulus of 1–2 GPa and in contrast to rubber that is much lower, 1–10 MPa. However, we know by observing microtubules and DNA that these molecules behave very differently in cells. In general, microtubules appear straight or curved, while DNA is highly coiled and compacted. The parameter that characterizes the fluctuation in shape of a flexible filament is its persistence length. Persistence length describes a filament’s resistance to thermal force and is the distance over which the correlation of the direction of the two ends of a polymer is lost (see inset B below). Fragments shorter than the persistence length of a material are most likely to be linear, while fragments longer than the persistence length are disordered (inset B, below). Microtubules have a persistence length on the order of 5–10 mm (Howard 2001), while DNA has a persistence length of 50 nm. Consider the consequences of microtubule polymers with persistence length in millimeters, while cellular dimensions are on the order of micrometers. One quickly realizes that microtubule-based motors likely influence the mechanical deformation state of the polymers to which they bind. Likewise, histone proteins significantly influence the trajectory of DNA (with a persistence length of 50 nm=~150 bp at 0.33 nm/bp) as DNA of one persistence length wraps one and a half times around a histone octamer. Histones (and other chromosomal proteins) dramatically alter the material properties of the chromosome, as noted in the large decrease in Young’s modulus relative to DNA (see Table 3).
Table 3.
Elastic modulus and persistence length of the major structural elements in the mitotic spindle
| Lp | E | |
|---|---|---|
| DNA | 50 nm | 1 GPa |
| Chromosome | 180 nm | 0.4 MPa |
| Microtubule | 6 mm | 2 GPa |
| Rubber | 1 MPa | |
| Plastic | 2.4 GPa |
Persistence length (Lp) is related to the Young’s modulus (E) in the following way, Lp=EI/kBT, where I = second moment of area, kB = Boltzmann constant, and T = roomtemperature (Kelvin). I is a measure of a material’s resistance to bending, kBT is the relevant energy scale for all molecular interactions inside a cell (at room temperature, kBT=4.1 pN nm). As a material’s flexural rigidity increases (EI), the longer the distance before thermally induced bending (Lp) becomes large.
For a molecule like DNA, where the persistence length is very short relative to the length of an average chromosome (in yeast, the average chromosome is 1 × 107/16 = ~ 0.2 mm; hence, L≫Lp), there are important mechanical consequences. Namely, DNA behaves as an entropic spring. The short rigid domains, being linked via flexible joints, will adopt a state of greatest disorder (entropy), as illustrated in inset B (below). From Hooke’s law, we know that F=κx, κ = spring constant (Newton/meter), x = change in distance. For small forces, F=3kBTx/n(2Lp)2. The spring constant of this freely jointed chain is equal to 3kBT/n(2Lp)2 (n = number of segments). For DNA length 10 kb, the spring constant = 0.036 fN/nm, small indeed. As force is applied to DNA, there is a corresponding decrease in the number of states of disorder, hence a decrease in entropy. In the absence of force, the chain will return to a state of highest disorder. This freely jointed chain has a number of mechanical properties in common with rubber-like materials, namely, that as temperature increases, the spring constant increases and the chains tend to shorten. A demonstration of an entropic spring can be found at the Department of Materials Science and Metallurgy (http://www.doitpoms.ac.uk/tlplib/stiffness-of-rubber/index.php).
Explanatory inset 1: polymer physics
References
- Baum M, Ngan VK, Clarke L. The centromeric K-type repeat and the central core are together sufficient to establish a functional Schizosaccharomyces pombe centromere. Mol Biol Cell. 1994;5:747–761. doi: 10.1091/mbc.5.7.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann C, Korner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 2007;128:101–114. doi: 10.1016/j.cell.2006.11.041. [DOI] [PubMed] [Google Scholar]
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Bloom K, Amaya E, Yeh E. Centromeric DNA structure in yeast. In: Borsey GG, Cleveland DW, Murphy DB, editors. Molecular biology of the cytoskeleton. Cold Spring Harbor Laboratory; New York: 1984. pp. 175–184. [Google Scholar]
- Bloom K, Sharma S, Dokholyan NV. The path of DNA in the kinetochore. Curr Biol. 2006;16:R276–R278. doi: 10.1016/j.cub.2006.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc Natl Acad Sci USA. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Cui Y, Bustamante C. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc Natl Acad Sci USA. 2000;97:127–132. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RB, Caro L, Purich DL. Force generation by cytoskeletal filament end-tracking proteins. Biophys J. 2004;87:2838–2854. doi: 10.1529/biophysj.104.045211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol. 1993;120:141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore J, Bryant Z, Nollmann M, Le MU, Cozzarelli NR, Bustamante C. DNA overwinds when stretched. Nature. 2006;442:836–839. doi: 10.1038/nature04974. [DOI] [PubMed] [Google Scholar]
- Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR. Force production by disassembling microtubules. Nature. 2005;438:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- Hays TS, Salmon ED. Poleward force at the kinetochore in metaphase depends on the number of kinetochore microtubules. J Cell Biol. 1990;110:391–404. doi: 10.1083/jcb.110.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D, Brinkley BR. Structure and dynamic organization of centromeres/prekinetochores in the nucleus of mammalian cells. J Cell Sci. 1996;109(Pt 11):2693–2704. doi: 10.1242/jcs.109.11.2693. [DOI] [PubMed] [Google Scholar]
- Howard J. Mechanics of motor proteins and the cytoskeleton. Sinauer; Sunderland: 2001. [Google Scholar]
- Jannink G, Duplantier B, Sikorav JL. Forces on chromosomal DNA during anaphase. Biophys J. 1996;71:451–465. doi: 10.1016/S0006-3495(96)79247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S, Mulder B. Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc Natl Acad Sci USA. 2006;103:12388–12393. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Marko JF, Agard DA, Sedat JW. Chromosome elasticity and mitotic polar ejection force measured in living Drosophila embryos by four-dimensional microscopy-based motion analysis. Curr Biol. 2001;11:569–578. doi: 10.1016/s0960-9822(01)00180-4. [DOI] [PubMed] [Google Scholar]
- McAinsh AD, Tytell JD, Sorger PK. Structure, function, and regulation of budding yeast kinetochores. Annu Rev Cell Dev Biol. 2003;19:519–539. doi: 10.1146/annurev.cellbio.19.111301.155607. [DOI] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Nicklas RB. A quantitative study of chromosomal elasticity and its influence on chromosome movement. Chromosoma. 1963;14:276–295. doi: 10.1007/BF00326816. [DOI] [PubMed] [Google Scholar]
- Nicklas RB. Chromosome velocity during mitosis as a function of chromosome size and position. J Cell Biol. 1965;25(Suppl):119–135. doi: 10.1083/jcb.25.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. Measurements of the force produced by the mitotic spindle in anaphase. J Cell Biol. 1983;97:542–548. doi: 10.1083/jcb.97.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. A quantitative comparison of cellular motile systems. Cell Motil. 1984;4:1–5. doi: 10.1002/cm.970040102. [DOI] [PubMed] [Google Scholar]
- Nicklas RB. The forces that move chromosomes in mitosis. Annu Rev Biophys Biophys Chem. 1988;17:431–449. doi: 10.1146/annurev.bb.17.060188.002243. [DOI] [PubMed] [Google Scholar]
- O’Toole ET, Winey M, McIntosh JR. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:2017–2031. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrasanta LI, Thrower D, Hsieh W, Rao S, Stemmann O, Lechner J, Carbon J, Hansma H. Probing the Saccharomyces cerevisiae centromeric DNA (CEN DNA)-binding factor 3 (CBF3) kinetochore complex by using atomic force microscopy. Proc Natl Acad Sci USA. 1999;96:3757–3762. doi: 10.1073/pnas.96.7.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG, Earnshaw WC. The centromere: hub of chromosomal activities. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- Purcell EM. Life at low Reynolds number. Am J Phys. 1977;45:3–11. [Google Scholar]
- Reynolds O. An experimental investigation of the circumstances which determine whether the motion of water shall be direct or sinuous, and the law of resistance in parallel channels. Philos Trans R Soc Lond. 1883;174:935–982. [Google Scholar]
- Sharp JA, Franco AA, Osley MA, Kaufman PD. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 2002;16:85–100. doi: 10.1101/gad.925302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci USA. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Skibbens RV, Salmon ED. Oscillating mitotic newt lung cell kinetochores are, on average, under tension and rarely push. J Cell Sci. 1996;109(Pt 12):2823–2831. doi: 10.1242/jcs.109.12.2823. [DOI] [PubMed] [Google Scholar]
- Willard HF. Centromeres of mammalian chromosomes. Trends Genet. 1990;6:410–416. doi: 10.1016/0168-9525(90)90302-m. [DOI] [PubMed] [Google Scholar]
- Yan J, Maresca TJ, Skoko D, Adams CD, Xiao B, Christensen MO, Heald R, Marko JF. Micromanipulation studies of chromatin fibers in Xenopus egg extracts reveal ATP-dependent chromatin assembly dynamics. Mol Biol Cell. 2006;18:464–474. doi: 10.1091/mbc.E06-09-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkowski RP, Meyne J, Brinkley BR. The centromere–kinetochore complex: a repeat subunit model. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]