Summary
Previous work on the DNA damage checkpoint in Saccharomyces cerevisiae has shown that two complexes independently sense DNA lesions: the kinase Mec1-Ddc2 and the PCNA-like 9-1-1 complex. To test whether colocalization of these components is sufficient for checkpoint activation, we fused these checkpoint proteins to the LacI repressor and artificially colocalized these fusions by expressing them in cells harboring Lac operator arrays. We observed Rad53 and Rad9 phosphorylation, Sml1 degradation, and metaphase delay, demonstrating that colocalization of these sensors is sufficient to activate the checkpoint in the absence of DNA damage. Our tethering system allowed us to establish that CDK functions in the checkpoint pathway downstream of damage processing and checkpoint protein recruitment. This CDK dependence is likely, at least in part, through Rad9, since mutation of CDK consensus sites compromised its checkpoint function.
Author Keywords: DNA, SIGNALING, CELLCYCLE
Introduction
Unrepaired DNA damage can lead to the inaccurate propagation of an organism's genome. When eukaryotic cells detect DNA damage, they activate a signal transduction pathway, called a checkpoint, to delay cell division and promote DNA repair. In response to double-strand breaks (DSBs), the DNA damage checkpoint in Saccharomyces cerevisiae arrests cells at the G2/M phase (Weinert and Hartwell, 1988). This response requires the function of at least four classes of checkpoint proteins: a clamp complex, sensor kinases, adaptor proteins, and effector kinases.
DSBs are processed by exonucleases that resect the 5′ strand, leaving a 3′ single-stranded DNA (ssDNA) overhang. This structure is thought to be a signal for the recruitment of a damage-specific DNA clamp, referred to as the 9-1-1 complex, that resembles the processivity factor for DNA replication, PCNA (Thelen et al., 1999). The 9-1-1 complex is a heterotrimer composed of three subunits, Ddc1, Mec3, and Rad17 (hRad9, hHus1, hRad1). Loading of the PCNA clamp at 3′ ssDNA/dsDNA junctions during replication is accomplished by the heteropentameric replication factor C (RFC) complex (Tsurimoto and Stillman, 1991). In contrast, the 9-1-1 complex is thought to be loaded at 5′ ssDNA/dsDNA junctions generated at damage sites by a modified form of RFC, in which one subunit, Rfc1, is replaced by a checkpoint-specific subunit called Rad24 ([Bermudez et al., 2003], [Ellison and Stillman, 2003], [Green et al., 2000], [Kondo et al., 2001], [Majka and Burgers, 2003], [Melo et al., 2001] and [Zou et al., 2003]).
Mec1 and Tel1 (ATR and ATM in mammals, respectively) are thought of as sensor kinases since they directly recognize DNA damage. These two kinases appear to function somewhat redundantly, Mec1 being the primary checkpoint signaling molecule in yeast. Mec1 associates with damaged chromatin through its partner, Ddc2, which binds RPA-coated ssDNA ([Paciotti et al., 2000], [Rouse and Jackson, 2002] and [Zou and Elledge, 2003]). One of the functions of the Mec1 kinase is to activate the effector kinase, Rad53 (hCHK2). Rad53 activation is mediated by either of two adaptor proteins, Rad9 or Mrc1. Mrc1 is thought to function as an adaptor during DNA replication, whereas Rad9 can also recognize damage that occurs outside of S phase. Rad9 is phosphorylated in a damage-dependent manner by Mec1 ([Aboussekhra et al., 1996], [Emili, 1998] and [Schwartz et al., 2002]). This promotes its association with Rad53, leading to Rad53 activation by Mec1 and subsequent autophosphorylation of Rad53 ([Schwartz et al., 2002] and [Sweeney et al., 2005]).
The exact mechanism by which Rad9, and its orthologs S. pombe Crb2 and mammalian 53BP1, is recruited to break sites remains unclear. Efficient Rad9 and Crb2 recruitment requires at least two histone modifications ([Huyen et al., 2004], [Nakamura et al., 2004], [Sanders et al., 2004], [Toh et al., 2006] and [Vidanes et al., 2005]). H2A is phosphorylated by Mec1 or Tel1 at its C terminus in response to DNA damage (Downs et al., 2000). This phosphorylation promotes an interaction between Rad9 and H2A. Similarly, vertebrate 53BP1 associates with the related histone H2A variant H2AX after phosphorylation by ATM or ATR ([Celeste et al., 2003] and [Ward et al., 2003]). In addition, a constitutive methylation on lysine 79 of H3 (H3K79) by the Dot1 methyltransferase mediates interactions with the Tudor domains in Rad9 (Huyen et al., 2004). Analogously, methylation of lysine 20 on H4 (H4K20) is required for the maintenance of Crb2 and 53BP1 (Botuyan et al., 2006). In summary, adaptor proteins from all three species use related mechanisms, H2A phosphorylation and methylations on the histone core, for their enrichment at chromatin adjacent to damage sites.
The observation that the Ddc2-Mec1 and the 9-1-1 complexes localize independently to sites of damage ([Kondo et al., 2001], [Melo et al., 2001] and [Zou et al., 2002]) suggested a model in which the DNA damage site serves as a platform to concentrate these molecules. To test this hypothesis, we artificially colocalized the Ddc2-Mec1 and 9-1-1 complexes by fusing one member of each complex to LacI and expressing these fusions in a strain with multimerized LacI-binding sites (LacO arrays). Using this system, we show that neither ssDNA nor the 5′ ssDNA/dsDNA junctions are directly required for the activation or function of checkpoint proteins, since colocalization of Ddc2-Mec1 kinase and the 9-1-1 complex is sufficient to activate Rad53 and delay cell-cycle progression. By altering the exact number of LacO sites, we were able to establish a correlation between the number of checkpoint molecules colocalized and the degree of Rad53 phosphorylation. Furthermore, we show that the Ddc1 subunit of 9-1-1 is sufficient for Rad53 activation. This activation functions in the context of chromatin, requiring H2A phosphorylation and H3K79 methylation for maximum signaling. Last, we show that CDK activity contributes to checkpoint activation through a mechanism independent from its established role in damage processing. Mutating the CDK consensus sites on Rad9 eliminates its cell-cycle-regulated electrophoretic shift and generates a checkpoint-deficient allele.
Results
Colocalization of Checkpoint Proteins Activates the Rad53 Kinase
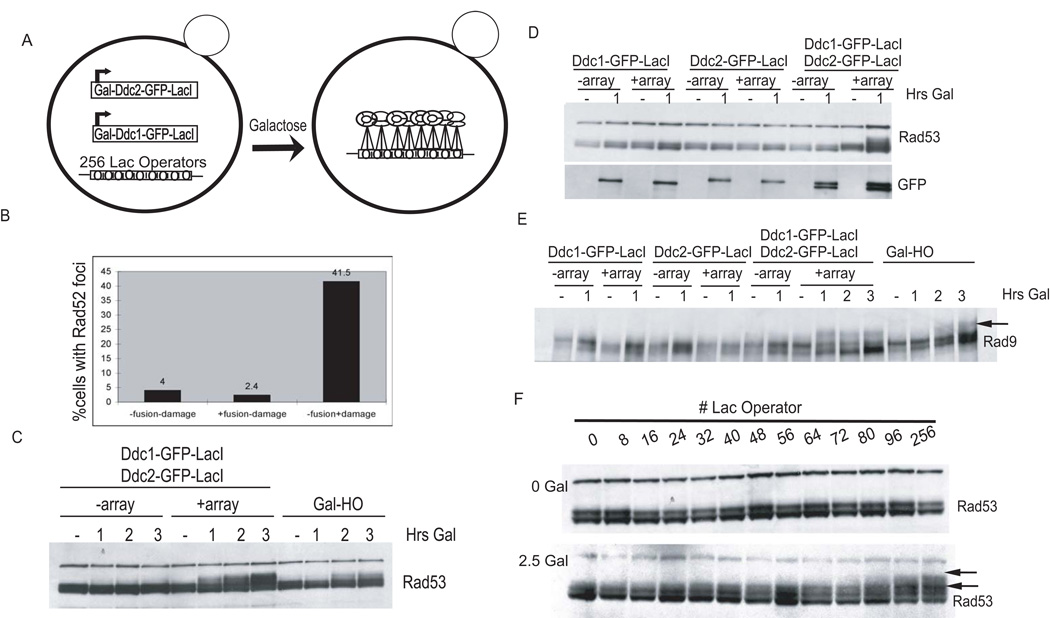
We set out to test the requirement for a DNA break in the initial activation step of the DNA damage checkpoint. Recruitment of Ddc2-Mec1 and the 9-1-1 complex to a DSB site is essential for activation of the DNA damage checkpoint. The association of Ddc2-Mec1 with ssDNA or RPA could stimulate conformational changes required for direct activation of kinase activity. Alternatively, if ssDNA serves strictly as a scaffold to concentrate Ddc2-Mec1 and the 9-1-1 complex, the requirement for a DSB could be bypassed by artificially colocalizing the two complexes. The prokaryotic repressor protein LacI binds with high affinity to the Lac operator sequence. GFP-LacI fusions have been used to recruit GFP to arrays of LacO repeats in order to visualize chromosome dynamics (Straight et al., 1996). We co-opted this approach to recruit Ddc2-Mec1 and the 9-1-1 complex to a region on chromosome IV containing 256 tandem copies of LacO, encompassing 10.5 Kb, which we will refer to as a LacO array. GFP-LacI fusions of DDC1 and DDC2 were placed under a galactose-inducible promoter and introduced into the LacO array strain (Figure 1A and see Figure S1 available online). Upon addition of galactose, each fusion was expressed at equivalent levels, as determined by western blot (Figure 1D, lower panel), and could complement deletions of DDC1 and DDC2, respectively (data not shown). To avoid the possibility that these fusions could disrupt DNA replication, we performed all experiments with cells that were first arrested in G2/M with nocodazole. We also confirmed that colocalization of checkpoint fusions did not create de novo DNA breaks by monitoring the formation of Rad52-RFP foci. Rad52 is required for homologous recombination and has been shown to localize to DSB (Lisby et al., 2003). DNA damage created by treatment with 100 ug/mL Zeocin for 3 hr resulted in Rad52 focus formation in 42% of cells. In contrast, only 4% of cells showed spontaneous Rad52 foci in the untreated sample, and this was not further increased by the expression of the checkpoint fusions (Figure 1B). In fact, only 1 out of the 167 fusion-expressing cells examined formed a spontaneous Rad52 focus that colocalized with the checkpoint protein fusions at the LacO array. This suggests that colocalization of Ddc1-GFP-LacI and Ddc2-GFP-LacI at the array does not induce DNA damage.
Figure 1. Colocalization of Checkpoint Fusions Promotes Rad53 and Rad9 Phosphorylation.
(A) Inducible checkpoint fusions' experimental design.
(B) Colocalization of Ddc1 and Ddc2 at a LacO array does not cause Rad52 foci formation. Cells containing Ddc1-GFP-LacI, Ddc2-GFP-LacI, LacO array, and Rad52-RFP were arrested with nocodazole and treated as follows: 100 ug/mL Zeocin for 3 hr in rich media plus dextrose, rich media plus dextrose, and rich media plus a 1 hr galactose pulse. Digital microscopy was used to measure the frequency of Rad52-RFP foci formation in all samples.
(C–E) Strains were arrested with nocodazole and maintained arrested while galactose was added to induce fusions or the HO endonuclease as a positive control. Rad53-HA (C and D) and Rad9-HA were visualized with anti-HA antibody (E). Checkpoint fusions were visualized with anti-GFP antibodies.
(F) Arrays of tandem LacO repeats were introduced in the strain coexpressing the checkpoint fusions. Strains were nocodazole arrested (top panel), and galactose was added to induce fusion expression for 2.5 hr (bottom panel).
Expression of both Ddc1-GFP-LacI and Ddc2-GFP-LacI in the presence of a LacO-array induced checkpoint activation (Figure 1C). In contrast, no Rad53 phosphorylation was seen induced by the localization of either single complex or by expression of both complexes in the absence of LacO arrays (Figure 1D). The Rad53 that we observed as shifted was activated, as judged by in situ kinase assay (data not shown), and was comparable to that induced by a single DSB (Figure 1C), suggesting that this system mimics the physiological levels of Rad53 activation observed upon DNA damage. Moreover, the Rad53 activation observed was independent of Mre11 (data not shown) arguing against ssDNA formation by Mre11's exonuclease activity (Nakada et al., 2004) leading to Rad53 phosphorylation during colocalization.
Mec1 phosphorylation of the adaptor protein Rad9 is required for its association with, and activation of, Rad53. Rad9 exhibits a damage-independent electrophoretic shift in G2/M that is supershifted upon DNA damage by Mec1/Tel1 phosphorylation ([Emili, 1998], [Sun et al., 1998] and [Vialard et al., 1998]). Rad9 was supershifted in a LacO array-dependent manner when both Ddc2-Mec1 and 9-1-1 were colocalized (Figure 1E). The observation that the localization of neither Ddc2-Mec1 nor 9-1-1 alone was sufficient to promote Rad9 or Rad53 phosphorylation further demonstrates that we have recapitulated the physiological DNA damage response.
In order to estimate the number of checkpoint complexes required for signaling, we tested the minimum number of LacO repeats required for Rad53 activation. To this end, we integrated LacO arrays of different sizes, ranging from 8 to 256 repeats in length, into a strain coexpressing Ddc1-GFP-LacI and Ddc2-GFP-LacI. As shown in Figure 1F, Rad53 phosphorylation is observed in strains with as few as 40 LacO sites. The shifted form of Rad53 continued to increase with an increasing number of LacO sites, whether they were continuous (Figure 1F) or integrated in small groups separated by 3 Kb (C.Y.B. and D.P.T., unpublished data). This suggests that the total number of corecruited molecules determined the extent of Rad53 phosphorylation.
Rad53 Properly Targets Downstream Substrates Following Artificial Sensor Localization
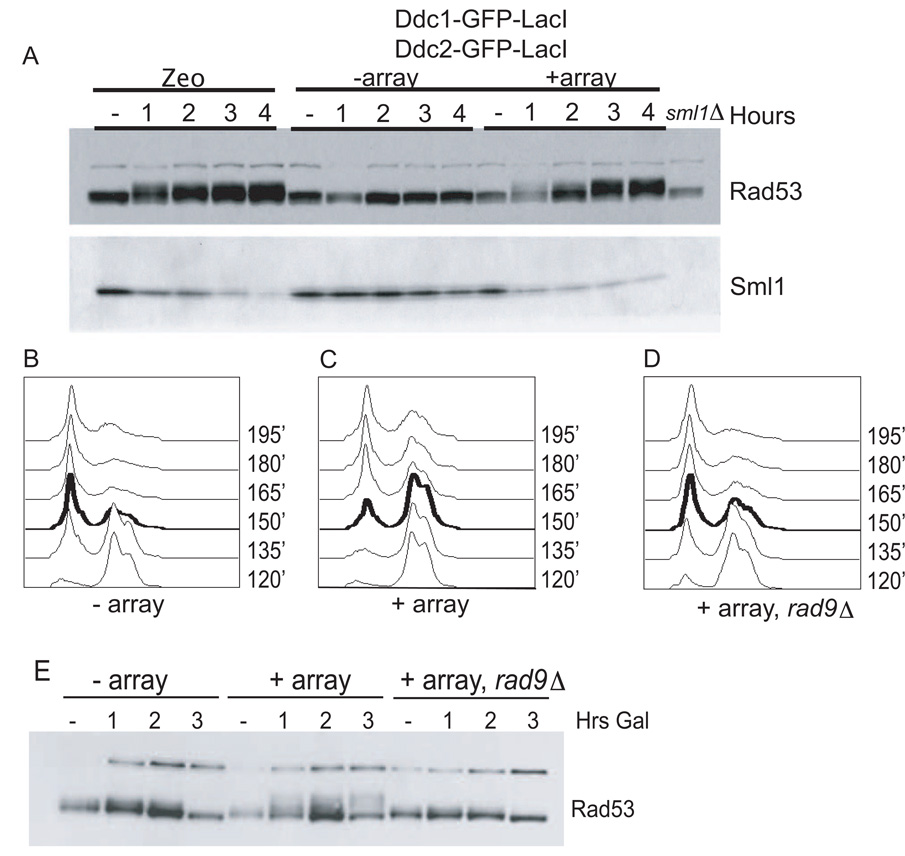
Having shown that Rad9 and Rad53 are phosphorylated after Ddc1 and Ddc2 colocalization, we wanted to test whether Rad53 kinase was active and led to downstream signaling. One direct target of Rad53 is the kinase Dun1, which is activated by Rad53 phosphorylation (Zhou and Elledge, 1993). Dun1 phosphorylates the ribonucleotide reductase inhibitor Sml1, inducing its degradation (Zhao and Rothstein, 2002). We were unable to observe Dun1 phosphorylation directly in our system or after a single DSB. Therefore, we used Sml1 protein levels as a readout of Dun1 activation. Induction of multiple DSBs with the bleomycin derivative Zeocin promoted Sml1 degradation (Figure 2A, lanes 2–5). Similarly, when both checkpoint fusions were induced in the presence of the LacO array, Sml1 levels consistently decreased (Figure 2A, lanes 11–15). Sml1 protein level decreased in an array-dependent manner on induction of the fusions, suggesting that the Rad53 phosphorylation seen in Figure 1C represents in vivo activation of the Rad53 kinase.
Figure 2. Colocalization Activates Multiple Checkpoint Readouts.
(A) Sml1 is degraded upon colocalization. A wild-type strain was left untreated or was treated 100 ug/ml Zeocin for 4 hr. Strains expressing checkpoint fusions −array and +array were arrested with nocodazole, and galactose was added to induce fusions. The westerns were blotted against endogenous Sml1 and Rad53-HA. A sml1 delete strain served as control for specificity (last lane).
(B–D) Strains containing fusions −array (B), +array (C), and +array, rad9 (D) were arrested in nocodazole, induced with galactose for 1 hr while arrested, and then released into media with α factor. A time course of FACS analysis is shown starting at 2 hr (120′) into galactose induction.
(E) Analysis of Rad53-HA by western blot of samples taken from (B)–(D).
The DNA damage checkpoint acts primarily at G2/M to arrest the cell cycle prior to chromosome segregation. We asked whether colocalization was sufficient to signal arrest at this cell-cycle stage upon release from nocodazole into α factor, which subsequently arrests cells in G1. Cell-cycle progression was monitored by flow cytometry (FACS). Cells without LacO arrays began to enter G1 90 min after release from a nocodazole arrest (150′ after galactose induction) (Figure 2B, Figure S2). The strain coexpressing Ddc2 and Ddc1 fusions in the presence of a LacO array maintained a 2N peak after nocodazole release for the duration of the experiment (most obvious at 150′), indicating maintenance of the G2/M arrest (Figure 2C). To ensure that this represented a checkpoint-mediated arrest, we examined an isogenic strain deleted for RAD9 and found that the cell-cycle delay was relieved (Figure 2D). Rad53 phosphorylation levels correlated with the observed delay in G2/M (Figure 2E).
The Rad24 Requirement Is Bypassed by Colocalization of Ddc2-Mec1 and 9-1-1
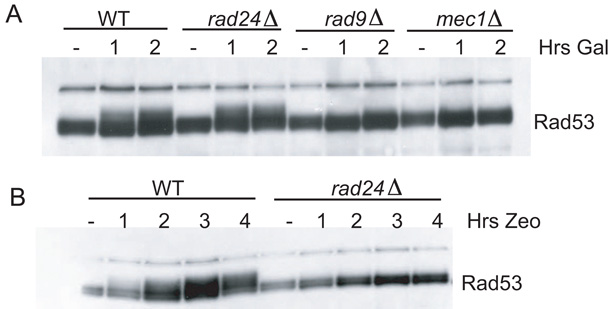
To further characterize the genetic requirements for checkpoint activation in our system, we examined which other checkpoint genes were required for Rad53 activation after Ddc2-Mec1 and 9-1-1 colocalization. We deleted MEC1, RAD9, and RAD24 in strains coexpressing both checkpoint fusions and carrying LacO arrays. Unlike wild-type strains, strains deleted for RAD9 and MEC1 did not display Rad53 phosphorylation upon coexpression, suggesting that both Rad9 and Mec1 are still needed to transduce the signal to Rad53 (Figure 3A). Importantly, this demonstrates that the Ddc2-GFP-LacI fusion was activating Rad53 through its association with Mec1.
Figure 3. 9-1-1 and Ddc2-Mec1 Colocalization Bypasses the Requirement for Rad24.
(A) Both checkpoint fusions were induced in nocodazole-arrested isogenic wild-type, rad24, rad9, and mec1 strains containing a LacO array.
(B) Checkpoint fusions were induced in nocodazole-arrested wild-type and rad24 strains. Galactose and Zeocin were added simultaneously in the presence of nocodazole.
Rad24-RFC has been shown to function as the clamp loader for the 9-1-1 complex. We observed that Rad24 was dispensable for Rad53 phosphorylation in the colocalization strain, suggesting that the role of Rad24 is restricted to localizing 9-1-1 to damage (Figure 3A). These data also suggest that the 9-1-1-LacI-clamp did not need to be loaded (i.e., it does not need to encircle DNA) at the array. Rad24 was still required for checkpoint activation when a Ddc1-GFP-LacI, Ddc2-GFP-LacI strain lacking a LacO array was treated with Zeocin (Figure 3B). This result verifies that the Ddc1 fusion is capable of interacting with Rad24 and can be loaded onto DNA damage sites. These data also confirm that expression of LacI fusions does not activate Rad53 by producing damage, since damage-induced activation requires Rad24 function.
Ddc1 Can Act Independently of the 9-1-1 Complex
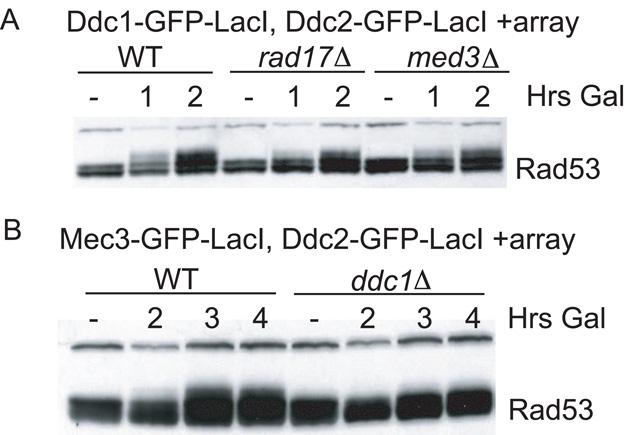
The Ddc1 subunit of the 9-1-1 complex was sufficient to activate Rad53 when colocalized with Ddc2-Mec1. We tested whether the other 9-1-1 subunits, Mec3 and Rad17, were required for checkpoint activation. Deletion of MEC3 and RAD17 in the strain coexpressing Ddc1-GFP-LacI and Ddc2-GFP-LacI containing a LacO array did not abolish Rad53 phosphorylation (Figure 4A). The Rad53 phosphorylation observed corresponded to a metaphase delay in the rad17Δ and mec3Δ strains as assessed by FACS (Figures S2A–S2D). Deletion of MEC3 and RAD17 did not prevent cells from delaying with a 2N peak like wild-type strains by 3 hr of galactose induction. This result suggested that Ddc1 is capable of activating Mec1 without the other two 9-1-1 subunits. If Ddc1 is the subunit that activates Mec1, it should be indispensable when the 9-1-1 complex is recruited through either of the other subunits, Mec3 or Rad17. To test this hypothesis, we fused Mec3 to GFP-LacI and recruited it to the array along with Ddc2-Mec1. Unlike the Ddc1-GFP-LacI fusion, the Mec3-GFP-LacI fusion was hypomorphic for DNA damage-induced checkpoint activation (Figure S2E). Still, the colocalization of Mec3-GFP-LacI and Ddc2-GFP-LacI at the array also resulted in Rad53 activation, although not to the degree seen for Ddc1-GFP-LacI. Mec3-GFP-LacI required the Ddc1 subunit to activate Mec1, as seen by the lack of Rad53 phosphorylation in the ddc1Δ strain, consistent with the hypothesis that Ddc1 mediates checkpoint activation (Figure 4B).
Figure 4. Ddc1 Is the Mec1-Activating Subunit.
(A) Ddc1-GFP-LacI and Ddc2-GFP-LacI checkpoint fusions were induced in nocodazole-arrested isogenic wild-type, mec3, and rad17 strains containing a LacO array.
(B) Mec3-GFP-LacI and Ddc2-GFP-LacI checkpoint fusions were induced in nocodazole-arrested isogenic wild-type and ddc1 strains containing a LacO array.
Chromatin Is Required for Checkpoint Activation
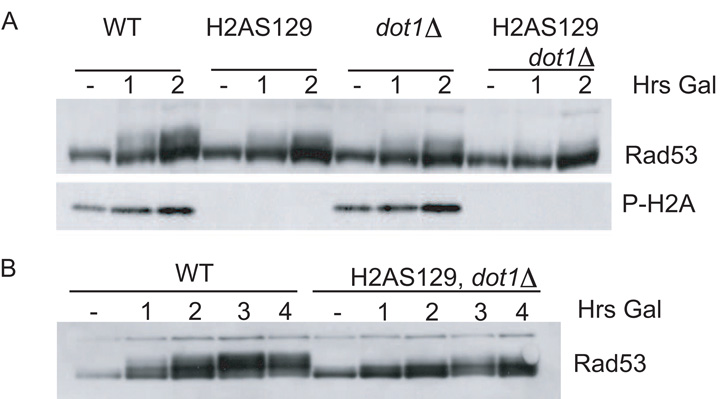
In recent years, histone modifications have been implicated in the DNA damage checkpoint. Phosphorylation of the H2A variant H2AX by ATM and ATR on S139 is a hallmark of DNA damage. While yeast does not possess the H2AX variant, the C-terminal tail of yeast H2A is phosphorylated by Mec1 and Tel1 on the analogous residue. This phosphorylation is thought to help recruit Rad9 to break sites by the physical interaction between Rad9 and phosphorylated H2A (Hammet et al., 2007). Methylation of histone H3 on lysine 79 (H3K79) by the Dot1 methyltransferase is also required for full Rad9 phosphorylation and localization ([Giannattasio et al., 2005] and [Toh et al., 2006]). Since our colocalization system brings Ddc2-Mec1 and 9-1-1 to DNA, we examined the role of neighboring chromatin in this checkpoint activation. Mutation of the phosphorylation sites on H2A (Figure 5A, bottom panel) or deletion of DOT1 resulted in a slight decrease in Rad53 phosphorylation upon colocalization. When both H2AS129 phosphorylation and H3K79 methylation were abolished, Rad53 phosphorylation decreased markedly (Figure 5A). At later time points (4 hr), the double mutant strain displayed some Rad53 phosphorylation, but not to the extent seen in the wild-type strain (Figure 5B). Thus, while we have bypassed the requirement for DNA damage in checkpoint activation, chromatin is still required for efficient signaling.
Figure 5. Rad53 Activation Requires H2AX-P and H3K79Me.
(A) Ddc1-GFP-LacI and Ddc2-GFP-LacI checkpoint fusions were induced in nocodazole-arrested isogenic wild-type, htaS129; dot1, and htaS129, dot1 strains containing a LacO array.
(B) Same as in (A) but for a longer time.
CDK Activity Promotes Rad53 Activation
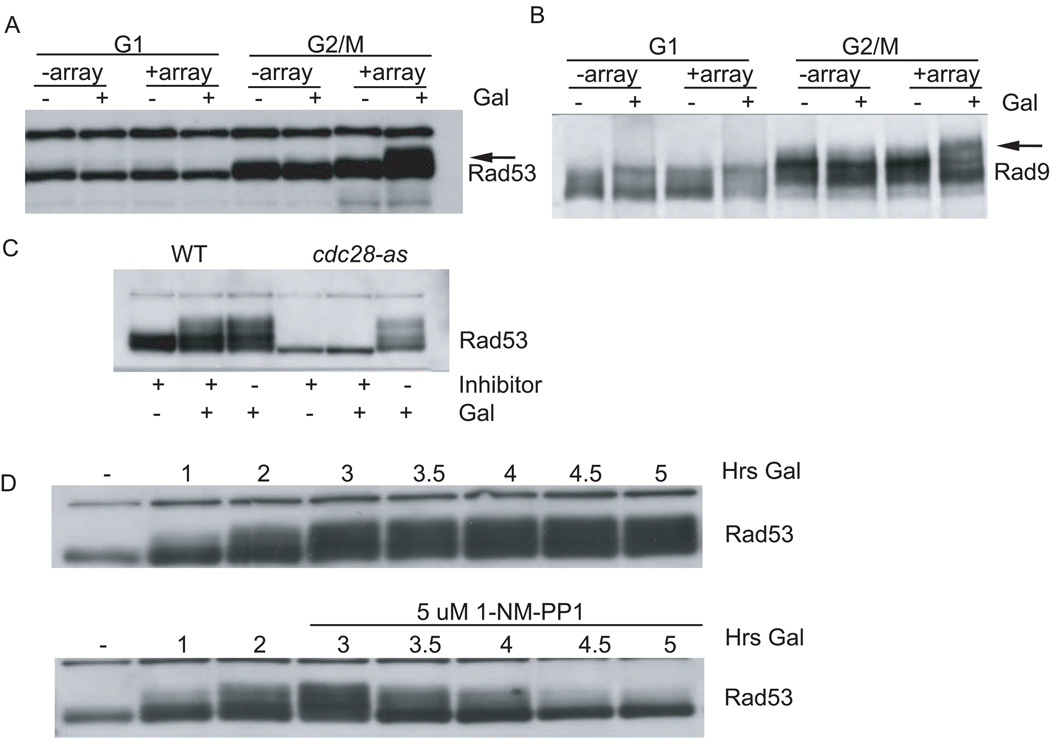
Several studies have suggested that checkpoint activation is less efficient in G1. In part, this effect can be attributed to a reported reduction in damage processing ([Clerici et al., 2004], [Ira et al., 2004] and [Pellicioli et al., 2001]). Efficient 5′-3′ resection is thought to require the cyclin-dependent kinase (CDK), Cdc28. Thus ssDNA, which is thought to be the intermediate recognized by the checkpoint machinery, is less abundant in G1 when CDK is inactive. We used our recruitment system to ask whether a reduction in damage processing, leading to reduced Ddc2-Mec1 and 9-1-1 recruitment, was the sole reason for low checkpoint activity in G1. Strains were arrested in G1 with α factor or in G2/M with nocodazole, and then both checkpoint fusions were induced with galactose. As expected from our previous results, colocalizing Ddc2-Mec1 and 9-1-1 resulted in Rad53 phosphorylation and Rad9 hyperphosphorylation in G2/M (Figures 6A and 6B). When cells were arrested in G1, induction of checkpoint fusions did not result in Rad53 or Rad9 phosphorylation (Figures 6A and 6B), suggesting that CDK activity is required for efficient checkpoint activation independent of its described role in damage processing.
Figure 6. CDK Promotes Rad53 Activation.
(A and B) Strains were arrested in G1 with α factor and G2/M with nocodazole, and galactose was added to induce checkpoint fusions.
(C) Checkpoint fusions were induced in nocodazole-arrested isogenic wild-type and analog (1NM-PP1)-sensitive, cdc28-as, strains containing a LacO array. Both strains were treated as follows: inhibitor alone (5 uM), galactose plus inhibitor, and galactose alone.
(D) The strain carrying the cdc28-as allele, checkpoint fusions, and array was arrested with nocodazole and induced with galactose. After 3 hr, the culture was split and inhibitor was added.
In order to address directly whether our inability to activate the checkpoint in G1 was due to low CDK activity, we used an analog-sensitive allele of CDC28 (cdc28-as) that renders the kinase inactive upon addition of the inhibitor 1NM-PP1 (Ubersax et al., 2003). Isogenic CDC28 and cdc28-as strains were arrested in G2/M with nocodazole. 1NM-PP1 was added at the initiation of galactose induction to inhibit Cdc28-as, and Rad53 phosphorylation was measured. When cells carrying the cdc28-as allele were treated with inhibitor, they failed to activate Rad53 (Figure 6C), suggesting that CDK activity was required even when both sensors were colocalized (Figure 6C). These data support the idea that CDK activity controls checkpoint activation downstream of damage processing.
CDK activity could be required for either the initiation or the maintenance of the checkpoint. We tested the requirement for CDK activity in checkpoint maintenance by inhibiting CDK after the checkpoint signal had been established. Checkpoint fusions were induced with galactose after a nocodazole arrest in the cdc28-as strain. After 3 hr of galactose induction (a time when the checkpoint signal is robust), the inhibitor 1NM-PP1 was added to half the culture (Figure 6D, bottom). Checkpoint inhibition was visible within 30 min, and by 2 hr the checkpoint signal was almost eliminated. Therefore, CDK activity is required to maintain Rad53 activation.
Rad9 CDK Consensus Sites Are Important for Checkpoint Function
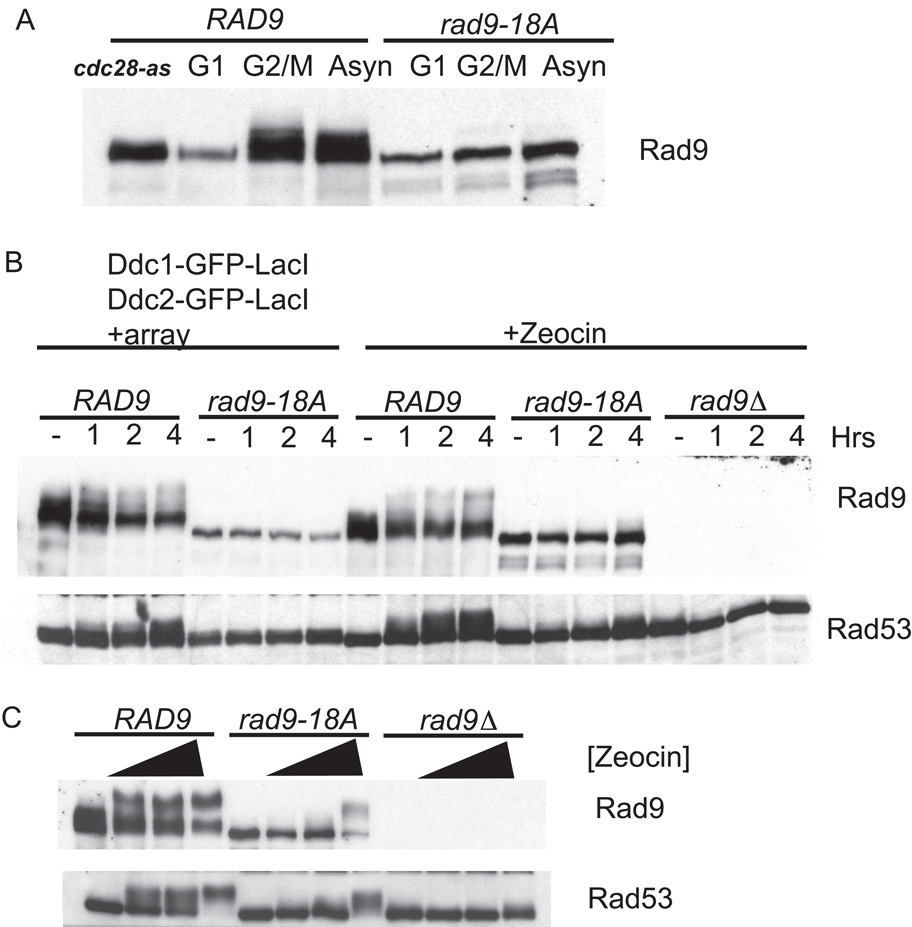
Having shown that CDK activity was important for the DNA damage checkpoint, we next wanted to identify the CDK target responsible for this requirement. The adaptor protein Rad9 has nine full (S/T-P-x-K/R) and 11 partial (S/T-P) CDK consensus sites and shows a cell-cycle-dependent mobility shift indicative of phosphorylation (Figure 7A). We made alanine substitutions of the N-terminal 18 sites, naming this allele rad9-18A. Upon initial examination, it was clear that Rad9-18A did not undergo cell-cycle-dependent phosphorylation compared to the wild-type Rad9 when cells were nocodazole arrested or allowed to cycle (Figure 7A). This observation suggests that we have eliminated the CDK-dependent phosphorylations on Rad9. The allele was then tested for its ability to activate the checkpoint in response to colocalization of sensors and induction of DSBs with Zeocin. As we have previously shown, colocalization of Ddc2-Mec1 and the 9-1-1 clamp at an array induced Rad53 and Rad9 phosphorylation (Figure 7B, left lanes). The Rad9-18A allele failed to undergo checkpoint-dependent phosphorylation upon colocalization of Ddc1-GFP-LacI and Ddc2-GFP-LacI. Moreover, the Rad9-18A allele did not support Rad53 phosphorylation. Similarly, when the Rad9-18A mutant was challenged with Zeocin, the rad9-18A strain did not show Rad53 phosphorylation. Therefore, CDK consensus sites on Rad9 are important for transducing the checkpoint signal from the sensors (Ddc2-Mec1 and 9-1-1) to the effector kinase Rad53.
Figure 7. Rad9 CDK Site Mutant Is Checkpoint Deficient.
(A) Isogenic strains carrying wild-type Rad9-Flag or Rad9-18A-Flag were α factor arrested, nodocazole arrested, allowed to cycle, or treated with cdc28-as inhibitor 1NM-PP1.
(B) Ddc1-GFP-LacI and Ddc2-GFP-LacI checkpoint fusions were induced in nocodazole-arrested Rad9-Flag and Rad9-18A-Flag strains. Zeocin was added to Rad9-Flag, Rad9-18A-Flag, and rad9Δ strains.
(C) Rad9-Flag, Rad9-18A-Flag, and rad9Δ cycling strains were left untreated or treated with Zeocin at 100 ug/ml, 200 ug/ml, and 2 mg/ml final concentration for 2 hr.
Mutating multiple serine and threonine residues on Rad9 could render the protein unstable, resulting in its checkpoint deficiency. However, we noticed that at long time points, slight Rad53 phosphorylation could be seen in the rad9-18A strain. We determined whether the CDK site requirement could be overridden with very high levels of damage. To test this, a range of Zeocin concentrations were used to damage wild-type and rad9-18A strains, and Rad9 and Rad53 phosphorylation was monitored (Figure 7C). At low Zeocin concentrations, Rad9-18A and Rad53 were not phosphorylated. At higher Zeocin concentrations, most of the Rad9-18A exhibited a damage-dependent mobility shift and Rad53 qualitatively activated. Thus, the rad9-18A allele is activated at very high damage doses, suggesting it is not misfolded.
Discussion
The DNA damage checkpoint employs two related protein kinases, ATR/Mec1 and ATM/Tel1, which must act on their substrates in a regulated manner: they do not become activated until a DNA lesion occurs. We set out to understand the mechanism by which the yeast checkpoint kinase Mec1 is activated and have shown that its colocalization with the 9-1-1 complex is sufficient to activate the checkpoint in vivo. By recruiting Ddc2-Mec1 and the 9-1-1 complex to LacO arrays, we were able to bypass the requirement for DNA damage, arguing against a strict damage-dependent activation step for Mec1/ATR. Having shown that colocalization activates the DNA damage checkpoint in vivo, we sought to understand the relationship between the number of sensors recruited and the extent of Rad53 phosphorylation. We consistently observed an array size-dependent increase in Rad53 phosphorylation and propose that checkpoint signaling correlates with the number of sensors corecruited. This result could explain the kinetics of Rad53 activation when a DSB is being processed: more checkpoint molecules on ssDNA result in more Rad53 phosphorylation.
Rad24-RFC is an essential component of the DNA damage checkpoint, presumably because it is required to recruit the damage-specific clamp (9-1-1) to a DSB ([Kondo et al., 2001] and [Melo et al., 2001]). rad24 mutants fail to form damage-induced Ddc1-GFP foci or fully activate Rad53. We found that the requirement for Rad24-RFC could be bypassed if we colocalized 9-1-1 with Ddc2-Mec1 through LacO arrays. Our result argues against the existence of any additional role of Rad24 in Rad53 activation. It also suggests that 9-1-1 activity does not require that it encircle DNA.
The Ddc1 Subunit of 9-1-1 Can Activate Mec1
Despite the similarities between the ATM and ATR kinases, the mechanisms used by cells to activate them appear quite different. Mammalian ATM has been shown to be activated by DNA in vitro. Dimeric ATM is directly activated by the MRN complex in the presence of DNA ([Dupre et al., 2006] and [Lee and Paull, 2005]). This activation leads to the formation of active monomers that act on their substrates ([Bakkenist and Kastan, 2003] and [Lee and Paull, 2005]). Mec1/ATR kinase is activated through an alternate mechanism that does not require a DNA break, as we have shown. Activation of Mec1 could take place directly through an intimate Mec1-Ddc1 interaction, or Ddc1 could promote the activation of Mec1 by recruiting a second factor. Mec1 activation by DNA damage outside of S phase requires the 9-1-1 complex, whereas its activation during DNA replication is less dependent on the 9-1-1 complex. Recently, in vitro data showed that the replication protein TopBP1 is able to activate ATR through an ATR-activating domain (AAD) (Kumagai et al., 2006). BRCT domains I and II of TopBP1 interact with the C terminus of the vertebrate Ddc1 homolog Rad9, suggesting that the 9-1-1 complex could recruit TopBP1 to ATR ([Delacroix et al., 2007], [Kumagai et al., 2006] and [Lee et al., 2007]). Both S. cerevisiae and S. pombe TopBP1 homologs, scDpb11 and spCut5, interact with scDdc1 and spRad9, respectively, suggesting a similar mechanism whereby Dbp11 could be recruited to Mec1 in vivo through the 9-1-1 complex in response to replication damage in S phase ([Furuya et al., 2004] and [Wang and Elledge, 2002]).
Alternatively, Ddc1 could activate Mec1 directly. It has recently been shown in vitro by Majka et al. that 9-1-1 loaded onto a DNA template mimicking a processed damage site could promote Mec1 activation (Majka et al., 2006). Our system furthers the understanding of this mechanism by showing that in vivo activation of Mec1 by the 9-1-1 complex does not require an association of either complex with DNA damage and that this colocalization is sufficient for checkpoint activation. The authors also reported that low-salt conditions allowed the Ddc1 subunit to activate Mec1 in vitro. We show that Ddc1 is able to activate Mec1 and recapitulate the whole checkpoint signaling pathway in vivo in the absence of other 9-1-1 members. The Mec3 and Rad17 subunits could serve to recognize the Rad24/RFC complex and allow loading of the 9-1-1 complex, which is important for maintaining Ddc1 in proximity to Mec1 at the DNA break. In support of this hypothesis, recruitment of Mec3 to Ddc2-Mec1 also activated the checkpoint, but it strictly required Ddc1, arguing that Ddc1 is the activating subunit.
Colocalization Requires Chromatin for Full Activation of Rad53
The role of chromatin in checkpoint signaling could be explained by its ability to maintain a pool of adaptor proteins close to the sensor kinases. This hypothesis is supported by the interaction between the BRCT domains of Rad9 and phosphorylated H2AX. Rad9 Tudor domains and methylated histone residues also provide a binding interface that contributes to adaptor protein localization. Elimination of both H2AX phosphorylation and H3K79 methylation decreases Rad9 phosphorylation upon IR treatment (Toh et al., 2006). Consistent with these data, removing both, H2A phosphorylation and H3K79 methylation decreased Rad53 phosphorylation but did not completely eliminate it in our system. Therefore, recruitment of Ddc2-Mec1 and the 9-1-1 complex alone is insufficient to fully activate the checkpoint. Thus, DNA is important not only as a scaffold for corecruitment of sensors but also as neighboring chromatin, where it may serve to amplify the signal by retaining other checkpoint components.
Activation and Maintenance of Checkpoint Signal Requires CDK
Efficient checkpoint signaling requires CDK activity, and recent experiments have suggested that this is due to a requirement for CDK in DSB processing ([Clerici et al., 2004], [Ira et al., 2004], [Jazayeri et al., 2006] and [Pellicioli et al., 2001]). We found that CDK activity is required for full checkpoint signaling even when colocalization was achieved by artificially concentrating Ddc2-Mec1 and the 9-1-1 complex. While resection does play an important role in signal amplification through damage processing, we have now shown that CDK is also required during additional steps downstream of sensor recruitment. Rad9 contains 9 full and 11 partial consensus CDK sites, more than other proteins in the yeast genome, and is phosphorylated by CDK in vitro (Ubersax et al., 2003). Rad9 is phosphorylated in the absence of damage in G2/M, when CDK activity is high (Figure 6B). If Rad9 phosphorylation by CDK promotes Rad9's activity as an adaptor, eliminating the CDK phosphorylation could impair its checkpoint function. By mutating 18 putative CDK sites on Rad9, we eliminated its cell-cycle-dependent electrophoretic shift and Rad9's ability to activate Rad53, suggesting that CDK also plays a role in controlling Rad9 as an adaptor of the DNA damage checkpoint. Importantly, a residual electrophoretic shift in Rad9 was often observed in G1-arrested cells. This, and the fact that the Rad9-18A defect can be overridden by high levels of damage, may explain the variation seen in the literature for the role of CDK in checkpoint signaling. The residual Rad9 damage-independent phosphorylation in G1 could be due to these sites being shielded from phosphatases or to their phosphorylation being mediated by an alternative proline-directed kinase. Rad9 activation involves multiple steps that include recruitment of Rad9 to a damage site, change in its oligomerization state, phosphorylation by Mec1/Tel1, and binding of Rad53 to allow for Rad53's activation ([Emili, 1998], [Gilbert et al., 2001] and [Schwartz et al., 2002]). Phosphorylation by CDK could be important for one or more of these steps. The Rad9-18A allele described in this manuscript will allow us to probe into the role of CDK in the checkpoint signaling cascade through a specific protein and avoid additional CDK effects.
Cells that encounter damage in G1 delay progression into S phase for only 20–30 min. This delay is significantly shorter than the checkpoint-mediated delay in G2/M, which lasts up to 8 hr. The difference between the two checkpoint responses in S. cerevisiae is consistent with the fact that cells do not require a strong checkpoint response in G1. The difference may reflect the fact that a wider range of repair options are available in G2/M, such as sister chromatid exchange. Previous experiments have suggested that the mechanisms of checkpoint activation are different in G1. The requirement for both H2AX phosphorylation and H3K79 methylation for checkpoint activation is made stronger in G1 ([Hammet et al., 2007], [Javaheri et al., 2006] and [Wysocki et al., 2005]). CDK phosphorylations on Rad9 may help promote Rad9 localization such that H2AX phosphorylation and H3K79 methylation are more important for the activation of Rad9 in G1, when Rad9 would have low levels of CDK phosphorylation. Similarly, Crb2, a S. pombe BRCT-containing checkpoint protein similar to Rad9, has also been shown to undergo CDK phosphorylation ([Caspari et al., 2002], [Du et al., 2006] and [Esashi and Yanagida, 1999]). Initial reports suggested a role for this phorphorylation in checkpoint adaptation and repair ([Caspari et al., 2002] and [Esashi and Yanagida, 1999]). More recently, it has been suggested that this phosphorylation has a redundant role in Crb2 localization (Du et al., 2006).
Many aspects of chromosome biology require the construction of complex protein assemblies seeded at loci dictated by their DNA sequence. In some cases, the sequences may simply be recognition sites for protein binding, whereas in other situations they may contribute in a more direct fashion. For example, transcriptional activation by the bacterial CAP protein requires its binding site to be bent (Gartenberg and Crothers, 1988). Some DNA sequences have significantly different structures that could profoundly affect their function, such as the G quartet structure found at telomeres. DNA damage sites do not differ from the rest of the genome in sequence, but instead the double helix itself is modified. DNA damage can take the form of covalent modifications or the disruption of the helix in the form of a DSB or ssDNA. Historically, studies in transcription have differentiated between structural versus recruitment contributions of a DNA target through the use of artificial tethering systems (Ptashne, 2005). Here, we have expanded upon this for use in systems that require large assemblies of proteins. In the future, this technology could be applied to other aspects of biology, such as the creation of artificial centromeres or telomeres.
Experimental Procedures
Plasmids and Strains
All strains are derivatives of CBY36 W303, mat a, ddc1Δ, ade2, leu2, trp1, his3, and ura3.
Plasmid pJAM150 was used to integrate the checkpoint fusion Gal-Ddc2-GFP-LacI at the his3 locus. Plasmid pCB5 was used to integrate GalS-Ddc1-GFP-LacI at the ura3 locus. Plasmid pCB10 was used to integrate GalS-Mec3-GFP-LacI at the ura3 locus. Integrations were checked by PCR and western blotting using antibodies against GFP. Endogenous RAD53 and RAD9 were C-terminally tagged with HA::LEU2. Plasmids pCB18 and pCB19 were used to integrate Rad9-Flag and Rad9-18A-Flag, respectively, into a rad9Δ strain at the RAD9 locus. Plasmid pAFS52 was digested with EcoRV to integrate 265 LacO arrays at the trp1 locus. Integration of the correct number of arrays was verified by southern blot by BglII digest, probing against a LacO array-specific sequence. Smaller array plasmids were integrated and verified similarly. MEC3 and RAD17 were deleted by gene replacement using the Kan-G418 cassette. DDC1, RAD24, RAD9, and SML1 were deleted by gene replacement using the hygromycin resistance cassette. MEC1 was deleted in the sml1::hygromycin strain by gene replacement with the Kan-G418 cassette. Serine 129 in HTA1 and HTA2 were deleted and replaced with the hygromycin resistance cassette and the KanMX-G418 cassette, respectively. DOT1 was deleted by gene replacement with the Nat resistance cassette. The cdc28-as allele was obtained from Dave Morgan and crossed into the required strain. The Rad52-mRFP allele was obtained from Rodney Rothstein and used to tag the endogenous Rad52.
Galactose Induction and Zeocin Treatment
Galactose induction experiments were conducted with nocodazole-arrested cells. Log cycling cells were arrested with nocodazole for 2 hr in rich media + raffinose. Galactose was added for 1 hr, at the end of which dextrose was added to prevent continued induction. Checkpoint fusions were stable for up to 6 hr after induction as determined by western blot. In the nocodazole release experiment, cells were arrested with nocodazole for 2 hr, induced with galactose for 1 hr in the presence of nocodazole, and released into rich media with dextrose and 8 ug/ml α factor. For experiments performed in G1, cells were first arrested in α factor for 2 hr, and then galactose was added for 2 hr in the presence of α factor. Experiments using the cdc28-as allele containing strain were done in nocodazole-arrested cells, and 5 uM 1-NM-PP1 was used as final concentration. For experiments performed with Zeocin, 100 ug/ml, was used commonly, unless otherwise stated.
Protein Detection
Cell pellets were collected and lysed in boiling SDS buffer for 3 min and loaded onto 10% SDS-PAGE for Rad53-HA detection and 8% SDS-PAGE for Rad9-HA detection and 6% for Rad9-Flag. Proteins were transferred to nitrocellulose and incubated with 16B12 anti-HA antibody or M2 anti-Flag antibody. For Sml1 detection, samples were loaded onto a 15% SDS-PAGE and incubated with an anti-Sml1 antibody obtained from Rodney Rothstein. Cdc28 was visualized with Santa Cruz antibody cY-20. H2A phosphorylation was assayed by using a yeast phosphor-H2A antibody obtained from William Bonner.
Supplementary Material
Acknowledgments
We would like to thank members of the Toczyski, Morgan, and Li labs for helpful discussions. Thanks to Dave Morgan, Joachim Li, Hiten Madahni, Wendell Lim, Bodo Stern, and Andrew Murray for intellectual contributions; Rodney Rothstein for the Sml1 antibody and Rad52-RFP strain; and William Bonner for the phospho-H2AXantibody. Funding was provided by National Institute of General Medical Sciences grant #1R25 GM56847 and National Institutes of Health grant #59691.
References
- Aboussekhra A, Vialard JE, Morrison DE, de la Torre-Ruiz MA, Cernakova L, Fabre F, Lowndes NF. A novel role for the budding yeast RAD9 checkpoint gene in DNA damage-dependent transcription. EMBO J. 1996;15:3912–3922. [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc. Natl. Acad. Sci. USA. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspari T, Murray JM, Carr AM. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 2002;16:1195–1208. doi: 10.1101/gad.221402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- Clerici M, Baldo V, Mantiero D, Lottersberger F, Lucchini G, Longhese MP. A Tel1/MRX-dependent checkpoint inhibits the metaphase-to-anaphase transition after UV irradiation in the absence of Mec1. Mol. Cell. Biol. 2004;24:10126–10144. doi: 10.1128/MCB.24.23.10126-10144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- Du LL, Nakamura TM, Russell P. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev. 2006;20:1583–1596. doi: 10.1101/gad.1422606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre A, Boyer-Chatenet L, Gautier J. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat. Struct. Mol. Biol. 2006;13:451–457. doi: 10.1038/nsmb1090. [DOI] [PubMed] [Google Scholar]
- Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell. 1998;2:183–189. doi: 10.1016/s1097-2765(00)80128-8. [DOI] [PubMed] [Google Scholar]
- Esashi F, Yanagida M. Cdc2 phosphorylation of Crb2 is required for reestablishing cell cycle progression after the damage checkpoint. Mol. Cell. 1999;4:167–174. doi: 10.1016/s1097-2765(00)80364-0. [DOI] [PubMed] [Google Scholar]
- Furuya K, Poitelea M, Guo L, Caspari T, Carr AM. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev. 2004;18:1154–1164. doi: 10.1101/gad.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg MR, Crothers DM. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature. 1988;333:824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- Gilbert CS, Green CM, Lowndes NF. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell. 2001;8:129–136. doi: 10.1016/s1097-2765(01)00267-2. [DOI] [PubMed] [Google Scholar]
- Green CM, Erdjument-Bromage H, Tempst P, Lowndes NF. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr. Biol. 2000;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- Hammet A, Magill C, Heierhorst J, Jackson SP. Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep. 2007;8:851–857. doi: 10.1038/sj.embor.7401036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaheri A, Wysocki R, Jobin-Robitaille O, Altaf M, Cote J, Kron SJ. Yeast G1 DNA damage checkpoint regulation by H2A phosphorylation is independent of chromatin remodeling. Proc. Natl. Acad. Sci. USA. 2006;103:13771–13776. doi: 10.1073/pnas.0511192103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Kondo T, Wakayama T, Naiki T, Matsumoto K, Sugimoto K. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science. 2001;294:867–870. doi: 10.1126/science.1063827. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J. Biol. Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat. Cell Biol. 2003;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- Majka J, Burgers PM. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc. Natl. Acad. Sci. USA. 2003;100:2249–2254. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka J, Niedziela-Majka A, Burgers PM. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol. Cell. 2006;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JA, Cohen J, Toczyski DP. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D, Hirano Y, Sugimoto K. Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol. Cell. Biol. 2004;24:10016–10025. doi: 10.1128/MCB.24.22.10016-10025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Du LL, Redon C, Russell P. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol. Cell. Biol. 2004;24:6215–6230. doi: 10.1128/MCB.24.14.6215-6230.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciotti V, Clerici M, Lucchini G, Longhese MP. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol. Cell. 2001;7:293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- Ptashne M. Regulation of transcription: from lambda to eukaryotes. Trends Biochem. Sci. 2005;30:275–279. doi: 10.1016/j.tibs.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Rouse J, Jackson SP. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell. 2002;9:857–869. doi: 10.1016/s1097-2765(02)00507-5. [DOI] [PubMed] [Google Scholar]
- Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Schwartz MF, Duong JK, Sun Z, Morrow JS, Pradhan D, Stern DF. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell. 2002;9:1055–1065. doi: 10.1016/s1097-2765(02)00532-4. [DOI] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Sun Z, Hsiao J, Fay DS, Stern DF. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- Sweeney FD, Yang F, Chi A, Shabanowitz J, Hunt DF, Durocher D. Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr. Biol. 2005;15:1364–1375. doi: 10.1016/j.cub.2005.06.063. [DOI] [PubMed] [Google Scholar]
- Thelen MP, Venclovas C, Fidelis K. A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins. Cell. 1999;96:769–770. doi: 10.1016/s0092-8674(00)80587-5. [DOI] [PubMed] [Google Scholar]
- Toh GW, O'Shaughnessy AM, Jimeno S, Dobbie IM, Grenon M, Maffini S, O'Rorke A, Lowndes NF. Histone H2A phosphorylation and H3 methylation are required for a novel Rad9 DSB repair function following checkpoint activation. DNA Repair (Amst.) 2006;5:693–703. doi: 10.1016/j.dnarep.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J. Biol. Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- Vialard JE, Gilbert CS, Green CM, Lowndes NF. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 1998;17:5679–5688. doi: 10.1093/emboj/17.19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidanes GM, Bonilla CY, Toczyski DP. Complicated tails: histone modifications and the DNA damage response. Cell. 2005;121:973–976. doi: 10.1016/j.cell.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Wang H, Elledge SJ. Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics. 2002;160:1295–1304. doi: 10.1093/genetics/160.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Minn K, Jorda KG, Chen J. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 2003;278:19579–19582. doi: 10.1074/jbc.C300117200. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell. Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Rothstein R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc. Natl. Acad. Sci. USA. 2002;99:3746–3751. doi: 10.1073/pnas.062502299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Elledge SJ. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell. 1993;75:1119–1127. doi: 10.1016/0092-8674(93)90321-g. [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. USA. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.