Abstract
The gene clusters responsible for the biosynthesis of two anti-tumor antibiotics, ravidomycin and chrysomycin, have been cloned from Streptomyces ravidus and Streptomyces albaduncus, respectively. Sequencing of the 33.28 kb DNA region of the cosmid cosRav32 and the 34.65 kb DNA region of cosChry1-1 and cosChryF2 revealed 36 and 35 open reading frames (ORFs), respectively, harboring tandem sets of type II polyketide synthase (PKS) genes, d-ravidosamine and d-virenose biosynthetic genes, post-PKS tailoring genes, regulatory genes, and genes of unknown function. The isolated ravidomycin gene cluster was confirmed to be involved in ravidomycin biosynthesis through the production of a new analogue of ravidomycin along with anticipated pathway intermediates and biosynthetic shunt products upon heterologous expression of the cosmid, cosRav32, in Streptomyces lividans TK24. The identity of the cluster was further verified through cross complementation of gilvocarcin V (GV) mutants. Similarly, the chrysomycin gene cluster was demonstrated to be indirectly involved in chrysomycin biosynthesis through cross-complementation of gilvocarcin mutants deficient in the oxygenases GilOII, GilOIII, and GilOIV with the respective chrysomycin monooxygenase homologues. The ravidomycin glycosyltransferase (RavGT) appears to be able to transfer both amino- and neutral sugars, exemplified through the structurally distinct 6-membered d-ravidosamine and 5-membered d-fucofuranose, to the coumarin-based polyketide derived backbone. These results expand the library of biosynthetic genes involved in the biosyntheses of gilvocarcin class compounds that can be used to generate novel analogues through combinatorial biosynthesis.
Keywords: biosynthesis, deoxysugars, gilvocarcins, glycosylation, polyketides, ravidomycin
Introduction
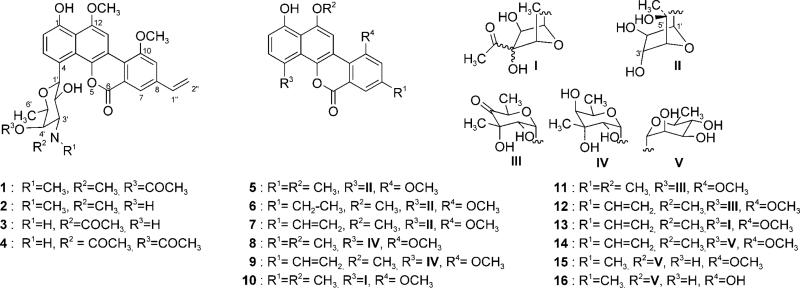
Ravidomycins (RMs; 1, 2) and chrysomycins (CMs; 8, 9) are potent antibacterial and antitumor antibiotics isolated from the culture broths of Streptomyces ravidus and Streptomyces albaduncus, respectively.[1–3] Both antibiotics share a polyketide derived tetracyclic coumarin-based benzo[d]naphtho[1,2-b]pyran-6-one moiety, also found in other analogues such as the gilvocarcins (5–7), FE35 A (3) and B (4),[4] BE 12 406A (15) and B (16),[5,6] Mer 1020 dA-D (10–13),[7] and the recently isolated polycarcin V (14).[8] However, they differ in their C-glycosidically linked sugar moieties: ravidomycin V (RMV, 1, also known as ravidomycin) possesses an amino pyranose, 4′-O-acetyl-d-ravidosamine, whereas chrysomycin V (CMV, 9, also known as chrysomycin A) possesses a branched neutral pyranose, d-virenose.[4,9–11] RMV shows a broad range of biological activities, such as antibacterial and antitumor both in vivo and in vitro.[11] Such activities primarily arise from the inhibitory effect of 1 on bacterial DNA and RNA synthesis.[12] In addition, excellent cytotoxicity of 1 was reported for human colon carcinoma cell lines.[1,11] Furthermore, 1 has been reported to act as potent inhibitor of human topoisomerase II.[13] Similarly, antitumor activities of CMV have been reported against various cancer cell lines.[2,3,13] Despite all these reports of biological activities, the exact mechanism of action of 1 and 9 at the molecular level still remains elusive. The members of this class of antibiotics are also often called gilvocarcin-type anticancer agents;[14] their biological activity is light-dependent. Mechanistic studies of gilvocarcin V (GV, 7) showed its equilibrium DNA binding and light-induced DNA adduct formation through [2+2] cycloaddition of the vinyl side chain with thymidine residues of DNA besides its strong DNA intercalation property.[15] GV-mediated selective cross-linking of DNA with histone H3 in the presence of light represents an unique mode of action.[16] The common coumarin-based structural backbone and light enhanced anti-tumor/antibacterial activities indicate that 1, 9 and 7 share a similar mode of action. (Scheme 1)
Scheme 1.
Ravidomycins, chrysomycins and related natural products.
Although there are no direct experimental reports on activity comparisons of RMs with the other analogues, the literature indicates 1 to be the most active compound against tumor cell lines.[17] This is a clear indication that the principally more hydrophilic amino sugar moiety plays a crucial role in the enhanced biological activities of RMV compared to its relatives. Such amino compounds also provide better formulation and delivery options, either as salt forms or for the loading into nanoparticles.[18–20] Chemical modifications of 1 and structure–activity relationship (SAR) studies suggested that the 2′- and 4′-hydroxyl groups of d-ravidosamine are essential for its biological activities. Further development of RM was hindered by its inherent cytotoxicity, thereby necessitating the generation of new analogues with better pharmacological profiles. More recently, total syntheses of 1, 5 and 6 have been reported.[21–24] However, complicated synthetic routes and poor yields hindered the generation of new analogues for further SAR studies. Future attempts to generate potentially valuable ravidomycin and chrysomycin analogues will require a detailed understanding of the role and orchestration of their biosynthetic enzymes. Glycodiversification provides a unique means to generate ravidomycin and chrysomycin analogues but it requires flexible glycosyltransferases (GTs) and a library of NDP-sugars.[25,26] Unfortunately, only a handful of C-GTs are known to possess marginal substrate promiscuity,[27–29] but no C-GTs capable of transferring amino or branched sugars have been reported except for Med-ORF8 (TDP-angolosamine).[30] In this context, characterization of the 4′-O-acetyl-NDP-d-ravidosamine (45) and NDP-d-virenose (37) biosynthetic enzymes and the corresponding GTs that establish the C-glycosidic linkage to the coumarin backbone is particularly intriguing. Incorporation experiments showed that the gilvocarcin, ravidomycin, and chrysomycin backbone derived from acetate and propionate, and it was suggested that the pathway involves an oxidative rearrangement of an angucyclinone intermediate.[31–33] We recently isolated the GV gene cluster and characterized various genes through inactivation and cross complementation experiments.[14,34–37] However, our efforts to address the biosynthetic sequence of key post-PKS tailoring events and their relevant intermediates were hampered by the resistant nature of the producer strain for genetic manipulations, and the inability of several mutant constructs to express in a heterologous host. Cloning and characterization of RM's and CM's gene clusters provide additional pathways to further explore these issues, while providing crucial toolkits to generate new gilvocarcin-type natural products through combinatorial biosynthesis.[38–45] In this context, we have cloned and sequenced the RM and CM biosynthetic loci, and also have produced a new ravidomycin derivative through the heterologous expression of its gene cluster. Also reported here is a C-GT that accepts a structurally most distinct NDP-d-fucofuranose in addition to its natural substrate NDP-d-ravidosamine.
Results and Discussion
Cloning, sequence analysis and organization of the ravidomycin and chrysomycin gene clusters
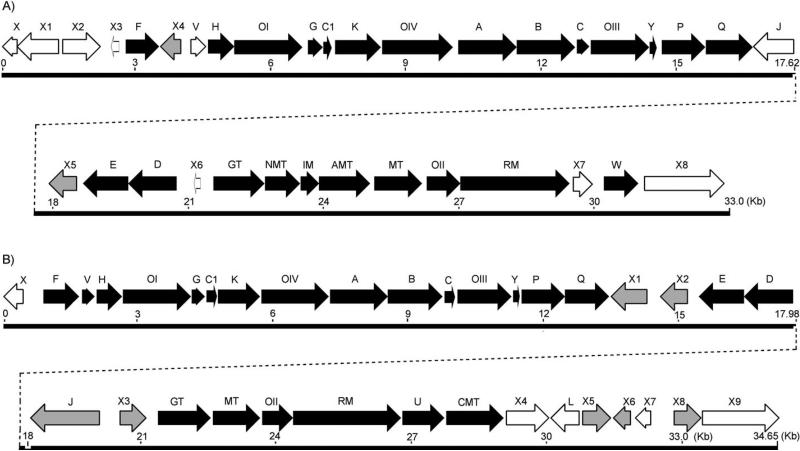
A library screening strategy was taken to clone the RMV and CMV biosynthetic gene clusters. Genomic libraries of S. ravidus and S. albaduncus were constructed in the pOJ446 E. coli-Streptomyces shuttle vector. Because it was obvious that the RMV and CMV polyketide cores derive from the activities of type II PKS enzymes[46–48] involving a ketosynthase α (KSα), and that their corresponding deoxysugar biosyntheses involve an NDP-sugar 4,6-dehydratase (4,6-DH), the corresponding genes were used to probe the genomic library. The primers based on the conserved residues of GilA (encodes the KSα in the GV pathway),[14] and its homologues in the database resulted in the expected amplifications from S. ravidus and S. albaduncus genomic DNA. Similarly, nucleotide fragments of 4,6-DH encoding gene were amplified using a previously reported forward primer,[49] but a modified reverse primer (Figure S3). Cosmid DNA was isolated from colonies hybridizing with the KSα and the 4,6-DH probes. Out of approximately 2000 colonies screened from each genomic library, four and three different colonies, respectively, from the S. ravidus and S. albaduncus library, respectively, were hybridized with both probes. One cosmid from each library (cosRav32 and cosChry1-1 from the S. ravidus and S. albaduncus library, respectively) was sequenced through a shotgun approach using the HyperMu™ MuA Transposase kit (Epicentre) following the reported protocol.[50–53] A third probe was constructed for screening S. albaduncus genomic DNA based on conserved residues from type II PKS-associated ketoacyl reductases, which resulted in three additional positively hybridized colonies. Bioinformatic analyses of the cosRav32 and the overlapping cosmids cosChry1-1 and cosChryF2 sequence using Frame plot and the NCBI BLAST database revealed clusters of 36 and 35 open reading frames (ORFs), respectively. Both the ravidomycin and chrysomycin gene clusters were flanked by ORFs with functions irrelevant to RMV and CMV biosynthesis, respectively. The organization of the clusters and the putative function of the genes are summarized in Figure 1 (Tables S1 and S2 in the Supporting Information). The sequence data have been deposited in the EMBL nucleotide database under the accession numbers FN565 485 (rav cluster) and FN565 166 (chry cluster).
Figure 1.
Gene clusters for A) ravidomycin and B) chrysomycin biosyntheses. The black filled arrows indicate biosynthetic genes, gray arrows represent regulatory genes, and blank arrows represent the genes with unknown functions.
Biosynthesis of the polyketide core
Nine genes in the RMV and CMV clusters were identified to be involved in the biosynthesis of the polyketide backbone. The encoded proteins of ravA, ravB and ravC show typical homologies to the various minimal PKS ketoacyl synthases (KSα), chain length determinants (CLF or KSβ), and the acyl carrier protein (ACP), respectively), enzymes typical of a type II PKS system.[46–48] Likewise, chryA, chryB, and chryC represent the CMV counterparts. Interestingly, both clusters possess an additional acyl carrier protein gene (ravC1 and chryC1, respectively) between two cyclase genes (that is, ravK/chryK and ravG/chryG). Despite their close homologies, the corresponding ACPs might be specific for the selection and priming of either the acetate or the propionate starter unit.[54–59] Typically only one ACP has been reported for well-characterized type II PKS clusters, such as the aclacinomycin A,[60] doxorubicin,[61] aranciamycin,[62] jadomycin B,[63] actinorhodin,[64] medermycin,[30] and tetracenomycin C[65] Even the closely related GV biosynthetic gene cluster[14] contains only one ACP, despite the fact that GV as well as the anthracyclines aclacinomycin and doxorubicin also use propionate starter units. However, two ACP encoding genes were found previously, e.g., in the gene clusters of the frenolicin[66] and the R1128[67] pathways, and it was discussed that the second ACP might serve as an intermediate anchor on the diketide stage.[66] The second ACP in the R1128 pathway was identified as part of an initiation module that is responsible for starter-unit selection and catalysis of the first chain elongation step, and the additional ACP was found to be indispensable for the incorporation of nonacetate primer units.[68] The products of ravP/chryP and ravQ/chryQ exhibit their closest homologies to gilP and gilQ, corresponding to malonyl CoA:ACP transacylases (MAT, also abbreviated as MCAT) and acyltransferases (AT) in the database, respectively. Based on sequence homologies and inactivation experiments of gilP and gilQ (unpublished results), we favor RavQ/ChryQ to be responsible for the selection of propionate as a starter unit which ends up as the C-8-bound vinyl side chain in 1 and 9. Given that type II PKSs often recruit the MAT from fatty acid biosynthesis, a process known as cross-talk,[69] the presence of such genes seemed unique. Although it has been shown that a number of type II polyketide ACPs undergo rapid self-acylation from malonyl-CoA in the absence of an MAT, this observation has been often ascribed to contamination with Escherichia coli MAT (FabD) rather than an intrinsic property of the ACP. However, recent work with a synthetic holo-ACP from the actinorhodin pathway of S. coelicolor showed that this ACP can undergo self-malonylation in the presence of malonyl-CoA, and careful controls ruled out the possibility of contamination with E. coli MAT, proving that self-malonylation is indeed an inherent activity of the ACP of the actinorhodin PKS.[70] The fact that we found MAT encoding genes in all three biosyntheses of gilvocarcin-type compounds suggests that the ACPs of their biosynthetic pathways cannot undergo self-malonylation. Two cyclases (encoded by ravK/chryK and ravG/chryG), and a ketoreductase (encoded by ravF/chryF) are the other PKS-associated enzymes that are hypothesized to catalyze multiple cyclizations and a ketoreduction step en route to homo-UWM6 (17).
Post-PKS tailoring enzymes
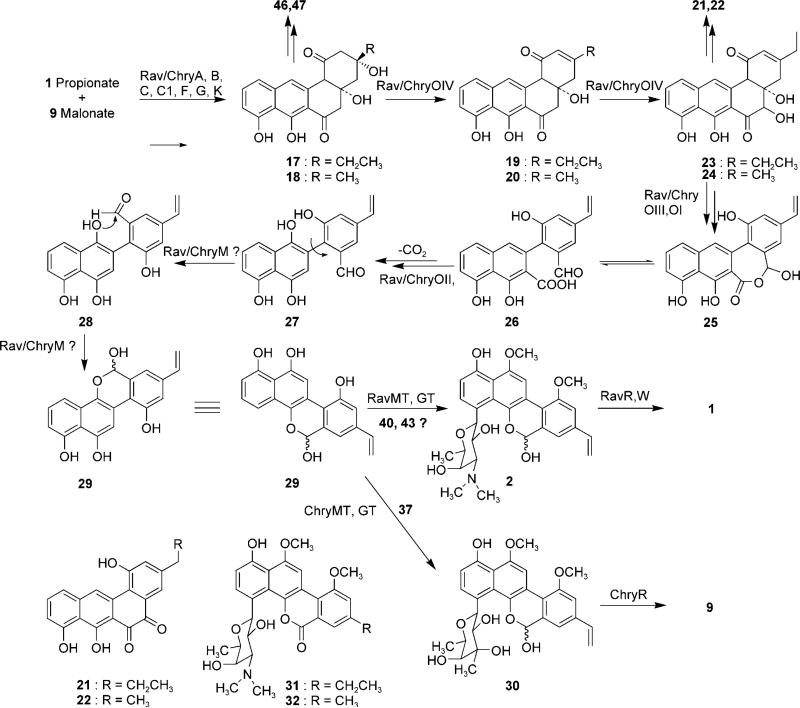
As expected, the post-PKS tailoring enzymes identified in the RMV and CMV clusters are similar to those of their GV counterpart. Four different oxygenase encoding genes (ravOI/chryOI, OII, OIII and OIV) were identified in the RMV and CMV clusters. Recent results showed that the homologues of RavOIV/ChryOIV, JadF and GilOIV, both catalyze identical reactions (i.e., 2,3-dehydration of 17 and oxidation at the C-5 position of 19) despite the different end products of their respective pathways (Scheme 2).[36]
Scheme 2.
Proposed pathways for the biosyntheses of ravidomycin V and chrysomycin V. For the NDP sugar structures 37, 40 and 43, see Scheme 3.
Similarly, RavOI/ChryOI homologues, JadH and GilOI, also show identical activities: both catalyze 4a-12b dehydration of 23, and might also be involved in C–C bond cleavage, presumably through Baeyer–Villiger monooxygenation.[71–75] Based on these similarity studies, we propose that RavOIV/ChryOIV and RavOI/ChryOI are functionally identical to GilOIV and GilOI of the GV cluster, respectively. The translated protein sequences of ravH/chryH exhibit similarity to various NADPH-dependent FMN reductases, including the recently characterized GV pathway counterpart GilH.[37] We assume RavH/ChryH generates the reduced coenzyme FADH from NADPH and FAD, which is necessary for RavOI,OIV/ChryOI,OIV during the oxygenation cascade.[19–25] The product sequences of ravOII/chryOII show their closest homologies to GilOII of the GV cluster (68%:77% and 65%:76% identity/similarity, respectively) and other putative anthrone oxygenases. Similar to GilOII of the GV cluster, RavOII/ChryOII might introduce a hydroxyl group at the naphthalene moiety of 26; this hydroxyl could later react with the aldehyde group resulting from the oxidative C–C bond cleavage to generate hemiacetal 29. The protein sequences encoded by ravOIII/chryOIII exhibit similarities to various cytochrome-P450 monooxygenases (CYPs), including the recently characterized GilOIII of the GV cluster. Through inactivation studies, we previously demonstrated the role of GilOIII in installing the C-8 vinyl side chain of GV.[35] Therefore, RavOIII/ChryOIII are most likely involved in C-8 vinyl side chain formation in their respective pathways. A gene encoding a homologue of ferrodoxin protein was identified in both clusters (ravY/chryY). Because most of the cytochrome-P450 monooxygenases require ferredoxin as an electron carrier to carry out their oxidative activities, we assume that the product of ravY/chryY might supply ferredoxin to RavOIII/ChryOIII. However, a homologous gene encoding a ferredoxin protein was not found in the GV cluster. But it is well known that such genes are often located outside the secondary metabolite biosynthetic gene clusters.
Sequence analyses showed that the stop codon of ravOII/chryOII was translationally coupled with the start codon of a relatively bigger ORF (ravRM/chryRM), the translated N-terminal protein sequence of which shows homology to FAD-dependent oxidoreductases, including the recently characterized GilR,[76] whereas the C-terminal region is similar to thiopurine-S-methyltransferase superfamily proteins from various organisms, GilM of the GV pathway being the closest homologue. The fact that GilR installs the lactone of GV at the last step of the biosynthetic pathway was recently confirmed both in vivo and in vitro.[76] Similarly, inactivation of GilM demonstrated its involvement in GV biosynthesis, but its exact role in the biosynthetic pathway remained elusive due to the instability of the accumulated product of the GilM− (unpublished results). It has been proposed that GilM might be involved in C–C bond rotation of 27 and formation of the hemiacetal moiety in 29 via 28 (Scheme 2). Therefore, the fusion of the genes encoding these functionally distinct Gil-homologues in the RM and CM clusters is particularly notable. Based on the homology analyses and comparison to the gilvocarcin biosynthetic genes, we believe ravMT/chryMT to encode O-methyltransferases responsible for possibly both O-methylations of 29.
Genes for deoxysugar biosynthesis and attachment
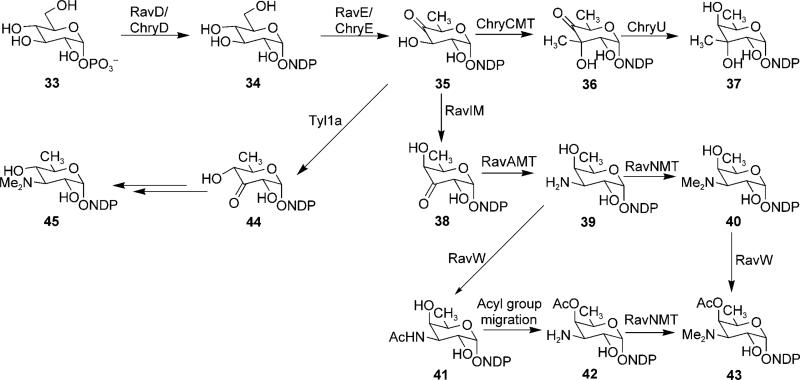
A total of five genes encoding deoxysugar biosynthetic enzymes were identified in the ravidomycin cluster. The products of ravD and ravE resemble various NDP-glucose synthases and NDP-glucose-4,6-dehydratases from Streptomyces sp., which generate NDP-4-keto-6-deoxy-d-glucose from TMP and glucose-1-phosphate (Scheme 3). The product of ravIM shows its closest homology to NDP-hexose-3,4-ketoisomerase ORF2[77] (51%:68% identity/similarity) and TyL1a[78] (62%:48%) from the spiramycin and mycaminose biosynthetic pathways, respectively. In addition, RavIM also exhibits remarkable identity (39%) to the TDP-4-keto-6-deoxy-d-glucose-3,4-ketoisomerase (FdtA) from Aneurinibacillus thermoaerophilus. FdtA has been demonstrated to catalyze the reaction of TDP-4-keto-6-deoxy-d-glucose (35) to TDP-3-keto-6-deoxy-d-galactose (38) during the biosynthesis of surface layer 3-acetamido-3,6-dideoxy-α-d-galactose moieties of the bacterial surface glycan chain,[79] whereas Ty L1a acts on the same substrate but alternatively generates the product TDP-3-keto-6-deoxy-d-glucose (44) with an equatorial-facing hydroxyl group at the C-4 position. Despite the closer similarity of RavIM with TyL1a, we propose that RavIM catalyzes an isomerization reaction identical to that of FdtA (Scheme 3) generating an axial hydroxyl group at the 4-position, which is later acetylated to furnish 4′-O-acetyl-d-ravidosamine. If this hypothesis is true, RavIM will be the first enzyme from a secondary metabolite biosynthetic pathway to have such an activity. ORFs ravAMT and ravNMT are homologous to various proposed/characterized deoxysugar amino-transferase encoding genes and to genes of N,N-dimethyltransferases, respectively.[80–85] In context with RM biosynthesis, we assume that RavAMT catalyzes the transamination of 38 to generate 39, which undergoes subsequent methylation of the amino-nitrogen to generate 40 through the activity of RavNMT.
Scheme 3.
Proposed pathways for the biosyntheses of 4′-O-acetyl-NDP-d-ravidosamine and NDP-d-virenose.
The product of ravW shows modest similarity to GCN-5 related N-acetyltransferase (GNAT)-family enzymes that catalyze the transfer of an acetyl group from acetyl-CoA to the primary amine of a variety of acceptor substrates. This suggests the possibility of RavW-mediated N-acetylation of 39 followed by the migration of the acetyl group to the 4′-hydroxyl group (41→42) during NDP-O-acetyl ravidosamine biosynthesis. However, the presence of significant amounts of deacetylravidomycin (2) in the culture broth of S. ravidus indicates that RavW might also act as an O-acetyltransferase, thereby directly acetylating the axial 4′-hydroxyl group of 40 completing the 4′-O-acetyl-ravidosamine moiety of RMV prior to or after the glycosyltransfer step (Scheme 2). Further experiments are necessary to clarify the details of these late steps in RM biosynthesis.
The GT responsible for the C-glycosidic linkage between the aromatic backbone and amino sugar of RMV is particularly interesting from a biosynthetic point of view as a unique tool for further combinatorial biosynthetic studies. We have identified a GT-encoding gene, ravGT, upstream of ravNMT. The deduced amino acid sequence of RavGT exhibits high identity/similarity to various O- and C-GTs involved in secondary metabolite biosyntheses such as LanGT2 (47%:67%), GilGT (46%:66%), Med-ORF8 (48%:65%) and UrdGT2 (42%:60%) from the landomycin, GV, medermycin and urdamycin biosynthetic pathways, respectively.[30,86,87] Unlike GilN of the GV pathway, there are no additional GT-encoding homologues in the RM cluster, the products of which could potentially be involved or assist C-glycosylation. Therefore, we assume that RavGT catalyzes the attachment of the NDP-ravidosamine to 29, establishing a C–C bond. Here, it is important to note that UrdGT2, GilGT and IroB are the only C-GTs to be characterized at the functional level.[88] The former two enzymes use NDP-deoxysugars as donor substrates whereas the latter uses UDP-d-glucose. There is obvious similarity of the gilvocarcin-type GTs regarding both the NDP-sugar donor substrate and the coumarin-like acceptor substrate to ChaGT1,[89] a GT that is presumably responsible for the O-glycosidic linkage of a d-fucopyranose to the chartreusin aglycone, which resembles the coumarin-like polyketide core of the gilvocarcins. The sequence comparison here yielded the strongest relation of ChaGT1 to RavGT (32%:48% aa identity/similarity), then to ChryGT (32%:46%) and GilGT (27%:42%).
Given the fact that the sugar donor substrate flexibility of a GT determines its potential applicability to generate novel gilvocarcin-type analogues through glycodiversification strategies, we were curious to preliminarily explore the flexibility of RavGT. The mutant strain of the GV pathway[47] (S. lividans TK24/cosG9B3-GilGT̄), in which gilGT was deleted in-frame, was the ideal host to test the glycosylation activity of ravGT.[35] This strain harbors the entire GV biosynthetic genes but lacks the GT (encoded by gilGT), thus providing the yet unidentified sugar acceptor substrate and the NDP-d-fucofuranose. Therefore, we attempted the cross-complementation of the GilGT− mutant with the ravGT expression construct (pRavGT). Interestingly, the complementation of the mutant restored gilvocarcin E (GE) production (Figure S1). Accumulation of GE instead of GV was not surprising as the activity of GilOIII, which is responsible for the vinyl group formation, was suppressed in the GilGT− mutant due to polar effects inherited by the inactivation.[35] UrdGT2 is reportedly the only known C-GT to possess flexibility for both sugar donor and sugar acceptor substrates.[27,28] More recently, the generation of 4′-hydroxygilvocarcins through the inactivation of the ketoreductase GilU has demonstrated the flexibility of GilGT towards its donor substrate in the GV pathway. However, GilGT could not accept a variety of NDP-aminosugar donor substrates tested in our laboratory (unpublished results). Our current report provides the first example of a C-GT that can utilize both a neutral 5-membered NDP-d-fucofuranose and a basic 6-membered NDP-aminopyranose (NDP-d-ravidosamine) as sugar donor substrates.
The C-glycosidically linked d-virenose is a unique structural feature of CMs. d-virenose has an equatorial methyl group at C-3′ position and an axial C-3′ hydroxyl group, otherwise it is identical to d-fucopyranose. Analysis of the CM cluster revealed a set of four genes that could potentially be involved in the biosynthesis of NDP-d-virenose. The products of chryD and chryE catalyze the first two biosynthetic steps to generate the common intermediate 35. ChryCMT is proposed to catalyze the equatorial C-3 methylation of this intermediate. The deduced sequence of ChryCMT shows typical homology to several putative/characterized deoxysugar SAM-dependent C-methyltransferases from various organisms. The product encoded by chryU reduces the 4-keto group of 36 to afford NDP-d-virenose (37). Gene chryGT is the only candidate C-GT gene identified in the CM cluster, the product of which might establish the C–C bond between the PKS-derived coumarin backbone and C-1 of NDP-d-virenose. In fact, this gene encodes the first C-GT that potentially utilizes a branched sugar (NDP-d-virenose) as a natural sugar donor substrate. The substrate specificity of this enzyme needs to be explored to determine its future potential for combinatorial biosynthetic attempts towards new GV analogues.
Genes encoding regulatory and resistance enzymes or enzymes of unknown function
RavX4 and ravX5 represent two regulatory genes, the corresponding amino acid sequences of which resemble transcriptional regulatory proteins from various Streptomyces sp. in the RMV cluster. The function of the corresponding enzymes of six other ORFs (ravX2, X3, J, X6, X7 and X8) could not be assigned through homology studies. The products of ravX and X1 represent homologues of phenylacetic acid degrading enzymes which might not be related to the RM biosynthesis. Located at the right end of the cluster, the product of ravX8 shows close similarity to several hypothetical proteins of unknown function not involved in biosyntheses of natural product secondary metabolites, thereby indicating the end of the cluster.
Similarly chryX2, X3, X4, X5, X6, and X7 represent the regulatory genes in the CM cluster. The deduced amino acid sequences of genes chryX, X8 and X9 show similarity to spore transformation proteins, hexokinase from glycolysis and acyl-CoA dehydrogenase from β-oxidation, respectively, and thus are likely not involved in CM biosynthesis, therefore providing the boundaries of the CM cluster. Like in case of the gilL product of the GV pathway, the role of the chryL product for CM biosynthesis could not be assigned through data bank searches. The protein encoded by chryJ likely serves as an efflux pump necessary for the self-protection of S. albaduncus from its own product chrysomycin, much like GilJ (aa-identity/similarity between ChryJ/GilJ is 62%:75%).
Heterologous production of deacetylravidomycins in S. lividans TK24
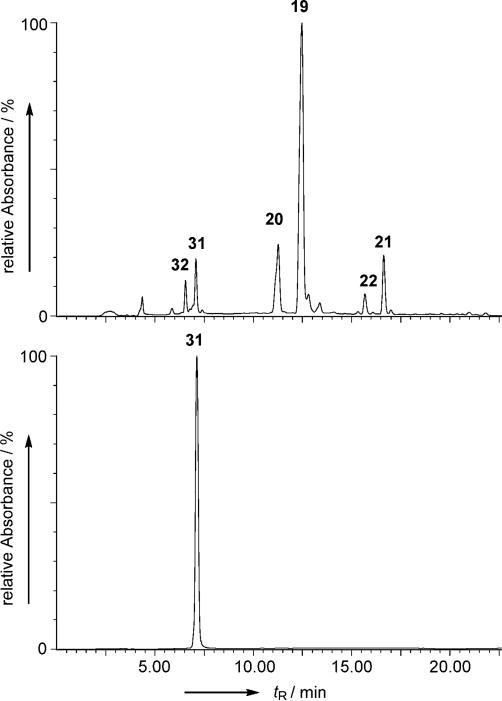
Analysis of the DNA sequence of cosRav32 indicated that the cosmid harbors all of the necessary genes for the production of RM. This prompted us to attempt a heterologous expression of the entire RM cluster in S. lividans TK24. Such expression would utilize all the naturally occurring promoters, regulatory, resistance and RM biosynthetic genes encoded within cosRav-32. The cosmid was introduced into S. lividans TK24 through a well-established E. coli: Streptomyces inter-generic conjugation protocol.[90] The yellowish green exconjugants obtained on the plate were subjected to liquid culture for metabolite production. HPLC analysis showed that there are at least six new metabolite peaks in the culture broth of the S. lividans TK24-(cosRav32) as compared to the control strain S. lividans TK24/pOJ446. UV spectra and retention time (tR) in HPLC/MS comparisons revealed that four of the six peaks represented 2,3-dehydroUWM6 (20, 2 mg L−1), 2,3-dehydro-homoUWM6 (19, 20 mg L−1), preGM-o-quinone (22, 0.2 mg L−1) and preGE-o-quinone (21, 1.3 mg L−1), see Figure 2.
Figure 2.
A) HPLC analysis of the metabolites of S. lividans TK24 (cosRav32); B) Standard deacetylravidomycin E (31)
The other two peaks (eluted at tR = 6.6 and 7.1 min) exhibited UV spectra typical to that of ravidomycin, however with a slight hypsochromic shift at one of the maxima from 392 nm to 380 nm for both compounds. This indicated that the C-8 vinyl side chain of 1 might be replaced with a saturated ethyl or methyl group. Low resolution ESI-MS analyses further showed that the mass of the first compound (m/z 510 [M+H]+ in positive APCI MS and 508 m/z [M–H]− in negative APCI MS) is reduced by 14 amu compared to that of the second (m/z 524 [M+H]+ in positive and m/z 522 [M–H]− in negative APCI MS); this indicates their real molecular weights to be 511 and 523, respectively. This suggested that these compounds differ from one another by one methylene group (-CH2-), preferably at the 8-sidechain, and that the C-4′-O-acetyl group is missing in both compounds. The molecular formula of the latter compound obtained through the high resolution EI-MS (523.2207 gmol−1) matched the predicted structure of deacetylravidomycin E (31, C29H33NO8). However, poor production (<50 μg L−1) of these metabolites did not allow a larger scale isolation necessary for NMR analysis. However, we reasoned that compound 31 could be obtained from S. ravidus by inhibiting its putative P450 oxygenase RavOIII with ancymidol, a known, effective CYP450 inhibitor.[91,92] Indeed, fermentation of S. ravidus in presence of ancymidol revealed a new metabolite in HPLC/MS, the HPLC retention time and mass of which seemed identical to the larger ravidomycin compound from S. lividans TK24(cosRav32).
About 4 mg of the compound was isolated using preparative HPLC and its structure was analyzed by NMR and HRMS. When comparing its 1H NMR with that of 1 (RMV), all signals of the RV protons were observed except for the vinyl and 4′-O-acetyl protons which typically appear at δ 5.5–7.0 and δ 2.0– 2.5, respectively. The presence of an additional triplet at δ 1.32 and quartet at δ 2.83 clearly indicated the presence of an 8-ethyl side chain instead of the 8-vinyl side chain, thereby confirming structure 31 as shown in Scheme 2. The structure of the compound was further confirmed by 13C NMR analysis (see NMR data in the Supporting Information). Having standard 31 in hand, we compared it to the S. lividans TK24(cosRav32) metabolites. Identical HPLC Rt, UV spectrum and MS fingerprints confirmed that the second ravidomycin compound mentioned above is indeed 31. Based on these observations, we also propose the structure of the minor compound to be 32.
The results clearly show that the isolated cluster covers most of the genes required for the biosynthesis of RV. However, the accumulation of a marginal amount of 31 (deacetyl-RME) in instead of 1, and the accumulation of 19–22 as major metabolites is surprising. Gene inactivation, complementation and in vitro assay results have suggested that 19 and 20 are biosynthetic intermediates of the GV and jadomycin pathways, respectively.[34,36,93,94] As in the GV pathway, hydroxylation of 19 followed by lactone formation (19→25) through the activities of RavOI and RavOIV are the suggested RV biosynthetic steps in which 21 and 22 are generated through further shunt oxidation of 23 (Scheme 2). The accumulation of these intermediates indicated that there are some disturbances in the C–C bond cleavage process (particularly 19→25). The lack of the acetyl group at 4-O′-position of the sugar residue and the missing 8-vinyl side chain in 31 suggests that the proposed acetyltransferase RavW and CYP450 RavOIII do not function in the heterologous host for unclear reasons. For example, these enzymes are functionally inactive, and the real functional enzymes are encoded by genes located somewhere outside of the sequenced region. Alternatively, the expression of RavOIII and RavW might be negatively regulated in the heterologous host. To check the first hypothesis, a RavOIII expression construct (pRavOIII) was generated based on plasmid pEM4, in which the inserted ravOIII gene is expressed under the control of the erythromycin resistance gene promoter ermEp*. This construct was used to complement the S. lividans TK24 (cosG9B3-GilOIII− ) mutant that harbors the entire GV cluster except gilOIII, and produces GE (6) along with other biosynthetic shunt products, such as rabelomycin and homorabelomycin. The expression of RavOIII in this GilOIII− mutant resulted in the complete reconstitution of the GV production (Figure S2); this demonstrates that ravOIII encodes a functionally active P450 enzyme, which is responsible for the vinyl side chain formation of RV. Therefore, we still suspect that the expression of RavOIII as well as RavW might have been negatively regulated during heterologous expression of cosRav32, by unknown mechanisms. However, because both P450 enzymes and acetyltransferases are commonly found in many organisms including Streptomycetes, various regulation mechanisms might exist. Further investigation, including the in vitro characterization of these enzymes, might help to resolve these ambiguities, but so far all our attempts to prepare RavW in vitro repeatedly failed due to the insolubility of the enzyme.
Cross complementation experiments using chrysomycin monooxygenases
Various attempts to heterologously produce CM in S. lividans TK24 through the expression of the cosmid cosChry1-1 were unsuccessful. Unfortunately, the CM producer strain S. albaduncus AD0819 turned out to be recalcitrant to direct manipulations of its CM biosynthetic genes. Therefore, cross-complementations of GV mutants with homologous CM genes were carried out to verify that the aforementioned cluster is responsible for CM biosynthesis. S. lividans TK24 (cosG9B3-GilOII−, cosG9B3-GilOIV− and cosG9B3-GilOIII−) mutants that are blocked in GV biosynthesis at various stages were complemented with overexpression plasmids containing their CM counterparts (pChryOII, pChryOIV and pChryOIII, pEM4-based plasmids containing the chry oxygenase genes under ermEp* promotion; Figure S2). All complementations successfully restored GV production, thus providing indirect evidence for their involvement in CM biosynthesis.
Conclusions
The above-mentioned experiments clearly proved that we cloned the gene clusters of the gilvocarcin-type anticancer antibiotics ravidomycin V and chrysomycin V; this significantly expands the repertoire of genes involved in gilvocarcin class compound biosyntheses that can be used to generate novel analogues through combinatorial biosynthesis.
The assignments of the product function of critical and related oxygenase-encoding genes were not only based on sequence data, but also on heterologous complementation (for example, of ravOIII, ravGT, chryOII, chryOIV, chryOIII). Furthermore, it was shown that P450 enzyme RavOIII, responsible for the generation of the vinyl side chain in RV, can be inhibited by the P450-inhibitor ancymidol, and that the aminosugar transferase RavGT possesses significant flexibility towards its sugar donor substrate. The experiments also yielded new gilvocarcin-type drugs, such as 31 and 32.
Experimental Section
Construction of plasmids and DNA manipulation
Plasmids and bacterial strains used in this study are summarized in Table S3. Routine DNA manipulations such as cloning, restriction analysis, ligation, transformation etc. were carried out following the standard protocols for E. coli[95] and Streptomyces.[90] High fidelity pfu DNA polymerase was used to amplify various DNA fragments throughout the experiments. PCR products were cloned into PCR-Blunt-II-TOPO cloning vectors and sequenced to confirm that no mutation had occurred during PCR amplification. For complementation studies, ravOIII, chryOIV, chryOIII, chryOII were amplified and ligated into the pEM4 vector at the XbaI and EcoRI sites, whereas ravGT was ligated at the NdeI and EcoRI sites of pUWL201PW.
Bacterial strains and culture conditions
Various strains of E. coli were cultured at 37 °C in Lauria Bertani (LB) broth or on the LB-agar medium supplemented with various antibiotics (kanamycin, 50 μg mL−1; chloramphenicol, 25 μg mL−1; apramycin, 50 μg mL−1 and ampicillin, 50 μg mL−1) whenever necessary. Streptomyces strains were grown on M2-agar or in SG liquid medium for the metabolites analyses as reported previously.[34] MS-agar medium was used for the transfer of the cosmids from E. coli into Streptomyces following the standard protocol.[90] Antibiotics (apramycin, 50 μg mL−1 and thiostrepton 25 μg mL−1) were supplemented in the medium whenever necessary.
Construction and screening of genomic cosmid libraries
Genomic DNA of S. ravidus and S. albaduncus were isolated following the standard protocol. The genomic DNA was partially digested with Sau3AI, dephosphorylated with calf intestinal phosphatase (CIP) and ligated to the pO J446 vector digested with BamHI and HpaI. The ligation sample was transduced into E. coli XL1-Blue MRF using Gigapack III XL packaging extract (Stratagene). More than 2300 colonies were obtained for each library. Two sets of degenerate primers (a standard KS-probe-for and KS-probe-rev, and a newly designed DH-probe-for and DH-probe-rev) were used to amplify the internal nucleotide sequences of ketoacyl synthase (KSα) and NDP-glucose-4,6-dehydratase from the genomic DNA of S. ravidus and S. albaduncus. The amplified KSα fragments were labeled with DIG and used to screen positive colonies. The positive colonies were further screened with the NDP-glucose-4,6-dehydratase probe. Colony hybridization and southern blot analyses revealed four and three different cosmid clones for S. ravidus and S. albaduncus genomic libraries, respectively. A probe suited to detect a type II PKS-associated ketoreductase gene (gilF homologue) was also constructed (CM_cons_F1 and CM_cons_R1) to probe for cosmids that contained chryF. Three positively hybridizing colonies yielded cosmids cosChryF1, cosChryF2, and cosChryF3, cosChryF2 with the most overlap with cosChry1–1 according to the restriction analysis map. Thus, cosmids cosRav32, cosChry1–1, and cosChryF2 were selected for sequencing. All primers used in this study are summarized in Table S4.
Sequencing and annotation of the gene clusters
Sequencing of the cosmids was carried out involving a standard shotgun approach using the HyperMu™ MuA Transposase kit (Epicentre) following the reported protocol[50–53] used by the sequencing facilities of the Advanced Genetic Technologies Center (AGTC) located in the College of Agricultural Sciences of the University of Kentucky (http://www.uky.edu/Centers/AGTC/). Phred/Phrap/Consed software package (http://www.phrap.org.) was used to process and assemble raw sequence data into larger contigs. The small gaps between contigs were filled by primer walking. Frame plot (http://www0.nih.go.jp/~jun/cgi-bin/frameplot.pl) and ORF finder (http://www.ncbi.nlm.nih.gov/projects/gorf/) were used to assign the open reading frames. Functional assignments of ORFs were performed through database comparison using various BLAST search tools on the server of the National Center for Biotechnology Information, Bethesda, Maryland, USA (http://www.ncbi.nlm.nih.gov).
Fermentation and isolation of metabolites
Streptomyces strains were grown on MS agar for 6 days at 30 °C. An agar chunk full of bacterial spores was inoculated into SG liquid (100 mL) medium in a Erlenmeyer flask (500 mL) and incubated for three days at 30 °C and 200 rpm to prepare seed cultures. For analyses of metabolites, the liquid seed culture (3%) was used to inoculate larger-scale SG liquid medium cultures (up to 4 L) and grown for five days. The cultures were harvested through centrifugation to obtain culture broth and pellet fractions. The broth was twice extracted with equal volumes of ethyl acetate. The organic layer was combined and dried in a rotary evaporator. Such prepared extracts were dissolved in methanol and filtered through syringe filter (0.2 μm) prior to High Performance Liquid Chromatography-Mass Spectrometry (HPLC-MS) analysis. The pellet fraction was re-suspended in acetone and sonicated for 10 min. The acetone extract was collected through centrifugation and dried under reduced pressure. The prepared extract was dissolved in methanol and subjected to HPLC-MS analysis.
The production of chrysomycins, ravidomycins and gilvocarcins by wild/mutant strains were analyzed by the aforementioned protocol. The production scale was increased to 4 L when isolating compounds for structural analysis. The culture was harvested through centrifugation, and the cell pellets were extracted with acetone as described above. The extract was combined with the broth and loaded onto a reverse phase silica (Rp18, 200 g) column (5 × 65 cm) pre-equilibrated with deionized water. The effluent was discarded and the column was washed with acetonitrile (1 L of 10%, in water). Elution was carried out using an increasing gradient of acetonitrile (20%, 40%, 60%, 80% and 100%, 1 L each). The fraction containing target compounds were dried in a lyophilizer. Final purification of the metabolites was carried out in preparative-scale HPLC (columns type and size, see below). The purity and authenticity of the compound was confirmed through 1H and 13C NMR analyses. A linear gradient of acetonitrile and acidified water (solvent A = 0.1% formic acid in H2O; solvent B = acetonitrile; 0–15 min 25% B to 100% B; 16–24 min 100% B; 25–26 min 100% to 25% B; 27–29 min 25% B) was used to separate compounds. Sun-Fire prepC18 column (19×150 mm, 5 μm) and Symmetry C18 (4.6 250 mm, 5 μm) columns were used for preparative and analytical scale separations, respectively. Flow rate was maintained to 10 mL min−1 and 1.0 mL min−1 for preparative and analytical scale separations, respectively. Micromass ZQ 2000 (Waters) equipped with HPLC (Waters alliance 2695 model) and photodiode array detector (Waters 2996) were used to analyze the compounds. Atmospheric pressure chemical ionization (APCI) and Electrospray ionization (ESI) probes were used to detect molecular ions. Electron ionization-High resolution mass (EI-HR-MS) of compounds were acquired using a high resolution electron impact (EI) ionization mass spectrometer. Data were recorded at 25 eV on a JEOL JMS-700T MStation at the University of Kentucky mass spectrometry facility. Samples were introduced through a heatable direct probe inlet. Perfluorokerosene (pfk) was used to produce reference masses.
To isolate deacetylravidomycin E, fully grown S. ravidus seed culture (20 mL) was cultured in SG medium (5 flasks × 100 mL) and of ancymidol solution (100 μL, in DMSO, 2 mm, final concentration) was added into the culture. Only DMSO was used in the control flask. The fermentation was carried out for 6 days and the metabolites were analyzed by HPLC-MS. Isolation and purification of 4′-O-deacetylravidomycin E (31) was carried out according to the aforementioned protocol. The structure of 31 was determined through comparative analysis of its experimental NMR data and reported NMR data of 2 (see the Supporting Information).
Supplementary Material
Acknowledgements
This work was supported by the US National Institutes of Health (grant CA 102 102 to J.R.). The University of Kentucky Advanced Genetic Technologies Center (AGTC) and Center for Mass Spectrometry are acknowledged for their services.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.200900673.
References
- 1.Sehgal SN, Czerkawski H, Kudelski A, Pandev K, Saucier R, Vezina C. J. Antibiot. 1983;35:355. doi: 10.7164/antibiotics.36.355. [DOI] [PubMed] [Google Scholar]
- 2.Matson JA, Rose WC, Bush JA, Myllymaki R, Bradner WT, Doyle TW. J. Antibiot. 1989;42:1446. doi: 10.7164/antibiotics.42.1446. [DOI] [PubMed] [Google Scholar]
- 3.Weiss U, Yoshihira K, Highet RJ, White RJ, Wei TT. J. Antibiot. 1982;35:1194. doi: 10.7164/antibiotics.35.1194. [DOI] [PubMed] [Google Scholar]
- 4.Yamashita N, Shin-ya K, Furihata K, Hayakawa Y, Seto H. J. Antibiot. 1998;51:1105. doi: 10.7164/antibiotics.51.1105. [DOI] [PubMed] [Google Scholar]
- 5.Nakajima S, Kojiri K, Suda H, Okanishi M, Antibiot J. 1991;44:1061. doi: 10.7164/antibiotics.44.1061. [DOI] [PubMed] [Google Scholar]
- 6.Kojiri K, Arakawa H, Satoh F, Kawamura K, Okura A, Suda H, Okanishi M. J. Antibiot. 1991;44:1054. doi: 10.7164/antibiotics.44.1054. [DOI] [PubMed] [Google Scholar]
- 7.Nakashima T, Fujii T, Sakai K, Tomohiro S, Kumagai H, Yoshioka T. United States Patent 6,030,951, Mercian Corporation (JP), United States. 2000. [Google Scholar]
- 8.Li YQ, Huang XS, Ishida K, Maier A, Kelter G, Jiang Y, Peschel G, Menzel KD, Li MG, Wen ML, Xu LH, Grabley S, Fiebig HH, Jiang CL, Hertweck C, Sattler I. Org. Biomol. Chem. 2008;6:3601. doi: 10.1039/b808633h. [DOI] [PubMed] [Google Scholar]
- 9.Brazhnikova MG, Kudinova MK, Kulyaeva VV, Potapova NP, Ponomarenko VI. Antibiotiki. 1977;22:967. [PubMed] [Google Scholar]
- 10.Strelitz F, Flon H, Ashenov IN. J. Bacteriol. 1955;69:280. doi: 10.1128/jb.69.3.280-283.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenstein M, Monji T, Yeung R, Maiese WM, White RJ. Antimicrob. Agents Chemother. 1986;29:861. doi: 10.1128/aac.29.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh K. J. Antibiot. 1984;37:71. doi: 10.7164/antibiotics.37.71. [DOI] [PubMed] [Google Scholar]
- 13.Lorico A, Long BH. Eur. J. Cancer. 1993;29 A:1985. doi: 10.1016/0959-8049(93)90459-s. [DOI] [PubMed] [Google Scholar]
- 14.Fischer C, Lipata F, Rohr J. J. Am. Chem. Soc. 2003;125:7818. doi: 10.1021/ja034781q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knobler RM, Radlwimmer FB, Lane MJ. Nucleic Acids Res. 1993;21:3920. doi: 10.1093/nar/20.17.4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumoto A, Hanawalt PC. Cancer Res. 2000;60:3921. [PubMed] [Google Scholar]
- 17.Rakhit S, Eng C, Baker H, Singh K. J. Antibiot. 1983;36:1490. doi: 10.7164/antibiotics.36.1490. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Li S, Li X. Recent Pat. Nanotechnol. 2009 doi: 10.2174/187221009789177803. in press. [DOI] [PubMed] [Google Scholar]
- 19.Fang JY. Chang Gung Med. J. 2006;29:358. [PubMed] [Google Scholar]
- 20.Lassalle V, Ferreira ML. Macromol. Biosci. 2007;7:767. doi: 10.1002/mabi.200700022. [DOI] [PubMed] [Google Scholar]
- 21.Hosoya T, Takashiro E, Matsumoto T, Suzuki K. J. Am. Chem. Soc. 1994;116:1004. [Google Scholar]
- 22.Futagami S, Ohashi Y, Imura K, Hosoya T, Ohmori K, Matsumoto T, Suzuki K. Tetrahedron Lett. 2000;41:1063. [Google Scholar]
- 23.Matsumoto T, Hosoya T, Suzuki K. J. Am. Chem. Soc. 1992;114:3568. [Google Scholar]
- 24.Hsu DS, Matsumoto T, Suzuki K. Synlett. 2005:801. [Google Scholar]
- 25.Griffith BR, Langenhan JM, Thorson JS. Curr. Opin. Biotechnol. 2005;16:622. doi: 10.1016/j.copbio.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Langenhan JM, Griffith BR, Thorson JS. J. Nat. Prod. 2005;68:1696. doi: 10.1021/np0502084. [DOI] [PubMed] [Google Scholar]
- 27.Baig I, Kharel M, Kobylyanskyy A, Zhu LL, Rebets Y, Ostash B, Luzhetskyy A, Bechthold A, Fedorenko VA, Rohr J. Angew. Chem. 2006;118:8006. doi: 10.1002/anie.200603176. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2006;45:7842. [Google Scholar]
- 28.Dürr C, Hoffmeister D, Wohlert SE, Ichinose K, Weber M, Von Mulert U, Thorson JS, Bechthold A. Angew. Chem. 2004;116:3022. doi: 10.1002/anie.200453758. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2004;43:2962. [Google Scholar]
- 29.Liu T, Kharel MK, Zhu L, Bright SA, Mattingly C, Adams VR, Rohr J. ChemBioChem. 2009;10:278. doi: 10.1002/cbic.200800348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ichinose K, Ozawa M, Itou K, Kunieda K, Ebizuka Y. Microbiol.-SGM. 2003;149:1633. doi: 10.1099/mic.0.26310-0. [DOI] [PubMed] [Google Scholar]
- 31.Carter GT, Fantini AA, James JC, Borders DB, White RJ. Tetrahedron Lett. 1984;25:255. [Google Scholar]
- 32.Takahashi K, Tomita F. J. Antibiot. 1983;36:1531. doi: 10.7164/antibiotics.36.1531. [DOI] [PubMed] [Google Scholar]
- 33.Carter GT, Fantini AA, James JC, Borders DB, White RJ. J. Antibiot. 1985;38:242. doi: 10.7164/antibiotics.38.242. [DOI] [PubMed] [Google Scholar]
- 34.Liu T, Fischer C, Beninga C, Rohr J. J. Am. Chem. Soc. 2004;126:12262. doi: 10.1021/ja0467521. [DOI] [PubMed] [Google Scholar]
- 35.Liu T, Kharel MK, Fischer C, McCormick A, Rohr J. ChemBioChem. 2006;7:1070. doi: 10.1002/cbic.200600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kharel MK, Zhu LL, Liu T, Rohr J. J. Am. Chem. Soc. 2007;129:3780. doi: 10.1021/ja0680515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu T, Kharel MK, Zhu L, Bright S, Rohr J. ChemBioChem. 2009;10:278. doi: 10.1002/cbic.200800348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menzella HG, Reeves CD. Curr. Opin. Microbiol. 2007;10:238. doi: 10.1016/j.mib.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Van Lanen SG, Shen B. Drug Discovery Today Technol. 2006;3:285. doi: 10.1016/j.ddtec.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 40.Rix U, Fischer C, Remsing LL, Rohr J. Nat. Prod. Rep. 2002;19:542. doi: 10.1039/b103920m. [DOI] [PubMed] [Google Scholar]
- 41.Baltz RH. Nat. Biotechnol. 2006;24:1533. doi: 10.1038/nbt1265. [DOI] [PubMed] [Google Scholar]
- 42.Wilkinson B, Micklefield J. Nat. Chem. Biol. 2007;3:379. doi: 10.1038/nchembio.2007.7. [DOI] [PubMed] [Google Scholar]
- 43.Salas JA, Mendez C. Trends Microbiol. 2007;15:219. doi: 10.1016/j.tim.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Salas JA, Mendez C. Curr. Opin. Chem. Biol. 2009;13:152. doi: 10.1016/j.cbpa.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 45.Weissman KJ, Leadlay PF. Nat. Rev. Microbiol. 2005;3:925. doi: 10.1038/nrmicro1287. [DOI] [PubMed] [Google Scholar]
- 46.Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Nat. Prod. Rep. 2007;24:162. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- 47.Rohr J, Hertweck C. In: Comprehensive Natural Products II–Chemistry and Biology, Vol. 2. Mander L, Liu H.-w., editors. Elsevier; Amsterdam: 2010. [Google Scholar]
- 48.Tang Y, Lee TS, Khosla C. PLoS Biol. 2004;2:e31. doi: 10.1371/journal.pbio.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Decker H, Gaisser S, Pelzer S, Schneider P, Westrich L, Wohlleben W, Bechthold A. FEMS Microbiol. Lett. 1996;141:195. doi: 10.1111/j.1574-6968.1996.tb08384.x. [DOI] [PubMed] [Google Scholar]
- 50.Kobryn K, Watson MA, Allison RG, Chaconas G. Mol. Cell. 2002;10:659. doi: 10.1016/s1097-2765(02)00596-8. [DOI] [PubMed] [Google Scholar]
- 51.Butterfield YSN, Marra MA, Asano JK, Chan SY, Guin R, Krzywinski MI, Lee SS, MacDonald KWK, Mathewson CA, Olson TE, Pandoh PK, Prabhu AL, Schnerch A, Skalska U, Smailus DE, Stott JM, Tsai MI, Yang GS, Zuyderduyn SD, Schein JE, Jones SJM. Nucleic Acids Res. 2002;30:2460. doi: 10.1093/nar/30.11.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Savilahti H, Rice PA, Mizuuchi K. EMBO J. 1995;14:4893. doi: 10.1002/j.1460-2075.1995.tb00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mizuuchi K. Cell. 1983;35:785. doi: 10.1016/0092-8674(83)90111-3. [DOI] [PubMed] [Google Scholar]
- 54.Bililign T, Hyun C-G, Williams JS, Czisny AM, Thorson JS. Chem. Biol. 2004;11:959. doi: 10.1016/j.chembiol.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 55.Crawford JM, Vagstad AL, Ehrlich KC, Townsend CA. Bioorg. Chem. 2008;36:16. doi: 10.1016/j.bioorg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerlitz M, Meurer G, Wendt-Pienkowski E, Madduri K, Hutchinson CR. J. Am. Chem. Soc. 1997;119:7392. [Google Scholar]
- 57.Moore BS, Hertweck C. Nat. Prod. Rep. 2002;19:70. doi: 10.1039/b003939j. [DOI] [PubMed] [Google Scholar]
- 58.Xu Z, Schenk A, Hertweck C. J. Am. Chem. Soc. 2007;129:6022. doi: 10.1021/ja069045b. [DOI] [PubMed] [Google Scholar]
- 59.Zhang W, Ames BD, Tsai SC, Tang Y. Appl. Environ. Microbiol. 2006;72:2573. doi: 10.1128/AEM.72.4.2573-2580.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Räty K, Kantola J, Hautala A, Hakala J, Ylihonko K, Mantsala P. Gene. 2002;293:115. doi: 10.1016/s0378-1119(02)00699-6. [DOI] [PubMed] [Google Scholar]
- 61.Hutchinson CR. Chem. Rev. 1997;97:2525. doi: 10.1021/cr960022x. [DOI] [PubMed] [Google Scholar]
- 62.Luzhetskyy A, Mayer A, Hoffmann J, Pelzer S, Holzenkamper M, Schmitt B, Wohlert SE, Vente A, Bechthold A. ChemBioChem. 2007;8:599. doi: 10.1002/cbic.200600529. [DOI] [PubMed] [Google Scholar]
- 63.Han L, Yang KQ, Ramalingam E, Mosher RH, Vining LC. Microbiology. 1994;140:3379. doi: 10.1099/13500872-140-12-3379. [DOI] [PubMed] [Google Scholar]
- 64.Fernández-Moreno MA, Martinez E, Boto L, Hopwood DA, Malpartida F. J. Biol. Chem. 1992;267:19 278. [PubMed] [Google Scholar]
- 65.Bibb MJ, Biro S, Motamedi H, Collins JF, Hutchinson CR. EMBO J. 1989;8:2727. doi: 10.1002/j.1460-2075.1989.tb08414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bibb MJ, Sherman DH, Omura S, Hopwood DA. Gene. 1994;142:31. doi: 10.1016/0378-1119(94)90351-4. [DOI] [PubMed] [Google Scholar]
- 67.Marti T, Hu Z, Pohl NL, Shah AN, Khosla C. J. Biol. Chem. 2000;275:33443. doi: 10.1074/jbc.M006766200. [DOI] [PubMed] [Google Scholar]
- 68.Tang Y, Lee TS, Kobayashi S, Khosla C. Biochemistry. 2003;42:6588. doi: 10.1021/bi0341962. [DOI] [PubMed] [Google Scholar]
- 69.Summers RG, Ali A, Shen B, Wessel WA, Hutchinson CR. Biochemistry. 1995;34:9389. doi: 10.1021/bi00029a015. [DOI] [PubMed] [Google Scholar]
- 70.Arthur CJ, Szafranska A, Evans SE, Findlow SC, Burston SG, Owen P, Clark-Lewis I, Simpson TJ, Crosby J, Crump MP. Biochemistry. 2005;44:15414. doi: 10.1021/bi051499i. [DOI] [PubMed] [Google Scholar]
- 71.Liu T, Fischer C, Beninga C, Rohr J. J. Am. Chem. Soc. 2004;126:12262. doi: 10.1021/ja0467521. [DOI] [PubMed] [Google Scholar]
- 72.Beam MP, Bosserman MA, Noinaj N, Wehenkel M, Rohr J. Biochemistry. 2009;48:4476. doi: 10.1021/bi8023509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang J, Tetzlaff CN, Takamatsu S, Iwatsuki M, Komatsu M, Ikeda H, Cane DE. Biochemistry. 2009;48:6431. doi: 10.1021/bi900766w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kamerbeek NM, Janssen DB, van Berkel WJH, Fraaije MW. Adv. Synth. Catal. 2003;345:667. [Google Scholar]
- 75.van Berkel WJH, Kamerbeek NM, Fraaije MW. J. Biotechnol. 2006;124:670. doi: 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 76.Kharel MK, Pahari P, Lian H, Rohr J. ChemBioChem. 2009;10:1305. doi: 10.1002/cbic.200900130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karray F, Darbon E, Oestreicher N, Dominguez H, Tuphile K, Gagnat J, Blondelet-Rouault MH, Pernodet C. Microbiol-Sgm. 2007;153:4111. doi: 10.1099/mic.0.2007/009746-0. [DOI] [PubMed] [Google Scholar]
- 78.Melançon CE, Hong L, White JA, Liu YN, Liu HW. Biochemistry. 2007;46:577. doi: 10.1021/bi061907y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pfoestl A, Hofinger A, Kosma P, Messner P. J. Biol. Chem. 2003;278:26410. doi: 10.1074/jbc.M300858200. [DOI] [PubMed] [Google Scholar]
- 80.Hong L, Zhao Z, Melancon CE, Zhang H, Liu HW. J. Am. Chem. Soc. 2008;130:4954. doi: 10.1021/ja0771383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Burgie ES, Holden HM. Biochemistry. 2008;47:3982. doi: 10.1021/bi800063j. [DOI] [PubMed] [Google Scholar]
- 82.Chen H, Yamase H, Murakami K, Chang CW, Zhao L, Zhao Z, Liu HW. Biochemistry. 2002;41:9165. doi: 10.1021/bi020245j. [DOI] [PubMed] [Google Scholar]
- 83.Pickens LB, Tang Y. Metab. Eng. 2009;11:69. doi: 10.1016/j.ymben.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang W, Watanabe K, Cai X, Jung ME, Tang Y, Zhan J. J. Am. Chem. Soc. 2008;130:6068. doi: 10.1021/ja800951e. [DOI] [PubMed] [Google Scholar]
- 85.Schenk A, Xu Z, Pfeiffer C, Steinbeck C, Hertweck C. Angew. Chem. 2007;119:7165. doi: 10.1002/anie.200702033. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2007;46:7035. [Google Scholar]
- 86.Luzhetskyy A, Taguchi T, Fedoryshyn M, Durr C, Wohlert SE, Novikov V, Bechthold A. ChemBioChem. 2005;6:1406. doi: 10.1002/cbic.200500018. [DOI] [PubMed] [Google Scholar]
- 87.Künzel E, Faust B, Oelkers C, Weissbach U, Bearden DW, Weitnauer G, Westrich L, Bechthold A, Rohr J. J. Am. Chem. Soc. 1999;121:11058. [Google Scholar]
- 88.Fischbach MA, Lin HN, Liu DR, Walsh CT. Proc. Natl. Acad. Sci. USA. 2005;102:571. doi: 10.1073/pnas.0408463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu Z, Jakobi K, Welzel K, Hertweck C. Chem. Biol. 2005;12:579. doi: 10.1016/j.chembiol.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 90.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics, The John Innes Foundation, Norwich, UK. 2000. [Google Scholar]
- 91.Oikawa H, Aihara Y, Ichihara A, Sakamura S. Biosci. Biotechnol. Biochem. 1992;56:684. doi: 10.1271/bbb.56.684. [DOI] [PubMed] [Google Scholar]
- 92.Rademacher W. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:501. doi: 10.1146/annurev.arplant.51.1.501. [DOI] [PubMed] [Google Scholar]
- 93.Chen YH, Wang CC, Greenwell L, Rix U, Hoffmeister D, Vining LC, Rohr JR, Yang KQ. J. Biol. Chem. 2005;280:22508. doi: 10.1074/jbc.M414229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rix U, Wang CC, Chen YH, Lipata FM, Rix LLR, Greenwell LM, Vining LC, Yang KQ, Rohr J. ChemBioChem. 2005;6:838. doi: 10.1002/cbic.200400395. [DOI] [PubMed] [Google Scholar]
- 95.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual 2001. Cold Spring Harbor Laboratory Press; New York: [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.