Abstract
There has been an explosion of interest in the human B cell response to HIV infection of late. Recent advances in techniques for isolation of human antibodies and antibody secreting cell lines have facilitated a rapid expansion in the number of antibodies available for study. Early analysis of these repertoires reveals interesting features of the HIV-specific antibody response. HIV-specific repertoires exhibit a high level of clonality in circulating cells, and high levels of somatic mutations within the antibody variable gene segments. It appears that many if not most antibodies in circulation bind to virus envelope conformations that are found only in complex oligomeric structures on virion particles or virus-like particles. The rapid isolation of large panels of novel human neutralizing antibodies promises to reveal new insights into the fundamental principles underlying antibody-mediated neutralization of HIV.
Introduction
The recent failure of the STEP study, which tested the efficacy of a vaccine able to induce HIV-specific T cell responses, suggests that vaccine-induced neutralizing antibody responses also are likely to be required for protection [1]. There has been a recent swell of interest in the role of antibodies (Abs) in controlling HIV infection in humans [2, 3]. Despite a large body of research on HIV immunity, relatively little is understood about the fundamental principles underlying inhibition of HIV by human Abs. Several groups have identified isolate-specific Abs that inhibit viral replication, however monoclonal antibodies able to neutralize diverse isolates are rare, and the molecular basis for broadly neutralizing antibody (Ab) responses to HIV is not completely clear [4, 5].
Obstacles to Study of HIV B Cell Responses
Antigenic variability
One of the principal obstacles to studying antibody repertoires in humans is the high level of variability of virus sequences, with concomitant variation in epitope structure, viral susceptibility to neutralization, and viral fitness that all contribute to the protean nature of the virus and its antigens. Genotypic and phenotypic variation in virus populations exists in any one person at all times, and accumulates within individuals over time. In a population of infected individuals, the potential virus variation is enormous. The reagents that are in common use for immune studies typically were derived from representative individuals in the past and have been collected into panels of virus strains that represent only a portion of the spectrum of variation known to exist in field strains. These reagents, such as the envelope (Env) proteins from the NIH AIDS Reagent Repository, may or may not be appropriate for the study of the repertoire in any one individual. It is crucial and more biologically relevant to study the repertoire of response to an individual's native circulating viral quasi-species.
Lack of structural information
Significant progress has been made in recent years with cryo-electron microscopy, crystallization studies, and other techniques for determining the structure of Env and antibody-Env complexes. Nevertheless, we still do not have a crystal clear appreciation of the structure of Env trimers in the virion membrane. There are suggestions that these protein trimers exhibit features that contribute to variation in structure, due to lack of a high level of stability of the trimeric head, high levels of glycosylation that may be variable, and the frequent occurrence of defective or shed trimers. The very reasons that this trimer is difficult to resolve structurally may correspond to features that contribute to the difficulty in developing broadly neutralizing antibodies.
Polyspecificity
Some antibodies that react with HIV virions or infected cells may overlap with autoreactive antibodies that bind to human proteins. The molecular basis for these interactions is still under study. First, a number of host proteins incorporate into virion particles as they bud from the cell, therefore it is expected that host proteins on the surface of virions might bind to autoreactive B cells or antibodies. Second, it appears that in some cases autoreactive antibodies acquire additional specificity for HIV proteins, resulting in antibodies that bind both host and viral proteins, although typically with differing affinities. The physiologic role of such cross-reactivity or polyspecificity is not clear at this time. However, interpretation of HIV-specific B cell repertoires ultimately must account for these complex specificities.
Emerging Techniques for Study of Human B Cell Repertoires
A large number of new or improved techniques for isolation of human antibodies has emerged in the last decade, and these techniques are being focused rapidly on the HIV antibody problem. Phage display of single chain or Fabs in large libraries has been used to isolate important and interesting antibodies from mRNAs from human B cells [6-10]. Such antibodies contain randomly associated heavy and light chains, so the isolated antibodies may or may not have existed in the donor. More recently, techniques that maintain the natural pairing of heavy and light chains have been developed. EBV transformation has long been used in the field, but was recently greatly improved with the finding that EBV transformation in the presence of CpG, through activation of survival and proliferation signals via Toll-like receptor 9, is much more efficient [10, 11]. This technique yields polyclonal collections of transformed lines that can be screened for antigen specificity, however cloning stable EBV lines can be challenging. We have combined this method with electrofusion to myeloma partners to yield stable human hybridomas in some cases [12, 13]. Isolation of antibody genes from single antigen-selected cells that are expanded in culture [14, 15] or cloned directly [16, 17] has yielded large panels of antibody sequences that are virus-specific, including HIV-specific repertoires. Typically most of the cloned cells are memory cells, but other subsets of cells can be selected with the appropriate markers. An important challenge to these studies is the fact that most recombinant antigens do not fully recapitulate the native antigenic structure of the Env protein on the virion, so that antibody repertoires obtained from selection of B cells with altered antigens may be misleading. A variation of this technique is to isolate plasmablasts that are reacting to a recent immunization or acute infection, with the presumption that blasting cells are mostly specific for the antigen present in the recent immunization [18]. It is not yet clear if the specificity of plasmablasts will be high enough for the antigens of chronic infections like HIV. Simple analysis of the antibody gene repertoire without Ab expression is of interest, but without expression of the antibodies and confirmation of binding to virions these studies are difficult to interpret.
In our studies of antigen-specific sorting of B cells, 10% at most of the Abs secreted by sorted cells can be shown definitively to bind to viral particles after expansion in culture (see Figure 1 for overview). This suggests that while physical sorting of single antigen-specific cells enriches their frequency, not every sorted cell produces antibodies able to bind HIV. Care is needed when drawing conclusions from studies on repertoire usage since the physical sorting process results in potentially a high level of non-specific binding. The way to overcome this concern is to express the antibody genes and produce antibody proteins for in vitro studies from the isolated genes.
Figure 1. Overview of method for generation of HIV-specific human antibodies.
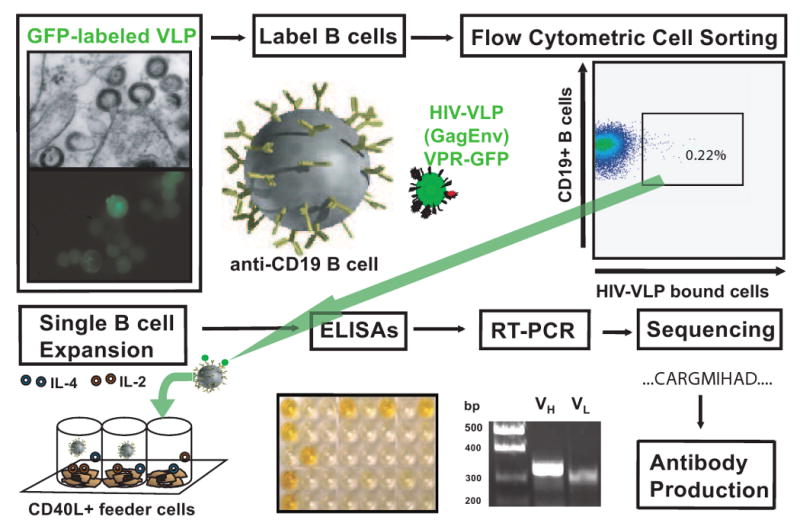
In the upper portion of the figure, HIV-VLPs are created in inducible cell lines and are shown here budding from the cell surface (by electron or fluorescent microscopy). B cells then are labeled with the resulting fluorescent HIV-VLPs and are shown here specifically labeling a subset of HIV-specific B cells from a HIV+ subject. These VLP-labeled B cells then are sorted into individual culture wells of a 96-well plate containing a stimulator cell line and cytokine mixture (shown in bottom portion of figure). ELISAs are used to identify clonal B cell populations HIV-specific immunoglobulins. Cells are collected from wells of interest, RNA isolation is performed and RT-PCR is used to clone the corresponding antibody variable gene segments of the heavy and light chains. Antibody genes of interest are sequenced and cloned into a plasmid based antibody expression system.
Critical Nature of the Antigen
It has been proposed that conformationally dependent trimer-specific binding of Abs is a critical component of viral neutralization [7, 19]. Conformational epitope specific Abs have been raised in mice by oligomeric vaccination [20], however these studies were performed with oligomeric forms of uncleaved gp140 constructs that exhibited heterogeneity in protein form. There have been reports of a broad neutralizing response following vaccination of oligomeric forms of gp140. Unfortunately, the reported breadth of activity consisted of mostly low serum titers, often with titers of <1:100, and not all gp140 trimeric Env preparations induce significant titers of neutralizing Abs able to recognize conformational epitopes [21]. Additionally, virus-like particles (VLPs) bearing disulfide-stabilized functional trimers (SOS-VLPs) and cleavage-site mutated Env VLPs predominantly induced Ab responses against non-functional forms of Env and not against the trimeric Env [22].
Other studies have shown that not all trimerized constructs hide irrelevant epitopes completely, such as studies characterizing a panel of 11 mAbs isolated from mice immunized with gp140 trimerized constructs [23]. The anti-gp41 Abs in this study exhibited improved binding in the context of the trimerized gp140 versus the monomeric gp140. These data imply that there is a conformational change upon trimerization with this construct in the gp41 region. However, this effect was not noted for the Abs that mapped to gp120 epitopes. The trimer in this study contains a mutation in the protease cleavage site between gp120 and gp41 [24], and such mutations may interfere with formation of the native epitope conformation during trimerization. The association of gp120 and gp41 is unstable in mature native virions [25, 26]. Many groups have utilized proteolytic cleavage site mutations to stabilize this interaction in their oligomeric or lipid-associated Env trimers [20, 22, 27, 28]. Similarly, immunization with HIV-1 Env gp140 trimerized constructs created by the addition of the trimerization motif of the T4 bacteriophage fibritin did not induce significant amounts of Abs to conformational epitopes [21].
Proteoliposomes also have been utilized to present a constrained gp160 trimer. Unfortunately, the proteoliposome platform of these constructs did not elicit a broad or as potent of a response as a GCN4-trimerized gp140 construct [29]. The neutralizing Abs that were raised were directed predominantly against non-V3 loop epitopes of gp120. Like other approaches, the proteoliposomes contain a proteolytic cleavage-site mutation in gp160 that may prevent them from representing the native trimerized Env conformation.
Single Cell Studies with Novel VLPs
In this section we summarize the findings from our own recent studies, which have focused on the use of novel VLPs for study of the human HIV-specific B cell repertoire [30]. It has been shown previously that Gag-only VLPs self assemble when the gag gene is expressed in an appropriate cell line. When gag and env genes are expressed together, VLPs are created in which Env protein self assembles in the lipid outer-membrane and forms trimeric Env (herein referred to as HIV-VLP), thus mimicking a natural HIV virion [25, 31]. A Vpr-GFP gene fusion construct that incorporates into the VLP was created in order to track the VLPs by fluorescence emission. Co-transfection with the gag and env genes produced fluorescently labeled HIV-VLPs. (Figure 1).
We have used a single B cell sorting method successfully in the past to clone human Ab genes that encode productive Abs reactive against other viruses such as rotavirus and respiratory syncytial virus [15, 18, 32-35]. For HIV repertoire studies we have labeled HIV-specific B cells with the purified fluorescently labeled HIV-VLPs. These studies have been successful in isolating a large number of antibody gene pairs for antibodies to HIV. Strikingly, the level of mutation in the variable genes encoding trimer-specific anti-HIV Abs is greater than that in randomly-selected B cells or other virus-specific B cells. Similar to recent reports on anti-HIV Abs [16], we have also found a dramatic number of circulating clonal populations of HIV-specific Abs in infected subjects.
An important aspect of this study was the development and usage of separate screening ELISAs to delineate specificity prior to cloning of the Ab gene segments. In this study, clonal supernatants were screened against: BaL strain-VLPs (which present gp120/gp41 as a trimer), BaL strain-gp120 monomer, or Gag-only VLPs that present all of the unrelated lipid membrane associated components that would be on the surface of the BaL-VLPs but have no gp41 or gp120. Minimal non-specific interaction was detected with the Gag-only VLPs in the flow cytometric sorting experiments
In order to study the Ab repertoire of HIV positive patients, GFP-labeled HIV-VLP reactive B cells were isolated from infected patient peripheral blood samples. These HIV-VLPs bound HIV-positive subject B cells but not those from HIV-negative controls (Figure 2). Generally, between 0.1% and 1% of circulating B cells from an HIV-positive subject bound HIV-VLP in the subjects we have studied to date, which were obtained from long-term non-progressors [30]. As part of the original characterization of these individuals, neutralization assays were performed to confirm the presence of BaL neutralizing circulating Abs, which is the parent strain of our HIV-VLP. Individual B cells were subjected to flow cytometric sorting into wells of a 96-well plate onto a feeder cell line. After culturing and expansion to small clones containing a few hundreds of cells, supernatants were screened for immunoglobulin production and antigen specificity by utilizing three distinct ELISAs (gp120 protein, HIV-VLP, or Gag-only VLP). Both the gp120 used in ELISA and the VLP Env used for sorting and in ELISA were from the clade B HIV virus isolate BaL.
Figure 2. Flow cytometric identification of HIV-specific B cells.
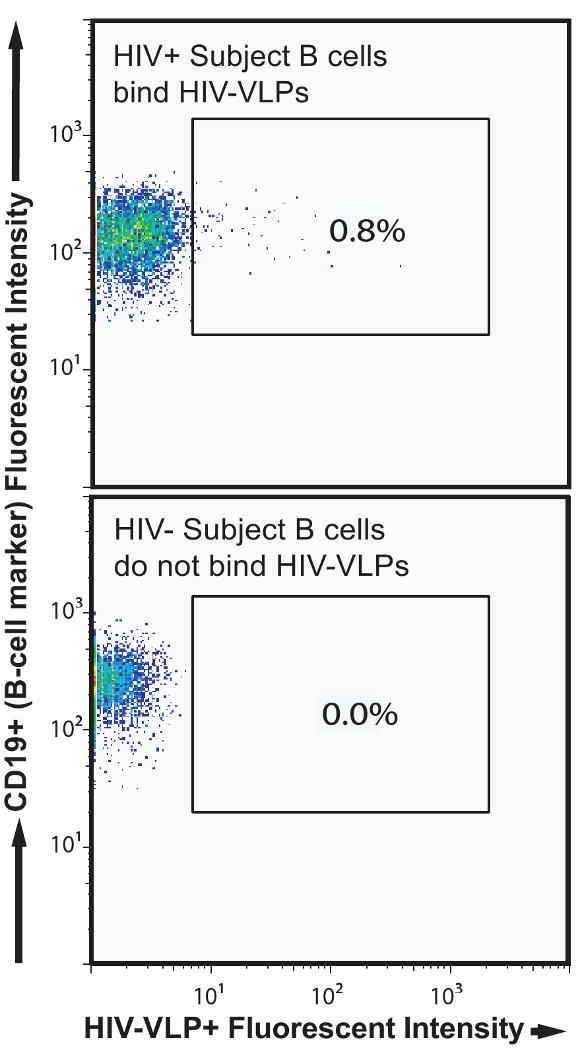
After paramagnetic bead enrichment of B cells from PBMCs, cells from an HIV+ subject (top flow panel) or an HIV- subject (bottom flow panel) were labeled separately with HIV-VLPs and appropriate cell markers. Detection of the anti-CD19 and HIV-VLP reagents are shown after prior gating (not shown) to exclude dead cells, CD3+ T cells, and CD14+ monocytes.
After supernatants were applied to the ELISA plates, HRP-conjugated goat anti-human Ig was used as a secondary antibody conjugate for detection. Photographs of ELISA plates from a representative assay were taken after acid stop (Figure 3A-C). Interestingly, most of the antibody responses detected were specific for the HIV-VLP ELISA, and not reactive with gp120 monomers nor reactive with the Gag-only VLP. Expanded HIV-VLP-reactive B cells were selected and RT-PCR was performed to clone the variable regions of the heavy and light chains of the expressed Abs. We have isolated a large number of heavy and light chain Ab sequences from over anti-HIV B cell clones.
Figure 3. Complex binding patterns of antibodies to HIV antigens.
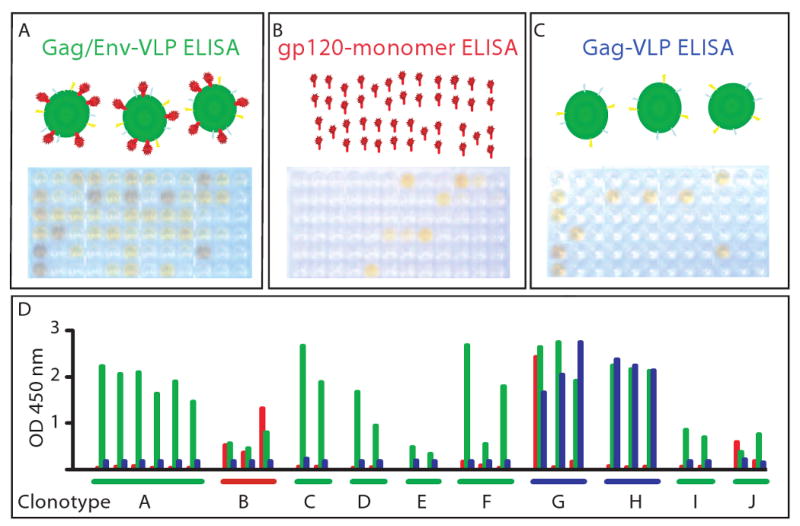
Binding of HIV-specific B cell culture supernates in enzyme linked immunosorbent assay to (A) HIV-VLPs containing Env protein, (B) monomeric gp120, or (C) VLPs containing Gag but not Env protein. Panels A through C show photographs of the colorimetric readout of the reactivity in plates tested with the same supernates arrayed in the same pattern. In (D) is shown the optical density (OD at 450 nm) of the same samples color-coded for reactivity as in the title of the ELISAs above (A=green, B=red, C=blue). In D, the wells are grouped by genetic clonotypes, determined by subsequent sequence analysis of the antibody genes of B cells from corresponding wells. The data in D show that cells within a clonotype (i.e., B cells sharing antibody genes) exhibit similar binding specificity for monomeric (red) or oligomeric (green) forms of Env. Interestingly, two clonotypes (designated G and H) exhibit binding to VLPs that do not contain Env.
The variable regions of Ab sequences were analyzed utilizing the IMGT database. Analysis revealed common usage of gene segments and significant homology in the CDR3 region indicating clonal populations (clonotypes). By gene segment usage and CDR3 similarity, the Abs from the cells shown in Figure 1 were divided into 10 different clonotypes. Interestingly, comparing the optical densities of the three different ELISA screens revealed that the pattern of reactivity for a particular antigen (monomer, HIV-VLP or Gag-only VLP) also was shared within each clonotype (Figure 3D). Once again, most of the Abs bound the trimeric Env presented on the VLP (shown in green), but did not bind monomeric gp120 (shown in red). However some clones were not specific to HIV Env containing antigens on ELISA (clonotypes G and H, exhibiting Gag-only VLP binding in blue). Clonotype B bound both gp120 monomer on ELISA and the HIV-VLP, which should have gp120/gp41 heterodimers presented on the surface, indicating the epitope of this antibody is likely a relatively linear epitope that does not change significantly upon trimeric conformation of the Env spike. So far in our studies, we have noted marked clonality in the B cell repertoire of all patients. Variable gene segments from each of the seven heavy chain variable gene families have been isolated, and the vast majority of our antibodies appear to bind epitopes present only in VLPs containing Env.
Clonality and repeated stimulation marked by extensive somatic hypermutation are principal features seen in the antibody genes in B cells from our cohort. In fact, these clones possess some of the most extensively mutated Ab genes described. Two of the most heavily mutated clones we have isolated fall into the same clonotype and contain unusual codon insertions that add to their diversity. Even without considering this insertion or the D region assignment and N and P type additions, these antibody variable genes average 73 of 350 (21%) nucleotides mutated [30]. This analysis does not even include the D segment, which encodes the middle of the heavy chain CDR3, generally the most mutated region of an Ab.
Interestingly, both clones within clonotype C, designated mAb 6B8 and mAb 2C6, showed homology in their CDR3 region to a known anti-HIV mAb 47e (also known as 4.7E) that binds to the CD4-inducible site on gp120. Comparison of the CDR3 regions of mAbs 47e, 2C6 and 6B8 reveal a shared DY*SDPFY motif. In this CDR3 region, the first DY these antibodies share has been shown in mAb 47e to be tyrosine sulfated thus mimicking CCR5 sulfation [36]. Strikingly, IMGT analysis predicts all three sequences to use VH1-69 and JH6 gene segments. However, both have 43 nucleic acid mutations from germline resulting in 19 amino acid changes compared to mAb 47e, which has 11 nucleic acid mutations from germline resulting in only 5 amino acid substitutions. In mAbs 2C6 and 6B8, the concentration of mutations is in the CDR1, CDR2 and FR3 Ab regions. Subsequently we have shown that mAb 2C6, like mAb 47e, neutralizes HIV in a CD4-induced fashion [30].
Studies identifying two novel broadly neutralizing monoclonal antibodies are consistent with these data [17]. Walker et al. separated over 30,000 B cells into clonal populations and found 97.7% of clonal supernatants that neutralized HIV-1JR-CSF and 46.5% that neutralized HIV-1SF-162 did not bind gp120JR-CSF nor gp41HxB2. This work led to the identification of a novel potentially broadly neutralizing epitope on the V2/V3 loop region that was dependent on trimeric envelope formation and was not available on monomeric gp120. This finding is consistent with studies in other recent publications that show the majority of neutralization activity targets epitopes that are not targeted by the known broadly neutralizing antibodies [37].
Summary
Emerging techniques for the study if human HIV-specific B cell repertoires promise to greatly expand our understanding of the molecular basis for antibody-mediated inhibition of HIV. Large panels of HIV-specific antibodies are being isolated, with a focus on isolating neutralizing antibodies. Care must be taken to use the most authentic antigen possible in order to recapitulate important native neutralizing epitopes. We and others increasingly find that most HIV-specific antibodies in humans bind to epitopes present only on complex oligomeric Env antigens, perhaps best represented currently by VLPs. Ultimately, knowledge of the fundamental principles underlying antibody-mediated neutralization of HIV will contribute to the rational design of immunogens to induce neutralizing responses.
Acknowledgments
This research was supported by U01 AI 078407 (NIAID, NIH), P30 AI 54999 (NIAID, NIH; the Vanderbilt-Meharry Center for AIDS Research), and P30 AI 50409 (Emory Center for AIDS Research). MDH was supported by a Pediatric Infectious Diseases Society-St. Jude Children's Research Hospital Fellowship Program in Basic Research and K08 AI 83078. JEC is a Burroughs Wellcome Clinical and Translational Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Watkins DI, Burton DR, Kallas EG, Moore JP, Koff WC. Nonhuman primate models and the failure of the Merck HIV-1 vaccine in humans. Nat Med. 2008;14:617–21. doi: 10.1038/nm.f.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fauci AS, Johnston MI, Dieffenbach CW, Burton DR, Hammer SM, Hoxie JA, et al. HIV vaccine research: the way forward. Science. 2008;321:530–2. doi: 10.1126/science.1161000. [DOI] [PubMed] [Google Scholar]
- 3.Alter G, Ananworanich J, Pantophlet R, Rybicki EP, Buonaguro L. Report on the AIDS Vaccine 2008 Conference. Hum Vaccin. 2009;5:119–25. doi: 10.4161/hv.5.3.7557. [DOI] [PubMed] [Google Scholar]
- 4.Johnston MI, Fauci AS. An HIV vaccine--challenges and prospects. N Engl J Med. 2008;359:888–90. doi: 10.1056/NEJMp0806162. [DOI] [PubMed] [Google Scholar]
- 5.Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med. 2009;15:866–70. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 6.Binley JM, Ditzel HJ, Barbas CF, 3rd, Sullivan N, Sodroski J, Parren PW, et al. Human antibody responses to HIV type 1 glycoprotein 41 cloned in phage display libraries suggest three major epitopes are recognized and give evidence for conserved antibody motifs in antigen binding. AIDS Res Hum Retroviruses. 1996;12:911–24. doi: 10.1089/aid.1996.12.911. [DOI] [PubMed] [Google Scholar]
- 7.Ditzel HJ, Parren PW, Binley JM, Sodroski J, Moore JP, Barbas CF, 3rd, et al. Mapping the protein surface of human immunodeficiency virus type 1 gp120 using human monoclonal antibodies from phage display libraries. J Mol Biol. 1997;267:684–95. doi: 10.1006/jmbi.1997.0912. [DOI] [PubMed] [Google Scholar]
- 8.Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75:10892–905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moulard M, Phogat SK, Shu Y, Labrijn AF, Xiao X, Binley JM, et al. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc Natl Acad Sci U S A. 2002;99:6913–8. doi: 10.1073/pnas.102562599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simmons CP, Bernasconi NL, Suguitan AL, Mills K, Ward JM, Chau NV, et al. Prophylactic and therapeutic efficacy of human monoclonal antibodies against H5N1 influenza. PLoS Med. 2007;4:e178. doi: 10.1371/journal.pmed.0040178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Traggiai E, Becker S, Subbarao K, Kolesnikova L, Uematsu Y, Gismondo MR, et al. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10:871–5. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu X, McGraw PA, House FS, Crowe JE., Jr An optimized electrofusion-based protocol for generating virus-specific human monoclonal antibodies. J Immunol Methods. 2008;336:142–51. doi: 10.1016/j.jim.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu X, Tsibane T, McGraw PA, House FS, Keefer CJ, Hicar MD, et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008;455:532–6. doi: 10.1038/nature07231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams JV, Weitkamp JH, Blum DL, LaFleur BJ, Crowe JE., Jr The human neonatal B cell response to respiratory syncytial virus uses a biased antibody variable gene repertoire that lacks somatic mutations. Mol Immunol. 2009;47:407–14. doi: 10.1016/j.molimm.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weitkamp JH, Kallewaard N, Kusuhara K, Feigelstock D, Feng N, Greenberg HB, et al. Generation of recombinant human monoclonal antibodies to rotavirus from single antigen-specific B cells selected with fluorescent virus-like particles. J Immunol Methods. 2003;275:223–37. doi: 10.1016/s0022-1759(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 16.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–40. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 17.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, et al. Broad and Potent Neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–9. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–71. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fouts TR, Binley JM, Trkola A, Robinson JE, Moore JP. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J Virol. 1997;71:2779–85. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Earl PL, Broder CC, Long D, Lee SA, Peterson J, Chakrabarti S, et al. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J Virol. 1994;68:3015–26. doi: 10.1128/jvi.68.5.3015-3026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bower JF, Li Y, Wyatt R, Ross TM. HIV-1 Envgp140 trimers elicit neutralizing antibodies without efficient induction of conformational antibodies. Vaccine. 2006;24:5442–51. doi: 10.1016/j.vaccine.2006.03.063. [DOI] [PubMed] [Google Scholar]
- 22.Crooks ET, Moore PL, Franti M, Cayanan CS, Zhu P, Jiang P, et al. A comparative immunogenicity study of HIV-1 virus-like particles bearing various forms of envelope proteins, particles bearing no envelope and soluble monomeric gp120. Virology. 2007;366:245–62. doi: 10.1016/j.virol.2007.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derby NR, Gray S, Wayner E, Campogan D, Vlahogiannis G, Kraft Z, et al. Isolation and characterization of monoclonal antibodies elicited by trimeric HIV-1 Env gp140 protein immunogens. Virology. 2007;366:433–45. doi: 10.1016/j.virol.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srivastava IK, Stamatatos L, Kan E, Vajdy M, Lian Y, Hilt S, et al. Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate. J Virol. 2003;77:11244–59. doi: 10.1128/JVI.77.20.11244-11259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammonds J, Chen X, Ding L, Fouts T, De Vico A, zur Megede J, et al. Gp120 stability on HIV-1 virions and Gag-Env pseudovirions is enhanced by an uncleaved Gag core. Virology. 2003;314:636–49. doi: 10.1016/s0042-6822(03)00467-7. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Wang S, Hoxie JA, LaBranche CC, Lu M. Mutations that destabilize the gp41 core are determinants for stabilizing the simian immunodeficiency virus-CPmac envelope glycoprotein complex. J Biol Chem. 2002;277:12891–900. doi: 10.1074/jbc.M110315200. [DOI] [PubMed] [Google Scholar]
- 27.Yang X, Farzan M, Wyatt R, Sodroski J. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 2000;74:5716–25. doi: 10.1128/jvi.74.12.5716-5725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Wyatt R, Sodroski J. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J Virol. 2001;75:1165–71. doi: 10.1128/JVI.75.3.1165-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grundner C, Li Y, Louder M, Mascola J, Yang X, Sodroski J, et al. Analysis of the neutralizing antibody response elicited in rabbits by repeated inoculation with trimeric HIV-1 envelope glycoproteins. Virology. 2005;331:33–46. doi: 10.1016/j.virol.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 30.Hicar MD, Chen X, Briney B, Hammonds J, Wang J, Kalams SA, et al. Pseudovirion particles bearing native HIV envelope trimers facilitate a novel method for generating human neutralizing monoclonal antibodies against HIV. doi: 10.1097/QAI.0b013e3181dc98a3. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammonds J, Chen X, Zhang X, Lee F, Spearman P. Advances in methods for the production, purification, and characterization of HIV-1 Gag-Env pseudovirion vaccines. Vaccine. 2007;25:8036–48. doi: 10.1016/j.vaccine.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 32.Weitkamp JH, Kallewaard NL, Bowen AL, Lafleur BJ, Greenberg HB, Crowe JE., Jr VH1-46 is the dominant immunoglobulin heavy chain gene segment in rotavirus-specific memory B cells expressing the intestinal homing receptor alpha4beta7. J Immunol. 2005;174:3454–60. doi: 10.4049/jimmunol.174.6.3454. [DOI] [PubMed] [Google Scholar]
- 33.Weitkamp JH, Lafleur BJ, Crowe JE., Jr Rotavirus-specific CD5+ B cells in young children exhibit a distinct antibody repertoire compared with CD5- B cells. Hum Immunol. 2006;67:33–42. doi: 10.1016/j.humimm.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 34.Weitkamp JH, Lafleur BJ, Greenberg HB, Crowe JE., Jr Natural evolution of a human virus-specific antibody gene repertoire by somatic hypermutation requires both hotspot-directed and randomly-directed processes. Hum Immunol. 2005;66:666–76. doi: 10.1016/j.humimm.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Tian C, Luskin GK, Dischert KM, Higginbotham JN, Shepherd BE, Crowe JE., Jr Evidence for preferential Ig gene usage and differential TdT and exonuclease activities in human naive and memory B cells. Mol Immunol. 2007;44:2173–83. doi: 10.1016/j.molimm.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U S A. 2004;101:2706–11. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, Davis KL, et al. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol. 2008;82:11651–68. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]


