Abstract
Integration of exogenous DNA into modified vaccinia Ankara (MVA) is often accomplished using mapped deletion sites in the viral genome. Since MVA has a large capacity (≥30kb) for foreign gene inserts and a limited number of unique integration sites, development of additional integration sites is needed to take full advantage of the extraordinary capacity for foreign gene insertion. In this report, we evaluate an alternative insertion site known as intergenic region 3 (IGR3). Recombinant MVA carrying the cytomegalovirus pp65 gene in IGR3 (rMVA-pp65-IGR3) demonstrated expression and genetic stability of the insert gene upon passage. Immunization of transgenic HLA-A2 mice with rMVA-pp65-IGR3 induced robust antigen-specific immune responses. Moreover, rMVA-pp65-IGR3-infected human EBV-transformed B cell lines were able to stimulate high levels of pp65-specific memory T cell responses in human PBMCs. These data support the usage of IGR3 for the development of highly immunogenic rMVA vaccines for clinical or veterinary use.
Keywords: modified vaccinia Ankara, recombinant vaccine, insertion site, deletion site, intergenic region, stability
Introduction
Enhancing the stability and immunogenic properties of transgenes inserted into MVA is vital for the generation of effective vaccines for use against infectious disease and cancer (Amato et al., 2009;Earl et al., 2007;Earl et al., 2009;Gomez et al., 2008;Hodge et al., 2003;Meyer et al., 2005). MVA itself is avirulent, even in immunocompromised individuals, yet it has the ability to induce significant immune responses to targeted transgene products (Gherardi & Esteban, 2005;Mayr & Danner, 1978;Moss, 1996;Ramirez et al., 2000;Wang et al., 2004a;Wang et al., 2008). Deletion (del) sites represent non-essential regions in the MVA genome whose deletion minimally impacts MVA viability in chicken embryo fibroblasts (CEFs). These regions formed during the process of 570 serial passages on CEFs that transformed the pathogenic vaccinia virus Ankara strain into MVA (Meyer et al., 1991). Del sites have been routinely used for insertion of recombinant genes encoding tumor- and virus-associated antigens to form MVA vaccines (Drexler et al., 2004). However, the stability of certain transgenes in del sites is variable depending on the toxicity of the expressed product during cellular and viral replication (Wyatt et al., 2008). In some cases, mutations or deletions that occur in transgenes after several passages of rMVA in culture may result in reduced or complete loss of expression (Wyatt et al., 2009). Therefore, alternative insertion sites known as intergenic regions (IGRs) within the MVA genome have been identified and studied for their support of stable transgene expression. A recent report described the stable insertion of HIV subunit genes into a site located between the essential I8R and GIL genes in the central conserved region of MVA (Wyatt et al., 2009). Recent studies have also looked at the genetic stability of transgenes in IGRs (Timm et al., 2006), but have not yet assessed their immunogenic properties. To this end, we have evaluated the stability and immunogenicity of a model cytomegalovirus (CMV) antigen (pp65) inserted specifically into intergenic region 3 (IGR3) (Howley P & Leyrer S, 2009) (Timm et al., 2006).
Results
Construction of rMVA-pp65-IGR3
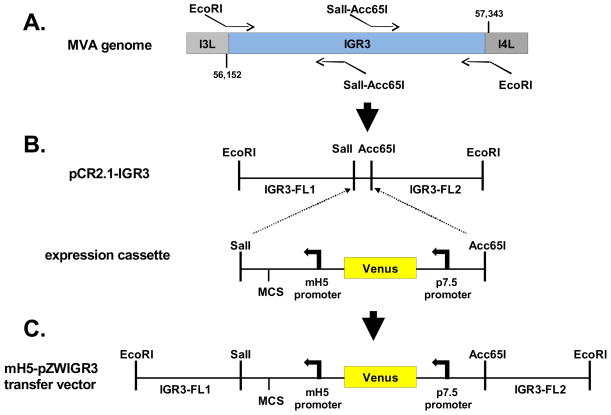
To investigate the properties of IGR3, a transfer vector was first generated to target the IGR3 site located between the essential I3L (Rochester & Traktman, 1998) and nonessential I4L genes (Howley et al., 1996b) spanning nucleotide positions 56,152 to 57,343 of the MVA genome for insertion of an expression cassette carrying the CMV-pp65 gene (McLaughlin-Taylor et al., 1994; Wang et al., 2007). As shown in Figure 1A, the IGR3 sequence of the MVA genome was first amplified by PCR using primers containing EcoRI restriction sites flanking the IGR3 site for insertion into the pCR2.1 vector (Invitrogen). SalI/Acc65I restriction enzyme sites were inserted into the IGR3 site in preparation to accept an expression cassette containing a modified H5 promoter (mH5) (Nemeckova et al., 2001;Wang et al., 2010), a multiple cloning site (MCS), a p7.5 promoter (Chakrabarti et al., 1985) and the Venus™ (Clontech) fluorescent marker gene (Nagai et al., 2002) used for screening of recombinant viral plaques (Figure 1B). In the final assembled transfer vector, mH5-pZWIGR3, the expression cassette is placed within the IGR3 site splitting it into two flanking segments (Figure 1C). For this study, the CMV-pp65 gene was inserted into the MCS of the transfer vector and then used to generate rMVA carrying the CMV-pp65 transgene in IGR3 (rMVA-pp65-IGR3) by homologous recombination in BHK-21 cells using established protocols (Moss & Earl, 1998;Wang et al., 2007;Yue et al., 2008).
Figure 1. Construction of recombinant MVA-pp65-IGR3.
(A) Schematic for mH5-pZWIGR3 transfer vector construction. Shown is a representation of the region of the MVA genome where IGR3 is located between the I3L (ORF 064) and I4L (ORF 065) genes. Two sets of primers were used to amplify individual regions of the IGR3 that would be used in the final transfer vector shown in C. The left most portion is approximately 596 base pairs and corresponds to IGR3-FL1, while the right fragment is approximately 607 base pairs and represents IGR3-FL2. Sequences of the primers and methods for amplification are described in Table 1 and Materials and Methods. The IGR3 region of the MVA genome was first amplified using primers containing EcoRI sites for cloning into pCR2.1 (pCR2.1-IGR3). (B) An expression cassette containing a multiple cloning site (MCS), modified H5 promoter (mH5) and fluorescent selection marker cassette (Venus™, Clontech) was inserted into pCR2.1-IGR3 using SalI and Acc65I restriction sites. The Sal1-Acc65I fragment bisects the IGR3 amplified region into two segments that are referred to as IGR3-FL1 and IGR3-FL2. (C) The expression cassette flanked by IGR3 (FL1 and FL2) was then inserted into the mH5-pZWIIA transfer vector (Wang et al, In Press) using EcoRI restriction sites (mH5-pZWIGR3). The mH5-pZWIGR3 transfer vector was used to generate recombinant MVA (rMVA-pp65-IGR3) by homologous recombination. Procedures and primer sequences are described in Table 1 and Materials and Methods.
Comparison of viral properties of Del II and IGR3 insertion site viruses
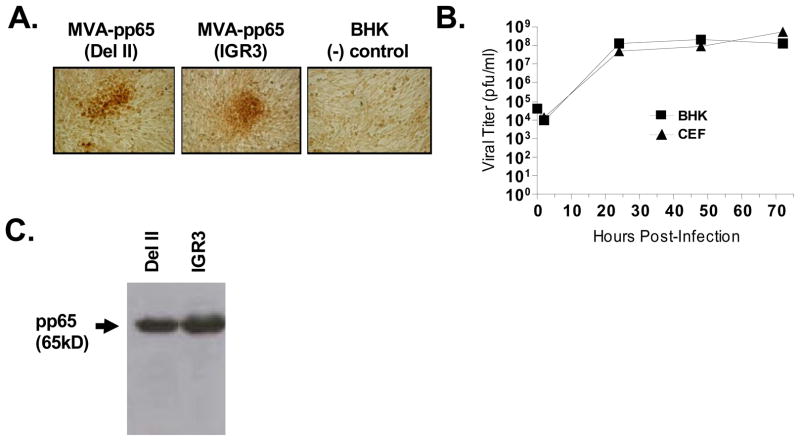
Although the Venus™ fluorescent marker gene allows for rapid screening of recombinant virus, it does not guarantee the presence of transgene products since it is not fused to the insert gene. Therefore, preliminary analysis of expression levels of CMV-pp65 was performed following 5 rounds of screening based on immunostaining of BHK-21 cells infected with isolates of rMVA-pp65-IGR3 or isolates containing CMV-pp65 in the del II insertion site (rMVA-pp65-del II) (Figure 2A). The size and degree of spread of rMVA-infected, pp65-positive BHK-21 cells, indicated by darker staining cells, was comparable between rMVA carrying pp65 in either IGR3 or del II insertion sites. These data suggest that insertion of CMV-pp65 is equally well tolerated in both del II and IGR3 sites.
Figure 2. Viability and expression levels of rMVA-pp65-IGR3.
(A) Expression of transgene product (CMV-pp65) during screening process of rMVA-pp65-IGR3. Following 5 rounds of screening, immunohistochemical staining specific for CMV-pp65 was done on BHK-21 cells infected with recombinant MVA containing the CMV-pp65 expression cassette inserted into either the del II site or the IGR3 site. Uninfected BHK-21 cells were used as a (−) staining control. (B) Growth kinetics of rMVA-pp65-IGR3 in BHK-21 and CEF cells. BHK-21 or CEF cells (4×106/100mm dish) were infected with 4×104 pfu (MOI=0.01) of rMVA-pp65-IGR3. Cell lysates were generated 2, 24, 48, and 72 hours after infection and used for viral titer by immunohistochemical staining. (C) Western blot analysis of transgene expression from a rMVA-pp65-IGR3 isolate after purification. Total cell lysates were generated from BHK-21 cells infected with either purified rMVA-pp65-del II or rMVA-pp65-IGR3 and 30 μg of total lysate were loaded for western blot analysis. Details of the approaches can be found in Materials and Methods.
Insertion of exogenous DNA into the MVA genome can potentially reduce the fitness of the recombinant virus and prevent optimal replication, making it difficult to obtain titers of virus sufficient for vaccine development. Thus, subsequent to purification, the viral growth kinetics of rMVA-pp65-IGR3 was evaluated in both BHK-21 and CEF cells, following infection with a starting concentration of 4.0 × 104 pfu/ml (multiplicity of infection (MOI) = 0.01), by determining viral titers at various time points through immunostaining against rMVA (Figure 2B). Viral replication reached maximal titers ranging from 108–109 pfu/ml in both BHK-21 and CEF cells, which is not significantly different from titers observed with wildtype or rMVAs carrying other transgenes in del II or del III sites (Drexler et al., 1998). These data strongly suggest that recombination of the CMV-pp65 expression cassette into IGR3 does not interrupt expression from the essential I3L gene, since doing so would have resulted in minimal production of recombinant virus(Howley et al., 1996a). Confirming unimpeded replication kinetics, we then proceeded to purify rMVA-pp65-IGR3 following 8 rounds of screening and performed western blot (WB) analysis for CMV-pp65 expression in lysates of BHK-21 cells infected with the purified virus. Using equal amounts of total protein lysate (Bradford assay), we observed CMV-pp65 expression from rMVA-pp65-IGR3-infected BHK-21 cells that was comparable to that of rMVA-pp65-del II-infected BHK-21 cells (Figure 2C). These data further validate the equivalence of insert expression in both viruses and the functional expression properties of rMVA carrying transgenes in the IGR3 site.
Genetic stability of rMVA-pp65-IGR3 upon serial passage
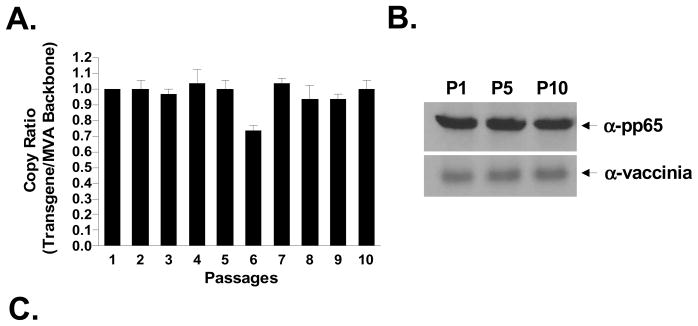
The genetic stability of a live rMVA vaccine is critical for maintaining potency and feasibility of manufacture. Verifying the expression of the transgene product over numerous passages, though important, will not necessarily indicate which fraction of the viral population has lost the inserted transgene. Therefore, we sought to determine the percentage of rMVA maintaining the CMV-pp65 transgene following multiple passages in vitro. We first measured the genetic stability of CMV-pp65 in IGR3 by performing real-time PCR analysis (ABI 7300 Real-Time PCR System) to detect the ratio of the CMV-pp65 transgene to MVA backbone over 10 passages of rMVA-pp65-IGR3 in BHK-21 cells. As shown in Figure 3A, the transgene to MVA backbone ratio (normalized to 1) at the majority of passage points were predominantly ≥1 through 10 passages, indicating that the CMV-pp65 transgene is likely retained in nearly 100% of viral isolates. However, since the primers used in the qPCR assay target only part of the CMV-pp65 transgene (Table 1) and can not detect mutations or deletions that would likely affect transgene expression, it was necessary to analyze the ratio of rMVA expressing CMV-pp65 to total MVA virus. By employing immunohistochemistry staining of BHK-21 cells infected with rMVA-pp65-IGR3 from passages 1, 5 and 10, and using CMV-pp65-specific mAb (for transgene expression) and vaccinia-specific mAb (for total MVA virus)(Schmelz et al., 1994), we found that greater than 97% of MVA expressed CMV-pp65 at each passage (Table 2).
Figure 3. Genetic stability during passage of recombinant MVA-pp65-IGR3.
(A) Following purification of rMVA-pp65-IGR3, transgene stability was determined by quantitative PCR (ABI 7300 Real-Time PCR System) during 10 passages on BHK-21 cells. Copy ratios are expressed as transgene copies (CMV-pp65) to MVA backbone copies. The analysis was repeated 3 times and shown is the mean of all 3 runs. Error bars represent standard error of the mean (SEM). Details of the approach are described in Materials and Methods. (B) Western blot analysis of CMV-pp65 at passages 1, 5, and 10. Equal amounts of total protein lysate (30 ug, Bradford assay) were loaded for each sample. Additional western blot against vaccinia protein using an anti-BR8 vaccinia mAb was performed as a loading control. (C) Using primers specific for the IGR3 flanking region (Table 1), PCR analysis was performed using a Perkin Elmer 9600 Gene Amp PCR system to detect for the presence or absence of wildtype (wt) IGR3 sequence (~300 bp) and the CMV-pp65 expression cassette (~2.7 kb) in viral genomic DNA isolated from rMVA-pp65-IGR3 at passages 1, 5, and 10.
Table 1.
Primers used in generation of IGR3 transfer vector and PCR analysis of virus stability during passage
| Target Gene | Primer Sequences |
|---|---|
| MVA IGR3 FL13 | Forward: 5′ CCGGAATTCCACCTTCTATAGATCTGAG 3′ |
| Reverse: 5′ GGTACCGCGCGTGTCGACTCAAATGAGATAAAGTGAAAATATATATC 3′ | |
| MVA IGR3 FL23 | Forward: 5′ CATTTGAGTCGACACGCGCGGTACCGAATAAAAATGTTTTTGTTTAACCACTGCA 3′ |
| Reverse: 5′ CGCGAATTCGGAATTGGGAACCTCTAAAAG 3′ | |
| MVA IGR3 (qPCR) | Forward: 5′ AACACCCGCACAAGTCTG 3′ |
| Reverse: 5′ TCCGCTCCCATTCAATTC 3′ | |
| CMV pp65 (qPCR) | Forward: 5′ A TCAAACCGGGCAAGATCTCGC 3′ |
| Reverse: 5′ ATCGTACTGACG CAGTTCCACG 3′ |
Table 2.
CMV-pp65 expression by serially passaged rMVA-pp65-IGR3
| Passage | % pp65 staining plaques out of total rMVA |
|---|---|
| 1 | 100 (n =83) |
| 5 | 98 (n =53) |
| 10 | 100 (n =95) |
n represents total number of anti-BR8 vaccinia staining plaques
To ensure that no wildtype MVA had been amplified during passage, we confirmed the presence of the entire CMV-pp65 expression cassette (~2.7 kb) in rMVA-pp65-IGR3 and also the absence of wildtype MVA (~300 bp) at passages 1, 5, and 10 by PCR analysis (Figure 3B) using primers specific for the IGR3 region (Table 1). At the same time, we also observed consistent CMV-pp65 protein expression by western blot analysis (Figure 3C). Lastly, and most importantly, sequencing of the CMV-pp65 transgene from purified DNA of rMVA-pp65-IGR3 viral isolates (n = 6) revealed no variation in transgene sequence at passages 1 or 5, and we found only a single nucleotide mutation that resulted in an amino acid change from Arg to His at residue position 494 in 1 isolate at passage 10 (Table 3). Altogether, these data further highlight the capacity for IGR3 to serve as a stable site of recombinant transgene insertion.
Table 3.
Sequence analysis of CMV-pp65 gene in passaged rMVA-pp65-IGR3
| Passage | Sequence Match to Input Virus |
|---|---|
| 1 | 6 of 6 isolates, 100% match |
| 5 | 6 of 6 isolates, 100% match |
| 10 | 5 of 6 isolates, 100% match; 1 of 6 contained 1 a.a. change* |
an amino acid (a.a.) change of Arg to His occurred at a.a. position 494 in this isolate.
Immunogenicity in HHDII transgenic mice
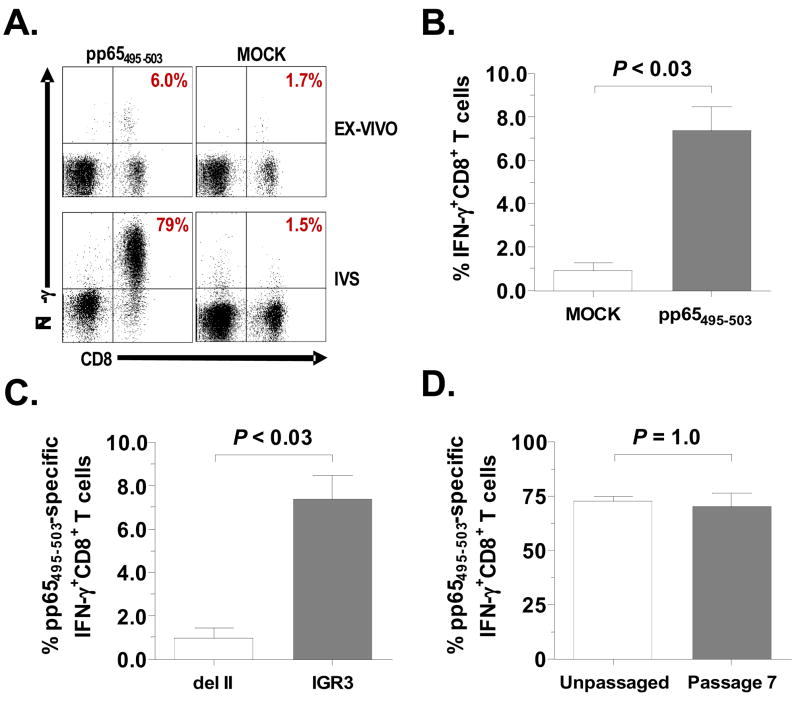
Although the genetic stability of various IGRs have been partially assessed in previous work (Timm et al., 2006), immunogenicity of genes inserted into these sites is not well known. We therefore evaluated the immunogenicity of purified rMVA-pp65-IGR3 in the transgenic HLA-A2 (HHDII) murine model system. HHDII mice (n=4) were inoculated intraperitoneally with 5.0 × 107 pfu of purified rMVA-pp65-IGR3. Spleens were harvested 3 weeks later and CD8+ splenocytes were assessed for IFN-γ production by intracellular cytokine staining (ICS) following ex vivo or 7 day in vitro stimulation (IVS) with a known HLA-A2-restricted CMV-pp65 epitope (pp65495–503) (Diamond et al., 1997;Wang et al., 2007;Wills et al., 1996). Following 7 day IVS of splenocytes with pp65495–503-loaded LCLs, we observed dramatic increases in CMV-pp65-specific CD8+IFN-γ+ responses, ranging from 70% to 79% (Figure 4A, bottom left) of the total CD8+ response with mock responses ranging from 0.7% to 1.5% (Figure 4A, bottom right) of the total CD8+ response. Surprisingly, splenocytes from rMVA-pp65-IGR3-vaccinated HHDII mice, stimulated ex-vivo with pp65495–503 peptide, consistently generated CD8+IFN-γ+ responses ranging from 6.0% to as high as 9.8% (Figure 4A, top left) of the total CD8+ response, which were significantly higher than mock (irrelevant peptide) stimulated responses that measured between 0.10% to 1.7% of the total CD8+ response (Figure 4A, top right and Figure 4B). Such ex-vivo responses are not always readily observed when immunizing with rMVA containing transgenes, such as CMV-pp65, inserted into del sites (Krishnan et al., 2008;Wang et al., 2007). We therefore compared ex-vivo CD8+IFN-γ+ splenocyte responses of HHDII mice immunized with rMVA containing CMV-pp65 in either del II or IGR3 (Figure 4C). We observed significantly higher CMV-pp65-specific responses in HHDII mice vaccinated with rMVA-pp65-IGR3 compared to those vaccinated with rMVA-pp65-del II (P< 0.03, Mann-Whitney). The consistent transgene-specific immune responses generated ex-vivo and after IVS by rMVA-pp65-IGR3 in the HHDII murine system further highlight the potential usefulness of IGR3 as an effective antigen gene insertion site.
Figure 4. Immunogenicity of rMVA-pp65-IGR3 in HHDII transgenic mice.
(A) HLA-A2 pp65-specific IFN-γ+ T cell responses generated by CD8+ splenocytes from transgenic HHDII mice immunized with purified rMVA-pp65-IGR3. HHDII mice (n=4) were inoculated intraperitoneally (i.p.) with 5×107 pfu of rMVA-pp65-IGR3. Splenocytes were harvested 3 weeks post-immunization and assessed for IFN-γ production ex-vivo and after in-vitro stimulation (IVS) to a known HLA-A2-restricted pp65 epitope (pp65495–503) by FACS analysis. Shown is a representative mouse for the ex-vivo and IVS groups. Mock groups represent re-stimulation with irrelevant peptide. (B) pp65495–503-specific IFN-γ+ responses generated ex-vivo by CD8+ splenocytes from HHDII mice (n=4) immunized i.p. with 5×107 pfu of purified rMVA-pp65-IGR3. Error bars represent SEM. Differences in pp65495–503-specific IFN-γ+ responses compared to mock stimulated responses were evaluated using the Mann-Whitney test and a P value < 0.05 is considered significant. Details of the approaches can be found in Materials and Methods. (C) pp65495–503-specific IFN-γ+ responses generated ex-vivo by CD8+ splenocytes from HHDII mice (n=4) immunized i.p. with 5×107 pfu of either purified rMVA-pp65-IGR3 or rMVA-pp65-del II. Error bars represent SEM. Differences in responses were evaluated using the Mann-Whitney test and a P value < 0.05 is considered significant. (D) pp65495–503-specific IFN-γ+ responses generated through IVS by CD8+ splenocytes from HHDII mice (n=4) immunized i.p. with 5×107 pfu of purified rMVA-pp65-IGR3 after 7 passages were compared to responses from mice immunized i.p. with 5×107 pfu unpassaged rMVA-pp65-IGR3. Error bars represent SEM. Differences in responses were evaluated using the Mann-Whitney test and a P value > 0.05 is not considered significant.
Stable immunogenicity after 7 passages assessed in HHDII transgenic mice
It is common to observe instability of transgene expression following multiple passages of rMVA in culture, which can ultimately result in decreased immunogenicity of purified vaccine stocks (Burgers et al., 2008; Wyatt et al., 2009). Therefore, to determine whether successive passage of rMVA-pp65-IGR3 changes its immunogenic properties, we immunized HHDII mice (n=4) with virus that had been passaged seven times in BHK-21 cells. IVS responses generated by this immunization (Figure 4D) were not significantly different from those seen in mice immunized with purified, unpassaged rMVA-pp65-IGR3 (P = 1.0, Mann-Whitney test), ranging from 58% to 81% of the total CD8+ response with mock responses ranging from 0.3% to 1.27% of the total CD8+ response. These data demonstrate the continued ability of rMVA-pp65-IGR3 to sustain robust immunogenicity following multiple passages in-vitro. Stable levels of immunogenicity is a valuable property during extended passage required to sustain the vaccine manufacturing process.
rMVA-pp65-IGR3 infection of human PBMC and expansion of a memory T cell repertoire
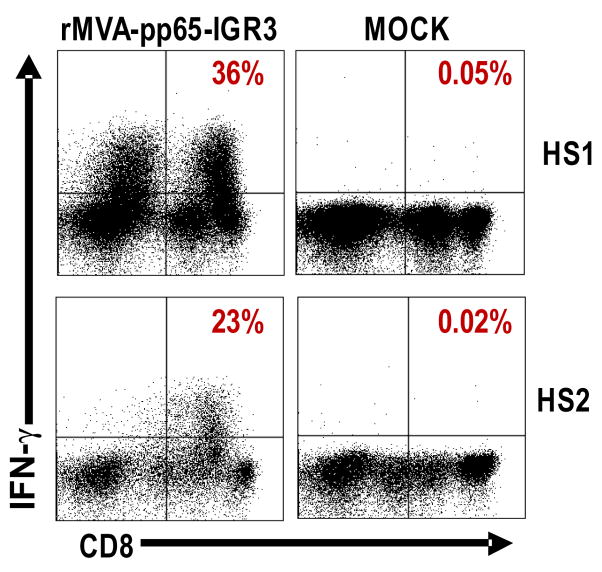
Following these preliminary experiments in the HHDII murine system, we next investigated the potential of rMVA-pp65-IGR3 to elicit human T cell memory responses using PBMC. Autologous PBMC were used as APC for IVS of PBMC from 2 healthy individuals after conditioning for 2 days with a cocktail of CpG ODN as described in Material and Methods. The heterogeneous population of APC becomes more capable of being infected with purified rMVA-pp65-IGR3. As shown in Figure 5, CD8+IFN-γ+ responses to CMV-pp65 ranged from 23% to 36% of the total CD8+ T cell response with responses from mock stimulation ranging from 0.02% to 0.05%. The significantly robust immune responses generated by rMVA-pp65-IGR3 in human PBMC further emphasizes the potential of IGR3 as an alternate, or an addition, to del sites for insertion of transgenes.
Figure 5. Memory T-cell responses to CMV-pp65 after infection of human PBMC with rMVA-pp65-IGR3.
CD8+ pp65-specific IFN-γ+ responses were generated from human PBMCs of 2 CMV-positive healthy subjects (HS1, HS2). The approach used in-vitro stimulation with rMVA-pp65-IGR3-infected autologous PBMC, after conditioning using a cocktail of CpG ODN. Details of the approaches can be found in Materials and Methods.
Discussion
Our results indicate that IGR3 defines a useful site of insertion in MVA because it maintains genetic stability of a wide variety of inserts including CMV-pp65, and allows for stable expression of proteins from transgenes that result in robust transgene-specific immune responses. Previous studies have assessed the stability of transgenes in IGRs (Timm et al., 2006), but have only in a single instance evaluated immunogenicity of genes in these insertion sites (Wyatt et al., 2009). The study of an HIV glycoprotein showed that a sequence and structural alteration of the membrane form of the glycoprotein expressed as a recombinant transgene was necessary to achieve stable expression during passage. For the first time we show that a transgene inserted into IGR3 of the MVA genome is able to consistently generate CD8+IFN-γ+ responses in both a transgenic HHDII murine model and in human PBMC. We also show that the inserted transgene (CMV-pp65) and expression levels are nearly 100% retained following multiple passages in-vitro, which in part may contribute to the enhanced magnitude and consistency of immune responses after serial passages of the recombinant virus during passage.
Transgene expression that results in adverse effects to MVA replication is often minimized by mutation or genetic truncation, to achieve optimal replication (Wyatt et al., 2009). Several consequences of inappropriate changes in antigen sequence include potential loss of diversity of MHC recognition, improper folding and processing of the truncated protein which may lead to poor immunogenicity, or more worrisome new regulatory properties generated by a change in primary sequence. Based on observations made in this study, IGR3 may serve as an alternate site for insertion and potentially improve expression and immunogenicity of unmodified transgenes that prove unstable or are shown to have poor immunogenicity in known del sites(Wang et al., 2004a;Wyatt et al., 2008). Under suboptimal treatment conditions that utilize only single antigens or epitopes in a vaccine regimen, certain cancers or infectious diseases may only be weakly controlled and have increased probability to evade immune surveillance. Therefore, IGR3 may serve as an additional site of insertion for multiple genes into MVA, where immunization against a diversity of tumor or viral antigens is necessary for greatest protection, such as is the case for HIV, CMV, and numerous cancers (Earl et al., 2009;Fischer et al., 2007;Gomez et al., 2007;Maruyama et al., 2000;Mooij et al., 2004;Wang et al., 2006;Wang et al., 2007;Young et al., 2001). Dual use of IGRs that are located between conserved MVA genes described in recent reports may serve as a more stable alternative to the use of dual insertion in del sites (Wyatt et al., 2009). Recently, we have continued to explore the usefulness of the IGR3 site for incorporation of additional inserts that include human and murine Survivin and murine p53. These additional gene products have proven to be well expressed and the viruses that contain them are stable throughout their purification and subsequent passage (data not shown). These preliminary results indicate that the prospects for using IGR3 for a wide array of genes is likely, and further confirms and extends its potential as an insertion site for a wide variety of antigens that can be incorporated into MVA vectors and explored as vaccines for infectious disease and cancer.
Materials and Methods
Cells, viruses, peptides, and mice
BHK-21 cells (ATCC CCL-10) were purchased from American Type Culture Collection (Manassas, VA) and maintained in minimal essential medium (MEM) supplemented with 10% fetal calf serum (ISC BioExpress, Kaysville, UT) in a 37° incubator containing 5% CO2. Chicken embryo fibroblasts (CEF) were purchased from Charles River Laboratories (Wilmington, MA) as a cell suspension that is freshly prepared weekly by the company. Cells are counted and frozen when they arrive, and are thawed and plated followed by 24 hour resting phase prior to infection with viruses. Wildtype (wt) MVA virus stock was kindly provided by Drs. Linda Wyatt and Bernard Moss (Laboratory of Viral Diseases, NIAID, NIH). mH5-pp65 inserted into deletion 2 (del II) of MVA is described in a recent publication from our laboratory(Wang et al., 2008;Wang et al., 2010). Peptide epitopes used in the mouse immunization studies and flow cytometry were prepared in our laboratory using standard Fmoc peptide synthesis techniques (Wang et al., 2008). HHDII mice are transgenic for HLA-A2 and have knockout of mouse MHC Class I and β2-microglobulin loci and have been used previously for evaluation of other rMVA expressing CMV genes in our laboratory (Krishnan et al., 2008;Pascolo et al., 1997).
Construction of MVA transfer plasmid and viruses containing IGR3 sequences
The first step in the process of generating a transfer plasmid incorporating the IGR3 segment of the MVA genome required amplification of the IGR3 genome from wildtype MVA genomic DNA. Primers were made to subdivide the IGR3 region into two segments that are flanked with Eco R1 and contain Sal1/Acc65I restriction enzyme sites. The sequence of the two pairs of primers are shown in Table 1. Conditions for PCR amplification from the MVA genome were 95°C for 5 min, 30 cycles of 95°C for 1 min, 55°C for 1 min, 72°C for 3 min, and ending with a final cycle at 72°C for 10 min. The modified fragment that contains Sal1/Acc65I sites and flanked by terminal EcoR1 sites was cloned into the plasmid transfer vector pCR 2.1 (Invitrogen) and referred to as pCR 2.1-IGR3. The IGR3 segment was further modified by insertion of an expression cassette that was previously described and contains a multiple cloning site (MCS) followed by a synthetically generated mH5 promoter sequence and a p7.5 promoter that drives the Venus™ (Clontech) fluorescent marker gene which was inserted into the transfer vector(Wang et al., 2010). The completed transfer vector is shown in Figure 1C and was further modified by cloning the CMV-pp65 gene into the MCS to complete the insertion vector for MVA. All plasmid vectors were sequenced on both strands using an ABI 3730 located in the DNA sequencing/Solexa Core facility of Beckman Research Institute of City of Hope (COH). rMVA-pp65-IGR3 was generated by transfecting the CMV-pp65 transfer vector into wt MVA-infected BHK-21 cells and screened based on the Venus™ fluorescent marker to eliminate wt MVA according to our published procedures (Wang et al., 2008). Cell debris were purified from final stocks of rMVA-pp65-IGR3 through ultracentrifugation on a sucrose gradient at 16,000 rpm for 80 min using a Sorvall Discovery 90 (SA-600 rotor). Viral pellets were then washed, ultracentrifuged for 60 min, and resuspended in aqueous 7.5% lactose/10 mM Tris (pH 7.5) and frozen at −80° C for long term storage.
Immunohistrochemical staining of rMVA-pp65-IGR3 in BHK-21 and CEF cells
For growth kinetic studies, BHK-21 and CEF monolayers were grown to a density of 4×106 cells/100mm dish and then infected with rMVA-pp65-IGR3 at an MOI of 0.01 (4×104 pfu/ml starting input). Infected 100mm dishes were then harvested at 2h, 24h, 48h, and 72h after initial infection and resuspended in 1.0 ml of MEM containing 2% fetal calf serum (MEM-2) and subjected to 3× freeze/thaw cycles followed by sonication to release the virus. Virus from each time point was subsequently titrated by immunostaining of virus-infected cell foci on BHK-21 monolayers using polyclonal vaccinia virus-specific antibody (Biogenesis Ltd., Poole, England, Cat. No. 9503-2057), reconstituted in saline buffer (120 mM NaCl, 10 mM TrisCL, pH 7.4) and used at a dilution of 1:2000. For genetic stability studies, BHK-21 monolayers were infected with a dilution (10−7) of purified, passaged rMVA-pp65-IGR3 (passages 1, 5, or 10). Immunohistochemistry staining was then performed as described above using purified mAb 28–103 (Wang et al., 2004b) against CMV-pp65 or anti-BR8-specific vaccinia mAb (Schmelz et al., 1994). The percentage of CMV-pp65-containing rMVA is calculated as: (CMV-pp65 staining plaques/BR8-specific vaccinia staining plaques) × 100%.
Western blot detection of rMVA protein expression
Protein expression levels by the CMV-pp65 transgene were measured by WB using the Amersham ECL Plus™ detection kit (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom). Cell lysates were separated by 10% SDS-PAGE. After electrotransfer of proteins from the gel onto PVDF membranes (Bio-Rad, Hercules, CA), the blots were incubated with purified mAb 28–103 (Wang et al., 2004b) against CMV-pp65, or anti-BR8-specific vaccinia antibody used for a loading control (Schmelz et al., 1994), then washed and further incubated with HRP-labeled goat anti-mouse polyclonal antibody according to the manufacturer instructions (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom). These methods closely follow our recent publications (Wang et al., 2008;Wang et al., 2010).
qPCR and sequencing analysis of rMVA-pp65-IGR3
rMVA-pp65-IGR3 viruses were serially passaged 10 times on BHK-21 cells. Briefly, BHK-21 cells growing on 150mm tissue culture dishes were infected with rMVA at MOI=0.1. rMVA viruses were harvested 48 hours after infection, resuspended in 1.0 ml of MEM containing 2% fetal calf serum (MEM-2) and subjected to 3× freeze/thaw cycles followed by sonication to release the virus. The virus from each passage was subsequently titrated on BHK-21 cells, and after adjustment to an MOI of 0.10, it was used for the next passage on BHK-21 cells. Viral DNA from each passage was purified for qPCR analysis using the Qiagen™ (Valencia, CA) column purification kit according to the manufacturers instructions. Cell lysates of each passage used for qPCR analysis were prepared from 100mm dishes of BHK-21 cells infected with the same amount of rMVA viruses from each serial passage. The plasmid DNA used to generate the standard curve was made by inserting the pp65 gene into the pSC11 vector containing the TK gene (Wang et al., 2010). The absolute concentration of the plasmid was measured by two independent means: OD260 by UV spectraphotometry and the fluorophor based method using Quant-iT™ Picogreen© DS DNA kit (Invitrogen, Carlsbad, CA). The concentration was converted to plasmid copy number using the molecular weight of the plasmid DNA. The primers to measure pp65 and MVA backbone copies were designed based on standard qPCR conditions using primer express software version 3.0 (Applied Biosystems Inc., Foster City, CA) and listed in Table 1. Primers for detection of CMV-pp65 using qPCR generate a ~190 bp fragment derived from the 1.70kb gene. qPCR was performed using an ABI 7300 real time PCR system and Power SYBR Green Master Mix Kit (ABI). Briefly, 5μL of MVA genomic DNA was amplified in a mixture of 25 μL containing 1μM forward, 1μM reverse primers and SYBR Green containing solution. The thermocycling conditions were 95°C for 10 min and 40 cycles at 95°C for 15 s and 60°C for 1 min. Gene copy numbers were calculated using ABI sequence detection software (SDS). The ratio of pp65 gene and MVA backbone (ratio equals pp65 gene copy number/MVA backbone copy number) was calculated for each passage. The copy numbers for each gene was normalized to 1.0 for passage 1 and subsequent passages were computed based upon the measured level at passage 1.
For PCR and sequencing analysis, rMVA-pp65-IGR3 genomic DNA isolated from infected BHK-21 cells at passages 1, 5, and 10 were subjected to PCR using IGR3 FL1 forward and IGR3 FL2 reverse primers shown in Table 1. A Perkin Elmer 9600 Gene Amp PCR system was used in conjunction with GoTaq Green Master Mix (Promega, San Luis Obispo, CA) for all PCR reactions. Fragments were then either run on a 1% agaraose ethidum bromide gel or cloned into pCR2.1 (Invitrogen) for sequencing using an ABI 3730 located in the DNA sequencing/Solexa Core facility of Beckman Research Institute of City of Hope (COH).
Immunogenicity of rMVA-pp65-IGR3 in HHDII transgenic mice
HHDII mice (transgenic HLA A2.1) were used at 10–12 weeks for immunization and were bred and maintained under SPF conditions (microisolator cages with top filtration and automatic water feed lines contained within a multi-rack suite) in the COH centralized animal core facility. HHDII mice were immunized with 5×107 pfu of purified rMVA by the intraperitoneal (i.p.) route. After 3 weeks post-immunization, spleens were removed and were stimulated in-vitro for 1 week using a simplified protocol with HLA-matched EBV-LCL (Krishnan et al., 2008) as antigen presenting cells (APC) loaded either with the relevant CMV-CTL epitope HLA-A*0201 pp65495–503 (pp65-A2) (Diamond et al., 1997) or an irrelevant HLA A*0201-specific epitope from HIV-gag (Johnson et al., 1991). For some experiments shown in Figure 4A, no IVS was used, and fresh splenocytes were harvested immediately after euthanasia and processed for ICC (intracellular cytokine) assays as described below. ICC was used to measure pp65 IFN-γ+/CD8+ T cells from the stimulated splenocytes according to previously described methods (Krishnan et al., 2008). 0.5 to 1.0 × 106 events were acquired for each sample on a FACSCanto™ flow analyzer (BD Biosciences, San Jose, CA). Analysis was performed using FCS Express version 2 software (De Novo, Ontario, Canada). The number of double positive cells is expressed as a percentage of the CD8+ T cell population.
Immunogenicity of rMVA-pp65-IGR3 in human PBMC
In-vitro stimulation using rMVA-pp65-IGR3 was modified from a published method (La Rosa et al., 2006). Briefly, cryopreserved PBMCs were rapidly thawed and immediately dispensed in a 12-well plate at a concentration of 2 × 106 cells/mL in RPMI 1640 medium (2.5 mL/well) supplemented with 10% human AB serum (COH Blood Bank) and were incubated with 5 μg/mL of both CpG-A ODN 2216 and CpG-B ODN 2006 (TriLink BioTechnologies, San Diego, CA, USA). After 3 days, ODN-treated PBMCs were infected with rMVA-pp65-IGR3 at a multiplicity of infection of 5, for 6 hours in RPMI 1640 medium with reduced (2%) human AB serum. Infected PBMCs were γ-irradiated (2500 rads) and used as APC. APC were plated in a 24-well plate (1.5 × 106/well), co-incubated with autologus PBMC (3 × 106/well), in a final volume consisting of 2ml/well RPMI 1640 medium with 10% human AB serum and human rIL-2 (NIH AIDS Research and Reference Reagent Program, 10 units/ml). Every 2 days, 50% of the culture medium was removed, and replaced with fresh medium containing rIL2. Cells were incubated for 8 days and split into additional wells when necessary. At day 8, cells were collected and washed with medium without rIL-2 and transferred into 15ml Falcon tubes in 1 mL medium without rIL-2 and starved overnight. PBMC were then incubated with a pp65 Pepmix™ of overlapping 15mers (JPT peptide technologies, Berlin, Germany) or peptide library diluent for 1.5 hrs and then GolgiPlug containing brefeldin A (BD Biosciences) was added. After 12 hours of incubation, PBMC were harvested, washed, labeled with PE–conjugated anti-CD8 antibody, fixed, and permeabilized (Cytofix-Cytoperm; Becton Dickinson Biosciences) before they were labeled with APC–conjugated antibody to IFN-γ. The stained cells were analyzed on a FACSCanto™ (BD Immunocytometry Systems, San Jose, CA), and data were analyzed using FCS Express (version 3.0; DeNovo Software). 0.5×106 events were acquired for each sample. Lymphocytes were initially gated using forward vs side scatter, and then CD8+ lymphocytes were gated separately. The number of IFN-γ expressing cells in the upper right quadrant is shown as a percentage of the CD8+ lymphocyte population.
Acknowledgments
Support for this work was from Public Health Service grants CA030206, CA077544, CA114889, and AI062496 to DJD and AI084019 to CLR. We thank Drs. Linda Wyatt and Bernard Moss for their wildtype MVA and plasmid vectors for construction of rMVA. We also thank Dr. F. Lemonnier for HHD II mice under MTA with the Institut Pasteur (Paris, France). We are grateful to Dr. William J. Britt (UAB, Birmingham, AL) for providing us with a hybridoma specific for the CMV-pp65 antigen. We’d like to acknowledge the technical assistance of Aparna Krishnan and Gideon Blumstein and the secretarial and administrative assistance of Donna Packer, Denise Marsano, and Peter Kwon. We gratefully acknowledge the support staff of City of Hope Donor/Apheresis Center for assistance in human sample preparation and the City of Hope Animal Research Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Amato RJ, Shingler W, Goonewardena M, de Belin J, Naylor S, Jac J, Willis J, Saxena S, Hernandez-McClain J, Harrop R. Vaccination of renal cell cancer patients with modified vaccinia Ankara delivering the tumor antigen 5T4 (TroVax) alone or administered in combination with interferon-alpha (IFN-alpha): a phase 2 trial. J Immunother. 2009;32:765–772. doi: 10.1097/CJI.0b013e3181ace876. [DOI] [PubMed] [Google Scholar]
- Burgers WA, Shephard E, Monroe JE, Greenhalgh T, Binder A, Hurter E, Van Harmelen JH, Williamson C, Williamson AL. Construction, characterization, and immunogenicity of a multigene modified vaccinia Ankara (MVA) vaccine based on HIV type 1 subtype C. AIDS Res Hum Retroviruses. 2008;24:195–206. doi: 10.1089/aid.2007.0205. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Brechling K, Moss B. Vaccinia virus expression vector: coexpression of beta-galactosidase provides visual screening of recombinant virus plaques. Mol Cell Biol. 1985;5:3403–3409. doi: 10.1128/mcb.5.12.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond DJ, York J, Sun J, Wright CL, Forman SJ. Development of a candidate HLA A*0201 restricted peptide-based vaccine against human cytomegalovirus infection. Blood. 1997;90:1751–1767. [PubMed] [Google Scholar]
- Drexler I, Heller K, Wahren B, Erfle V, Sutter G. Highly attenuated modified vaccinia virus Ankara replicates in baby hamster kidney cells, a potential host for virus propagation, but not in various human transformed and primary cells. J Gen Virol. 1998;79 (Pt 2):347–352. doi: 10.1099/0022-1317-79-2-347. [DOI] [PubMed] [Google Scholar]
- Drexler I, Staib C, Sutter G. Modified vaccinia virus Ankara as antigen delivery system: how can we best use its potential? Curr Opin Biotechnol. 2004;15:506–512. doi: 10.1016/j.copbio.2004.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl PL, Americo JL, Wyatt LS, Eller LA, Montefiori DC, Byrum R, Piatak M, Lifson JD, Amara RR, Robinson HL, Huggins JW, Moss B. Recombinant modified vaccinia virus Ankara provides durable protection against disease caused by an immunodeficiency virus as well as long-term immunity to an orthopoxvirus in a non-human primate. Virology. 2007;366:84–97. doi: 10.1016/j.virol.2007.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl PL, Cotter C, Moss B, Vancott T, Currier J, Eller LA, McCutchan F, Birx DL, Michael NL, Marovich MA, Robb M, Cox JH. Design and evaluation of multi-gene, multi-clade HIV-1 MVA vaccines. Vaccine. 2009;27:5885–5895. doi: 10.1016/j.vaccine.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W, Perkins S, Theiler J, Bhattacharya T, Yusim K, Funkhouser R, Kuiken C, Haynes B, Letvin NL, Walker BD, Hahn BH, Korber BT. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med. 2007;13:100–106. doi: 10.1038/nm1461. [DOI] [PubMed] [Google Scholar]
- Gherardi MM, Esteban M. Recombinant poxviruses as mucosal vaccine vectors. J Gen Virol. 2005;86:2925–2936. doi: 10.1099/vir.0.81181-0. [DOI] [PubMed] [Google Scholar]
- Gomez CE, Najera JL, Jimenez EP, Jimenez V, Wagner R, Graf M, Frachette MJ, Liljestrom P, Pantaleo G, Esteban M. Head-to-head comparison on the immunogenicity of two HIV/AIDS vaccine candidates based on the attenuated poxvirus strains MVA and NYVAC co-expressing in a single locus the HIV-1BX08 gp120 and HIV-1(IIIB) Gag-Pol-Nef proteins of clade B. Vaccine. 2007;25:2863–2885. doi: 10.1016/j.vaccine.2006.09.090. [DOI] [PubMed] [Google Scholar]
- Gomez CE, Najera JL, Krupa M, Esteban M. The poxvirus vectors MVA and NYVAC as gene delivery systems for vaccination against infectious diseases and cancer. Curr Gene Ther. 2008;8:97–120. doi: 10.2174/156652308784049363. [DOI] [PubMed] [Google Scholar]
- Hodge JW, Poole DJ, Aarts WM, Gomez YA, Gritz L, Schlom J. Modified vaccinia virus ankara recombinants are as potent as vaccinia recombinants in diversified prime and boost vaccine regimens to elicit therapeutic antitumor responses. Cancer Res. 2003;63:7942–7949. [PubMed] [Google Scholar]
- Howley P, Leyrer S. Intergenic regions as insertion sites in the genome of modified vaccinia ankara (MVA) Bavarian Nordic A/S. 10/514,761(7,550,147 B2) :1–40. 6–23-2009. Kvistgaard/DK. 5–14-2003. [Google Scholar]
- Howley PM, Spehner D, Drillien R. A vaccinia virus transfer vector using a GUS reporter gene inserted into the I4L locus. Gene. 1996b;172:233–237. doi: 10.1016/0378-1119(96)00192-8. [DOI] [PubMed] [Google Scholar]
- Howley PM, Spehner D, Drillien R. A vaccinia virus transfer vector using a GUS reporter gene inserted into the I4L locus. Gene. 1996a;172:233–237. doi: 10.1016/0378-1119(96)00192-8. [DOI] [PubMed] [Google Scholar]
- Johnson RP, Trocha A, Yang L, Mazzara GP, Panicali DL, Buchanan TM, Walker BD. HIV-1 gag-specific cytotoxic T lymphocytes recognize multiple highly conserved epitopes. Fine specificity of the gag-specific response defined by using unstimulated peripheral blood mononuclear cells and cloned effector cells. J Immunol. 1991;147:1512–1521. [PubMed] [Google Scholar]
- Krishnan A, Wang Z, Srivastava T, Rawal R, Manchanda P, Diamond DJ, La Rosa C. A novel approach to evaluate the immunogenicity of viral antigens of clinical importance in HLA transgenic murine models. Immunol Lett. 2008;120:108–116. doi: 10.1016/j.imlet.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa C, Wang Z, Lacey SF, Lalimarmo MM, Krishnan A, Longmate J, Diamond DJ. In vitro expansion of polyclonal T-cell subsets for adoptive immunotherapy by recombinant modified vaccinia Ankara. Exp Hematol. 2006;34:497–507. doi: 10.1016/j.exphem.2005.12.018. [DOI] [PubMed] [Google Scholar]
- Maruyama H, Zaloudik J, Li W, Sperlagh M, Koido T, Somasundaram R, Scheck S, Prewett M, Herlyn D. Cancer vaccines: single-epitope anti-idiotype vaccine versus multiple-epitope antigen vaccine. Cancer Immunol Immunother. 2000;49:123–132. doi: 10.1007/s002620050611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr A, Danner K. Vaccination against pox diseases under immunosuppressive conditions. Dev Biol Stand. 1978;41:225–34. 225–234. [PubMed] [Google Scholar]
- McLaughlin-Taylor E, Pande H, Forman SJ, Tanamachi B, Li CR, Zaia JA, Greenberg PD, Riddell SR. Identification of the major late human cytomegalovirus matrix protein pp65 as a target antigen for CD8+ virus-specific cytotoxic T lymphocytes. J Med Virol. 1994;43:103–110. doi: 10.1002/jmv.1890430119. [DOI] [PubMed] [Google Scholar]
- Meyer H, Sutter G, Mayr A. Mapping of deletions in the genome of the highly attenuated vaccinia virus MVA and their influence on virulence. J Gen Virol. 1991;72 (Pt 5):1031–1038. doi: 10.1099/0022-1317-72-5-1031. [DOI] [PubMed] [Google Scholar]
- Meyer RG, Britten CM, Siepmann U, Petzold B, Sagban TA, Lehr HA, Weigle B, Schmitz M, Mateo L, Schmidt B, Bernhard H, Jakob T, Hein R, Schuler G, Schuler-Thurner B, Wagner SN, Drexler I, Sutter G, Arndtz N, Chaplin P, Metz J, Enk A, Huber C, Wolfel T. A phase I vaccination study with tyrosinase in patients with stage II melanoma using recombinant modified vaccinia virus Ankara (MVA-hTyr) Cancer Immunol Immunother. 2005;54:453–467. doi: 10.1007/s00262-004-0616-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooij P, Nieuwenhuis IG, Knoop CJ, Doms RW, Bogers WM, Ten Haaft PJ, Niphuis H, Koornstra W, Bieler K, Kostler J, Morein B, Cafaro A, Ensoli B, Wagner R, Heeney JL. Qualitative T-helper responses to multiple viral antigens correlate with vaccine-induced immunity to simian/human immunodeficiency virus infection. The Journal of Virology. 2004;78:3333–3342. doi: 10.1128/JVI.78.7.3333-3342.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B. Genetically engineered poxviruses for recombinant gene expression, vaccination, and safety. Proc Natl Acad Sci USA. 1996;93:11341–11348. doi: 10.1073/pnas.93.21.11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B, Earl PL. Expression of proteins in mammalian cells using vaccinia virus vectors. In: Coligan JE, Kruisbeek AM, Margulies DH, Shevach E, Strober W, editors. Protocols in Immunology. Greene Publishing; New York: 1998. pp. 16.15.1–16.21.9. [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nemeckova S, Hainz P, Otahal P, Gabriel P, Sroller V, Kutinova L. Early gene expression of vaccinia virus strains replicating (Praha) and non-replicating (modified vaccinia virus strain Ankara, MVA) in mammalian cells. Acta Virol. 2001;45:243–247. [PubMed] [Google Scholar]
- Pascolo S, Bervas N, Ure JM, Smith AG, Lemonnier FA, Perarnau B. HLA-A2.1-restricted education and cytolytic activity of CD8(+) T lymphocytes from beta2 microglobulin (beta2m) HLA-A2.1 monochain transgenic H-2Db beta2m double knockout mice. The Journal of Experimental Medicine. 1997;185:2043–2051. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez JC, Gherardi MM, Esteban M. Biology of attenuated modified vaccinia virus Ankara recombinant vector in mice: virus fate and activation. The Journal of Virology. 2000;74:923–933. doi: 10.1128/jvi.74.2.923-933.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochester SC, Traktman P. Characterization of the single-stranded DNA binding protein encoded by the vaccinia virus I3 gene. J Virol. 1998;72:2917–2926. doi: 10.1128/jvi.72.4.2917-2926.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz M, Sodeik B, Ericsson M, Wolffe EJ, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. The Journal of Virology. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm A, Enzinger C, Felder E, Chaplin P. Genetic stability of recombinant MVA-BN. Vaccine. 2006;24:4618–4621. doi: 10.1016/j.vaccine.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Wang Z, La Rosa C, Lacey SF, Maas R, Mekhoubad S, Britt WJ, Diamond DJ. Attenuated poxvirus expressing three immunodominant CMV antigens as a vaccine strategy for CMV infection. J Clin Virol. 2006;35:324–331. doi: 10.1016/j.jcv.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Wang Z, La Rosa C, Li Z, Ly H, Krishnan A, Martinez J, Britt WJ, Diamond DJ. Vaccine properties of a novel marker gene-free recombinant modified vaccinia Ankara expressing immunodominant CMV antigens pp65 and IE1. Vaccine. 2007;25:1132–1141. doi: 10.1016/j.vaccine.2006.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, La Rosa C, Maas R, Ly H, Brewer J, Mekhoubad S, Daftarian P, Longmate J, Britt WJ, Diamond DJ. Recombinant modified vaccinia virus Ankara expressing a soluble form of glycoprotein B causes durable immunity and neutralizing antibodies against multiple strains of human cytomegalovirus. The Journal of Virology. 2004a;78:3965–3976. doi: 10.1128/JVI.78.8.3965-3976.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, La Rosa C, Mekhoubad S, Lacey SF, Villacres MC, Markel S, Longmate J, Ellenhorn JD, Siliciano RF, Buck C, Britt WJ, Diamond DJ. Attenuated Poxviruses Generate Clinically Relevant Frequencies of CMV-Specific T cells. Blood. 2004b;104:847–856. doi: 10.1182/blood-2003-10-3469. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhou W, Srivastava T, La Rosa C, Mandarino A, Forman SJ, Zaia JA, Britt WJ, Diamond DJ. A fusion protein of HCMV IE1 exon4 and IE2 exon5 stimulates potent cellular immunity in an MVA vaccine vector. Virology. 2008;377:379–390. doi: 10.1016/j.virol.2008.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Martinez J, Zhou W, La Rosa C, Srivastava T, Dasgupta A, Rawal R, Li Z, Britt WJ, Diamond D. Modified H5 promoter improves stability of insert genes while maintaining immunogenicity during extended passage of genetically engineered MVA vaccines. Vaccine. 2010;28:1547–1557. doi: 10.1016/j.vaccine.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills MR, Carmichael AJ, Mynard K, Jin X, Weekes MP, Plachter B, Sissons JG. The human CTL response to cytomegalovirus is dominanted by structural protein pp65: Frequency, specificity, and T Cell Receptor usage of pp65-Specific CTL. The Journal of Virology. 1996;70:7569–7579. doi: 10.1128/jvi.70.11.7569-7579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt LS, Belyakov IM, Earl PL, Berzofsky JA, Moss B. Enhanced cell surface expression, immunogenicity and genetic stability resulting from a spontaneous truncation of HIV Env expressed by a recombinant MVA. Virology. 2008;372:260–272. doi: 10.1016/j.virol.2007.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt LS, Earl PL, Xiao W, Americo JL, Cotter CA, Vogt J, Moss B. Elucidating and minimizing the loss by recombinant vaccinia virus of human immunodeficiency virus gene expression resulting from spontaneous mutations and positive selection. The Journal of Virology. 2009;83:7176–7184. doi: 10.1128/JVI.00687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Schneider DL, Zuckerman AJ, Du W, Dickson B, Maddrey WC. Adult hepatitis B vaccination using a novel triple antigen recombinant vaccine. Hepatology. 2001;34:372–376. doi: 10.1053/jhep.2001.26167. [DOI] [PubMed] [Google Scholar]
- Yue Y, Wang Z, Abel K, Li J, Strelow L, Mandarino A, Eberhardt MK, Schmidt KA, Diamond DJ, Barry PA. Evaluation of recombinant modified vaccinia Ankara virus-based rhesus cytomegalovirus vaccines in rhesus macaques. Med Microbiol Immunol. 2008;197:117–123. doi: 10.1007/s00430-008-0074-5. [DOI] [PubMed] [Google Scholar]