Abstract
The human immunodeficiency virus type 1 (HIV-1) regulator Tat is essential for viral replication because it achieves complete elongation of viral transcripts. Tat can be released to the extracellular space and taken up by adjacent cells, exerting profound cytoskeleton rearrangements that lead to apoptosis. In contrast, intracellular Tat has been described as protector from apoptosis. Tat gene is composed by two coding exons that yield a protein of 101 amino acids (aa). First exon (1–72aa) is sufficient for viral transcript elongation and second exon (73–101 aa) appears to contribute to non-transcriptional functions. We observed that Jurkat cells stably expressing intracellular Tat101 showed gene expression deregulation 4-fold higher than cells expressing Tat72. Functional experiments were performed to evaluate the effect of this deregulation. First, NF-κB-, NF-AT- and Sp1-dependent transcriptional activities were greatly enhanced in Jurkat-Tat101, whereas Tat72 induced milder but efficient activation. Second, cytoskeleton-related functions as cell morphology, proliferation, chemotaxis, polarization and actin polymerization were deeply altered in Jurkat-Tat101, but not in Jurkat-Tat72. Finally, expression of several cell surface receptors was dramatically impaired by intracellular Tat101 but not by Tat72. Consequently, these modifications were greatly dependent on Tat second exon and they could be related to the anergy observed in HIV-1-infected T cells.
INTRODUCTION
The human immunodeficiency virus type 1 (HIV-1) transcriptional activator (Tat) is essential for viral gene expression and replication (1). Tat fulfils an efficient viral transcript elongation through the interaction with the Tat response element (TAR), a stem-loop RNA located at the 5′-end of nascent viral transcripts that enables the recruitment of cellular factors as P-TEFb to increase the functional capacity of the RNA polymerase II (RNAPII) (2). P-TEFb is composed of cyclin T1, which directly interacts with Tat and permits the binding to TAR (3), and CDK9, which hyperphosphorylates RNAPII (4). Other functions have also been attributed to Tat as enhancing viral reverse transcription (5), mimicking chemokine functions (6), suppressing antigen specific CD8+ T-cell immune response (7), and regulating host cell gene expression through binding to canonical enhancer sequences of cellular transcription factors (8) or creating a complex interactome formed by several host cell proteins involved in gene expression regulation (9). Tat can also be released from infected cells and taken up by adjacent non-infected cells through receptor-mediated or adsorptive endocytosis (10,11), acting then as a secretable growth factor, T-cell activator, and modulator of apoptosis (12,13). However, Tat also shows non-transcriptional activities as the modulation of cellular protein synthesis, thereby affecting the metabolism of host cells (12,14).
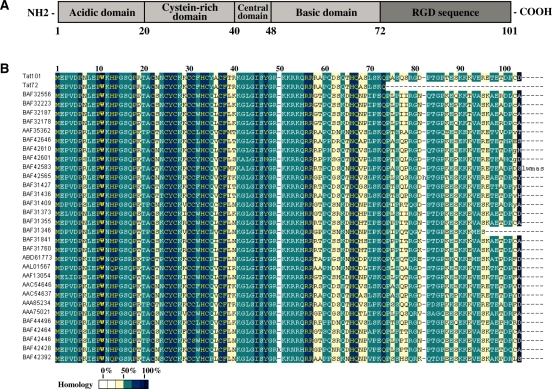
Tat coding gene consists of two spliced exons separated in the HIV-1 genome by more than 2300 nts. After complete splicing of the viral pre-mRNA, a highly conserved protein of 101 residues is synthesized (15). Tat101 is the most common protein in clinical HIV-1 isolates but several laboratory virus strains (LAI, HXB2, BRU, NL4.3) encode a Tat86 protein (3), a non-natural truncated form that appears to be fully functional (15). Tat is composed by several domains (Figure 1A): the cysteine-rich domain [amino acid (aa) 22–37] is required for Tat transcriptional activity (16); the central domain (aa 38–48) contains the conserved 36VCFT39 motif involved in tubulin binding and apoptosis (17–19), and the 41KGLGI45 motif that constitutes the minimal activation domain together with the cysteine-rich domain (20); the basic domain (aa 49–57) contains the nuclear localization signal 49RKKRRQRRR57 necessary for binding to TAR (21,22), Tat cellular uptake (23), and nuclear translocation (24), and this region is the minimal neurotoxic region causing cell death (25); the glutamine-rich region (aa 60–72) has been involved in Tat-mediated apoptosis of T-cells (26); and finally, the second exon (aa 73–101) is the least conserved region between different isolates, with homology below 50% (Figure 1B). This region contains two main motifs: an 78RGD80 sequence absent in HIV-2 and SIV (simian immunodeficiency virus) Tat that is involved in cell adhesion (27); and the 86ESKKKVE92 motif that has been described as critical for NF-κB transactivation (28). The second exon has been involved in several activities as the cellular uptake of the exogenous Tat (29), apoptosis through increasing activity of caspase-8 (30,31), improvement of HIV-1 replication in tissue culture and primary PBLs (2,28,32), and IL-2 superinduction in HIV-1-infected T cells after CD3/CD28 co-stimulation (33). The contribution of the second exon to HIV-1 in vivo replication has also been demonstrated after the accidental infection of three laboratory workers with the HXB2 HIV-1 isolate that shows a premature stop codon at the aa 89. In two of these patients, this mutation reverted spontaneously yielding a more pathogenic virus (34,35). These results were also confirmed in four macaques infected with SIVtat1ex virus that reverted to wild-type SIV in two of them, correlating with increased viral load and decreased CD4+ T-cell count (35).
Figure 1.
HIV-1 Tat protein structure. (A) Full-length Tat protein consists of 101aa. Tat first exon expands from 1 to 72 residues, while second exon expands from 73 to 101 residues. Tat complete sequence can be divided into five conserved domains. A highly conserved motif 41KGLGI45 located at the central domain constitutes the minimal activation domain together with both N-terminal acidic and cysteine-rich domains. The highly conserved motif 49RKKRRQRRR57 (NLS) located at the basic domain confers Tat the ability to bind TAR and translocate to the nucleus. The second exon region contains the rather conserved motif 78RGD80 involved in cell adhesion signal and a partially conserved 86ESKKKVE92 motif involved in NF-κB transactivation. (B) Alignment of Tat protein sequences from several clinical isolates shows that the first exon is highly conserved amongst different HIV-1 isolates, whereas the second exon shows homology lower than 50%. Sequence names provided correspond to the GenBank accession number for each sequence. Sequences of Tat101 and Tat72 proteins used in this study are also included.
Both extra and intracellular Tat display the same structure, but this protein acts differently when is synthesized inside the cell during HIV-1 infection than when is released from infected cells. Intracellular Tat is able to protect cells from apoptosis (14,36,37), whereas extracellular Tat is considered a dose-dependent pro-apoptotic factor (38–41) that contributes to the acquired immunodeficiency syndrome (AIDS)-associated immunosuppresion and pathologies as Kaposi’s sarcoma, dementia and degenerative diseases (17,42–44). One pro-apoptotic mechanism described for extracellular Tat involves its attachment to microtubules and to the actin cytoskeleton (18,19,26,44–46), altering the cytoskeleton dynamics. Cytoskeleton is involved in cell motility, proliferation and intracellular trafficking, as well as in cell shape maintenance and polarity. Actin filaments, actin-binding proteins and microtubules are not only essential for regulating cell death, but also play a major role in HIV-1 infection. Viral trafficking is mediated through microtubules by retrograde transport, the viral core uses dynein to move toward the nuclear pore complex (NPC) (47), and the cytoskeleton reorganization is necessary to initiate HIV-1-induced membrane fusion, viral assembly and budding (48). In this context, although intracellular Tat101 induces down-regulation of several cytoskeletal proteins, it does not cause cell death (14), suggesting that intracellular Tat has a role in the modification of the cytoskeleton structure for adapting the host cell to HIV-1 infection (49).
In the present work, the contribution of intracellular Tat to changes in the T-cell cytoskeleton structure and function and the specific role of the second exon in these modifications were analysed by microarrays and functional assays. Deregulation of gene expression was 4-fold higher in T cells expressing intracellular Tat101 than Tat truncated at the second exon (Tat72). We prove that the C-terminus is important for modifying cytoskeleton functions as cell morphology, growth, proliferation, and chemotaxis, as well as T-cell signalling and activation. Furthermore, in silico modelling of Tat72 suggests that the second exon can induce higher affinity or strength in the binding of Tat to its target sequences or protein partners.
MATERIALS AND METHODS
Cells
Jurkat cell lines were cultured in RPMI 1640 medium (Biowhitaker, Walkersville, MD, USA) with 10% fetal calf serum (PAN Biotech GmbH, Aidenbach, Germany), 2 mM l-glutamine, 100 µg/ml streptomycin and 100 U/ml penicillin, at 37°C. Jurkat TetOff cell line was purchased from BD Biosciences Clontech (Mountain View, CA, USA) and cultured according to manufacturer’s instructions. Jurkat TetOff cells stably transfected with HIV-1 pTRE2hyg-tat101 (Jurkat-Tat101) or pTRE2hyg-tat72 (Jurkat-Tat72) expression vectors were maintained in 200 µg/ml geneticin/G418 (Sigma-Aldrich, St. Louis, MO, USA) and 200 µg/ml hygromycin B (BD Biosciences Clontech). Tat expression could be stopped by adding 1 µg/ml doxycycline (BD Biosciences Clontech) in the culture medium for 2 days.
Vectors
Plasmid pCMV-tat101 has already been described by Arenzana-Seisdedos et al. (50). Complementary DNA (cDNA) from tat first exon (1-219 nt; 1–72aa) was obtained from pCMV-tat101 using specific oligonucleotides and cloned in pTRE2hyg vector (BD Biosciences Clontech) using BamHI/NheI cloning sites (pTRE2hyg-tat72 vector). Empty pTRE2hyg vector and pcDNA3.1 vector (Invitrogen, Carlsbad, CA, USA) were used as negative controls. Long terminal repeat (LTR)-LUC vector containing the luciferase (LUC) reporter gene under the control of HIV-1 LTR U3+R region (LAI strain) was described by Bachelerie et al. (51). LTR-EGFP vector was generated by cloning the enhanced green fluorescence protein (EGFP) gene from pEGFP plasmid (BD Bioscience) into LTR-LUC vector using AgeI/NotI cloning sites after removing LUC gene. 3κB-LUC vector that contains a LUC gene under the control of three −κB consensus motifs of the immunoglobulin κ-chain promoter was also described by Arenzana-Seisdedos et al. (50). NF-AT-LUC has been described by Northrop et al. (52) and encompasses three tandem copies of the NF-AT-1 binding site in the interleukin-2 (IL-2) enhancer. The SP1-LUC plasmid contains two consensus sequences for SP1 cloned into the p19LUC vector (53). pSV-β-galactosidase vector, used as control of transfection efficiency, was purchased from Promega (Madison, WI, USA). Plasmids were purified using Qiagen Plasmid Maxi Kit (Qiagen, CA, USA), following the manufacturer’s instructions.
Reagents and antibodies
Monoclonal antibody against HIV-1 Tat (aa 2–9) was obtained from Advanced Biotechnologies Inc. (Columbia, MD, USA). Specific antibody against the β-isoform of actin was obtained from Sigma Aldrich (St. Louis, MO, USA). Antibodies against the chemokine (C-X-C motif) receptor 4 (CXCR4) conjugated with phycoerythrin (PE) and against CD4 and CD1a conjugated with fluorescein isothiocyanate (FITC) were purchased from BD Biosciences. Antibody against ezrin-radixin-moesin (ERM) protein family was kindly gifted by Dr. Francisco Sánchez-Madrid from Hospital de la Princesa, Madrid, Spain (54,55). Polyclonal antibodies against p65/RelA (clone C-20) and against IκBα (clone C-21) were supplied by Santa Cruz Biotechnology (Santa Cruz, CA, USA). Polyclonal antibodies against phosphorylated-NF-κB p65/RelA (Ser536) and against phosphorylated-IκBα (Ser32) were supplied by Cell Signaling Technology (Danvers, MA, USA). Secondary antibodies conjugated with horseradish peroxidase (HRP) were purchased from GE Healthcare (Uppsala, Sweden). Secondary antibodies conjugated to Alexa 488 and TexasRed were purchased from Molecular Probes (Eugene, OR, USA). Stromal cell-derived factor-1 alpha (SDF-1α)/CXCL12 was used at 50 nM and it was kindly provided by Dr. Françoise Baleux (Institut Pasteur, Paris, France). Phytohemagglutinin (PHA) was used at 5 µg/ml. Phalloidin conjugated with FITC was purchased from Invitrogen. Propidium iodide and 4′,6-diamidino-2-phenylindole (Dapi) were obtained from Sigma Aldrich.
Transfection assays
Stable transfection of Jurkat TetOff cells with pTRE2hyg-tat72 and pTRE2hyg expression vectors was performed by electroporation with an Easyjet Plus Electroporator (Equibio, Middlesex, UK). In brief, 5 × 106 cells were collected in 250 µl of RPMI without supplement and mixed with 5 µg of plasmid DNA in an electroporation cuvette with 2 mm electrode gap (Equibio). Cells were transfected by two pulses at 280V, 150 µF and 330Ω. After transfection, cells were incubated in supplemental RPMI at 37°C for 18 h and then geneticin and hygromycin B were added to the culture medium at 50 µg/ml each per day until selection. For transient transfections, 5 × 106 cells were resuspended in 350 µl of RPMI without supplements and mixed with 5 µg of plasmid DNA in an electroporation cuvette with 4 mm electrode gap (Equibio). Cells were transfected by one pulse at 280V and 1500 µF. After transfection, cells were incubated in supplemental RPMI at 37°C for 18 h. LUC and β-Galactosidase activities were assayed using Luciferase Assay System and β-Galactosidase Enzyme Assay System (Promega), respectively, according to manufacturer’s instructions. Relative LUC units (RLUs) were measured in supernatants with a Sirius luminometer (Berthold Detection Systems, Oak Ridge, TN, USA) after the addition of the appropriate substrate; β-Galactosidase activity was measured at 420 nm in a microplate reader Sunrise (Tecan Group Ltd., Männedorf, Switzerland). RLUs were normalized by measuring both β-Galactosidase activity and total protein concentration by the method of Bradford (56) using a bovine serum albumin (BSA) standard curve.
HIV-1 infection
Briefly, 10 × 106 of MT-2 cells were incubated with 0.5 µg/ml of p24-gag of NL4.3 virus for 2 h at gentle rotation and room temperature. Cells were then centrifuged at 2500 rpm for 30 min at 25°C, and washed five times with PBS1x. After incubation for 2 or 7 days post-infection, nuclear protein extracts were obtained as described by Laín de Lera et al. (57) and protein concentration was determined by the method of Bradford using a BSA standard curve (56).
RT–PCR assays
Total RNA from Jurkat cells was isolated with RNeasy Mini kit (Qiagen) following manufacturer’s instructions. Specific primers for amplifying full-length HIV-1 tat gene (306bp) were as follows: Tat-s, 5′-ATGGAGCCAGTAGATCCTA-3′; and Tat-as, 5′-ATCGCACGGATCTGTCTCT-3′. Primers for amplifying tat first exon (219bp) were the former Tat-s and Tat72-as, containing a stop codon (underlined text), 5′-CTATGCTCTGATAGAGAAGCT-3. Primers for amplifying human CD4 gene were as follows: CD4-s, 5′-GCACTTGCTTCTGGTGCTGCAACT-3′, and CD4-as, 5′-ACTGAGGAGTCTCTTGATCTGAGA-3′, yielding a fragment of 1,296bp. Primers for generic amplification of human CD1 genes (A, B, C, D, E) were as follows: CD1-s, 5′-AAGTGAAGCCCGAGGCCTGGCTGT-3′, and CD1-as, 5′-GTCCTGGCCCTCTA GACTGCTGTG-3′, yielding a fragment of 265 bp. Primers for amplifying human β-actin gene were as follows: β-actin-s, 5′-TCACCCACACTGTGCCCATCTA-3′, and β-actin-as, 5′-AGTTGAAGGTAGTTTCGTGGAT-3′, yielding a fragment of 360 bp. RT–PCR assay was performed by using Qiagen OneStep RT–PCR Kit (Qiagen). Briefly, the reaction mix contained 10 µl of buffer 5X, 2 mM MgCl2, 0.1 mM dNTPs, 0.4 µM of each primer pair, and 1 µl of Qiagen OneStep RT-PCR enzyme mix in a final volume of 50 µl. A total of 5 µg of RNA was added and thermal cycling conditions were as follows: an initial cycle of 48°C for 45 min and 95°C for 10 min; 45 cycles of 95°C for 30 s, 55°C for 30 sec, 72°C for 30 s; and a final extension cycle of 72°C for 10 min. Amplified products were analysed by electrophoresis on 3% Seakem agarose gel containing 5 µg/ml of ethidium bromide in 0.5x Tris-acetate buffer. DNA Molecular Weight Marker XIV (Roche Applied Science, Barcelona, Spain) was used as 100 bp ladder. BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) was used for sequencing the amplified DNA fragments according to manufacturer’s instructions by using an ABI Prism 3700 DNA Analyzer (Applied Biosystems). Densitometry analysis was performed using Quantity One software (Bio-Rad). Gel bands were quantified and background noise was subtracted from the images. Relative ratio of optical density units was calculated regarding to the gel band corresponding to the internal control for each lane and each type of RNA sample.
Sequence alignment
Thirty Tat protein sequences obtained from different HIV-1 isolates were obtained at random from GenBank database (58), National Center for Biotechnology Information (NCBI, Rockville Pike, Bethesda, MD, USA). These sequences were aligned with those corresponding to Tat101 and Tat72 proteins used in this study, previously amplified with specific primers and sequenced with BigDye Terminator Cycle Sequencing Kit (Applied Biosystems). Alignment was performed by using the Multiple Alignment Construction & Analysis Workbench (Macaw) software v2.05 (59).
Immunofluorescence assays
For immunofluorescence assays, cells were immobilized in PolyPrep slides (Sigma-Aldrich) for 15 min and then fixed with 2% paraformaldehyde (PFA)–0.025% glutaraldehyde in PBS for 10 min at room temperature. After washing twice with 0.1% glycine/PBS, cells were permeabilized with 0.1% Triton X-100/PBS for 10 min. Cells were then treated with 1 mg/ml NaBH4 for 10 min. Incubation for 1 h at room temperature with each primary and secondary antibodies and subsequent washes were performed with PBS1x-2% BSA-0.05% saponine buffer. Coverslips were immobilized with 70% glycerol/PBS. Images were obtained with a Radiance 2100 confocal microscope (BioRad, Hercules, CA, USA), Leica TCS-SP confocal microscope (Leica Microsystems, Wetzlar, Germany) or a Leica DMI 4000B Inverted Microscope (Leica Microsystems).
Flow cytometry
Cells transiently transfected with vector LTR-EGFP were analysed on a FACScalibur Flow Cytometer (BD Biosciences), using CellQuest software (BD Biosciences). Analysis was performed by gating live (propidium iodide-negative) cells that also showed the highest fluorescent on FL1 green channel (60). For studying changes in cell morphology, live (propidium iodide-negative) cells were analysed by their relative size and complexity in sideward scatter (SSC)/forward scatter (FSC) dot plots. For staining of cell surface antigens, cells were washed with PBS1x and then incubated for 20 min at 4°C with specific antibodies or the appropriate isotype control.
Cell proliferation assays
Cell proliferation was measured with CellTiter-96 Aqueous Non-Radioactive Cell Proliferation Assay (Promega) accordingly to manufacture’s instructions. Briefly, 2 × 104 cells per well were cultured in a 96-well plate in the absence of any stimuli or with 5 µg/ml PHA for 72 h. Compound 3-(4,5-dimethylthiazol-2-yl)-5(3-carboxy methonyphenol)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) was mixed with phenazine methosulfate solution and then added to the culture medium. Absorbance was measured at 490 nm after 3 h incubation. Proliferation rate for each cell type in response to PHA was compared with their proliferation rate in the absence of stimulus.
Analysis of cytoskeleton structure and function
Migration assays were performed in Transwell plates (Costar, Cambridge, MA, USA) of 6.5-mm diameter with 5 µm-pore filters. Briefly, 105 cells diluted in complete medium were plated in the upper compartment, filling the lower compartment with medium alone or with SDF-1α/CXCL12 at 50 nM. Plates were then incubated for 2 h at 37°C with 5% CO2. Migrated cells were collected from the lower compartment and counted by flow cytometry for 1 min sharp. Migration ratio represents the ratio between the migration input in the presence or absence of SDF-1α/CXCL12. For cell polarization assays, Jurkat cells adhered to fibronectin (75 µg/ml) pre-treated coverslips were cultured for 45 min at 37°C in the absence or presence of SDF-1α/CXCL12. Cells were stained with an antibody against ERM proteins and a secondary antibody conjugated with TexasRed. Images were acquired with a Leica TCS-SP confocal microscope (Leica Microsystems). Polarization rate represents the ratio percentage between the number of polarized cells and the number of total cells counted in 15 different microscope fields. For F-actin polymerization assays, cells cultured during 15 s at 37°C in the absence or presence of SDF-1α/CXCL12 were simultaneously fixed and permeabilized with 2% PFA and 0.1% Tritón X-100. Phalloidin conjugated with FITC (6 U/ml) was then added during 15 min at 37°C. Phalloidin binds at the interface between F-actin subunits, avoiding its depolymerization. The amount of polymerized F-actin was determined by flow cytometry.
Immunoblotting assays
Cytosolic and nuclear protein extracts were obtained as described by Laín de Lera et al. (57) and protein concentration was determined by the Bradford method using a BSA standard curve (56). Ten micrograms were fractionated by sodium dodecyl sulfate-polyacrilamide gel electrophoresis (SDS-PAGE) and transferred onto Hybond-ECL nitrocellulose paper (GE Healthcare). After blocking and incubation with primary and secondary antibodies, proteins were detected with SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL, USA). Antibodies against histone H1 (clone AE-4) and NF-κB p105 (clone H-119) provided by Santa Cruz Biotechnology were used as internal control for purity of nuclear and cytosolic extracts, respectively. Images were acquired in a BioRad Geldoc 2000 (BioRad) and densitometry analysis was performed using Quantity One software (Bio-Rad). Relative ratio of optical density units was calculated regarding to the gel band corresponding to the internal control for each lane and each type of protein extracts after subtracting the background noise.
DNA affinity immunoblotting assay
The DNA affinity immunoblotting (DAI) assay was performed as previously described by Liu et al. (61) with minor modifications. Briefly, 100 µg of nuclear protein extracts from Jurkat TetOff, Jurkat-Tat72 and Jurkat-Tat101 cells were incubated at 4°C for 30 min with 25 nM of a 5′-end-labelled biotin probe that contains the double −κB consensus motif located in the HIV-1 LTR promoter (5′-AGCTTACAAGGGACTTTCCGCTGGGGACTTTCCAGGGA-3′), with this probe mutated at the −κB consensus sites (5′AGCTTACAACTCACTTTCCGCTGCTCACTTTCCAGGGA) or with a probe that contains the estrogen receptor (ER) DNA element, a non-related NF-κB gene (62). Biotin-probe/protein complexes were captured by incubating with streptavidin agarose resins (Pierce) at 4°C for 30 min and then collected by centrifugation at 3000g for 10 min. Protein complexes were fractionated by SDS-PAGE and the presence of p65/RelA protein bound to DNA probes was detected by immunoblotting with a specific antibody (clone C-20; Santa Cruz Biotechnology). As internal control 10 µg of protein were separated before adding each probe and resolved by Western-blot using an antibody against β-actin (Sigma-Aldrich).
Microarrays assays
Total RNA from Jurkat cells was isolated with RNeasy Maxi kit (Qiagen) following manufacturer’s instructions. One-Color Microarray-Based Gene Expression Analysis Protocol (Agilent Technologies, Palo Alto, CA, USA) was used to amplify and label RNA. Briefly, 1 µg of total RNA was reverse transcribed using T7 promoter primer and the Moloney murine leukemia virus (MMLV) reverse transcriptase (RT). cDNA was then converted to antisense RNA (aRNA) by using T7 RNA polymerase that amplifies target material and incorporates cyanine 3 (Cy3)-labelled CTP simultaneously. Samples containing 1.65 µg of Cy3-labeled aRNA were hybridized to a Whole Human Genome Microarray 4 × 44K (G4112F, Agilent Technologies) for 17 h at 65°C in a Agilent hybridization oven (G2545A, Agilent Technologies) set to 10 rpm in a final concentration of GEx Hybridization Buffer HI-RPM 1x (Agilent Technologies). Arrays were washed and dried out using a centrifuge according to manufacturer's; instructions (One-Color Microarray-Based Gene Expression Analysis, Agilent Technologies). Arrays were scanned at 5 µm resolution on an Agilent DNA Microarray Scanner (G2565BA, Agilent Technologies) using the default settings for 4 × 44K format one-color arrays. Images provided were analysed using Feature Extraction software v9.5.3.1 (Agilent Technologies).
Statistical analysis
Data files from Feature Extraction software were imported into GeneSpring GX software v9.0 (Agilent Technologies). Quantile normalization was performed and expression values (log2 transformed) were obtained for each probe. Probes were also flagged as Present, Marginal or Absent using GeneSpring default settings. Probes that were flagged as Present or Marginal in all three replicates for the two experimental conditions to be compared on each contrast were selected for further analysis. These filtered data were loaded into SAM (Significance Analysis of Microarrays) software for genomic expression data mining (63). SAM uses the false discovery rate (FDR) and q-value method as described by Storey (64). Expression ratios (log2) were calculated using control cells values as baseline. For considering a fold change as statistically significant, the q-value cutoff was set at 5%.
Functional and canonical pathway analyses
Functional and canonical pathway analyses of specific gene datasets coming from SAM analysis were performed by using Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA, USA). Functional analysis was performed to identify functions and/or diseases that were most significant to the dataset. All genes from the dataset that were associated with biological functions and/or diseases in the Ingenuity knowledge database were considered for the analysis. B-H Multiple Testing Correction p-value test (65) was used to calculate the p-value for determining the probability that each biological function and/or disease assigned to the dataset was due to chance alone. Canonical pathway analysis identified from the Ingenuity Pathway Analysis library those pathways that were more significant to the dataset. All genes associated with a canonical pathway in the Ingenuity knowledge base were considered for the analysis. The significance of the association between the dataset and the canonical pathway was measured in two ways: first, the ratio of the number of genes from the dataset that map to the pathway divided by the total number of molecules that exist in the canonical pathway; and second, the B-H Multiple Testing Correction p-value test was used to calculate a p-value to determine the probability that the association between the genes in the dataset and the canonical pathway was due to chance alone.
Tat in silico modelling
Tat72 protein model was constructed by homology modelling based on the nuclear magnetic resonance (NMR) spectra structure of unbound HIV-1 Tat with 86 residues from HIV-1 BRU isolate (PDB code: 1jfw) (66) plus residues 49–68 from EIAV Tat protein bound to TAR/P-TEFb complex (PDB code: 2w2h) (67) using Modeller 9v4 software (68). Electrostatic potentials were calculated using the Adaptive Poisson–Boltzmann Solver (APBS) software (69) and displayed at the solvation accessible surface with Pymol software (http://www.pymol.org/). Tat interaction with TAR/cyclin T1/CDK9 complex was modelled by substituting EIAV Tat from 2w2h structure by Tat72 model, after structural alignment with Matching Molecular Models obtained from Theory (MAMMOTH) software (70,http://ub.cbm.uam.es/software.php?lang=en). CDK9 structure was incorporated to the complex, mimicking the interaction with cyclin T1 observed in PDB structure 3blh (71), after superposition of cyclinT1 from both complexes with MAMMOTH software. It was necessary to apply subtle deformations to modelled Tat72 using the sculpting tool of Pymol software in order to avoid collisions with cyclin T1. The complex was energy minimized with the SANDER module from the Assisted Model Building with Energy Refinement (AMBER) v8 package of molecular simulation programs (http://ambermd.org/). Hydrogen atom positions and standard atomic charges and radii were assigned according to Amber ff99 force field.
RESULTS
Analysis of Tat72 expression in stably transfected Jurkat cells
Jurkat TetOff cells stably transfected with gene codifying Tat72 protein were generated as described previously by Coiras et al. (14) for the stable Jurkat-Tat101 cell line . All figures show the means of data or representative experiments performed with two independent populations of Jurkat-Tat72 and Jurkat-Tat101 cells. No clones were used. Due to the high mortality induced by long-term treatment with doxycycline, Jurkat TetOff stably transfected with empty plasmid pTRE2hyg were mostly used as negative control.
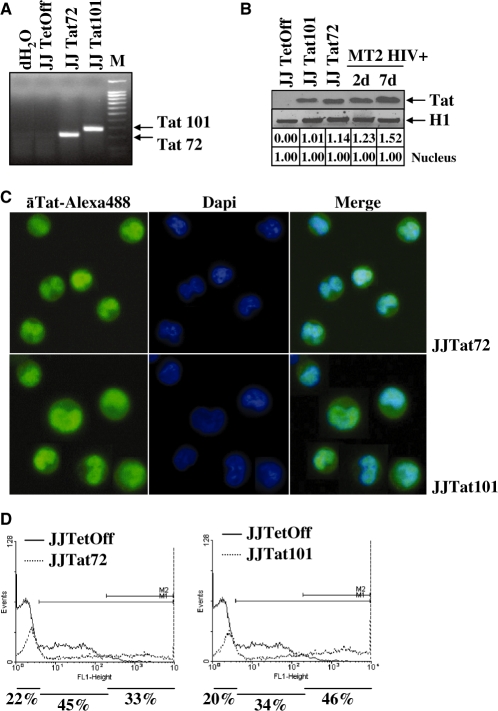
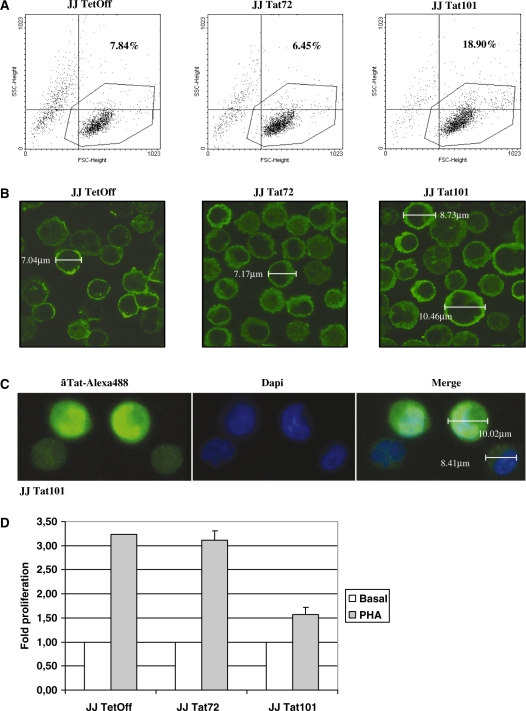
Total RNA from stably transfected Jurkat-Tat72 cells was analysed by RT–PCR. A single band of the expected size (219 nt) was observed, in comparison with the full-length Tat gene (306 nt) detected in Jurkat-Tat101 cells (Figure 2A). RT–PCR assay specificity was proved by automatic sequencing. Expression of Tat protein in both Jurkat-Tat101 and Jurkat-Tat72 cells was highly stable, with comparable half-life, and very similar to a real infection (Figure 2B), as was determined previously by Coiras et al. (14). Jurkat-Tat72 cells were also analysed by immunofluorescence to determine Tat subcellular localization. This truncated protein showed a predominant nuclear localization, similarly to Tat101 (Figure 2C). The percentage of cells that were expressing Tat72 or Tat101 within the whole population was calculated by transient transfection of vector LTR-EGFP and flow cytometry analysis. Jurkat-Tat72 cells were a mixed population in which 33% of cells showed very high expression of Tat (Figure 2D). The role of Tat in LTR transactivation was also confirmed by transfecting LTR-EGFP in the presence of doxycycline (data not shown).
Figure 2.
Characterization of Jurkat-Tat72 cell line. (A) Analysis of mRNA expression for tat gene in Jurkat-Tat101 cells (306 nt) and Jurkat-Tat72 cells (219 nt) by RT–PCR. Jurkat TetOff-pTRE2hyg cells were used as negative control. 100 bp DNA ladder was used as molecular marker. (B) Tat protein expression was analysed in 150 μg of nuclear protein extracts from Jurkat-Tat101 (lane 2) and Jurkat-Tat72 (lane 3) by immunoblotting using a monoclonal antibody against Tat (aa 2–9) and compared with the expression of Tat in MT2 infected with NL4.3 for 2 and 7 days (lanes 4 and 5). Jurkat TetOff-pTRE2hyg cells were used as negative control. Histone H1 was used as internal loading control. Gel bands were quantified by densitometry and background noise was subtracted from the images. Relative ratio of optical density units was calculated regarding to the gel band corresponding to the internal control for each lane and for each protein extract. (C) Subcellular localization of Tat was analysed by immunofluorescence assay in Jurkat-Tat72 and Jurkat-Tat101 using an antibody against Tat and a secondary antibody conjugated with Alexa 488. Dapi was used for nuclear staining. Images were captured by fluorescence microscopy. (D) Analysis of percentage of cells within the whole population expressing high amounts of Tat was performed by flow cytometry 18 h after transfection of Jurkat-Tat72 and Jurkat-Tat101 cells with LTR-EGFP vector. Histograms show one representative experiment selected from three independent experiments. EGFP expression from control cells was used as basal fluorescence (continuous line) compared to EGFP expression from Jurkat-Tat72 or Jurkat-Tat101 (discontinuous line). Percentage of Tat activity was calculated by subtracting the EGFP expression corresponding to control cells from the EGFP expression for each Tat-expressing cell line.
Transcriptional activity induced by Tat101 or Tat72 intracellular expression
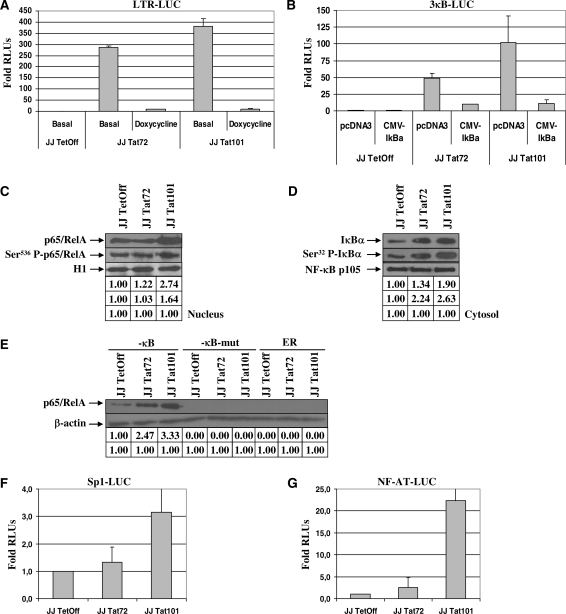
Jurkat-Tat101 and Jurkat-Tat72 were transiently transfected with vector LTR-LUC and LUC activity was measured after 18 h. Both Tat72 and Tat101 were able to promote the transactivation of the HIV-1 LTR, although full-length Tat was slightly more efficient in LTR transactivation than Tat72 (Figure 3A). Role of Tat in LTR transactivation was assessed by treatment with doxycycline. Basal transcriptional activity due to NF-κB exclusively was also evaluated by transient transfection of vector 3κB-LUC. κB-dependent activity was also higher in Jurkat-Tat101 than in Jurkat-Tat72 cells (Figure 3B). NF-κB-dependent synthesis of LUC was confirmed by simultaneous co-transfection of vector CMV-IκBα to produce the over-expression of IκBα, the main inhibitor of NF-κB. Analysis by immunoblotting of nuclear localization of the main active NF-κB protein, p65/RelA, showed that while this protein remained unchanged in Jurkat-Tat72, total p65/RelA was 2.7-fold increased in Jurkat-Tat101 and Ser536-phosphorylated p65/RelA was 1.6-fold increased (Figure 3C). Cytosolic IκBα was mainly increased in Jurkat-Tat101 cells, but Ser32-phosphorylated IκBα was more than 2-fold increased in both cell types (Figure 3D). Because IκBα resynthesis is dependent on the NF-κB activity, higher quantity of IκBα proved the existence of high NF-κB activity in Jurkat-Tat101 and Jurkat-Tat72. Besides, IκBα phosphorylated at Ser32 is susceptible to be degraded in the proteasome, letting active NF-κB free to be translocated to the nucleus. NF-κB activity in Jurkat-Tat72 and Jurkat-Tat101 was measured by DAI assay using a specific probe containing both -κB consensus sites located at the HIV-1 LTR promoter. Free NF-κB able to bind the promoter was more than 2-fold increased in Jurkat-Tat72 and more than 3-fold increased in Jurkat-Tat101 when compared with control cells (Figure 3E). NF-AT- and Sp1-dependent transcriptional activities were also analysed by transient transfection of vectors Sp1-LUC (Figure 3F) or NF-AT-LUC (Figure 3G). Both NF-AT and Sp1 activity were greatly enhanced in Jurkat-Tat101 cells, whereas Jurkat-Tat72 showed similar activity than control cells. Same results were obtained when Jurkat-Tat cells were treated with doxycycline immediately after transfection (data not shown).
Figure 3.
HIV-1 LTR transactivation by Tat intracellular expression. (A) Basal LTR-dependent transactivation activity was measured in Jurkat-Tat72 and Jurkat-Tat101 by transient transfection of LTR-LUC in the absence of stimulus or after treatment with 1 µg/ml doxycycline for 18 h to inhibit Tat expression. Fold mean of RLUs corresponding to three independent experiments was represented as a bar diagram. Lines on the top of the bars correspond to the standard deviation (SD). (B) NF-κB-dependent transactivation activity was measured by transient transfection of Jurkat-Tat72 and Jurkat-Tat101 with 3κB-LUC in the absence of stimulus. Specificity of NF-κB-dependent transactivation was assessed by co-transfection of CMV-IκBα. Co-transfection of pcDNA3 vector was used as negative control. Fold mean of RLUs corresponding to three independent experiments is represented and lines on the top of the bars correspond to SD. (C and D) NF-κB activation was also analysed by immunoblotting of 30 μg of nuclear protein extracts, using an antibody against total p65/RelA or Ser536-phosphorylated p65/RelA (C); and by immunoblotting of 30 μg of cytosolic protein extracts, using an antibody against total IκBα or Ser32-phosphorylated IκBα (D). Histone H1 was used as internal loading control of nuclear protein extracts and NF-κB p105 was used as internal loading control of cytosolic protein extracts. Gel bands were quantified by densitometry and background noise was subtracted from the images. Relative ratio of optical density units was calculated regarding to the gel band corresponding to the internal control for each lane and for each protein extract. (E) NF-κB activity was further analysed in Jurkat-Tat72 and Jurkat-Tat101 in comparison with control cells by using DAI assay. Nuclear protein extracts were incubated with a specific probe containing two -κB consensus sites labelled with biotin. After immunoprecipitation with an antibody against p65/RelA, protein/DNA complexes were purified, analysed by SDS-PAGE and immunoblotted with an antibody against p65/RelA. Non-related ER DNA element and mutated -κB probes were used as control of specificity. Gel bands were quantified by densitometry and background noise was subtracted from the images. Relative ratio of optical density units was calculated regarding to the gel band corresponding to the internal control for each lane and for each sample. (F and G) Sp1- and NF-AT-dependent transactivation activity was measured by transient transfection of either Sp1-LUC or NF-AT-LUC plasmid. Fold mean of RLUs corresponding to three independent experiments is represented and lines on the top of the bars correspond to SD. pSV-β-galactosidase vector was used as transfection internal control.
Bioinformatics analysis and microarray data clustering
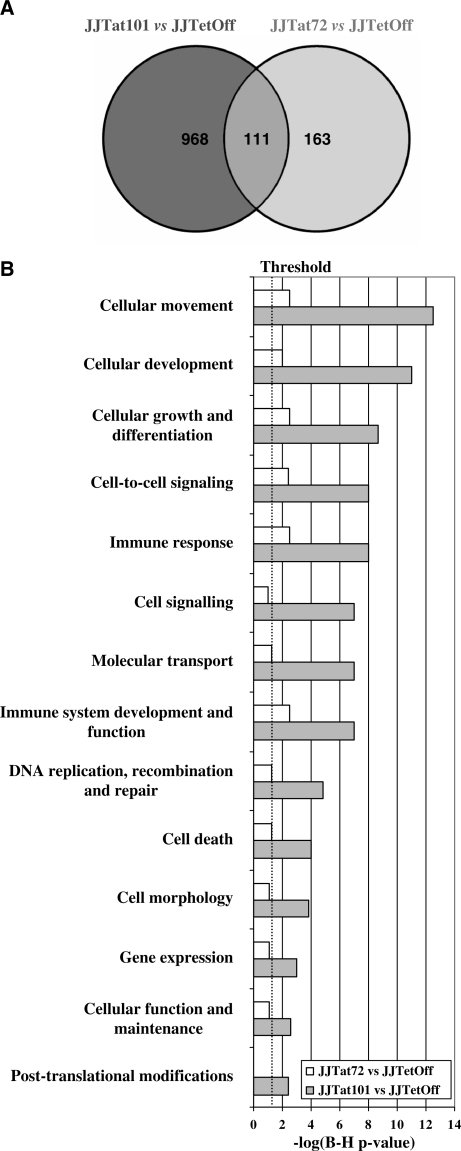
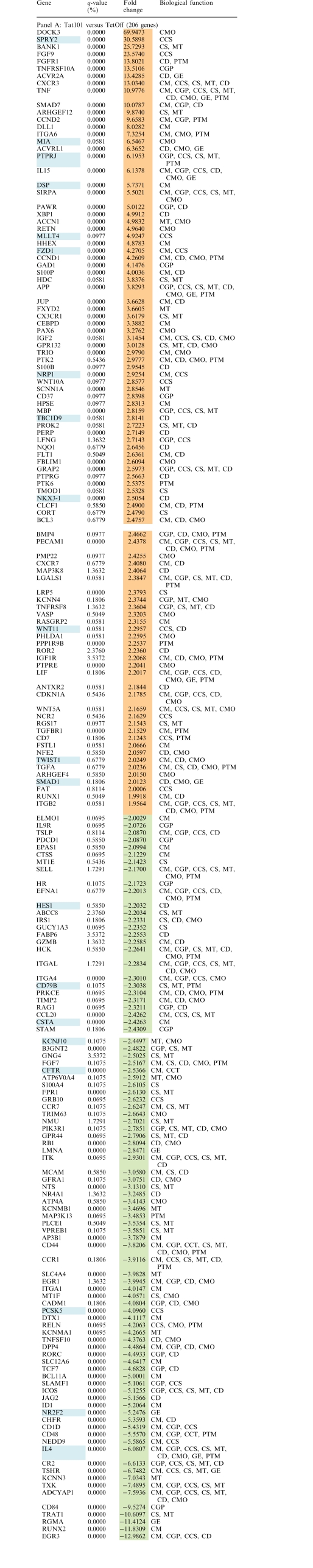
Gene expression patterns of total RNA from Jurkat-Tat101 and Jurkat-Tat72 cell lines from three independent extractions were examined using whole human genome microarrays containing 44 000 probes representing 41 000 human genes and transcripts. Due to the high toxicity of long-term treatment with doxycycline, Jurkat TetOff cells stably transfected with empty vector pTRE2hyg were used as control of basal expression. After filtering the scanned images, 25 069 and 25 863 gene probes were considered for statistical analysis for the Jurkat-Tat101 versus control comparison and the Jurkat-Tat72 versus control comparison, respectively. Expression values (log2 transformed) were obtained for each probe in three replicates for all cell types. Expression ratios (log2) were calculated using control cells values as baseline. Only probes with q-value <5% and fold change >2 or <−2 were considered as statistically significant. These statistical analysis criteria yielded 1079 and 274 gene probes for Jurkat-Tat101 versus control and Jurkat-Tat72 versus control comparisons, respectively. These results were summarized in a Venn diagram (Figure 4A). Within these significant gene probes, 111 genes located at the overlapped area were deregulated in both Jurkat-Tat101 and Jurkat-Tat72 cell lines. Ontological analysis of gene expression data suggested that some essential cellular functions were changed in Jurkat-Tat101, being this change significantly lower in Jurkat-Tat72 (Figure 4B). Abbreviated grouping of deregulated genes shorted by fold change and indicating their biological functions, is given in Table 1, panel A (Jurkat-Tat101 versus control) and panel B (Jurkat-Tat72 versus control).
Figure 4.
Gene expression and bioinformatics analysis. (A) Gene expression patterns of Jurkat-Tat101 and Jurkat-Tat72 were compared with control cells. Data were considered statistically significant when q-value was <5% and fold change >2 or <−2. These filtering criteria yielded 1079 analysable gene spots for Jurkat-Tat101 versus control, and 274 analysable gene spots for Jurkat-Tat72 versus control. These results are summarized in a Venn diagram. The overlapped area represents 111 genes that were deregulated in both cell lines. (B) Analysis of functional and canonical pathways for specific gene datasets generated with the B-H Multiple Testing Correction p-value determined that all cellular functions were mostly altered in Jurkat-Tat101. Data representing the −log(B–H p-value) are shown in a histogram for Jurkat-Tat101 versus control and Jurkat-Tat72 versus control comparisons.
Table 1.
Deregulated genes in Jurkat-Tat101 and Jurkat-Tat72 cells
 |
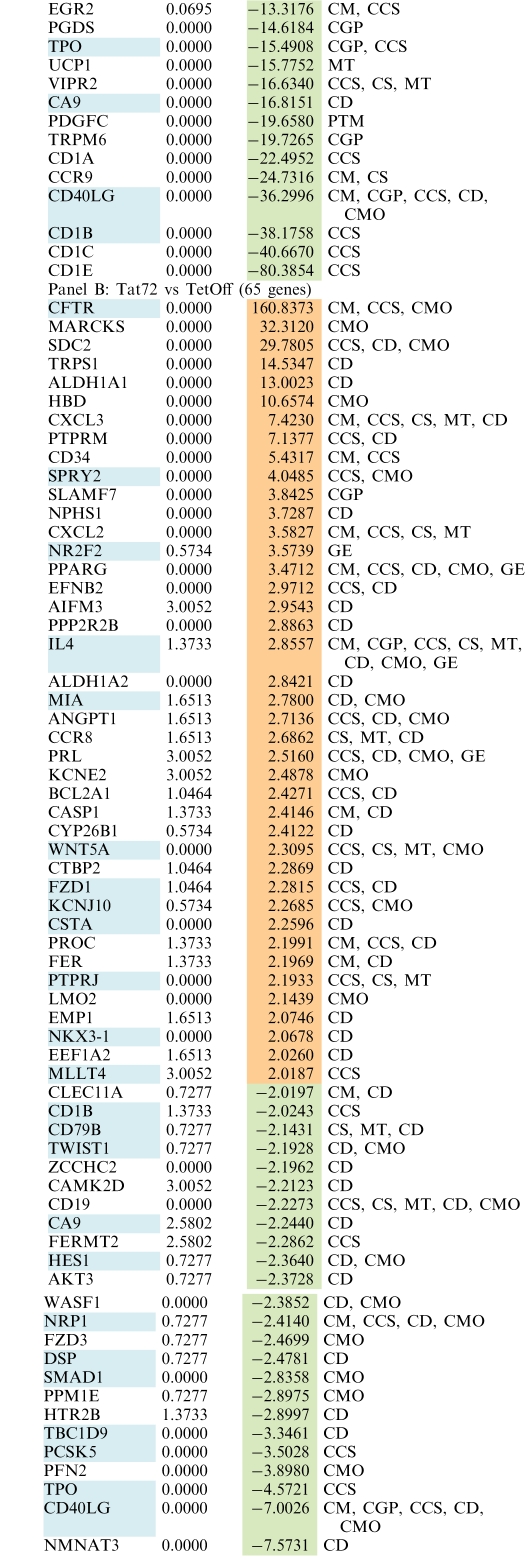 |
Fold change expression values for genes deregulated in Jurkat-Tat101 versus control (Panel A) and Jurkat-Tat72 versus control (Panel B) are listed according to higher (orange) and lower (green) expression. q-value <5% was fulfilled by all genes and fold change was >2 or <−2. Genes marked in blue represent those genes that are deregulated in both comparisons. Biological functions in which these gene could be involved are abbreviated as follows: CCS, Cell-to-Cell Signaling; CD, Cell Death; CGP, Cell Growth and Proliferation; CM, Cell Movement; CMO, Cell Morphology; CS, Cell Signaling; GE, Gene Expression; MT, Molecular Transport; PTM, Postraductional Modifications.
Within the essential functions potentially modified by the stable intracellular expression of Tat are noticeable those related to the cytoskeletal structure, as cellular movement, growth and proliferation, apoptosis and cell morphology. However, differences in gene expression only reflect differences in RNA levels. Accordingly, several experiments were performed to prove that the related cellular functions were modified.
Cellular morphology and growth of Jurkat-Tat101 and Jurkat-Tat72 cells
Jurkat-Tat101 and Jurkat-Tat72 cells were analysed by flow cytometry to determine differences in cell shape. Jurkat-Tat72 showed similar size (FSC) and complexity (SSC) than control cells, but Jurkat-Tat101 population was scattered, being more than 11% of the population bigger and more complex (Figure 5A). These results were confirmed by immunofluorescence analysis using an antibody against β-actin (Figure 5B). Jurkat-Tat101 cells presenting bigger size were also expressing higher quantity of nuclear Tat (Figure 5C).
Figure 5.
Changes in cell morphology and proliferation induced by Tat intracellular expression. (A) Relative cell size (forward scatter, FSC) and complexity (sideward scatter, SSC) of Jurkat-Tat72 and Jurkat-Tat101 was analysed by flow cytometry after staining with propidium iodide. Representative SSC/FSC dot plots for each cell type were selected from three independent experiments. Percentage of cells inside the dot plot upper right quadrant corresponds to cells with higher size and complexity than control cells. (B) Jurkat-Tat72 and Jurkat-Tat101 were analysed by confocal microscopy after staining with an antibody against β-actin and a secondary antibody conjugated with Alexa 488. Cell diameter was calculated in some selected cells. (C) Analysis of Jurkat-Tat101 by confocal microscopy after staining with a monoclonal antibody against Tat and a secondary antibody conjugated with Alexa 488 confirmed that cells with higher size were also expressing higher amounts of intranuclear Tat. Dapi was used for nuclear staining. Cell diameter was measured in cells expressing high or low levels of Tat. (D) PHA-induced proliferation was measured in both Jurkat-Tat72 and Jurkat-Tat101 in comparison with control cells. The histogram shows the fold-change mean of three different experiments and lines on the top of the bars represent the SD.
It was hypothesized that the modification in cell shape could impair the migration capacity of these cells. Accordingly, cell proliferation assays were performed in response to the mitogen phytohemagglutinin (PHA). All cell types showed a similar growth rate in the absence of stimulus, but proliferation upon PHA treatment was rather impaired in Jurkat-Tat101 cells in comparison with both Jurkat-Tat72 and control cells, which showed a similar proliferation rate (Figure 5D).
Cell movement and cytoskeleton structure of Jurkat-Tat101 and Jurkat-Tat72 cells
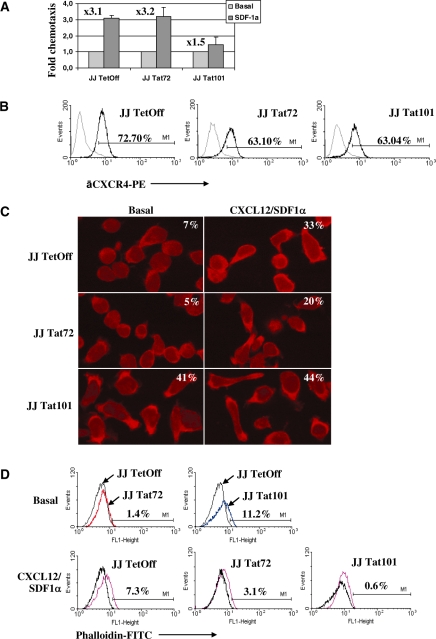
Jurkat-Tat72 showed similar ability as control cells for migrating in response to SDF-1α/CXCL12, but this capacity was impaired in Jurkat-Tat101 (Figure 6A). This chemotaxis impairment was also observed in human peripheral blood lymphocytes (PBLs) from healthy donors transiently transfected with Tat101 (data not shown). This impairment was not due to a significant deficient expression of CXCR4—the major receptor for SDF-1α/CXCL12 (Figure 6B). Extracellular Tat can inhibit SDF-1/CXCL12 signalling through binding to CXCR4 (72) but Tat was not detected in the culture medium of Jurkat-Tat101 or Jurkat-Tat72 as a soluble form (data not shown). Basal and SDF-1α/CXCL12-induced polarization capacity was also measured in Jurkat-Tat101 and Jurkat-Tat72 in comparison with control cells by confocal microscopy after staining with an antibody against ERM proteins (Figure 6C), which are involved in the linking of transmembrane proteins to the actin cytoskeleton (73). More than 40% of Jurkat-Tat101 cells showed a basal polarized morphology in the absence of chemoattractant stimulus, in comparison with less than 10% of polarized cells in both Jurkat-Tat72 and control cell populations. Treatment of Jurkat-Tat101 with SDF-1α/CXCL12 did not significantly modify the polarized morphology of these cells that seemed unable to respond to the stimulus. These data correlate with the chemotaxis impairment observed in Figure 6A. Analysis by flow cytometry after staining with phalloidin conjugated to FITC showed that basal F-actin polymerization was more than 10% increased in Jurkat-Tat101 (Figure 6D) and could not be further induced by SDF-1α/CXCL12, in contrast with control cells. Jurkat-Tat72 showed a moderate ability to respond to SDF-1α/CXCL12.
Figure 6.
Impairment of cytoskeleton functions by the intracellular expression of Tat. (A) Analysis of SDF-1α/CXCL12-induced migration capacity in Jurkat-Tat72 and Jurkat-Tat101 in comparison with control cells. Histogram shows the fold-change mean relative to three different experiments and lines on the top of the bars represent the SD. (B) Flow cytometry analysis of CXCR4 expression on the cell surface of Jurkat-Tat72 and Jurkat-Tat101 after staining with a specific antibody conjugated with PE (line in bold). An antibody against whole IgG2a conjugated with PE was used as isotype control (discontinuous line). (C) Jurkat-Tat72, Jurkat-Tat101 and control cells treated or not with SDF-1α/CXCL12 were adhered to fibronectin coverslips and stained with an antibody against ERM proteins and a secondary antibody conjugated with TexasRed. Images were acquired by confocal microscopy and one representative experiment selected from three independent experiments is shown. Percentage of polarized cells was determined by calculating the ratio between the number of polarized cells and the number of total cells counted in 15 different microscope fields. (D) Basal F-actin polymerization in Jurkat-Tat72 (red line) and Jurkat-Tat101 (blue line) was analysed by flow cytometry after labelling with phalloidin-FITC in comparison with control cells (black line). F-actin polymerization in response to SDF-1α/CXCL12 was also measured in control cells (purple line), Jurkat-Tat72 and Jurkat-Tat101 (pink lines) using each cell background as basal control (black line). One representative experiment selected from three independent experiments is shown.
Expression of cell surface receptors in Jurkat-Tat101 and Jurkat-Tat72 cells
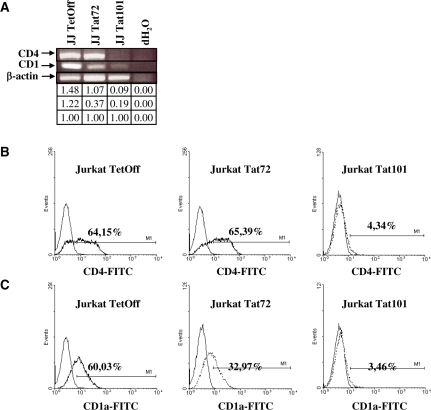
Microarrays showed decrease of CD4 and CD1 (CD1a, b, c, d, e) gene expression in both Jurkat-Tat101 and Jurkat-Tat72. Parameters of statistical significance were fulfilled for all CD1 genes but not for CD4 gene, which showed q-value >5%. However, analysis by RT–PCR of mRNA expression for CD4 gene showed that it was severely reduced in Jurkat-Tat101, as well as CD1 (Figure 7A). Flow cytometry analysis showed that CD4 and CD1a surface expression was greatly diminished in Jurkat-Tat101 cells. CD4 surface expression in Jurkat-Tat72 remained similar to control cells, but CD1a diminished nearly 2-fold (Figure 7B and C). Both CD4 and CD1a expression was annulled in both populations of Jurkat-Tat101, proving that those changes in CD4 expression were not due to random fluctuations. Moreover, treatment with doxycycline partially restored CD4 and CD1a expression in Jurkat-Tat101 (data not shown).
Figure 7.
Expression of CD1 and CD4 in Jurkat-Tat72 and Jurkat-Tat101. (A) mRNA expression for CD1 (generic) and CD4 in Jurkat-Tat101 and Jurkat-Tat72 was analysed by semiquantitative RT–PCR. β-actin gene was used as housekeeping gene. Gel bands were quantified by densitometry and relative ratio of optical density units was calculated regarding to the internal control for each lane and each RNA sample. (B and C) Flow cytometry analysis of CD4 (B) and CD1a (C) expression on the cell surface of Jurkat-Tat72 and Jurkat-Tat101 after staining with specific antibodies conjugated with FITC (discontinuous line). An antibody against whole IgG1 conjugated with FITC was used as isotype control (continuous line). One representative experiment out of three independent experiments is shown.
In silico modelling of Tat86 and Tat72 proteins
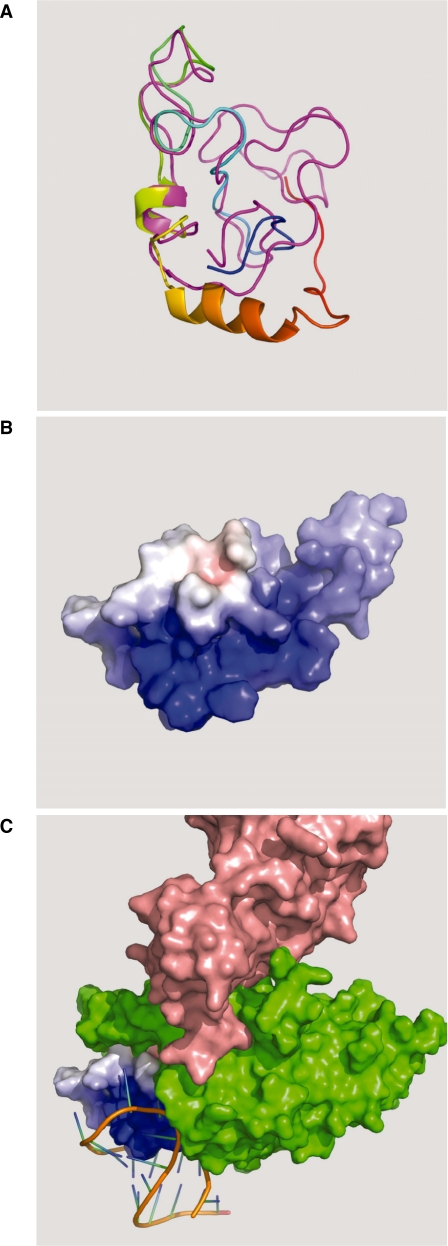
Lack of the second exon (29aa) could imply some structural rearrangements inside Tat protein that would explain the functional and structural differences observed in Jurkat-Tat72 in comparison with Jurkat-Tat101. However, there are few studies about HIV-1 Tat crystallization that could permit in silico modelling to evaluate the effect on Tat function after truncating the second exon. PDB structure 1jfw corresponding to the NMR spectra of a chemically synthesized Tat86 protein from BRU isolate (66) has been used to develop a reliable homology modelling of Tat72, with which shows an identity of 91%. The absence of published structural data for residues 86–101 makes impossible to perform a reliable modelling of Tat101. Results from modelling Tat72 showed that this protein is highly flexible and displays extended regions and connecting loops, although it did not show highly defined α-helixes or β-sheets (Figure 8A). Tat72 model showed very polarized electrostatic charges, with a high-positive charged TAR-interacting region in the N-terminus, whereas the C-terminus was severely reduced and showed quite diminished positive charging (Figure 8B). This end would likely interact with several host cell proteins and structural targets, providing Tat101 with 29aa residues that would strength these interactions. In order to infer the contribution of the second exon to this interaction, modelling of Tat72/cyclin T1/TAR complex was attempted. Because HIV-1 Tat structure has not been crystallized in complex with TAR and cyclin T1, a crystallization of Tat from equine infectious anaemia virus (EIAV) in complex with TAR and cyclin T1 (PDB structure 2w2h) was used as model (67). Probably due to the high flexibility of the protein, only one α-helix interacting with TAR was resolved in the structure 2w2h. EIAV and HIV-1 are distantly related, with less that 30% sequence identity, but the TAR interacting region is relatively conserved and both human and horse cyclin T1 are almost identical. EIAV TAR is formed by two distinct structural regions and although the activation domains of both HIV-1 and EIAV Tat contain a conserved core element, the cysteine-rich domain is absent from EIAV Tat. As a result, HIV-1 Tat requires the presence of additional structural regions to completely fulfil its functions (20). Based on this assumption, Tat72 was modelled using as template ijfw structure except for the TAR-interacting region, for which the resolved α-helix from 2w2h was used (Figure 8C). Although 2w2h diffracting crystal was generated by using a 57 residues-EIAV Tat (13-69aa) (67), quite good volumetric fitting in the space comprised between TAR and cyclin T1 was obtained for Tat72 after slight manually adjustment to avoid molecular collisions, despite 1jfw had been resolved in an unbound conformation. Accordingly, folding of cyclin T1 around Tat should be a highly conserved three-dimensional structure. In this model, the C-terminal domain of Tat72 is deeply enfolded in the cyclin T1 structure, leaving no physical space in the inner part of cyclin T1 for accommodating the C-terminus of 29aa that constitute the second exon. This would imply that the second exon of Tat101 in an extended conformation should be interacting not only with the internal core of cyclin T1 but with its external surface or with other protein/s bound to the complex.
Figure 8.
Tat72 in silico modelling. (A) Tat72 (coloured ribbon: N-terminus in blue, C-terminus in red) and Tat86 (magenta ribbon) proteins were modelled using the PDB structure 1jfw according to their fitting in TAR/P-TEFb complex (theoretical bound conformation). Both proteins were highly flexible and displayed extended regions and connecting loops, although they did not show highly defined α-helixes or β-sheets. (B) Modelled Tat72 (surface view) showed that the electrostatic charges were very polarized, with a highly positive charged TAR-interacting region in the amino-terminal extreme of the protein (dark blue) and a quite diminished positive charging in the severely reduced C-terminal extreme (light blue). (C) Modelling of Tat72 (surface view) in complex with cyclin T1/TAR was attempted by using a crystallized EIAV Tat/cyclin T1/TAR complex (PDB structure 2w2h). Tat72 was modelled using ijfw structure as template except for the TAR-interacting region, for which a resolved α-helix from 2w2h was used. The C-terminal interface of Tat72 perfectly adjusted inside the cyclin T1 structure, with quite good volumetric fitting in the space comprised between TAR (orange) and cyclin T1 (green), where CDK9 (pink) is bound to cyclin T1. Tat72 was completely buried inside the cyclin T1 structure, with no space left for accommodating the C-terminal domain of Tat101 (29aa).
DISCUSSION
HIV-1 Tat acts in a different way when it is synthesized inside the cell during HIV-1 infection than when it is released to the extracellular medium and taken up by adjacent cells. As a secretable factor, Tat induces dramatic cytoskeleton rearrangements (74) that lead to apoptotic events related to HIV-1-induced pathogenesis (6,43,44,75). As an intracellular protein, Tat is a regulator involved in LTR transcriptional activation and HIV-1 replication (1). Despite the different behaviour, the protein structure is the same. Tat first exon (1–72aa) contains the minimal functional domain to generate a protein competent in HIV-1 replication (20), whereas the second exon (73–101aa) has often been described as dispensable for Tat activity. However, most primate lentiviruses conserve the second exon despite the high error rate of the RT, suggesting it must have an essential biological function (12,29,30,76–78). Moreover, an HIV or SIV virus carrying one stop codon at the end of the first exon can spontaneously revert to full-length Tat in vivo, yielding a more pathogenic virus (34,35). This higher pathogenicity could be related to a higher efficiency for initiating the LTR transactivation or to non-transcriptional activities. In order to study the role of the second exon of Tat in the modification of T-cell functionality, two lines of Jurkat cells stably expressing Tat101 or Tat72 have been obtained. Because the 86ESKKKVE92 motif is critical for NF-κB transactivation (28), it was assumed that Tat72 would be less efficient to transactivate the HIV-1 LTR promoter in our cellular system. This was confirmed when Tat72 proved to be 25% less efficient than Tat101 to initiate the LTR-dependent transactivation. This lower activity was related to lower NF-κB-, NF-AT- and Sp1-dependent activities, in accordance with previous reports (79–84). Due to Tat concentration in Jurkat-Tat101 and Jurkat-Tat72 was very similar and highly stable after long-term culture, differences in the transactivation activity could not be caused by different quantities of Tat-expressing cells in each population. Therefore, Tat72 was less efficient to transactivate the LTR but nevertheless, active.
The main activities attributed to Tat are the LTR transactivation and the efficient elongation of viral transcripts, but other functions have also been related to Tat as the modification of the cytoskeleton dynamics (6,12,14,43,44,75). The cytoskeleton integrity is indispensable for maintaining the cellular shape and survival and for an appropriate immune response. Actually, T cells become motile cells upon activation, switching from a spherical to a polarized shape and orienting their components towards one pole of the cell. This mechanism relies on the cytoskeletal elements, being the microfilament system remodelling essential for T-cell polarization (85,86). It has been described that extracellular Tat can induce dramatic cytoskeleton rearrangements that lead to apoptosis (6,43,44,75). Conversely, although the cytoskeleton can also be greatly modified by Tat intracellular expression, these changes do not decrease cell viability but even prevent apoptosis induced by different stimuli (14,17). Accordingly, the role of intracellular Tat in the T-cell cytoskeleton disorganization was investigated. To avoid interferences caused by extracellular Tat, the possibility that Tat could be solubilized in the culture medium of Jurkat-Tat101 or Jurkat-Tat72 was previously ruled out.
A global functional analysis of changes in the T-cell gene expression profile induced by intracellular Tat and the role of the second exon in these changes was performed by using microarrays of whole human genome. This wide approach has previously allowed the identification of cellular functions modified by the intracellular expression of Tat, as those related to cell survival, proliferation, transcription, immune response, metabolism and chemokine release (36,87). De la Fuente et al. (88) also determined that Tat86 deregulates many cellular genes involved in cell signalling, translation and control of proliferative and differentiation signals. However, Tat86 is a non-natural truncated form that lacks the 86ESKKKVE92 motif critical for NF-κB transactivation (28) and therefore, it should not be fully functional. In fact, our results proved that Tat72 was not fully functional either because the intracellular expression of Tat101 induced up- or down-regulation of nearly 300 genes, whereas less than 70 genes were deregulated in Jurkat-Tat72. Within these impaired functions, we focused on those related to the cytoskeleton functionality as cell morphology, growth and proliferation, movement or apoptosis, as well as cell signalling and T-cell activation. All these functions are related to HIV-1 infectivity and virulence. These data prove that although the second coding exon of Tat may be dispensable for the transcriptional activity of this protein, it is essential for non-transcriptional functions.
Several functional analyses were performed to evaluate the consequences of the gene expression deregulation caused by intracellular Tat in the cytoskeleton activity and the role of the second exon in these modifications. First, we observed that intracellular Tat101 caused modifications in the morphology and shape of Jurkat cells and a strong inhibition of PHA-induced proliferation, whereas Tat72 did not produce significant changes. These results were valuable because the role of Tat in cell proliferation is controversial. Our data agree with previous reports that demonstrate the inhibition of T-cell proliferation by extracellular Tat (89–91), but in other studies Tat did not show effect on PHA-induced T-cell proliferation (92,93). Moreover, Gibellini et al. (36) suggested that Tat could be involved in the positive regulation of T-cell proliferation, although no functional studies were performed in this report. In our study, intracellular Tat101 not only caused the inhibition of PHA-induced proliferation but also of SDF-1/CXCL12-induced chemotaxis. This inhibition was not observed in Jurkat-Tat72 either. The chemotaxis impairment in Jurkat-Tat101 was not due to changes in CXCR4 surface expression or to the binding of extracellular Tat to CXCR4 (72) because soluble Tat was not detected in the culture medium. Instead, Tat-mediated mechanism underlying the impairment of chemotaxis and proliferation could be related to the deregulation of essential genes related to T-cell growth, infiltration and development as CADM1 (94), Bcl11a (95), or SMAD7—antagonist of TGF-β signalling (96). Other genes related to migration were up-regulated in Jurkat-Tat101 as DOCK3, which is involved in cytoskeleton reorganization (97), modification of cell morphology (98), impairment of cell migration, and regulation of the Wnt signalling pathway—related to cell growth, proliferation and polarity (99–101). There are other proteins required for cell motility that are down-regulated as ELMO1 that acts together with DOCK1 and DOCK2 to activate Rac/Rho small GTPases (102,103). This mechanism could be similar to the Nef-dependent inhibition of T-cell chemotaxis induced by the activation of Rac through the binding of Nef to the complex DOCK2–ELMO1 (103). Besides, intracellular Tat induced the up-regulation of Spry2 -which can inhibit cell growth, migration and invasion (104), as well as the down-regulation of CD40LG, which modulates proliferation and movement (105), and CD44, which mediates T-cell migration, recirculation and homing through Rac-mediated cytoskeletal rearrangements (106). Other receptors involved in cell migration as CCR1, CCR7 and MCAM/CD146 (107–109) were also down-regulated. Within all these genes modified in Jurkat-Tat101, only Wnt5a, Spry2 and CD40LG were also deregulated in Jurkat-Tat72.
Impairment of chemotaxis and proliferation are related to changes in the cytoskeleton structure mediated by Tat. It has been demonstrated that extracellular Tat increases the rate of tubulin polymerization in vitro (18,26). Now we determined that intracellular Tat increased the rate of basal F-actin polymerization in vivo and that this effect was dependent on the second exon. The higher basal F-actin polymerization and polarization showed by Jurkat-Tat101 could be responsible for the impairment of migration because it would impede further response to chemotactic stimuli. However, it has been described that full-length Tat is not necessary to enhance the tubulin polymerization that leads to apoptosis but only peptides containing the glutamine-rich region are required (18,26). These results were obtained by adding high concentrations of exogenous Tat to an in vitro system of purified lamb brain tubulin. Our results indicated that the second exon was essential to induce extensive modifications in the T-cell actin cytoskeleton in vivo mediated by intracellular Tat. These modifications can not be due exclusively to the direct interaction of Tat with cytoskeleton elements as tubulin or actin but to other indirect mechanisms as the up-regulation of DOCK3 and Spry2 or the down-regulation of ELMO1, CD40LG and CD44. These results increase the differences between the behaviour of extra and intracellular Tat. Other possible explanation is that Tat localizes predominantly in the nucleus when is synthesized inside the cell, but it remains mainly at the cytosol after being endocytosed (17), where it could be retained through the binding to cytoskeletal proteins as actin, tubulin, Lis1 and Hic (19,110,111), causing higher toxicity.
Alterations in the cytoskeleton structure could impair the HIV-1 infection because HIV-1 uses dynein and the microtubule network to move towards the NPC (47). Besides, the nucleocapsid core interacts with actin filaments to allow the intracellular transport of Gag, virus assembly and budding (112), and actin and other cytoskeletal proteins as ERM could act as structural component of HIV-1 particles (113). These mechanisms could mean that re-infection of cells already infected and expressing intracellular Tat could be hindered. In fact, Tat down-regulates surface receptors as CD4 (114) and TCR/CD3 (115). Accordingly, Jurkat-Tat101 lacked of several essential receptors on the cell surface. Absence of CD4 and CCR9—an alternative receptor to CD4 (116) —would also mean that these cells could not be re-infected. Down-regulation of other surface receptors as CD1 (CD1a, b, c, d, e) and CD2 was also dramatic in Jurkat-Tat101. It has been described that endogenous HIV-1 Nef down-regulates CD1a selectively among CD1 molecules in dendritic cells (117), but we observed that intracellular Tat101 down-regulated all CD1 molecules in Jurkat. Because CD1 receptors act as antigen presenting molecules (118), their down-regulation would contribute to the impairment of the immune response. In fact, Tat has been described as an inhibitor of antigen-specific responsiveness of T cells, inducing anergy (93). Down-regulation of these receptors was dependent on the second exon because it was not observed in Jurkat-Tat72.
In summary, Tat second exon was responsible for altering the cytoskeleton structure and functions as proliferation, chemotaxis and migration, and for the down-expression of surface receptors, when Tat is synthesized inside the cell. These changes suggest that intracellular Tat could be essential for adapting the host cell to HIV-1 infection, for hindering re-infections, and for the impairment of T-cell functionality during the immune response. Structurally, the second exon is a high-positive charged region of 29aa that would likely enhance the binding strength or affinity to host cell proteins and structural targets, as was described for the HIV-2 Tat second exon (119–121). To test this statement, in silico modelling of Tat72 was performed and adjusted within the Tat/TAR/P-TEFb complex, leaving Tat72 C-terminus deeply buried in the cyclin T1 structure. It would be improbable to fit Tat101 in the same space, implying that the C-terminal domain must interact not only with the internal core of cyclin T1 but with its external surface. Accordingly, the presence of the second exon would explain why Tat101 was more competent than Tat72 to cause gene expression deregulation and cytoskeleton modifications. More analyses will be necessary to evaluate how these changes in the cytoskeleton structure and functionality would affect HIV-1 infection. Data obtained by microarrays provide important clues about the pathways impaired by Tat and will be used as the basis for more extensive studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Plan Nacional del SIDA (MVI 1434/05–5), FIPSE 36584/06 and 36633/07, VIRHORST Network from Comunidad de Madrid (Spain), FIS PI040614 and PI0808752, ISCIII-RETIC RD06/0006, EUROPRISE Network of Excellence of the EU (Grant no. LSHP CT-2006-037611), and BIO2008-04384 from the Ministerio de Ciencia e Innovación, España. Funding for open access charge: Instituto de Salud Carlos III, Ministry of Science and Technology, Spain.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors acknowledge excellent technical assistance of Almudena Cascajero and brilliant secretarial assistance from Olga Palao. They would like to thank Dr. Francisco Sánchez-Madrid and Dr. Mónica Gordón-Alonso (Hospital Universitario de La Princesa, Madrid, Spain) for their great support with the cytoskeleton studies. They also thank Dr. Carles Suñé (Instituto de Parasitología y Biomedicina López Neyra, Granada, Spain) for kindly providing the pCMV-Tat vector and Dr Solís-Herruzo (Hospital 12 de Octubre, Madrid, Spain) for the kind gift of NF-AT-LUC and Sp1-LUC plasmids. They also thank Dr. Carlota Dominguez (Centro Nacional de Microbiología, Instituto de Salud Carlos III, Madrid, Spain) for great support with DAI assays.
REFERENCES
- 1.Gaynor RB. Regulation of HIV-1 gene expression by the transactivator protein Tat. Curr. Top. Microbiol. Immunol. 1995;193:51–77. doi: 10.1007/978-3-642-78929-8_3. [DOI] [PubMed] [Google Scholar]
- 2.Berkhout B, Jeang KT. Trans activation of human immunodeficiency virus type 1 is sequence specific for both the single-stranded bulge and loop of the trans-acting-responsive hairpin: a quantitative analysis. J. Virol. 1989;63:5501–5504. doi: 10.1128/jvi.63.12.5501-5504.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcello A, Zoppe M, Giacca M. Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator. IUBMB Life. 2001;51:175–181. doi: 10.1080/152165401753544241. [DOI] [PubMed] [Google Scholar]
- 4.Zhou M, Halanski MA, Radonovich MF, Kashanchi F, Peng J, Price DH, Brady JN. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 2000;20:5077–5086. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrich D, Ulich C, Garcia-Martinez LF, Gaynor RB. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 1997;16:1224–1235. doi: 10.1093/emboj/16.6.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albini A, Ferrini S, Benelli R, Sforzini S, Giunciuglio D, Aluigi MG, Proudfoot AE, Alouani S, Wells TN, Mariani G, et al. HIV-1 Tat protein mimicry of chemokines. Proc. Natl Acad. Sci USA. 1998;95:13153–13158. doi: 10.1073/pnas.95.22.13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agwale SM, Shata MT, Reitz M.S., Jr, Kalyanaraman VS, Gallo RC, Popovic M, Hone DM. A Tat subunit vaccine confers protective immunity against the immune-modulating activity of the human immunodeficiency virus type-1 Tat protein in mice. Proc. Natl Acad. Sci. USA. 2002;99:10037–10041. doi: 10.1073/pnas.152313899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dandekar DH, Ganesh KN, Mitra D. HIV-1 Tat directly binds to NFkappaB enhancer sequence: role in viral and cellular gene expression. Nucleic Acids Res. 2004;32:1270–1278. doi: 10.1093/nar/gkh289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gautier VW, Gu L, O’Donoghue N, Pennington S, Sheehy N, Hall WW. In vitro nuclear interactome of the HIV-1 Tat protein. Retrovirology. 2009;6:47. doi: 10.1186/1742-4690-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ensoli B, Buonaguro L, Barillari G, Fiorelli V, Gendelman R, Morgan RA, Wingfield P, Gallo RC. Release, uptake, and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J. Virol. 1993;67:277–287. doi: 10.1128/jvi.67.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magnuson DS, Knudsen BE, Geiger JD, Brownstone RM, Nath A. Human immunodeficiency virus type 1 tat activates non-N-methyl-D-aspartate excitatory amino acid receptors and causes neurotoxicity. Ann. Neurol. 1995;37:373–380. doi: 10.1002/ana.410370314. [DOI] [PubMed] [Google Scholar]
- 12.Xiao H, Neuveut C, Benkirane M, Jeang KT. Interaction of the second coding exon of Tat with human EF-1 delta delineates a mechanism for HIV-1-mediated shut-off of host mRNA translation. Biochem. Biophys. Res. Commun. 1998;244:384–389. doi: 10.1006/bbrc.1998.8274. [DOI] [PubMed] [Google Scholar]
- 13.Campioni D, Corallini A, Zauli G, Possati L, Altavilla G, Barbanti-Brodano G. HIV type 1 extracellular Tat protein stimulates growth and protects cells of BK virus/tat transgenic mice from apoptosis. AIDS Res. Hum. Retroviruses. 1995;11:1039–1048. doi: 10.1089/aid.1995.11.1039. [DOI] [PubMed] [Google Scholar]
- 14.Coiras M, Camafeita E, Urena T, Lopez JA, Caballero F, Fernandez B, Lopez-Huertas MR, Perez-Olmeda M, Alcami J. Modifications in the human T cell proteome induced by intracellular HIV-1 Tat protein expression. Proteomics. 2006;6(Suppl. 1):S63–S73. doi: 10.1002/pmic.200500437. [DOI] [PubMed] [Google Scholar]
- 15.Jeang KT, Xiao H, Rich EA. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J. Biol. Chem. 1999;274:28837–28840. doi: 10.1074/jbc.274.41.28837. [DOI] [PubMed] [Google Scholar]
- 16.Garcia JA, Harrich D, Pearson L, Mitsuyasu R, Gaynor RB. Functional domains required for tat-induced transcriptional activation of the HIV-1 long terminal repeat. EMBO J. 1988;7:3143–3147. doi: 10.1002/j.1460-2075.1988.tb03181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aprea S, Del Valle L, Mameli G, Sawaya BE, Khalili K, Peruzzi F. Tubulin-mediated binding of human immunodeficiency virus-1 Tat to the cytoskeleton causes proteasomal-dependent degradation of microtubule-associated protein 2 and neuronal damage. J. Neurosci. 2006;26:4054–4062. doi: 10.1523/JNEUROSCI.0603-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egele C, Barbier P, Didier P, Piemont E, Allegro D, Chaloin O, Muller S, Peyrot V, Mely Y. Modulation of microtubule assembly by the HIV-1 Tat protein is strongly dependent on zinc binding to Tat. Retrovirology. 2008;5:62. doi: 10.1186/1742-4690-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen D, Wang M, Zhou S, Zhou Q. HIV-1 Tat targets microtubules to induce apoptosis, a process promoted by the pro-apoptotic Bcl-2 relative Bim. EMBO J. 2002;21:6801–6810. doi: 10.1093/emboj/cdf683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carroll R, Martarano L, Derse D. Identification of lentivirus tat functional domains through generation of equine infectious anemia virus/human immunodeficiency virus type 1 tat gene chimeras. J. Virol. 1991;65:3460–3467. doi: 10.1128/jvi.65.7.3460-3467.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang YN, Jeang KT. The basic RNA-binding domain of HIV-2 Tat contributes to preferential trans-activation of a TAR2-containing LTR. Nucleic Acids Res. 1992;20:5465–5472. doi: 10.1093/nar/20.20.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Churcher MJ, Lamont C, Hamy F, Dingwall C, Green SM, Lowe AD, Butler JG, Gait MJ, Karn J. High affinity binding of TAR RNA by the human immunodeficiency virus type-1 tat protein requires base-pairs in the RNA stem and amino acid residues flanking the basic region. J. Mol. Biol. 1993;230:90–110. doi: 10.1006/jmbi.1993.1128. [DOI] [PubMed] [Google Scholar]
- 23.Vives E, Brodin P, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 24.Hauber J, Malim MH, Cullen BR. Mutational analysis of the conserved basic domain of human immunodeficiency virus tat protein. J. Virol. 1989;63:1181–1187. doi: 10.1128/jvi.63.3.1181-1187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabatier JM, Vives E, Mabrouk K, Benjouad A, Rochat H, Duval A, Hue B, Bahraoui E. Evidence for neurotoxic activity of tat from human immunodeficiency virus type 1. J. Virol. 1991;65:961–967. doi: 10.1128/jvi.65.2.961-967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Mareuil J, Carre M, Barbier P, Campbell GR, Lancelot S, Opi S, Esquieu D, Watkins JD, Prevot C, Braguer D, et al. HIV-1 Tat protein enhances microtubule polymerization. Retrovirology. 2005;2:5. doi: 10.1186/1742-4690-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barillari G, Gendelman R, Gallo RC, Ensoli B. The Tat protein of human immunodeficiency virus type 1, a growth factor for AIDS Kaposi sarcoma and cytokine-activated vascular cells, induces adhesion of the same cell types by using integrin receptors recognizing the RGD amino acid sequence. Proc. Natl Acad. Sci. USA. 1993;90:7941–7945. doi: 10.1073/pnas.90.17.7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahlknecht U, Dichamp I, Varin A, Van Lint C, Herbein G. NF-kappaB-dependent control of HIV-1 transcription by the second coding exon of Tat in T cells. J. Leukoc. Biol. 2008;83:718–727. doi: 10.1189/jlb.0607405. [DOI] [PubMed] [Google Scholar]
- 29.Ma M, Nath A. Molecular determinants for cellular uptake of Tat protein of human immunodeficiency virus type 1 in brain cells. J. Virol. 1997;71:2495–2499. doi: 10.1128/jvi.71.3.2495-2499.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartz SR, Emerman M. Human immunodeficiency virus type 1 Tat induces apoptosis and increases sensitivity to apoptotic signals by up-regulating FLICE/caspase-8. J. Virol. 1999;73:1956–1963. doi: 10.1128/jvi.73.3.1956-1963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campbell GR, Watkins JD, Esquieu D, Pasquier E, Loret EP, Spector SA. The C terminus of HIV-1 Tat modulates the extent of CD178-mediated apoptosis of T cells. J. Biol. Chem. 2005;280:38376–38382. doi: 10.1074/jbc.M506630200. [DOI] [PubMed] [Google Scholar]
- 32.Verhoef K, Bauer M, Meyerhans A, Berkhout B. On the role of the second coding exon of the HIV-1 Tat protein in virus replication and MHC class I downregulation. AIDS Res. Hum. Retroviruses. 1998;14:1553–1559. doi: 10.1089/aid.1998.14.1553. [DOI] [PubMed] [Google Scholar]
- 33.Ott M, Emiliani S, Van Lint C, Herbein G, Lovett J, Chirmule N, McCloskey T, Pahwa S, Verdin E. Immune hyperactivation of HIV-1-infected T cells mediated by Tat and the CD28 pathway. Science. 1997;275:1481–1485. doi: 10.1126/science.275.5305.1481. [DOI] [PubMed] [Google Scholar]
- 34.Pincus SH, Messer KG, Nara PL, Blattner WA, Colclough G, Reitz M. Temporal analysis of the antibody response to HIV envelope protein in HIV-infected laboratory workers. J. Clin. Invest. 1994;93:2505–2513. doi: 10.1172/JCI117260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith SM, Pentlicky S, Klase Z, Singh M, Neuveut C, Lu CY, Reitz M.S., Jr, Yarchoan R, Marx PA, Jeang KT. An in vivo replication-important function in the second coding exon of Tat is constrained against mutation despite cytotoxic T lymphocyte selection. J. Biol. Chem. 2003;278:44816–44825. doi: 10.1074/jbc.M307546200. [DOI] [PubMed] [Google Scholar]
- 36.Gibellini D, Vitone F, Schiavone P, Re MC. HIV-1 tat protein and cell proliferation and survival: a brief review. New Microbiol. 2005;28:95–109. [PubMed] [Google Scholar]
- 37.Zauli G, Gibellini D. The human immunodeficiency virus type-1 (HIV-1) Tat protein and Bcl-2 gene expression. Leuk. Lymphoma. 1996;23:551–560. doi: 10.3109/10428199609054864. [DOI] [PubMed] [Google Scholar]
- 38.Borgatti P, Zauli G, Colamussi ML, Gibellini D, Previati M, Cantley LL, Capitani S. Extracellular HIV-1 Tat protein activates phosphatidylinositol 3- and Akt/PKB kinases in CD4+ T lymphoblastoid Jurkat cells. Eur. J. Immunol. 1997;27:2805–2811. doi: 10.1002/eji.1830271110. [DOI] [PubMed] [Google Scholar]
- 39.Jones M, Olafson K, Del Bigio MR, Peeling J, Nath A. Intraventricular injection of human immunodeficiency virus type 1 (HIV-1) tat protein causes inflammation, gliosis, apoptosis, and ventricular enlargement. J. Neuropathol. Exp. Neurol. 1998;57:563–570. doi: 10.1097/00005072-199806000-00004. [DOI] [PubMed] [Google Scholar]
- 40.Misumi S, Takamune N, Ohtsubo Y, Waniguchi K, Shoji S. Zn2+ binding to cysteine-rich domain of extracellular human immunodeficiency virus type 1 Tat protein is associated with Tat protein-induced apoptosis. AIDS Res. Hum. Retroviruses. 2004;20:297–304. doi: 10.1089/088922204322996536. [DOI] [PubMed] [Google Scholar]
- 41.Zhu X, Yao H, Peng F, Callen S, Buch S. PDGF-mediated protection of SH-SY5Y cells against Tat toxin involves regulation of extracellular glutamate and intracellular calcium. Toxicol. Appl. Pharmacol. 2009;240:286–291. doi: 10.1016/j.taap.2009.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Battaglia PA, Zito S, Macchini A, Gigliani F. A Drosophila model of HIV-Tat-related pathogenicity. J. Cell. Sci. 2001;114:2787–2794. doi: 10.1242/jcs.114.15.2787. [DOI] [PubMed] [Google Scholar]
- 43.Huigen MC, Kamp W, Nottet HS. Multiple effects of HIV-1 trans-activator protein on the pathogenesis of HIV-1 infection. Eur. J. Clin. Invest. 2004;34:57–66. doi: 10.1111/j.1365-2362.2004.01282.x. [DOI] [PubMed] [Google Scholar]
- 44.McCloskey TW, Ott M, Tribble E, Khan SA, Teichberg S, Paul MO, Pahwa S, Verdin E, Chirmule N. Dual role of HIV Tat in regulation of apoptosis in T cells. J. Immunol. 1997;158:1014–1019. [PubMed] [Google Scholar]
- 45.Campbell GR, Pasquier E, Watkins J, Bourgarel-Rey V, Peyrot V, Esquieu D, Barbier P, de Mareuil J, Braguer D, Kaleebu P, et al. The glutamine-rich region of the HIV-1 Tat protein is involved in T-cell apoptosis. J. Biol. Chem. 2004;279:48197–48204. doi: 10.1074/jbc.M406195200. [DOI] [PubMed] [Google Scholar]
- 46.Wu RF, Gu Y, Xu YC, Mitola S, Bussolino F, Terada LS. Human immunodeficiency virus type 1 Tat regulates endothelial cell actin cytoskeletal dynamics through PAK1 activation and oxidant production. J. Virol. 2004;78:779–789. doi: 10.1128/JVI.78.2.779-789.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Damsky CH, Sheffield JB, Tuszynski GP, Warren L. Is there a role for actin in virus budding? J. Cell Biol. 1977;75:593–605. doi: 10.1083/jcb.75.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matarrese P, Malorni W. Human immunodeficiency virus (HIV)-1 proteins and cytoskeleton: partners in viral life and host cell death. Cell Death Differ. 2005;12(Suppl. 1):932–941. doi: 10.1038/sj.cdd.4401582. [DOI] [PubMed] [Google Scholar]
- 50.Arenzana-Seisdedos F, Fernandez B, Dominguez I, Jacque JM, Thomas D, Diaz-Meco MT, Moscat J, Virelizier JL. Phosphatidylcholine hydrolysis activates NF-kappa B and increases human immunodeficiency virus replication in human monocytes and T lymphocytes. J. Virol. 1993;67:6596–6604. doi: 10.1128/jvi.67.11.6596-6604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bachelerie F, Alcami J, Arenzana-Seisdedos F, Virelizier JL. HIV enhancer activity perpetuated by NF-kappa B induction on infection of monocytes. Nature. 1991;350:709–712. doi: 10.1038/350709a0. [DOI] [PubMed] [Google Scholar]
- 52.Northrop JP, Ullman KS, Crabtree GR. Characterization of the nuclear and cytoplasmic components of the lymphoid-specific nuclear factor of activated T cells (NF-AT) complex. J. Biol. Chem. 1993;268:2917–2923. [PubMed] [Google Scholar]
- 53.Garcia-Ruiz I, de la TP, Diaz T, Esteban E, Fernandez I, Munoz-Yague T, Solis-Herruzo JA. Sp1 and Sp3 transcription factors mediate malondialdehyde-induced collagen alpha 1(I) gene expression in cultured hepatic stellate cells. J. Biol. Chem. 2002;277:30551–30558. doi: 10.1074/jbc.M203368200. [DOI] [PubMed] [Google Scholar]
- 54.Amieva MR, Furthmayr H. Subcellular localization of moesin in dynamic filopodia, retraction fibers, and other structures involved in substrate exploration, attachment, and cell–cell contacts. Exp. Cell Res. 1995;219:180–196. doi: 10.1006/excr.1995.1218. [DOI] [PubMed] [Google Scholar]
- 55.Serrador JM, Vicente-Manzanares M, Calvo J, Barreiro O, Montoya MC, Schwartz-Albiez R, Furthmayr H, Lozano F, Sanchez-Madrid F. A novel serine-rich motif in the intercellular adhesion molecule 3 is critical for its ezrin/radixin/moesin-directed subcellular targeting. J. Biol. Chem. 2002;277:10400–10409. doi: 10.1074/jbc.M110694200. [DOI] [PubMed] [Google Scholar]
- 56.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 57.Laín de Lera T, Folgueira L, Martin AG, Dargemont C, Pedraza MA, Bermejo M, Bonay P, Fresno M, Alcami J. Expression of IkappaBalpha in the nucleus of human peripheral blood T lymphocytes. Oncogene. 1999;18:1581–1588. doi: 10.1038/sj.onc.1202455. [DOI] [PubMed] [Google Scholar]
- 58.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schuler GD, Altschul SF, Lipman DJ. A workbench for multiple alignment construction and analysis. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 60.Ormerod MG. Flow Cytometry: A Practical Approach. New York: Oxford University Press Inc; 1992. [Google Scholar]
- 61.Liu Y, Asch H, Kulesz-Martin MF. Functional quantification of DNA-binding proteins p53 and estrogen receptor in cells and tumor tissues by DNA affinity immunoblotting. Cancer Res. 2001;61:5402–5406. [PubMed] [Google Scholar]
- 62.Klein-Hitpass L, Schorpp M, Wagner U, Ryffel GU. An estrogen-responsive element derived from the 5′ flanking region of the Xenopus vitellogenin A2 gene functions in transfected human cells. Cell. 1986;46:1053–1061. doi: 10.1016/0092-8674(86)90705-1. [DOI] [PubMed] [Google Scholar]
- 63.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Storey JD. A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B Stat. Methodol. 2002;64:479–498. [Google Scholar]
- 65.Klipper-Aurbach Y, Wasserman M, Braunspiegel-Weintrob N, Borstein D, Peleg S, Assa S, Karp M, Benjamini Y, Hochberg Y, Laron Z. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med. Hypotheses. 1995;45:486–490. doi: 10.1016/0306-9877(95)90228-7. [DOI] [PubMed] [Google Scholar]
- 66.Peloponese JM, Jr, Gregoire C, Opi S, Esquieu D, Sturgis J, Lebrun E, Meurs E, Collette Y, Olive D, Aubertin AM, et al. 1H-13C nuclear magnetic resonance assignment and structural characterization of HIV-1 Tat protein. C. R. Acad. Sci. III, Sci. Vie. 2000;323:883–894. doi: 10.1016/s0764-4469(00)01228-2. [DOI] [PubMed] [Google Scholar]
- 67.Anand K, Schulte A, Vogel-Bachmayr K, Scheffzek K, Geyer M. Structural insights into the cyclin T1-Tat-TAR RNA transcription activation complex from EIAV. Nat. Struct. Mol. Biol. 2008;15:1287–1292. doi: 10.1038/nsmb.1513. [DOI] [PubMed] [Google Scholar]
- 68.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 69.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ortiz AR, Strauss CE, Olmea O. MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison. Protein Sci. 2002;11:2606–2621. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baumli S, Lolli G, Lowe ED, Troiani S, Rusconi L, Bullock AN, Debreczeni JE, Knapp S, Johnson LN. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 2008;27:1907–1918. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiao H, Neuveut C, Tiffany HL, Benkirane M, Rich EA, Murphy PM, Jeang KT. Selective CXCR4 antagonism by Tat: implications for in vivo expansion of coreceptor use by HIV-1. Proc. Natl Acad. Sci. USA. 2000;97:11466–11471. doi: 10.1073/pnas.97.21.11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fais S, Luciani F, Logozzi M, Parlato S, Lozupone F. Linkage between cell membrane proteins and actin-based cytoskeleton: the cytoskeletal-driven cellular functions. Histol. Histopathol. 2000;15:539–549. doi: 10.14670/HH-15.539. [DOI] [PubMed] [Google Scholar]
- 74.Schwarze SR, Dowdy SF. In vivo protein transduction: intracellular delivery of biologically active proteins, compounds and DNA. Trends Pharmacol. Sci. 2000;21:45–48. doi: 10.1016/s0165-6147(99)01429-7. [DOI] [PubMed] [Google Scholar]
- 75.Mitola S, Soldi R, Zanon I, Barra L, Gutierrez MI, Berkhout B, Giacca M, Bussolino F. Identification of specific molecular structures of human immunodeficiency virus type 1 Tat relevant for its biological effects on vascular endothelial cells. J. Virol. 2000;74:344–353. doi: 10.1128/jvi.74.1.344-353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo X, Kameoka M, Wei X, Roques B, Gotte M, Liang C, Wainberg MA. Suppression of an intrinsic strand transfer activity of HIV-1 Tat protein by its second-exon sequences. Virology. 2003;307:154–163. doi: 10.1016/s0042-6822(02)00068-5. [DOI] [PubMed] [Google Scholar]
- 77.Kim YS, Panganiban AT. The full-length Tat protein is required for TAR-independent, posttranscriptional trans activation of human immunodeficiency virus type 1 env gene expression. J. Virol. 1993;67:3739–3747. doi: 10.1128/jvi.67.7.3739-3747.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Westendorp MO, Shatrov VA, Schulze-Osthoff K, Frank R, Kraft M, Los M, Krammer PH, Droge W, Lehmann V. HIV-1 Tat potentiates TNF-induced NF-kappa B activation and cytotoxicity by altering the cellular redox state. EMBO J. 1995;14:546–554. doi: 10.1002/j.1460-2075.1995.tb07030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Biswas P, Poli G, Kinter AL, Justement JS, Stanley SK, Maury WJ, Bressler P, Orenstein JM, Fauci AS. Interferon gamma induces the expression of human immunodeficiency virus in persistently infected promonocytic cells (U1) and redirects the production of virions to intracytoplasmic vacuoles in phorbol myristate acetate-differentiated U1 cells. J. Exp. Med. 1992;176:739–750. doi: 10.1084/jem.176.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu J, Perkins ND, Schmid RM, Nabel GJ. Specific NF-kappa B subunits act in concert with Tat to stimulate human immunodeficiency virus type 1 transcription. J. Virol. 1992;66:3883–3887. doi: 10.1128/jvi.66.6.3883-3887.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alcami J, Lain dL, Folgueira L, Pedraza MA, Jacque JM, Bachelerie F, Noriega AR, Hay RT, Harrich D, Gaynor RB, et al. Absolute dependence on kappa B responsive elements for initiation and Tat-mediated amplification of HIV transcription in blood CD4 T lymphocytes. EMBO J. 1995;14:1552–1560. doi: 10.1002/j.1460-2075.1995.tb07141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Demarchi F, Gutierrez MI, Giacca M. Human immunodeficiency virus type 1 tat protein activates transcription factor NF-kappaB through the cellular interferon-inducible, double-stranded RNA-dependent protein kinase, PKR. J. Virol. 1999;73:7080–7086. doi: 10.1128/jvi.73.8.7080-7086.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Macian F, Rao A. Reciprocal modulatory interaction between human immunodeficiency virus type 1 Tat and transcription factor NFAT1. Mol. Cell. Biol. 1999;19:3645–3653. doi: 10.1128/mcb.19.5.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hidalgo-Estevez AM, Gonzalez E, Punzon C, Fresno M. Human immunodeficiency virus type 1 Tat increases cooperation between AP-1 and NFAT transcription factors in T cells. J. Gen. Virol. 2006;87:1603–1612. doi: 10.1099/vir.0.81637-0. [DOI] [PubMed] [Google Scholar]
- 85.Serrador JM, Alonso-Lebrero JL, del Pozo MA, Furthmayr H, Schwartz-Albiez R, Calvo J, Lozano F, Sanchez-Madrid F. Moesin interacts with the cytoplasmic region of intercellular adhesion molecule-3 and is redistributed to the uropod of T lymphocytes during cell polarization. J. Cell Biol. 1997;138:1409–1423. doi: 10.1083/jcb.138.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fais S, Malorni W. Leukocyte uropod formation and membrane/cytoskeleton linkage in immune interactions. J. Leukoc. Biol. 2003;73:556–563. doi: 10.1189/jlb.1102568. [DOI] [PubMed] [Google Scholar]
- 87.Izmailova E, Bertley FM, Huang Q, Makori N, Miller CJ, Young RA, Aldovini A. HIV-1 Tat reprograms immature dendritic cells to express chemoattractants for activated T cells and macrophages. Nat. Med. 2003;9:191–197. doi: 10.1038/nm822. [DOI] [PubMed] [Google Scholar]
- 88.de la Fuente C, Santiago F, Deng L, Eadie C, Zilberman I, Kehn K, Maddukuri A, Baylor S, Wu K, Lee CG, et al. Gene expression profile of HIV-1 Tat expressing cells: a close interplay between proliferative and differentiation signals. BMC Biochem. 2002;3:14. doi: 10.1186/1471-2091-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meyaard L, Otto SA, Schuitemaker H, Miedema F. Effects of HIV-1 Tat protein on human T cell proliferation. Eur. J. Immunol. 1992;22:2729–2732. doi: 10.1002/eji.1830221038. [DOI] [PubMed] [Google Scholar]
- 90.Li CJ, Friedman DJ, Wang C, Metelev V, Pardee AB. Induction of apoptosis in uninfected lymphocytes by HIV-1 Tat protein. Science. 1995;268:429–431. doi: 10.1126/science.7716549. [DOI] [PubMed] [Google Scholar]
- 91.Westendorp MO, Frank R, Ochsenbauer C, Stricker K, Dhein J, Walczak H, Debatin KM, Krammer PH. Sensitization of T cells to CD95-mediated apoptosis by HIV-1 Tat and gp120. Nature. 1995;375:497–500. doi: 10.1038/375497a0. [DOI] [PubMed] [Google Scholar]
- 92.Viscidi RP, Mayur K, Lederman HM, Frankel AD. Inhibition of antigen-induced lymphocyte proliferation by Tat protein from HIV-1. Science. 1989;246:1606–1608. doi: 10.1126/science.2556795. [DOI] [PubMed] [Google Scholar]
- 93.Subramanyam M, Gutheil WG, Bachovchin WW, Huber BT. Mechanism of HIV-1 Tat induced inhibition of antigen-specific T cell responsiveness. J. Immunol. 1993;150:2544–2553. [PubMed] [Google Scholar]
- 94.Dewan MZ, Takamatsu N, Hidaka T, Hatakeyama K, Nakahata S, Fujisawa J, Katano H, Yamamoto N, Morishita K. Critical role for TSLC1 expression in the growth and organ infiltration of adult T-cell leukemia cells in vivo. J. Virol. 2008;82:11958–11963. doi: 10.1128/JVI.01149-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu P, Keller JR, Ortiz M, Tessarollo L, Rachel RA, Nakamura T, Jenkins NA, Copeland NG. Bcl11a is essential for normal lymphoid development. Nat. Immunol. 2003;4:525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- 96.Nakao A, Miike S, Hatano M, Okumura K, Tokuhisa T, Ra C, Iwamoto I. Blockade of transforming growth factor beta/Smad signaling in T cells by overexpression of Smad7 enhances antigen-induced airway inflammation and airway reactivity. J. Exp. Med. 2000;192:151–158. doi: 10.1084/jem.192.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kashiwa A, Yoshida H, Lee S, Paladino T, Liu Y, Chen Q, Dargusch R, Schubert D, Kimura H. Isolation and characterization of novel presenilin binding protein. J. Neurochem. 2000;75:109–116. doi: 10.1046/j.1471-4159.2000.0750109.x. [DOI] [PubMed] [Google Scholar]
- 98.Namekata K, Enokido Y, Iwasawa K, Kimura H. MOCA induces membrane spreading by activating Rac1. J. Biol. Chem. 2004;279:14331–14337. doi: 10.1074/jbc.M311275200. [DOI] [PubMed] [Google Scholar]
- 99.Caspi E, Rosin-Arbesfeld R. A novel functional screen in human cells identifies MOCA as a negative regulator of Wnt signaling. Mol. Biol. Cell. 2008;19:4660–4674. doi: 10.1091/mbc.E07-10-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.He F, Xiong W, Yu X, Espinoza-Lewis R, Liu C, Gu S, Nishita M, Suzuki K, Yamada G, Minami Y, et al. Wnt5a regulates directional cell migration and cell proliferation via Ror2-mediated noncanonical pathway in mammalian palate development. Development. 2008;135:3871–3879. doi: 10.1242/dev.025767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kurayoshi M, Oue N, Yamamoto H, Kishida M, Inoue A, Asahara T, Yasui W, Kikuchi A. Expression of Wnt-5a is correlated with aggressiveness of gastric cancer by stimulating cell migration and invasion. Cancer Res. 2006;66:10439–10448. doi: 10.1158/0008-5472.CAN-06-2359. [DOI] [PubMed] [Google Scholar]
- 102.Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, Walk SF, Nemergut ME, Macara IG, Francis R, et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- 103.Janardhan A, Swigut T, Hill B, Myers MP, Skowronski J. HIV-1 Nef binds the DOCK2-ELMO1 complex to activate rac and inhibit lymphocyte chemotaxis. PLoS Biol. 2004;2:E6. doi: 10.1371/journal.pbio.0020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee CC, Putnam AJ, Miranti CK, Gustafson M, Wang LM, Vande Woude GF, Gao CF. Overexpression of sprouty 2 inhibits HGF/SF-mediated cell growth, invasion, migration, and cytokinesis. Oncogene. 2004;23:5193–5202. doi: 10.1038/sj.onc.1207646. [DOI] [PubMed] [Google Scholar]
- 105.Foy TM, Aruffo A, Bajorath J, Buhlmann JE, Noelle RJ. Immune regulation by CD40 and its ligand GP39. Annu. Rev. Immunol. 1996;14:591–617. doi: 10.1146/annurev.immunol.14.1.591. [DOI] [PubMed] [Google Scholar]
- 106.Murai T, Miyazaki Y, Nishinakamura H, Sugahara KN, Miyauchi T, Sako Y, Yanagida T, Miyasaka M. Engagement of CD44 promotes Rac activation and CD44 cleavage during tumor cell migration. J. Biol. Chem. 2004;279:4541–4550. doi: 10.1074/jbc.M307356200. [DOI] [PubMed] [Google Scholar]
- 107.Sallusto F, Lenig D, Mackay CR, Lanzavecchia A. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J. Exp. Med. 1998;187:875–883. doi: 10.1084/jem.187.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yoshida R, Imai T, Hieshima K, Kusuda J, Baba M, Kitaura M, Nishimura M, Kakizaki M, Nomiyama H, Yoshie O. Molecular cloning of a novel human CC chemokine EBI1-ligand chemokine that is a specific functional ligand for EBI1, CCR7. J. Biol. Chem. 1997;272:13803–13809. doi: 10.1074/jbc.272.21.13803. [DOI] [PubMed] [Google Scholar]
- 109.Pickl WF, Majdic O, Fischer GF, Petzelbauer P, Fae I, Waclavicek M, Stockl J, Scheinecker C, Vidicki T, Aschauer H, et al. MUC18/MCAM (CD146), an activation antigen of human T lymphocytes. J. Immunol. 1997;158:2107–2115. [PubMed] [Google Scholar]
- 110.Epie N, Ammosova T, Sapir T, Voloshin Y, Lane WS, Turner W, Reiner O, Nekhai S. HIV-1 Tat interacts with LIS1 protein. Retrovirology. 2005;2:6. doi: 10.1186/1742-4690-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gautier VW, Sheehy N, Duffy M, Hashimoto K, Hall WW. Direct interaction of the human I-mfa domain-containing protein, HIC, with HIV-1 Tat results in cytoplasmic sequestration and control of Tat activity. Proc. Natl Acad. Sci. USA. 2005;102:16362–16367. doi: 10.1073/pnas.0503519102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sasaki H, Nakamura M, Ohno T, Matsuda Y, Yuda Y, Nonomura Y. Myosin-actin interaction plays an important role in human immunodeficiency virus type 1 release from host cells. Proc. Natl Acad. Sci. USA. 1995;92:2026–2030. doi: 10.1073/pnas.92.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ott DE, Coren LV, Kane BP, Busch LK, Johnson DG, Sowder RC, Chertova EN, Arthur LO, Henderson LE. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J. Virol. 1996;70:7734–7743. doi: 10.1128/jvi.70.11.7734-7743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nagy G, Ward J, Mosser DD, Koncz A, Gergely P., Jr, Stancato C, Qian Y, Fernandez D, Niland B, Grossman CE, et al. Regulation of CD4 expression via recycling by HRES-1/RAB4 controls susceptibility to HIV infection. J. Biol. Chem. 2006;281:34574–34591. doi: 10.1074/jbc.M606301200. [DOI] [PubMed] [Google Scholar]
- 115.Willard-Gallo KE, Furtado M, Burny A, Wolinsky SM. Down-modulation of TCR/CD3 surface complexes after HIV-1 infection is associated with differential expression of the viral regulatory genes. Eur. J. Immunol. 2001;31:969–979. doi: 10.1002/1521-4141(200104)31:4<969::aid-immu969>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 116.Choe H, Farzan M, Konkel M, Martin K, Sun Y, Marcon L, Cayabyab M, Berman M, Dorf ME, Gerard N, et al. The orphan seven-transmembrane receptor apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J. Virol. 1998;72:6113–6118. doi: 10.1128/jvi.72.7.6113-6118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shinya E, Owaki A, Shimizu M, Takeuchi J, Kawashima T, Hidaka C, Satomi M, Watari E, Sugita M, Takahashi H. Endogenously expressed HIV-1 nef down-regulates antigen-presenting molecules, not only class I MHC but also CD1a, in immature dendritic cells. Virology. 2004;326:79–89. doi: 10.1016/j.virol.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 118.Janeway CA, Travers P, Walport M, Schlomchik M. Immunobiology 5: The Immune System in Health and Disease. New York: Garland Publishing; 2001. [Google Scholar]
- 119.Jeang KT, Berkhout B, Dropulic B. Effects of integration and replication on transcription of the HIV-1 long terminal repeat. J. Biol. Chem. 1993;268:24940–24949. [PubMed] [Google Scholar]
- 120.Rhim H, Rice AP. Exon2 of HIV-2 Tat contributes to transactivation of the HIV-2 LTR by increasing binding affinity to HIV-2 TAR RNA. Nucleic Acids Res. 1994;22:4405–4413. doi: 10.1093/nar/22.21.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tong-Starksen SE, Baur A, Lu XB, Peck E, Peterlin BM. Second exon of Tat of HIV-2 is required for optimal trans-activation of HIV-1 and HIV-2 LTRs. Virology. 1993;195:826–830. doi: 10.1006/viro.1993.1438. [DOI] [PubMed] [Google Scholar]