Abstract
POLN is a nuclear A-family DNA polymerase encoded in vertebrate genomes. POLN has unusual fidelity and DNA lesion bypass properties, including strong strand displacement activity, low fidelity favoring incorporation of T for template G and accurate translesion synthesis past a 5S-thymine glycol (5S-Tg). We searched for conserved features of the polymerase domain that distinguish it from prokaryotic pol I-type DNA polymerases. A Lys residue (679 in human POLN) of particular interest was identified in the conserved ‘O-helix’ of motif 4 in the fingers sub-domain. The corresponding residue is one of the most important for controlling fidelity of prokaryotic pol I and is a nonpolar Ala or Thr in those enzymes. Kinetic measurements show that K679A or K679T POLN mutant DNA polymerases have full activity on nondamaged templates, but poorly incorporate T opposite template G and do not bypass 5S-Tg efficiently. We also found that a conserved Tyr residue in the same motif not only affects sensitivity to dideoxynucleotides, but also greatly influences enzyme activity, fidelity and bypass. Protein sequence alignment reveals that POLN has three specific insertions in the DNA polymerase domain. The results demonstrate that residues have been strictly retained during evolution that confer unique bypass and fidelity properties on POLN.
INTRODUCTION
The human genome encodes 15 DNA polymerases specialized for different functions, including DNA replication, repair, recombination and bypass of DNA damage. The nuclear A-family DNA polymerase POLN (polν) is a 100 kDa enzyme with moderate processivity. POLN has several unique properties, including efficient strand displacement activity. It is a low fidelity enzyme lacking an intrinsic proofreading 3′–5′ exonuclease, and favors incorporation of T almost as often as C for template G, resulting in frequent GC to AT transitions (1,2). POLN can catalyze accurate translesion synthesis past a 5S-thymine glycol (Tg), a major DNA lesion generated by reactive oxygen species that blocks the progression of replicative DNA polymerases (1).
POLN’s low fidelity and ability to efficiently bypass lesions is in contrast to prokaryotic A-family DNA polymerases, which generally have high fidelity. Another nuclear DNA polymerase in the A-family encoded by the human genome is designated as POLQ (polθ), but it has properties very different from POLN. The 290 kDa POLQ harbors a superfamily-2 helicase-like domain as well as an A-family DNA polymerase domain (3). POLQ has a high frameshift error rate (in contrast to POLN) (4). POLQ, but not POLN, can efficiently bypass an apurinic (AP) site in DNA. POLQ is broadly expressed in several tissues and cell lines, and disruption of POLQ causes phenotypes associated with hypersensitivity to ionizing radiation (5,6). In contrast, POLN is preferentially expressed in testis (7), and POLN protein expression is very difficult to detect in cultured cell lines. It is important to understand why POLN has functionally unique activities that distinguish it from prototypical A-family DNA polymerases as well as from POLQ. Here, we report an analysis of primary protein sequence features that distinguish vertebrate POLN. We identify key residues in the O-helix that confer reduced fidelity and are evolutionarily conserved in POLN.
MATERIALS AND METHODS
Production of POLN derivatives
Wild-type POLN (WT) was amplified by PCR from the plasmid pEGFP-C1 containing full-length POLN (7) using the primers 5′- CACCCTGGAAGTTCTGTTCCAGGGGCCCGAAAATTATGAGGCATTGGTAGGC-3' (for the 5′-end) and 5′-ATATATGAATTCCTACTTGTCGTCATCGTCTTTGTAGTCCATCAGACAAAATGAAGGCGAAAAATGC-3' (for the 3′-end), and cloned into plasmid pENTR/D-TOPO (Invitrogen, Carlsbad, CA, USA). After DNA sequencing, the cDNA was transfered into plasmid pDEST17 (Invitrogen), resulting in a protein tagged with six His residues at the N-terminus (contributed by the pDEST17 vector), and a FLAG tag at the C-terminus. The following primers were chemically synthesized to create POLN point mutations (altered DNA sequences are underlined): 5′-GAGCAAACCAAGAGGGTGGTGTACGCG-3′ and 5′-CGCGTACACCACCCTCTTGGTTTGCTC-3′ (for K679R), 5′-GAGCAAACCAAGACGGTGGTGTACGC-3′ and 5′-GCGTACACCACCGTCTTGGTTTGCTC-3′ (for K679T), 5′-GAGCAAACCAAGGCGGTGGTGTACG-3′ and 5′-CGTACACCACCGCCTTGGTTTGCTC-3′ (for K679A), 5′-GAGCAAACCAAGCAGGTGGTGTACGC-3′ and 5′-GCGTACACCACCTGCTTGGTTTGCTC-3' (for K679Q), 5′-CAAGAAGGTGGTGTTCGCGGTGGTCTATG-3′ and 5′-CATAGACCACCGCGAACACCACCTTCTTG-3′ (for Y682F). Mutagenesis was performed by using a Quick Change II site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). To generate K679R, K679T, K679Q and Y682F, the pDEST17 vector carrying wild-type POLN was used as a template. To generate K679A, the K679T vector was used as a template. Recombinant POLN derivatives were bacterially expressed and protein from the soluble fraction was purified (1), yielding ∼50 µg from 1 l of culture. The fraction of soluble protein was <1% and was similar for all derivatives. These proteins were concentrated by NANOSEP 30 K (PALL, Ann Arbor, MI, USA), and stored in buffer containing 50 mM sodium phosphate (pH 7.0), 300 mM NaCl, 10% glycerol, and 0.01% NP-40. Sequence-verified DNA polymerase Q (POLQ) was purified as reported (8). RB69 gp43 was purified as reported (9), and Kf (exo−) (exonuclease-deficient Klenow fragment of Escherichia coli pol I) was obtained from Promega, San Luis Obispo, CA, USA.
Oligonucleotide substrates
Primer oligonucleotides were purchased from Bio-Synthesis or Sigma GenoSys, purified by HPLC or gel extraction, and 5′-labeled using polynucleotide kinase and [γ-32P] dATP. Oligonucleotides containing a 5S-thymine glycol were synthesized as described (10). The phosphoramidite precursors for an AP site analog and a 5R-thymine glycol were purchased from Glen Research (Sterling, VA, USA), synthesized into 30-mer oligonucleotides and gel-purified. 5′-[32P]-labeled primers were annealed to these oligonucleotides to create substrates for bypass assays (lesions highlighted by bold letters) as follows:
Non-damaged substrate
5′-CACTGACTGTATGATG-3′
3′-GTGACTGACATACTACXTCTACGACTGCTC-5′
The first template base denoted by X was A, C, G or T. 5′-AAGATGCTGACGAG was annealed to the nondamaged substrate (X was T) to create the nicked substrate.
Tg-substrate (5S-Tg or 5R-Tg is denoted by Tg)
5′-CACTGACTGTATGATG-3'
3′-GTGACTGACATACTAC(Tg)TCTACGACTGCTC-5'
AP site-substrate (site of the AP-analog denoted by X)
5′-CACTGACTGTATGATG-3'
3′-GTGACTGACATACTACXTCTACGACTGCTC-5'
For processivity assays, the 24-mer primer oligonucleotide 5′-[32P]-TCTTCTTCTGTGCACTCTTCTTCT was annealed to M13mp18GTGx single-stranded DNA (11). For strand displacement assays, a 60-mer oligonucleotide blocked at the 3′-end from priming using TdT and ddATP, with a 5′-phosphoryl, 5′-CCCCAGGAATTCGGTCATAGCTGTTTCCTGCTCGAGGGCGCCAGGGTGGTTTTTCTTTTC was annealed to the 24-mer oligomer to M13mp18GTGx ssDNA to generate a nicked substrate.
DNA polymerase assays
A 5′-32P-labeled 16-mer primer and a 30-mer template (sequences given above) were annealed at a molar ratio of 1:1 to detect DNA polymerase activity (1). 5′-32P-labeled 16-mer primer, a downstream oligomer (5′-AAGATGCTGACGAG), and the 30-mer template were used at a molar ratio of 1:5:2 for the nicked substrate. Singly primed DNA templates were prepared by annealing the 5′-32P-labeled 24-mer primer to single-stranded circular M13mp18GTGx DNA at a molar ratio of 2:1 to detect processivity. To measure strand displacement, substrates were prepared by mixing the 5′-32P-labeled 24-mer primer, a downstream 60-mer oligomer with a 5′-phosphoryl, and single-stranded M13mp18GTGx at a molar ratio of 1:5:2. Primer templates were heated for 5 min at 65°C and cooled down slowly for annealing. Reaction mixtures (10 µl) in buffer A [20 mM Tris–HCl pH 8.8, 4% glycerol, 2 mM dithiothreitol (DTT), 80 µg/ml bovine serum albumin (BSA), 8 mM Mg acetate], 100 µM of each dNTP, 30 nM of the primer-template (unless otherwise indicated), and the indicated amount of POLN derivatives. Mixtures with POLQ (10 µl) contained buffer A and 0.1 mM EDTA. RB69 gp43 reaction mixtures (10 µl) contained 10 mM Tris–HCl pH 7.9, 50 mM NaCl, 1 mM DTT, 200 µg/ml BSA, 10 mM MgCl2 and 100 µM of each dNTP. After incubation at 37°C for 10 min (unless otherwise indicated), reactions were terminated by adding 10 µl of formamide stop buffer and boiling at 95°C for 3 min. Products were electrophoresed on a denaturing 20% polyacrylamide–7 M urea gel and exposed to BioMax MS film or analyzed with a Fuji FLA3000 Phosphor Imager. For translesion synthesis, the same amounts of templates containing specific lesions were used. For steady-state kinetics, 1 pmol of primer-template (100 nM) was used and the procedure was as previously described (12). This procedure utilizes extensive titration of deoxynucleotide concentration (0.1, 1, 5, 10, 50, 100, 200, 500 and 1000 µM) and four time points (0, 2.5, 5 and 10 min), producing results valid for a wide range of enzyme concentrations. Vmax and Km were determined from a Hanes–Woolf plot of [dNTP]/velocity versus [dNTP]. The nucleotide misincorporation ratio, finc was determined by dividing (kcat/Km)incorrect by (kcat/Km)correct. Less than 10% of the primers were extended under steady-state conditions, ensuring single hit conditions. In the steady state, Vmax values were proportional to enzyme concentration (data not shown). Here, kcat was presented by utilizing the equation kcat = [Vmax (mol of primer-template)]/ [(mol of polymerase) min].
To test sensitivity to ddNTP, ddTTP (2′,3′-dideoxythymidine-5′-triphosphate, Amersham, Piscataway, NJ, USA) and a poly(dA)-oligo(dT)10:1 template were used. Reaction mixtures (25 µl) in buffer A contained 8 µg/ml of poly(dA)-oligo(dT)10:1, 10 µM of dTTP, the indicated amount of ddTTP, 1 µCi of [α-32P] dTTP, 23 nM of WT (POLN) or delP, or 115 nM of Y682F, or 1 pM of Kf (exo−). After incubation at 37°C for 20 min, reactions were stopped by adding 25 µl of 40 mM EDTA and placed on ice. A 10 µl aliquot of each mixture was spotted onto DE81 paper (Whatman), and washed three times with 0.5 M Na2HPO4 for 5 min and twice with ethanol. The paper was dried and radioactivity was quantified with a Fuji Phosphor Imager.
M13mp2 fidelity assay
Reaction mixtures (25 µl) contained buffer A (at pH 8.4), 1 mM each of dATP, dCTP, dGTP and dTTP and 0.2 nM of gapped M13mp2 DNA substrate. Reactions were initiated by adding POLN (240 ng, 90 nM), incubated at 37°C for 30 min, and terminated by adding EDTA to 20 mM. Half of the reaction mixture was mixed 1:1 with SDS buffer (20 mM Tris pH 8.0, 5 mM EDTA, 5% SDS, 0.5 % bromophenol blue and 25% glycerol) and analyzed by agarose gel electrophoresis as described (13). POLN wild-type and mutant proteins completely filled the gap [data not shown, but for a typical result, see ref. (2)]. Each gap-filled DNA product was examined for the frequency of lacZ mutants by electroporation and plating onto indicator plates as described (13). Errors were scored as light blue or colorless mutant plaques, while correct synthesis yielded plaques that were dark blue. DNA from independent mutants was sequenced in order to identify the errors made during gap-filling synthesis. For sequence changes that yield light blue and colorless plaques, error rates per detectable nucleotide incorporated were calculated (13).
Bypass efficiency reactions
The assays were performed as reported (14,15). 5′-32P-labeled 14- and 15-mer primers (Supplementary Figure S5), 1 or 2 nt shorter than the similar primer used for DNA polymerase assays in Figure 4, were annealed to a 30-mer template at a molar ratio of 1:1. Primer extension reactions with POLN, POLQ and RB69 gp43 were as described above. Reaction mixtures (10 µl) were incubated at 37°C for 2, 4 or 6 min and diluted in 10 µl of formamide stop buffer. Products were heated at 95°C for 3 min and separated on a denaturing 20% polyacrylamide–7 M urea gel. Product bands were quantified with a phosphorimager and the values used to calculate the probability of termination of processive synthesis and the insertion efficiencies at each template nucleotide (14). The termination probability at position (X) is the band intensity at (X)/the summed intensity for all bands ≥[X]. The insertion probability at position (X) is the intensity at bands ≥[X]/the summed intensity at bands ≥[X–1]. The extension probability at position (X) is the band intensity ≥[X + 1]/ the intensity at bands ≥[X]. The bypass probability at position (X) is the summed band density ≥[X + 1] / the intensity of ≥[N1]. Bypass efficiency is the bypass probability (damaged) divided by the bypass probability (undamaged).
Figure 4.
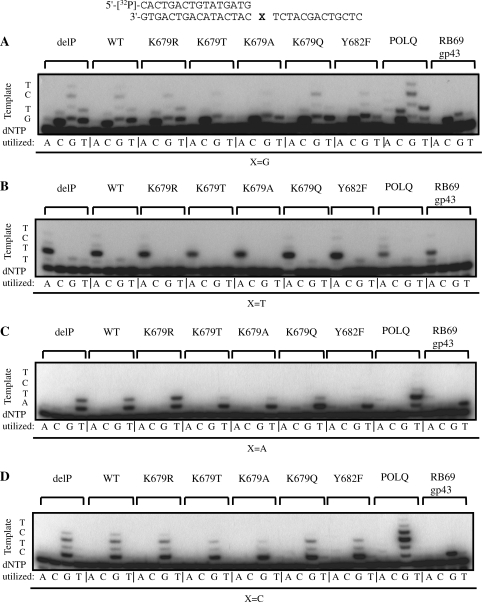
Nucleotide selectivities of POLN derivatives. Twenty-three nanomolars of delP, WT, K679R, K679T, K679A and K679Q, 115 nM of Y682F, 12 nM of POLQ and 10 pM of RB69 gp43 were incubated with 300 fmol of 5′-32P-labeled 16-mer primer annealed to a 30-mer DNA template in the presence of one of the indicated dNTPs (100 µM) for 10 min. The first template base denoted by X was G, T, A or C in (A, B, C and D), respectively. Template sequences are indicated to the left of each panel.
RESULTS
Unusual residue conservation and sequence insertions in the POLN DNA polymerase domain
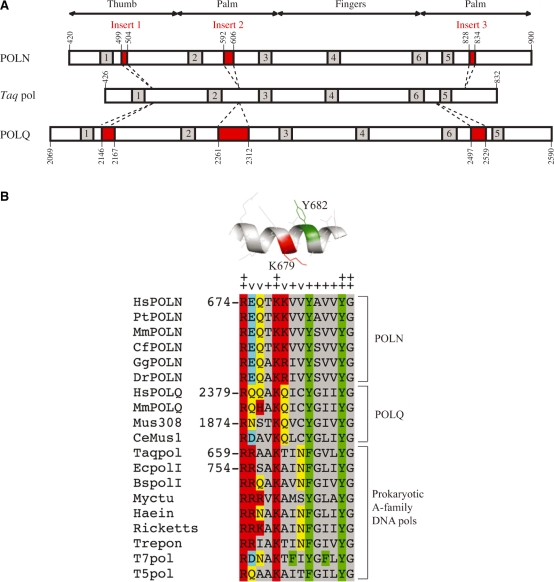
Using recently available complete sequences for vertebrate genomes, an alignment of the polymerase domain of POLN sequences was made with the equivalent region of prokaryotic A-family DNA polymerases. We found that POLN has three previously unrecognized extra sequence insertions compared to the prokaryotic enzymes (denoted Insert 1, 2 and 3) (Figure 1A and Supplementary Figure S1). Insert 1 maps to the tip of the ‘thumb’ subdomain of known pol I structures, and Inserts 2 and 3 to the ‘palm’ subdomain. Inserts 1 and 2 correspond in position to two longer sequence inserts present in the DNA polymerase domain of POLQ (8). In contrast, Insert 3 of POLN (7 aa) lies in a position C-terminal of motif 5, entirely distinct from that of the 33 aa Insert 3 of POLQ (Figure 1A). The functions of these inserts are currently unknown, but they provide a further means to distinguish POLN from POLQ in vertebrate genomes.
Figure 1.
Distinct sequence insertions in POLN. (A) The DNA polymerase catalytic region of human POLN is shown in relation to Taq pol and human POLQ. The locations of the three inserts found in POLN and POLQ are shown in red and the conserved DNA polymerase motifs are shown in gray. (B) Sequence alignment of motif 4 of POLN, POLQ and prokaryotic A-family DNA polymerases. Similarity groups for colored residues are (K, R, H), (D, E), (F, Y, W) and (Q, N). Perfectly conserved residues are denoted by ++, relatively conserved residues are denoted by +, flexible amino acid residues are denoted by ∨. K679 and Y682 are shown in red and green in a modeled structure for motif 4 of HsPOLN. Hs, Homo sapiens; Pt, Pan troglodytes; Mm, Mus musculus; Cf, Canis familiaris; Gg, Gallus gallus; Dr, Danio rerio; Mus308, Drosophila melanogaster Mus308; CeMus1, Caenorhabditis elegans Mus1; Taqpol, Thermus aquaticus DNA polymerase; EcpolI, Escherichia coli; BspolI, Bacillus subtilis; Myctu, Mycobacterium tuberculosis pol I; Haein, Haemophilus influenzae pol I; Ricketts, Rickettsia prowazekii pol I; Trepon, Treponema pallidum pol I; T7pol, T7 DNA polymerase; T5pol, T5 DNA polymerase. PtPOLN, CfPOLN and GgPOLN are predicted from their genomic DNA sequences.
In the six recognized A family DNA polymerase motifs, amino acid residues are generally highly conserved. We focused on Motifs 3 and 4, which form structures positioned to contribute critically to DNA polymerase fidelity and substrate specificity (16). Motif 3 residues are well-conserved between POLN and prokaryotic A-family DNA polymerases, but the 14 residues of POLN Motif 4 display an important difference from the prokaryotic homologs (Figure 1B). Four amino acid residues (R674, K678, Y686 and G687 of POLN) are perfectly conserved among A-family DNA polymerases and the first two correspond to ‘immutable’ positions in the screen of Loeb and co-workers (17) of permitted base changes in the O-Helix of Taq pol I. Six other residues are highly conserved in the alignment, and four residues (E675, Q676, K679 and V681 of POLN) show poor conservation (Figure 1B). Position 679 is of particular interest. It is consistently a positively charged Lys or Arg in POLN, and an uncharged Ala or Thr in the prokaryotic enzymes (Figure 1B). This residue stands out for its exceptional importance in extensive screens for fidelity mutants of prokaryotic pol I. The single mutant with the lowest fidelity isolated by mutagenesis of this motif in Taq pol I was a change of a Thr residue at this position to Arg, T664R (18). In E. coli DNA polymerase I, the single mutant isolated with the lowest fidelity was the substitution of an Ala at this position for Arg (19). The mutation rate was increased by 14- to 25-fold by this single amino acid substitution in both cases. It is intriguing that POLN homologs consistently have Lys or Arg at this position as in the low fidelity prokaryotic pol I mutants, and we reasoned that this amino acid difference might be responsible for much of the low fidelity of POLN.
K679 and Y682 in the O-helix are critical for fidelity in POLN
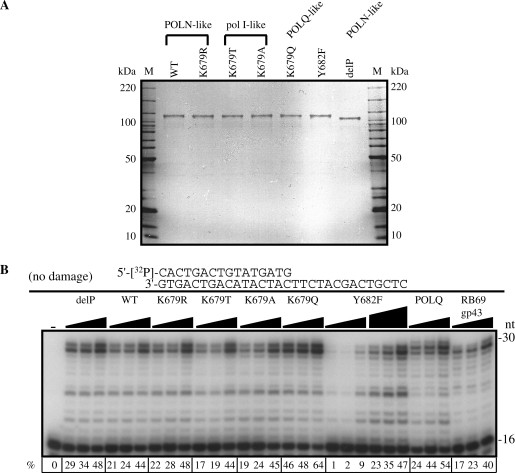
To test this proposal, several recombinant POLN derivatives were expressed in bacteria and purified (Figure 2A). K679T and K679A mutants were constructed to correspond to the prokaryotic enzymes, and a K679Q because a Gln residue occurs in this position of POLQ. In motif 4, a single aromatic residue that is either Phe or Tyr is highly conserved among A-family DNA polymerases (Y682 in HsPOLN) (Figure 1B). A Y682F mutant was made to determine whether this residue is critical for ddNTP selectivity as it is for other A-family DNA polymerases. DNA polymerases with Phe at this position are not sensitive to ddNTP, while DNA polymerases carrying Tyr (including POLN) are sensitive (1,20). A control POLN was also purified with a truncation of the nonessential proline-rich C-terminal tail (delP) (1). Further controls were the high fidelity B-family gp43 DNA polymerase from phage RB69, and human POLQ.
Figure 2.
Activity of wild-type (WT) and mutant POLN on a nondamaged DNA template. (A) Purified POLN derivatives (300 ng) and molecular weight markers were separated by electrophoresis in a 4–15% SDS–polyacrylamide gradient gel and stained with colloidal Coomassie Brilliant Blue G-250. Substituted residues in POLN derivatives are shown in Figure 1. delP is POLN with a short truncation of the C-terminal proline-rich tail. (B) DNA polymerase activities of POLN derivatives. Increasing amounts of delP, WT, K679R, K679T, K679A and K679Q (6, 12 and 23 nM), Y682F (6, 12, 23, 29, 58 and 115 nM), POLQ (3, 6 and 12 nM), RB69 gp43 (2.5, 5 and 10 pM) were incubated with the 5′-32P-labeled primer templates indicated beside the panel in the presence of all 4 nt at 37°C for 10 min. The first lane contained no enzyme. The percentage (%) of the product extension from the primer is shown below each lane. The specific activity of POLN is 127 U/mg, delP is 125 U/mg, K679R is 116 U/mg, K679T is 112 U/mg, K679A is 124 U/mg, K679Q is 153 U/mg and Y682F is 32 U/mg. One unit is defined as 10 nmol of dTTP incorporated into poly(dA)/oligo(dT)10:1 template at 37°C for 30 min.
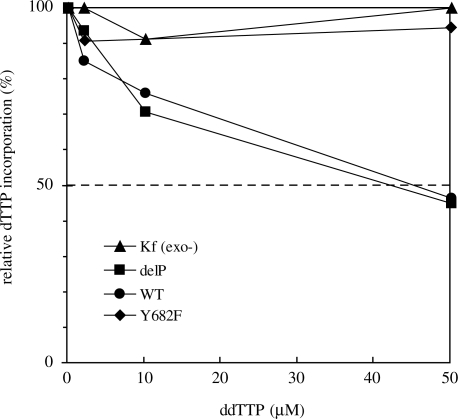
On a nondamaged DNA template, all POLN derivatives showed similar DNA polymerase activities except Y682F, which had a specific activity about 5-fold lower (Figure 2). Consistent with data on other A-family DNA polymerases, the WT and delP proteins harboring Tyr at position 682 were sensitive to ddTTP on a poly(dA)-oligo(dT) template, whereas POLN carrying Phe at the corresponding position (Y682F) was not sensitive (Figure 3).
Figure 3.
Substitution of Phe for Tyr in POLN increases discrimination against dideoxynucleotide. Escherichia coli Kf (exo−), delP, WT and Y682F were incubated with ddTTP in a reaction mixture containing radioactively labeled dTTP and poly(dA)/oligo(dT) template.
Fidelities of POLN derivatives were examined qualitatively by gel electrophoresis (Figure 4) and quantitatively by measuring kinetics (Table 1). Substrates were analyzed with first template base G, T, A or C (Figure 4A, B, C and D, respectively). All enzymes efficiently incorporated the correct C deoxynucleotide when the first template base was G (Figure 4A). Frequent incorporation of T deoxynucleotide opposite G was noted for POLN delP, WT, K679R, K679Q and POLQ (Figure 4A, lanes utilizing T), consistent with previous results (1,8). POLQ was the most promiscuous enzyme on this template. However, K679T, K679A and Y682F incorporated T opposite G less efficiently (Figure 4A, lanes utilizing T).
Table 1.
Steady-state kinetic constants of POLN and mutant derivatives
| DNA substrate | enzyme | dNTP | Km (µM) | kcat (min−1 × 10−4) | kcat/Km (µM−1 min−1 × 10−4) | finc |
|---|---|---|---|---|---|---|
| Insertion opposite G | WT | dCTP | 7.5 ± 0.3 | 959 ± 50 | 128 ± 12 | 1 |
| 5′-ATG-TACGTC | dTTP | 18.5 ± 1.5 | 899 ± 101 | 49.3 ± 9.4 | 3.9 × 10−1 | |
| delPa | dCTP | 8.2 ± 0.6 | 987 ± 28 | 124 ± 43 | 1 | |
| dTTP | 18.8 ± 4.6 | 928 ± 20 | 55.2 ± 2.4 | 4.5 × 10−1 | ||
| K679R | dCTP | 8.1 ± 0.2 | 549 ± 23 | 67.5 ± 1.0 | 1 | |
| dTTP | 19.8 ± 1.7 | 507 ± 19 | 25.6 ± 5.2 | 3.8 × 10−1 | ||
| K679T | dCTP | 5.7 ± 0.6 | 473 ± 67 | 83.5 ± 8.8 | 1 | |
| dTTP | 43.2 ± 1.5 | 171 ± 30 | 4.0 ± 0.8 | 4.8 × 10−2 | ||
| K679A | dCTP | 7.8 ± 0.8 | 588 ± 104 | 78.1 ± 26 | 1 | |
| dTTP | 52.1 ± 3.3 | 227 ± 47 | 4.4 ± 1.2 | 5.6 × 10−2 | ||
| Y682F | dCTP | 32.8 ± 3.5 | 157 ± 12 | 4.9 ± 0.9 | 1 | |
| dTTP | 98.6 ± 11.6 | 42.5 ± 8.0 | 0.4 ± 0.0 | 8.2 × 10−2 | ||
| DNA substrate | enzyme | dNTP | Km (µM) | kcat (min−1 × 10−4) | kcat/Km (µM−1 min−1 × 10−4) | Relative efficiencyb |
| Insertion opposite T | WT | dATP | 8.5 ± 1.4 | 1265 ± 91 | 152 ± 14 | |
| 5′-ATG-TACTTC | delPa | dATP | 7.0 ± 1.4 | 1385 ± 212 | 201 ± 9 | |
| K679R | dATP | 8.8 ± 1.0 | 1402 ± 219 | 165 ± 43 | ||
| K679T | dATP | 6.8 ± 0.1 | 410 ± 10 | 60.2 ± 2.6 | ||
| K679A | dATP | 7.4 ± 0.4 | 695 ± 96 | 92.8 ± 7.9 | ||
| Y682F | dATP | 26.2 ± 1.3 | 220 ± 38 | 8.4 ± 0.6 | ||
| Insertion opposite 5S-Tg | WT | dATP | 14.2 ± 1.5 | 346 ± 24 | 24.7 ± 4.3 | 1.6 × 10−1 |
| 5′-ATG-TACTgTC | delPa | dATP | 11.8 ± 1.1 | 392 ± 15 | 33.5 ± 2 | 1.7 × 10−1 |
| K679R | dATP | 13.0 ± 0.1 | 361 ± 9 | 27.8 ± 0.8 | 1.7 × 10−1 | |
| K679T | dATP | 28.6 ± 1.1 | 132 ± 10 | 4.6 ± 0.5 | 7.6 × 10−2 | |
| K679A | dATP | 36.2 ± 1.0 | 167 ± 11 | 4.6 ± 0.2 | 5.0 × 10−2 | |
| Y682F | dATP | 126 ± 20 | 156 ± 14 | 1.2 ± 0.2 | 1.4 × 10−1 | |
aData were taken from ref. (1).
bkcat/Km values for incorporation of dATP were normalized to 1 and the kcat/Km values were given relative to these values.
Similarly, with first template base T, POLN mutants K679T and K679A were less efficient at incorporating G deoxynucleotide (Figure 4B). When the first template base was A (Figure 4C) and only dTTP was provided, each enzyme efficiently incorporated T deoxynucleotide. Wild-type POLN (and delP, K679Q) also incorporated a further T, but the K679T, K679A and Y682F mutants rarely did so. When the first template base was C (Figure 4D), each enzyme efficiently inserted a G. The POLN mutants K679T and K679A were the least able to extend further by incorporating a G opposite the second T template base. POLQ efficiently incorporated G opposite the second template base T and was able to extend the furthest; RB69 gp43 hardly incorporated G opposite T.
From kinetic measurements Km and kcat were determined, and nucleotide incorporation frequencies by POLN derivatives for template base G were calculated from kcat/Km. This enabled calculation of the frequency of misincorporation of T opposite G (ƒinc). The ƒinc values reveal that insertion fidelity was increased about 10-fold by the K679T and K679A substitutions, and increased about 5-fold in Y682F compared to WT, delP or K679R proteins (Table 1).
The DNA products of complete gap filling by POLN and POLN derivatives [data not shown, but for example see Figure 3 in ref. (13)] yielded a lacZ mutant frequency of 12% for POLN, 2.3% for K679T and 3.4% for K679A (Table 2). These results confirm the above findings that both POLN mutants exhibit increased fidelity. To determine the types and positions of errors made by POLN and POLN derivatives, the sequence of the 407 template bases within the gap was determined for sets of independent lacZ mutants (Table 2). The single base changes were distributed throughout the target sequence (Supplementary Figure S2). As previously noted (2), the predominant error observed was misincorporation of dTMP opposite G, resulting in G to A transitions (Supplementary Table S1). Error rates were calculated from these data (Table 2). There was a 10- and 5-fold decrease in G-dTMP error rate for K679T and K679A, respectively compared to WT POLN and the observed mutations were in similar hotspots [Table 1 and ref. (2)].
Table 2.
Summary of error rates for POLN and POLN derivatives
| WTa | K679Ta | K679Aa | ||||
|---|---|---|---|---|---|---|
| LacZ mutant frequency | 0.12 | 0.023 | 0.034 | |||
| Total mutants sequenced | 47 | 45 | 44 | |||
| Total bases sequenced | 19 129 | 18 315 | 17 908 | |||
| Detectable changesb | No. of events | Error rate (×10−4) | No. of events | Error rate (×10−4) | No. of events | Error Rate (×10−4) |
| Base substitutions | 54 | 18.0 | 28 | 1.9 | 43 | 4.4 |
| Frameshifts (−1) | 12 | 2.5 | 8 | 0.3 | 3 | 0.2 |
| Frameshifts (+1) | 2 | 1.3 | 5 | 0.2 | 1 | 0.1 |
| G:dTMP | 46 | 86.0 | 20 | 8.0 | 34 | 20.0 |
aFor WT, the total number of plaques scored was 6793, among which 789 were scored as mutants. For K679T the total number of plaques scored was 3380, among which 78 were scored as mutants. For K679A, the total number of plaques scored was 2552, among which 87 were scored as mutants.
bError rates were calculated for phenotypically detectable changes, as described in ‘Materials and Methods’ section.
K679 and Y682 are critical for bypass activity in POLN
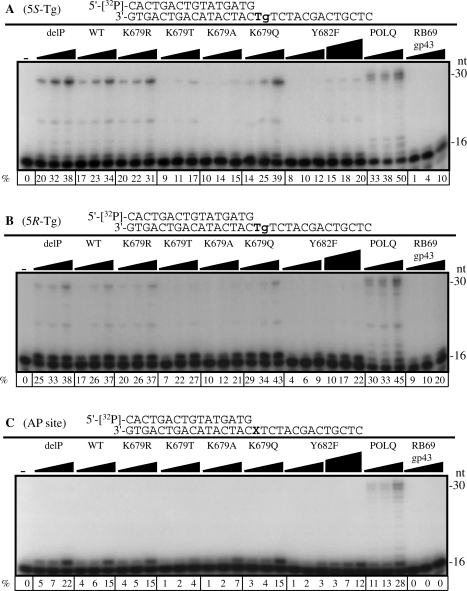
We examined whether the POLN mutants with increased fidelity had an altered ability to bypass single sites of DNA template damage. In comparison to WT POLN, the K679T, K679A and Y682F mutants showed reduced bypass of both a 5S-Tg (Figure 5A) and a 5R-Tg (Figure 5B) which is more difficult to bypass (1). The efficiency of A incorporation opposite 5S-Tg was calculated from kcat/Km (Table 1). Relative efficiencies of bypass (compared to utilization of dATP opposite an undamaged T) of K679T and K679A were reduced to about half compared with delP, WT, K679R and Y682F POLN. The relative bypass efficiency of Y682F was similar to WT, although its absolute frequency of A incorporation opposite 5S-Tg (kcat/Km) was lowest among all the derivatives, because of the pronounced effect of the Y682F mutation on overall DNA polymerase activity.
Figure 5.
Translesion synthesis activities of POLN derivatives. Increasing amounts of delP, WT, K679R, K679T, K679A and K679Q (6, 12 and 23 nM), Y682F (6, 12, 23, 29, 58 and 115 nM), POLQ (3, 6 and 12 nM) and RB69 gp43 (2.5, 5 and 10 pM) were incubated with the 5′-32P-labeled primer templates indicated, in the presence of all four dNTPs for 10 min. The first lane contained no enzyme. The first base templates were 5S-Tg (A), 5R-Tg (B) or AP analog (C). The percentage (%) extension of the primer is shown below each lane.
Only POLQ could fully bypass an AP site (Figure 5C). POLN inserted a nucleotide opposite an AP-site but could not extend further and this insertion was reduced in the K679T and K679A derivatives (Figure 5C).
For successful translesion DNA synthesis, a primer-template must be extended after a nucleotide is inserted opposite a lesion in DNA. The K679T, K679A and Y682F mutants extended less efficiently from A:5S-Tg than did the wild-type group of enzymes (Supplementary Figure S3A). No POLN derivative could efficiently extend a C, G or T terminus opposite a 5S-Tg (Supplementary Figure S3B–D). Thus, K679 is important in efficient A insertion opposite a 5S-Tg, and both K679 and Y682 are essential for efficient extension after A incorporation opposite the lesion. We also tested whether K679 and Y682 of POLN are important for the extension from a T:G mismatch. Unlike the extension from A:5S-Tg, K679T, K679A and Y682F extended from a T:G mismatch as efficiently as wild-type POLN (Supplementary Figure S4).
To further quantify the abilities of POLN and the low fidelity mutant derivatives to bypass a 5S-Tg or an undamaged T in the same sequence context, primers 1 or 2 bases shorter than in Figure 4 were used so that bypass probabilities, insertion probabilities, and extension probabilities could be calculated (Supplementary Figure S5C and D). Each of these probabilities was reduced in K679T and K679A for 5S-Tg. Insertion and extension probabilities were reduced to about half in K679T and K679A, in agreement with the steady-state kinetics measurements (Table 1) and the result observed in Supplementary Figure S3A. RB69 gp43 completely stopped at the lesion, as in Figure 5 and Supplementary Figure S3A.
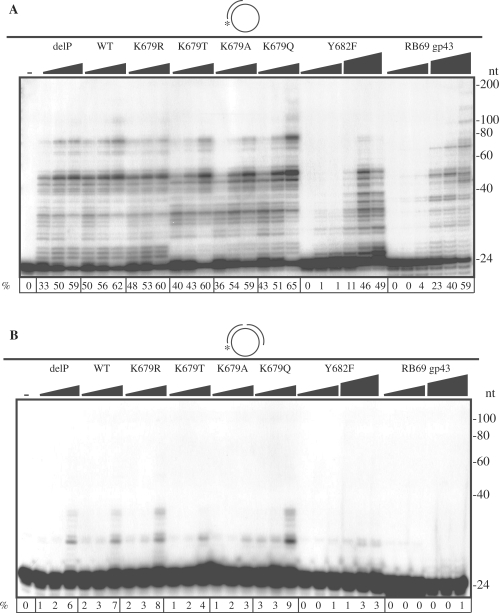
Processivity and strand displacement activities in mutant POLN
Moderate processivity was previously noted for POLN. We found that all POLN derivatives could elongate DNA chains up to ∼100 nt (1), except Y682F which elongated up to ∼50 nt and showed pronounced stalling at the first template base (Figure 6A). The first four template bases of the substrate were G, and much more stalling at these positions occurred with wild-type enzymes than with K679A and K679T. This is probably because the wild-type enzymes incorporated T opposite G more often than the mutant derivatives, and extension from a T:G mismatch is less efficient that from a matched C:G terminus.
Figure 6.
Mutation of Lys 679 to Ala or Thr reduces strand displacement. Increasing amounts of delP, WT, K679R, K679T, K679A and K679Q (12, 23 and 46 nM), and Y682F (12, 23, 46, 58, 115 and 230 nM) and RB69 gp43 (5, 10, 20, 103, 205 and 410 pM) were incubated at 37°C for 15 min with 100 fmol primer annealed to M13mp18GTGx (A) or nicked substrate, 100 fmol primer and downstream oligomer were annealed to M13mp18GTGx (B). The substrates are schematically diagrammed and were radiolabeled at the 5′-end of the upstream 24-mer primer (asterisk). The 5′-end of the downstream 60-mer oligonucleotide was phosphorylated, and its 3′-end was blocked with ddATP. The first lane contained no enzyme. The percentage (%) extension of the primer is shown below each lane.
Human POLN has a strong strand displacement activity (1). Strand displacement of 10 bp or more was observed with the wild type enzymes, and was still present but reduced to about half with the K679T, K679A and Y682F mutant derivatives (Figure 6B). Control RB69 gp43 did not show strand displacement activity, as expected (Figure 6B).
DISCUSSION
Conservation of a low fidelity residue in vertebrate POLN
The most unusual and striking property of POLN is its very high G to A base substitution rate, in comparison to other A-family DNA polymerases. It is important to understand why POLN has this biochemical property. In this study, we focused on the residues of conserved motif 4, which comprise the ‘O-helix’ in the polymerase structure. A main finding is that a residue in this domain previously identified as critical for fidelity of A-family DNA polymerases was uniformly conserved as a low fidelity ‘mutant’ residue in POLN. We demonstrated that this residue contributes significantly to the error-rate by affecting both fidelity of insertion and extension. The strong conservation of the residue 679 position as Lys or Arg and its effect on fidelity and bypass strongly indicates that it is critical for the specialized cellular function of POLN.
Structural information is not currently available for wild-type and mutant POLN complexed with DNA, and so we can only speculate briefly on possible mechanisms. In the pol I family, residues of the O-helix of the fingers subdomain interacts with the incoming dNTP and changes its conformation to facilitate base pairing. In the structures of prokaryotic pol I the residue corresponding to K678 directly interacts with the incoming dNTP. In Taq pol, alteration of the following residue from Thr to Arg (Thr664, corresponding to POLN 679) was proposed to stabilize the closed conformation of the polymerase, even without the correct pairing (16). This could occur by mediating a subtle conformational change in the O-helix (18), or perhaps by direct interaction of K679 to stabilize base pairs formed in the active site. The most frequent change by POLN is a G to A transition, while a T to C transition is the most frequently observed event with T664R Taq pol (18). Both changes involve G:T base mismatches, indicating that a basic residue at this position in the O-helix can stabilize the polymerase active site with such a mismatch.
A residue critical for ddNTP selectivity also modulates fidelity
A second major finding in this study is that the aromatic Tyr residue present at position 682 of POLN is important not only for ddNTP selectivity, but also for fidelity, translesion synthesis activity, processivity and overall DNA polymerase activity. The importance of this residue for ddNTP selectivity was expected because the absence of a hydroxyl group on Phe is known to better exclude ddNTP from the active site of A-family enzymes (20). However, the presence of Tyr at this position in POLN gives higher overall DNA polymerase activity. A Y682F mutation in POLN decreased DNA polymerase activity, increasing Km(dCTP) and Km(dATP) about 5- and 3-fold, respectively compared with wild-type POLN. This is comparable to an inverse experiment with E. coli Kf (exo−), where a F762Y mutation increased DNA polymerase activity compared to wild-type Kf (exo−), attributed to a 5-fold decrease in Km(dTTP) (21). It is notable that low fidelity A-family enzymes including POLN and POLQ only have Tyr at the corresponding position, suggesting that it is functionally conserved to maintain the properties of these enzymes. Single amino acid substitutions in K679 and Y682 of POLN influenced the fidelity 10- and 5-fold respectively. In prokaryotic pol I, the residue corresponding to K679 influences the fidelity 14- to 25-fold, but the influence of the residue corresponding to Y682 is not well-characterized.
A basic residue at position 679 aids in bypass and strand displacement
Thymine glycol (Tg) presents a strong block to both repair and replicative DNA polymerases. Replicative DNA polymerases generally can insert A opposite Tg but cannot extend further. The crystal structure of a binary complex of the replicative RB69 gp43 with DNA shows that extension past the A:Tg pair, rather than insertion opposite the Tg, constitutes the block to the replicative polymerases (22). In contrast, POLN both efficiently inserts the correct nucleotide A opposite 5S-Tg and extend. In POLN, K679 is important for both steps: efficient insertion of A opposite a 5S-Tg, and extension after the insertion. This supports the notion that the presence of a basic residue at position 679 stabilizes a closed conformation even with non-standard base pairing. Bypass of 5S-Tg seems particularly important biologically as the lesion is removed less efficiently than 5R-Tg in mammalian cells (23). The 5S-Tg bypass reaction by POLN is accurate and is possibly used in certain cell types as part of defense against reactive oxygen attack.
Strand displacement activities were also reduced in K679T, K679A, and Y682F mutants, but these activities were still stronger than RB69 gp43. Interactions of the O-helix with a strand-displacing primer terminus thus also appears to have some importance in the mechanism of strand displacement activity. Strand displacement in E. coli pol I is proposed to also involve the three residues S769, F771 and R841 in the fingers subdomain (24). Substitutions of S769, F771 and R841 of Kf to Ala reduced strand displacement activity. The corresponding residues of POLN are G689, E691 and R761. These are conserved in POLN orthologs (Supplementary Figure S1) and Gly and Glu are unique in POLN. The residues may be important for the especially effective strand displacement activity of POLN. The biological function of POLN is currently under investigation, with contrasting roles suggested in translesion synthesis past an unhooked interstrand crosslink (25), or in homologous recombination (26). Efficient strand displacement could be important for either of these functions, or for additional ones.
Basis for differences in properties of POLN and POLQ
Substitution of Lys 679 in POLN for Gln, the residue normally found in POLQ, did not confer key properties of POLQ on the mutant POLN; for example, no bypass or extension of an AP site occurred in this mutant. This study demonstrates that some of the unique characteristics of human POLN are highly dependent on a K679 residue in the fingers subdomain. Enzymes with basic Lys or Arg sidechains at residue 679 had similar properties, quite different from neutral Thr, Ala or Gln sidechains.
Nucleotide selectivities of K679Q were also different from that of POLQ (Figure 4). Although the corresponding Q2384 may be important for the fidelity of POLQ, additional mechanisms are necessary to explain the unique activities and high +1 frameshift error rate of POLQ. The three insertions in the DNA polymerase domain of POLQ are suggested to be important in this respect (8). Among them, insertion 1 of POLQ is located at the tip of the thumb domain, an area important for DNA binding and frameshift fidelity (27,28). Insertion 1 is 22 amino acids long and contains seven positively charged residues, which possibly contact with duplex DNA. This insertion may be important for AP-site bypass and the unique fidelity observed in POLQ.
POLN also has three conserved insertions in the DNA polymerase domain, with marked differences from those in POLQ (Figure 1A and Supplementary Figure S1). Insertions 1 and 2 are in the same positions as those in POLQ, but are shorter. Insert 3 is in a different position of the palm subdomain (which forms a substrate-binding pocket for incoming dNTP) in the two enzymes. These results indicate that the K679 of POLN is not the only residue determining its unique properties, but also that insertions 1–3 and some other conserved amino acid differences also influence the unique properties of POLN.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (CA101980 to R.D.W.); the University of Pittsburgh Cancer Institute; and the Grady F. Saunders, Ph.D. Endowed Professorship; Division of Intramural Research of the National Institutes of Health Project (Z01 ES065070 to T.A.K. and M.A.) in part; National Institute of Environmental Health Sciences. Funding for open access charge: MD Anderson Endowed Professorship.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Karen Zima and Anthony Schuffert for assistance with the fidelity measurements, and members of our laboratories for discussion and comments. We thank Tatsuhiko Shimizu and Shigenori Iwai (Osaka University, Japan) for providing the oligonucleotide containing a 5S-Tg. The NIEHS DNA Sequencing and Molecular Genetics Cores provided expert assistance in the DNA sequence analysis of lacZ mutants. We appreciate insightful comments on the article from Katarzyna Bebenek, Alan B. Clark, Sabine S. Lange, Matthew Yousefzadeh and Ella Bedford.
REFERENCES
- 1.Takata K, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low-fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J. Biol. Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 2.Arana ME, Takata K, Garcia-Diaz M, Wood RD, Kunkel TA. A unique error signature for human DNA polymerase ν. DNA Repair. 2007;6:213–223. doi: 10.1016/j.dnarep.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seki M, Marini F, Wood RD. POLQ (Pol θ), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36:3847–3856. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shima N, Munroe RJ, Schimenti JC. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol. Cell Biol. 2004;24:10381–10389. doi: 10.1128/MCB.24.23.10381-10389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goff JP, Shields DS, Seki M, Choi S, Epperly MW, Dixon T, Wang H, Bakkenist CJ, Dertinger SD, Torous DK, et al. Lack of DNA polymerase theta (POLQ) radiosensitizes bone marrow stromal cells in vitro and increases reticulocyte micronuclei after total-body irradiation. Radiat. Res. 2009;172:165–174. doi: 10.1667/RR1598.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marini F, Kim N, Schuffert A, Wood RD. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 2003;278:32014–32019. doi: 10.1074/jbc.M305646200. [DOI] [PubMed] [Google Scholar]
- 8.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang G, Franklin M, Li J, Lin TC, Konigsberg W. Correlation of the kinetics of finger domain mutants in RB69 DNA polymerase with its structure. Biochemistry. 2002;41:2526–2534. doi: 10.1021/bi0119924. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu T, Manabe K, Yoshikawa S, Kawasaki Y, Iwai S. Preferential formation of (5S,6R)-thymine glycol for oligodeoxyribonucleotide synthesis and analysis of drug binding to thymine glycol-containing DNA. Nucleic Acids Res. 2006;34:313–321. doi: 10.1093/nar/gkj443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moggs JG, Yarema KJ, Essigmann JM, Wood RD. Analysis of incision sites produced by human cell extracts and purified proteins during nucleotide excision repair of a 1,3-intrastrand d(GpTpG)-cisplatin adduct. J. Biol. Chem. 1996;271:7177–7186. doi: 10.1074/jbc.271.12.7177. [DOI] [PubMed] [Google Scholar]
- 12.Creighton S, Goodman MF. Gel kinetic analysis of DNA polymerase fidelity in the presence of proofreading using bacteriophage T4 DNA polymerase. J. Biol. Chem. 1995;270:4759–4774. doi: 10.1074/jbc.270.9.4759. [DOI] [PubMed] [Google Scholar]
- 13.Bebenek K, Kunkel TA. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 14.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 15.McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 16.Patel PH, Suzuki M, Adman E, Shinkai A, Loeb LA. Prokaryotic DNA polymerase I: evolution, structure, and “base flipping” mechanism for nucleotide selection. J. Mol. Biol. 2001;308:823–837. doi: 10.1006/jmbi.2001.4619. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki M, Baskin D, Hood L, Loeb LA. Random mutagenesis of Thermus aquaticus DNA polymerase I: concordance of immutable sites in vivo with the crystal structure. Proc. Natl Acad. Sci. USA. 1996;93:9670–9675. doi: 10.1073/pnas.93.18.9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki M, Avicola AK, Hood L, Loeb LA. Low fidelity mutants in the O-helix of Thermus aquaticus DNA polymerase I. J. Biol. Chem. 1997;272:11228–11235. doi: 10.1074/jbc.272.17.11228. [DOI] [PubMed] [Google Scholar]
- 19.Camps M, Naukkarinen J, Johnson BP, Loeb LA. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I. Proc. Natl Acad. Sci. USA. 2003;100:9727–9732. doi: 10.1073/pnas.1333928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tabor S, Richardson CC. A single residue in DNA polymerases of the Escherichia coli DNA polymerase I family is critical for distinguishing between deoxy- and dideoxyribonucleotides. Proc. Natl Acad. Sci. USA. 1995;92:6339–6343. doi: 10.1073/pnas.92.14.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Astatke M, Ng K, Grindley ND, Joyce CM. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc. Natl Acad. Sci. USA. 1998;95:3402–3407. doi: 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aller P, Rould MA, Hogg M, Wallace SS, Doublie S. A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc. Natl Acad. Sci. USA. 2007;104:814–818. doi: 10.1073/pnas.0606648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katafuchi A, Nakano T, Masaoka A, Terato H, Iwai S, Hanaoka F, Ide H. Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. J. Biol. Chem. 2004;279:14464–14471. doi: 10.1074/jbc.M400393200. [DOI] [PubMed] [Google Scholar]
- 24.Singh K, Srivastava A, Patel SS, Modak MJ. Participation of the fingers subdomain of Escherichia coli DNA polymerase I in the strand displacement synthesis of DNA. J. Biol. Chem. 2007;282:10594–10604. doi: 10.1074/jbc.M611242200. [DOI] [PubMed] [Google Scholar]
- 25.Zietlow L, Smith LA, Bessho M, Bessho T. Evidence for the involvement of human DNA polymerase N in the repair of DNA interstrand cross-links. Biochemistry. 2009;48:11817–11824. doi: 10.1021/bi9015346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moldovan GL, Madhavan MV, Mirchandani KD, McCaffrey RM, Vinciguerra P, D'A;ndrea AD. DNA polymerase POLN participates in crosslink repair and homologous recombination. Mol. Cell. Biol. 2010;30:1088–1096. doi: 10.1128/MCB.01124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson JF, Fox R, Harris DD, Lyons-Abbott S, Loeb LA. Insertion of the T3 DNA polymerase thioredoxin binding domain enhances the processivity and fidelity of Taq DNA polymerase. Nucleic Acids Res. 2003;31:4702–4709. doi: 10.1093/nar/gkg667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minnick DT, Astatke M, Joyce CM, Kunkel TA. A thumb subdomain mutant of the large fragment of Escherichia coli DNA polymerase I with reduced DNA binding affinity, processivity, and frameshift fidelity. J. Biol. Chem. 1996;271:24954–24961. doi: 10.1074/jbc.271.40.24954. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.