Abstract
Human THAP1 is the prototype of a large family of cellular factors sharing an original THAP zinc-finger motif responsible for DNA binding. Human THAP1 regulates endothelial cell proliferation and G1/S cell-cycle progression, through modulation of pRb/E2F cell-cycle target genes including rrm1. Recently, mutations in THAP1 have been found to cause DYT6 primary torsion dystonia, a human neurological disease. We report here the first 3D structure of the complex formed by the DNA-binding domain of THAP1 and its specific DNA target (THABS) found within the rrm1 target gene. The THAP zinc finger uses its double-stranded β-sheet to fill the DNA major groove and provides a unique combination of contacts from the β-sheet, the N-terminal tail and surrounding loops toward the five invariant base pairs of the THABS sequence. Our studies reveal unprecedented insights into the specific DNA recognition mechanisms within this large family of proteins controlling cell proliferation, cell cycle and pluripotency.
INTRODUCTION
Gene expression is tightly modulated by the interplay of sequence-specific transcription factors that recruit direct transcription effectors in vivo. In this context, the thermodynamic, structural and kinetic strategies adopted by a DNA-binding protein to locate and bind to its specific DNA target among a huge excess of non-specific DNA in the cell are of considerable interest and are still under investigation (1–3). During the last decade, structural studies performed on a number of DNA-binding domains bound to their DNA target provided some molecular details about specific DNA recognition (4–6). Understanding the molecular mechanisms of DNA specific recognition also requires the issue of binding to non-specific DNA target to be tackled, as recently reported for the dimeric lac repressor, highlighting the importance of structural flexibility and plasticity in DNA recognition (2).
The THanatos-Associated protein (THAP) DNA-binding domain is an evolutionary conserved C2CH zinc-finger motif shared between a large family of cellular factors with functions associated to cell-proliferation and cell-cycle control (7,8). Human THAP1, the prototype member of the family, is described as a novel transcription factor involved in endothelial cell proliferation and G1/S cell-cycle control, regulating expression of several pRb/E2F cell-cycle target genes (9). The DNA-binding domain of THAP1 recognizes a consensus DNA target of 11 nt (THABS) comprising a core of five invariant base pairs 5′TxxxGGCA3′ (7). An unexpected finding was recently reported concerning the DNA-binding function of THAP1 associated with DYT6 primary torsion dystonia, a neurological disease characterized by twisting movements and abnormal postures (10). It was proposed that transcriptional dysregulation associated with mutations in the DNA-binding domain of THAP1 might contribute to the DYT6 disease (10,11). We have previously reported the solution structure of the DNA-binding module (THAP zinc finger) of THAP1 by NMR showing that the core fold consists of an anti-parallel two-stranded β-sheet with the two strands separated by a long loop-helix-loop motif (12). Using NMR and mutagenesis data, we provided the first structure-activity analysis of a functional DNA-binding THAP domain with demonstrated sequence-specific DNA-binding activity. Furthermore, we have shown that recombinant THAP domains from human THAP2 and THAP3 and from Caenorhabditis elegans CTBP and GON-14 do not exhibit sequence-specific DNA binding toward the THABS sequence recognized by THAP1, suggesting that although the different THAP zinc fingers share some structural homologies, they may recognize their own specific DNA sequence (12). This hypothesis was confirmed with the recent identification of Ronin, the mouse ortholog of THAP11 that underlies embryogenesis and Embryonic Stem cell pluripotency (13). Ronin exhibits DNA-binding activity toward a DNA sequence that is clearly distinct from the THABS consensus motif recognized by THAP1 (13).
In an attempt to get some clues regarding the molecular mechanisms by which the THAP zinc finger recognizes a specific DNA sequence, we determined the solution structure of the complex between the THAP zinc finger of THAP1 and a 16-bp oligonucleotide containing the THABS sequence identified in the natural rrm1 responsive element. The latter is a G1-S regulated gene coding for the Ribonucleotide Reductase M1 subunit essential for S-phase DNA synthesis, that was recently identified as the first direct transcriptional target of endogenous THAP1 (9). The rrm1 promoter contains two THABS-binding sites approximately 100-nt upstream of the 5′-end of the mRNA to which endogenous THAP1 binds in vivo (9).
By solving the first structure of a functional THAP protein–DNA complex, we show in the present article that the THAP zinc finger of THAP1 contacts the DNA major groove using its two-stranded β-sheet. The association relies on numerous non-specific contacts to the sugar phosphate backbone, allowing efficient positioning of the protein onto the DNA before setting up base-specific contacts. The DNA recognition specificity resides in a combination of crucial contacts provided by poorly conserved residues among the THAP members, that are located in the β-sheet, the N-terminal tail and surrounding loops and that cover the five invariant base pairs of the consensus THABS sequence. To increase the DNA-binding specificity, a loop in the C-terminal region of the THAP zinc finger gives additional contacts to the DNA minor groove. We also report structural and fluorescence studies on the binding of the THAP zinc finger of THAP1 to non-specific DNA. Our work provides new insights into the structural determinants controlling the DNA recognition specificity within this large family of cellular factors with major roles in cell proliferation, cell-cycle control and pluripotency.
MATERIALS AND METHODS
Sample preparation
The plasmid coding for the THAP zinc-finger domain of hTHAP1 (Met1-Phe81) with a double mutation C62SC67S was generated by PCR. The expression and purification protocols have been described previously (12). The 16-bp rrm1 DNA duplex was reconstituted by hybridizing oligonucleotides, 5′GCTTGTGTGGGCAGCG3′ and 5′CGCTGCCCACACAAGC3′ (Eurofins MWG) in a 1:1 ratio. The DNA–protein complex (∼1 mM) was formed by mixing either unlabeled protein or uniformly 15N- or 15N13C labeled protein with unlabeled duplex rrm1 DNA under high-salt conditions (50 mM Tris, pH 6.8, 250 mM NaCl, 5 mM DTT). The DNA duplex with an unrelated sequence was reconstituted by hybridizing oligonucleotides, 5′CGATTTGAATTTTAAC3′ and 5′GTTAAAATTCAAATCG3′, and mixed with the THAP zinc finger following the same protocol. All protein–DNA samples were exchanged against 50 mM Tris (pH 6.8), 30 mM NaCl, 5 mM DTT, 0.01% sodium azide and 10% or 100% 2H2O before NMR experiments.
NMR spectroscopy
NMR experiments were performed at 296K on cryo-probed Bruker DRX950 and DRX600 spectrometers. Protein (1H, 15N and 13C) backbone and side-chain resonances were assigned from analysis of standard 3D experiments (14). Distance restraints were extracted from 3D 15N HSQC NOESY (Tm 100 ms), 3D 13Cali HSQC-NOESY (Tm 80 ms) and 3D 13Caro HSQC-NOESY (Tm 120 ms) recorded at 950 MHz. DNA 1H resonances were assigned for the free rrm1 oligonucleotide using a combination of 2D TOCSY and NOESY recorded in 2H2O and H2O. DNA assignments in the protein–DNA complex were obtained from TOCSY and NOESY spectra recorded on the unlabeled sample at 950 MHz. Intermolecular protein–DNA NOEs were assigned from 15N- and 13C-edited NOESY spectra. Protein backbone ϕ and Ψ angle constraints were predicted with TALOS software using chemical shift assignments (15). Slow exchanging amide protons were identified from 2D 1H-15N HSQC spectra collected following resuspension of freeze-dried protein–DNA samples in 2H2O.
A number of 1DNH RDCs were collected at 600 MHz with the uniformly 15N13C-labeled protein in DNA-bound state oriented in Pf1 bacteriophage medium (15 mg/ml) from 2D IPAP 1H -15N HSQC (16). The data were processed using the NMRPipe suite (17). The magnitude of the axial and rhombic components of the alignment tensor was determined with the Module 1.0 software (18). Heteronuclear 15N relaxation parameters (T1, T2, NOE) were recorded at 600 MHz using standard pulse sequences on the protein–DNA sample and analyzed with NMRView (19). The overall and internal mobility parameters were determined using the Tensorv2.0 software (20).
Cross-saturation experiments were performed on the DNA–protein complex. Saturation of the DNA imino proton resonances was achieved by means of a pulse train of adiabatic inversion pulses centered at 13 ppm. This cross-saturation transfer period was introduced prior to the classical 1H-15N HSQC sequence as previously described (21). The 2D 1H-15N HSQC experiments were recorded with different saturation periods up to 1.8 s. The peak intensities were extracted from the 2D 1H-15N HSQC spectra using NMRView (19) and analyzed using GOSA (22).
Structure calculation
Structures of the rrm1-bound protein were calculated using torsion angle dynamics simulated annealing protocol using the CNSv1.21 software suite (23). From 500 structures, 20 were selected as acceptable with no NOE violations higher than 0.4 Å and no dihedral angle violations higher than 5°. The protein was then docked to rrm1 B-DNA using HADDOCK 2.0 (24). The docking protocol consists of three stages, rigid-body docking, semi-flexible simulated annealing and refinement in explicit solvent, as already described for protein–DNA docking (25). An ensemble of 20 protein NMR structures together with models of canonical B-DNA were used as starting structures in the rigid-body docking with intermolecular NOEs as docking restraints, generating 1000 models. 200 lowest-energy structures were selected for semi-flexible refinement stage with all NMR experimental restraints including the 39 intermolecular NOEs and the intramolecular restraints (for the protein: hbonds, dihedral angles, RDCs and NOEs and for the DNA: hbonds, B-form canonical dihedral angle restraints, planarity restraints and NOEs). Residues displaying high solvent accessibility, that were affected in the cross-saturation experiments and that showed large chemical shift changes upon DNA binding and for which no intermolecular NOE could be identified were defined as active (Gln3, Lys24, Lys46, Ser52, Arg65). The protein side chains of the active residues were allowed to move in a semi-flexible simulated annealing stage (25). The DNA bases encompassing the five invariant base pairs (from T6 to A13 and T20 to A27) were defined as active and 12 ambiguous interaction restraints between suitable atoms of protein and DNA were used in the calculation. Intra-residual DNA NOEs quantitative analysis allowed us to define C2′-endo conformation for all of the assigned riboses (26) and inter-residual DNA NOEs analysis could unambiguously confirm Watson–Crick base pairings. Additional restraints were introduced to maintain DNA base planarity and Watson–Crick bonds. During the first calculation, DNA was considered as fully flexible during the semi flexible simulated annealing stage. The structures were further refined in an explicit solvent with all NMR experimental restraints. Then, an ensemble of 10 DNA structures issued from the first calculation were analyzed and selected as initial pre-bent DNA structures for a final complete run with all NMR experimental data and in which only DNA base pairs located at the protein DNA interface were allowed to move in a semi-flexible simulated annealing stage. Finally, solution analysis was performed using HADDOCK2.0 package scripts and best structures were selected on the basis of lower unambiguous restraints violations. Intermolecular contacts analysis was performed using HADDOCK2.0 package scripts with an upper hydrogen bond cut-off at 2.5 Å. Finally, geometrical analysis was done using PROCHECK software.
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays were performed as previously described (7), using a 16-bp rrm1 oligonucleotide (∼7.6 µM) and increasing amount (1, 2.5 and 5 µM) of the recombinant THAP zinc finger of THAP1 containing the double mutation (C62SC67S). Binding reactions were performed for 10 min at room temperature in 20 µl of binding buffer [20 mM Tris-HCl (pH 7.5)/100 mM KCl/0.1% Nonidet P-40/100 µg/ml BSA/2.5 mM DTT and 5% glycerol].
Fluorescence measurements
Steady-state fluorescence anisotropy binding titrations were performed on a PTI Model QM-4 spectrofluorimeter at 25°C following the intrinsic fluorescence of the single tryptophan residue (λexc 295 nm and λem 324 nm). To measure the affinity of the protein toward rrm1, the THAP zinc finger was diluted to 0.5 µM in a volume of 4 ml and the 16-bp rrm1 DNA duplex (100 µM) was prepared in a buffer consisting of 50 mM Tris, 30 mM NaCl, pH 6.8. The rrm1 solution was progressively added to the protein sample with protein:DNA ratios ranging from 1:0 to 1:6. To study the influence of the ionic strength on the non-specific binding, samples with different protein:DNA ratios ranging from 1:0 to 1:6 were initially prepared in 250 mM NaCl (100 µl of THAP zinc finger at 3 µM) and were then exchanged in buffer containing suitable NaCl concentrations (30 or 150 mM). Fluorescence anisotropy was calculated including a correction factor as previously described (27) and the data were fitted from a previously described equation (28) using a non-linear fit with GOSA software (22).
RESULTS
Monitoring DNA binding by NMR and fluorescence anisotropy
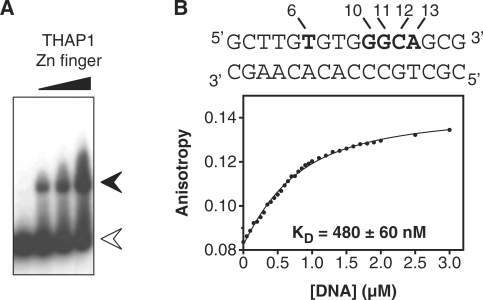
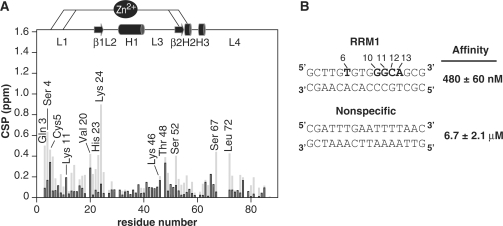
In a previous work, we solved the NMR structure of the THAP zinc finger of human THAP1 (residues 1–81) for which demonstrated sequence-specific THABS DNA-binding activity was known (12). But, initial attempts failed to produce a stable DNA–protein complex with limited conformational exchange. In order to improve the quality of the NMR spectra, we constructed two Cys-Ser mutations at positions 62 and 67. The doubly mutated THAP domain is a stable folded protein as judged by the quality and chemical shift dispersion of the 1H-15N HSQC spectrum, that is highly similar to the one recorded for the wild type THAP domain, showing that the two mutations do not induce major structural changes. A 16-bp oligonucleotide containing the THABS motif identified in the natural rrm1 responsive element (referred to rrm1) was chosen for further structural and biophysical characterisation of the specific DNA–protein complex (20 kDa). The THAP mutant retains its rrm1-binding activity as shown by electrophoresis mobility shift assay (Figure 1A); a dissociation constant of 480 ± 60 nM was determined by fluorescence anisotropy (Figure 1B).
Figure 1.
The rrm1-DNA binding of the THAP zinc finger of hTHAP1 observed by EMSA and fluorescence anisotropy. (A) EMSA experiments were performed using a 16-bp oligonucleotide found in the rrm1 DNA target and increasing amounts (1, 2.5 and 5 µM) of the recombinant THAP zinc finger of THAP1. White arrow, free rrm1 and black arrow, rrm1-THAP zinc-finger complex (B) top, sequence of the 16-bp rrm1 oligonucleotide. Invariant bases of the THABS core motif are highlighted in bold bottom, plot showing the fluorescence anisotropy of the single tryptophan residue (Trp36) as a function of increased DNA concentrations. The protein concentration was 0.5 µM and the DNA concentration ranged from 0 to 3 µM.
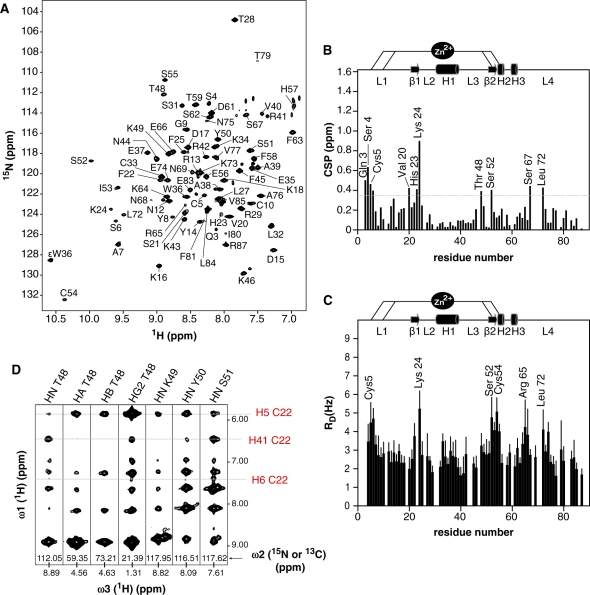
The quality of the NMR spectra allowed us to unambiguously identify residues that exhibit chemical shift changes of their backbone amide nitrogen resonances upon rrm1 DNA binding (Figure 2A and B). The regions showing important chemical shift perturbation (CSP) in the complex (Δδ >Δδaverage +SD ∼0.35 ppm) include the N-terminal tail close to the zinc ion (Gln3-Ser6), the double-stranded β-sheet (residues Val20 to Lys24 and residue Ser52) and the loop L3 encompassing Thr48 (Figure 2B). Additional strong CSP were observed for two residues Ser67 and Leu72 located in loop L4. The DNA–protein interface was further defined by means of cross-saturation experiments. Upon saturation of DNA imino proton resonances, large reduction rates of peak intensities were observed for residues Cys5-Ser6, Lys24, Ser52-Ser55, Arg65 and Leu72 (Figure 2C). Finally, solvent exchange experiments were performed on the rrm1-protein complex to identify protected residues upon DNA binding. In particular, the amide protons of Thr48, Tyr50 and Ser51 remain protected from hydrogen exchange after several hours while they exchange in less than an hour in the free protein (data not shown).
Figure 2.
The rrm1-DNA binding of the THAP zinc finger of hTHAP1 observed by NMR. (A) The 1H-15N HSQC spectrum of the THAP zinc finger in the presence of an equimolar amount of the 16-bp rrm1 DNA. (B) Histogram of chemical shift changes upon DNA binding as a function of the residue number. Reported chemical shift changes represent combined 15N and 1H chemical shift changes as (Δδ = [(ΔδHN)2 +(ΔδN × 0.154)2]½). Secondary structure elements are shown above the panel. (C) Plot showing reduction rates of peak intensities as function of residue number upon saturation of the DNA imino proton resonances (cross-saturation experiments). The peak intensities were extracted from 2D 1H-15N HSQC spectra recorded with different saturation periods up to 1.8 s. (D) Strip views extracted from 3D 15N and 13C NOESY spectra showing selected intermolecular NOEs observed between the doubly 15N, 13C-labeled THAP zinc finger of hTHAP1 and unlabeled rrm1-DNA.
Structure determination of the complex
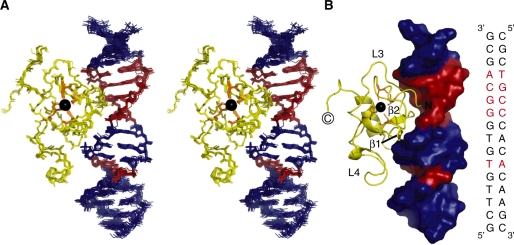
NMR spectra collected at 950 MHz allowed us to assign most of the protein and DNA resonances in the complex and to identify 39 intermolecular NOEs involving nine residues of the THAP zinc finger and seven bases of the rrm1 DNA duplex (Figure 2D and Supplementary Table 1), that were sufficient to unambiguously determine the protein orientation with respect to the DNA (Figure 3). The solution structure of the complex was determined using the data-driven biomolecular docking HADDOCK approach (29) including NMR restraints. Structure calculations for the THAP zinc finger in the DNA-bound state were performed by simulated annealing on the basis of experimental restraints including 1796 NOEs, 12 hydrogen bonds, 156 dihedral angles and 55 1DNH residual dipolar couplings (RDC) (Table 1). The 20 lowest-energy NMR structures of the THAP zinc-finger domain in its DNA-bound form were used as initial structures for the HADDOCK calculations of the DNA–protein complex. Most of the bound DNA resonance frequencies were unambiguously assigned except for bases G10-G11 and 679 DNA intramolecular NOEs were identified, unambiguously establishing that rrm1 adopts a B-DNA conformation in the complex, with standard base pairings. The structural ensemble presented a root mean square (r.m.s.) deviation of 1.22 ± 0.32 Å over all backbone atoms of both protein and DNA (Table 1). 15N relaxation analysis gave a correlation time of 5.6 ± 0.1 ns and 10.3 ± 0.1 ns, for the free and bound protein respectively, consistent with a monomeric form in both states (Supplementary Figure S1).
Figure 3.
The NMR solution structure of the complex between the THAP zinc finger of hTHAP1 and its specific rrm1 DNA target. (A) Stereo-view of the NMR ensemble for the 15 lowest energy structures. Colour code: protein, yellow and DNA, blue and red. The five invariant base pairs of the DNA THABS core motif are coloured red. The zinc atom is shown in black and the four ligands are depicted in orange. (B) Ribbon diagram of the protein bound to the DNA molecular surface with the same orientation as in (A). For clarity, some secondary structure elements are indicated on the protein ribbon. The sequence of the 16-bp rrm1 oligonucleotide is shown in the right panel. The five invariant base pairs of the DNA THABS core motif are highlighted in red.
Table 1.
Structural statistics of the THAP zinc finger of hTHAP1 in complex with its specific rrm1 target
| Protein | Nucleic acid | |
|---|---|---|
| NMR distance and dihedral constraints | ||
| Distance restraints | ||
| Total NOE | 1796 | 679 |
| Intra-residue | 0 | 517 |
| Inter-residue | 1796 | 152 |
| Sequential (|i – j| = 1) | 851 | 123 |
| Non-sequential (|i – j| > 1) | 945 | 29 |
| Hydrogen bonds | 12 | 43a |
| Zinc coordination | 14 | |
| Protein–nucleic acid intermolecular | 39 | |
| Ambiguous intermolecular restraints | 12 | |
| RDC 1DHN-N | 55 | |
| Total dihedral angle restraints | 156 | 256 |
| Protein | ||
| ϕ | 78 | |
| Ψ | 78 | |
| Nucleic acid (B-DNA conformation) | ||
| Sugar puckerb | 96 | |
| Backboneb | 160 | |
| Planarity restraints | 16 | |
| Structure statistics | ||
| Violations (mean ± SD) | ||
| Distance constraint violations >0.25Å | 1.20 ± 0.74 | |
| Dihedral angle constraints violation >5° | 0.14 ± 0.34 | |
| Max. dihedral angle violation (°) | 5.71 ± 0.62c | |
| Max. distance constraint violation (Å) | 0.28 ± 0.05 | |
| Deviations from idealized geometry | ||
| Bond lengths (Å) | 0.0046 | |
| Bond angles (°) | 0.75 | |
| Impropers (°) | 0.79 | |
| Average pairwise r.m.s. deviation (Å)d | ||
| Protein | ||
| Heavy | 0.88 ± 0.30 | |
| Backbone | 0.48 ± 0.16 | |
| DNA | ||
| DNA heavy atoms | 0.66 ± 0.25 | |
| DNA heavy atoms at the binding interfacee | 0.41 ± 0.11 | |
| Complex | ||
| Heavy atoms (C, N, O, P) | 1.22 ± 0.32 | |
| Non-terminal heavy atomsf | 0.86 ± 0.25 | |
aThe DNA intramolecular hydrogen bonds were deduced from base pairings.
bBased on a B-form geometry derived from NOE analysis, where α = −63 ± 15°, β = 176 ± 15°, γ = 51 ± 15°, ε = 171 ± 15°, ζ = −103 ± 15° and ν1 (C1′-C2′-C3′-C4′) = 37.5 ± 5°, ν2 (C5′-C4′-C3′-C2′) = –155 ± 5° and ν3 (C5′-C4′-C3′-C2′) = 144 ± 5°.
cDouble violation of a Ψ angle of residue Glu83 locate in the highly dynamic C-terminal tail of the protein.
dPairwise r.m.s. deviation was calculated among 15 refined structures.
eRegion T6–A13 and T20–A27 encompassing the invariant base pairs of the THABS motif.
fExcluding protein residues Leu82–Arg87 and terminal DNA base pairs (positions 1–4 and 15–16).
The complex structure reveals a DNA-binding interface using the double-stranded β-sheet
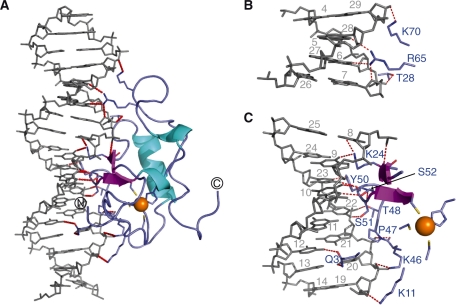
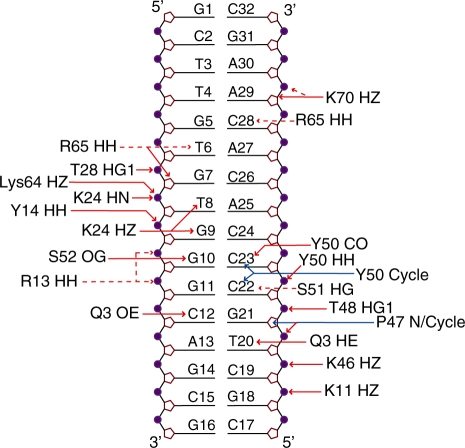
The THAP zinc-finger contacts the rrm1 DNA by filling the major groove with its side containing the double-stranded β-sheet giving rise to a buried area of 2120 Å2. The two strands insert into the major groove with an orientation perpendicular to the DNA axis (Figure 3A and B). The N-terminal tail and loop L3 that connects the α-helix to the β2 strand contribute to the DNA-binding surface in the major groove (Figure 3). In particular, the double-stranded β-sheet contacts two backbone phosphates at positions T8 and G9 and three bases T8, G9 and G10 in the coding strand. Two residues, Lys24 and Ser52 from the β-sheet mediate base-specific contacts (Figure 4). Bidentate hydrogen bonds are formed between the side-chain amino group of Lys24 and both atoms O6 of G9 and O4 of T8 while the side-chain HG proton of Ser52 contacts N7 of the invariant base G10. Loop L3 preceding the β2 strand is also involved in DNA recognition as residues Lys46 to Ser51 provide several contacts with the complementary half of the DNA duplex, either by contacting backbone phosphates at position 20–23 or by giving base-specific contacts with C22 and C23 or by maintaining Van der Waals contacts with the major groove (Figure 4). The protein backbone at Pro47 gives polar contacts with G21 phosphate and the side-chain amino terminal group of Lys46 points toward the phosphate group of T20 while Thr48 and Tyr50 contact phosphate groups of C22 and C23, respectively (Figure 5). The carboxyl group of Tyr50 interacts with the two DNA strands simultaneously as it could give polar contacts to the O6 of G10 in the coding strand and to the N4 of C23 in the complementary strand. In addition, the aromatic side chain of Tyr50 makes extensive hydrophobic contacts with bases and sugar rings of C22 and C23. Finally, the OG atom of Ser51 participates in hydrogen bonding with the amino group of C22. In the vicinity of Ser51, the N-terminal tail of the protein participates in interactions with both DNA strands within the major groove. In particular, Gln3 uses its carboxyl side chain to contact the N4 of C12 in the coding strand while its side chain amino group can be hydrogen bonded simultaneously with the O4 of T20 (Figure 5).
Figure 4.
DNA recognition by the THAP zinc finger (A) Ribbon representation indicating the polar contacts (red dotted lines) observed at the DNA–protein interface. DNA is shown in grey and the protein is coloured blue with the exception of the β-strands and the helices, which are coloured magenta and cyan, respectively. The zinc ion is depicted in orange. The four zinc ligands together with the amino acid side chains at the DNA–protein interface are shown as sticks. (B) Close-up view of the minor groove interface with the same orientation as in (A). (C) Close-up view of the major groove interface with the same orientation as in (A).
Figure 5.
Schematic representation illustrating the protein–DNA contacts in the structure of the rrm1-THAP zinc-finger complex. Red and blue arrows indicate hydrogen bonds and hydrophobic contacts, respectively. A hydrogen bond is considered to occur when potential donor and acceptor are <2.5 Å apart. Dotted red arrows indicate polar contacts observed between potential donors and acceptors that do not satisfy the hydrogen bond distance criteria. The plot was generated by NucPlot (46). The contacts represented here are summarized in Supplementary Tables S2 and S3.
In addition to the contacts observed toward the DNA major groove, the structure of the complex reveals few additional contacts to bases within the minor groove, which are achieved by loop L4 from the C-terminus of the THAP domain. In particular, polar contacts are made between the guanidine group of Arg65 and both the O2 of C28 and the atoms O4 of T6. Simultaneously, the O4′ ribose atom of G7 could be hydrogen bonded to the guanidine group of Arg65 (Figure 4).
Structural and dynamic modifications upon binding
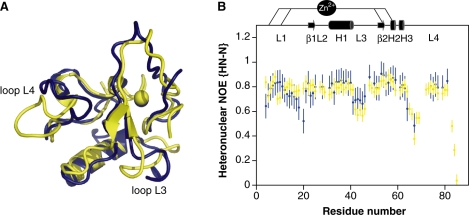
The protein in the complex adopts a βαβ fold consisting of a double-stranded anti-parallel β-sheet with a long loop-helix-loop motif (L2-H1-L3) inserted between the two strands. Despite a similar topology to the one previously described for the THAP zinc finger in its DNA-free form (12), binding to specific DNA is accompanied by remarkable structural changes (Figure 6A). The greatest change occurs in loop L4 from residues Arg65 to Leu72 in order to allow contacts to the DNA minor groove. The loop displacement pulls Asn68 away from the DNA by 15 Å while Arg65 is pushed toward the DNA by almost 6 Å. The flip is accompanied by large ps-ns timescale motions, observed for residues Arg65 to Lys71, allowed to pivot around two rigid residues Phe63 and Leu72 (Figure 6B). The C-terminal region of loop L3 preceding the second β-strand undergoes a displacement of residues Thr48 to Ser51 of 6-7 Å, providing favourable contacts with the DNA complementary strand. This part of the loop is not disordered as it displays restricted mobility (Figure 6B) and as several NOEs were identified between residues Thr48, Lys49 and the methyl group of Ile53 (data not shown). Residues 42–46 (beginning of loop L3) and residues 66–69 (beginning of loop L4) that exhibit mobility in the free protein remain mobile in the complex, as seen from heteronuclear NOE values (Figure 6B). In contrast, residues 16–21 (end of loop L1) are immobilized upon DNA binding, presumably via electrostatic interactions between the DNA phosphates and the side chains of Lys11, Arg13 and Tyr14 that might anchor the entire loop L1 to the DNA. Notably, the amide proton of Lys18 is hydrogen bonded to the carboxyl group of Asp15 and remains protected from hydrogen exchange (data not shown), contributing to the reduced mobility of this part of loop L1.
Figure 6.
Structural and dynamic modifications of the THAP zinc finger of hTHAP1 upon DNA binding. (A) Expanded view showing superposition of the THAP zinc finger in its free state (blue) (Protein Data Bank entry 2jtg) (12), and in the presence of its specific DNA target (yellow). Loops L3 and L4, which undergo structural changes upon DNA binding, are indicated. (B) Histogram showing the heteronuclear NOE values for the protein in the absence (12) (blue) and in the presence of DNA (yellow) as a function of residue number. The secondary structure elements are depicted on the top.
From the DNA point of view, the binding does not change the overall conformation of the rrm1 target, which remains that of a standard B-form as confirmed by NOE analysis (see Materials and methods section). However, a moderate degree of bending (15°) starting at the G9/C24 base pair and a slight enlargement of 3 Å for the major groove width at the G10/C23 base pair are observed (data not shown).
Recognition specificity
The structure of the complex shows that most of the DNA–protein contacts cover the bases from the invariant base T6 on one strand to the last invariant base T20 on the DNA complementary strand (Figure 5). The side chains of two residues Lys24 and Ser52 from the double-stranded β-sheet, donate base-specific contacts to the DNA major groove. Lys24 is relatively well conserved and mostly replaced by an arginine in other THAP proteins. It contacts the two bases T8 and G9 that do not contribute to the specificity of the THABS sequence (7). The structure of the complex explains why a guanine in position 9 can be substituted by a thymine (7) since they both have a carboxyl group in the major groove as an acceptor of hydrogen bonds from the amino side-chain group of Lys24. In the present work, binding experiments combining NMR and fluorescence anisotropy were performed in the presence of a 16-bp oligonucleotide containing an unrelated sequence (non-specific DNA, Figure 7A and B). The Lys24 HN chemical shift is clearly not affected in the presence of non-specific DNA while it displays the largest chemical shift change upon rrm1 binding (Figure 7A). This is presumably due to the loss in base-specific contacts, as the two bases T8 and G9 contacted by Lys24 in the rrm1-THAP complex are replaced by adenines in the non-specific sequence (Figure 7B). A single-point mutant K24A retains its capacity to bind to non-specific DNA, as monitored by fluorescence anisotropy (data not shown) whereas it abrogates specific DNA-binding activity (12). Similarly, the chemical shift perturbation of Ser52 HN proton within the β2 strand is clearly reduced upon addition of non-specific DNA compared to its chemical shift change in the presence of rrm1. But in contrast to Lys24, Ser 52 is poorly conserved among the THAP family proteins, and it creates a hydrogen bond with G10 inside the GGCA core recognition motif. Therefore, Ser52 in the β2-strand must play a crucial role in specific DNA recognition. Just preceding the β-sheet, two poorly conserved residues, namely Tyr50 and Ser51 from loop L3 provide additional base-specific contacts to two bases (C22-C23) at positions 1 and 2 inside the GGCA recognition site, helping to increase specificity (Figure 5). Finally, Gln3 in the N-terminal tail of the DNA-binding domain gives hydrogen bonds to two invariant bases at positions 3 (C12) and 4 (T20) simultaneously (Figure 5). Given that Gln3 is poorly conserved among the THAP members, these two contacts are likely to affect DNA-binding specificity. Remarkably, its neighbouring amino acid Ser4, which is also poorly conserved displays notable changes in amide resonance chemical shift in the specific complex while it is only slightly disturbed by addition of the non-specific DNA, confirming the importance of the N-terminal tail in specific DNA recognition. In the opposite direction, loop L4 points toward the minor groove contacting the invariant base T6 inside the recognition 5′TxxxGGCA3′ motif, using the guanidine group of Arg65, another poorly conserved residue that is likely to play a crucial role in specificity.
Figure 7.
Comparison of specific and non-specific recognition (A) Backbone amide nitrogen chemical shift perturbation upon addition of rrm1 specific (grey) and non-specific (black) DNA sequences. (B) Specific rrm1 and non-specific DNA sequences together with the respective binding affinity values are indicated. Measurements were performed at 30 mM NaCl.
Importance of non-specific interactions on the overall affinity
At 30 mM NaCl, the protein binds to non-specific DNA with a significantly lower affinity compared to specific DNA (dissociation constant values of 6.7 ± 2 µM versus 480 ± 60 nM, (Figure 5B). In the presence of non-specific DNA, only slight chemical shift perturbations (Δδ <0.4 (ppm) were observed for a small number of residues. Affected backbone amide nitrogen resonances (Δδ > Δδaverage + SD ∼0.15 ppm) correspond to Cys5 (from the N-terminus), Lys11 and Val20 (loop L1), Lys46 and Thr48 (loop L3) and Arg65 (loop L4) (Figure 7A). Our data show that the regions affected in the presence of non-specific DNA are similar to those described in the rrm1-THAP zinc-finger complex, suggesting that the DNA orientation relative to the protein should not be much different. A number of non-specific contacts between the protein and DNA phosphate groups were identified in the structure of the rrm1-THAP zinc-finger complex (see above). In particular, residues Lys46 and Thr48 from loop L3 that point toward DNA phosphate groups in the specific complex are affected in the non-specific complex, consistent with the idea that they contribute to positioning the protein onto the DNA. As the salt concentration increases, the affinity of the protein toward non-specific DNA decreases (Supplementary Figure S2). At 250 mM NaCl, the dissociation constant is 33.5 ± 5 µM and the 2D 1H-15N HSQC spectrum of the protein in the presence of non-specific DNA looks similar to the one recorded in the absence of DNA (Supplementary Figure S2).
DISCUSSION
We solved the first 3D structure of a THAP zinc finger bound to its DNA target and compared the binding characteristics to specific and non-specific DNA sequences, in terms of binding affinities and protein positioning. On its rrm1 specific target, the protein contacts the DNA major groove by presenting its double stranded β-sheet as secondary structure element with the amino terminus tail and loop L3 contributing significantly to form the molecular interface. We previously demonstrated the originality of the THAP zinc finger characterized by particular features such as a βαβ topology and the long loop-helix-loop (L2-H1-L3) motif inserted into the atypical spacing between the two pairs of zinc ligands (12). The structure of the complex reveals an important role for the two-stranded β-sheet while evidencing that the helix H1 is not the primary structural element used to recognize DNA. From this finding, the THAP zinc finger clearly differs from classical zinc-finger motifs that mainly use residues in α-helices to specifically contact the DNA bases. Among the vast number of DNA-binding proteins, few have been shown to contact DNA using a β-sheet (30). In the case of the prokaryotic MetJ-Arc repressor (31,32), a double stranded β-sheet, formed upon homo-dimerization of the protein, is used to recognize the major groove. In the lambda integrase protein (33,34) and the plant GCC box-binding protein (35), DNA recognition is mediated by a triple stranded β-sheet that anchors into the major groove by providing contacts with the DNA sugar-phosphate backbone. Larger β-sheets can also play a central role in DNA recognition, mostly by inducing intricate recognition mechanisms associated with DNA bending, as previously described for the Tata-Binding Protein (36) and for the Integration Host Factor (37). However, very few examples of zinc fingers using a β-sheet as secondary structure element to recognize DNA have been described so far. The crystal structure of the zinc-coordinating GCM domain, bound to its octameric DNA target revealed the involvement of a five-stranded beta-sheet and three surrounding helices to contact the DNA major groove (38). Contrary to the proposed classification for the CtBP-THAP domain to belong to the treble clef finger superfamily (39), the DNA-binding mode by the THAP-zinc finger of THAP1 differs from the one described for the treble clef motif in which the α-helix is engaged in the DNA major groove while a β-strand interacts with the sugar phosphate backbone of the DNA (40). In the case of the THAP-zinc finger, the double-stranded β-sheet fills the DNA major groove with remarkably good complementarity and in a specific-sequence manner; however, it is only a piece of the binding interface, as other regions of the domain contribute to DNA base-pair contacts. To cope with the relatively small size of its double-stranded β-sheet, the THAP-zinc finger has increased the number of contacts to DNA by using its N-terminal tail and additional loops.
Recognition of the rrm1 sequence resides in a number of specific side chain interactions with the five invariant base pairs (T6/A27, G10/C23, G11/C22, C12/G21 and A13/T20) of the THABS motif and a number of non-specific contacts with the sugar-phosphate DNA backbone. Four amino acids located within the β-sheet (Lys24, Ser52), the N-terminal tail (Gln3) and loop L4 (Arg65) confer specific DNA recognition. Two additional residues Tyr50 and Ser51 from loop L3 preceding the β-sheet also contact two invariant bases of the motif. Interestingly, the combination of these six residues is only found in the THAP1 protein and may explain the recognition specificity toward the THABS motif.
Our data show that the N-terminal tail of the domain contributes to binding specificity and could explain why most of the THAP domains are located at the N-terminal position of the THAP family (8). Another interesting feature involves loop L4 and in particular the side chain of Arg65 that provides specific contacts to T6 and C28 bases in the minor groove, stabilizing DNA interaction as previously observed in a number of protein–DNA complexes (41). Notably, loop L4 is poorly conserved among the THAP domains and insertions or deletions in this loop are notable in the sequences among the family of THAP proteins. For example, loop L4 is not present in the recently identified THAP domain of the Ronin protein, which binds a DNA sequence clearly different from the THABS consensus sequence recognized by THAP1 (13).
Loop L3 located between helix H1 and the β2 strand is critical for both specific and non-specific DNA recognition. We show that residue Thr48 plays a crucial role in DNA binding and that it contributes to positioning the protein onto the DNA duplex allowing further specific side chain contacts to occur. This would allow post-translational modification such as site-specific phosphorylation of its hydroxyl group, to efficiently regulate DNA interaction, as previously observed for other transcription factors (42).
We find that the THAP zinc finger binds DNA as a monomer with a relatively low affinity as previously observed for isolated domains such as the lac repressor (6). In vivo, the recognition might require dimerization of the THAP zinc finger in order to enhance binding affinity and specificity. It is noteworthy that the rrm1 DNA sequence used in the present study corresponds to the first THABS-binding site, while two THABS-binding sequences are located approximately 100-nt upstream of the 5′-end of the mRNA to which endogenous THAP1 associates in vivo (9). By solving the structure of the complex, we show that the helix, which contains several highly conserved residues, is not directly involved in DNA recognition and is instead exposed. It could mediate homodimerization with another THAP domain bound to the second THABS-binding sequence within the rrm1 gene or it might be involved in the formation of protein–protein complexes. Furthermore, it should be kept in mind that the full-length THAP proteins, beyond their DNA-binding domain, exhibit other functional regions such as the coiled coil domains frequently involved in protein–protein interactions. The Ronin protein (mTHAP11) interacts with host cell factor-1 HCF-1, a key transcriptional regulator associated to chromatin remodelling (13). Two other THAP members involved in complexes associated to chromatin modification were previously identified, namely THAP7 and HIM17 (43–45). Overall, these studies suggest that the THAP proteins could play a major role in targeting genes to promote transcription regulation through interactions with protein complexes associated to chromatin remodelling.
In this regard, the data presented here provide the first 3D structure of a protein–DNA complex within the THAP-zinc-finger family and give unique clues to understanding the structural determinants of specific DNA recognition by this previously uncharacterized family of transcription factors.
While our manuscript was in the reviewing process, the crystal structure of the THAP domain from the D. melanogaster P-element transposase (dmTHAP) in complex with a naturally occuring 10-bp DNA site has been published [Sabogal et al., Nat. Struct. Mol. Biol. (2010) 17, 117–123; accession code 3KDE]. This structure shows that the THAP domain binds to DNA in a bipartite manner using both the DNA major and minor grooves. The DNA sequence-specific recognition is achieved by the insertion of the dmTHAP central β-sheet into the major groove while the basic loop L4 provides contacts with the DNA minor groove. Our NMR study also reveals this bipartite recognition mechanism. Both studies performed on two distinct THAP domains and DNA targets and using different approaches (NMR versus X-ray cristallography) are consistent and complementary and provide clues to understand the mechanism of specific DNA recognition by the THAP proteins.
ACCESSION NUMBERS
The 1H, 13C and 15N chemical shifts, NMR restraints and coordinates have been deposited in the BioMagResBank (BMRB) and Protein Data Bank (PDB) with the accession codes 16485 and 2ko0, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French Research Ministry; Centre National de la Recherche Scientifique; Université Paul Sabatier; Région Midi-Pyrénées and European structural funds. Extended access to the EU-NMR facility in Frankfurt (6th Framework Program of the EC [contract number RII3-026145]) is duly acknowledged. Financial support from the TGE RMN THC Fr3050 for conducting the research is gratefully acknowledged. Funding for open access charge: CNRS and Université Paul Sabatier.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to J.P. Girard for initiating the project and for critically reading the manuscript. They thank their collaborators at the IPBS, L. Mourey and V. Guillet for useful discussions and S. Mazere for technical assistance with fluorescence measurements. They acknowledge their colleagues, P. Demange, I. Muller and J. Czaplicki for help with biochemistry, fluorescence and data analysis.
REFERENCES
- 1.Jen-Jacobson L, Engler LE, Jacobson LA. Structural and thermodynamic strategies for site-specific DNA binding proteins. Structure. 2000;8:1015–1023. doi: 10.1016/s0969-2126(00)00501-3. [DOI] [PubMed] [Google Scholar]
- 2.Kalodimos CG, Biris N, Bonvin AM, Levandoski MM, Guennuegues M, Boelens R, Kaptein R. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 3.Iwahara J, Zweckstetter M, Clore GM. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc. Natl Acad. Sci. USA. 2006;103:15062–15067. doi: 10.1073/pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schildbach JF, Karzai AW, Raumann BE, Sauer RT. Origins of DNA-binding specificity: role of protein contacts with the DNA backbone. Proc. Natl Acad. Sci. USA. 1999;96:811–817. doi: 10.1073/pnas.96.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalodimos CG, Boelens R, Kaptein R. Toward an integrated model of protein-DNA recognition as inferred from NMR studies on the Lac repressor system. Chem. Rev. 2004;104:3567–3586. doi: 10.1021/cr0304065. [DOI] [PubMed] [Google Scholar]
- 7.Clouaire T, Roussigne M, Ecochard V, Mathe C, Amalric F, Girard JP. The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc. Natl Acad. Sci. USA. 2005;102:6907–6912. doi: 10.1073/pnas.0406882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roussigne M, Kossida S, Lavigne AC, Clouaire T, Ecochard V, Glories A, Amalric F, Girard JP. The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem. Sci. 2003;28:66–69. doi: 10.1016/S0968-0004(02)00013-0. [DOI] [PubMed] [Google Scholar]
- 9.Cayrol C, Lacroix C, Mathe C, Ecochard V, Ceribelli M, Loreau E, Lazar V, Dessen P, Mantovani R, Aguilar L, et al. The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood. 2007;109:584–594. doi: 10.1182/blood-2006-03-012013. [DOI] [PubMed] [Google Scholar]
- 10.Djarmati A, Schneider SA, Lohmann K, Winkler S, Pawlack H, Hagenah J, Bruggemann N, Zittel S, Fuchs T, Rakovic A, et al. Mutations in THAP1 (DYT6) and generalised dystonia with prominent spasmodic dysphonia: a genetic screening study. Lancet Neurol. 2009;8:447–452. doi: 10.1016/S1474-4422(09)70083-3. [DOI] [PubMed] [Google Scholar]
- 11.Bressman SB, Raymond D, Fuchs T, Heiman GA, Ozelius LJ, Saunders-Pullman R. Mutations in THAP1 (DYT6) in early-onset dystonia: a genetic screening study. Lancet Neurol. 2009;8:441–446. doi: 10.1016/S1474-4422(09)70081-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bessiere D, Lacroix C, Campagne S, Ecochard V, Guillet V, Mourey L, Lopez F, Czaplicki J, Demange P, Milon A, et al. Structure-function analysis of the THAP zinc finger of THAP1, a large C2CH DNA-binding module linked to Rb/E2F pathways. J. Biol. Chem. 2008;283:4352–4363. doi: 10.1074/jbc.M707537200. [DOI] [PubMed] [Google Scholar]
- 13.Dejosez M, Krumenacker JS, Zitur LJ, Passeri M, Chu LF, Songyang Z, Thomson JA, Zwaka TP. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133:1162–1174. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sattler M, Schleucher J, Griedinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Progr. Nucl. Magn. Reson. Spectr. 1999;34:93–158. [Google Scholar]
- 15.Cornilescu G, delaglio F, Bax A. TALOS: torsion angle likelihood obtained from shift and sequence similarity. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 16.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 17.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 18.Dosset P, Hus JC, Marion D, Blackledge M. A novel interactive tool for rigid-body modeling of multi-domain macromolecules using residual dipolar couplings. J. Biomol. NMR. 2001;20:223–231. doi: 10.1023/a:1011206132740. [DOI] [PubMed] [Google Scholar]
- 19.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 20.Dosset P, Hus JC, Blackledge M, Marion D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J. Biomol. NMR. 2000;16:23–28. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- 21.Ramos A, Kelly G, Hollingworth D, Pastore A, Frenkiel T. Mapping the Interfaces of Protein−Nucleic Acid Complexes Using Cross-Saturation. JACS. 2000;122:11311–11314. [Google Scholar]
- 22.Czaplicki J, Cornélissen G, Halberg F. GOSA, a simulated annealing-based program for global optimization of nonlinear problems, also reveals transyears. J. Appl. Biomed. 2006;4:87–94. [PMC free article] [PubMed] [Google Scholar]
- 23.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination [In Process Citation] Acta Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 24.de Vries SJ, van Dijk AD, Krzeminski M, van Dijk M, Thureau A, Hsu V, Wassenaar T, Bonvin AM. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 25.van Dijk M, van Dijk AD, Hsu V, Boelens R, Bonvin AM. Information-driven protein-DNA docking using HADDOCK: it is a matter of flexibility. Nucleic Acids Res. 2006;34:3317–3325. doi: 10.1093/nar/gkl412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuniasse P, Sowers LC, Eritja R, Kaplan B, Goodman MF, Cognet JA, Le Bret M, Guschlbauer W, Fazakerley GV. Abasic frameshift in DNA. Solution conformation determined by proton NMR and molecular mechanics calculations. Biochemistry. 1989;28:2018–2026. doi: 10.1021/bi00431a009. [DOI] [PubMed] [Google Scholar]
- 27.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd edn. New York: Springer; 2006. [Google Scholar]
- 28.Lundblad JR, Laurance M, Goodman RH. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol. Endocrinol. 1996;10:607–612. doi: 10.1210/mend.10.6.8776720. [DOI] [PubMed] [Google Scholar]
- 29.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 30.Tateno M, Yamasaki K, Amano N, Kakinuma J, Koike H, Allen MD, Suzuki M. DNA recognition by beta-sheets. Biopolymers. 1997;44:335–359. doi: 10.1002/(SICI)1097-0282(1997)44:4<335::AID-BIP3>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 31.Somers WS, Phillips SE. Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands. Nature. 1992;359:387–393. doi: 10.1038/359387a0. [DOI] [PubMed] [Google Scholar]
- 32.Raumann BE, Rould MA, Pabo CO, Sauer RT. DNA recognition by beta-sheets in the Arc repressor-operator crystal structure. Nature. 1994;367:754–757. doi: 10.1038/367754a0. [DOI] [PubMed] [Google Scholar]
- 33.Fadeev EA, Sam MD, Clubb RT. NMR structure of the amino-terminal domain of the lambda integrase protein in complex with DNA: immobilization of a flexible tail facilitates beta-sheet recognition of the major groove. J. Mol. Biol. 2009;388:682–690. doi: 10.1016/j.jmb.2009.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wojciak JM, Connolly KM, Clubb RT. NMR structure of the Tn916 integrase-DNA complex. Nat. Struct. Biol. 1999;6:366–373. doi: 10.1038/7603. [DOI] [PubMed] [Google Scholar]
- 35.Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M. A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998;17:5484–5496. doi: 10.1093/emboj/17.18.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 37.Lynch TW, Read EK, Mattis AN, Gardner JF, Rice PA. Integration host factor: putting a twist on protein-DNA recognition. J. Mol. Biol. 2003;330:493–502. doi: 10.1016/s0022-2836(03)00529-1. [DOI] [PubMed] [Google Scholar]
- 38.Cohen SX, Moulin M, Hashemolhosseini S, Kilian K, Wegner M, Muller CW. Structure of the GCM domain-DNA complex: a DNA-binding domain with a novel fold and mode of target site recognition. EMBO J. 2003;22:1835–1845. doi: 10.1093/emboj/cdg182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liew CK, Crossley M, Mackay JP, Nicholas HR. Solution Structure of the THAP Domain from Caenorhabditis elegans C-terminal Binding Protein (CtBP) J. Mol. Biol. 2007;366:382–390. doi: 10.1016/j.jmb.2006.11.058. [DOI] [PubMed] [Google Scholar]
- 40.Grishin NV. Treble clef finger–a functionally diverse zinc-binding structural motif. Nucleic Acids Res. 2001;29:1703–1714. doi: 10.1093/nar/29.8.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jean A, Gutierrez-Hartmann A, Duval DL. A Pit-1 Threonine 220 phosphomimic reduces binding to monomeric DNA sites to inhibit Ras and estrogen stimulation of the prolactin gene promoter. Mol. Endocrinol. 2009;24:91–103. doi: 10.1210/me.2009-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miele A, Medina R, van Wijnen AJ, Stein GS, Stein JL. The interactome of the histone gene regulatory factor HiNF-P suggests novel cell cycle related roles in transcriptional control and RNA processing. J. Cell Biochem. 2007;102:136–148. doi: 10.1002/jcb.21284. [DOI] [PubMed] [Google Scholar]
- 44.Macfarlan T, Kutney S, Altman B, Montross R, Yu J, Chakravarti D. Human THAP7 is a chromatin-associated, histone tail-binding protein that represses transcription via recruitment of HDAC3 and nuclear hormone receptor corepressor. J. Biol. Chem. 2005;280:7346–7358. doi: 10.1074/jbc.M411675200. [DOI] [PubMed] [Google Scholar]
- 45.Reddy KC, Villeneuve AM. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell. 2004;118:439–452. doi: 10.1016/j.cell.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 46.Luscombe NM, Laskowski RA, Thornton JM. NUCPLOT: a program to generate schematic diagrams of protein-nucleic acid interactions. Nucleic Acids Res. 1997;25:4940–4945. doi: 10.1093/nar/25.24.4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.