Summary
The 9.5-megadalton human pyruvate dehydrogenase complex (PDC) utilizes the specific dihydrolipoamide dehydrogenase (E3)-binding protein (E3BP) to tether the essential E3 component to the 60-meric core of the complex. Here, we report crystal structures of the binding domain (E3BD) of human E3BP alone and in complex with human E3 at 1.6 Å and 2.2 Å, respectively. The latter structure shows that residues from E3BD contact E3 across its 2-fold axis, resulting in one E3BD-binding site on the E3 homodimer. Negligible conformational changes occur in E3BD upon its high-affinity binding to E3. Modifications of E3BD residues at the center of the E3BD/E3 interface impede E3 binding far more severely than those of residues on the periphery, validating the “hot spot” paradigm for protein interactions. A cluster of disease-causing E3 mutations located near the center of the E3BD/E3 interface prevents the efficient recruitment of these E3 variants by E3BP into the PDC, leading to the dysfunction of the PDC catalytic machine.
Introduction
The mammalian mitochondrial pyruvate dehydrogenase complex (PDC) catalyzes the oxidative decarboxylation of pyruvate to give rise to acetyl-CoA, NADH and CO2 (Reaction 1).
| (1) |
FAD and lipoyl moieties are used as a coenzyme and a prosthetic group, respectively, by PDC. The overall reaction links glycolysis to the energy-producing Krebs cycle and lipogenic pathways. PDC is a member of the highly conserved mitochondrial α-ketoacid dehydrogenase complexes, which comprise the PDC, the branched-chain α-ketoacid dehydrogenase complex (BCKDC), and the α-ketoglutarate dehydrogenase complex (KGDC) (Patel and Roche, 1990; Reed, 2001; Reed et al., 1985). The mammalian PDC is a 9.5 × 106-dalton nuclear-encoded macromolecular machine organized around an icosahedral 60-meric transacetylase (E2p) core. To the E2p core, 30 copies of heterotetrameric pyruvate dehydrogenase (E1p), 12 copies of homodimeric dihydrolipoamide dehydrogenase (E3) and 12 copies of monomeric E3-binding protein (E3BP) are attached via noncovalent protein-protein interactions (Reed, 2001). Each E2p subunit consists of two lipoyl-bearing domains (LBD’s), one subunit-binding domain (SBDp) that binds E1p, and one inner-core domain that contains the E2p active site and is responsible for the self-assembly of the core. Of the above PDC components, E1p, E2p, and E3BP are specific for PDC, whereas E3 is common to all α-ketoacid dehydrogenase complexes. The overall reaction of PDC (Reaction 1) is achieved by the substrate-channeled summation of the individual reactions of the three catalytic components. In addition, unspecified copies of pyruvate dehydrogenase kinase and pyruvate dehydrogenase phosphatase, which are associated primarily with the second lipoyl-bearing domain of the E2p core, regulate PDC activity by reversible phosphorylation (Reed et al., 1985; Roche et al., 2003).
Because of its central importance in intermediary metabolism, mutations in the genes coding for subunits of human PDC produce severe clinical phenotypes (Robinson, 2001). Congenital defects in the gene for human E1p result in neonatal lactic acidemias and encephalopathies (Robinson, 2001). Mutations in the gene encoding human E3BP have also been reported (Aral et al., 1997; Dey et al., 2002; Dey et al., 2003; Ling et al., 1998). These mutations invariably result in the absence of the E3BP protein. The null phenotype for E3BP leads to congenital PDC deficiency as characterized by the above clinical manifestations (Robinson, 2001). Errors in the gene coding human E3 cause combined deficiencies in α-ketoacid dehydrogenase complexes manifested by lactic acidemias and Maple Syrup Urine Disease (Chuang and Shih, 2001; Hengeveld and de Kok, 2002; Odievre et al., 2005). A subset of the human E3 mutations has been suggested to occur at the homodimer interface (Hong et al., 1996; Hong et al., 1997; Shany et al., 1999) or at the putative E3/E3BP (Odievre et al., 2005) interaction surface. The clinical consequences resulting from these mutations are similar to those caused by the E3BP-null mutations. Additionally, the aberrant down-regulation of PDC activity by reversible phosphorylation contributes to hyperglycemic states observed in type-2 diabetes (Holness and Sugden, 2003), raising the prospect that the PDC represents a therapeutic target for a disease that afflicts over 150 million people worldwide (Wild et al., 2004).
The fundamental metabolic and clinical importance of PDC has prompted the examination of its macromolecular structure. Three-dimensional reconstructions of yeast PDC derived from cryo-electron microscopy show that 12 copies of E3BP are buried inside the 12 pentagonal openings of the truncated E2p scaffold, with each of the 12 E3 homodimers bound to a single E3BP monomer (Reed, 2001; Stoops et al., 1997). In contrast, a recent study has suggested that the inner-core domain of bovine E3BP, which has a significantly longer C-terminal extension than its yeast counterpart, is integrated into the 60-meric core of the bovine PDC with an E2p : E3BP = 48 : 12 subunit stoichiometry (Hiromasa et al., 2004). The structural basis for the fundamental differences in the organization of E3BP between yeast and mammalian PDC is unknown.
Only a few high-resolution studies of the structural details of the vital inter-subunit interactions of PDC are available. The structure of the SBDp of E2p from the Geobacillus stearothermophilus (formerly Bacillus stearothermophilus) PDC bound to its cognate E3 shows that a single SBDp binds near to the twofold axis of the E3 homodimer (Mande et al., 1995). This mode of binding sterically excludes the binding of a second SBDp. In this Geobacillus SBDp/E3 complex, the interaction between the two proteins consists largely of electrostatic interactions between one α-helix of the SBDp and surface residues of E3. SBDp is also responsible for securing the E1p to the Geobacillus PDC, and E1p as well as E3 bind to SBDp in a mutually exclusive mode (Reed, 2001; Perham, 2000). A recent SBDp/E1p complex structure of Geobacillus PDC demonstrates that the electrostatic nature of the SBDp/E3 interface is conserved, but much less surface area is buried at the SBP/E1p interface than that of the SBDp/E3 (Frank et al., 2005).
Unlike Geobacillus stearothermophilus and other prokaryotes, eukaryotes require a separate protein, E3BP, to tether E3 to the PDC. E3BP was previously designated “protein X” because its biological function was unknown when it was first discovered (De Marcucci and Lindsay, 1985; Jilka et al., 1986). The identity and the E3-tethering function of E3BP was established by limited proteolysis (Gopalakrishnan et al., 1989; Powers-Greenwood et al., 1989) and later confirmed by disruption and mutagenesis of the gene encoding yeast E3BP (Lawson et al., 1991). The domain structure of human E3BP is similar to that of human E2p (Aral et al., 1997; Harris et al., 1997; Ling et al., 1998); at the N-terminus is a single LBD, followed by the E3-binding domain (E3BD) and the C-terminal domain. The C-terminal domain of E3BP shows about 50% sequence identity to its counterpart in human E2p. The covalently attached lipoyl moiety in E3BP (Neagle et al., 1989; Powers-Greenwood et al., 1989), like those of E2p, is competent to serve as a coenzyme in the PDC reaction (Hodgson et al., 1986; Jilka et al., 1986; Rahmatullah and Roche, 1987). The inner-core domain of E2p harbors a histidine residue that is essential for its transacetylase activity. This residue is not conserved in E3BP, and this protein therefore does not possess transacetylase activity.
In this study, we present crystal structures of human E3BD alone and in complex with E3 of the human PDC at resolutions of 1.6 Å and 2.2 Å, respectively. These structures show that E3BD is structurally similar to SBD’s and that it rigidly associates with E3 using a complex network of interactions. Alanine-scanning mutagenesis of E3BD residues involved in the E3BD/E3 interactions and subsequent binding studies further validate the nature of the association between E3BD and E3. Moreover, we show that a subset of disease-causing human E3 mutations, which are located at the E3BD/E3 interface, disrupt the binding of E3 to the E2p core, resulting in dysfunction of the PDC.
Results & Discussion
The Crystal Structure of E3BD: Similarity to the SBD Fold
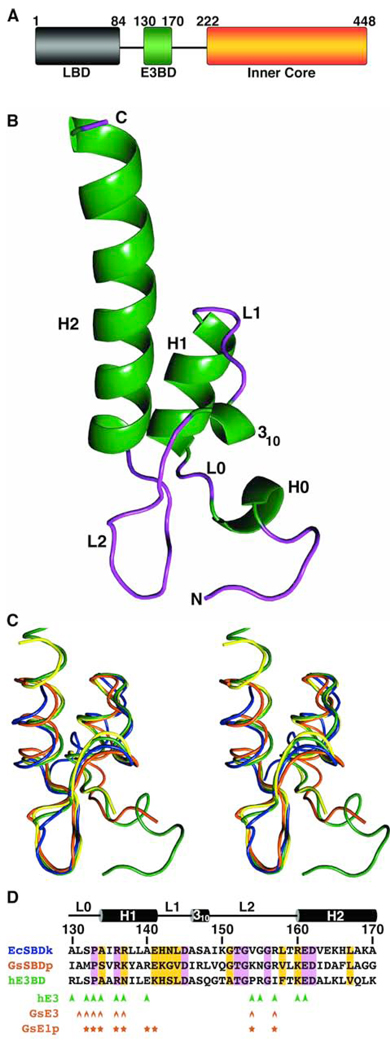
The human E3BP comprises one copy each of an LBD (residues 1–84), E3BD (residues 130–170) and the inner-core (residues 222–448) domains (Figure. 1A). These domains are connected by flexible linker regions. The 1.55-Å crystal structure of the E3BP region (residues 121–175) harboring the E3BD has been determined. This was accomplished via molecular replacement using the structure of 43 amino acids of human E3BD known from the bound structure (see below) as the search model (Table 1; Figure 1B). In the model, residues 121–126 are ordered but do not form regular secondary structure. Residues 127–130 form a small α-helix (H0), which is followed by a loop (L0) comprising residues 131–132 of E3BD. The α-helix H1 of the binding domain ranges from residues 133 to 141. After H1, there is a short region devoid of regular secondary structure (residues 142–145, termed L1). A small 310 helix (residues 146–148) is followed by a second loop (L2; residues 149–159) with an abrupt turn. The remainder of E3BD is an α-helix (H2; residues 160–175). Additional vector-derived amino acids, labeled residues 176–180 in the construct (see Experimental Procedures), are visible at the C-terminus of H2. H1 and H2 helices are parallel to each other and form extensive packing interactions.
Figure 1. Crystal Structure of E3BD and Comparison to SBD Structures.
(A) The domain structure of human E3BD. The single lipoyl domain (LBD, gray), the E3-binding domain (E3BD, green) and the inner core domain (orange) are connected by flexible linkers (solid lines). The boxes depict individual domains with numbers of the first and last residues for each domain indicated. (B) Ribbon drawing of the crystal structure of E3BD, with helices in green and areas without regular secondary structure in purple. Parts of the structure mentioned in the text are labeled. (C) A stereo diagram of the comparison of the structures of SBD’s to E3BD. Each tube represents a trace through the Cα’s of the respective structure. Shown are human E3BD (green), human E3BD from the E3BD/E3 structure (yellow), the NMR structure of the SBDp from the E2p of Geobacillus PDC (orange; 1W3D; Allen et al., 2005), and the NMR structure of the SBDk from the E2k of E. coli KGDC (blue; 1BBL; Robien et al., 1992). (D) Sequence alignment of the three structures. The names of the sequences retain the color of the structure shown in part (C). The numbering is that of human E3BD. Abbreviations: hE3BD, human E3BD; GsSBDp, Geobacillus SBD from the PDC, EcSBDk, E. coli SBDk from the E2k of KGDC; hE3, human E3; GsE3, Geobacillus E3; GsE1p; E1 of the Geobacillus PDC. The residues boxed in purple are identical in all three structures; regions of similarity identified by CLUSTALW are boxed in yellow. Above the alignment is a cartoon representation of the secondary structure of E3BD. Below are marked the positions of side chains of the binding proteins that contact hE3 (arrowheads), GsE3 (circumflexes), or GsE1p (stars). The color of the symbols matches the color of the name of the protein to which the E3 or E1p binds (C).
Table 1.
Data Collection and Refinement Statistics
| Structure | E3BD | E3BD/E3 |
|---|---|---|
| Space Group | P6522 | P212121 |
| a, Å | 36.8 | 171.1 |
| b, Å | 36.8 | 187.7 |
| c, Å | 191.5 | 224.4 |
| α, β, γ, ° | 90, 90, 120 | 90, 90, 90 |
| Resolution, Å* | 28.5-1.55 (1.61-1.55) | 37.4-2.18 (2.26-2.18) |
| No. reflections | 165,906 | 1,412,815 |
| Unique reflections (F≥0) | 12,109 (1117) | 367,237 (31492) |
| Completeness, % | 98.9 (95.4) | 98.2 (84.8) |
| Avg. Redundancy | 13.7 (7.9) | 3.9 (2.5) |
| Avg. I/σ | 67.8 (8.6) | 21.9 (1.9) |
| Rsym† | 0.032 (0.148) | 0.054 (0.401) |
| Refinement Statistics | ||
| Resolution range, Å | 28.5-1.55 | 37.4-2.18 |
| No. reflections used | 12,034 | 367,068 |
| No. protein atoms | 561 | 39,790 |
| No. waters | 59 | 1,415 |
| Rwork‡ | 0.203 | 0.211 |
| Rfree§ | 0.220 (584) | 0.247 (4518) |
| Average B-factors (Å2) | ||
| Protein | 23.5 | 40.3 |
| E3 | N/A** | 41.4 |
| E3BD | 23.5 | 49.1 |
| Ligand/Ion | 52.9 | 41.1 |
| Water | 39.8 | 38.7 |
| r.m.s. deviations | ||
| bond lengths, Å | 0.034 | 0.011 |
| bond angles, ° | 2.8 | 1.5 |
| Ramachandran distribution¶ | ||
| favored, % | 100 | 96.1 |
| allowed, % | 100 | 98.6 |
Numbers in parentheses are for the highest-resolution shell of the data.
Rsym = ΣhΣi |Î(h) – Ii(h)| / ΣhΣiIi(h), where Ii(h) is the ith measurement of the intensity for Miller indices h and Î(h) represents the mean intensity value of the symmetry- (or Friedel-) equivalent reflections of Miller indices h.
Rwork = Σh∥Fobs | - |Fcalc ∥ / Σh|Fobs |
The formula for Rfree is the same as that for Rwork, except it is calculated with a portion of the structure factors that had not been used for refinement. The number of reflections used for calculating Rfree is indicated in parentheses.
Using a modified Ramachandran plot (Lovell et al., 2002).
Not applicable.
The structure of E3BD is similar to those of SBD’s from E2’s of other α-ketoacid dehydrogenase complexes. The solution structures of the E2k SBD’s from E. coli KGDC (SBDk) and the Geopbacillus SBDp are known (Allen et al., 2005; Robien et al., 1992). In addition, the crystal structures of Geobacillus SBDp in complex with either the cognate E1p or E3 have been determined (Frank et al., 2004; Mande et al., 1995). Pairwise comparisons of the structures reveal that the three domains are structurally homologues (Figure 1C); the largest r.m.s. deviation between the approximately 40 comparable Cα atoms in any of the structures when compared to that of E3BD is 1.6 Å. An alignment of the three sequences is shown in Figure 1D.
The Crystal Structure of the E3BD/E3 Complex: Rigid Structural Association
The binding of E3BD to E3 was characterized by isothermal titration calorimetry (ITC). The LBD-E3BD construct (the LBD of human BCKDC fused to E3BD from human E3BP; see “Experimental Procedures” for details) was used in the present binding experiments. LBD-E3BD binds tightly to E3, exhibiting a dissociation constant (Kd) of 7.8 × 10−10 M as measured in the competitive binding experiment (Figure 2). For high-affinity binding with Kd < 10−8 M, the presence of a weaker-binding competitor ligand such as the SBDb of human BCKDC in this ITC measurement is necessary to obtain an accurate dissociation constant from the binding isotherm (Sigurskjold, 2000). The LBD moiety of human BCKDC in the fusion construct does not interact with E3, since similar Kd values were obtained when either isolated E3BD or LBD-E3BD was titrated against E3 (see above). The binding model with the best fit to the observed binding isotherm demonstrates one binding site for E3BD per E3 homodimer. In contrast, the binding of the SBDp from human E2p to E3 in the absence of the competitor ligand is not measurable due to negligible enthalpy changes (Figure 2). These results establish the specialized role of E3BD in binding human E3.
Figure 2. Binding of E3BD and SBDp to Human E3 Measured by Isothermal Titration Calorimetry.
The upper panel shows raw data of heat changes for the titration of LBD-E3BD (blue tracing) or LBD-SBDp (red tracing) into E3 in the cell in the presence and absence of a competitor ligand LBD-SBDb (from human BCKDC), respectively. The lower panel represents the binding isotherms for each injection versus the molar ratio of the LBD-E3BD monomer (blue circle) or the LBD-SBDp monomer (red circle) to the E3 dimer. The solid line in the lower panel depicts the fitting of the binding isotherms for LBD-E3BD and LBD-SBDp, based on a single-site binding model using the ORIGIN v. 7.0 software package. A dissociation constant (Kd) of 7.8 × 10−10 M is obtained for E3BD binding to E3 as derived from the competitive binding data (see Supplemental “Experimental Procedures”). The absence of heat changes with LBD-SBDp is consistent with the inability of SBDp to bind E3.
The 2.2-Å crystal structure of E3BD in complex with human E3 (the E3BD/E3 complex) reveals the interactions between the binding domain and its larger homodimeric partner (Figure 3). The structure was determined using the human E3 homodimer (1ZWD; Brautigam et al., 2005) as a molecular-replacement search model (Table 1). Five homodimers were located in the asymmetric unit of the E3BD/E3 crystals. Following rigid-body refinement, electron-density maps revealed density for E3BD bound to all five homodimers of E3. In most cases, this density was difficult to interpret. In four out of the five dimers, E3BD apparently binds in two orientations with respect to the homodimer, and each orientation is observed in about 50% of the asymmetric units in the crystal (see below). However, on one homodimer, the "AB dimer", unambiguous electron density is present for only one orientation of E3BD, which allowed the tracing of 43 amino acids of E3BD. Using this information, two E3BD’s were positioned on each of the other four homodimers at overlapping binding sites and refined as alternate conformations of one another, with occupancies of approximately 0.5 (the occupancies were refined). It is important to note that the two conformations therefore represent a single E3BD binding to these E3 homodimers in two possible orientations. The occupancy of the lone E3BD bound to the AB dimer is 1.0. Although we limit our discussion below to the AB-dimer/E3BD interface, the observations hold for all copies of E3 and E3BD in the asymmetric unit. As modeled above, the structure refined to an Rfree of 0.247 and has good geometry (Table 1). The E3BD/E3 interface buries approximately 1,490 Å2 of the E3 and E3BD surfaces. Residues 130–172 of E3BD (Figures 3B and 3C) and nearly all of the E3 residues are visible in electron-density maps. The well-ordered residues 121–129 and 173–180 in the structure of the E3BD construct alone are not visible in the E3BD/E3 complex and are assumed disordered. Based on proteolysis results and homology to SBD’s, the human E3BD had previously been surmised to consist of residues 131–163 (Harris et al., 1997). In this study, residues 130–170 were chosen as the boundaries of E3BD, because residue 130 makes contact to E3 and the side chain of residue 170 is the last to interact with a neighboring secondary structural element of E3BD. At merely 41 residues, E3BD and the structurally homologous SBD’s represent one of the smallest known folding units (Perham, 2000).
Figure 3. Crystal Structure of the E3BD/E3 Complex.
(A) Ribbon representation of the structure. Shown are the monomers of E3 (blue, tan), E3BD (green), and FAD (black sticks). (B) Electron density for E3BD. A close-up of the structure in the same orientation as in part (A) is shown. The ball-and-stick model represents the refined coordinates of E3BD. Superposed on this is a 2Fo-Fc electron-density map contoured at the 1-σ level. (C) A closer, stereo view of the map shown in part (B). (D) The overlap of the two E3BD-binding sites. A surface representation of E3 is shown in an orientation rotated 90° around the horizontal axis with respect to that of part (A). A ribbon diagram of E3BD (green) is shown, and a second theoretical molecule (E3BD*), generated by rotating the green monomer about the dyad axis of E3, is shown in pink. The surface is colored as above, except for the surface in contact with the green E3BD (purple), that contacting the yellow E3BD (blue), and that contacting both (orange).
Because E3 is a homodimer, two putative binding sites are available for E3BD (Figure 3D). However, only one E3BD binds to the E3 homodimer in the E3BD/E3 structure (Figure 3A and 3B), which confirms the ITC data. This stoichiometry results from the fact that portions of E3BD bind across the local two-fold axis present in E3, thereby contacting residues in both monomers of the homodimer non-equivalently (Figure 4A). With an E3BD bound in this configuration, a second E3BD attempting to access the second binding site on E3 would be sterically excluded by the first. This method of enforcing the stoichiometry of binding is similar to that observed in Geobacillus SBDp bound to its cognate E1p or E3 (Frank et al., 2004; Mande et al., 1995). The location of the E3BD-binding site on human E3 does not obstruct the binding sites for the enzyme’s substrates; indeed, it is possible to soak NADH into the E3BD/E3 crystals and observe electron density (data not shown) for this substrate bound at its normal binding site (Brautigam et al., 2005). The surface area on E3BD buried at the E3BD/E3 interface is approximately 785 Å2. About 27% of the total surface of E3BD (2900 Å2) is therefore involved in this interaction.
Figure 4. Interactions between E3 and E3BD.
(A) Stereo diagram of selected interactions. Carbons from E3 are colored tan or light blue, depending on their monomer of origin. Secondary structure is shown semi-transparently. The local two-fold axis of E3 is shown as a black cylinder. (B) The interactions of residues in helix H2 of E3BD (numbers with bd) with those from E3. (C) Dissociation constants (Kd) for the binding of wild-type and mutant E3BD to E3 determined by ITC. ΔΔGA is also shown. NM, not measurable. (D) Location of site-directed mutations in E3BD. A surface representation of E3BD with residues color-coded according to the magnitude of ΔΔGA (in units of kcal/mol) caused by their mutations to alanine (or methionine, in the case of A134bd). Red, ΔΔGA > 4.8; yellow, 4.8 > ΔΔGA > 4.0; green, 4.0 > ΔΔGA > 3.0; blue, 3.0 > ΔΔGA > 2.0. The total surface for E3 binding is outlined in orange. Locations of residues are labeled on the surface.
Comparison of the structures of E3 in the E3BD/E3 complex versus human E3 alone (Brautigam et al., 2005) reveals that E3 is essentially unperturbed when bound to E3BD; the r.m.s. deviation of the Cα atoms of the two structures is only 0.3 Å. Likewise, E3BD maintains essentially the same fold in crystal structures alone and when bound to E3 (Figure 1B); the r.m.s. deviation of the two sets of Cα atoms is 0.7 Å. The secondary structure of E3BD undergoes a small change when bound to E3. L0 is slightly larger (residues 130–134) and H1 slightly smaller (residues 135–141) in the bound form of E3BD compared to E3BD alone. The other local changes in both E3 and E3BD are the movements of side chains to accommodate and facilitate binding. In contrast to the Geobacillus SBDP/E3 structure (Mande et al., 1995), there is no change in the active site of E3 when E3BD binds. Both E3 and E3BD appear to be rigid structural units that associate as a consequence of their surface, electrostatic, and hydrogen-bonding compatibility.
Many of the interactions between human E3 and the side chains of E3BD are made between residues in the L0 and L2 regions of E3BD and residues from the interface domain (Brautigam et al., 2005) of E3 (Figures 4A & 4B). Both helices H1 and H2 of E3BD are also involved in the interaction. A complex network of interactions between the E3 homodimer and E3BD occurs very close to the dyad axis of E3. Y438 (one monomer) and Y438’ (the other monomer) stack on one another, forming a platform onto which residues from E3BD (designated P133bd, P154bd and I157bd) stack (Figure 4A). The side chain of R130bd makes a hydrogen bond to Y438’ and an ionic contact to D444. The equivalent D444’ is in a position to hydrogen bond with S132bd and the main-chain oxygen of P133bd. The guanidinium moiety of R447’ stacks on the side chain of N137bd instead of forming a hydrogen bond with it, as might be expected. Also, two residues from H2 of E3BD contact E3; the side chain K160bd forms hydrogen bonds with the side chains of E443’ and D413’, while the side chains of E161bd and T412’ stack on one another (Figure 4B). These interactions are noteworthy because in the Geobacillus SBDP/E3 structure, H2 does not interact with E3 (Mande et al., 1995).
Binding Studies of E3BD’s Mutated at the E3BD/E3 Interface
Further characterization of the E3BD/E3 interface was carried out by site-directed mutagenesis of interfacial residues in the LBD-E3BD construct. The structural integrity of all of the mutant proteins was ascertained using CD spectroscopy; all had spectra that were similar to wild-type LBD-E3BD (data not shown). The binding of the mutant LBD-E3BD proteins to human E3 was measured using ITC. The binding of E3BD’s containing the mutations S132Abd, P133Abd, A134Mbd, N137Abd, and I157Abd to E3 could not be measured by ITC, because heat changes upon their titration into solutions of E3 were too small (Figure 4C). Presumably, this loss of enthalpy is due to a severe diminution in the ability of these mutants to bind E3. All of these residues contact the E3 homodimer near to its dyad axis, and they make either van der Waals or uncharged polar interactions with E3 (Figure 4A). The binding of K160Abd to E3 is measurable; its dissociation constant is raised four orders of magnitude compared to that of wild-type E3BD (Figure 4C). This residue is not as close to the twofold axis of E3 as those mentioned above, but it is the only basic residue of E3BD to interact with two acidic residues of E3 (Figure 4B). Other residues at the interface, R130bd, R136bd, E140bd, R155bd, and E161bd were mutated to alanine, and each mutation diminishes the binding affinity by 2 to 3 orders of magnitude (Figure 4C), indicating that they are also important to the interaction. In sum, these mutational data validate the E3BD/E3 interface observed crystallographically.
It may seem counterintuitive that the mutation of residues involved in modest hydrogen-bonding or stacking interactions apparently causes abolition of E3BD binding (Figures 4A–4C). Nonetheless, similar behavior has been detected at other protein-protein interfaces (Chen and Shapiro, 1997; Clackson and Wells, 1995; Schreiber and Fersht, 1995). Many protein-protein interfaces feature mutational “hot spots.” A hot spot is generally defined as a residue whose mutation to alanine causes a large change in the association free energy (ΔΔGA = RT ln (Kd(mutant)/Kd(wild-type))) of two proteins (Clackson and Wells, 1995). Figure 4D shows that the residues causing the largest ΔΔGA’s upon mutation (ΔΔGA > 4.8 kcal/mol; i.e. those whose binding characteristics is not measurable) are located in a swath along the middle of the E3BD/E3 interface. Mutation of residues on the periphery of the interface has a smaller effect on ΔΔGA. Thus, the present binding results validate the “hot-spot” paradigm previously proposed for other protein-protein interfaces: the mutations of residues in the center of the interface cause the largest changes in binding energy (Bogan and Thorn, 1998; Clackson and Wells, 1995). The theoretical underpinnings of this phenomenon are not well understood; they may involve the necessity of excluding solvent from the center of the interaction (Bogan and Thorn, 1998) or the ease with which water may substitute for a lost interaction (DeLano, 2002).
Comparisons between E3BD/E3 and Geobacillus SBDp/E3 Interfaces
A comparison of the human E3BD/E3 complex to the Geobacillus SBDp/E3 complex reveals that, in both cases, the H1 helices of the binding domains intimately contact the respective E3’s (Figure 5). Both H1’s interact using polar and apolar contacts. The sequences of the binding domains and their binding sites on the E3’s are not well conserved (Figure 1D); thus, the identity of the residues that form the aforementioned interactions varies between the two species. The positions of the H2 helices of the binding domains with respect to the respective E3’s are divergent. The H2 of the Geobacillus SBDp/E3 complex is oriented away from the surface of E3, but in the human E3BD/E3 complex, H2 is closer to E3 (Figure 5). Indeed, as noted above, residues from H2 of E3BD contact human E3 (Figure 4B). The orientation of E3BD with respect to E3 is not influenced by crystal contacts. This fact is evidenced by the observation that, in the E3BD/E3 asymmetric unit, all nine E3BD’s are nearly identically oriented despite differences in their crystalline environments.
Figure 5. Comparison of the Geobacillus SBDp/E3 and E3BD/E3 Structures.
The Cα atoms of Geobacillus E3 and human E3 (not the binding domains) were used for this superposition. The coloration is that of Figure 3, and the Geobacillus SBDp is orange.
The binding sites for the binding domains on the respective E3’s also differ. In the Geobacillus SBDp/E3 complex, SBDp comes close to the dyad axis of E3, but does not cross it. Pairs of residues related by this axis are not contacted by Geobacillus SBDp. However, parts of E3BD do cross the dyad axis of E3, and they do contact twofold-related pairs of residues non-equivalently (Figures 3C and 4A). In this respect, the E3BD/E3 complex is like the Geobacillus SBDp/E1 complex (Frank et al., 2005). The extent of contact for the binding domains on human E3, Geobacillus E3, and Geobacillus E1p varies. Calculations performed using CNS show that the total surface area buried by the coupling of the Geobacillus SBDp and E1p is only 1030 Å2, compared to 1240 Å2 for the Geobacillus SBDp/E3 complex and 1490 Å2 for the human E3BD/E3 complex.
Figure 1D shows an alignment of human E3BD and Geobacillus SBDp, with residues that contact the respective E3’s (and Geobacillus E1p) marked with different symbols. This analysis demonstrates that the positions, if not the identities, of many of the residues that contact E3’s are conserved in these diverse species; the positions of 7 out of 12 human E3BD/E3 contacts are recapitulated at the Geobacillus E3/SBDp interface. As noted before (Frank et al., 2005), many of the same residues used to contact Geobacillus E3 are used to associate with Geobacillus E1p (Figure 1D). These interactions explain the mutually exclusive mode in which Geobacillus E3 and E1p bind to the SBDp. Moreover, in this sequence alignment, T152bd, G153bd, and D162bd are invariant, but do not make contact with a binding partner. Instead, these residues play important structural roles within the three binding domains, explaining their conservation.
Analysis of Disease-causing Mutations in Human E3
Several mutations in human E3 are known to result in combined enzyme deficiencies in α-ketoacid dehydrogenase complexes PDC, BCKDC and KGDC (Hengeveld and de Kok, 2002). Among the E3 missense mutations reported to date, E340K (Hong et al., 1997), D444V (Shany et al., 1999), R447G (Odievre et al., 2005), and R460G (Hong et al., 1996) are close to the E3BD-binding site (Figure 6A). All four residues make inter-monomer contacts in the human E3 homodimer. It has been speculated that these mutations diminish E3 activity by disrupting the ability of the enzyme to form homodimers (Brautigam et al., 2005; Hengeveld and de Kok, 2002) or by interfering with E3-E3BP interactions (Odievre et al., 2005).
Figure 6. Human Mutations Causing E3 Deficiency.
(A) The surface of the human E3 homodimer is shown as in Figure 3. One of the E3BD-binding sites is outlined in orange. Disease-causing mutations are colored dark blue (E340), red (D444), yellow (447), and magenta (R460). (B) Enzyme activities of wild-type and mutant human E3’s. The black bars represent E3 activity, the speckled bars show PDC activity reconstituted with wild-type and mutant E3. The identities of the E3’s used are shown on the x-axis. Activities are normalized to that of the wild-type enzymes, which at 100% are 314 s−1 for human E3 and 401 min−1 for the overall activity of the reconstituted PDC. (C) Dissociation constants (Kd) for the binding of wild-type and mutant E3 to LBD-E3BD as determined by ITC. NM, not measurable.
We examined the effects of these mutations on E3BD binding to E3. Analyses of the proteins carrying the E340K, D444V, R447G and R460G mutations using analytical ultracentrifugation show that they have no effect on the dimerization of human E3. The wild-type and mutant E3’s have similar dissociation constants (Kd = 2–3 × 10−7 M) at 20°C for the monomer-dimer equilibrium (data not shown). Except for R460G, the recombinant mutant proteins expressed at 28°C exhibit essentially wild-type E3 activity (Figure 6B). These results differ from those measured with cultured cells from E3-deficient patients carrying these mutations, which showed markedly reduced E3 activity (Hong et al., 1997; Odievre et al., 2005; Shany et al., 1999). A plausible explanation of this discrepancy is that these missense mutations render these E3 variants unstable in patient’s cells at 37°C. PDC activity reconstituted with E340K, D444V and R447G E3 variants is reduced to 38%, 12% and 18% of the wild-type, respectively (Figure 6D). Moreover, the activity of human BCKDC is impaired when reconstituted with human E3 variants carrying the D444V and E340K mutations (data not shown). The results suggest that these E3 variants interact with binding domains poorly, thereby impeding the recruitment of E3 to the PDC or the BCKDC. This notion is confirmed by ITC measurements, which show that the affinities of the mutant E3 proteins for E3BD are severely reduced (Figure 6C). The Kd for R447G is two orders of magnitude higher than that of the wild-type protein. The heat changes for the interactions between E3BD and E340K and D444V are too weak to be detected. It is therefore likely that poor recruitment of E3 into PDC accounts at least partially for the poor PDC activity detected in human patients carrying these mutations. For the R460G mutant E3, the Kd is three orders of magnitude higher than that of wild-type E3. The weak binding of this mutant E3 to E3BD is compounded by its low dihydrolipoamide dehydrogenase activity, yielding no detectable PDC activity. The side chain of R460 contacts the side chain of E333’, and both residues are mostly buried in the human E3 structure (Figure 6A). Removal of the bulky, charge-compensating argininyl side chain, as in the R460G mutant, may therefore cause a significant structural rearrangement that diminishes both E3 activity and E3BD binding.
Concluding Remarks
The conserved folds of E3BD, Geobacillus SBDp and E. coli SBDk (Figure 1C) suggest that the E3BP arose from its cognate E2p through gene duplication; in support of this is the fact that the genes for both human E3BP and E2p are located on chromosome 11 (Harris et al., 1997). The structural role of E3BP in tethering E3 to the icosahedral 60-meric E2p/E3BP core of the eukaryotic PDC (Harmych et al., 2002; Maeng et al., 1996; Seyda and Robinson, 2000) is confirmed by the binding studies presented herein (Figure 2). Three different strategies are employed by α-ketoacid dehydrogenase complexes to sequester the essential E1 and E3 components to the E2 core. Eukaryotic PDC uses E3BP to secure E3, and the SBDp of E2p binds E1p (Reed, 2000). In the mammalian KGDC, there is biochemical evidence that the E3 binds to the amino-terminal region of the cognate E1 homodimer, allowing the separation of a stable E1-E3 subcomplex from the octahedral E2 core (McCartney et al., 1998; Rice et al., 1992; Sanderson et al., 1996). In the Geobacillus PDC, both E1p and E3 sterically compete for essentially the same binding site on the SBDp domain of the 60-meric E2p scaffold (Figure 1D). Similarly, the single SBD in the octahedral 24-meric E2 core of the mammalian BCKDC, which is devoid of E3BP, also binds both of its respective E1 and E3 components, albeit with markedly lower affinity for E3 than E1 (Pettit et al, 1978). Thus, while the overall molecular architecture and mechanism of α-ketoacid dehydrogenase complexes is conserved, the mode of binding for the common E3 component has diverged to accommodate the special organizational and functional requirements of each catalytic machine.
Experimental Procedures
Protein Expression, Purification, and Site-directed Mutagenesis
Recombinant human E3 was expressed and purified as previously described (Brautigam et al., 2005). To facilitate expression, a recombinant fusion protein LBD-E3BD was produced. This construct comprises the LBD from the E2b of human BCKDC, followed by residues 121–175 of human E3BP, which harbors the entire E3BD sequence (residues 130–170), and eight vector-derived residues including the 6 × His-tag at its C-terminus. A tobacco-etch virus (TEV) protease recognition sequence (LENLYFQ ↓G) was introduced into the connecting region immediately 5’ to residue 121. The SBDp protein (E2p residues 260–315) of human PDC was also expressed as a C-terminal fusion protein to LBD of human BCKDC (LBD-SBDp), containing a TEV-cleavage site similar to LBD-E3BD. LBD-E3BD and LBD-SBDp proteins were isolated from E. coli lysates by Ni-NTA (Qiagen) extraction and Sephadex S-200 gel filtration.
The LBD-E3BD fusion protein was digested with TEV protease followed by re-extraction with Ni-NTA resin. The resultant E3BD protein was further purified on a Sephacryl S-100 size-exclusion column that had been equilibrated with a buffer containing 50 mM potassium phosphate, pH 7.5, 150 mM KCl, and 5% (v/v) glycerol.
To prepare the E3BD/E3 complex for crystallization, purified human E3 and E3BD proteins were mixed at the ratio of one E3 dimer to two E3BD monomers. The mixture was transferred to a buffer consisting of 20 mM HEPES, pH 7.5 and 20 mM β-mercaptoethanol, and concentrated to 25 mg/ml concentration.
Mutations in E3 and LBD-E3BD were introduced using the QuickChange kit (Stratagene), according to the manufacturer’s protocols.
Crystallization and Structure Determination
Crystals of E3BD were obtained using the vapor diffusion method at 20° C by mixing 2 µl E3BD (33 mg/ml) with 2 µl well solution containing 1.35M sodium citrate, pH 6.3 and 0.05–0.075% (w/v) β-octyl-glucopyranoside. Microseeding was used to induce crystal growth. Cryostabilization was accomplished by transferring the crystals to well solution containing 10% (v/v) glycerol. The 300–500 µm pyramidal crystals have the symmetry of space group P6522. Crystals were flash-cooled by plunging them into liquid propane.
Hanging drops of the E3BD/E3 complex consisted of 3 µl of the protein solution and 3 µl of the well solution containing 14% (w/v) polyethylene glycol 4000, 0.2M (NH4)2SO4, 0.1M sodium acetate, pH 4.6, 0.03M spermidine and 6–8% glycerol. Crystallization was induced by streak seeding after the drops were equilibrated for 24 h at 20° C. The rod-like crystals have typical dimensions of approximately 500 × 50 × 50 µm and have the symmetry of space group P212121. Cryo-cooling of the crystals was carried out at 4°C with 20% (v/v) glycerol added to the well solution to serve as the cryoprotectant. After this step, the crystals were flash-cooled in liquid propane. X-ray diffraction data from crystals of E3BD and the E3BD/E3 complex were collected at beamline 19-ID at the Structural Biology Center (SBC) of the Advanced Photon Source (APS) at Argonne National Laboratory (ANL). The data were integrated and scaled using the HKL2000 package of programs (Otwinowski and Minor, 1997). The E3BD/E3 complex structure was determined using the molecular-replacement protocols available in CNS version 1.1, as outlined above (Brunger et al., 1998). Refinement of the E3BD/E3 model proceeded using the simulated annealing, conjugate-gradient minimization, and individual B-factor refinement protocols available in CNS. The model was manipulated using the program XtalView (McRee, 1992). The final model contains ten nearly complete chains of human E3 and nine chains of E3BD (eight of which have occupancy < 1). The positions of the E3BD molecules have been verified using simulated-annealing omit maps.
The structure of E3BD alone was determined using molecular replacement. The search model utilized was that of E3BD associated with the AB dimer, as discussed above. Phaser (Read, 2001; Storoni et al., 2004) was used to locate a single monomer of E3BD in the asymmetric unit of these crystals. The search model represented only 75% of the residues present in the E3BD protein construct; electron density was evident for additional residues after rigid-body refinement in CNS of the molecular replacement model. These residues were added to the model, which was subjected to refinement in CNS as outlined above. Alternate conformations of several residues were included in the refinement. The final model of E3BD alone contains 60 amino-acid residues. In addition, elongated electron density is present along a crystallographic two-fold axis in Fo - Fc maps, with a maximum extent of 6 σ. Attempts to model buffer constituents into this density raised Rfree; it therefore remains unmodeled. Final statistics of the model are in Table 1.
Supplementary Material
Acknowledgments
The authors thank Dr. Kirill Popov (University of Alabama at Birmingham) for kindly providing the E1p and E2p/E3BP plasmids of human PDC used in this study. This paper is dedicated to the memory of Debbie Skaggs. This work was supported by Grants DK26758 and DK62306 from the National Institutes of Health and Grant I-1286 from the Welch Foundation. The use of the Argonne National Laboratory Structural Biology Center beam lines at the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Energy Research under Contract No. W-31-109-ENG-38.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data
Supplemental Data including additional “Experimental Procedures” are available at http://www.structure.org/cgi/content/full/.
Accession Numbers
The model coordinates and structural factor amplitudes have been deposited in the Protein Data Bank for structures of human E3BD (2F60) and the human E3BD/E3 complex (2F5Z).
References
- Allen MD, Broadhurst RW, Solomon RG, Perham RN. Interaction of the E2 and E3 components of the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus: use of a truncated protein domain in NMR spectroscopy. FEBS Journal. 2005;272:259–268. doi: 10.1111/j.1432-1033.2004.04405.x. [DOI] [PubMed] [Google Scholar]
- Aral B, Benelli C, Ait-Ghezala G, Amessou M, Fouque F, Maunoury C, Creau N, Kamoun P, Marsac C. Mutations in PDX1, the human lipoyl-containing component X of the pyruvate dehydrogenase-complex gene on chromosome 11p1, in congenital lactic acidosis. American Journal of Human Genetics. 1997;61:1318–1326. doi: 10.1086/301653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. Journal of Molecular Biology. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- Brautigam CA, Chuang JL, Tomchick DR, Machius M, Chuang DT. Crystal structure of human dihydrolipoamide dehydrogenase: NAD+/NADH binding and the structural basis of disease-causing mutations. Journal of Molecular Biology. 2005;350:543–552. doi: 10.1016/j.jmb.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallographica. Section D: Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chen C-Z, Shapiro R. Site-specific mutagenesis reveals differences in the structural bases for tight binding of RNase inhibitor to angiogenin and RNase A. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:1761–1766. doi: 10.1073/pnas.94.5.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang DT, Shih VE. Maple syrup urine disease (branched-chain ketoaciduria) In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. The Metabolic and Molecular Bases of Inherited Disease. Vol. 2. New York: McGraw-Hill; 2001. pp. 1971–2005. [Google Scholar]
- Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- De Marcucci O, Lindsay JG. Component X. An immunologically distinct polypeptide associated with mammalian pyruvate dehydrogenase multi-enzyme complex. European Journal of Biochemistry. 1985;149:641–648. doi: 10.1111/j.1432-1033.1985.tb08972.x. [DOI] [PubMed] [Google Scholar]
- DeLano WL. Unraveling hot spots in binding interfaces: progress and challenges. Current Opinion in Structural Biology. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- Dey R, Aral B, Abitbol M, Marsac C. Pyruvate dehydrogenase deficiency as a result of splice-site mutations in the PDX1 gene. Molecular Genetics and Metabolism. 2002;76:344–347. doi: 10.1016/s1096-7192(02)00104-x. [DOI] [PubMed] [Google Scholar]
- Dey R, Mine M, Desguerre I, Slama A, Van Den Berghe L, Brivet M, Aral B, Marsac C. A new case of pyruvate dehydrogenase deficiency due to a novel mutation in the PDX1 gene. Annals of Neurology. 2003;53:273–277. doi: 10.1002/ana.10478. [DOI] [PubMed] [Google Scholar]
- Frank RAW, Pratap JV, Pei XY, Perham RN, Luisi BF. The molecular origins of specificity in the assembly of a multienzyme complex. Structure. 2005;13:1119–1130. doi: 10.1016/j.str.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Frank RAW, Titman CM, Pratap JV, Luisi BF, Perham RN. A molecular switch and proton wire synchronize the active sites in thiamine enzymes. Science. 2004;306:872–876. doi: 10.1126/science.1101030. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan S, Rahmatullah M, Radke GA, Powers-Greenwood SL, Roche TE. Role of protein X in the function of the mammalian pyruvate dehydrogenase complex. Biochemical and Biophysical Research Communications. 1989;160:715–721. doi: 10.1016/0006-291x(89)92492-3. [DOI] [PubMed] [Google Scholar]
- Harmych S, Arnette R, Komuniecki R. Role of dihydrolipoyl dehydrogenase (E3) and a novel E3-binding protein in the NADH sensitivity of the pyruvate dehydrogenase complex from anaerobic mitochondria of the parasitic nematode Ascaris suum. Molecular and Biochemical Parasitology. 2002;125:135–146. doi: 10.1016/s0166-6851(02)00221-9. [DOI] [PubMed] [Google Scholar]
- Harris RA, Bowker-Kinley MM, Wu P, Jeng J, Popov KM. Dihydrolipoamide dehydrogenase-binding protein of the human pyruvate dehydrogenase complex: DNA-derived amino acid sequence, expression, and reconstitution of the pyruvate dehydrogenase complex. Journal of Biological Chemistry. 1997;272:19746–19751. doi: 10.1074/jbc.272.32.19746. [DOI] [PubMed] [Google Scholar]
- Hengeveld AF, de Kok A. Structural basis of the dysfunctioning of human 2-oxo acid dehydrogenase complexes. Current Medicinal Chemistry. 2002;9:499–520. doi: 10.2174/0929867023370996. [DOI] [PubMed] [Google Scholar]
- Hiromasa Y, Fujisawa T, Aso Y, Roche TE. Organization of the cores of the mammalian pyruvate dehydrogenase complex formed by E2 and E2 plus E3-binding protein and their capacities to bind the E1 and E3 components. Journal of Biological Chemistry. 2004;279:6921–6933. doi: 10.1074/jbc.M308172200. [DOI] [PubMed] [Google Scholar]
- Hodgson JA, De Marcucci OG, Lindsay JG. Lipoic acid is the site of substrate-dependent acetylation of component X in ox heart pyruvate dehydrogenase multienzyme complex. European Journal of Biochemistry. 1986;158:595–600. doi: 10.1111/j.1432-1033.1986.tb09796.x. [DOI] [PubMed] [Google Scholar]
- Holness MJ, Sugden MC. Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochemical Society Transactions. 2003;31:1143–1151. doi: 10.1042/bst0311143. [DOI] [PubMed] [Google Scholar]
- Hong YS, Kerr DS, Craigen WJ, Tan J, Pan Y, Lusk M, Patel MS. Identification of two mutations in a compound heterozygous child with dihydrolipoamide dehydrogenase deficiency. Human Molecular Genetics. 1996;5:1925–1930. doi: 10.1093/hmg/5.12.1925. [DOI] [PubMed] [Google Scholar]
- Hong YS, Kerr DS, Liu T-C, Lusk M, Powell BR, Patel MS. Deficiency of dihydrolipoamide dehydrogenase due to two mutant alleles (E340K and G101del): analysis of a family and prenatal testing. Biochimica et Biophysica Acta. 1997;1362:160–168. doi: 10.1016/s0925-4439(97)00073-2. [DOI] [PubMed] [Google Scholar]
- Jilka JM, Rahmatullah M, Kazemi M, Roche TE. Properties of a newly characterized protein of the bovine kidney pyruvate dehydrogenase complex. Journal of Biological Chemistry. 1986;261:1858–1867. [PubMed] [Google Scholar]
- Lawson JE, Behal RH, Reed LJ. Disruption and mutagenesis of the Saccharomyces cerevisiae PDX1 gene encoding the protein X component of the pyruvate dehydrogenase complex. Biochemistry. 1991;30:2834–2839. doi: 10.1021/bi00225a015. [DOI] [PubMed] [Google Scholar]
- Ling M, McEachern G, Seyda A, MacKay N, Scherer SW, Bratinova S, Beatty B, Giovannucci-Uzielli ML, Robinson BH. Detection of a homozygous four bas pair deletion in the protein X gene in a case of pyruvate dehydrogenase complex deficiency. Human Molecular Genetics. 1998;7:501–505. doi: 10.1093/hmg/7.3.501. [DOI] [PubMed] [Google Scholar]
- Liu S, Baker JC, Andrews PC, Roche TE. Recombinant expression and evaluation of the lipoyl domains of the dihydrolipoyl acetyltransferase component of the human pyruvate dehydrogenase complex. Archives of Biochemistry and Biophysics. 1995;316:926–940. doi: 10.1006/abbi.1995.1124. [DOI] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB, III, de Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Ca geometry: f, y, and Cb deviation. Proteins. 2002;Vol. 50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Maeng C-Y, Yazdi MA, Reed LJ. Stoichiometry of binding of mature and truncated forms of the dihydrolipoamide dehydrogenase-binding protein to the dihydrolipoamide acetyltransferase core of the pyruvate dehydrogenase complex from Saccharomyces cerevisiae. Biochemistry. 1996;35:5879–5882. doi: 10.1021/bi9600254. [DOI] [PubMed] [Google Scholar]
- Mande SS, Sarfaty S, Allen MD, Perham RN, Hol WGJ. Protein-protein interactions in the pyruvate dehydrogenase multienzyme complex: dihydrolipoamide dehydrogenase complexed with the binding domain of dihydrolipoamide acetyltransferase. Structure. 1995;4:277–286. doi: 10.1016/s0969-2126(96)00032-9. [DOI] [PubMed] [Google Scholar]
- McCartney RG, Rice JE, Sanderson SJ, Bunik V, Lindsay H, Lindsay JG. Subunit interactions in the mammalian alpha-ketoglutarate dehydrogenase complex. Evidence for direct association of the alpha-ketoglutarate dehydrogenase and dihydrolipoamide dehydrogenase components. Journal of Biological Chemistry. 1998;273:24158–24164. doi: 10.1074/jbc.273.37.24158. [DOI] [PubMed] [Google Scholar]
- McRee DE. A visual protein crystallographic software system for X11/Xview. Journal of Molecular Graphics. 1992;10:44–46. [Google Scholar]
- Neagle J, De Marcucci OG, Dunbar B, Lindsay JG. Component X of mammalian pyruvate dehydrogenase complex: structural and functional relationship to the lipoate acetyltransferase (E2) component. FEBS Letters. 1989;253:11–15. doi: 10.1016/0014-5793(89)80919-6. [DOI] [PubMed] [Google Scholar]
- Odievre M-H, Chretien D, Munnich A, Robinson BH, Dumoulin R, Masmoudi S, Kadhom N, Rotig A, Rustin P, Bonnefont J-P. A novel mutation in the dihydrolipoamide dehydrogenase E3 subunit gene (DLD) resulting in an atypical form of a-ketoglutarate dehydrogenase deficiency. Human Mutation. 2005 doi: 10.1002/humu.9319. in press. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Patel MS, Roche TE. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB Journal. 1990;4:3224–3233. doi: 10.1096/fasebj.4.14.2227213. [DOI] [PubMed] [Google Scholar]
- Perham RN. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annual Review of Biochemistry. 2000;69:961–1004. doi: 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- Powers-Greenwood SL, Rahmatullah M, Radke GA, Roche TE. Separation of protein X from the dihydrolipoyl transacetylase component of the mammalian pyruvate dehydrogenase complex and function of protein X. Journal of Biological Chemistry. 1989;264:3655–3657. [PubMed] [Google Scholar]
- Rahmatullah M, Roche TE. The ctatlytic requirements for reduction and acetylation of protein X and the related regulation of various forms of resolved pyruvate dehydrogenase kinase. Journal of Biological Chemistry. 1987;262:10265–10271. [PubMed] [Google Scholar]
- Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallographica. Section D: Biological Crystallography. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- Reed LJ. A trail of research from lipoic acid to alpha-keto acid dehydrogenase complexes. J Biol Chem. 2001;276:38329–38336. doi: 10.1074/jbc.R100026200. [DOI] [PubMed] [Google Scholar]
- Reed LJ, Damuni Z, Merryfield ML. Regulation of mammalian pyruvate alpha-keto acid dehydrogenase complexes by phosphorylation-dephosphorylation. Current Topics in Cellular Regulation. 1985;27:41–49. doi: 10.1016/b978-0-12-152827-0.50011-6. [DOI] [PubMed] [Google Scholar]
- Rice JE, Dunbar B, Lindsay JG. Sequences directing dihydrolipoamide dehydrogenase (E3) binding are located on the 2-oxoglutarate dehydrogenase (E1) component of the mammalian 2-oxoglutarate dehydrogenase multienzyme complex. EMBO Journal. 1992;11:3229–3235. doi: 10.1002/j.1460-2075.1992.tb05400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robien MA, Clore GM, Omichinski JG, Perham RN, Appella E, Sakaguchi K, Gronenborn AM. Three-dimensional solution structure of the E3-binding domain of the dihydrolipoamide succinyltransferase core from the 2-oxoglutarate dehydrogenase multienzyme complex of Escherichia coli. Biochemistry. 1992;31:3463–3471. doi: 10.1021/bi00128a021. [DOI] [PubMed] [Google Scholar]
- Robinson BH. Lactic acidemia: disorders of pyruvate decarboxylase and pyruvate dehydrogenase. In: Scriver CR, Beaudet AL, Valle D, Sly WS, Childs B, Kinzler KW, Vogelstein B, editors. The Metabolic and Molecular Bases of Inherited Disease. Vol. 2. New York: McGraw-Hill; 2001. pp. 2275–2295. [Google Scholar]
- Roche TE, Hiromasa Y, Turkan A, Gong X, Peng T, Yan X, Kasten SA, Bao H, Dong J. Essential roles of lipoyl domains in the activated function and control of pyruvate dehydrogenase kinases and phosphatase isoform 1. European Journal of Biochemistry. 2003;270:1050–1056. doi: 10.1046/j.1432-1033.2003.03468.x. [DOI] [PubMed] [Google Scholar]
- Sanderson SJ, Khan SS, McCartney RG, Miller C, Lindsay JG. Reconstitution of mammalian pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase complexes: analysis of protein X involvement and interaction of homologous and heterologous dihydrolipoamide dehydrogenases. Biochemical Journal. 1996;319:109–116. doi: 10.1042/bj3190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G, Fersht AR. Energetics of protein-protein interactions: analysis of the barnase-barstar interface by single mutations and couble mutant cycles. Journal of Molecular Biology. 1995;248:478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- Seyda A, Robinson BH. Expression and functional characterization of human protein X variants in SV40-immortalized protein X-deficient and E2-deficient human skin fibroblasts. Archives of Biochemistry and Biophysics. 2000;382:219–223. doi: 10.1006/abbi.2000.2026. [DOI] [PubMed] [Google Scholar]
- Shany E, Saada A, Landau D, Shaag A, Hershkovitz E, Elpeleg ON. Lipoamide dehydrogenase deficiency due to a novel mutation in the interface domain. Biochemical and Biophysical Research Communications. 1999;262:163–166. doi: 10.1006/bbrc.1999.1133. [DOI] [PubMed] [Google Scholar]
- Sigurskjold BW. Exact analysis of competition ligand binding by displacement isothermal titration calorimetry. Anal Biochem. 2000;277:260–266. doi: 10.1006/abio.1999.4402. [DOI] [PubMed] [Google Scholar]
- Stoops JK, Cheng RH, Yazdi MA, Maeng C-Y, Schroeter JP, Klueppelberg U, Kolodziej SJ, Baker TS, Reed LJ. On the unique structural organization of the Saccharomyces cerevisiae pyruvate dehydrogenase complex. Journal of Biological Chemistry. 1997;272:5757–5764. doi: 10.1074/jbc.272.9.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallographica. Section D: Biological Crystallography. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.