This work examines the interaction between jasmonate (JA) and light signaling. It finds that attenuation of shade responses by low red/far-red light requires the JA signal component COI1 and that some responses to JA are partly dependent on the light signal component phyA. The JA and phyA pathways are integrated through stability of the repressor protein JAZ1.
Abstract
Jasmonate (JA) activates plant defense, promotes pollen maturation, and suppresses plant growth. An emerging theme in JA biology is its involvement in light responses; here, we examine the interdependence of the JA- and light-signaling pathways in Arabidopsis thaliana. We demonstrate that mutants deficient in JA biosynthesis and signaling are deficient in a subset of high irradiance responses in far-red (FR) light. These mutants display exaggerated shade responses to low, but not high, R/FR ratio light, suggesting a role for JA in phytochrome A (phyA) signaling. Additionally, we demonstrate that the FR light–induced expression of transcription factor genes is dependent on CORONATINE INSENSITIVE1 (COI1), a central component of JA signaling, and is suppressed by JA. phyA mutants had reduced JA-regulated growth inhibition and VSP expression and increased content of cis-(+)-12-oxophytodienoic acid, an intermediate in JA biosynthesis. Significantly, COI1-mediated degradation of JASMONATE ZIM DOMAIN1-β-glucuronidase (JAZ1-GUS) in response to mechanical wounding and JA treatment required phyA, and ectopic expression of JAZ1-GUS resulted in exaggerated shade responses. Together, these results indicate that JA and phyA signaling are integrated through degradation of the JAZ1 protein, and both are required for plant responses to light and stress.
INTRODUCTION
The group of signaling molecules known collectively as the jasmonates (JAs) are plant hormones that regulate many physiological processes, including defense, growth, development, fertility, and senescence, throughout the life cycle of higher plants (Devoto and Turner, 2003; Wasternack, 2007; Balbi and Devoto, 2008; Reinbothe et al., 2009). JAs inhibit growth by suppressing mitosis in apical meristems (Zhang and Turner, 2008), inhibit photosynthesis and energy-generating processes (Ueda and Kato, 1980; Reinbothe et al., 1993; Jung et al., 2007), and activate plant defense responses to a variety of biotic and abiotic stresses (Rao et al., 2000; Glazebrook, 2005; Howe and Jander, 2008; Frenkel et al., 2009; Clarke et al., 2009), suggesting that JAs regulate the balance between growth and defense in response to a dynamic environment.
In response to biotic and abiotic stress, α-linolenic acid is released from plastid membranes by phospholipases, providing the precursor for JA synthesis via the octadecanoid pathway (Mueller et al., 1993). Regulation of JA responses occurs via a global reprogramming of transcription that results in the upregulation of genes involved in defense, secondary metabolism, hormone biosynthesis, and, in a feedback loop, JA synthesis (Sasaki et al., 2001; Devoto et al., 2005; Cho et al., 2007; Dombrecht et al., 2007; Jung et al., 2007; Pauwels et al., 2008; Wang et al., 2008). Arabidopsis thaliana mutants in JA biosynthesis, allene oxide synthase (aos; Park et al., 2002) and 12-oxo-phytodienoic acid reductase (opr3; Stintzi and Browse, 2000), are male sterile, highlighting the role of JAs in fertility.
A number of key players in JA signal transduction have been identified in Arabidopsis. The gene for the F-box protein COI1 was identified in a mutant screen for plants insensitive to growth inhibition by the JA analog, coronatine, a phytotoxin produced by some pathogenic strains of Pseudomonas syringae (Feys et al., 1994). Mutations in the COI1 gene result in plants that are compromised in all known JA responses: defense against biotic and abiotic stresses, growth inhibition, and fertility (Xie et al., 1998; Zhang and Turner, 2008). COI1 binds to the Arabidopsis homologs of S-PHASE KINASE-ASSOCIATED PROTEIN1 and CULLIN to form the SCFCOI1 complex, an E3 ubiquitin ligase that degrades target proteins in response to JA via the 26S proteosome (Devoto et al., 2002; Xu et al., 2002). Targets of SCFCOI1 include the JAZ transcriptional repressors (Chini et al., 2007; Thines et al., 2007). JAZ proteins interact with and repress activators of JA gene expression, including a transcription factor, the Arabidopsis homolog of myelocytomatosis viral oncogene (MYC), MYC2 (Boter et al., 2004; Lorenzo et al., 2004). In response to a JA signal, JAZ proteins bind to COI1 and are degraded, relieving the repression of MYC2 that leads to expression of JA-regulated genes (Chini et al., 2007; Thines et al., 2007). COI1 has been identified as a receptor for the bioactive JA derivative (+)-7-iso-jasmonoyl-l-isoleucine (JA-Ile; Fonseca et al., 2009; Yan et al., 2009). All interactions of COI1 with JAZ proteins tested so far are mediated by JA-Ile (Thines et al., 2007; Katsir et al., 2008; Melotto et al., 2008; Chini et al., 2009). The amino acid conjugate synthase, JA-RESISTANT1 (JAR1), the enzyme responsible for conjugating JA to Ile and other amino acids, is a crucial component of the JA signal transduction cascade for a subset of JA responses (Staswick et al., 2002; Staswick and Tiryaki, 2004; Chung et al., 2008).
As well as providing the primary source of energy to support photosynthesis, light is one of the most important environmental signals governing the growth and development of plants throughout their life cycle (Chen et al., 2004). The phytochromes, which absorb light primarily in the red (R) and far-red (FR) part of the spectrum (600 to 750 nm), regulate seed germination, seedling establishment, circadian entrainment, and flowering time (Chory et al., 1996). Phytochromes are also responsible for monitoring the change in R/FR ratio that occurs when plants are shaded by neighbors due to the selective absorbance of red light by photosynthetic pigments (Ballaré et al., 1990). This allows shade-intolerant plants like Arabidopsis to preempt the threat of crowding by competitors and initiate escape mechanisms, collectively known as shade avoidance responses, to reach light and so maximize photosynthesis. Changes in R/FR ratio light are mainly detected by phyB that acts to suppress shade avoidance in high R/FR light (Franklin et al., 2003). Consequently, the phyB mutant displays a constitutive shade avoidance response and, in all light conditions, has elongated hypocotyls, petioles, and stems, elevated leaves, and early flowering (Whitelam and Johnson, 1982; Somers et al., 1991). However, in low R/FR ratio light, such as occurs under dense canopies, phyA signaling limits excessive shade avoidance, and plant survival is enhanced. The principal evidence is that the phyA mutant is impaired in deetiolation under extreme canopy shade and displays exaggerated shade avoidance responses only in low, but not high, R/FR ratio light (Johnson et al., 1994; Yanovsky et al., 1995; Smith et al., 1997). Moreover, that the phyA phyB double mutant displays more exaggerated shade responses than the phyB mutant alone suggests that in very low R/FR ratio light, phyA antagonizes phyB.
One of the first indications of a link between JA and phyA signaling came from the discovery that a mutation in the Arabidopsis gene FAR-RED-INSENSITIVE219, which was originally identified as a suppressor of the constitutive photomorphogenesis1 mutation (Hsieh et al., 2000), is allelic to jar1 (Staswick et al., 2002). Although jar1 was at first thought not to have a photomorphogenic deficiency, a subsequent investigation reported that both fin219 and the jar1 alleles are insensitive to FR light with respect to hypocotyl growth inhibition (Chen et al., 2007). Other screens have also identified Arabidopsis mutants that are involved in both light and JA signaling. For example, the mutant jasmonate insensitive1 (jin1) was found to be an allele of MYC2 (Lorenzo et al., 2004), which was also identified as the gene for Z-BOX BINDING FACTOR1 (ZBF1) on the basis of its binding to the Z-box of light-regulated promoters (Yadav et al., 2005). Mutations in MYC2/JIN1/ZBF1 lead to hypersensitivity to blue light with respect to hypocotyl growth inhibition and altered blue and FR light gene expression. In a screen for mutants with increased sensitivity to JA, a mutation in LONG HYPOCOTYL1 (HY1), which encodes a plastid heme oxygenase required for phytochrome chromophore biosynthesis, was recovered. Subsequently, it was demonstrated that mutations in either HY1 or HY2, which encodes a phytochromobilin synthase, cause elevated JA production and sensitivity (Zhai et al., 2007). The ecological significance of an interaction between phytochrome and JA signaling was revealed in a study of Arabidopsis plants grown at high density or in illumination supplemented with FR light. In these plants, phyB signaling led to shade avoidance phenotypes and suppressed both JA-mediated gene expression and JA-dependent defenses against an insect herbivore (Moreno et al., 2009).
In rice (Oryza sativa), characterization of the hebiba mutant provided the first evidence of a link between JA signaling and photomorphogenesis in monocots (Riemann et al., 2003). This mutant was isolated in a screen for reduced sensitivity to red light. Significantly, hebiba is male sterile, compromised in JA accumulation, and has been reported to map close to a known JA biosynthesis gene. The light-regulated turnover of the phyA protein is delayed in hebiba and restored by exogenous JA, suggesting a specific role for JA in a key facet of phytochrome A signaling (Sineshchekov et al., 2004; Riemann et al., 2009). In addition, a knockout mutation in the rice homolog of JAR1 is insensitive to FR and blue light inhibition of coleoptile elongation (Riemann, et al., 2008). These two rice mutants point to a key role for JA biosynthesis and signaling in photomorphogenesis, which is largely absent from our current understanding of photomorphogenesis in Arabidopsis. It is possible that photomorphogenic phenotypes in Arabidopsis JA mutants have been overlooked because many of the JA biosynthesis mutants and the JA receptor coi1 are male sterile and may have been lost during screens to identify mutants with altered light perception.
We previously noticed that coi1-16 mutants flower early, and in some growth conditions display elongated, hyponastic petioles, and elongated hypocotyls, phenotypes that are classic indicators of deficient responses to light quality (Whitelam and Johnson, 1982). An upright growth habit had also been reported for the coi1-20 mutant that was isolated in a screen for resistance to P. syringae (Kloek et al., 2001). We have therefore investigated the interaction of JA and phytochrome signaling in light-regulated growth responses. We demonstrate that COI1 and JA signaling is required for some aspects of photomorphogenesis and shade avoidance growth responses, that phy A is required for full sensitivity to growth inhibition by JA, and that phyA and JA signaling are integrated at the level of degradation of a target of the COI1 E3 ligase, the JAZ1 protein.
RESULTS
COI1 Modulates Responses to Light
COI1 Regulates Flowering and Shade Responses
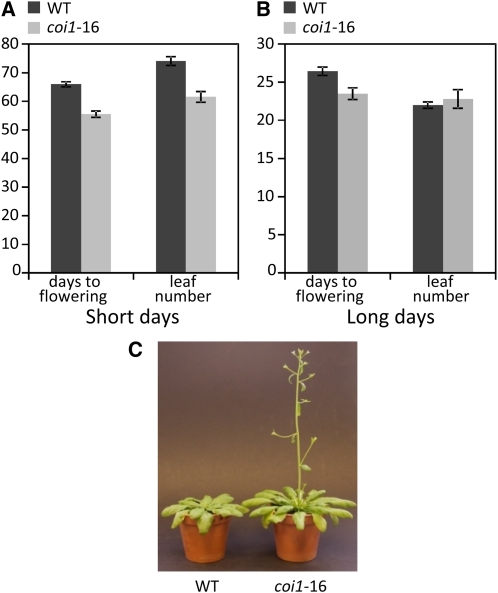
The COI1 gene controls most of the known JA responses in Arabidopsis (Xie et al., 1998). We reasoned that if JA regulates some light responses, then COI1 would be required for at least some of these. We first compared time to flower in the partial loss-of-function mutant coi1-16 and its wild-type background Columbia (Col)-gl1, to which we refer hereafter as the wild type. In short days (SDs), coi1-16 plants flowered earlier than wild-type plants and did so with fewer leaves (Figures 1A and 1C). In long days (LDs), the coi1-16 mutant also flowered earlier, although with a similar number of leaves to the wild type (Figure 1B). However, sensitivity to daylength was retained in respect of both flowering time and leaf number. In this regard, the coi1-16 mutant displays the early flowering response that is reminiscent of phyB mutants (Goto et al., 1991).
Figure 1.
coi1-16 Mutants Flower Early.
(A) Flowering time in SD.
(B) Flowering time in LD. Days to flowering: number of days from germination to first appearance of buds. Leaf number: total number of rosette and cauline leaves on the primary inflorescence, counted after bolting. The wild type and coi1-16 are significantly different with respect to the number of days to flowering and the total leaf number in SD and with respect to days to flowering in LD (Student's t test; P < 0.01). Total leaf numbers in LD are not significantly different. n = 20 (mean ± se).
(C) Eight-week-old wild type (left) and coi1-16 plants in SD.
[See online article for color version of this figure.]
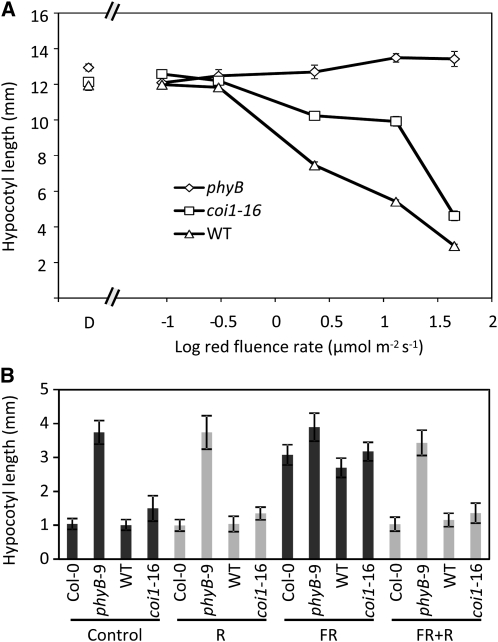
When Arabidopsis plants are shaded by neighboring vegetation, the incident light they receive has a reduced ratio of R/FR because of the selective absorption of red light by the photosynthetic pigments of plants forming the canopy (Ballaré et al., 1990). Under these conditions, Arabidopsis displays shade avoidance responses (Whitelam and Johnson, 1982). To test whether COI1 is required for these phytochrome-dependent responses, we compared the shade avoidance phenotypes of coi1-16 and the wild type. Under low R/FR ratio light, the hypocotyls of coi1-16 were >30% longer than those of the wild type (Figures 2A and 2B). By contrast, when plants were grown under a high R/FR ratio, the hypocotyls of both coi1-16 and the wild type were shorter than under low R/FR and were not significantly different in length (Figures 2A and 2B). In this respect, coi1-16 mutants are reminiscent of phyA mutants, which, when grown in low, but not high, R/FR, display enhanced hypocotyl elongation compared with wild-type controls (Figure 2C; Johnson et al., 1994; Yanovsky et al., 1995; Smith et al., 1997). Our results are consistent with the hypothesis that phytochrome A (phyA) modulates sensitivity to reductions in R/FR perceived by phyB under FR-enriched light conditions, such as those found underneath dense canopies. Moreover, that both coi1-16 and phyA mutants display enhanced shade avoidance only under low R/FR ratio light further suggests that COI1 may be required for the phyA-mediated antagonism of phyB signaling in shade responses. In contrast with coi1-16, however, phyA mutants flower slightly later in LDs and significantly later in SDs than the wild type (Johnson et al., 1994). This suggests that, in addition to its role in phyA-mediated shade avoidance, COI1 is involved in flowering responses mediated by a different mechanism, possibly involving phyB.
Figure 2.
coi1-16 Mutants Display an Exaggerated Shade Avoidance Response to Low R/FR.
(A) Hypocotyl elongation response to low R/FR ratio light of wild-type and coi1-16 seedlings. Average hypocotyl lengths of 30 12-d-old seedlings per genotype/treatment (mean ± sd) are shown. Differences between the wild type and coi1-16 in low R/FR are significant (Student's t test; P < 0.0001).
(B) Photographs of representative 10-d-old wild-type and coi1-16 seedlings under high and low R/FR ratios. A 10-mm ruler is shown.
(C) Hypocotyl elongation response to low R/FR ratio light of Col-0 and phyA-211 seedlings. Average hypocotyl lengths of 30 12-d-old seedlings per genotype/treatment (mean ± sd) are shown. Differences between Col-0 and phyA-211 in low R/FR are significant (Student's t test; P = 0.002).
[See online article for color version of this figure.]
COI1 Is Required during Early Seedling Development for Photomorphogenic Responses to FR Light
Light is an important signal during early seedling development, promoting photomorphogenic responses such as cotyledon expansion and chloroplast development while inhibiting hypocotyl growth, processes that are collectively referred to as seedling deetiolation (Chen et al., 2004). We examined these responses in monochromatic R, FR, and blue (B) light to further dissect the light responses regulated by COI1.
Elongation of Arabidopsis hypocotyls is inhibited in continuous FR light (FRc) in a fluence-dependent, high irradiance response (HIR) mediated by phyA (Whitelam et al., 1993; Shinomura et al., 2000). The seminal evidence for the role of phyA in this response is that null mutants, including phyA-211 (Reed et al., 1994), lack inhibition of hypocotyl elongation in FRc (Whitelam et al., 1993). We therefore tested whether COI1 is also required for this phyA-mediated response. coi1-16 seedlings grown under FRc light (1 μmol m−2 s−1) had significantly longer hypocotyls than wild-type seedlings (Figures 3A and 3C). This key observation indicates that COI1 is necessary for the inhibition of hypocotyl elongation that occurs in FRc light. However, unlike the phyA mutants, which are deficient in all seedling deetiolation responses to FRc light, coi1-16 seedlings, like the wild type, had open, expanded cotyledons and no apical hook. Thus, COI1 is not required for all FR responses. To confirm the requirement for COI1 in the inhibition of hypocotyl elongation, we also tested the coi1-1 null mutant. This mutant, like coi1-16, is recessive, but in addition is infertile and therefore can only be maintained in segregating F2 populations. An F2 population collected from a COI1/coi1-1 plant was grown in FRc light (1 μmol m−2 s−1). This population segregated in a ratio of 18:4 for seedlings with short (≤8 mm) versus long (>8 mm) hypocotyls, whereas wild-type seedlings all had short hypocotyls (≤8 mm) under this condition (Figures 3B and 3C). We presumed that seedlings with the longer hypocotyls in the segregating population were homozygous coi1-1 seedlings and that the seedlings with shorter hypocotyls were heterozygous and wild type. This indicates that the observed phenotypes of the coi1-16 mutants are not allele specific. Together, these results identify a novel role for COI1 in hypocotyl growth inhibition in response to FRc light. Like the coi1-16 mutants, coi1-1 mutants displayed wild-type apical hook opening and cotyledon expansion in response to FR light.
Figure 3.
coi1 Mutants Are Partially Insensitive to FR Light.
(A) Average hypocotyl lengths of 4-d-old wild-type and coi1-16 seedlings grown for 2 d in the dark and transferred for 2 d to FRc at 1 μmol m−2 s−1. n = 10 (mean ± sd). Differences are significant (Student's t test; P < 0.0001).
(B) Frequency distribution of hypocotyl lengths of a wild-type population and a coi1-1+/− segregating population grown as in (A).
(C) Photographs of 4-d-old wild-type, coi1-16, and coi1-1 seedlings grown as in (A) and (B).
(D) Fluence rate response to FRc light. Average hypocotyl lengths of 6-d-old wild-type, coi1-16, and phyA-211 seedlings grown in difference fluence rates of FRc light (730 ± 10 nm). Forty seedlings per genotype/treatment were measured (mean ± se). The wild type and coi1-16 were significantly different at all fluences (Student's t test; P < 0.01). The wild-type background of phyA-211 (Col-0) was not significantly different from the wild-type background of the coi1-16 mutant and was omitted for clarity. Most error bars are smaller than symbols.
We examined the fluence rate dependence of the requirement for COI1 for inhibition of hypocotyl elongation in the range previously demonstrated to be regulated by the HIR. As expected, FRc-grown wild-type seedlings exhibited a fluence-dependent inhibition of hypocotyl growth, whereas the phyA-211 hypocotyls were the same length as dark-grown hypocotyls at all fluences tested (Figure 3D). coi1-16 hypocotyls were significantly longer (by 10 to 25%) than wild-type hypocotyls at FR light fluence rates from 0.01 to 120 μmol m−2 s−1. However, coi1-16 exhibited only a partial defect in FR hypocotyl responses, retaining sensitivity in particular to higher fluences. These results indicated that COI1 is required for part of the FR- and fluence rate–dependent inhibition of hypocotyl elongation.
A Role for COI1 in Photomorphogenesis Extends to Red Light Responses
phyB is the primary photoreceptor for the inhibition of elongation of hypocotyls of Arabidopsis seedlings exposed to continuous R (Rc) light (McCormac et al., 1993). To determine whether COI1 is also required for phyB-mediated seedling responses, we grew seedlings of coi1-16, wild type, and the loss-of-function phyB mutant, phyB-9 (Reed et al., 1994) in Rc light at different fluence rates and compared hypocotyl lengths. Again, as anticipated, the hypocotyl length of the wild type was reduced in approximate proportion to the log of the fluence rate, while at all fluence rates, hypocotyls of phyB-9 were as fully elongated as those of dark-grown seedlings. By contrast, coi1-16 hypocotyls were significantly longer than those of the wild type at fluence rates >0.3 μmol m−2 s−1 (Figure 4A). Similar to the responses in FRc, coi1-16 mutants displayed only partial insensitivity to Rc-mediated inhibition of hypocotyl growth. Additionally, coi1-16 seedlings, similar to wild-type seedlings, had open, expanded, and green cotyledons and no apical hook, indicating that COI1 is not required for all R light responses. Significantly, hypocotyls of dark-grown coi1-16 seedlings were not different in length from those of their wild-type counterparts (Figure 4A), indicating that the altered hypocotyl length of light-grown seedlings is light dependent and does not result from a general defect in growth.
Figure 4.
COI1 Is Required for Hypocotyl Growth Inhibition Responses to Rc Light but Is Not Required for the phyB-Mediated Low Fluence Response to EOD-FR.
(A) Fluence rate response to Rc light. Average hypocotyl lengths of 6-d-old wild-type, coi1-16, and phyB-9 seedlings grown in difference fluence rates of Rc light (660 ± 10 nm). Forty seedlings per genotype/treatment were measured (mean ± se). The wild type and coi1-16 were significantly different at the three highest fluences of red light tested only (45, 13, and 2.3 μmol m−2 s−1; Student's t test; P < 0.01) but not at the lower fluences (0.3 and 0.09 μmol m−2 s−1). The wild-type background of phyB-9 (Col-0) was not significantly different from the wild-type background of the coi1-16 mutant and was omitted for clarity. Error bars are smaller than symbols.
(B) EOD-FR hypocotyl response. Hypocotyl lengths of 3-d-old white light/SD-grown Col-0, phyB-9, wild-type, and coi1-16 plants given 5 min red, 10 min FR, or 10 min FR followed by 5 min red light at the end of every light cycle. n = 10 (mean ± sd).
These data indicate that COI1 is necessary for light-induced suppression of hypocotyl elongation in response to both Rc and FRc light and may therefore be required for both phyA and phyB signaling during early seedling development. However, coi1-16 was only slightly insensitive to suppression of hypocotyl elongation by continuous blue (Bc) light (see Supplemental Figure 1 online). While the major receptors for blue light responses are the cryptochromes (Ahmad and Cashmore, 1993), suppression of hypocotyl elongation by Bc light is also partly mediated by phyA (Johnson et al., 1994). The slight insensitivity to Bc in coi1-16 could also reflect loss of COI1-dependent phyA activity.
Low Fluence Responses to Red Light Do Not Involve COI1
We showed in the preceding section that COI1 is required for Rc light inhibition of hypocotyl elongation, a classic HIR mediated by phyB. PhyB acts as a molecular switch that is activated by R light and inactivated by FR light, to regulate R/FR reversible low fluence responses (Nagatani et al., 1991). We tested whether COI1 was also required for phyB-mediated low fluence responses by monitoring the response to end-of-day (EOD) FR. Wild-type, coi1-16, and phyB-9 seedlings were grown in white light (SD) and at the end of each day were exposed to FR light for 10 min for three consecutive days. As anticipated, wild-type, but not phyB-9, seedlings showed FR-mediated elongation of hypocotyls (Nagatani et al., 1991) (Figure 4B). A subsequent 5-min red light treatment resulted in hypocotyl growth inhibition of wild-type seedlings similar to that of white light or white light plus EOD red light–grown control seedlings. Significantly, coi1-16 seedlings responded like wild-type seedlings to the EOD-FR treatment.
Although the previous section identified a role for COI1 in HIR to Rc light, our data above show that COI1 is not required for the R/FR reversible phyB-mediated low fluence EOD-FR response. Because we show here that COI1 is required for shade responses to low, but not high, R/FR and additionally for HIR to FRc light, both of which are mediated by phyA, we investigated further the interaction of COI1 and phyA signaling.
JA Regulates the Expression of the FR-Inducible Transcription Factors HFR1 and PIL1 in a COI1-Dependent Manner
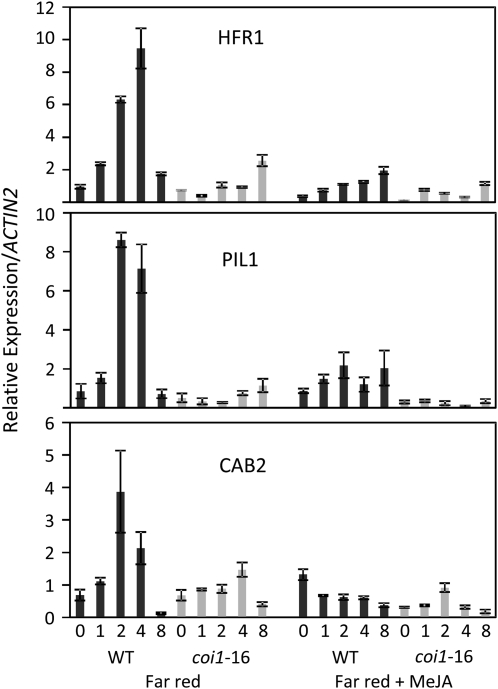
We showed in the previous section that COI1 is required for several FR-dependent responses, including inhibition of hypocotyl elongation and shade avoidance. We reasoned that COI1 might also be required for regulation of expression of genes involved in FR signaling. FR induces expression of the basic helix-loop-helix (bHLH) transcription factors encoded by HFR1 and PIL1, which regulate responses to FRc and low R/FR ratio light (Fairchild et al., 2000; Salter et al., 2003; Sessa et al., 2005), and the chlorophyll a/b binding protein gene, CAB2, which is often employed as a marker of light signaling (Karlin-Neumann et al., 1988). We grew the wild-type and coi1-16 seedlings in the dark and transferred them to FRc light and used quantitative RT-PCR (qRT-PCR) to compare levels of transcripts of HFR1, PIL1, and CAB2 (Figure 5). As expected, the expression of HFR1, PIL1, and CAB2 was elevated in wild-type seedlings transferred to FR light, but this induction was significantly attenuated in coi1-16 seedlings. Notably, the COI1-dependent FR induction of HFR1, PIL1, and CAB2 was itself suppressed by JA treatment. We conclude that COI1 is required for both the FR induction and for the JA suppression of transcription of HFR1, PIL1, and CAB2.
Figure 5.
Light-Regulated Genes Are Induced by FR in a COI1-Dependent Manner and Are Suppressed by Exogenous JA.
Expression of HFR1, PIL1, and CAB2 genes in 6-d-old etiolated wild-type or coi1-16 seedlings exposed to FR or FR + 20 μM methyl jasmonate (meJA) for the number of hours indicated. Expression levels (mean ± sd) of three technical replicate qRT-PCR reactions are expressed relative to ACTIN2.
COI1 Is Required for the FR Light–Preconditioned Block of Greening
We examined whether other phyA responses required COI1. Wild-type Arabidopsis seedlings raised in FRc light and subsequently transferred to white light fail to produce chlorophyll and they die. This preconditioned block of greening by FR (Barnes et al., 1996) is in part a consequence of the phyA-mediated repression of the light-dependent NADPH-protochlorophyllide oxidoreductase A gene (PORA). This repression causes hyperaccumulation of the chlorophyll precursor, protochlorophyllide-F632, and irreversible photooxidative damage on transfer of seedlings to white light (Runge et al., 1996). The phyA mutants do not exhibit FR repression of PORA and are therefore resistant to FR-mediated block of greening. To examine whether COI1 was required for this FR response, we raised wild-type, phyA-211, and coi1-16 seedlings in FRc light (10 μmol m−2 s−1) for 6 d, transferred these to white light for 2 d, and then measured their chlorophyll content. Wild-type seedlings had bleached cotyledons at harvest and did not survive, whereas both phyA-211 and coi1-16 cotyledons were detectably green following transfer from FR to white light, and their chlorophyll contents were significantly higher than the wild type (Figures 6A and 6B).
Figure 6.
COI1 Is Required for the FR Light–Preconditioned Block of Greening Response.
(A) Wild-type, coi1-16, and phyA-211 seedlings grown in FRc (10 μmol m−2 s−1) for 6 d and transferred to white light/LD (100 μmol m−2 s−1) for 2 d.
(B) Chlorophyll accumulation (μg/mg tissue) in plants grown as in (A).
(C) Expression of the PORA gene in 6-d-old etiolated wild-type, coi1-16, and phyA-211 seedlings exposed to FR (10 μmol m−2 s−1) for the number of hours indicated. Expression levels (mean ± sd) of three technical replicate qRT-PCR reactions are expressed relative to ACTIN2.
We tested whether the resistance to FR block of greening in coi1-16 was associated with failure to repress PORA, as it is in phyA-211. Wild-type, phyA-211, and coi1-16 seedlings were grown in the dark for 6 d and transferred into FRc light for 4 or 24 h. mRNA was extracted and PORA expression was measured by qRT-PCR. As expected, PORA was depleted in wild-type seedlings illuminated with FRc for 4 h and was undetectable after 24 h (Figure 6C). PORA was not depleted in phyA-211 and was still detectable after 24 h of FRc light. Significantly, PORA expression in coi1-16 was higher than in the wild type, both in the dark and after exposure to FRc light. The elevated basal level of PORA in coi1-16 may explain its survival of FR light–preconditioned block of greening and indicates that that COI1 contributes to repression of PORA during skotomorphogenesis and during FR light–mediated photomorphogenesis.
JA Biosynthesis and Signaling Regulate Responses to FR Light
JA Biosynthesis and Signaling Are Required for FR-Dependent Photomorphogenesis
In the preceding sections, we showed that COI1 is required for the phyA-mediated FR-dependent inhibition of hypocotyl elongation. Because COI1 is a component of the JA signal pathway, we wished to determine whether COI1 in particular, or JA signaling in general, is required for the inhibition of hypocotyl elongation by FR. Mutants defective in synthesis of JA (aos and opr3) were grown in FRc light (1 μmol m−2 s−1) and in darkness, and their hypocotyl lengths were measured. We represent hypocotyl growth as a percentage of dark-grown controls. The average length of Col-0 wild-type hypocotyls in FRc was 56.5% (±5.2) of that in darkness, while hypocotyls of the phyA-211 mutants in FR light were 98.5% ± 6.3% of their length in darkness (Figure 7A). The hypocotyls of the aos mutant were inhibited by FR light to 78.4% (±9.4) of their length in darkness, which was significantly less (P < 0.0001) than the inhibition of Col-0 wild-type hypocotyls. At variance with this, the null opr3 mutant was not significantly different from its wild type (Wassilewskija) with respect to percentage of hypocotyl growth inhibition by FRc light (wild type, 72.9% ± 4.6%; opr3, 72.3% ± 4.3%).
Figure 7.
JA Biosynthesis and Signaling Genes Are Involved in FR Responses.
(A) Average percentage of hypocotyl growth inhibition by FR of 4-d-old (2 d dark, 2 d FR at 1 μmol m−2 s−1) JA and JA-Ile biosynthesis mutants aos, jar1-1/fin219, and opr3, along with their wild-type parental lines (Col-0 and Wassilewskija) and the phyA-211 and coi1-16 mutants for comparison. The aos and opr3 mutants are taller than the wild type in the dark so bars represent percentage of dark-grown (4 d dark) hypocotyl length. The differences in percentage of growth inhibition between the wild type and the mutants were tested by Student's t test, and the significant results (P < 0.05) are indicated by asterisks. n = 20 (mean ± sd).
(B) Average percentage of hypocotyl growth inhibition by FR of 4-d-old (2 d dark, 2 d FR at 1 μmol m−2 s−1) JA signaling mutants jin1-1 and jai3, the wild type (Col-0), and the phyA-211 mutant.
(C) Expression of AOC1, MYC2/JIN1/ZBF1, JAZ1, and VSP1 genes in 6-d-old wild-type and coi1-16 etiolated seedlings exposed to FR (10 μmol m−2 s−1) for the number of hours indicated. Expression levels (mean ± sd) of three technical replicate qRT-PCR reactions are expressed relative to ACTIN2.
We also analyzed the FRc growth phenotype of the mutants fin219 and jar1-1. Our results are consistent with former studies (Hsieh et al., 2000; Chen et al., 2007): in FRc light, fin-219 and jar-1 hypocotyl lengths were inhibited 84.3% ± 4.6% and 62.4% ± 4.3%, respectively, of their dark-grown lengths and significantly less than the inhibition of hypocotyls of their wild type, Col-0 (fin219, P < 0.0001; jar1-1, P < 0.01).
We investigated the role of signaling components that act downstream of JA biosynthesis. COI1 is involved in JA perception, and Figure 3 shows that coi1-16 is less sensitive to FRc inhibition of hypocotyl length than the wild type. Similarly, Figure 7A shows that in FRc, coi1-16 hypocotyls were 79% of the length of dark-grown hypocotyls. These results indicate that COI1 is required for FRc inhibition of hypocotyl elongation. We also examined the response to FRc of lines with mutations in genes for negative (JASMONATE INSENSITIVE3 [JAI3]/JAZ3) and positive (MYC2/JIN1/ZBF1) regulators of JA signaling. Mutants of these genes, jai3 and jin1-1, respectively, were originally isolated by virtue of their insensitivity to growth inhibition by JA (Lorenzo et al., 2004).
In FRc light, wild-type Col-0 hypocotyls were 66.2% (±7.5) of their dark-grown length, but the jai3 and jin1-1 mutant hypocotyls were less strongly inhibited to 78.5% (±6.6) and 76.6% (±9.1) of their dark-grown length, respectively (Figure 7B), and the reduced sensitivity of the mutants compared with the wild type was significant (P < 0.0001 and P = 0.011, respectively). It was previously reported that there was no difference between the hypocotyl growth response of the wild type and myc2 (jin1) mutants to FRc light (Yadav et al., 2005). The difference we report here may be due to the lower fluence rate, 1 μmol m−2 s−1 we have used, compared with the 90 μmol m−2 s−1 fluence rate employed by Yadav et al. (2005). In support of this explanation, others (Staswick et al., 2002; Chen et al., 2007) report that hypocotyl length of jar1-1 mutants differ from the wild type in FRc only at fluences <10 μmol m−2 s−1. Alternatively, Yadav et al. (2005) used T-DNA mutants myc2-2 and myc2-3 rather than the jin1-1 allele, and there may be allele-specific responses that could explain the difference in our results. Our experiments demonstrate that wild-type alleles of genes JAI3/JAZ3 and MYC2/JIN1/ZBF1, whose products act downstream of COI1, are necessary components of the JA signaling required for FR light inhibition of hypocotyl elongation. These data indicate that JA synthesis and signaling contribute to the activation of phyA-mediated inhibition of hypocotyl elongation by FR light. OPR3, however, is not required for this response.
Expression of JA Biosynthesis and Signaling Genes Is Regulated by FR Light
The transcription of JA biosynthesis genes is activated by JA in a COI1-dependent manner, apparently in a feedback loop (Devoto et al., 2005). We sought to establish whether genes involved in JA biosynthesis and signaling are regulated transcriptionally by FR light, as has been demonstrated in rice (Haga and Iino, 2004). For this, seedlings of the wild type and coi1-16 were grown in darkness and transferred to FRc, and samples taken at intervals were assayed by qRT-PCR for transcription of the JA biosynthesis gene ALLENE OXIDE CYCLASE1 (AOC1), the signaling genes JAZ1 and MYC2/JIN1/ZBF1, and the response gene VEGETATIVE STORAGE PROTEIN1 (VSP1), which is commonly used as a marker of JA responses (Berger et al., 1995). FR caused transient induction of all genes, and this induction was attenuated in the coi1-16 mutants (Figure 7C). Evidently, FRc induces COI1-dependent transcription of genes involved in JA biosynthesis and signaling.
JAR1, JAZ3, and MYC2/JIN1/ZBF1 Are Required for Shade Responses
We show above that JAR1, COI1, JAZ3, and MYC2 were required for the FRc inhibition of hypocotyl elongation by phyA. We therefore tested whether these genes were also required for shade responses in low R/FR ratio light. coi1-16, jar1-1, jin1-1, and jai3 (Figures 8A and 8B), and coi1-1 and fin219 (see Supplemental Figure 2 online) showed enhance shade avoidance at low, but not high, R/FR ratio light compared with the wild type. Together, these results indicate that JA biosynthesis and signaling are involved in the adaptive response of plants to deep canopy shade.
Figure 8.
JA Biosynthesis and Response Mutants Show Exaggerated Shade Responses to Low R/FR.
(A) Photographs of representative 10-d-old wild-type, jin1-1, jai3, and jar1-1 seedlings grown in high (left) and low (right) R/FR. Arrows denote the ends of the hypocotyls.
(B) Average percentage of increase in hypocotyl length (mm) by low R/FR compared with high R/FR of seedlings grown as in (A). The differences between the wild type and the mutants were tested by Student's t test and are significant (P < 0.05). n = 20 (mean ± sd).
phyA Regulates Sensitivity to JAs
phyA Is Required for JA-Mediated Root Growth Inhibition
The foregoing demonstrates that JA is required for phyA signaling. We therefore examined whether this is reciprocated and whether phyA is required for JA signaling. In Arabidopsis, root growth is inhibited by exogenous JA in a dose-dependent manner (Staswick et al., 1992; Feys et al., 1994; Lorenzo et al., 2004). We therefore grew the wild type, coi1-16, Col-0, and phyA-211 in LD for 8 d on media containing different concentrations of MeJA. As expected, wild-type roots were progressively inhibited by increasing MeJA concentration, whereas the coi1-16 mutant was significantly less sensitive to root growth inhibition by MeJA (Figures 9A and 9B). The phyA-211 mutant was also significantly less sensitive to root growth inhibition by MeJA than its wild type (P < 0.001) at all concentrations, though less so than coi1-16 (Figures 9A and 9C). Col-0 and phyA-211 roots were not significantly different in length in the absence of JA, indicating that the altered root phenotype of phyA-211 was JA dependent and not due to a general defect in root growth. These data indicate that a functional phyA gene is required for part of the root growth inhibition by exogenous JA.
Figure 9.
phyA Mutants Display Altered JA-Related Phenotypes.
(A) From the left: 8-d-old white light–grown wild type, coi1-16, Col-0, and phyA-211 plants grown on media containing 0, 10, or 50 μM MeJA.
(B) Dose–response curves. Root lengths of 8-d-old wild-type and coi1-16 plants (left) and Col-0 and phyA-211 plants (right) grown on different MeJA concentrations. n = 20 (mean ± se). Col-0 and phyA-211 are not significantly different from each other in the absence of MeJA (Student's t test; P = 0.77) but are significantly different at all concentrations of MeJA (1, 5, 10, and 25 μM; Student's t test; P < 0.01, 50 μM; P < 0.05). Error bars are smaller than symbols.
(C) Fold induction of VSP1 in wild-type (Col-0) and phyA seedlings in response to treatment for 8 h with 20 μM MeJA. Expression levels (mean ± sd) of three technical replicate qRT-PCR reactions are expressed relative to ACTIN2.
(D) OPDA content of 3-d-old (2 d dark, 1 d FRc 10 μmol m−2 s−1 or 3 d dark) phyA-211, coi1-16, and control seedlings. phyA-211 seedlings contain significantly more OPDA than Col-0 in both growth conditions (Student's t test; P < 0.01). n = 4 (mean ± sd).
[See online article for color version of this figure.]
phyA Is Required for JA-Mediated Induction of VSP1
Arabidopsis VSP1 is transcriptionally activated by JA and wounding in a COI1-dependent manner (Benedetti et al., 1995; Berger et al., 1995). Transcription of VSP1 is transiently induced by FR light and is COI1 dependent (Figure 7C). We tested whether phyA was required for the transcription of VSP1. For this, 6-d-old Col-0 and phyA-211 seedlings were transferred to media containing 20 μM MeJA or fresh control media and incubated in the dark or in FRc light for 8 h. RNA was extracted and VSP1 was measured by qRT-PCR. In Col-0, VSP1 MeJA treatment increased VSP1 expression over the untreated control by 226-fold in the dark and 314-fold in FRc light (Figure 9C). By contrast, MeJA increased VSP1 expression phyA mutants, over the untreated control, by 3.9-fold in the dark and by 30-fold in FR light (Figure 9C). Therefore, in young seedlings, transcriptional activation of VSP1 by MeJA requires phyA.
phyA Mutants Have Altered OPDA Levels
Treatment of etiolated rice seedlings with red light resulted in accumulation of JA (Riemann et al., 2003). We therefore investigated whether in Arabidopsis FRc treatments that promote photomorphogenesis also result in accumulation of JA or its derivatives. We grew the wild type, coi1-16, Col-0, and phyA-211 for 3 d in the dark or 2 d dark followed by 24 h of FR light (10 μmol m−2 s−1). Seedlings were flash-frozen and analyzed for JA content. JA and JA-Ile were undetectable in two independent experiments in both dark and FR, in all genotypes and all replicates. Cis-(+)-12-oxophytodienoic acid (OPDA), however, was abundant in both dark-grown and FR-treated seedlings (Figure 9D), consistent with a former study (Brüx et al., 2008). FRc treatment did not affect the OPDA content of seedlings, but the OPDA content of phyA-211 (400 to 500 pmol/g fresh weight) was significantly higher than that of Col-0 (235 to 240 pmol/g fresh weight, P < 0.01). Interestingly, the coi1-16 plants had significantly reduced OPDA content, supporting the hypothesis that there is feedback regulation of oxylipin biosynthesis downstream of JA perception (Stintzi and Browse, 2000).
phyA Is Not Required for Wound- and JA-Mediated Aerial Growth Inhibition, Anthocyanin Accumulation, or Chlorophyll Degradation
Responses of wild-type Arabidopsis to exogenous JA include inhibition of root growth, inhibition of aerial growth, accumulation of anthocyanin, and degradation of chlorophyll, and these responses are absent from coi1 mutants (Feys et al., 1994; Ellis and Turner, 2002; He et al., 2002). To establish whether phyA involvement in JA signaling is specific to particular JA-mediated processes, we examined additional JA responses in phyA-221. There were no significant differences between MeJA-treated phyA-211 and the wild type with respect to aerial growth inhibition, anthocyanin accumulation, or chlorophyll degradation, indicating that phyA is not required for these responses to JA (see Supplemental Figure 3 online).
Exogenous and wound-induced JAs inhibit mitosis, resulting in growth inhibition (Zhang and Turner, 2008). To test whether phyA was required for wound-induced growth inhibition, we repeatedly wounded wild-type, coi1-16, and phyA-211 plants over 10 d and recorded their size. Both the wild type and phyA-211 showed a similar level of growth inhibition, whereas coi1-16 was not significantly inhibited, indicating that growth inhibition by endogenous wound-induced JA does not require phyA (see Supplemental Figure 3 online).
Light and JA Pathways Converge at JAZ1
phyA Regulates Wound- and JA-Mediated JAZ1 Degradation
The JAZ proteins are repressors of JA-responsive gene expression, and a critical step in JA signaling is their COI1-mediated degradation (Chini et al., 2007; Thines et al., 2007). We show that JAZ1 transcription is enhanced by FR light (Figure 7C) and that the jai3 mutant hypocotyl was insensitive to inhibition by FRc and by low R/FR ratio light (Figures 7C and 8), demonstrating a role for JAZ genes in phyA-mediated responses. We therefore investigated whether phyA was required for JAZ degradation. The 35S:JAZ1-β-glucuronidase (GUS) transgene (Thines et al., 2007) was introduced into the phyA-211 mutant by crossing. Seedlings containing 35S:JAZ1-GUS in the wild type, in plants segregating for COI1/coi1-1, and in phyA-211 were grown in axenic culture for 10 d. In control plants, GUS activity was detected in all genotypes (Figure 10A, left panel). A single wound to a leaf caused the disappearance within 1 h of JAZ1-GUS from the aerial parts and roots of wild-type plants, as previously reported (Zhang and Turner, 2008) but not from one-quarter of the segregating COI1/coi1-1 population, which we predict to be coi1-1 homozygotes (Figure 10A, middle panel). Significantly, JAZ1-GUS was not degraded in the aerial parts of phyA-211 mutants, which resembled coi1-1 mutants in this respect. JAZ1-GUS was degraded, however, in the roots of phyA-211 plants.
Figure 10.
phyA Is Required for JA- and Wound-Mediated JAZ1 Degradation.
(A) Photographs of representative control, wounded (1 h), and 50 μM MeJA-grown 10-d-old seedlings of Col-gl1/35S:JAZ1-GUS (wild type), 35S:JAZ1-GUS/phyA-211, and 35S:JAZ1-GUS/coi1. Seedlings were grown on sucrose-containing media in LD conditions.
(B) On media lacking sucrose, JAZ1 is stabilized in aerial parts and degraded in roots of phyA-211 seedlings in the absence of any deliberately applied stress or hormone treatment. Photographs of 12-d-old seedlings of 35S:GUS, 35S:JAZ-GUS, and 35S:JAZ-GUS/phyA-211.
(C) Hypocotyl elongation response to low R/FR ratio light in 35S:JAZ1-GUS and control 35:GUS seedlings. Hypocotyl lengths (mean ± sd) of ≥15 seedlings per genotype/treatment are shown. Differences in low R/FR are significant (Student's t test; P < 0.0001).
The same genotypes were grown for 10 d on media containing 50 μM MeJA. Wild-type plants were very stunted, as expected, and JAZ1-GUS was completely degraded in these conditions (Figure 10A, right panel). The coi1-1 homozygotes showed complete insensitivity to growth inhibition as expected for the null allele, and JAZ1-GUS was not degraded in this genotype, as was shown previously (Thines et al., 2007). phyA-211 was as stunted as the wild type at this concentration of MeJA, as previously found (Figures 9A and 9B; see Supplemental Figure 3 online). However, similar to phyA-211 wounded plants, JAZ1-GUS was not degraded in the aerial parts but was degraded in the roots (Figure 10A, right panel). These results indicate that both phyA and COI1 are required for JAZ1 degradation in green tissues of wounded or MeJA-treated plants but that phyA is not required for JAZ1 destabilization in the root. Significantly, the strong GUS activity in the green tissues of the MeJA-treated, and severely stunted, phyA-211 mutants indicates that stabilization of JAZ1 does not result in insensitivity to growth inhibition by MeJA.
The preceding wound and JA experiments were on seedlings grown in standard experimental axenic conditions on sucrose-containing media in LDs. We previously noticed that JAZ1-GUS is less stable in seedlings raise in conditions we typically use for R/FR light experiments: media without sucrose and in continuous light. Consequently, we grew lines containing 35S:JAZ1-GUS in the wild type and in phyA backgrounds in media lacking sucrose, in continuous light, and compared GUS expression with lines containing 35S:GUS as control. In these conditions, JAZ-GUS was destabilized in the aerial parts of wild-type plants in the absence of any deliberate stress or JA, and GUS activity was detectable, although at reduced level, in roots (Figure 10B). In the phyA background, however, JAZ-GUS was present at high level in the aerial parts but was undetectable in roots, suggesting that phyA regulates JAZ1 stability differently in the shoot and the root.
JAZ1 Plays a Role in Shade Responses
Having established that JAZ1 stability is in part governed by phyA, we examined the shade response of 35S:JAZ1-GUS seedlings. We previously noted that, like most JA loss-of-function mutants, this line in which JAZ-GUS is expressed ectopically had elongated petioles in low light (H. Whitfield and J.G. Turner, unpublished data). Therefore, we grew seedlings containing 35S:JAZ1-GUS and control 35S:GUS seedlings in high or low R/FR ratio light as before. 35S:JAZ1-GUS seedlings, like coi1 and other JA mutants, and phyA had exaggerated shade responses in low, but not high, R/FR ratio light (Figure 10C). Therefore, the 35S:JAZ1-GUS seedlings had an exaggerated shade avoidance phenotype similar to that of the JA and FR light mutants, including jar1, fin219, coi1-16, coi1-1, jai3, jin1-1, and phyA.
DISCUSSION
We demonstrate roles for JA signaling in phyA-regulated hypocotyl growth inhibition and chlorophyll biosynthesis in Arabidopsis during FR-induced photomorphogenesis and in suppression of elongation growth in low R/FR ratio light when shade avoidance responses are attenuated. The JA receptor COI1 was required for the FR-induced expression of the light-regulated genes HFR1, PIL1, and CAB2, and exogenous JA suppressed this process. The influence of JA signaling on phyA-mediated responses was reciprocated: phyA was partially insensitive to JA-induced root growth inhibition and had altered oxylipin content and attenuated induction of the JA response gene VSP1.
Therefore, our work supports previous reports that Arabidopsis mutants defective in JA signaling (fin219/jar1, Hsieh et al., 2000; Staswick et al., 2002; Chen et al., 2007; myc2/jin1, Lorenzo et al., 2004; Yadav et al., 2005) express an altered light phenotype and that Arabidopsis mutants defective in light signaling (hy1/hy2, Zhai et al., 2007; det3, Brüx et al., 2008; phyB, Moreno et al., 2009) display JA-related phenotypes. In this context, we show that FR light regulated the expression of the JA-induced genes AOC1, JIN1, JAZ1, and VSP1. Crucially, integration of the JA and phyA signal pathways was at the level of JAZ1 stability: phyA was required for JAZ1 destabilization by wounding or exogenous JA. The photodestruction of phyA is in part controlled by JA, as was demonstrated in the hebiba mutant of rice (Sineshchekov et al., 2004; Riemann et al., 2009). Together, our results establish that the control of protein stability is central to the integration of these two signal pathways.
JA Signaling Regulates Diverse phyA Responses
phyA has a profound influence on seedling development, particularly in adverse environmental conditions. Thus, phyA seedlings exhibit exaggerated elongation growth under increasing canopy density of wheat (Triticum aestivum; Yanovsky et al., 1995). In extreme conditions, seedlings fail to deetiolate properly and show reduced survival rates. phyA therefore suppresses exaggerated growth responses in conditions in which a plant must balance limited resource between extension growth and physiological and developmental processes such as photosynthesis and seed production that allow survival.
Our work shows that JA mutants, similar to phyA, display exaggerated shade responses in low, but not high, R/FR ratio light. JAs also downregulate photosynthetic gene expression (Ueda and Kato, 1980; Reinbothe et al., 1993; Jung et al., 2007). We speculate that plants growing in photosynthetically poor radiation regulate JA action through the phyA photoreceptor. They do so to prevent inappropriate extension growth, decrease energy-costly defense mechanisms, and increase photosynthetic output.
coi1 mutant seedlings were also compromised in a subset of FR light–mediated HIRs during photomorphogenesis, including hypocotyl growth inhibition and the preconditioned block of greening. However, they showed normal responses with respect to opening of the apical hook and cotyledon expansion. Like coi1, most of the JA biosynthesis and response mutants displayed reduced sensitivity to FR light especially in hypocotyl growth inhibition. The opr3 mutant, which is blocked in JA biosynthesis (Stintzi and Browse, 2000; Stintzi et al., 2001), displayed a wild-type response to FR light. This observation may instead implicate an intermediate in the JA biosynthesis pathway as the oxylipin(s) directly responsible for hypocotyl growth inhibition, as was suggested by Brüx et al. (2008). Exogenous JA in growth media has a dramatic effect on growth inhibition of roots and aerial parts of light-grown plants, but the effect on hypocotyl growth is mild (Brüx et al., 2008). This suggests the possibility that the JA component of hypocotyl growth inhibition is regulated downstream of biosynthesis and perception of JA. In that case, the role of COI1 and the downstream signaling components in phyA-mediated hypocotyl deetiolation might be in regulating a feedback loop for JA synthesis.
JA responses extend also to phyB signaling in Arabidopsis (Moreno et al., 2009) and in rice, as demonstrated by the hebiba mutant, which is compromised in red light coleoptile growth responses (Riemann et al., 2003). We show that coi1 mutants are partially insensitive to red light–mediated hypocotyl growth inhibition. Thus, JA signaling contributes to all major phytochrome responses during seedling deetiolation.
Chlorophyll Biosynthesis Is Suppressed by COI1
JAs downregulate expression of genes involved in photosynthesis and reduce photosynthetic output (Ueda and Kato, 1980; Reinbothe et al., 1993; Jung et al., 2007). They also promote turnover of proteins involved in light capture and energy-generating processes during dark-induced senescence (Tsuchiya et al., 1999; He et al., 2002). We show that COI1 suppresses chlorophyll biosynthesis: PORA gene expression is higher in the coi1-16 mutant, and coi1 seedlings, like phyA, are resistant to the FR preconditioned block of greening. Intriguingly, coi1 seedlings also accumulate higher levels of chlorophyll on transfer from dark to white light (F. Robson and J.G. Turner, unpublished data). The hebiba mutant seedlings also have higher levels of active protochlorophyllide than their wild-type counterparts in the dark and in FR (Sineshchekov et al., 2004). This indicates that JA signaling regulates chlorophyll biosynthesis during skotomorphogenesis and during photomorphogenesis in FR-enriched light, in both monocots and dicots.
phyA Controls JA Sensitivity and Response to Mechanical Wounding
phyA mutants had reduced JA-mediated root growth inhibition. The phenotype was mild, however, and only apparent at low concentrations of JA, which may explain why phyA has not been identified in previous screens for JA-insensitive root growth. Interestingly, we demonstrate that OPDA is increased in phyA mutants and markedly reduced in coi1-16 mutants. We interpret this in terms of feedback regulation of oxylipin biosynthesis downstream of JA perception (Stintzi and Browse, 2000; Sasaki et al., 2001).
phyA mutants also displayed reduced JA induction of VSP1 (Figure 9C). VSP proteins are acid phosphatases (Thaller et al., 1998) that may have a role in defense against herbivores (Berger et al., 2002; Liu et al., 2005). phyB signaling suppresses responses to herbivory (Ballaré, 2009; Moreno et al., 2009). Additionally, plants exposed to low R/FR have reduced resistance to insect pests, suggesting that in light conditions that favor extension growth, defenses may be compromised (Izaguirre et al., 2006). The phytochromes are known also to modulate the SA pathway. For example, the phyA phyB double mutant was compromised in its defense response to the bacterial pathogen P. syringae (Genoud et al., 2002); moreover, full activation of systemic acquired resistance was dependent on a prolonged light period during the early stage of plant–pathogen interaction (Griebel and Zeier, 2008).
phyA regulates transcription in response to FR light (Tepperman et al., 2001; Wang et al., 2002; Devlin et al., 2003). The defense-related genes Thionin2.2 and PR5 and a disease resistance protein of the TIR-NBS-LRR class are regulated by phyA during shade responses (Devlin et al., 2003). It will be interesting, therefore, to compare the transcript profiles of phyA and the wild type with respect to responses to mechanical wounding, herbivory, or plant diseases.
We observed altered JA-regulated VSP1 expression and altered OPDA content of phyA mutants in the light and in the dark. The latter is at odds with conventional understanding that light is a requirement for phyA action. The most likely explanation of phyA activity in our dark-grown seedlings is that phyA protein is reporting a very low fluence response to our green safelight.
The Role of JAZ1
We show that JAZ1 destabilization requires signals from the phyA and JA signal pathways. A crucial question arises: What is the role of JAZ1 in phytochrome and JA signaling? Most knockout mutations in JAZ genes do not manifestly show JA-regulated growth inhibition phenotypes, suggesting that there is functional redundancy among genes in this family (Chini et al., 2007; Thines et al., 2007). The functions of JAZ proteins are revealed in the phenotypes of a dominant JAZ mutant (jai3/jaz3; Lorenzo et al., 2004) or from transgenic plants (35S:JAZ3Δ and 35S:JAZ1Δ) that express JAZ proteins lacking the C-terminal motif required for JAZ destabilization (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Chung et al., 2008). It is proposed that these gain-of-function mutant proteins interfere with COI1 signaling by blocking the degradation of other JAZ proteins (Chini et al., 2009; Chung and Howe, 2009). Although it is difficult to attribute particular phenotypes to individual JAZ proteins, dominant 35S:JAZ1Δ transgenic plants display a range of JA-related phenotypes: they are male sterile, have reduced defense against P. syringae, and show altered JA-mediated root growth inhibition and gene expression (Thines et al., 2007; Melotto et al., 2008). They also display reduced resistance to the generalist herbivore Spodoptera exigua (Chung et al., 2008). Transgenic lines that have reduced JAZ1 expression have increased sensitivity to root growth inhibition by MeJA (Grunewald et al., 2009). This is consistent with JAZ1 suppressing JA-mediated root growth inhibition. The phyA mutant also displayed slightly reduced sensitivity to JA-mediated root growth inhibition. However, in this mutant, JAZ1-GUS was constitutively degraded in the roots (Figures 10A and 10B). Although phyA clearly regulates JAZ1 stability, phyA may also regulate other components of JA signaling that contribute to the overall mutant phenotype.
The most striking effect of the phyA mutation was that JAZ1-GUS was stabilized in green tissues of seedlings that had either been wounded or grown on 50 μM MeJA for an extended period. JAZ1-GUS is degraded in wild-type plants by these treatments within 1 h (Thines et al., 2007; Zhang and Turner, 2008). Significantly, the aerial parts of the phyA mutant displayed wild-type sensitivity to JA-mediated growth inhibition, despite JAZ1-GUS stabilization. This suggests that JAZ1 destabilization is not required for JA-mediated inhibition of growth of green tissues. The 35S:JAZ1-GUS transgenic plants had exaggerated shade avoidance responses in low, but not high, R/FR ratio light (Figure 10C), similar to that of coi1-16, other JA mutants, and phyA mutants. Similar phenotypes have been reported in plants overexpressing members of a JAZ-related family of proteins: the Arabidopsis ZIM/TIFYs (Shikata et al., 2004). These results, taken together with our findings, highlight a role for the JAZ and related ZIM/TIFY families of transcriptional repressors in shade responses. Looking ahead, it is possible that JAZ proteins regulate responses beyond those presently associated with JA or phytochromes revealed here.
Integration of Signal Pathways through Common bHLH Transcription Factors
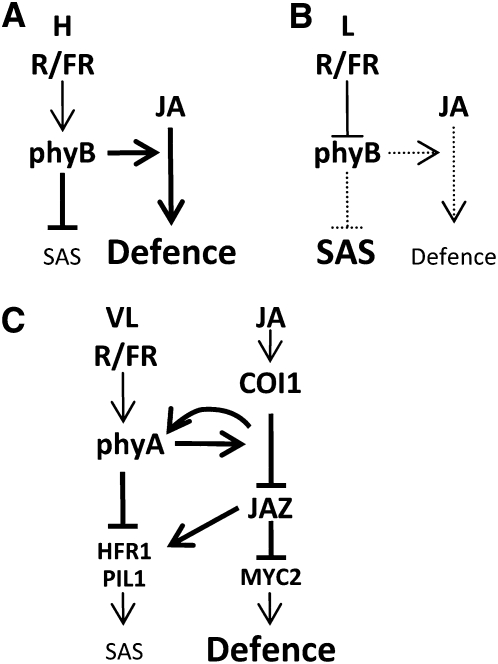
Shade and defense responses involve a common set of bHLH transcription factors. MYC2 is a bHLH transcription factor that is induced by JA in a COI1-dependent manner and regulates a diverse set of JA responses (Dombrecht et al., 2007). HFR1, another bHLH transcription factor, is a negative master regulator of shade responses. HFR1 attenuates elongation growth responses and flowering of plants that have been unsuccessful in escaping canopy shade (Sessa et al., 2005). The hfr1 mutant is remarkably similar to phyA and coi1 in that it displayed an exaggerated shade response only in low R/FR ratio light, for which it is known as slender in canopy shade1. With regard to shade, we have found that HFR1, and the positive regulator PIL1, are both, in part at least, under the control of COI1. Paradoxically, JA suppressed this FR induction, suggesting that COI1 either up- or downregulates individual genes depending on the environmental signal. The involvement of COI1 in positive (FR) and negative (JA) regulation of the same target demonstrates that shade and defense pathways may share common components and are integrative rather than mutually exclusive. We propose a regulatory pathway by which phyA and COI1 control the stability of negative regulators, the JAZ proteins, depending on the input signal and that the JAZ proteins dictate the specificity of the response by regulating a set of bHLH transcription factor genes, including MYC2, HFR1, and PIL1 (Figure 11).
Figure 11.
Model Showing the Proposed Integration of JA and Phytochrome Signaling in Shade and Defense Responses.
(A) In high R/FR light, phyB suppresses the shade avoidance syndrome (SAS) and enhances sensitivity to JA.
(B) In low R/FR light, phyB is inactivated, the suppression of shade responses is released, and sensitivity to JA is reduced (Moreno et al., 2009).
(C) In very low or extended R/FR light, phyA signaling suppresses exaggerated shade responses by recruiting a JA signal. Some responses to a JA signal in photosynthetic tissues require phyA.
To Grow or Defend?
Our study adds to the literature that describes the physiological and ecological implications of neighbor competition and responses to canopy shade on the one hand and defense responses to herbivores or other biotic and abiotic stresses on the other (Herms and Mattson, 1992; Izaguirre et al., 2006; Ballaré, 2009; Moreno et al., 2009). Modern agriculture requires both efficient land use and defense against pests and diseases. Dense sowing of crops can lead to reduced productivity due to neighbor competition that reduces plant yield and increased susceptibility to herbivory and mechanical damage. Elucidation of the molecular mechanisms that control the balance of these processes may benefit agriculture.
We propose that phyA and JA signaling cooperate to regulate the balance between shade avoidance responses in FR-enriched light and defense responses to mechanical damage or herbivores. Our work indicates that the interaction is at the level of degradation of JAZ proteins that are targets of both signaling pathways. These two signaling pathways are integrated, mutually dependent and reciprocal in their influence on the developmental processes that formerly have been assigned to each other.
METHODS
Plant Materials
The coi1-16 and coi1-1 mutants were identified in this lab (Feys et al., 1994; Ellis and Turner, 2002). coi1-16 is in a transgenic Col-gl1 background containing the VSP1:luciferase reporter gene. The coi1-16 parental line is referred to as the wild type in this article. coi1-16 seeds were propagated by maintenance of flowering plants at 16°C. coi1-1 is in the Col-gl1 background and is maintained as a heterozygous segregating population. Seeds of the aos-TJ1180, phyA-211, and phyB-9 mutants were obtained from the Nottingham Arabidopsis Stock Centre (Reed et al., 1994; Park et al., 2002). The opr3 mutant (Stintzi and Browse, 2000), the 35S-JAZ1-GUS line in COI1/coi1-1 (Thines et al., 2007), jai3, jin1-1, (Lorenzo et al., 2004), cry1-304 (Bruggemann et al., 1996), fin219 (Hsieh et al., 2000), and jar1-1 (Staswick et al., 2002) were gifts from the author labs stated here. All mutants are in Col-0 except for aos-TJ1180, which is in Col-gl1, and opr3, which is in Wassilewskija. Aos and opr3 seeds were propagated by repeatedly dipping inflorescences in 500 μM MeJA throughout the flowering period.
Growth Conditions
For flowering time experiments, wound-mediated growth inhibition, general propagation, crossing, and genetic analysis of plants, seeds were planted on damp soil (Levington's F2 compost plus fine grit at a ratio of 6:1), incubated at 4°C in the dark for 48 h to synchronize germination, and then transferred to growth chambers set at long (16 h light/8 h dark) or short (8 h light/16 h dark) days as appropriate, at 22°C with a 3/2 mix of cool white/gro-lux fluorescent bulbs (100 μmol m−2 s−1).
Seeds for axenic growth were surface sterilized by incubating in 70% ethanol for 2 min and then 10% bleach/0.01% triton for 10 min followed by five washes with sterilized deionized water. For seedling morphological and gene expression responses to specific light regimes, sterilized seeds were sown on 0.8% plant agar plates containing half-strength Murashige and Skoog salts plus vitamins and 0.5 g/L MES buffer, pH 5.7. For the analysis of seedling JA responses, seeds were sown on the same media plus 2% sucrose. MeJA (Sigma-Aldrich; 100 mM stock in ethanol) was added to cooled molten media to the required final concentration as appropriate. All plated seeds were stratified at 4°C in the dark for 72 h followed by 24 h white light (cool white fluorescent bulbs, 70 μmol m−2 s−1) at 22°C to synchronize germination. All subsequent manipulations were done in an inner dark room with a dim green safe light obtained by wrapping a fluorescent lamp (Sylvania standard white; OSRAM-Sylvania) with two layers of green filters (LEE Filters).
For hypocotyl length measurements, seeds prepared as above were transferred into the dark for 24 h at 22°C by which time the radicles had emerged. Germinated seedlings were then exposed to FRc, Rc, or Bc light for 2 to 6 d (see below for light sources). Control plates were kept in the dark.
For gene expression, seeds prepared as above were transferred into the dark for 6 d. The seedlings were then exposed to FRc for the required length of time before harvesting into liquid N2.
For seedling responses to high and low R/FR light, see below for light sources and ratios. Seeds prepared as above were transferred to a Sanyo growth cabinet (cool white fluorescent bulbs, 70 μmol m−2 s−1, continuous light, high R/FR) for 3 d before being transferred into low R/FR conditions for a further 9 d. Control plants were kept in high R/FR for a further 9 d.
For EOD-FR, seeds prepared as above were transferred to SD (cool white fluorescent bulbs, 70 μmol m−2 s−1) for 3 d with a 10-min FR and/or 5-min R treatment at the end of every light cycle.
For seedling JA responses, seeds prepared as above were transferred to LD growth cabinets with cool white fluorescent bulbs (70 μmol m−2 s−1) for the required number of days. Seedlings for root growth inhibition measurements were grown vertically.
Light Sources
Plants examined for their responses to continuous monochromatic FR (730 nm ± 10 nm), R (660 nm ± 10 nm), or B (470 nm ± 10 nm) light were grown in a growth chamber equipped with light emitting diodes (E30-LED; Percival Scientific) at 21°C. FR light was filtered through a layer each of #116 and #172 (LEE Filters) to cut off wavelengths shorter than 700 nm. This cabinet was also used for EOD-FR and R light treatments.
High and low R/FR experiments were conducted essentially as described by Keiller and Smith (1989) and Devlin et al. (1999) where continuous high R/FR light was provided by cool white fluorescent bulbs in a Sanyo growth cabinet (W) and low R/FR light was provided by adding supplementary FR light (W + FR; see Supplemental Figure 4 online). The high R/FR cabinet provided a photon irradiance, 400 to 700 nm, of 70 μmol m−2 s−1 and a R/FR of 10.8. The low R/FR cabinet provided the same photon irradiance but an R/FR of 0.068. The supplementary FR light was provided by prototype LED arrays (G. Whitelam, Leicester University) and was filtered through one layer of 0.3-mm black plexiglass to remove wavelengths lower than 700 nm. Light intensities and qualities were measured using a spectrometer (USB2000; Ocean Optics; see Supplemental Figure 4 online) and a SKP 200 photometer (Sky Instruments).
Measurement of Hypocotyls and Roots
Seedlings were placed flat on agar plates and scanned using a flatbed scanner. Hypocotyls and roots from scanned images were measured using the ImageJ program (Abramoff et al., 2004).
Flowering Time
Flowering time of soil-grown plants in LD and SD was scored as the number of days from germination to the first appearance of buds in the rosette center. The total number of rosette and cauline leaves on the primary inflorescence was counted after bolting.
RNA Extraction and Real-Time PCR
Total RNA was extracted from frozen plant material using an RNeasy plant mini kit (Qiagen). An RNase-free DNase step was incorporated according to the manufacturer's protocol for preparation of RNA for real-time PCR expression analysis. RNA was quantified using a NanoDrop 1000 (Thermo Scientific). A total of 1 μg of RNA was used per sample for cDNA synthesis using random hexamer primers and Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's protocol. Real-time PCR was performed to quantify cDNA using either inventoried small-scale TaqMan gene expression assays, each containing a FAM dye-labeled TaqMan MGB probe and a PCR primer pair (Applied Biosystems), or manually designed PCR primers and FAM/TAMRA probes (Sigma-Genosys). Each reaction was performed in 25 μL and contained the equivalent of 5 ng of reverse transcribed RNA, 33% TaqMan PCR Master Mix (Applied Biosystems), 200 nM each of the forward and reverse primer, and 100 nM of probe using the ABI Prism 7900 Fast real-time PCR system (Applied Biosystems), according to the manufacturer's protocol. To determine relative RNA levels within the samples, standard curves for the PCR were prepared using a mix of cDNA from all samples and making twofold serial dilutions covering the range equivalent to 20 to 0.625 ng RNA. All expression levels were quantified relative to the housekeeping gene ACTIN2, and reactions were performed in triplicate for each cDNA sample. Details of the TaqMan Gene Expression Assays and manually designed primers and probes used are in Supplemental Tables 1 and 2 online.
Histochemical GUS Assay
Seedlings were incubated in ice-cold 90% acetone for 20 min. The solution was changed to GUS staining solution (100 mM sodium phosphate buffer, 20% methanol, 5% Triton X-100, 10 mM EDTA, 500 μM potassium ferrocyanide, 500 μM potassium ferricyanide, and 500 μg/mL X-Gluc). The seedlings were infiltrated on ice by briefly applying and releasing a vacuum twice and then incubated overnight at 37°C in the dark followed by destaining and clearing in several changes of 70% ethanol.
Chlorophyll Extraction and Measurement
Approximately 3 to 5 mg tissue per sample was extracted overnight in 800 μL dimethylformamide at 4°C in the dark. Absorbance at 664 and 647 nm was measured in a SmartSpec Plus spectrophotometer (Bio-Rad) and total chlorophyll content determined in μg/mg tissue according to the extinction coefficients calculated by Porra et al. (1989).
Anthocyanin Extraction and Measurement
Anthocyanin was extracted and measured according to Neff and Chory (1998) with the following modifications. Approximately 20 mg frozen and ground plant tissue per sample was incubated overnight in 300 μL methanol:HCl (99:1) at 4°C in the dark. After addition of 200 μL deionized waterd, anthocyanins were separated from chlorophylls by the addition of 500 μL chloroform. Total anthocyanins were determined by measuring the absorbance at 530 and 657 nm using a spectrophotometer. Relative anthocyanin levels (abs 530 to abs 657) were calculated per milligram of tissue.
JA, JA-Ile, and OPDA Extraction and Measurement
Frozen plant tissue samples were extracted with methanol and acetylated before preparative HPLC (Miersch et al., 2008). Fractions obtained were analyzed by gas chromatography–mass spectrometry. JA and OPDA were analyzed as pentafluorobenzyl esters on an Rtx-5ms column in chemical ionization mode with NH3 as collision gas (Miersch et al., 2008). JA-Ile was quantified by gas chromatography–mass spectrometry on a DB-5ht column after derivitization with (trimethylsilyl)diazomethane (Guranowski et al., 2007).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: COI1 (At2G39940), phyA (At1G09570), phyB (At2G18790), CRY1 (At4G08920), JAZ1 (At1G19180), MYC2 (At1G32640), FIN219/JAR1 (At2G46370), JAI3/JAZ3 (At3G17860), AOS (At5G42650), and OPR3 (At2G06050). Accession numbers for genes used in qPCR are listed in Supplemental Tables 1 and 2 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. coi1-16 Mutants Are Slightly Insensitive to Blue Light.
Supplemental Figure 2. coi1-1 and fin219 Mutants Show Exaggerated Shade Responses to Low R/FR.
Supplemental Figure 3. phyA Mutants Have Wild-Type Responses to JA and Wounding with Respect to Aerial Growth Inhibition, Anthocyanin Accumulation, and Chlorophyll Degradation.
Supplemental Figure 4. Light Conditions of Sanyo Growth Cabinet Supplemented (Red and Yellow Traces) or Not (Black Trace) with FR.
Supplemental Table 1. Taqman Gene Expression Assays Used for Real-Time qPCR.
Supplemental Table 2. Sequences of Manually Designed Primers and Probes Used for qPCR.
Supplementary Material
Acknowledgments
This article is dedicated to the memory of the late Garry Whitelam to acknowledge his enormous contribution to the field of phytochrome biology and his enthusiasm for science. Garry also generously lent us his hand-built prototype FR, red, and blue LED arrays that allowed us to perform light experiments at the University of East Anglia. We thank the following people: Birgit Ortel for oxylipin measurements, John Baker for photography, Caroline Pennington for help with real-time qRT-PCR, John Browse, Roberto Solano, Chentao Lin, Paul Staswick, and Xing Wang Deng, and their colleagues for gifts of seeds, and Andrew Mayes for assistance with spectrum measurements in high/low R/FR growth conditions. F.R. is funded by Biotechnology and Biological Science Research Council (BBSRC) Grant BBD013429/1; H.O. is a BBSRC David Phillips Research Fellow (43JF20606); and S.-R.H. is funded by the Electro Magnetic Field Biological Trust.
References
- Abramoff M.D., Magelhaes P.J., Ram S.J. (2004). Image Processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Ahmad M., Cashmore A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162–166 [DOI] [PubMed] [Google Scholar]
- Balbi V., Devoto A. (2008). JA signalling network in Arabidopsis thaliana: Crucial regulatory nodes and new physiological scenarios. New Phytol. 177: 301–318 [DOI] [PubMed] [Google Scholar]
- Ballaré C.L. (2009). Illuminated behavior. Phytochrome as a key regulator of light foraging and plant anti-herbivore defense. Plant Cell Environ. 32: 713–725 [DOI] [PubMed] [Google Scholar]
- Ballaré C.L., Scopel A.L., Sanchez R.A. (1990). Far-red radiation reflected from adjacent leaves: An early signal of competition in plant canopies. Science 247: 329–332 [DOI] [PubMed] [Google Scholar]
- Barnes S.A., Nishizawa N.K., Quaggio R.B., Whitelam G.C., Chua N.H. (1996). Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell 8: 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti C.E., Xie D., Turner J.G. (1995). COI1-dependent expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine or methyl JA. Plant Physiol. 109: 567–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S., Mitchell-Olds T., Stotz H.U. (2002). Local and differential control of vegetative storage protein expression in response to herbivore damage in Arabidopsis thaliana. Physiol. Plant. 114: 85–91 [DOI] [PubMed] [Google Scholar]
- Berger S., Bell E., Sadka A., Mullet J.E. (1995). Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl JA, wounding, sugars, light and phosphate. Plant Mol. Biol. 27: 933–942 [DOI] [PubMed] [Google Scholar]
- Boter M., Ruiz-Rivero O., Abdeen A., Prat S. (2004). Conserved MYC transcription factors play a key role in JA signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggemann E., Handwerger K., Essex C., Storz G. (1996). Analysis of fast neutron-generated mutants at the Arabidopsis thaliana HY4 locus. Plant J. 10: 755–760 [DOI] [PubMed] [Google Scholar]
- Brüx A., Liu T.-Y., Krebs M., Stierhof Y.-D., Lohmann J.U., Miersch O., Wasternack C., Schumacher K. (2008). Reduced V-ATPase activity in the trans-Golgi network causes oxylipin-dependent hypocotyl growth inhibition in Arabidopsis. Plant Cell 20: 1088–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.-C., Huang I.C., Liu M.-J., Wang Z.-G., Chung S.-S., Hsieh H.-L. (2007). Glutathione S-transferase interacting with far-red insensitive 219 is involved in phytochrome A-mediated signaling in Arabidopsis. Plant Physiol. 143: 1189–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Chico J.M., Fernández-Calvo P., Solano R. (2009). The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 59: 77–87 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., Garcia-Casado G., Lopez-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in JA signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Cho K., Agrawal G.K., Shibato J., Jung Y.-H., Kim Y.-K., Nahm B.H., Jwa N.-S., Tamogami S., Han O., Kohda K., Iwahashi H., Rakwal R. (2007). Survey of differentially expressed proteins and genes in jasmonic acid treated rice seedling shoot and root at the proteomics and transcriptomics levels. J. Proteome Res. 6: 3581–3603 [DOI] [PubMed] [Google Scholar]
- Chory J., Chatterjee M., Cook R.K., Elich T., Fankhauser C., Li J., Nagpal P., Neff M., Pepper A., Poole D., Reed J., Vitart V. (1996). From seed germination to flowering, light controls plant development via the pigment phytochrome. Proc. Natl. Acad. Sci. USA 93: 12066–12071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Howe G.A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Koo A.J.K., Gao X., Jayanty S., Thines B., Jones A.D., Howe G.A. (2008). Regulation and function of Arabidopsis JA ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S.M., Cristescu S.M., Miersch O., Harren F.J., Wasternack C., Mur L.A. (2009). JAs act with salicylic acid to confer basal thermotolerance in Arabidopsis thaliana. New Phytol. 182: 175–187 [DOI] [PubMed] [Google Scholar]
- Devlin P.F., Robson P.R., Patel S.R., Goosey L., Sharrock R.A., Whitelam G.C. (1999). Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol. 119: 909–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin P.F., Yanovsky M.J., Kay S.A. (2003). A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol. 133: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A., Ellis C., Magusin A., Chang H.-S., Chilcott C., Zhu T., Turner J. (2005). Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl JA-induced secondary metabolism, defence, and hormone interactions. Plant Mol. Biol. 58: 497–513 [DOI] [PubMed] [Google Scholar]
- Devoto A., Nieto-Rostro M., Xie D.X., Ellis C., Harmston R., Patrick E., Davis J., Sherratt L., Coleman M., Turner J.G. (2002). COI1 links JA signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32: 457–466 [DOI] [PubMed] [Google Scholar]
- Devoto A., Turner J.G. (2003). Regulation of JA-mediated plant responses in Arabidopsis. Ann. Bot. (Lond.) 92: 329–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse JA-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C., Turner J. (2002). A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215: 549–556 [DOI] [PubMed] [Google Scholar]
- Fairchild C.D., Schumaker M.A., Quail P.H. (2000). HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14: 2377–2391 [PMC free article] [PubMed] [Google Scholar]
- Feys B.J.F., Benedetti C.E., Penfold C.N., Turner J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male-sterile, insensitive to methyl JA, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009). (+)-7-Iso-jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Franklin K.A., Praekelt U., Stoddart W.M., Billingham O.E., Halliday K.J., Whitelam G.C. (2003). Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol. 131: 1340–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel M., Kulheim C., Jankanpaa H., Skogstrom O., Dall'Osto L., Agren J., Bassi R., Moritz T., Moen J., Jansson S. (2009). Improper excess light energy dissipation in Arabidopsis results in a metabolic reprogramming. BMC Plant Biol. 9: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud T., Buchala A.J., Chua N.H., Métraux J.P. (2002). Phytochrome signalling modulates the SA-perceptive pathway in Arabidopsis. Plant J. 31: 87–95 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Goto N., Kumagai T., Koornneef M. (1991). Flowering responses to light-breaks in photomorphogenic mutants of Arabidopsis thaliana, a long-day plant. Physiol. Plant. 83: 209–215 [Google Scholar]
- Griebel T., Zeier J. (2008). Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: Phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol. 147: 790–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W., Vanholme B., Pauwels L., Plovie E., Inzé D., Gheysen G., Goossens A. (2009). Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep. 10: 923–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guranowski A., Miersch O., Staswick P.E., Suza W., Wasternack C. (2007). Substrate specificity and products of side-reactions catalyzed by JA:amino acid synthetase (JAR1). FEBS Lett. 581: 815–820 [DOI] [PubMed] [Google Scholar]
- Haga K., Iino M. (2004). Phytochrome-mediated transcriptional up-regulation of ALLENE OXIDE SYNTHASE in rice seedlings. Plant Cell Physiol. 45: 119–128 [DOI] [PubMed] [Google Scholar]
- He Y., Fukushige H., Hildebrand D.F., Gan S. (2002). Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 128: 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herms D.A., Mattson W.J. (1992). The dilemma of plants: To grow or defend. Q. Rev. Biol. 67: 283 [Google Scholar]
- Howe G.A., Jander G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Hsieh H.-L., Okamoto H., Wang M., Ang L.-H., Matsui M., Goodman H., Deng X.W. (2000). FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev. 14: 1958–1970 [PMC free article] [PubMed] [Google Scholar]
- Izaguirre M.M., Mazza C.A., Biondini M., Baldwin I.T., Ballaré C.L. (2006). Remote sensing of future competitors: Impacts on plant defenses. Proc. Natl. Acad. Sci. USA 103: 7170–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E., Bradley M., Harberd N.P., Whitelam G.C. (1994). Photoresponses of light-grown phyA mutants of Arabidopsis (phytochrome A is required for the perception of daylength extensions). Plant Physiol. 105: 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C., Lyou S., Yeu S., Kim M., Rhee S., Kim M., Lee J., Choi Y., Cheong J.-J. (2007). Microarray-based screening of JA-responsive genes in Arabidopsis thaliana. Plant Cell Rep. 26: 1053–1063 [DOI] [PubMed] [Google Scholar]
- Karlin-Neumann G.A., Sun L., Tobin E.M. (1988). Expression of light-harvesting chlorophyll a/b-protein genes is phytochrome-regulated in etiolated Arabidopsis thaliana seedlings. Plant Physiol. 88: 1323–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008). COI1 is a critical component of a receptor for JA and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiller D., Smith H. (1989). Control of carbon partitioning by light quality mediated by phytochrome. Plant Sci. 63: 25–29 [Google Scholar]
- Kloek A.P., Verbsky M.L., Sharma S.B., Schoelz J.E., Vogel J., Klessig D.F., Kunkel B.N. (2001). Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26: 509–522 [DOI] [PubMed] [Google Scholar]
- Liu Y., Ahn J.-E., Datta S., Salzman R.A., Moon J., Huyghues-Despointes B., Pittendrigh B., Murdock L.L., Koiwa H., Zhu-Salzman K. (2005). Arabidopsis vegetative storage protein is an anti-insect acid phosphatase. Plant Physiol. 139: 1545–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sanchez-Serrano J.J., Solano R. (2004). JA-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different JA-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac A.C., Wagner D., Boylan M.T., Quail P.H., Smith H., Garry C., Whitelam G.C. (1993). Photoresponses of transgenic Arabidopsis seedlings expressing introduced phytochrome B-encoding cDNAs: Evidence that phytochrome A and phytochrome B have distinct photoregulatory functions. Plant J. 4: 19–27 [Google Scholar]
- Melotto M., Mecey C., Niu Y., Chung H.S., Katsir L., Yao J., Zeng W., Thines B., Staswick P., Browse J., Howe G.A., He S.Y. (2008). A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miersch O., Neumerkel J., Dippe M., Stenzel I., Wasternack C. (2008). Hydroxylated JAs are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in JA signaling. New Phytol. 177: 114–127 [DOI] [PubMed] [Google Scholar]
- Moreno J.E., Tao Y., Chory J., Ballaré C.L. (2009). Ecological modulation of plant defense via phytochrome control of JA sensitivity. Proc. Natl. Acad. Sci. USA 106: 4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller M.J., Brodschelm W., Spannagl E., Zenk M.H. (1993). Signaling in the elicitation process is mediated through the octadecanoid pathway leading to jasmonic acid. Proc. Natl. Acad. Sci. USA 90: 7490–7494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A., Chory J., Furuya M. (1991). Phytochrome B is not detectable in the hy3 mutant of Arabidopsis, which is deficient in responding to end-of-day far-red light treatments. Plant Cell Physiol. 32: 1119–1122 [Google Scholar]
- Neff M.M., Chory J. (1998). Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-H., Halitschke R., Kim H.B., Baldwin I.T., Feldmann K.A., Feyereisen R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Pauwels L., Morreel K., De Witte E., Lammertyn F., Van Montagu M., Boerjan W., Inzé D., Goossens A. (2008). Mapping methyl JA-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 105: 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394 [Google Scholar]
- Rao M.V., Lee H.-i., Creelman R.A., Mullet J.E., Davis K.R. (2000). Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12: 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W., Nagatani A., Elich T.D., Fagan M., Chory J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S., Reinbothe C., Parthier B. (1993). Methyl JA-regulated translation of nuclear-encoded chloroplast proteins in barley (Hordeum vulgare L. cv. salome). J. Biol. Chem. 268: 10606–10611 [PubMed] [Google Scholar]
- Reinbothe C., Springer A., Samol I., Reinbothe S. (2009). Plant oxylipins: Role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J. 276: 4666–4681 [DOI] [PubMed] [Google Scholar]
- Riemann M., Bouyer D., Hisada A., Müller A., Yatou O., Weiler E., Takano M., Furuya M., Nick P. (2009). Phytochrome A requires JA for photodestruction. Planta 229: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Riemann M., Muller A., Korte A., Furuya M., Weiler E.W., Nick P. (2003). Impaired induction of the JA pathway in the rice mutant hebiba. Plant Physiol. 133: 1820–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemann M., Riemann M., Takano M. (2008). Rice JA RESISTANT 1 is involved in phytochrome and JA signalling. Plant Cell Environ. 31: 783–792 [DOI] [PubMed] [Google Scholar]
- Runge S., Sperling U., Frick G., Apel K., Armstrong G.A. (1996). Distinct roles for light-dependent NADPH:protochlorophyllide oxidoreductases (POR) A and B during greening in higher plants. Plant J. 9: 513–523 [DOI] [PubMed] [Google Scholar]
- Salter M.G., Franklin K.A., Whitelam G.C. (2003). Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426: 680–683 [DOI] [PubMed] [Google Scholar]
- Sasaki Y., et al. (2001). Monitoring of methyl JA-responsive genes in Arabidopsis by cDNA macroarray: Self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res. 8: 153–161 [DOI] [PubMed] [Google Scholar]
- Sessa G., Carabelli M., Sassi M., Ciolfi A., Possenti M., Mittempergher F., Becker J., Morelli G., Ruberti I. (2005). A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 19: 2811–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata M., Matsuda Y., Ando K., Nishii A., Takemura M., Yokota A., Kohchi T. (2004). Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J. Exp. Bot. 55: 631–639 [DOI] [PubMed] [Google Scholar]
- Shinomura T., Uchida K., Furuya M. (2000). Elementary processes of photoperception by phytochrome A for high-irradiance response of hypocotyl elongation in Arabidopsis. Plant Physiol. 122: 147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sineshchekov V.A., Loskovich A.V., Riemann M., Nick P. (2004). The JA-free rice mutant hebiba is affected in the response of phyA'/phyA'' pools and protochlorophyllide biosynthesis to far-red light. Photochem. Photobiol. Sci. 3: 1058–1062 [DOI] [PubMed] [Google Scholar]
- Smith H., Xu Y., Quail P.H. (1997). Antagonistic but complementary actions of phytochromes A and B allow optimum seedling de-etiolation. Plant Physiol. 114: 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D.E., Sharrock R.A., Tepperman J.M., Quail P.H. (1991). The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. Plant Cell 3: 1263–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Su W., Howell S.H. (1992). Methyl JA inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89: 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Tiryaki I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16: 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Tiryaki I., Rowe M.L. (2002). JA response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14: 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A., Browse J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for JA synthesis. Proc. Natl. Acad. Sci. USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A., Weber H., Reymond P., Browse J., Farmer E.E. (2001). Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 98: 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman J.M., Zhu T., Chang H.-S., Wang X., Quail P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98: 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaller M.C., Schippa S., Rossolini G.M. (1998). Conserved sequence motifs among bacterial, eukaryotic, and archaeal phosphatases that define a new phosphohydrolase superfamily. Protein Sci. 7: 1647–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCFCOI1 complex during JA signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Tsuchiya T., Ohta H., Okawa K., Iwamatsu A., Shimada H., Masuda T., Takamiya K. (1999). Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: Finding of a lipase motif and the induction by methyl jasmonate. Proc. Natl. Acad. Sci. USA 96: 15362–15367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda J., Kato J. (1980). Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.). Plant Physiol. 66: 246–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ma L., Habashi J., Li J., Zhao H., Deng X.W. (2002). Analysis of far-red light-regulated genome expression profiles of phytochrome A pathway mutants in Arabidopsis. Plant J. 32: 723–733 [DOI] [PubMed] [Google Scholar]
- Wang Z., Cao G., Wang X., Miao J., Liu X., Chen Z., Qu L.-J., Gu H. (2008). Identification and characterization of COI1-dependent transcription factor genes involved in JA-mediated response to wounding in Arabidopsis plants. Plant Cell Rep. 27: 125–135 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007). JAs: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.) 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam G.C., Johnson E., Peng J.R., Carol P., Anderson M.L., Cowl J.S., Harberd N.P. (1993). Phytochrome-a null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam G.C., Johnson C.B. (1982). Photomorphogenesis in Impatiens parviflora and other plant species under simulated natural canopy radiations. New Phytol. 90: 611–618 [Google Scholar]
- Xie D.-X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for JA-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xu L.H., Liu F.Q., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D.F., Xie D.X. (2002). The SCFCOl1 ubiquitin-ligase complexes are required for JA response in Arabidopsis. Plant Cell 14: 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V., Mallappa C., Gangappa S.N., Bhatia S., Chattopadhyay S. (2005). A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic Growth. Plant Cell 17: 1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chetelat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the JA pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Zhang C., Gu M., Bai Z., Zhang W., Qi T., Cheng Z., Peng W., Luo H., Nan F., Wang Z., Xie D. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky M.J., Casal J.J., Whitelam G.C. (1995). Phytochrome A, phytochrome B and HY4 are involved in hypocotyl growth responses to natural radiation in Arabidopsis: Weak de-etiolation of the phyA mutant under dense canopies. Plant Cell Environ. 18: 788–794 [Google Scholar]
- Zhai Q.Z., Li C.B., Zheng W.G., Wu X.Y., Zhao J.H., Zhou G.X., Jiang H.L., Sun J.Q., Lou Y.G., Li C.Y. (2007). Phytochrome chromophore deficiency leads to overproduction of jasmonic acid and elevated expression of JA-responsive genes in Arabidopsis. Plant Cell Physiol. 48: 1061–1071 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Turner J.G. (2008). Wound-induced endogenous JAs stunt plant growth by inhibiting mitosis. PLoS One 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.