This study exploits the recent production of a draft nuclear genome sequence for Arabidopsis lyrata to examine evolution of plant MIRNA and p4-siRNA loci between two congenic Brassicaceae species. The results indicate that many MIRNAs and most p4-siRNA hot spots are rapidly changing and evolutionarily transient within the Arabidopsis genus.
Abstract
Twenty-one-nucleotide microRNAs (miRNAs) and 24-nucleotide Pol IV-dependent small interfering RNAs (p4-siRNAs) are the most abundant types of small RNAs in angiosperms. Some miRNAs are well conserved among different plant lineages, whereas others are less conserved, and it is not clear whether less-conserved miRNAs have the same functionality as the well conserved ones. p4-siRNAs are broadly produced in the Arabidopsis genome, sometimes from active hot spot loci, but it is unknown whether individual p4-siRNA hot spots are retained as hot spots between plant species. In this study, we compare small RNAs in two closely related species (Arabidopsis thaliana and Arabidopsis lyrata) and find that less-conserved miRNAs have high rates of divergence in MIRNA hairpin structures, mature miRNA sequences, and target-complementary sites in the other species. The fidelity of miRNA biogenesis from many less-conserved MIRNA hairpins frequently deteriorates in the sister species relative to the species of first discovery. We also observe that p4-siRNA occupied loci have a slight tendency to be retained as p4-siRNA loci between species, but the most active A. lyrata p4-siRNA hot spots are generally not syntenic to the most active p4-siRNA hot spots of A. thaliana. Altogether, our findings indicate that many MIRNAs and most p4-siRNA hot spots are rapidly changing and evolutionarily transient within the Arabidopsis genus.
INTRODUCTION
Plant transcriptomes include a multitude of small RNAs produced by the action of Dicer-Like (DCL) proteins. These endogenous small RNAs function as specificity determinants bound to Argonaute (AGO) proteins within complexes that effect transcriptional and/or posttranscriptional regulation of RNA targets. MicroRNAs (miRNAs) are an abundant subset of the plant small RNA population. They are defined by precise, DCL-catalyzed excision from the helical stems of hairpin-forming single-stranded precursor RNAs (Meyers et al., 2008; Voinnet, 2009). Many plant miRNAs negatively regulate multiple target mRNAs at the posttranscriptional level, promote the formation of short interfering RNAs (siRNAs) from their RNA targets, and/or interact with naturally occurring target mimics (Mallory and Bouche, 2008). Through these regulatory mechanisms, plant miRNAs are critical for multiple processes, including diverse developmental events, meristem identity, abiotic stress responses, nutrient homeostasis, and pathogen responses.
Plant MIRNA loci are most often independent RNA Polymerase II (Pol II)–transcribed units whose expression patterns are individually regulated and consequently display tissue- or condition-specific accumulation patterns (Xie et al., 2005; Valoczi et al., 2006; Sieber et al., 2007). Identical or nearly identical mature miRNAs can be encoded by large families of paralogous MIRNA loci. The evolution of individual MIRNA loci (Warthmann et al., 2008) and patterns of MIRNA family expansion and contraction (Maher et al., 2006) can be tracked using comparative genomics. Some plant MIRNA families are quite conserved: Over 20 families are expressed in both monocots and eudicots, and at least seven of these families are also expressed in bryophytes (Axtell and Bowman, 2008). Compared with less-conserved miRNAs, well-conserved miRNAs tend to have higher expression levels, more paralogous loci per family, and RNA targets that are easier to computationally predict (using currently understood parameters for miRNA/target interactions in plants) and experimentally verify (chiefly by detecting remnants of AGO-catalyzed target cleavage) (Rajagopalan et al., 2006; Axtell et al., 2007; Fahlgren et al., 2007). These observations have led to the hypothesis that many less-conserved miRNA families may be nonfunctional and evolutionarily transient (Rajagopalan et al., 2006; Fahlgren et al., 2007; Axtell, 2008; Fenselau de Felippes et al., 2008). A second hypothesis, which explains the difficulty of predicting and validating targets of less conserved miRNAs, suggests that less conserved miRNAs are indeed often functional as target regulators but tend to interact with targets in configurations generally not captured by current target prediction methods and with molecular outcomes that do not often include readily detectable cleavage remnants (Brodersen and Voinnet, 2009). The less conserved miR834 partially conforms to this hypothesis because target regulation occurs without easily detected RNA cleavage (Brodersen et al., 2008). However, in this case, the target site itself was readily predicted by existing methods. A third hypothesis, which explains the lack of conservation and generally low expression levels, posits that less conserved miRNAs often perform regulatory tasks in restricted numbers of cells within a single family or genus. The regulation of AGL16 transcripts by the less conserved miR824 specifically within the stomatal precursor cells of the Brassicaceae provides an example conforming to this idea (Kutter et al., 2007).
In most angiosperm tissues that have been analyzed, the majority of small RNAs are not miRNAs, but instead are 24-nucleotide Pol IV–dependent siRNAs (p4-siRNAs) that arise from DCL processing of long, perfectly double-stranded RNA templated by genomic sequences. In Arabidopsis thaliana, most 24-nucleotide siRNAs are p4-siRNAs produced and used by the Pol IV/Pol V system, which uses them to direct RNA-directed DNA methylation and repressive histone modifications to target chromatin (Matzke et al., 2009). Production of p4-siRNAs is broadly distributed throughout the A. thaliana genome, with concentrations in pericentromeric regions, avoidance of protein-coding loci, and a tendency toward repetitive sequences (Lu et al., 2005; Rajagopalan et al., 2006; Kasschau et al., 2007). Nonetheless, there are clearly hot spots of p4-siRNA production from certain loci (Rajagopalan et al., 2006; Kasschau et al., 2007; Zhang et al., 2007; Mosher et al., 2008). Some A. thaliana p4-siRNA loci are active in all developmental stages (type II loci), while many others produce p4-siRNAs specifically in floral and reproductive tissues (type I loci; Mosher et al., 2009). Loci marked by cytosine methylation, some of which is likely directed by p4-siRNAs, can vary among A. thaliana ecotypes (Vaughn et al., 2007) as do the activities of some p4-siRNA loci (Vaughn et al., 2007; Zhai et al., 2008). However, it is not known if individual p4-siRNA hot spots are frequently retained as hot spots between species.
In this study, we exploit the recent production of a draft nuclear genome sequence for Arabidopsis lyrata to examine evolution of plant MIRNA and p4-siRNA loci between two congenic Brassicaceae species. We find that many less-conserved miRNA families have high rates of sequence divergence in MIRNA hairpin structures, mature miRNA sequences, and in the complementary sites of predicted targets between the two species. High-throughput identification of sliced miRNA targets in both A. lyrata and A. thaliana was generally unsuccessful for targets of less conserved miRNAs. We also observe that the most active p4-siRNA hot spots expressed within A. lyrata leaves are not syntenic to the p4-siRNA hot spots in multiple tissues of A. thaliana.
RESULTS
Identification and Annotation of A. lyrata miRNAs
A. lyrata MIRNAs were identified using two complementary methods: Identification of A. lyrata genomic regions syntenic to annotated A. thaliana MIRNAs and by analysis of sequenced A. lyrata small RNA populations. For synteny-based identification, A. thaliana MIRNAs annotated in miRBase 14.0 were filtered according to updated criteria for annotation of plant MIRNAs (Meyers et al., 2008) in combination with ∼1.6 × 107 sequenced small RNAs from nine publically available small RNA seq (sRNA-seq) data sets from various wild-type tissues (Table 1). Authentic MIRNA loci are defined by precise excision of one or more transient duplexes (consisting of the eventual miRNA and the miRNA*) from a stem-loop RNA precursor. A. thaliana loci that lacked clear evidence for precise excision of an authentic miRNA/miRNA* duplex from a qualifying hairpin structure were discarded, leaving a total of 157 loci. Syntenic A. lyrata loci were identified for 143 out of the 157 queries using a microsynteny-based method (see Supplemental Data Set 1 online). This procedure identified regions of similarity to the MIRNA queries that were flanked by upstream and downstream protein-coding loci that were highly similar to the upstream and downstream genes in A. thaliana (see Methods). In parallel, three sRNAseq libraries from A. lyrata were obtained and randomly sequenced: A library prepared from rosette leaf tissue yielded ∼5.8 × 105 reads, while two biological replicate sRNAseq samples from inflorescences yielded ∼2.6 × 107 and ∼2.2 × 107 reads, respectively (Table 1). A. lyrata MIRNA were identified de novo from these small RNA data using a combination of MIRcheck (Jones-Rhoades and Bartel, 2004) and expression-based filters to ensure conformity to current criteria of plant MIRNA annotation (Meyers et al., 2008). A total of 154 A. lyrata MIRNA loci were identified based on the sRNAseq data (see Supplemental Data Set 1 online). Many of these loci (106) were identical to the A. lyrata loci found by the synteny-based approach. Two of the A. lyrata loci were found to be syntenic to annotated A. thaliana MIRNA loci that had been missed in the initial homology search because the corresponding A. thaliana loci had not met our expression-based criteria for inclusion as queries. Five of the A. lyrata MIRNA loci found based on expression were members of known A. thaliana miRNA families but seemed to lack syntenic homologs in A. thaliana. Interestingly, these five A. lyrata loci were found in two genomic clusters: two clustered miR395 loci and three clustered miR399 loci. The remaining 41 A. lyrata MIRNA loci all produced mature miRNAs lacking appreciable similarity to previously annotated miRNAs in any species (based on miRBase 14). The microsynteny method found syntenic A. thaliana loci for about half (22) of these 41 new A. lyrata MIRNA loci. Four of these 22 A. thaliana syntenic regions passed the Meyers et al. (2008) expression criteria for confident annotation as a MIRNA based on our reference A. thaliana small RNA data set but had not been previously noticed in A. thaliana. Details on all A. thaliana and A. lyrata MIRNAs may be found in Supplemental Data Sets 1 to 3 online.
Table 1.
sRNAseq and Degradome Data Sets
| Name | Species/Tissue | Type | Sequencing Instrument | Number of Reads | Unique Reads | Reference(s) | Accession(s) (NCBI GEO) |
| AL-L1-sRNA | A. lyrata/rosette leaves | sRNAseq | Illumina/SBS | 583,895a | 382,520a | This study | GSM451894 |
| AL-F1-sRNA | A. lyrata/inflorescences | sRNAseq | SOLiD | 26,192,231a | 61,287,122b | This study | GSM512644 |
| AL-F2-sRNA | A. lyrata/inflorescences | sRNAseq | SOLiD | 21,620,398a | 52,184,197b | This study | GSM512645 |
| AT-deg1 | A. thaliana/inflorescences | Degradome | SOLiD | 9,031,213c | 671,981d | This study | GSM512878 |
| AT-deg2 | A. thaliana/inflorescences | Degradome | SOLiD | 9,612,258c | 878,714d | This study | GSM512879 |
| AL-deg1 | A. lyrata/inflorescences | Degradome | SOLiD | 7,087,227c | 739,992d | This study | GSM512880 |
| AL-deg2 | A. lyrata/inflorescences | Degradome | SOLiD | 15,665,420c | 743,889d | This study | GSM512881 |
| AT-F1-sRNA | A. thaliana/inflorescences | sRNAseq | Roche/454 | 205,649e | 100,658e | Rajagopalan et al. (2006) | GSM118372 |
| AT-F2-sRNA | A. thaliana/inflorescences | sRNAseq | Roche/454 | 78,596e | 57,966e | Kasschau et al. (2007) | GSM154336 |
| AT-F3-sRNA | A. thaliana/inflorescences | sRNAseq | Illumina/SBS | 7,686,781e | 2,841,896e | Lister et al. (2008) | GSM227608 |
| AT-F4-sRNA | A. thaliana/inflorescences | sRNAseq | Illumina/SBS | 7,576,080e | 1,482,150e | Montgomery et al. (2008) | GSM342999, GSM343000, GSM343001 |
| AT-Sq1-sRNA | A. thaliana/siliques | sRNAseq | Roche/454 | 305,764e | 141,539e | Rajagopalan et al. (2006) | GSM118375 |
| AT-Se1-sRNA | A. thaliana/seedlings | sRNAseq | Roche/454 | 188,954e | 77,937e | Rajagopalan et al. (2006) | GSM118374 |
| AT-Se2-sRNA | A. thaliana/seedlings | sRNAseq | Roche/454 | 22,467e | 12,718e | Kasschau et al. (2007) | GSM154375 |
| AT-L1-sRNA | A. thaliana/rosette leaves | sRNAseq | Roche/454 | 186,899e | 67,663e | Rajagopalan et al. (2006) | GSM118373 |
| AT-L2-sRNA | A. thaliana/rosette leaves | sRNAseq | Roche/454 | 15,833e | 8,112e | Kasschau et al. (2007) | GSM154370 |
Number of reads mapped to the genome.
Number of distinct genomic positions matched by one or more reads. Because SOLiD data were mapped allowing for errors, it is not possible to tally the number of unique reads that were mapped.
Number of reads mapped to the sense strand of the transcriptome.
Number of distinct 5′ ends matched by one or more reads. Because SOLiD data were mapped allowing for errors, it is not possible to tally the number of unique reads that were mapped.
Number of reads present within the GEO-deposited accession.
Our MIRNA annotation efforts led to a list of 205 MIRNA loci that, using the sRNAseq data sets referenced above, met the Meyers et al. (2008) criteria for annotation of plant MIRNAs in either A. thaliana, A. lyrata, or both (see Supplemental Data Set 1 online). These loci were classified based on the apparent conservation level of the mature miRNA families: The 26 families (encoded by 99 loci) that were also annotated in one or more non-Brassicaceae species in miRBase 14 were termed MC (for more conserved). The 104 families (encoded by 106 loci) that were not annotated as present in any non-Brassicaceae species in miRBase 14 were termed LC (for less conserved).
Less Conserved MIRNAs Are Often Species Specific, Weakly Expressed, Encoded by Single Loci, and More Likely to Produce 22-Nucleotide RNAs
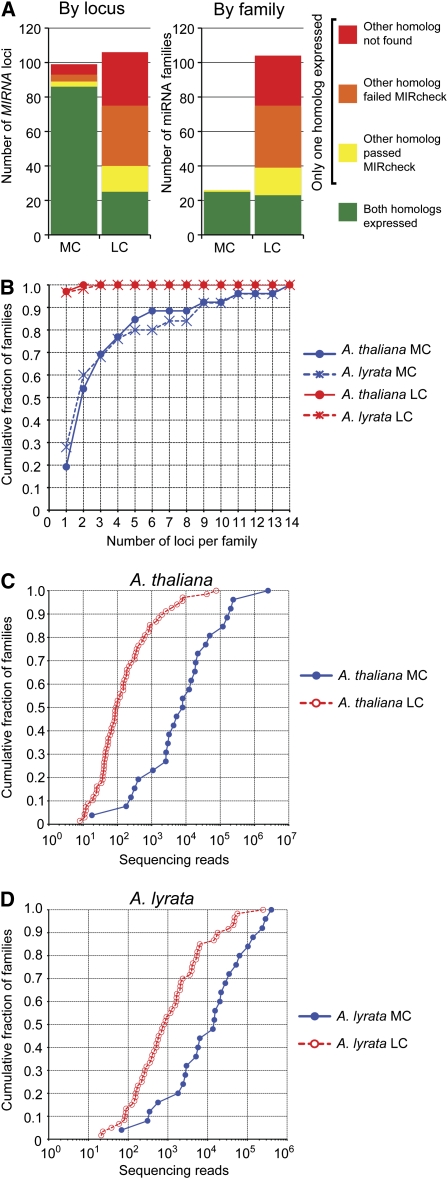
For most MIRNA loci corresponding to MC families, syntenic homologs were identified in both species that, with reference to our sRNAseq data, expressed microRNAs conforming to the Meyers et al. (2008) expression criteria (Figure 1A). By contrast, this situation was rare for LC families (Figure 1A). Instead, many homologs of LC miRNAs expressed in one species were not found to be expressed in the other species; only a minority of these homologs retained a putative hairpin structure capable of passing MIRcheck (which assesses secondary structures but does not incorporate expression criteria; Figure 1A). Most MC miRNA families were encoded by two or more loci, while nearly all LC miRNA families were encoded by just a single locus (Figure 1B). As inferred by sRNAseq read coverage, accumulation levels of both MC and LC miRNAs spanned several orders of magnitude in both A. thaliana and A. lyrata (Figures 1C and 1D). However, as a group, MC miRNA accumulation levels were clearly higher than those for LC miRNA families (Figures 1C and 1D).
Figure 1.
Less Conserved MIRNAs Are Often Species Specific, Weakly Expressed, and Encoded by Single Loci.
(A) Identification of homologous MIRNA loci (left panel) and families (right panel) in A. thaliana and A. lyrata. Only loci/families that passed the expression criteria in at least one species were considered. MC, more conserved; LC, less conserved.
(B) Cumulative distributions of the number of paralogous loci per miRNA family.
(C) Cumulative distributions of the number of sequencing reads per A. thaliana miRNA family (based on nine sRNAseq data sets totaling ∼1.6 × 107 reads; Table 1).
(D) As in (C) for A. lyrata miRNA families (based on ∼4.8 × 107 reads; Table 1).
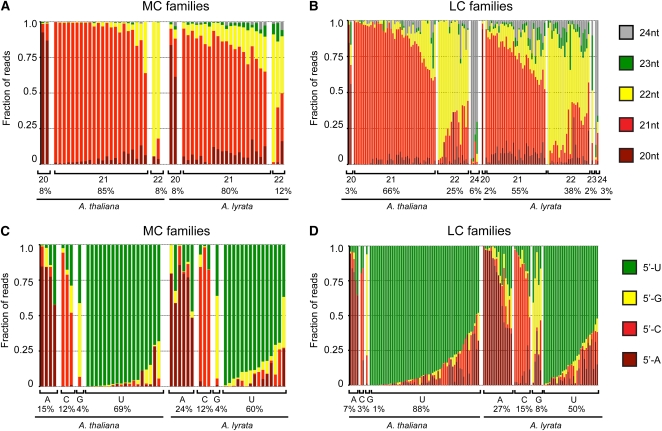
We next analyzed properties of the observed MIRNA hairpin-derived small RNAs themselves (which included annotated mature miRNAs, miRNA*s, as well as any length and positional variants). Most MC families were dominated by expression of 21-nucleotide small RNAs in both A. thaliana and A. lyrata (Figure 2A). However, some MC families were dominated by expression of either 20-nucleotide small RNAs or by 22-nucleotide small RNAs. The proportion of LC families dominated by 22-nucleotide small RNA expression was higher than in MC families in both A. thaliana and A. lyrata, although 21-nucleotide dominant families still accounted for the majority of LC families (Figure 2B). Small RNAs with a 5′-U accounted for most small RNAs produced by both MC and LC families in both species (Figures 2C and 2D). However, we noted that the number of families in which small RNA expression was not dominated by 5′-U RNAs was higher for A. lyrata miRNAs than for A. thaliana miRNAs. We do not understand the reason for this result, but we suspect it might be due to differential biases caused the different library construction and sequencing methods used for the different sRNAseq data sets (Table 1).
Figure 2.
Predominant Lengths and 5′ Nucleotides Produced by A. thaliana and A. lyrata MIRNA Hairpins.
(A) Proportions of sRNAseq reads of the indicated lengths from more conserved (MC) families. Families are grouped according to the most abundant small RNA length, as indicated below the chart. Percentages indicate the percentage of families dominated by small RNAs of the indicated length for a given species.
(B) As in (A) for less conserved (LC) families.
(C) As in (A) for 5′ nucleotides of reads from MC families.
(D) As in (A) for 5′ nucleotides of reads from LC families.
High Levels of MIRNA Hairpin and Mature miRNA Sequence Divergence between A. thaliana and A. lyrata
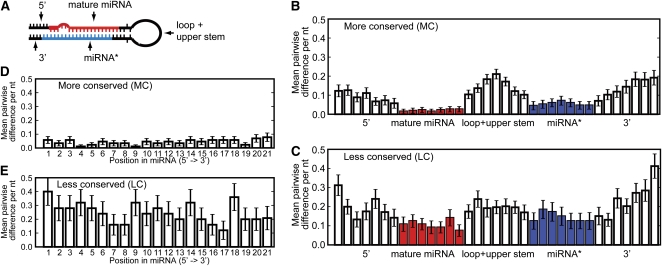
We next compared the sequences of syntenic A. thaliana and A. lyrata MIRNA hairpins. Analysis of sequence divergence was limited only to syntenic pairs for which both members passed the Meyers et al. (2008) expression criteria or pairs in which one member failed the expression criteria but still had a putative hairpin capable of passing MIRcheck in the expression-negative species (i.e., the green and yellow regions of Figure 1A). Sequence divergence between these 128 syntenic A. thaliana and A. lyrata loci (88 from MC families and 40 from LC families) was calculated by scoring each position of all pairwise alignments. Hairpins were divided into five regions (Figure 3A); regions were scaled and divided into seven bins each to account for variations in length among the loci. As expected for conserved, functional MIRNA hairpins (Ehrenreich and Purugganan, 2008; Warthmann et al., 2008), divergence was lowest within the mature miRNA itself for both the MC and the LC groups (Figures 3B and 3C); this likely reflects purifying selection on the mature miRNA sequences to maintain complementarity with target mRNAs. The miRNA*s also showed low levels of divergence, most likely reflecting the requirement to maintain base pairs with the constrained miRNAs in the context of the stem-loop secondary structure. By contrast, the 5′, loop, and 3′ regions were relatively unconstrained (Figures 3B and 3C). Although the divergence profiles of MC and LC MIRNAs were qualitatively similar, there was clearly more divergence in mature miRNA sequences among the LC families, even after discarding loci whose predicted secondary structures were highly aberrant in one of the two species (Figures 3B and 3C). This indicates that the mature miRNA sequences of LC families often have less constraint in mature miRNA and miRNA* sequences over short evolutionary distances. The mature MC miRNAs had a slight tendency toward higher divergence at their 3′ ends, although diversity was low throughout the MC miRNAs (Figure 3D). By contrast, mature LC miRNAs showed high levels of divergence at all sequence positions (Figure 3E).
Figure 3.
Less Conserved miRNAs Diverge More between A. thaliana and A. lyrata Than Do More Conserved miRNAs.
(A) A sketch showing the five regions of MIRNA hairpins that were analyzed. For convenience, the mature miRNA is shown on the 5′ arm, although in reality it can be either on the 5′ arm or 3′ arm.
(B) Average sequence divergence between more conserved A. thaliana and A. lyrata MIRNA hairpins. Both 5′-arm and 3′-arm mature miRNAs were tallied and displayed together. To account for differences in lengths among the population of hairpins, the five regions were each scaled to seven bins. Bars indicate the standard errors of the means.
(C) As in (B) for less conserved MIRNAs.
(D) As in (B) for each nucleotide position within mature miRNAs from more conserved families.
(E) As in (D) for less conserved families.
Imprecise and Inconsistent Processing of Less Conserved MIRNAs
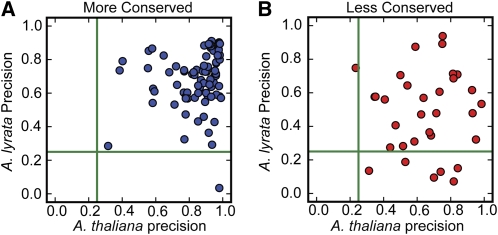
Plant MIRNAs are defined by precise processing of a single-stranded stem-loop precursor RNA to release one or more specific miRNA/miRNA* duplexes (Ambros et al., 2003; Meyers et al., 2008). In practice, stem-loop derived small RNAs fall into a continuous spectrum of processing precisions, from very imprecisely processed inverted repeats (whose products are often classified as a form of endogenous siRNA; Lu et al., 2006; Zhang et al., 2007) to canonical MIRNAs producing almost exclusively a single miRNA/miRNA* duplex. Some less conserved MIRNAs are imprecisely processed in A. thaliana, and this inaccuracy is sometimes correlated with their reliance upon DCL4 instead of DCL1 for processing (Rajagopalan et al., 2006). To examine MIRNA processing precision, the ∼4.8 × 107 A. lyrata sRNAseq reads from our three libraries (Table 1) were mapped to the A. lyrata MIRNA hairpins. In parallel, the ∼1.6 × 107 publicly available A. thaliana sRNAseq reads (all nine A. thaliana sRNAseq libraries in Table 1) from several wild-type tissues were mapped to the A. thaliana MIRNA hairpins. We defined processing precision at each locus as the abundance of reads corresponding exactly to the mature miRNA or miRNA* divided by the total abundance of all reads mapping to the hairpin. Thus, values close to one indicate very high precision, while values close to zero indicate processing which produces only small amounts of any given miRNA/miRNA* duplex. To compare how MIRNA processing precision differed between A. thaliana and A. lyrata homologs, we considered only those syntenic pairs for which both members passed the Meyers et al. (2008) expression criteria or pairs in which one member failed the expression criteria but still had a putative hairpin that both passed MIRcheck and expressed at least 10 sRNAseq reads. The processing precisions of hairpins from MC miRNA families were typically high in both species (Figure 4A). Some processing precisions from LC hairpins also had similar precisions in both species (Figure 4B). However, several of the LC hairpins were processed quite imprecisely in A. lyrata; some of these were also imprecisely processed in A. thaliana, where most of them were first described, while others had higher precisions in A. thaliana (Figure 4B). We conclude that many LC MIRNAs are processed very imprecisely, especially outside of the species in which they were first observed.
Figure 4.
Less Conserved MIRNAs Tend to Be Processed Imprecisely.
(A) Scatterplot of A. lyrata versus A. thaliana MIRNA processing precisions for more conserved MIRNAs. Green lines show the precision value of 0.25, which we used as a cutoff for determining miRNA-like expression patterns (Meyers et al., 2008).
(B) As in (A) for less conserved MIRNAs.
High Levels of miRNA Target Divergence between A. thaliana and A. lyrata
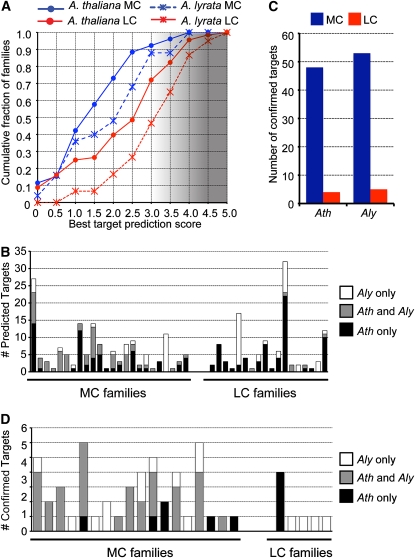
We next examined predicted targets of A. thaliana and A. lyrata miRNAs. Targets were predicted only for miRNAs whose precursors passed the Meyers et al. (2008) expression criteria. Potential miRNA target sites were scored according to the criteria of Allen et al. (2005); higher scores indicated less confidence in the predictions. In both species, lower-scoring (and thus higher confidence) targets were more frequently predicted for MC families and less frequently predicted for LC families (Figure 5A). Based on previous results (Allen et al., 2005; Rajagopalan et al., 2006), we used a score of three as the upper limit for confident target prediction (Figure 5A). All target predictions meeting this cutoff for A. lyrata miRNAs are given in Supplemental Data Set 4 online. The overlap in confident target predictions (score ≤ 3) between A. thaliana and A. lyrata was examined. Importantly, this analysis was limited to the subset of miRNA families that had at least one hairpin that passed the Meyers et al. (2008) expression criteria in both species; in other words, we examined miRNA target overlap only for miRNA families that actually existed in both species. Syntenic homologs were frequently predicted targets of MC miRNA families, whereas this was rarely the case for the predicted targets of LC miRNA families (Figure 5B). We conclude that target predictions using standard criteria are more unreliable and more inconsistent between species for LC miRNA families than for MC families.
Figure 5.
Targets of Less Conserved miRNAs Are Difficult to Identify and Inconsistent between A. thaliana and A. lyrata.
(A) Cumulative distributions of the number of miRNA families with the indicated target prediction scores. The lowest scoring prediction (i.e., the most confident prediction) for each family was used. MC, more conserved; LC, less conserved. The gradient of shading indicates increasingly less confident predictions, beginning at a score of 3.
(B) miRNA target predictions by family. The number of predicted targets found only in A. thaliana (Ath), only in A. lyrata (Aly), or syntenic homologs predicted in both species are shown. Families without any predicted targets in either species are omitted, as are families that were expressed only in a single species.
(C) Sliced targets confidently found by degradome sequencing. Sliced targets were those that were found in both biological replicate degradome libraries for the given species.
(D) As in (B) for degradome-confirmed targets.
To determine experimentally miRNA targets, four degradome libraries were prepared: two biological replicates from A. thaliana inflorescences and two biological replicates from A. lyrata inflorescences. Each library consisted of ∼1 × 107 reads that mapped to the sense strand of one or more annotated transcripts (Table 1). Degradome sequencing (synonymous with parallel analysis of RNA ends and genome-wide mapping of uncapped and cleaved transcripts) determines the 5′ ends of RNAs with a 5′-monophospate (Addo-Quaye et al., 2008; German et al., 2008; Gregory et al., 2008). This RNA population includes the sliced remnants of many miRNA-targeted transcripts. Sliced miRNA targets were identified from these data using an updated version of the CleaveLand software (Addo-Quaye et al., 2009a), which calculates empirically estimated P values for each possible sliced miRNA target (see Methods). Targets with a P value ≤ 0.05 in both biological replicates were considered verified; all verified targets along with supporting information are found in Supplemental Data Sets 5 to 8 online. Nearly all verified targets in both species were those of MC miRNA families (Figure 5C), and many of these were syntenic homologs that were validated in both species (Figure 5D). Taken together, these observations suggested that many of the putative targets of LC miRNA families were inconsistent between A. thaliana and A. lyrata and difficult to verify by looking for evidence of slicing.
PoI IV siRNA Occupancy and Hot Spots Differ between A. thaliana and A. lyrata
We next turned our attention to comparisons between A. thaliana and A. lyrata 24-nucleotide siRNAs, most of which are likely due to the action of the Pol IV/Pol V pathway. A whole-genome alignment between A. thaliana and A. lyrata was performed. The A. thaliana genome was divided into 1000-nucleotide bins (119,184 in total), and A. lyrata genomic regions aligned to each of the bins were identified. In total, 82.5% of the bins (98,357) were confidently paired with an A. lyrata syntenic region; the rest were ambiguous, largely due to small-scale inversions (data not shown).
Repeat-normalized small RNA abundances from nine publicly available A. thaliana sRNAseq data sets (Table 1) were tabulated for all A. thaliana bins. Importantly, these data sets represented diverse wild-type tissues (inflorescences, leaves, seedlings, and siliques) and were produced with multiple technologies (Roche/454 pyrosequencing and Illumina sequencing by synthesis) by different investigators. Abundances derived from our three A. lyrata sRNAseq samples were also calculated for all aligned A. lyrata bins. Occupancies of small RNAs of a given size were determined by applying a minimum threshold that corrects for loci that produce multiple sizes of small RNAs (see Methods). Only bins confidently aligned between A. thaliana and A. lyrata were used for analysis. The number of bins occupied by 24-nucleotide RNAs varied from 1378 to 11,431 (1.4 to 11.6% of 98,357 bins); variations in occupancy were directly correlated with the sequencing depths of the samples. The number of bins that were co-occupied in each pairwise combination of all data sets was calculated (O: observed overlap). Additionally, the number of co-occupied bins expected by random chance (E: expected overlap) was also calculated for each pairwise combination. In this scheme, positive values of O/E indicate more co-occupancy than would be expected by chance. Importantly, this metric can be compared across samples of differing sequencing depths. The observed co-occupancies for 24-nucleotide RNAs between different A. thaliana samples was always greater than expected by chance alone, typically between four- and eightfold higher (Figure 6A). These data indicate that 24-nucleotide RNA producing loci are somewhat consistent across different tissues of A. thaliana whose small RNAs were sampled at different depths and using different sequencing methodologies. Comparisons to the 24-nucleotide RNA accumulation pattern of A. lyrata also showed that co-occupancy of syntenic bins occurred more often than expected by chance. However, the enrichments relative to chance alone were very modest, with all O/E ratios less than twofold for all interspecies comparisons (Figure 6A). As a control, the same analysis was performed for 21-nucleotide occupied bins from each sample, with the expectation that because many of these bins contain conserved MIRNAs or TAS loci, they would be consistently occupied both in the various A. thaliana samples and in A. lyrata. Indeed, the number of 21- nucleotide RNA co-occupied bins in A. thaliana intraspecies comparisons and in the A. lyrata–A. thaliana interspecies comparisons greatly exceeded the values expected by chance alone and exceeded the values seen for 24-nucleotide RNA co-occupancy (Figures 6A and 6B). We conclude that only a small percentage of the genomic regions producing 24-nucleotide small RNAs in one species also produce 24-nucleotide RNAs from the syntenic region in a closely related species. This contrasts with 21-nucleotide RNA expressing loci, which are more consistently found to be 21-nucleotide expressers in both species.
Figure 6.
24-Nucleotide RNA Expression and Hot Spots Frequently Differ between A. thaliana and A. lyrata.
(A) Log2 ratios of observed to expected overlaps between 24-nucleotide small RNA occupied 1-kb bins for all pairwise comparisons between various A. thaliana and A. lyrata small RNA samples. F1-F4, floral; Sq1, silique; Se1-Se2, seedlings; L1-L2, rosette leaves.
(B) As in (A) for 21-nucleotide small RNA occupied bins.
(C) Overlaps between the top 100 24-nucleotide small RNA expressing 1-kb bins for all pairwise comparisons between various A. thaliana and A. lyrata small RNA samples.
(D) As in (C) for the top 100 21-nucleotide small RNA expressing loci.
(E) Percentages of A. thaliana 21- and 24-nucleotide small RNA occupied bins that were ambiguously aligned in the A. thaliana–A. lyrata whole genome alignment. Dashed line indicates the percentage of the entire A. thaliana genome that was ambiguously aligned.
(F) As in (E) for the top 100 A. thaliana 21- and 24-nucleotide small RNA hot spots.
(G) Fractions of 24-nucleotide small RNAs that mapped to annotated genes, compared with the overall fraction of genomic nucleotides overlapping with gene annotations in A. thaliana and A. lyrata. A. thaliana sRNAseq data were the combination of all nine sRNAseq libraries, and A. lyrata sRNAseq data were the combination of all three sRNAseq libraries (Table 1).
Next, we examined whether the most active hot spots of small RNA expression behaved similarly to the global patterns for all occupied loci. The top 100 24-nucleotide expressing 1-kb bins, ranked in order of the abundance of mapped 24mers, were obtained for each A. thaliana and A. lyrata data set within the 98,357 confidently aligned 1-kb regions. The raw observed overlap between all of the data sets was analyzed. The top 100 21-nucleotide expressing bins from each sample were analyzed in the same way, as a control. Similar to the global patterns of occupancy, the consistency of 24-nucleotide RNA hot spots between different A. thaliana samples was generally lower than that of 21-nucleotide hot spots (Figures 6C and 6D). In all A. thaliana-only pairwise combinations, between one and 63 24-nucleotide hot spots were in the top 100 in both samples, with a mean of 19.0, while values for 21-nucleotide hot spot overlap between A. thaliana samples ranged from 32 to 76 with a mean of 54.4. Some of the variation in A. thaliana 24-nucleotide RNA hot spots was due to tissue-specific expression patterns. Particularly striking were the highly consistent 24-nucleotide RNA hot spots from the inflorescence and silique samples (Figure 6C). These are likely to be type I p4-siRNA loci, defined by their specific expression in reproductive tissues (Mosher et al., 2009). By contrast, 24-nucleotide siRNA hot spots from A. thaliana leaves were not as consistent, with 14 out of 100 overlapping between the two A. thaliana leaf samples examined (Figure 6C). The leaf hot spots are likely type II p4-siRNA loci, which are defined by their broad expression patterns, particularly in nonreproductive tissues (Mosher et al., 2009). Other probable sources of the within-species variation in small RNA hot spots include sampling error due to nonsaturating sequencing depths, variations in small RNA library construction and particularly in the level of contamination by degraded RNA fragments, and artifacts arising from differing sequencing technologies. Pairwise comparisons of all nine A. thaliana hot spot lists to all three A. lyrata hot spot lists revealed very low overlap in the top 100 24-nucleotide RNA hot spots, with between zero and two shared syntenic bins (mean = 0.52; Figure 6C). This low to nonexistent overlap in 24-nucleotide RNA hot spots was much lower than the 14 out of 100 overlap seen when comparing two A. thaliana leaf samples, but higher than the values expected from random chance (∼1 E−6). By contrast, many more of the top 100 21-nucleotide hot spots overlapped between A. thaliana and A. lyrata; values ranged from 21 to 34 with a mean of 28.3 (Figure 6D). Most of these shared 21-nucleotide hot spots were likely to have been abundant miRNAs or trans-acting siRNAs.
It is possible that the failure to identify more 24-nucleotide RNA hot spots shared between A. thaliana and A. lyrata was because syntenic hot spots disproportionately fell into genomic regions that were not confidently aligned. To test this, we examined both co-occupancy and the top 100 small RNA hot spots from all 119,184 1kb bins, including the ambiguously aligned bins, for the nine A. thaliana data sets. Both for global occupancy, and for hot spots, 24-nucleotide expressing regions were indeed more likely to fall into nonaligned bins than were the control 21-nucleotide hot spots (Figures 6E and 6F). In all cases, the percentages of 24-nucleotide occupied bins or hot spots exceeded the genome-wide percentage of all nonalignable bins. By contrast, the 21-nucleotide occupied bins or hot spots fell into ambiguously aligned bins with consistently lower frequencies that were roughly centered upon the genome-wide value (Figures 6E and 6F). Thus, A. thaliana 24-nucleotide RNA hot spots are indeed more likely to arise from genomic regions difficult to align with A. lyrata. However, it should be noted that this effect is modest; most A. thaliana 24-nucleotide RNA hot spots and occupied loci were in confidently aligned bins but almost none of these were also 24-nucleotide RNA hot spots in A. lyrata. Like their A. thaliana counterparts, A. lyrata 24-nucleotide small RNAs tended to emanate from regions of the genome devoid of annotated protein-coding capacity (Figure 6G). Altogether, we observe a slight tendency of 24-nucleotide RNA expressing loci to be retained as 24-nucleotide expressers between species, but our data provide little evidence for retention of individual 24-nucleotide RNA hot spots between A. thaliana and A. lyrata. This contrasts strongly with the most active 21-nucleotide expressing loci, which are often highly expressed in both species.
DISCUSSION
Emergence or Degeneration of MIRNAs at the Species Level
The notion that many plant MIRNAs are lineage specific has been clearly supported by comparisons of MIRNA inventories between different plant families (Rajagopalan et al., 2006; Fahlgren et al., 2007). MIRNA emergence must be fairly rapid, as there are several examples of MIRNAs that probably arose specifically in the species A. thaliana, as judged by their absence in the closest relative A. lyrata (Fenselau de Felippes et al., 2008). Our sampling of A. lyrata small RNA expression also indicated that there are many A. lyrata–specific MIRNAs that arose after the divergence of the A. thaliana lineage. We found that, as a group, homologs of young A. thaliana or A. lyrata MIRNA loci frequently do not conform to classical ideas of MIRNA biogenesis and function. Specifically, syntenic homologs of young MIRNAs in the closest related species (1) frequently lack the capacity to form a MIRNA-like stem-loop, (2) have high divergence rates in mature miRNA sequences, (3) tend to lose complementarity with homologs of the known/predicted targets, and (4) are processed with diminished accuracy in the sister species. Misannotations of MIRNA loci were unlikely to have confounded these results, as we restricted our analyses to a subset of annotated MIRNAs for which there is unambiguous experimental evidence for miRNA biogenesis in at least one of the two species being analyzed. Thus, we conclude that many of the less conserved MIRNAs either degenerated in one species from a functional common ancestor or were specifically refined from a nonfunctional ancestor in a given lineage. Either option entails relatively rapid changes in MIRNA sequences, processing accuracies, and target repertoires.
At least two hypotheses are consistent with the differences in properties between young MIRNA loci and their syntenic homologs in closely related species. The first is that these homologs are performing biologically meaningful regulatory roles in both species. Because of the noncanonical sequence conservation, diminished target site conservation, and reduced processing accuracies, this hypothesis necessitates that the young MIRNAs exert biological effects in a manner that is quite different than for canonical MIRNAs. For instance, it could be that these young miRNAs interact with targets with pairing geometries not captured by commonly used prediction methods (Brodersen and Voinnet, 2009). Such pairing configurations could potentially be more tolerant of positional and sequence heterogeneity (although this is not the case for the seed targeting that has been extensively documented in animals). Another possibility is that some of these young MIRNAs could exert regulatory roles on host transcripts in cis simply by being processed by DCL proteins, similar to the suspected functions of A. thaliana MIR838 and Physcomitrella patens MIR1047 (Rajagopalan et al., 2006; Axtell et al., 2007). A second hypothesis that cannot be excluded with currently available data is that many homologs of young MIRNAs are simply degenerate and do not function in any biologically relevant role in the sister species. This would in turn imply that these young MIRNAs are truly species specific, in that they exist and function only in a single species but not in its closest relative.
Our analysis highlighted broad trends that differentiated more conserved and less conserved MIRNAs between A. thaliana and A. lyrata. However, it is important to point out that not all less conserved MIRNAs followed the overall trends. Some homologs of less conserved MIRNAs had conserved mature miRNA sequences within MIRNA-like hairpins, maintained high levels of complementarity to their targets, and were processed accurately in both species. Thus, there are certainly some MIRNAs that are restricted to the Brassicaceae but also have the properties of more conserved, canonical MIRNA loci.
Pol IV siRNA Hot Spots Can Be Evolutionarily Transient
P4-siRNAs, which are 24 nucleotides in length, are hypothesized to counteract productive Pol II transcription of intergenic regions by directing chromatin modifications to Pol V transcribed areas (Wierzbicki et al., 2009). Much of the genome is involved in p4-siRNA production at low levels, and there are clear hot spots of production that account for a disproportionate amount of p4-siRNA production (Zhang et al., 2007). We find that global expression patterns of 24-nucleotide RNAs, which we presume to be p4-siRNA loci, are relatively consistent across different tissue samples derived from A. thaliana, with pairwise overlaps between different samples consistently four- to eightfold higher than expected by chance alone. By contrast, 24-nucleotide RNA hot spots are consistent between different inflorescence and silique samples of A. thaliana, but often differ between vegetative (leaves and seedlings) and reproductive (inflorescences and siliques) tissues of A. thaliana. These data imply that type I p4-siRNAs (defined by their absence from vegetative tissues) have more reproducible hot spots than do type II p4-siRNAs (defined by their presence in vegetative tissues). Despite the higher intraspecies variability in the vegetative p4-siRNA hot spots, there are many that are reproducibly active in different samples: For instance, 14 out of the top 100 p4-siRNA hot spots are shared between two independently derived A. thaliana leaf samples (Figure 5C). However, there is essentially no significant overlap between the top 100 p4-siRNA loci expressed in A. lyrata leaves or inflorescences and the top 100 p4-siRNA loci from any A. thaliana tissue. Similarly, there is only a slight trend toward overlap in the global patterns of p4-siRNA accumulation between A. lyrata and A. thaliana regardless of expression level. Thus, the loci which produce p4-siRNAs often differ between A. thaliana and A. lyrata.
The significance of most individual p4-siRNA hot spots within plant genomes is unknown. In Drosophila melanogaster, Piwi-interacting RNAs (piRNAs) have some functional analogies to plant p4-siRNAs. Like plant p4-siRNAs, fly piRNAs also function to silence transposable element expression by the use of Watson-Crick interactions between the small RNA and target (Aravin et al., 2007). In germline cells and surrounding somatic support cells, piRNA expression disproportionately emanates from just a few master regulator loci, the most prominent of which is flamenco (Brennecke et al., 2007; Lau et al., 2009; Malone et al., 2009). The flamenco locus is an ∼180-kb region dominated by transposon fragments that are arranged almost exclusively in a single orientation (Brennecke et al., 2007). The abundant piRNAs produced from the flamenco locus function to initiate posttranscriptional silencing of active transposon copies elsewhere in the genome. The flamenco piRNA hot spot is critical for suppression of gypsy retroelements in the female germline (Prud'homme et al., 1995). Importantly, flamenco is conserved with respect to high production of piRNAs and single-stranded transposon orientation in both Drosophila yakuba and Drosophila erecta (Malone et al., 2009). By analogy with flamenco, one potential function for plant p4-siRNA hot spots could be as master loci that produce siRNAs with the capacity to silence expression of many unlinked transposons with sequence similarity in trans. However, unlike in the flamenco analogy, we did not find evidence for maintenance of high expression for any p4-siRNA hot spots between two closely related plant species. This implies that highly active individual p4-siRNA loci can, like less-conserved MIRNAs, be evolutionarily transient in plants.
METHODS
Small RNA Sequencing and Data Analysis
Total RNA was extracted using Tri-Reagent (Sigma-Aldrich). Illumina sequencing of the Arabidopsis lyrata leaf sample was as follows: Small RNA-enriched fractions were purified from 20% PAGE gels by recovering the 20- to 30-nucleotide area from the total RNA sample. A pre-adenylated 3′ adapter (IDT) linker 1 (5′-AppCTGTAGGCACCATCAATddC-3′) was added using T4 RNA ligase without exogenous ATP. The 3′-ligated products were gel eluted and then ligated to a 5′ adapter composed of RNA (5′-GUUCAGAGUUCUACAGUCCGACGAUC-3′) using T4 RNA ligase with ATP. Gel purification of the ligated product was followed by reverse transcription using an oligo (5′-ATTGATGGTGCCTACAG-3′) specific to the 3′ linker. The cDNA library was then amplified using a 5′ adapter oligo (5′-AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGA-3′) and a 3′ adapter oligo (5′-CAAGCAGAAGACGGCATACGAATTGATGGTGCCTACAG-3′). The amplified library was then gel purified and sequenced using an Illumina genome analyzer by Fasteris. The A. lyrata data (described here) and another library embedded within the raw data were computationally separated by parsing the 3′ adapter sequences. Reads between 19 and 26 nucleotides in length were retained for analysis. Construction of A. lyrata inflorescence-derived small RNAs was performed using the SOLiD Small RNA Expression Kit per the manufacturer's instructions, followed by sequencing on the SOLiD 2 instrument at the Penn State/Huck Institutes Genomics Core Facility.
Identification of MIRNAs in Arabidopsis thaliana and A. lyrata
A. thaliana small RNA reads combined from nine publicly available sRNAseq data sets (Table 1) were mapped to the TAIR9 genome, and reads mapped to the genome fewer than 15 times were used then aligned to 190 annotated A. thaliana MIRNA loci retrieved from miRBase 14.0 (Griffiths-Jones et al., 2008). Loci were then filtered based on a conservative interpretation of the updated criteria for plant MIRNA annotation (Meyers et al., 2008), which required at least 10 raw small RNA sequencing reads matching the hairpin, and an miRNA/miRNA* duplex processing precision >0.25 (calculated as the proportion of the raw read abundance mapping exactly to the mature miRNA and miRNA* out of the total abundance of reads mapping anywhere on the hairpin). A. lyrata MIRNA homologs of those A. thaliana MIRNA loci that passed the filtering were computationally identified using a microsynteny-based method. For each A. thaliana MIRNA considered, the two flanking protein coding loci were retrieved from the TAIR9 annotation set. The top five hits in the A. lyrata filtered gene models generated by the Joint Genome Initiative (JGI; http://genome.jgi-psf.org/Araly1/Araly1.home.html) of the A. thaliana MIRNA and the flanking loci were identified using BLASTn. Flanking loci and MIRNA hits were compared to identify microsyntenic regions. The MIRNA hit with maximally preserved synteny (i.e., between two hits to the respective A. thaliana flanking loci) was identified as the predicted A. lyrata MIRNA hairpin homolog. If none of the top five hits of the MIRNA maintained the same synteny of MIRNA and two flanking genes in A. lyrata, a MIRNA hit that maintained the synteny with only one of the flanking genes was identified as the predicted A. lyrata MIRNA homolog. Meanwhile, A. lyrata MIRNA loci were identified de novo from the three sRNAseq data sets produced in this study (Table 1). Reads from each data set were mapped to the A. lyrata genome assembly generated by the JGI (http://genome.jgi-psf.org/Araly1/Araly1.home.html), and each 300-nucleotide flanking genomic region was prefiltered based on polarity of small RNA accumulation (≥75% from the dominant strand), 21-22mer abundance relative to other size classes (amount of 21-22mers more than double the amount of non-21-22mers), and number of hits for individual small RNAs in the genome (no greater than 15 genome matches) and examined by MIRcheck (Jones-Rhoades and Bartel, 2004). Candidates that survived this prescreening were then subject to the following additional filters: The most abundant small RNA on the hairpin had to have more than 15 reads in at least one of the three libraries; the processing precision had to be greater than 0.25; the hairpin had to pass MIRcheck (Jones-Rhoades and Bartel, 2004) with the parameters “-mir_bulge”,3, “-ass”, 2,”-unpair”; the miRNA* had to be expressed, or the mature miRNA had to be the most abundant small RNA in two or more libraries. A. thaliana genomic loci syntenic to previously unknown A. lyrata MIRNAs were identified as described above.
MIRNA Divergence Analysis
Syntenic A. thaliana and A. lyrata MIRNA hairpins were divided into five regions: the loop/upper stem, mature miRNA, miRNA*, and 5′ and 3′ regions. Each region was further divided into seven equal-length bins (rounded to the closest integer). Divergence was calculated as the pairwise difference per nucleotide of each bin for each MIRNA based on the pairwise alignment using MUSCLE (Edgar, 2004) between the MIRNA hairpins in both species. Mature miRNAs in both species were identified as the small RNA in the most precisely processed miRNA/miRNA* duplexes in a family calculated from the aforementioned sRNAseq data sets. Thus, the mature miRNA sequences were not necessarily the same as annotated miRNAs in miRBase 14.0.
miRNA Target Prediction and Validation
All mature miRNAs derived from hairpins that passed our expression criteria were used to predict targets from the TAIR9 transcriptome (for A. thaliana) or from the JGI FM3 transcriptome (A. lyrata). Predictions were accomplished with the PERL script axtell_targetfinder.pl. This program first uses rmapper-ls (from the SHRiMP package; Rumble et al., 2009) to find a large set of alignments with very low stringency. These initial alignments are then parsed into RNA-RNA alignments and scored using the scheme of Allen et al. (2005), retaining only those alignments scoring seven or better. Target prediction with this method also includes annotation of the predicted cleavage site as well as randomizations. This program is available as part of the CleaveLand 2.0 package on our lab's website (http://homes.bio.psu.edu/people/faculty/Axtell/AxtellLab/Software.html).
Construction of degradome libraries differed considerably from our past efforts (Addo-Quaye et al., 2008, 2009b). Approximately 150 ng of poly(A)+ RNA was used as input to the SOLiD whole transcriptome analysis kit, following the manufacturer's instructions except that (1) the initial RNaseIII-catalyzed RNA fragmentation was omitted and (2) a large size range was gel isolated after the RT-PCR. Omitting the RNaseIII fragmentation step restricts the initial adapter ligation to only those RNAs containing 5′-monophosphates. Libraries were sequenced using the P1 (5′) adapter only, resulting in the sequencing of the first 35 nucleotides of the inserts that represented the 5′ ends of the original RNAs.
Raw degradome data, in colorspace format, were mapped to the appropriate transcriptomes using rmapper-cs (part of the SHRiMP package, version 1.3.1; Rumble et al., 2009) using the nondefault settings: M 35bp,fast -o 10,000 -F. Initial mappings were then filtered to retain only the best scoring alignment(s) for each read with less than six mismatches total and those that had perfect alignments to nucleotides one through six (to allow confident detection of 5′ ends) and whose alignments extended at least to position 29 (to exclude any potential miRNAs or siRNAs that might have contaminated the libraries). The filtered map data were compacted into a standard degradome format that gives the number of 5′ ends observed at each position in the transcriptome, along with a peak categorization score between zero and four. For each miRNA query, axtell_targetfinder.pl was used to predict all targets with alignment scores of seven or less, along with 1000 identical target predictions for randomly permuted versions of the query miRNA. The significance of any degradome signatures that matched the cleavage sites of a predicted miRNA target was assessed by examining the frequencies with which the randomized queries also matched degradome information. Specifically, for each peak category, a cumulative distribution representing the frequency with which the random queries had one or matches at a given miRNA alignment score was calculated. The likelihood that a given cleavage fragment was observed by chance was estimated by retrieving the frequency with which the random queries gave hits of the given peak category at the given alignment score or better; we interpreted this frequency as a P value. The cutoff for confident target identification was P ≤ 0.05 in both biological replicates. A series of PERL scripts that accomplish these calculations is available from our lab website as the CleaveLand 2.0 package (http://homes.bio.psu.edu/people/faculty/Axtell/AxtellLab/Software.html).
Small RNA Occupancy Calculation and Hot Spot Identification
A whole genome alignment between A. thaliana and A. lyrata was performed using lastz (http://www.bx.psu.edu/miller_lab/dist/README.lastz-1.01.50/README.lastz-1.01.50.html; B. Harris and C. Riemer, unpublished data) and further processed to retain a one-to-one best alignment using chainnet (Kent et al., 2003). The A. thaliana genome (TAIR9 release) was divided into 119,184 1-kb bins and the A. lyrata syntenic regions to 98,357 of the bins were confidently obtained via the whole-genome alignment. Small RNAs from 12 data sets (AT-F1, AT-F2, AT-F3, AT-F4, AT-Sq1, AT-Se1, AT-Se2, AT-L1, AT-L2, AL-L1, AL-F1, and AL-F2; Table 1) were mapped to their respective genomes, and repeat-normalized small RNA abundances were tabulated for each confidently aligned bin (98,357 bins). All bins with abundance greater than 10 reads per million, calculated by the small RNA abundance of a certain length (21 or 24 nucleotides) minus the abundance of small RNAs of all other lengths, were considered “occupied.” The overlap of the occupied bins between each pair of the data sets was analyzed, and the normalized overlap (calculated by log2 transformation of the observed overlap divided by the expected overlap) was reported. Expected overlap was the product of the fractions occupied in the two data sets being compared. For example, given a data set with 2000/98,357 (2.03%) bins occupied and a second data set with 10,000/98,357 (10.17%) bins occupied, the percentage of bins occupied in both data sets expected by random chance is 0.0203 × 0.1017 = 0.00206 (0.206%; ∼203 bins). For hot spot identification, the bins in each data set were ranked by the abundance of small RNAs of a certain length (21 or 24 nucleotides) minus the abundance of small RNAs of all other lengths. The top 100 ranking bins were considered hot spots. The number of overlapping bins out of the 100 top-ranking bins from every pair of the data sets was calculated. Overlap of 24-nucleotide small RNAs with annotated genes was calculated by comparing the mapping positions of 24-nucleotide small RNAs with genomic annotations (TAIR9 transcripts for A. thaliana and FM3 transcripts for A. lyrata). Reads falling within introns were also counted as mapping to genes. Genomic fractions annotated as genes simply scored every genomic nucleotide as either genic (exonic or intronic annotated) or intergenic.
Accession Numbers
Newly generated A. lyrata small RNA data have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) (GSE18077 and GSE20442). A. lyrata and A. thaliana degradome data have also been deposited at NCBI GEO (GSE20451). Accession numbers for all data sets used in this study are listed in Table 1.
Author Contributions
Z.M. and M.J.A. designed the research, Z.M., C.C., and M.J.A. performed the research, and Z.M. and M.J.A. analyzed the data and wrote the article.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Data Set 1. A. thaliana and A. lyrata MIRNA Loci Examined in This Study.
Supplemental Data Set 2. Expression Details of A. thaliana MIRNA Loci That Passed the Meyers et al. (2008) Criteria.
Supplemental Data Set 3. Expression Details of A. lyrata MIRNA Loci That Passed the Meyers et al. (2008) Criteria.
Supplemental Data Set 4. Predicted Targets of A. lyrata miRNAs.
Supplemental Data Set 5. Degradome Information for A. thaliana Sliced Targets Confidently Identified in Both A. thaliana Biological Replicates: Data from the AT-deg1 Sample.
Supplemental Data Set 6. Degradome Information for A. thaliana Sliced Targets Confidently Identified in Both A. thaliana Biological Replicates: Data from the AT-deg2 Sample.
Supplemental Data Set 7. Degradome Information for A. lyrata Sliced Targets Confidently Identified in Both A. lyrata Biological Replicates: Data from the AL-deg1 Sample.
Supplemental Data Set 8. Degradome Information for A. lyrata Sliced Targets Confidently Identified in Both A. lyrata Biological Replicates: Data from the AL-deg2 Sample.
Acknowledgments
We thank Webb Miller and Bob Harris for access to and advice on the LASTZ program, Craig Praul for SOLiD sequencing services, and James Carrington and Noah Fahlgren for sharing data and materials prior to publication. The A. lyrata draft genome assembly was kindly provided by the Department of Energy-Joint Genome Initiative's Community Sequencing Program, through a proposal coordinated by Detlef Weigel (MPI Tübingen, Germany). This study was supported by a grant from the National Science Foundation (Award MCB-0718051) to M.J.A.
References
- Addo-Quaye C., Eshoo T.W., Bartel D.P., Axtell M.J. (2008). Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18: 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo-Quaye C., Miller W., Axtell M.J. (2009a). CleaveLand: A pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25: 130–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo-Quaye C., Snyder J.A., Park Y.B., Li Y.F., Sunkar R., Axtell M.J. (2009b). Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA 15: 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Ambros V., et al. (2003). A uniform system for microRNA annotation. RNA 9: 277–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J. (2007). Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 [DOI] [PubMed] [Google Scholar]
- Axtell M.J. (2008). Evolution of microRNAs and their targets: Are all microRNAs biologically relevant? Biochim. Biophys. Acta 1779: 725–734 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Bowman J.L. (2008). Evolution of plant microRNAs and their targets. Trends Plant Sci. 13: 343–349 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Snyder J.A., Bartel D.P. (2007). Common functions for diverse small RNAs of land plants. Plant Cell 19: 1750–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Brodersen P., Voinnet O. (2009). Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 10: 141–148 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich I.M., Purugganan M.D. (2008). Sequence variation of microRNAs and their binding sites in Arabidopsis. Plant Physiol. 146: 1974–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N., Howell M.D., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Carrington J.C. (2007). High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS One 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenselau de Felippes F., Schneeberger K., Dezulian T., Huson D.H., Weigel D. (2008). Evolution of Arabidopsis thaliana microRNAs from random sequences. RNA 14: 2455–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- German M.A., et al. (2008). Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26: 941–946 [DOI] [PubMed] [Google Scholar]
- Gregory B.D., O'Malley R.C., Lister R., Urich M.A., Tonti-Filippini J., Chen H., Millar A.H., Ecker J.R. (2008). A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell 14: 854–866 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. (2008). miRBase: tools for microRNA genomics. Nucleic Acids Res. 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Kasschau K.D., Fahlgren N., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Carrington J.C. (2007). Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Baertsch R., Hinrichs A., Miller W., Haussler D. (2003). Evolution's cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. USA 100: 11484–11489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter C., Schob H., Stadler M., Meins F., Jr, Si-Ammour A. (2007). MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell 19: 2417–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.C., Robine N., Martin R., Chung W.J., Niki Y., Berezikov E., Lai E.C. (2009). Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 19: 1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., O'Malley R.C., Tonti-Filippini J., Gregory B.D., Berry C.C., Millar A.H., Ecker J.R. (2008). Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Kulkarni K., Souret F.F.MuthuValliappan R. Tej S.S. Poethig R.S. Henderson I.R. Jacobsen S.E. Wang W. Green P.J. Meyers B.C. (2006). MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 16: 1276–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Tej S.S., Luo S., Haudenschild C.D., Meyers B.C., Green P.J. (2005). Elucidation of the small RNA component of the transcriptome. Science 309: 1567–1569 [DOI] [PubMed] [Google Scholar]
- Maher C., Stein L., Ware D. (2006). Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 16: 510–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Bouche N. (2008). MicroRNA-directed regulation: To cleave or not to cleave. Trends Plant Sci. 13: 359–367 [DOI] [PubMed] [Google Scholar]
- Malone C.D., Brennecke J., Dus M., Stark A., McCombie W.R., Sachidanandam R., Hannon G.J. (2009). Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M., Kanno T., Daxinger L., Huettel B., Matzke A.J. (2009). RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 21: 367–376 [DOI] [PubMed] [Google Scholar]
- Meyers B.C., et al. (2008). Criteria for annotation of plant microRNAs. Plant Cell 20: 3186–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery T.A., Yoo S.J., Fahlgren N., Gilbert S.D., Howell M.D., Sullivan C.M., Alexander A., Nguyen G., Allen E., Ahn J.H., Carrington J.C. (2008). Inaugural Article: AGO1-miR173 complex initiates phased siRNA formation in plants. Proc. Natl. Acad. Sci. USA 105: 20055–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher R.A., Melnyk C.W., Kelly K.A., Dunn R.M., Studholme D.J., Baulcombe D.C. (2009). Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460: 283–286 [DOI] [PubMed] [Google Scholar]
- Mosher R.A., Schwach F., Studholme D., Baulcombe D.C. (2008). PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc. Natl. Acad. Sci. USA 105: 3145–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme N., Gans M., Masson M., Terzian C., Bucheton A. (1995). Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics 139: 697–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumble S.M., Lacroute P., Dalca A.V., Fiume M., Sidow A., Brudno M. (2009). SHRiMP: Accurate mapping of short color-space reads. PLOS Comput. Biol. 5: e1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber P., Wellmer F., Gheyselinck J., Riechmann J.L., Meyerowitz E.M. (2007). Redundancy and specialization among plant microRNAs: Role of the MIR164 family in developmental robustness. Development 134: 1051–1060 [DOI] [PubMed] [Google Scholar]
- Valoczi A., Varallyay E., Kauppinen S., Burgyan J., Havelda Z. (2006). Spatio-temporal accumulation of microRNAs is highly coordinated in developing plant tissues. Plant J. 47: 140–151 [DOI] [PubMed] [Google Scholar]
- Vaughn M.W., et al. (2007). Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 5: e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Warthmann N., Das S., Lanz C., Weigel D. (2008). Comparative analysis of the MIR319a microRNA locus in Arabidopsis and related Brassicaceae. Mol. Biol. Evol. 25: 892–902 [DOI] [PubMed] [Google Scholar]
- Wierzbicki A.T., Ream T.S., Haag J.R., Pikaard C.S. (2009). RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 41: 630–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138: 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., Liu J., Liu B., Li P., Meyers B.C., Chen X., Cao X. (2008). Small RNA-directed epigenetic natural variation in Arabidopsis thaliana. PLoS Genet. 4: e1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henderson I.R., Lu C., Green P.J., Jacobsen S.E. (2007). Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. USA 104: 4536–4541 [DOI] [PMC free article] [PubMed] [Google Scholar]