This article describes the localization and organization of several members of a family of proteins known as the reticulons that reside in the membrane of the endoplasmic reticulum (ER), which is responsible for synthesizing proteins for export out of the cell. The reticulons reside in the ER membrane, interact with each other, and induce curvature to make these ER compartments tubular in structure.
Abstract
The cortical endoplasmic reticulum (ER) in tobacco (Nicotiana tabacum) epidermal cells is a network of tubules and cisternae undergoing dramatic rearrangements. Reticulons are integral membrane proteins involved in shaping ER tubules. Here, we characterized the localization, topology, effect, and interactions of five Arabidopsis thaliana reticulons (RTNs), isoforms 1-4 and 13, in the cortical ER. Our results indicate that RTNLB13 and RTNLB1-4 colocate to and constrict the tubular ER membrane. All five RTNs preferentially accumulate on ER tubules and are excluded from ER cisternae. All isoforms share the same transmembrane topology, with N and C termini facing the cytosol and four transmembrane domains. We show by Förster resonance energy transfer and fluorescence lifetime imaging microscopy that several RTNs have the capacity to interact with themselves and each other, and we suggest that oligomerization is responsible for their residence in the ER membrane. We also show that a complete reticulon homology domain is required for both RTN residence in high-curvature ER membranes and ER tubule constriction, yet it is not necessary for homotypic interactions.
INTRODUCTION
The endoplasmic reticulum (ER) is the site of synthesis for many cellular proteins and the port of entry into the secretory pathway. The plant ER is an extremely dynamic organelle with a unique architecture consisting of a network of membrane tubules and sheets (cisternae) connected by three-way junctions (for a recent review, see Sparkes et al., 2009b). In cells of plant vegetative tissues, the cortical ER is predominantly tubular with small cisternal patches. This shape is independent of the ER being attached to a functional cytoskeleton (Boevink et al., 1998; Dreier and Rapoport, 2000), indicating that factors within the membrane itself must be responsible for conferring its curvature. Recently, a class of ER membrane proteins named the reticulons (RTNs) was shown to be sufficient to induce ER tubulation in yeast cells, mammalian cells (Voeltz et al., 2006), and in artificial proteoliposomes (Hu et al., 2008). It is assumed that these curvature-inducing properties result from the wedge-like transmembrane topology of the conserved structural motif of these proteins (the reticulon homology domain [RHD]), which includes two large hydropobic segments (Shibata et al., 2009).
The RTN gene family is much larger in higher plants than it is in mammals, with Arabidopsis thaliana having 21 RTN genes (Oertle et al., 2003; Nziengui et al., 2007; Nziengui and Schoefs, 2009; Sparkes et al., 2009b). This suggests that the numerous RTN isoforms may perform plant-specific functions. However, very little information is available regarding plant RTNs and their potential functions. RTNLB (for Reticulon Like Protein subfamily B) 1, 2, and 4 were isolated in a screen for agrobacterial pilin-interacting proteins, and a direct correlation between the level of RTNLB1 and plant susceptibility to Agrobacterium tumefaciens transformation was documented (Hwang and Gelvin, 2004). We have recently shown that the smallest plant reticulon isoform, RTNLB13, localizes to the ER membrane and, when overexpressed, can dramatically affect ER morphology by causing constrictions in the ER tubules (Tolley et al., 2008). These constrictions reduce diffusion within the ER lumen but do not cause major anterograde (from the ER toward the Golgi) trafficking defects.
The ability of a surfeit of RTN to cause constrictions fits the hypothesis that RTNs not only individually bend the membrane but can also form multimeric, arc-like structures (Shibata et al., 2008, 2009), which may ultimately shape and determine the diameter of ER tubules (Hu et al., 2008). It is not known whether the properties of RTNLB13, which contains the conserved RHD but has small soluble N- and C-terminal domains and is predicted to be expressed in seeds (see Supplemental Figure 1 online; Winter et al., 2007), are shared by the more ubiquitously expressed members of the RTN family, such as RTNLB1, 2, 3, and 4 (see Supplemental Figure 1 online). Here, we characterized the location, topology, ER-shaping function, and interactions of these five isoforms. We report that all five proteins share the same topology and general properties, indicating that these are likely to be shared by the other, less abundant isoforms. We also show that homo- and heterotypic interactions between RTN are responsible for their residence in the ER membrane.
RESULTS
RTNLB1-4 Localize to the ER Membrane and Remodel the ER Lumen
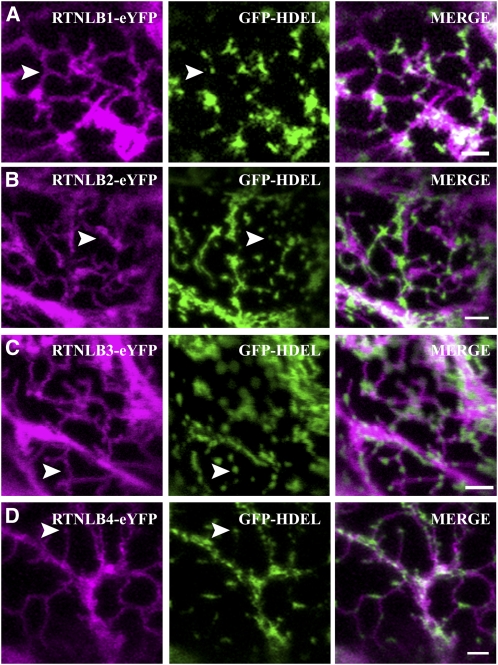
RTNLB1-4 were cloned from Arabidopsis seedling cDNA, and fluorescent fusions were transiently expressed in tobacco (Nicotiana tabacum) epidermal cells by agroinfiltration (Sparkes et al., 2006), owing to the extremely high transformation efficiency of this system and its prior use to characterize RTNLB13 (Tolley et al., 2008). Both N- and C-terminal fusions to full-length RTNLB1-4 genes resulted in localization patterns similar to ER networks. Coexpression of these fusions with an ER marker green fluorescent protein (GFP-HDEL), confirmed their ER location and indicated that all isoforms can induce constriction of ER tubules, similar to that previously observed with both untagged and yellow fluorescent protein (YFP)-tagged RTNLB13 overexpression (Figures 1A to 1D).
Figure 1.
RTNLB1-4 Colocalize to and Constrict Tubular ER.
RTNLB1-eYFP (A), RTNLB2-eYFP (B), RTNLB3-eYFP (C), and RTNLB4-eYFP (D) were transiently coexpressed with the ER lumenal marker GFP-HDEL (green) in tobacco leaf epidermal cells. All RTN isoforms constrict the ER, resulting in pockets of lumenal content (arrowheads). Bars = 2 μm.
Reticulons Are Enriched in the Tubular ER
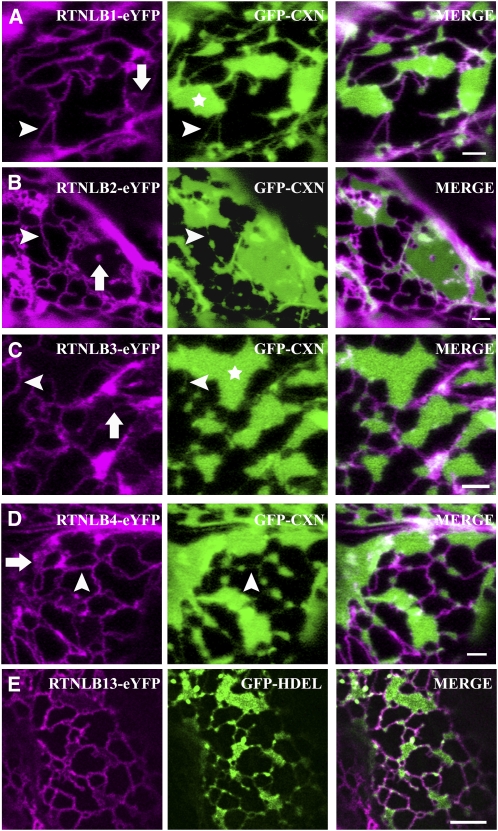
By virtue of their topology, mammalian and yeast reticulon/YOP (YIP 1 partner) proteins are predicted to insert preferentially into high-curvature membranes (i.e., tubules rather than cisternae) (Shibata et al., 2009). We tested whether this is also true for our selected plant RTN isoforms. As the cortical ER in leaf epidermal cells exists predominantly in tubular rather than cisternal form, we used two strategies to induce the formation of ER sheets. First, it has been previously shown that the overexpression of the transmembrane domain of GFP-calnexin (GFP-CXN) (Runions et al., 2006), a single-pass transmembrane ER protein, causes dilation of ER membranes and induces formation of cisternae. When RTNLB1-eYFP was coexpressed with GFP-CXN, RTNLB1 localization appeared to be limited to the ER tubules and cisternal rims (Figure 2A), while GFP-CXN was enriched in the cisternal regions of the membrane (Figure 2A, asterisk). Similar results were observed for RTNLB2, 3, and 4 fusions (Figures 2B to 2D). RTN-eYFP fusions occasionally located to tubular structures that traversed and perforated ER cisternae, indicating substructure within cisternae (Figure 2, arrows). Additionally, all full-length RTN fusions colocated to the ER contiguous with the nuclear envelope (see Supplemental Figure 2 online).
Figure 2.
RTNLB1-4 and RTNLB13 Preferentially Localize to High-Curvature ER Membranes.
(A) to (D) RTNLB1-eYFP (A), RTNLB2-eYFP (B), RTNLB3-eYFP (C), and RTNLB4-eYFP (D) were transiently coexpressed with GFP-CXN (green) in tobacco leaf epidermal cells. Whereas some RTNLB was present on ER tubules (arrowheads in magenta panels), the vast majority was confined to the rims of ER cisternae (asterisks). On occasion there seemed to be low levels and/or parallel tubules of RTNLB over the sheets (arrows). Unlike coexpression with an ER luminal marker, occasional fine tubular connections containing the ER membrane marker (GFP-CXN) were observed (arrowheads in green panels). Bars = 2 μm.
(E) Tobacco epidermal cells coexpressing RTNLB13-YFP (magenta) and GFP-HDEL were treated with 50 μg/mL BFA for 30 min. Note that RTNLB13 is enriched in ER tubules and in the rim of the BFA-induced ER sheets. Bar = 5 μm.
Treatment with the secretory inhibitor brefeldin A (BFA) causes reabsorption of Golgi membranes into the ER and induction of extensive ER cisternae (Boevink et al., 1999). Figure 2E shows that when cells coexpressing RTNLB13-eYFP and GFP-HDEL were treated with BFA, extensive cisternal sheets were produced but RTNLB13-eYFP remained mostly excluded from them and inhabited only the rims of the sheets and the rest of the tubular network (Figure 2E). Taken together, these results indicate that the five plant reticulons in this study localize to tubular ER.
RTNLB1-4 and -13 Share the Same Transmembrane Topology
Topology prediction with TOPCONS (Bernsel et al., 2008) indicates that all 21 Arabidopsis RTN genes have the same W topology (i.e., N and C termini in the cytosol and the central loop between transmembrane domains [TMDs] 2 and 3 also in the cytosol) (see Supplemental Figure 3 online). The only experimental determination of RTN topology available to date was performed on mammalian Rtn4c from rat (Voeltz et al., 2006), and it is not known whether this topology is shared by the whole family of RTN proteins. We experimentally assessed the topology of Arabidopsis RTNLB1-4 and -13 using three methods: redox-sensitive GFP (roGFP2; Schwarzlander et al., 2008; Brach et al., 2009), protease protection assays, and bimolecular fluorescence complementation (Zamyatnin et al., 2006).
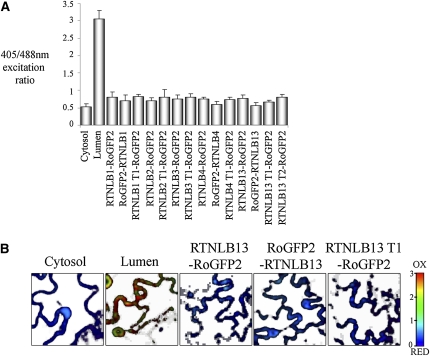
RoGFP2, a mutagenized form of GFP, interacts with the intracellular glutathione pool and detects differences in the glutathione redox potential (EGSH), which is generated by reduced glutathione (GSH) and its oxidized form GSSG. roGFP2 has differing excitation spectra dependent on the local EGSH and allows ratiometric analyses, which can be exploited for determination of membrane protein topology in membranes separating a steep EGSH gradient (Meyer et al., 2007; Brach et al., 2009). A more negative EGSH, as found in the cytosol, results in higher excitation at 488 nm, whereas 405 nm is more efficient at exciting roGFP2 in an environment with less negative EGSH, such as the ER lumen. Therefore, RTN N- and C-terminal fusions to roGFP2 indicate whether termini face the ER lumen or the cytosol.
To prevent movement and remodeling of the ER during sequential 405/488-nm image capture, samples were incubated in latrunculin B to disrupt the actin cytoskeleton (Sparkes et al., 2009a). Fast scans, at lower resolution, of untreated cells expressing the roGFP2 RTN fusions were also taken to minimize the effects of ER movement on ratiometric analysis. Both sets of data gave similar results: the N- and C-terminal fusions each showed low 405/488-nm fluorescence ratios, indicating a reducing environment close to that measured for the cytosolic control. Therefore, we concluded that both N and C termini of RTNLB1, 4, and 13 and the C termini of RTNLB2 and 3 face the cytosol (Figure 3). Data for roGFP2-RTNLB2/3 were not taken as the fusions were not expressed.
Figure 3.
Determination of RTNLB1-4 and -13 Topology through Ratiometric Imaging of roGFP2 Fusions.
RoGFP2 fusions to RTNLB1-4 and -13 were transiently expressed in tobacco leaf epidermal cells and imaged at 405 and 488 nm to detect oxidized and reduced forms, respectively. Example of ratiometric analysis from a single experiment, including standard deviations where n ≥ 6. The reduced/oxidized forms are presented in the histogram and controls for ER lumen and cytosol are indicated (A). Representative ratiometric images for controls and RTNLB13 fusions are shown, with red representing more oxidized and blue representing more reduced (B). Note that T1 and T2 refer to truncations 1 and 2 (see Figure 6A): RTNLB1 T1 (1 to 153 amino acids), RTNLB2 T1 (1 to 150 amino acids), RTNLB3 T1 (1 to 127 amino acids), RTNLB4 T1 (1 to 132 amino acids), RTNLB13 T1(1 to 93 amino acids), and RTNLB13 T2 (1 to 131 amino acids).
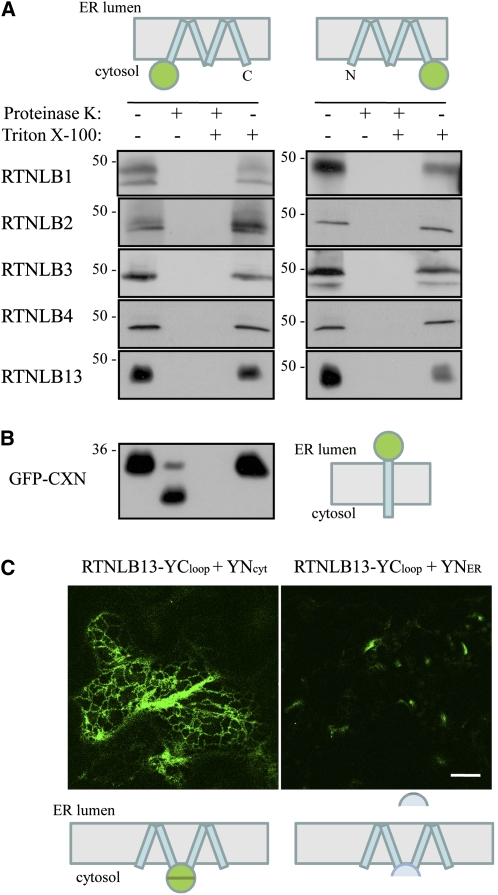
In parallel with these experiments, we performed protease protection assays on microsomes purified from leaf sectors infiltrated with eYFP-tagged versions of RTNLB1-4 and -13 (Figure 4). Incubation of microsomes with proteinase K resulted in the loss of the eYFP moiety from all RTN constructs, as revealed by immunoblotting with GFP antisera (Figure 4A). Both N- and C-terminally appended eYFP were susceptible to protease digestion, indicating that the N and C termini of all the RTN isoforms analyzed were exposed to the cytosol. As a control, in microsomes from leaf sectors expressing GFP-CXN (in which GFP faces the ER lumen), the GFP moiety was indeed protected from protease digestion (Figure 4B). Protection was lost when the microsomal membranes were solubilized with the detergent Triton X-100 (Figures 4A and 4B).
Figure 4.
Determination of RTNLB1-4 and -13 Topology through Protease Protection and BiFC.
(A) and (B) Microsomes from tobacco epidermal leaves expressing the indicated constructs were subjected to proteinase K treatment in the presence or absence of Triton X-100. Microsomal proteins were solubilized, resolved by SDS-PAGE, and subjected to immunoblot with GFP antiserum. Numbers indicate molecular size in kilodaltons.
(C) Tobacco epidermal cells were agroinfiltrated with the indicated expression construct combinations and visualized by confocal microscopy. Bar = 20 μm
The cartoons depict topological orientations of N- and C-terminal fusions for the protein fusion(s) under study. Blue rectangles represent transmembrane domains, and green circles represent GFP.
Both sets of results confirm that N and C termini of RTNLB1-4 and -13 are cytosolic but do not discriminate between a V configuration of TMDs, in which the large hydrophobic segments of the RHD span the membrane twice, with a connecting hydrophilic luminal loop, and a W configuration, in which each hydrophobic region forms a hairpin structure resulting in four membrane-spanning regions and a cytosolic connecting loop. The latter topology is predicted by TOPCONS (see Supplemental Figure 3 online). To discriminate between these two possibilities, we adopted two strategies: (1) expression of roGFP2 fusions to RTN truncations lacking the second hydrophobic stretches in the RHD (RTN truncation 1, schematized in Figure 6A), and (2) bimolecular fluorescence complementation (BiFC). This technique is based on the coexpression of proteins fused to either the N- or C-terminal half of YFP. It has been recently shown that, when overexpressed, the two halves interact and therefore yield a fluorescent YFP, if they are located in the same subcellular compartment (Zamyatnin et al., 2006). Thus, this technique is useful for the determination of plant membrane protein topology.
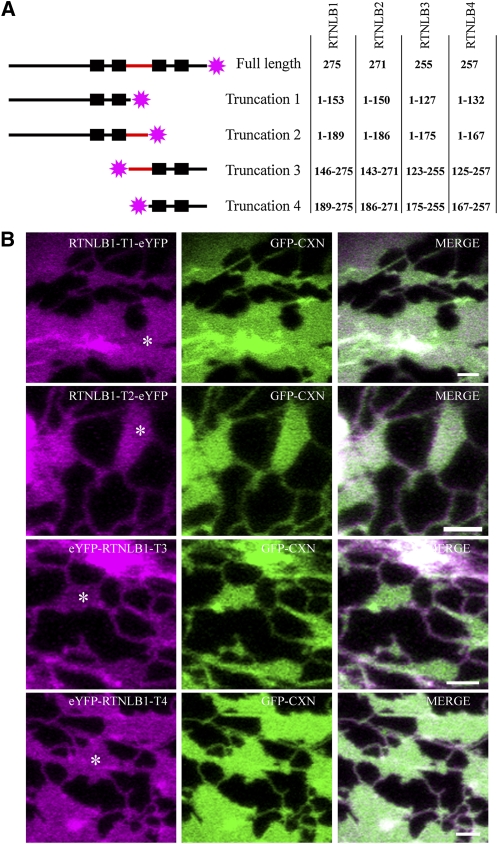
Figure 6.
RTNLBs Require a Fully Functional RHD to Constrict the ER and Constrain It to Cisternal Rims.
Truncations of RTNLB1-4 were generated containing either the N terminus with the first stretch of hydrophobic sequence (two black boxes) without (truncation 1) or with (truncation 2) the predicted loop domain (red) or only the C terminus with the second stretch of hydrophobic sequence with (truncation 3) or without (truncation 4) the predicted loop domain. The truncations were fused to eYFP (pink stars), and the amino acids of each RTNLB fusion are shown (A). Note that each stretch of hydrophobic sequence is predicted to contain two TMDs (black boxes) and that this has been verified experimentally. Images of tobacco leaf epidermal cells coexpressing RTNLB1 truncations 1-4 (T1-T4) with GFP-CXN are depicted (B). Bars = 2 μm.
RoGFP2 fusions to the N-terminal half of RTNLB1-4 lacking the second hydrophobic segment were generated (Figure 3A; also see Figure 6A schematic truncation 1). Ratiometric imaging indicated a cytosolic location for the roGFP2, thus suggesting a W shaped topology. The alternative V topology would have resulted in the probe residing on the luminal face of the ER membrane, and this was not observed.
For BiFC, we generated an expression construct where the C-terminal half of YFP was cotranslationally fused between Val-91 and Val-92 of the predicted central loop of RTNLB13 (RTNLB13YCloop) (Figure 4C). We then coexpressed RTNLB13YCloop in tobacco leaf cells with the complementary N-terminal half of YFP, targeted either to the cytosol (YNcyt) or to the ER lumen (YNER) (Zamyatnin et al., 2006). Figure 4C shows that complementation of YFP and, therefore, fluorescence was clearly detectable with the coexpression of YNcyt. Moreover, the interaction of the two YFP halves clearly highlighted the ER network (Figure 4C). This result corroborates the roGFP2 results and strongly suggests that the RHD loop is located in the cytosol, therefore confirming the TOPCONS prediction of a W topology. This prediction is also maintained for RTNLB13YCloop (see Supplemental Figure 3 online), indicating that the addition of the C-terminal half-barrel of YFP is unlikely to affect membrane insertion of this construct. We also used BiFC to further test the topology of the N and C termini of RTNLB13. The results confirm the protease protection assays and roGFP2 fusion data and indicate that both ends of RTNLB13 are cytosolic (see Supplemental Figure 4 online).
The Dilysine ER Retrieval Signal Is Not Required for ER Residence of RTNLB13
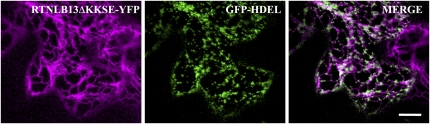
Eighteen out of 21 Arabidopsis RTN isoforms are predicted to contain the C-terminal ER retrieval dilysine motif (KKXX; Nziengui et al., 2007). To test whether the dilysine motif is necessary in the context of the full-length protein, we generated a deletion mutant of RTNLB13 lacking the four C-terminal residues (ΔKKSE). When expressed in tobacco epidermal cells, this mutant both showed ER location and maintained the ability to constrict the ER lumen (Figure 5). We therefore conclude that this motif is not essential for ER retention. In addition, expression of the first half of RTNLB1-4 as an eYFP fusion (truncation 1, Figure 6A), shows that the first large hydrophobic region (i.e., the first two predicted transmembrane domains) alone is sufficient to afford ER residence (Figure 6B). This further indicates that an alternative mechanism is responsible for the ER retention of the reticulons under study.
Figure 5.
RTNLB13 Location to the ER Is Not Dependent on Its Dilysine ER Retention Signal.
Tobacco epidermal cells were agroinfiltrated with RTNLB13ΔKKSE (magenta) and GFP-HDEL (green). Bar = 20 μm.
RTNLB1-4 Require Complete RHDs to Efficiently Constrict the ER and Constrain RTN to Tubules and Cisternal Rims
To determine the minimal structure of RTN that is capable of constricting ER tubules, we tested the requirement for the number of transmembrane domains. The topology experiments described above indicate that all the five RTN proteins under study contain four TMDs. Therefore, we generated RTN truncations containing two rather than four TMDs and monitored both their location and effect on tubular ER morphology (Figure 6).
Truncations containing either the first two TMDs, the first two TMDs plus cytosolic loop, the last two TMDs, or the last two TMDs plus cytosolic loop were generated (Figure 6A). In general, RTN truncation fusions lacking two of the four TMDs still localized to the ER but failed to induce tubule constrictions (Figure 6B, RTNLB1; RTNLB2-4 data not shown). This was irrespective of whether the N-proximal or the C-proximal TMD pair was removed and also independent of the presence of a full-length cytosolic loop domain. Remarkably, however, the location of truncated RTNs on the ER membrane was no longer limited to tubules and cisternal rims (as shown in Figure 2) but became evenly spread over the ER membrane, including cisternal surfaces (Figure 6B). Similar results were obtained for a RTNLB13 fusion lacking the C-proximal TMD pair (data not shown).
We therefore conclude that both localization of RTN to high-curvature ER regions and their capacity to constrict ER tubules requires a complete RHD (i.e., all four TMDs and intervening loop). Targeting to the ER is, however, independent of the presence of the complete RHD, with only two TMD being sufficient for ER localization.
Occasionally, eYFP-RTNLB2 truncations 3 and 4 were observed to constrict the ER in very few cells (see Supplemental Figure 5 online).The resulting phenotype indicated that the cisternal ER was undergoing constriction producing small pockets of the luminal ER marker, GFP-HDEL, on cisternae (see Supplemental Figure 5 online). This was not observed with an ER membrane marker.
RTNLB1 and RTNLB13 Can Form Both Homo- and Heterotypic Interactions
If deletion of the C-terminal KKXX does not result in loss of RTN, what then is anchoring the protein in the ER membrane? Shibata et al. (2008) showed that mammalian Rtn4a and yeast Rtn1 have the ability to form oligomers in the ER membrane. It is possible such oligomerization prevents the proteins from escaping from the ER while enhancing their ability to induce membrane curvature. We therefore decided to test homo- and heterotypic RTN association using Förster resonance energy transfer (FRET) measured by donor-excited state fluorescence lifetime imaging (FLIM). FRET-FLIM measures the reduction in the life time of GFP (donor) fluorescence when an acceptor fluorophore, in this case monomeric red fluorescent protein (mRFP), is close enough for FRET to occur, thus indicating physical interactions between the protein fusions (Osterrieder et al., 2009). Limitations in the speed of photon counting in the FLIM system required us to monitor a region of the ER with relatively low mobility, such as ER associated with the nuclear envelope (see Supplemental Figures 2A and 2B online).
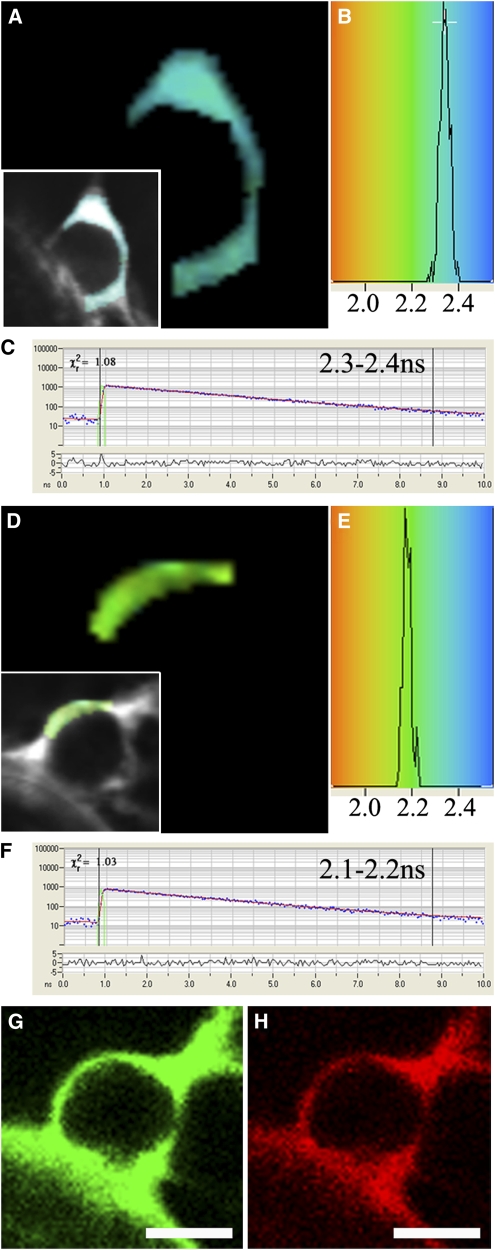
Combinations of eGFP (donor) and mRFP (acceptor) RTN fusions were transiently coexpressed in tobacco leaf epidermal cells and interactions assessed through FRET-FLIM. Previous FRET-FLIM studies have shown that Golgi-targeted fusion pairs STGFP/STmRFP (ST being the signal anchor sequence of a rat sialyl transferase; Boevink et al., 1998) do not interact, whereas At GRIP-GFP/mRFP-ARL1 do interact (At GRIP is a trans-Golgi matrix protein, and ARL1 is a small Rab-like GTPase). These pairs showed respective lifetimes for GFP of 2.5 to 2.4 ns and 1.8 to 2.0 ns and were used as negative and positive controls, respectively, for the system (Osterrieder et al., 2009). Figure 7 shows lifetime images of RTNLB1-eGFP expressed alone (Figures 7A to 7C) and RTNLB1-eGFP coexpressed with mRFP-RTNLB1 (Figures 7D to 7H). The pseudocolored lifetime map (Figures 7A and 7D) reflects the lifetime values of each point within the region of interest. The distribution of lifetimes within the region of interest is depicted (Figures 7B and 7E) where blue shades represent higher eGFP fluorescence lifetimes than those in green. Representative decay curves (Figures 7C and 7F) of a single point highlight an optimal single exponential fit (where points with χ2 values from 0.9 to 1.4 were taken). Hence, we concluded that RTNLB1 homodimerizes because the lifetime values for the eGFP/mRFP fusion pair (Figure 7) are lower than those for the eGFP fusion alone. The range of lifetimes for each RTN pair is shown in Table 1. The average of the range indicates at least a 0.2-ns decrease in lifetime of the pairs compared with the control donor expressed alone. Therefore, all combinations of full-length RTN and truncations tested appeared to oligomerize (Table 1). The fact that homotypic interactions (at least the ones we tested for RTNLB1 and RTNLB13; Table 1) were also observed for the truncated constructs indicates that the two C-terminal TMDs are not necessary for RTN to interact.
Figure 7.
FRET-FLIM Analysis of RTNLB1 Dimerization in Tobacco Leaf Epidermal Cells.
RTNLB1-eGFP ([A] to [C]) and coexpression with mRFP-RTNLB1 ([D] to [H]) in tobacco leaf epidermal cells were tested for interaction through FRET-FLIM. A region of interest on the ER continuous with the nuclear envelope was selected ([A] and [D], see insets) based on the lifetime decay curves of points within the region having a χ2 value between 0.9 and 1.4 ([C] and [F]). The lifetimes of the points within the selected region are displayed ([B] and [E]) and psuedocolored accordingly in (A) and (D). Confirmation of coexpression of eGFP and mRFP RTNLB1 fusions are shown ([G] and [H]). The lifetime of the combination is >0.1 ns lower than the donor fusion alone, indicating RTNLB1 dimerization. Bars = 5 μm.
Table 1.
FRET-FLIM Lifetimes of RTNLB Pairs
| Donor | Acceptor | Range of FLIM Lifetimes (ns) | Average FLIM Lifetime (ns) |
| RTNLB1-eGFP | – | 2.3–2.4 | 2.35 |
| RTNLB1-eGFP | mRFP-RTNLB1 | 2.1–2.2 | 2.15 |
| RTNLB1-eGFP | RTNLB1-mRFP | 2.1–2.2 | 2.15 |
| eGFP-RTNLB1 | – | 2.3–2.5 | 2.40 |
| eGFP-RTNLB1 | mRFP-RTNLB1 | 2.1–2.2 | 2.15 |
| eGFP-RTNLB1 | RTNLB1-mRFP | 2.1–2.3 | 2.20 |
| RTNLB1 T1-eGFP | – | 2.3–2.5 | 2.40 |
| RTNLB1 T1-eGFP | RTNLB1 T1-mRFP | 2.1–2.2 | 2.15 |
| eGFP-RTNLB13 | – | 2.2–2.5 | 2.30 |
| eGFP-RTNLB13 | mRFP-RTNLB13 | 1.9–2.0 | 1.95 |
| RTNLB13 T1-eGFP | – | 2.3–2.4 | 2.35 |
| RTNLB13 T1-eGFP | RTNLB13 T1-mRFP | 2.0–2.2 | 2.10 |
| eGFP-RTNLB1 | mRFP-RTNLB3 | 2.1–2.3 | 2.20 |
| eGFP-RTNLB1 | mRFP-RTNLB4 | 2.1–2.3 | 2.20 |
| eGFP-RTNLB13 | mRFP-RTNLB3 | 2.1–2.2 | 2.15 |
| eGFP-RTNLB13 | mRFP-RTNLB4 | 2.0–2.1 | 2.05 |
mRFP and eGFP pairs of RTNLB fusions were coexpressed in tobacco epidermal cells and interactions tested through FRET-FLIM. Comparisons with control values without acceptor indicate that all pairs tested interact with one another.
DISCUSSION
The cortical ER network in plants is extremely dynamic with regular tubule outgrowth and transitions between tubules and cisternae being common. Based on several observations, a functional role of ER remodeling in secretion has been proposed. Actively growing and dividing cells have been shown to contain more cisternal ER than nondividing mature cells (Ridge et al., 1999), and heavily secreting root cap cells show an abundance of cisternal ER (Stephenson and Hawes, 1986). Hence, cisternalization may increase the surface area and consequently the capacity of the ER for secretion. Therefore, characterization of the key cellular players in ER remodeling, in particular those controlling tubulation versus cisternalization of the ER could have important biotechnological implications for protein production.
The relative roles of the cytoskeleton, several myosins (XIK and XIJ myosin tail domains) and Golgi bodies in these processes were recently addressed (Sparkes et al., 2009a, 2009c). To further understand the factors that control the physical constrictive process resulting in ER tubule formation over cisternae, we performed this comprehensive study on RTNs. Previously, we have shown that RTNLB13 locates to the ER membrane and, on transient expression, constricts the cortical ER (Tolley et al., 2008). Here, we have shown that other members of the family, RTNLB1-4, have a similar effect on the ER and locate to ER tubules, but are constrained to the rims of ER cisternae. The ER location for RTNLB2 and 4 is in agreement with previous studies (Nziengui et al., 2007); however, these authors also reported that RTNLB4 labeled motile puncta, which could reflect a difference between the tissue types (mesophyll derived protoplasts versus epidermal cells) under study. As truncated forms of the reticulons are no longer confined to tubules but label the whole ER network, including the cisternae, we conclude that a complete RHD, comprising all four transmembrane domains, is required for both efficient ER constriction and for partitioning to tubules and cisternal rims. This indicates that these transmembrane regions, and not the N- and C-terminal cytosolic regions, may be directly implicated in conferring the wedge-like shape that is likely to promote membrane curvature. Thus, given that all 21 Arabidopsis RTN isoforms share a highly conserved RHD (Nziengui et al., 2007; Nziengui and Schoefs, 2009; Sparkes et al., 2009b), the remaining members of this family are also likely to be ER tubule-localized proteins. However, in the case of RTNLB2 RHD truncations 3 and 4, which have a similar localization pattern to the other RTN truncations and overlie the entire ER cisternae, they also, on rare occasions, have a propensity to constrict the cisternal ER into nodular clumps (see Supplemental Figure 5 online). Since we have not tested RTNLB2 homotypic interactions, this could reflect differing levels of RTN interaction in the cell.
The requirement for a high level of membrane curvature is not restricted to the ER. Many organelles can have tubular extensions; this is seen in the case of tubular extensions on plastids known as stromules (Kwok and Hanson, 2004) and similarly in peroxules and matrixules on peroxisomes and mitochondria (Scott et al., 2007; Sinclair et al., 2009, reviewed in Shibata et al., 2009). Golgi cisternae also demonstrate a high degrees of curvature. Such curvature can be generated by a number of mechanisms (Shibata et al., 2009), including the action of enzymes that bend membranes into tubules as suggested for plant dynamins at the phragmoplast (Fujimoto et al., 2008). Proteins lying on the surface of a membrane, such as Bar domain proteins, can form crescent-shaped dimers on the bilayer, thereby curving the membrane (Suetsugu et al., 2010). Others, such as phospholipase A, are thought to modify phospholipids into a wedge shape, and insertion of these into in the outer leaflet of membranes, such as the rims of Golgi cisternae, increase the surface area of the outer leaflet as opposed to the inner, thus inducing curvature (San Pietro et al., 2009). It could be envisaged that the reticulons work by a similar mechanism (Voeltz et al., 2006; Shibata et al., 2009). This could result from the unusual length of each of their four TMDs. Each of the four TMDs in the RHD most likely exceeds 20 amino acid residues in length (e.g., 24 residues for RTNLB13), which differs from the observed average TMD length of 17 residues for proteins that locate to the ER membrane (Brandizzi et al., 2002). It is tempting to speculate that the extra length of the RTN membrane-spanning helices causes them to insert at an angle, ultimately conferring RTN the wedge-like topology that results in curvature.
We have shown that, in the proteins studied here, the C-terminal ER retention motif (which is necessary for the retrieval of many ER membrane proteins) is not required for ER residence of RTN. We have further shown that the presence of the N-terminal half of the RHD is sufficient for ER localization. Our FRET-FLIM data suggest a general mechanism for ER retention that may depend on interactions between reticulons. The capacity of RTNLB1 to interact with itself, RTNLB2, and RTNLB4 has been previously inferred by yeast two-hybrid assays and was shown to require an intact RHD (Hwang and Gelvin, 2004). Our FRET/FLIM data extend and rationalize these findings in vivo in the context of RTN function and localization. Homotypic interactions of RTNLB1 and RTNLB13 do not require a complete RHD, indicating that the presence of only a subset of TMDs is sufficient for interaction and ER residence. However, as only full-length reticulons are restricted to areas of high curvature, but truncations, even if they oligomerize, can insert into flat sheets, we hypothesize that the W topology combined with homo- or heterotypic interactions are necessary for the induction of membrane curvature. However, the full role of RTN oligomerization in plants remains to be addressed.
In summary, our data distinguish between the capacity of RTN to localize to the ER, which does not require a complete RHD but may depend on RTN–RTN interactions, and the membrane-shaping properties of RTN, which depend on the full RHD and also result in RTN partitioning to high-curvature ER membranes. Our data indicate a conserved location, topology, function, and mechanism of residence for the five most abundantly expressed members of the Arabidopsis RTN gene family. Downregulation of RTNLB1, 2, or 4 by T-DNA insertion, antisense RNA, or RNA interference resulted in plants being less susceptible to Agrobacterium infection, but no obvious plant growth phenotype was observed, likely indicating high functional redundancy between these isoforms (Hwang and Gelvin, 2004). Accordingly, only a triple knockout of Rtn1p, Rtn2p (the two yeast RTNs), and the structurally similar Yop1p resulted in detectable loss of ER tubules in yeast (Shibata et al., 2008). Even by invoking redundancy, however, it is somewhat difficult to explain the unique expansion of the RTN family in higher plants (Sparkes et al., 2009b). It is tempting to speculate that, aside from the abundant, ubiquitously expressed RTNs we described in this work, less abundant isoforms are involved in shaping specific ER subdomains in particular tissues or cell types. Only the detailed analysis of the expression of these isoforms will shed light on this issue.
METHODS
Generation of RTN Clones
The complete list of primers used in this study is shown in Supplemental Table 1 online.
RTNLB1-4 were cloned by RT-PCR using a Superscript III one-step RT-PCR platinum Taq HiFi kit (Invitrogen) from total RNA that was extracted from various Arabidopsis thaliana tissues using the Nucleospin RNA II kit (Macherey-Nagel). Using Gateway technology (Invitrogen) all cloned products were subsequently cloned into pDONR 207 and then into their respective binary vectors; eYFP fusions required pCAMBIA 1300 derived vectors (Sparkes et al., 2005), and roGFP2 fusions required pSS01 (Brach et al., 2009) and pCM01 (described below) for N- and C-terminal fusions, respectively. A new destination vector for N-terminal fusions to target proteins was created by PCR amplifying roGFP2, thereby adding a 5′-SpeI and a 3′-EcoRV restriction site. eGFP was replaced by roGFP2 in pK7WGF2,0 to create pCM01 (Karimi et al., 2005). N- and C-terminal mRFP (pB7WGR2 and pB7RWG2) and eGFP (pB7WGF2 and pB7FWG2) fusions required the respective vectors as listed (Karimi et al., 2002). RTNLB1-4 truncations were amplified from the respective pDONR clone using primers detailed in Supplemental Table 1 online and then subsequently recombined into pDONR 207 and then the destination vector. Binary clones were transformed into Agrobacterium tumefaciens GV3101. A GFP-HDEL protein fusion (containing the HDEL ER retention signal) was used as an ER marker for localization experiments.
The construction of RTNLB13-YFP has already been described (Tolley et al., 2008). Deletion of the ER retention signal KKSE to generate RTNLB13ΔKKSE was performed by QuickChange mutagenesis using the primers described in Supplemental Table 1 online. All RTNLB13 constructs for BiFC analysis were generated in the vectors described by Zamyatnin et al. (2006) using the primers indicated in Supplemental Table 1 online.
Plant Material and Transient Expression System
Nicotiana tabacum (cv Petit Havana SR1) was grown as described (Sparkes et al., 2005). Transient expression was performed as described (Sparkes et al., 2006). The final optical densities at OD600 for GV3101-containing Agrobacterium cultures were as follows: GFP-HDEL, 0.04; N- and C-terminal eYFP fusions to RTN, 0.05; roGFP2 RTN fusions, 0.05 and 0.1; roGFP2 cytosolic and HDEL controls, both at 0.03 and 0.05; GFP-CXN, 0.04; and CFP-SKL, 0.04. All BiFC constructs were infiltrated at OD600 = 0.05.
Sample Preparation and Imaging
Imaging was performed on a Zeiss LSM 510 (Meta) and on a Leica TCS SP5 using ×63 or ×100 oil immersion objective lenses. eYFP and GFP dual imaging was performed as described (Sparkes et al., 2009a).
The roGFP2 used was as described by Schwarzlander et al. (2008) and Brach et al. (2009). Its oxidized and reduced form excited differently at 405 and 488 nm, respectively, corresponding to the intracellular redox environment generated by glutathione. Ratiometric imaging was performed using a Zeiss LSM510 Meta confocal microscope with a ×63 oil immersion objective lens. The roGFP2 was excited at 488 and 405 nm in multitrack mode with frame switching. Ratiometric analysis was performed using a custom written MATLAB script (kindly provided by Mark Fricker, Oxford University). The signal of each pixel from 405-nm images was divided by the 488-nm signal, and the ratio was pseudocolor coded to enable direct binary readout of the respective localization of the roGFP2-tag in the cytosol or the ER lumen. As it is known that roGFP2 is reduced in the cytosol and oxidized in the lumen of ER, the calculated ratio of roGFP2-HDEL and free cytoplasmic roGFP2 was used to calibrate the scale (Meyer et al., 2007; Brach et al., 2009).
Leaf tissue was excised, and ∼5-mm2 pieces were incubated in latrunculin B (2.5 μM, 1 M stock dissolved in DMSO) or BFA (50 μg/mL, 5 mg/mL stock dissolved in DMSO) as stated.
Protease Protection Assay
Four to eight Agrobacterium-infiltrated tobacco leaf sectors expressing the relevant construct were ground with a chilled mortar and pestle in 4 mL extraction buffer (10 mM KCl, 1 mM MgCl2, 0.4 mM sucrose, 0.4% PVP, and 40 mM HEPES-KOH, pH 7.5) for 3 min. The homogenate was transferred to two prechilled 2-mL Eppendorf tubes. The samples were centrifuged at 1000g for 5 min at 4°C, and the supernatants transferred to a fresh tube on ice. A sucrose pad was set up in a 4-mL ultracentrifuge tube. Sucrose (750 μL; 20% [w/v]) in extraction buffer was layered on top of 750 μL of 60% (w/v) sucrose in extraction buffer. Finally, 2 mL of sample was carefully added to the sucrose pad and centrifuged at 55,000 rpm for 30 min at 4°C in a Beckman TL-100 ultracentrifuge.
The microsomes form an interphase between the 20 and 60% sucrose layers. The majority of the upper phase was removed by aspiration, and 200 μL of the microsome fraction was recovered and gently resuspended in a fresh tube. The sample (25 μL) was preincubated with 20 mM CaCl2, PK buffer (50 mM Tris.HCl, pH 8.0, and 1 mM CaCl2) and 1% Triton X-100 for 15 min on ice to ensure membranes were disrupted (Ma et al., 2006) before adding the Proteinase K (Sigma-Aldrich) and incubating at 30°C for 30 min. To terminate the reaction, one Complete-mini protease inhibitor cocktail tablet (Roche) was dissolved in 1 mL of sterile distilled water, and 10 μL of this was added to each reaction tube. Sixty microliters of loading dye (0.5 M Tris.HCl, pH 6.8, 4.4% [w/v] SDS, 2% β-mercaptoethanol, 20% glycerol, and 0.036% bromophenol blue) was added, and the samples were boiled for 5 min before loading on a 15% SDS-PAGE gel. Immunoblot analysis was performed with rabbit polyclonal anti-GFP antibodies (Clontech).
BiFC
Fusions were made of either residues 1 to 154 of YFP (termed YN) or residues 155 to 239 (termed YC) to either the N or C terminus or the loop of RTNLB13 using conventional PCR, with pLH-YN, pLH-YC (Zamyatnin et al., 2006), and myc-RTNLB13 (Tolley et al., 2008) as templates. Primers were designed to add the restriction sites XhoI to the 5′ end and XbaI to the 3′ end of each construct (see Supplemental Table 1 online). For the insertion of YC in the loop region of RTNLB13, a fusion was made of residues 155 to 239 of YFP (termed YC) and was inserted between Val-91 and Val-92 of the central loop region of RTNLB13 using fusion PCR, using pLH-YC or myc-RTNLB13 as a template, respectively. Amplified fragments were cloned into the plant binary vector pLH7000 (Hausmann and Töpfer, 1999). The binary vector was then transformed into Agrobacterium strain C58 harboring the pSoup helper plasmid (Hellens et al., 2000). Reciprocal clones of each BiFC construct were also created to test the efficiency of expression, yielding YN-RTN, YC-RTN, RTN-YN, RTN-YC, and RTN-YC-RTN.
FRET-FLIM Data Acquisition
Tobacco leaf epidermal samples were excised, and FRET-FLIM data capture was performed according to Osterrieder et al. (2009) using a two-photon microscope at the Central Laser Facility of the Rutherford Appleton Laboratory with slight modifications. Briefly, a two-photon microscope was constructred around a Nikon TE2000-U inverted microscope using custom-made XY galvanometers (GSI Lumonics) for the scanning system. Laser light at a wavelength of 920 ± 5 nm was obtained from a mode-locked titanium sapphire laser (Mira; Coherent Lasers), producing 180-fs pulses at 75 MHz, pumped by a solid-state continuous-wave 532-nm laser (Verdi V18; Coherent Laser). The laser beam was focused to a diffraction-limited spot through a water immersion objective (Nikon VC ×60, numerical aperture of 1.2) and specimens illuminated at the microscope stage. Fluorescence emission was collected without descanning, bypassing the scanning system, and passed through a BG39 (Comar) filter to block the near infrared laser light. Line, frame, and pixel clock signals were generated and synchronized with an external fast microchannel plate photomultiplier tube (MCP-PMT; Hamamatsu R3809U) used as the detector. These were linked via a time-correlated single-photon-counting PC module SPC830 (Becker and Hickl) to generate the raw FLIM data. Prior to FLIM data collection, the eGFP and mRFP expression levels in the plant samples within the region of interest were confirmed using a Nikon eC1 confocal microscope with excitation at 488 and 543 nm, respectively. A 633-nm interference filter was used to significantly minimize the contaminating effect of chlorophyll autofluorescence emission that would otherwise obscure the mRFP emission as well as that of eGFP.
Data were analyzed by obtaining excited state lifetime values of a region of interest on the nucleus, and calculations were made using the SPCImage analysis software (Becker and Hickl). The distribution of lifetime values within the ROI were generated and displayed as a curve. Only values that had a χ2 between 0.9 and 1.4 were taken. The median lifetime value and minimum and maximum values for a quarter of the median lifetime values from the curve were taken to generate the range of lifetimes per sample. At least eight nuclei per RTN combination were analyzed, and the average of the ranges taken. Results are from two independent experiments.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RTNLB1, At4g23630; RTNLB2, At4g11220; RTNLB3, At1g64090; RTNLB4, At5g41600; and RTNLB13, At2g23640.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. eFP Browser Expression Predictions for RTNLB1-4 and -13.
Supplemental Figure 2. RTNLB1-4 and -13 Colocate to the ER Continuous with the Nuclear Envelope.
Supplemental Figure 3. Bioinformatic Topology Predictions for the RTN Isoforms under Study.
Supplemental Figure 4. BiFC Controls and N- and C-Terminal Topology.
Supplemental Figure 5. eYFP-RTNLB2 Truncation 4 Occasionally Affects the Cortical ER.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank E.I. Savenkov for the gift of his BiFC vector collection and M. Fricker for the MATLAB script. I.S. was funded by a Leverhulme grant (F/00 382/G), I.A. and J.S. were funded by an Erasmus scheme from Heidelberg University, and A.O. by Biotechnology and Biological Science Research Council (BBSRC) Grant BB/F008147/1. Work in the L.F. lab was supported in part by the European Union (LSH-2002-1.2.5-2 “Recombinant Pharmaceuticals from Plant for Human Health–Pharma-Planta”) and by the Leverhulme Trust (Grant F/00215/AP). N.T. was supported by a BBSRC studentship. Access to the Confocal Microscopy Laboratory at the Central Laser Facility, Rutherford Appleton Laboratory, was made possible through an Science and Technology Facilities Council Programme Access grant to C.H.
References
- Bernsel A., Viklund H., Falk J., Lindahl E., von Heijne G., Elofsson A. (2008). Prediction of membrane-protein topology from first principles. Proc. Natl. Acad. Sci. USA 105: 7177–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P., Martin B., Oparka K., Santa Cruz S., Hawes C. (1999). Transport of virally expressed green fluorescent protein through the secretory pathway in tobacco leaves is inhibited by cold shock and brefeldin A. Planta 208: 392–400 [Google Scholar]
- Boevink P., Oparka K., Santa Cruz S., Martin B., Batteridge A., Hawes C. (1998). Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 15: 441–447 [DOI] [PubMed] [Google Scholar]
- Brach T., Soyk S., Muller C., Hinz G., Hell R., Brandizzi F., Meyer A.J. (2009). Non-invasive topology analysis of membrane proteins in the secretory pathway. Plant J. 57: 534–541 [DOI] [PubMed] [Google Scholar]
- Brandizzi F., Frangne N., Marc-Martin S., Hawes C., Neuhaus J.M., Paris N. (2002). The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 14: 1077–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreier L., Rapoport T.A. (2000). In vitro formation of the endoplasmic reticulum occurs independently of microtubules by a controlled fusion reaction. J. Cell Biol. 148: 883–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M., Arimura S., Nakazono M., Tsutsumi N. (2008). Arabidopsis dynamin-related protein DRP2B is co-localized with DRP1A on the leading edge of the forming cell plate. Plant Cell Rep. 27: 1581–1586 [DOI] [PubMed] [Google Scholar]
- Hausmann L., Töpfer R. (1999). Entwicklung von Plasmid-Vektoren. BioEngineering für Rapssorten nach Maß. Vortr. Pflanzenzüchtg. 45: 155–173 [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hu J., Shibata Y., Voss C., Shemesh T., Li Z., Coughlin M., Kozlov M.M., Rapoport T.A., Prinz W.A. (2008). Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science 319: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Hwang H.H., Gelvin S.B. (2004). Plant proteins that interact with VirB2, the Agrobacterium tumefaciens pilin protein, mediate plant transformation. Plant Cell 16: 3148–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., De Meyer B., Hilson P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inze D., Depicker A. (2002). Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kwok E.Y., Hanson M.R. (2004). Stromules and the dynamic nature of plastid morphology. J. Microsc. 214: 124–137 [DOI] [PubMed] [Google Scholar]
- Ma B., Cui M.L., Sun H.J., Takada K., Mori H., Kamada H., Ezura H. (2006). Subcellular localization and membrane topology of the melon ethylene receptor CmERS1. Plant Physiol. 141: 587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A.J., Brach T., Marty L., Kreye S., Rouhier N., Jacquot J.P., Hell R. (2007). Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 52: 973–986 [DOI] [PubMed] [Google Scholar]
- Nziengui H., Bouhidel K., Pillon D., Der C., Marty F., Schoefs B. (2007). Reticulon-like proteins in Arabidopsis thaliana: Structural organization and ER localization. FEBS Lett. 581: 3356–3362 [DOI] [PubMed] [Google Scholar]
- Nziengui H., Schoefs B. (2009). Functions of reticulons in plants: What we can learn from animals and yeasts. Cell. Mol. Life Sci. 66: 584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertle T., Klinger M., Stuermer C.A.O., Schwab M.E. (2003). A reticular rhapsody: Phylogenic evolution and nomenclature of the RTN/Nogo gene family. FASEB J. 17: 1238–1247 [DOI] [PubMed] [Google Scholar]
- Osterrieder A., Carvalho C.M., Latijnhouwers M., Johansen J.N., Stubbs C., Bothchway S., Hawes C. (2009). Fluorescence lifetime imaging of interactions between Golgi tethering factors and small GTPases in plants. Traffic 10: 1034–1046 [DOI] [PubMed] [Google Scholar]
- Ridge R.W., Uozumi Y., Plazinski J., Hurley U.A., Williamson R.E. (1999). Developmental transitions and dynamics of the cortical ER of Arabidopsis cells seen with green fluorescent protein. Plant Cell Physiol. 40: 1253–1261 [DOI] [PubMed] [Google Scholar]
- Runions J., Brach T., Kuhner S., Hawes C. (2006). Photoactivation of GFP reveals protein dynamics within the endoplasmic reticulum membrane. J. Exp. Bot. 57: 43–50 [DOI] [PubMed] [Google Scholar]
- San Pietro E., et al. (2009). Group IV phospholipase A(2)alpha controls the formation of inter-cisternal continuities involved in intra-Golgi transport. PLoS Biol. 7: e1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzlander M., Fricker M.D., Muller C., Marty L., Brach T., Novak J., Sweetlove L.J., Hell R., Meyer A.J. (2008). Confocal imaging of glutathione redox potential in living plant cells. J. Microsc. 231: 299–316 [DOI] [PubMed] [Google Scholar]
- Scott I., Sparkes I.A., Logan D.C. (2007). The missing link: Inter-organellar connections in mitochondria and peroxisomes? Trends Plant Sci. 12: 380–381 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Hu J., Kozlov M.M., Rapoport T.A. (2009). Mechanisms shaping the membranes of cellular organelles. Annu. Rev. Cell Dev. Biol. 25: 329–354 [DOI] [PubMed] [Google Scholar]
- Shibata Y., Voss C., Rist J.M., Hu J., Rapoport T.A., Prinz W.A., Voeltz G.K. (2008). The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 283: 18892–18904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair A.M., Trobacher C.P., Mathur N., Greenwood J.S., Mathur J. (2009). Peroxule extension over ER-defined paths constitutes a rapid subcellular response to hydroxyl stress. Plant J. 59: 231–242 [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Frigerio L., Tolley N., Hawes C. (2009b). The plant endoplasmic reticulum: A cell-wide web. Biochem. J. 423: 145–155 [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Hawes C., Baker A. (2005). AtPEX2 and AtPEX10 are targeted to peroxisomes independently of known endoplasmic reticulum trafficking routes. Plant Physiol. 139: 690–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Ketelaar T., Ruijter N.C., Hawes C. (2009c). Grab a Golgi: Laser trapping of Golgi bodies reveals in vivo interactions with the endoplasmic reticulum. Traffic 10: 567–571 [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Hawes C., Griffing L. (2009a). Movement and remodelling of the endoplasmic reticulum in nondividing cells of tobacco leaves. Plant Cell 21: 3937–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Stephenson J.L.M., Hawes C.R. (1986). Stereology and stereometry of endoplasmic reticulum during differentiation in the maize root cap. Protoplasma 131: 32–46 [Google Scholar]
- Suetsugu S., Toyooka K., Senju Y. (2010). Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin. Cell Dev. Biol., 21: 340–349 [DOI] [PubMed] [Google Scholar]
- Tolley N., Sparkes I.A., Hunter P.R., Craddock C.P., Nuttall J., Roberts L.M., Hawes C., Pedrazzini E., Frigerio L. (2008). Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport. Traffic 9: 94–102 [DOI] [PubMed] [Google Scholar]
- Voeltz G.K., Prinz W.A., Shibata Y., Rist J.M., Rapoport T.A. (2006). A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124: 573–586 [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An 'Electronic fluorescent pictograph' browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamyatnin A.A., Jr., Solovyev A.G., Bozhkov P.V., Valkonen J.P., Morozov S.Y., Savenkov E.I. (2006). Assessment of the integral membrane protein topology in living cells. Plant J. 46: 145–154 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.