This work examines role of sulfite reductase (SiR) in assimilatory reduction of inorganic sulfate to sulfide. Reduced sulfite reductase activity results in growth retardation and severe perturbations of sulfur, nitrogen, and carbon metabolism, demonstrating that, surprisingly, SiR plays a role in controlling flux in the assimilatory sulfate reduction pathway.
Abstract
The role of sulfite reductase (SiR) in assimilatory reduction of inorganic sulfate to sulfide has long been regarded as insignificant for control of flux in this pathway. Two independent Arabidopsis thaliana T-DNA insertion lines (sir1-1 and sir1-2), each with an insertion in the promoter region of SiR, were isolated. sir1-2 seedlings had 14% SiR transcript levels compared with the wild type and were early seedling lethal. sir1-1 seedlings had 44% SiR transcript levels and were viable but strongly retarded in growth. In mature leaves of sir1-1 plants, the levels of SiR transcript, protein, and enzymatic activity ranged between 17 and 28% compared with the wild type. The 28-fold decrease of incorporation of 35S label into Cys, glutathione, and protein in sir1-1 showed that the decreased activity of SiR generated a severe bottleneck in the assimilatory sulfate reduction pathway. Root sulfate uptake was strongly enhanced, and steady state levels of most of the sulfur-related metabolites, as well as the expression of many primary metabolism genes, were changed in leaves of sir1-1. Hexose and starch contents were decreased, while free amino acids increased. Inorganic carbon, nitrogen, and sulfur composition was also severely altered, demonstrating strong perturbations in metabolism that differed markedly from known sulfate deficiency responses. The results support that SiR is the only gene with this function in the Arabidopsis genome, that optimal activity of SiR is essential for normal growth, and that its downregulation causes severe adaptive reactions of primary and secondary metabolism.
INTRODUCTION
Plants take up the essential macronutrient sulfur from the soil in the form of sulfate. The uptake of sulfate and its subsequent assimilatory reduction into organic sulfur compounds proceed through a highly coordinated mechanism. First, uptake of sulfate is catalyzed by specific proton cotransporters in root epidermal cells. They belong to the group of high affinity sulfate transporters (SULTR group 1) and are inducible by external sulfate deprivation (Smith et al., 1997; Takahashi et al., 2000). Internal allocation of sulfate is catalyzed by members of the low affinity SULTR groups 2 and 3 (Hawkesford, 2008; Takahashi and Saito, 2008). Next, assimilatory reduction of sulfate is initiated by ATP-dependent activation of sulfate to adenosine 5′-phosphosulfate (APS) catalyzed by ATP sulfurylase (ATPS). Further activation with ATP is catalyzed by APS kinase and yields 3′-phosphoadenosyl-5′-phosphosulfate (PAPS). APS kinase is present in plastids and the cytosol to provide PAPS for sulfation reactions by sulfotransferases (Mugford et al., 2009).
APS reductase (APR) in plastids from Arabidopsis thaliana and other plants strongly prefers APS instead of PAPS as a substrate, its expression responds to sulfate and nitrate availability, and a number of stress factors result in regulation of its activity (Leustek et al., 2000). In addition, flux analysis using 35S-labeled sulfate hinted that APR, after sulfate uptake, exerts the strongest control over flux through the sulfate reduction pathway in Arabidopsis (Vauclare et al., 2002) and is responsible for genetically determined variation in sulfate content in Arabidopsis ecotypes (Loudet et al., 2007).
In contrast with APR, the second enzyme of the free reduction pathway, sulfite reductase (SiR), has received little attention. Plant SiR is a plastid-localized soluble enzyme of two 65-kD subunits, contains a single siroheme and (4Fe-4S) cluster as prosthetic groups, and has a high affinity (Kmsulfite ∼10 μM) for sulfite (Krüger and Siegel, 1982; Nakayama et al., 2000). Ferredoxin acts as the physiological donor of the six electrons required for sulfite reduction, whereas bacterial SiR uses NADPH (Yonekura-Sakakibara et al., 2000). The structure, sequence, and ligands of SiR in bacteria, archea, and eukaryotes are similar to those of nitrite reductase, which catalyzes an equivalent reduction step in nitrate assimilation (i.e., a six-electron reduction of nitrite to ammonia) (Crane et al., 1995; Swamy et al., 2005). In Arabidopsis, SiR shows 19% identity with nitrite reductase (NiR) at the amino acid level. Phylogenetic analysis showed that both SiR and NiR arose from an ancient gene duplication in eubacteria, before the primary endosymbiosis that gave rise to plastids (Patron et al., 2008). SiR is able to reduce nitrite as well as sulfite, and substrate preferences can be converted by a single amino acid mutation (Nakayama et al., 2000). NiR has been suggested to operate similarly and to accept sulfite as substrate at a low rate (Schmidt and Jäger, 1992).
SiR is encoded by the only single-copy gene in primary sulfur metabolism in Arabidopsis, whereas the rice (Oryza sativa) and poplar (Populus spp) genomes each contain two copies (Bork et al., 1998; Kopriva, 2006). It is expressed in nearly all tissue types and shows the least transcriptional responses among sulfur-related genes in Arabidopsis in classical sulfate starvation experiments or under other stress conditions, according to a survey in microarray databases (Zimmermann et al., 2004). Expression changes were observed after treatment with SO2 (Brychkova et al., 2007), but these were not translated into significant changes of SiR enzyme activity under similar conditions (Lang et al., 2007). Activity of SiR is generally believed to be maintained in excess to scavenge potentially toxic sulfite (Leustek, 2002; Kopriva, 2006), based on flux control and APR overexpression experiments in Arabidopsis and maize (Zea mays; Tsakraklides et al., 2002; Vauclare et al., 2002; Martin et al., 2005). SiR potentially competes with two other enzymes for sulfite: UDP-sulfoquinosovyl synthase (SQD1) for sulfolipid synthesis in plastids (Benning, 2007) and sulfite oxidase (SO), a molybdoenzyme, in peroxisomes (Eilers et al., 2001; Hänsch and Mendel, 2005; Brychkova et al., 2007). However, the bulk of sulfite is normally channeled into the assimilatory reduction pathway for sulfur amino acid and protein biosynthesis.
Assimilatory sulfate reduction is completed by the two-step process of Cys synthesis that is catalyzed by serine acetyltransferase (SAT) and O-acetylserine (thiol) lyase (OAS-TL) with O-acetylserine (OAS) as the intermediate. Both enzymes form a regulatory complex, the Cys synthase complex that controls SAT activity in reaction to intracellular changes of sulfide, OAS, and Cys concentrations (for review, see Hell and Wirtz, 2008). The Cys synthase complex is present in plastids, cytosol, and mitochondria with sulfide, OAS, and Cys being freely exchanged between the compartments in Arabidopsis (Heeg et al., 2008; Watanabe et al., 2008a, 2008b; Krueger et al., 2009). Metabolic repression by Cys and GSH and activation by OAS of genes encoding sulfate transporters, ATPS, and APR are major mechanisms discussed for regulation of primary sulfur metabolism (Leustek et al., 2000; Kopriva, 2006).
Here, we investigated two Arabidopsis lines with T-DNA insertions in the promoter region of SiR. Mutant line sir1-2 is early seedling lethal and unequivocally demonstrates that the free sulfate reduction pathway is essential for survival and cannot be compensated for by any other enzymatic process. In mature leaves, mutant sir1-1 has 28% of SiR activity and 3.6% of flux in the assimilatory reduction pathway in vivo compared with the wild type. sir1-1 has a strongly retarded growth phenotype, showing that in contrast with general assumptions, SiR can easily become limiting for growth. sir1-1 mutant plants are sensitive to cadmium due to lack of GSH for phytochelatin synthesis. Carbon, nitrogen, and sulfur composition are severely altered with a shift toward carbon-bound reduced nitrogen, indicating that lowered sulfite reduction leads to comprehensive reprogramming of primary metabolism.
RESULTS
Identification and Characterization of sir Mutants
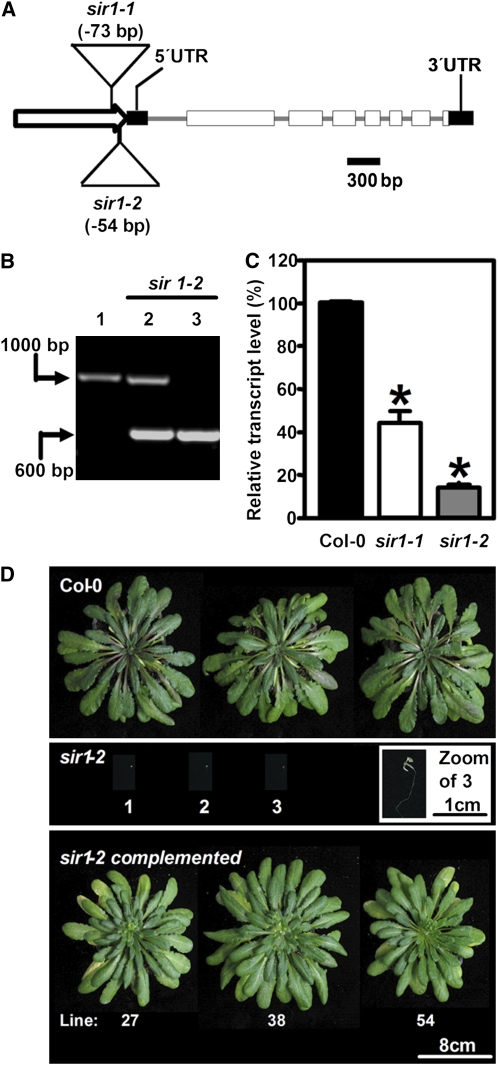
Two independent Arabidopsis T-DNA insertion lines, further annotated as sir1-1 and sir1-2, were identified in the GABI-Kat collection center. In both lines, the T-DNA was inserted in the promoter region of SiR (Figure 1A). PCR-verified heterozygous sir1-2 plants segregated in two distinct classes of seedlings after germination in the absence of selection marker: (1) wild type-like seedlings (401/533, 75%; χ2 = 0.016) and (2) bleached seedlings that died at the two cotyledon stage after germination (132/533, 25%). Genotyping of these seedlings (Figure 1B) revealed that bleached sir1-2 seedlings were homozygous for the T-DNA insertion. On these plates, wild-type-like plants represented a pool of heterozygous sir1-2 (1/2) and segregating Columbia-0 (Col-0) plants (1/4), suggesting a single insertion of a recessive allele. Thus, the homozygous sir1-2 line is seedling lethal. Flanking sequences of the T-DNA were PCR amplified and sequenced to characterize the insertion sites. Alignment with the genomic sequence of SiR revealed the beginning of the left border of the T-DNA at position −54 bp in sir1-2 relative to the transcription start site in the promoter region of SiR (Figure 1A). Quantitative real-time PCR (qRT-PCR) detected successful transcription initiation corresponding to 14% of mature SiR transcript in the early seedling stage that may account for embryo development and germination of sir1-2 (Figure 1C). For genetic complementation, the heterozygous sir1-2 plants were transformed with a construct that expressed the SiR cDNA along with its plastid transit peptide under control of the constitutive 35S cauliflower mosaic virus promoter. The phenotype of sir1-2 was completely restored (Figure 1D), demonstrating that the loss of function of SiR was the cause of the early seedling lethality observed in the homozygous sir1-2 plants.
Figure 1.
Molecular Identification of sir Mutants and Phenotype of sir1-2.
(A) Structure of the SiR locus with the T-DNA insertion sites in sir1-1 and sir1-2. The putative promoter is marked by a white arrow, exons are indicated as white boxes, and untranslated regions by black boxes.
(B) Genomic characterization of sir1-2 plants. Wild-type (1), heterozygous (2), and homozygous (3) sir1-2 plants were tested for the presence of wild-type and T-DNA insertion alleles of SiR by PCR.
(C) Transcript levels of SiR in the wild type, sir1-1, and sir1-2 determined by qRT-PCR at the developmental stage of five to six leaves of the wild type. The homozygous sir1-2 plants arrested at the two cotyledon stage. Bars represent the mean of pooled individuals (n = 6), while error bars show standard errors of technical replicates (n = 3). Asterisks indicate statistically significant (P < 0.05) differences from wild-type values.
(D) Growth phenotype and genetic complementation of sir1-2. For genetic complementation, the cDNA of SiR was fused with the 35S promoter and introduced in sir1-1 plants by Agrobacterium tumefaciens–mediated transformation.
[See online article for color version of this figure.]
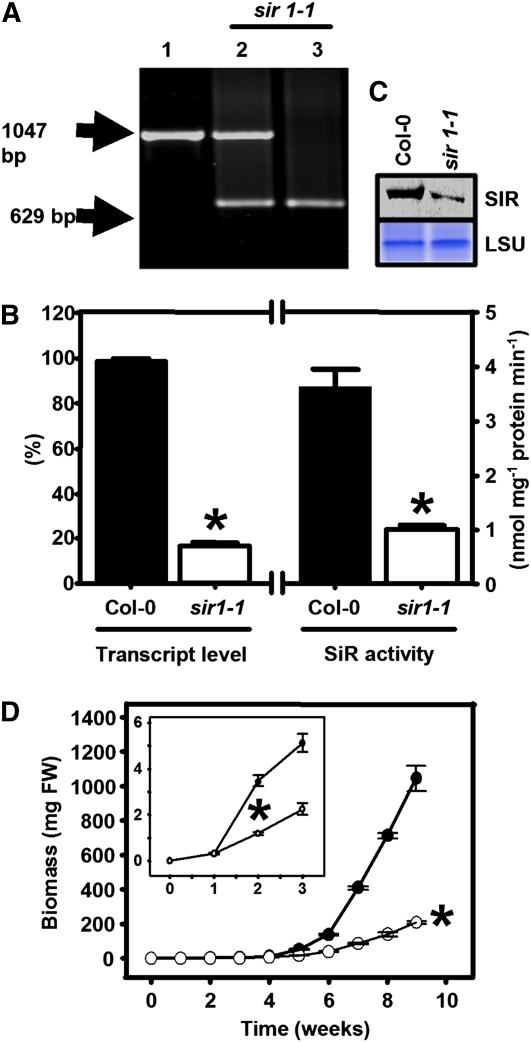
Like sir1-2, heterozygous sir1-1 plants also gave rise to two distinct classes of seedlings. Besides wild-type-like seedlings (352/471, 75%; χ2 = 0.018), severely growth-retarded and pale-looking seedlings (119/471, 25%) could be identified after germination in the absence of selection marker. The growth-retarded sir1-1 seedlings were found to be homozygous for the T-DNA insertion by PCR analysis (Figure 2A), again pointing to a single insertion site. The insertion site of the T-DNA in sir1-1 was determined by PCR amplification of the flanking sequences and revealed the insertion of the T-DNA at position −73 bp in the promoter region (Figure 1A). The viability of homozygous sir1-1 seedlings was in contrast with the seedling lethal phenotype of the sir1-2 mutant. In the early seedling stage of sir1-1, the abundance of SiR mRNA was 44% (Figure 1C) compared with wild-type seedlings of the same developmental stage. Thus, the levels of 44 and 14% of mature SiR mRNA made the difference between survival and death of the early seedlings of sir1-1 and sir1-2, respectively. It is remarkable that both T-DNA insertions in the SiR promoter allowed transcription of intact mRNA and the 19 bp between the insertion sites were responsible for a twofold difference in steady state mRNA of SiR. Transcript levels of SiR in mature leaves of 7-week-old soil-grown homozygous sir1-1 plants were decreased to 17% compared with the wild type (Figure 2B). Accordingly, the amount of SiR protein (Figure 2C) was significantly reduced, and SiR activity also was lowered to 28% of the wild-type level (Figure 2B). This provides an explanation for the slower vegetative growth in comparison to the wild type that became more pronounced with time (Figure 2D). The homozygous sir1-1 line had the same number of leaves compared with the wild type of the same age, but they were clearly paler and smaller (Figure 3A). An approximately fivefold reduction in total biomass was observed between sir1-1 and Col-0 plants grown for 8 weeks on soil under short-day conditions (Figure 3B). Short-day conditions delayed flowering of soil-grown homozygous sir1-1 plants by ∼4 weeks compared with Col-0. In spite of the retarded growth phenotype and delayed flowering, the homozygous sir1-1 plants were able to set seeds leading to viable offspring in the next generation. Thus, reduced expression of SiR causes a severe growth limitation. These results provide functional evidence that there is only a single annotated gene for SiR in the Arabidopsis genome and that its function is indispensible for survival. Nevertheless, the viability of sir1-1 allows investigation of the physiological role of SiR activity in plant metabolism. For that reason, all further experiments were performed with sir1-1.
Figure 2.
Abundance and Activity of SiR in the T-DNA Insertion Line sir1-1.
(A) Genomic characterization of sir1-1 plants. Wild-type (1), heterozygous (2), and homozygous (3) sir1-1 plants were tested for the presence of wild-type and T-DNA insertion alleles of SiR by PCR.
(B) Determination of SiR activity (n = 5, mean ± se) and relative SiR transcript levels by qRT-PCR (n = 3, mean ± se) in leaves of 7-week-old plants. Amplification of Ef1α from the same cDNA preparations was used as a control for qRT-PCR.
(C) Immunoblot of soluble leaf proteins from wild-type and sir1-1 plants was performed with a polyclonal antibody against SiR from Arabidopsis. Staining intensities of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (LSU) in the same sample confirms equal loading.
(D) Growth curve of wild-type (black circles) and sir1-1 plants (white circles). Growth retardation of sir1-1 was statistically significant from week 2 on, as detailed in the inset. All plants were grown on soil in a growth chamber under short-day conditions (n = 5). Means ± se are shown. Asterisks indicate statistically significant (P < 0.05) differences from wild-type values.
[See online article for color version of this figure.]
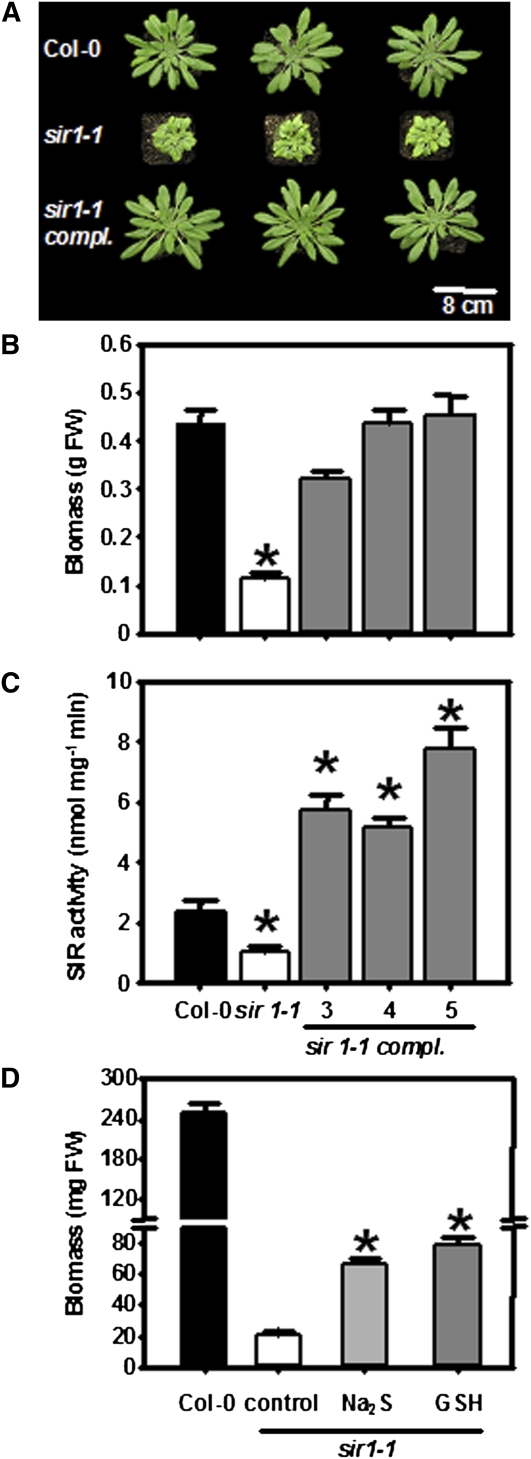
Figure 3.
Phenotype and Complementation of sir1-1 Plants.
(A) Top view of 7-week-old wild-type, sir1-1, and genetically complemented sir1-1 plants. Genetic complementation of sir1-1 was achieved by transformation of the wild-type SiR gene under control of the 35S promoter from the Cauliflower mosaic virus.
(B) Fresh weight of 7-week-old wild type (black), sir1-1 (white), and three complemented sir1-1 lines (gray) grown on soil under short-day conditions (n = 5).
(C) Specific activity of SiR in extracts from leaves shown in (B) (n = 5).
(D) FW of 6-week-old hydroponically grown wild-type (black), sir1-1 (white), and sir1-1 plants that were supplemented with 0.1 mM sodium sulfide (gray) and 1 mM GSH (dark gray) in the growth medium (n = 5). Means ± se are shown. Asterisks indicate statistically significant (P < 0.05) differences from wild-type values.
Genetic and Chemical Complementation of sir1-1
Stable transformation of homozygous sir1-1 with the 35S:SiR overexpression construct completely restored the phenotype of sir1-1, including the pale low chlorophyll view (Figures 3A and 3B). A 2.5-fold higher enzymatic activity of SiR was observed as a result of the high expression rate of SiR driven by the 35S promoter (Figure 3C). This resulted in wild-type-like thiol levels (see Supplemental Figure 1 online) and normal growth of the complemented sir1-1 lines under growth chamber conditions (Figure 3B). The latter results indicate an adequate SiR activity in Col-0 for optimal growth. Chemical complementation by exogenous supply of the limiting product of the SiR reaction, sulfide, and GSH, the major low molecular weight thiol in plants, partially rescued the phenotype (Figure 3D). The total shoot biomass of homozygous sir1-1 plants was increased twofold by feeding of 0.1 mM Na2S and 2.7-fold by feeding of 1 mM GSH in comparison to untreated sir1-1. However, the differences between chemically complemented homozygous sir1-1 and Col-0 control plants were still significant (Figure 3D).
Shifts of C, N, and S Metabolites in sir1-1
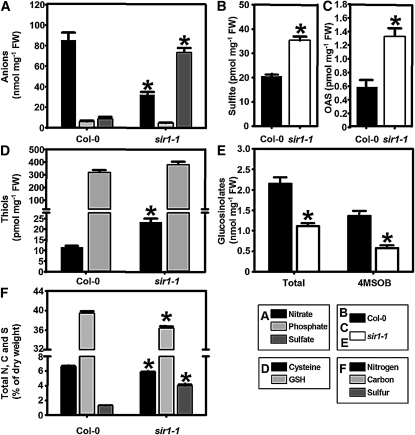
The consequences of reduced SiR activity for metabolic homeostasis were analyzed by determination of the steady state levels of primary and secondary metabolites in leaves of 7-week-old soil-grown wild-type and sir1-1 plants. Measurements of anion levels in homozygous sir1-1 plants revealed a ninefold accumulation of free sulfate, while the steady state level of nitrate was reduced threefold in comparison to the wild type, indicating a deregulation of nitrate and sulfate uptake and assimilation pathways (Figure 4A). Indeed, the accumulation of sulfate in leaves of sir1-1 was found to correlate with strongly increased uptake rates of sulfate in roots. Feeding of 35SO42− to roots of Arabidopsis wild-type and sir1-1 mutant plants revealed a 13-fold increased rate of sulfate uptake in sir1-1 (2 ± 0.8 fmol 35S min−1 mg−1 fresh weight [FW] in Col-0 compared with 26.6 ± 7.6 fmol 35S min−1 mg−1 FW in sir1-1).
Figure 4.
Impact of Reduced SiR Activity on Metabolism in Leaves of sir1-1 Plants.
(A) to (D) Metabolites were extracted from 7-week-old wild-type and sir1-1 plants and quantified by HPLC (n = 5).
(E) Total glucosinolates were extracted and quantified from the same samples (4MSOB, glucoraphanin, n = 10). Other glucosinolates are shown in Supplemental Figure 2 online.
(F) Aerial parts of 8-week-old wild-type and sir1-1 plants (n = 10) were used to determine the total content of nitrogen (black), carbon (gray), and sulfur (dark gray) twice for each sample. All plants were grown on soil under short-day conditions. Means ± se are shown. Asterisks indicate statistically significant (P < 0.05) differences from wild-type values.
In agreement with reduced SiR activity, a 1.7-fold accumulation of sulfite levels was evident (Figure 4B). OAS, the activated amino acid for synthesis of Cys, was more than twofold accumulated due to decreased formation of sulfide (Figure 4C). Unexpectedly, the steady state level of Cys was 1.8-fold upregulated, while the level of GSH was not significantly affected (Figure 4D). Both thiols had been assumed to be downregulated in sir1-1, as a result of lower SiR activity. The analysis of glucosinolates, the major sulfur-containing secondary compounds in Arabidopsis revealed strong differences between sir1-1 and Col-0. The profiles of aliphatic glucosinolates, especially the major glucosinolate, glucoraphanin, were reduced (Figure 4E; see Supplemental Figure 2 online), causing a significant twofold reduction of the total glucosinolate content either by decreased biosynthesis or by enhanced turnover.
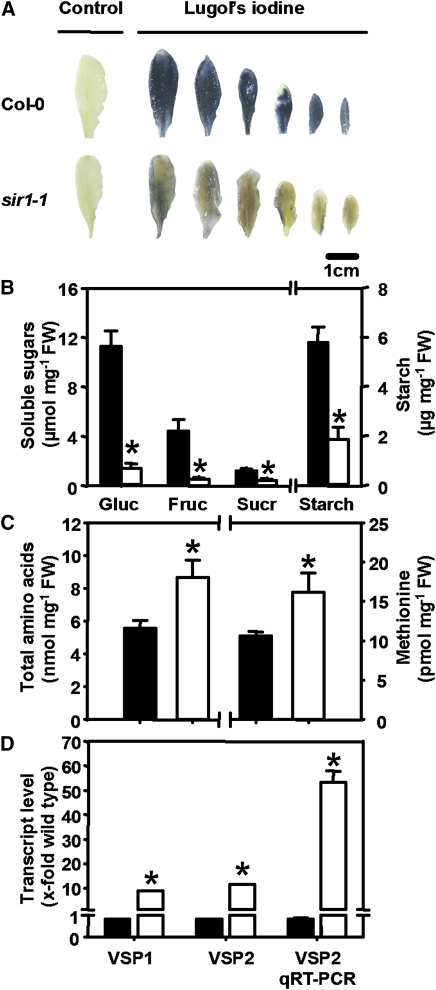
The analysis of total carbon (C) nitrogen (N) and sulfur (S) demonstrated significant reductions of total C (8%) and total N (11%) in sir1-1. A striking increase of total S of more than threefold was observed in the sir1-1 mutant (Figure 4F). This increase can be almost entirely attributed to the observed rise of free sulfate (Figure 4A) and not to organically bound sulfur (see Supplemental Figure 3A online). By contrast, the decrease in nitrate content accounted only partially for the lowered total nitrogen share in sir1-1 and strongly suggests that the organically bound nitrogen fraction was increased (see Supplemental Figure 3B online). These severe changes in the reduced carbon and nitrogen fractions prompted us to dissect the major C and N metabolites. In situ staining of leaves with iodine solution showed a clearly visible significant reduction in starch content 3 h after onset of light in source as well as sink leaves of rosettes of sir1-1 plants compared with the wild type (Figure 5A). On average, leaf starch content was reduced nearly threefold after extraction of starch from young leaves (Figure 5B). Sucrose contents were decreased by the same range and glucose and fructose were down by ∼90% in sir1-1 (Figure 5B). While the levels of major free sugars were reduced, the opposite was observed for amino acids (Figure 5C). Contents of 14 out of 18 determined amino acids were higher in sir1-1 (see Supplemental Figure 4 online), including that of Met (Figure 5C). In total, the amount of free amino acids increased significantly by 55% in sir1-1 compared with the wild type. These findings suggest an enhanced fixation of reduced nitrogen into carbon, possibly on cost of sugars. Concomitantly, the mRNA levels of VEGETATIVE STORAGE PROTEIN1 (VSP1) and VSP2 in leaves of sir-1 plants were at least 10-fold enhanced according to expression analyses (Figure 5D). Taken together, these results indicate massive and far-reaching shifts between carbon, nitrogen, and sulfur fractions in response to the partial block in sulfite reduction.
Figure 5.
Deregulation of Carbon- and Reduced Nitrogen-Containing Compounds in sir1-1 Plants.
(A) Starch (blue) was determined using Lugol's iodine staining. Leaves after ethanol treatment but without Lugol staining are shown as control.
(B) and (C) Quantification of amino acids (n = 5), glucose (Gluc), fructose (Fruc) sucrose (Sucr), and starch (mean ± se; n = 3) after extraction from leaves of wild-type (black) and sir1-1 (white) plants.
(D) Transcript levels of VSP1 and VSP2 in leaves of wild-type (black) and sir1-1 (white) plants were determined by microarray hybridization (VSP1 and VSP2, n = 12) and qRT-PCR (VSP2 qRT-PCR, n = 3). In all cases, material was harvested from wild-type and sir1-1 plants that were grown for 7 to 8 weeks on soil under short-day conditions. Asterisks indicate statistically significant (P < 0.05) differences from wild-type values.
[See online article for color version of this figure.]
S-Related Enzymatic Activities in sir1-1 Plants
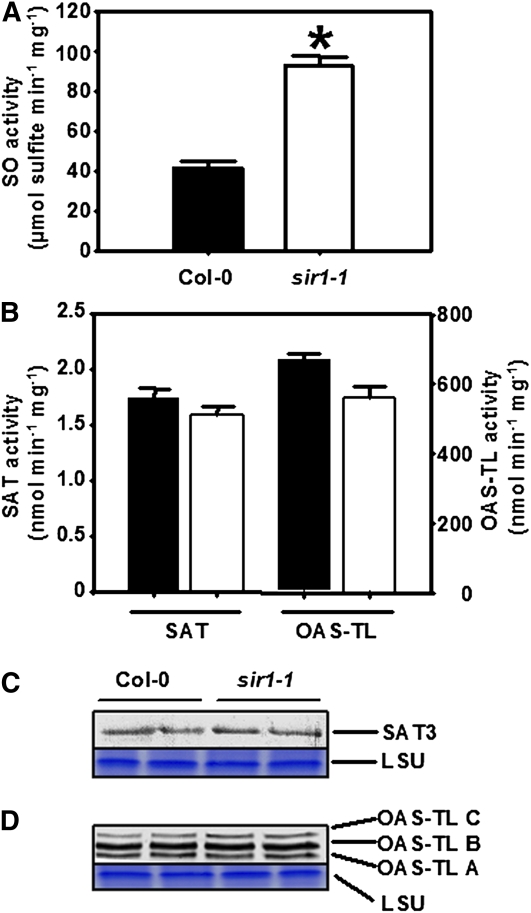
The significantly higher sulfite contents in leaves of sir1-1 prompted us to test the activity of SO in these plants. The activity of SO in leaves of hydroponically grown sir1-1 plants was found to be 2.4-fold higher in comparison to Col-0 (Figure 6A). This observation points to the theoretical possibility of reoxidation of excess sulfite to sulfate in peroxisomes of sir1-1. The ninefold higher accumulation of foliar sulfate may therefore result from insufficient SiR activity but also reoxidation of toxic sulfite to sulfate by SO. The amount of SO protein was not increased in sir1-1 mutants according to immunoblotting using an SO-specific antiserum (see Supplemental Figure 5 online), suggesting a posttranslational upregulation of SO activity. SAT catalyzes the production of OAS that is converted by OAS-TL to Cys under consumption of sulfide. Despite twofold increases in OAS and Cys steady state-levels, the total activities of SAT and OAS-TL were not significantly affected in sir1-1 (Figure 6B). Accordingly, the amount of mitochondrial SAT3 protein, which functions as pacemaker of Cys synthesis in Arabidopsis by providing OAS (Haas et al., 2008), did not change, as shown by immunological detection using a SAT3 specific antiserum (Figure 6C). Mitochondrial OAS-TL C, which is supposed to function as a regulator of SAT3 activity in the Cys synthase complex (Wirtz and Hell, 2006), showed no changes in its abundance and neither did cytosolic OAS-TL A, which is responsible for the majority of Cys synthesis (Heeg et al., 2008). Furthermore, the amount of plastid OAS-TL B was not altered, despite presumably strongly decreased sulfide production in the plastids of sir1-1 plants (Figure 6D).
Figure 6.
Abundance and Activity of SO, SAT, and OAS-TL in Leaves of sir1-1 Plants.
(A) Specific activity of SO in protein extracts of 7-week-old wild-type (black) and sir1-1 (white) plants that were grown on soil under short-day conditions.
(B) Specific activity of SAT and OAS-TL from the same extracts as in (A). The specific activities of SO, SAT, and OAS-TL were determined for each extract in triplicates with varying amounts of proteins to prove time and protein linearity of measurement (n = 5 to 7). Means ± se are shown. The asterisk indicates a statistically significant (P < 0.05) difference from the wild-type value.
(C) An immunoblot loaded with soluble protein from two extractions each of the leaves from the wild type and sir1-1 was decorated with At-SAT3 polyclonal antiserum.
(D) Same experimental set up as in (C), but a polyclonal antiserum against At-OAS-TLC was used, which also detects At-OAS-TL A and B. Staining of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (LSU) with Coomassie blue was used to confirm equal loading of lanes.
[See online article for color version of this figure.]
Incorporation of Radioactively Labeled Sulfate into Thiols and Protein
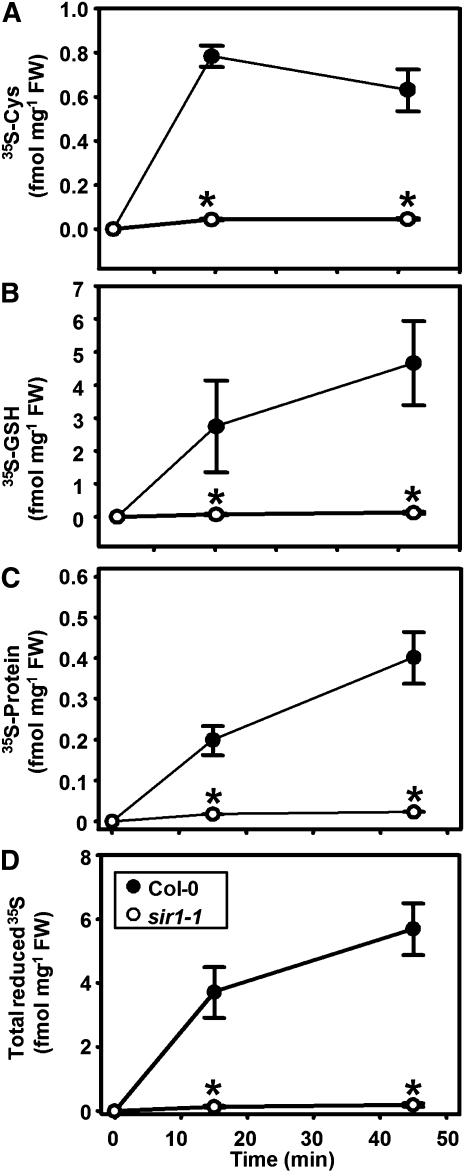
The higher steady state level of Cys in combination with unaffected SAT and OAS-TL activities are in contrast with the retarded growth phenotype and the assumed reduced sulfide production in sir1-1 plants. To unravel the underlying fluxes of sulfur through the assimilatory sulfate reduction pathway of sir1-1 plants, the incorporation of radiolabeled sulfate (35SO42−) into Cys and GSH was determined by feeding of leaf pieces according to Heeg et al. (2008). Time-resolved analysis of incorporation of the 35S label into Cys and GSH demonstrated a very strong reduction of flux through the sulfate assimilation pathways in sir1-1 plants compared with the wild type. Only 5.5% of incorporation of 35S label into Cys compared with the wild type was observed in leaves of sir1-1 after the 15-min feeding period. The incorporation was lower at the end of the 30-min chase period at time point 45 min, indicating the active turnover of Cys (Figure 7A). Most of the 35S-Cys went into the GSH pool (Figure 7B), and a smaller part was found in the protein fraction (Figure 7C), documenting active protein biosynthesis under these conditions. Thus, the total incorporation into organic sulfur compounds in sir1-1 after 15 min amounted to a 28-fold reduction compared with the wild type. The experiment was independently repeated using soil-grown plants instead of hydroponically grown plants to exclude influences of nutritional status of the plants. The distribution patterns were quite similar, albeit incorporation rates of the 35S-label were somewhat lower (see Supplemental Figure 6 online). The results from feeding experiments thus confirm a strongly reduced rate of sulfide formation and corroborate the unique function of SiR for the assimilatory sulfate reduction pathway.
Figure 7.
Incorporation Rates of Sulfate in Wild-Type and sir1-1 Plants.
(A) to (C) Leaf pieces of 7-week-old wild-type (black circles) and sir1-1 (white circles) plants, which were grown hydroponically under short-day conditions, were incubated with 35SO42--spiked half-strength Hoagland solution for 15 min (pulse, beginning at time 0) and subsequently kept in the same medium without radiolabel for 30 min (chase). Cys (A), GSH (B), and proteins (C) were extracted and separated by HPLC or specific precipitation. The incorporated 35S label was quantified by scintillation counting.
(D) Total sum of incorporated 35S label in Cys, GSH and proteins. The mean ± se from four independent extractions of the wild type and sir1-1 are shown. Asterisks indicate statistically significant (P < 0.05) differences from wild-type values.
Response of sir1-1 Seedlings toward Cadmium Stress
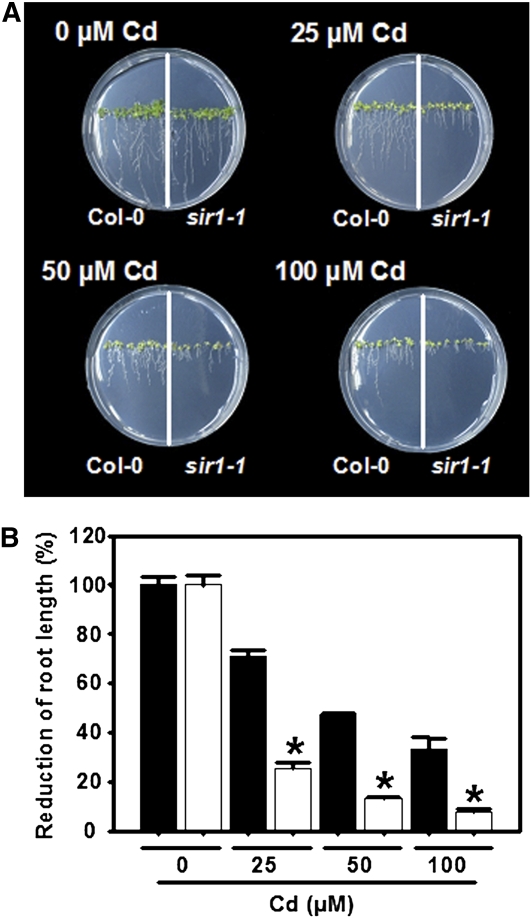
Phytochelatins are enzymatically synthesized from GSH by cytosolic phytochelatin synthase for detoxification of heavy metal ions (Ha et al., 1999). For that reason, the reduced capacity of the sir1-1 plants to incorporate sulfur for synthesis of GSH was expected to result in a higher sensitivity toward cadmium. Wild-type Arabidopsis and sir1-1 seeds were germinated on solid medium containing increasing concentrations of Cd (0 to 100 μM CdCl2). After 14 d of treatment both Col-0 and sir1-1 showed a pale leaf phenotype and shortened primary roots (Figure 8A) as typical symptoms of Cd stress (Cobbett et al., 1998). The degree of root shortening was used for quantification of Cd tolerance. Since the root length of sir1-1 seedlings was already strongly reduced in comparison to Col-0 in absence of Cd, the reduction of root length was expressed as a percentage, whereby the root length of untreated plants of the respective genotype was set to 100% (Figure 8B). Upon exposure to 25, 50, and 100 μM CdCl2, reductions of 75, 87, and 92%, respectively, were observed in the root length of sir1-1 seedlings. By contrast, the magnitudes of reduction at the same CdCl2 concentrations for Col-0 were only 29, 53, and 67%, demonstrating that root growth of sir 1-1 was 2.6-fold more sensitive than Col-0 toward 25 μM CdCl2, for instance. Taken together, the data demonstrate that reduced sulfur assimilation leads to higher Cd sensitivity in Arabidopsis.
Figure 8.
Cadmium Sensitivity of sir1-1 Plants.
(A) Top views of wild-type (Col-0, left) and sir 1-1 (right side) plants grown for 15 d on At-medium supplemented with varying concentrations of cadmium (0 to 0.1 mM CdCl2).
(B) Quantification of root growth of wild-type (black) and sir1-1 (white) plants grown as in (A). To allow comparison between wild-type and sir1-1 plants, the root growth is shown as percentage of growth of the respective line under nonstress conditions. Mean ± se are shown (n = 6 to 10). Asterisks indicate statistically significant (P < 0.05) differences from wild-type plants grown at the same concentration of Cd.
[See online article for color version of this figure.]
Response of Sulfur Metabolism at the Transcriptional Level
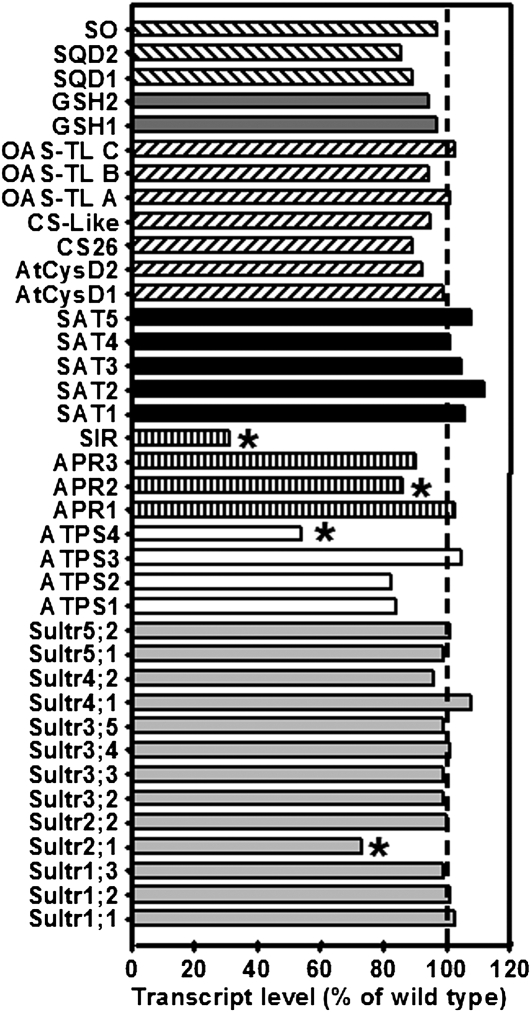
The impact of reduced SiR activity on the transcription of sulfur metabolism-related genes in leaves of 7-week-old soil-grown plants was investigated with a microarray carrying 920 genes related to primary metabolism and stress responses as described by Haas et al. (2008). Based on three biological repetitions of the wild type and sir1-1 with four technical replicates, each including dye swaps for each set, 67 genes were found to be significantly up- or downregulated in the leaves of hydroponically grown sir1-1 plants compared with Col-0 according to P values of <0.05. Most regulated genes were related to redox homeostasis (20), while genes of sulfur metabolism (11), pathogen resistance (11), glucosinolate synthesis (10), hormone synthesis and signaling (5), GSH transfer activity (4), sulfur-induced nonsulfur genes (3), and amino acid synthesis (3) were also found to be significantly changed in abundance (see Supplemental Table 1 online). The microarray analysis confirmed independently the downregulation of SiR transcript in mature leaves of sir1-1 (Figure 2C). Besides SiR, three genes of the primary sulfur assimilation pathway were significantly downregulated: ATPS4, APS REDUCTASE2 (APR2), and SULFATE TRANSPORTER 2.1 (SULTR2.1; Figure 9). SULTR2.1 is known to be specific for the vasculature and downregulated in leaves upon sulfur deficiency (Takahashi et al., 2000). ATPS4 catalyzes the activation of sulfate in plastids, which leads to formation of APS. APR2 is the key APR isoform in leaves for reduction of activated sulfate in APS to sulfite that is further reduced by SiR. Downregulation of APR2 was independently confirmed by real-time PCR and revealed 58% mRNA content in sir1-1 compared with wild-type plants (see Supplemental Figure 7 online). Most likely ATPS4 and APR2 are downregulated to avoid extensive accumulation of toxic sulfite, which cannot be incorporated into Cys as a result of reduced SiR activity in sir1-1. The stable total SAT and OAS-TL enzymatic activities and abundances of analyzed SAT and OAS-TL isoforms shown in leaves of sir1-1 (Figure 6) were further supported by unchanged SAT and OAS-TL transcript levels in sir1-1 (Figure 9). In accordance with unchanged SO protein contents, SO was not upregulated at the transcriptional level despite clearly increased enzymatic activity, leaving the possibility of posttranslational activation of SO in peroxisomes of sir1-1. Multiple genes involved in pathogen defense, such as PATHOGENESIS-RELATED1 (PR1), PR2, PR5, RECOGNITION OF PERONOSPORA PARASITICA1 (RPP1), RPP4, RPP5 as well as ETHYLENE INSENSITIVE3 (EIN3), and PENETRATION3 (PEN3) were significantly downregulated along with the sulfur assimilation pathway, indicating a coregulation of both processes, as predicted by the sulfur-enhanced defense hypothesis (Kruse et al., 2007).
Figure 9.
Transcript Levels of Sulfur Metabolism-Related Genes in Leaves of sir1-1 Plants.
The transcript levels of sulfur metabolism related genes in leaves of sir1-1 and wild-type plants (Col-0) grown on soil under short-day conditions for 7 weeks were compared using a targeted microarray approach. Total mRNA was extracted from three individuals of each plant line, labeled independently two times with Cy3 and Cy5, and cohybridized with the microarray twice (n = 12). From bottom to top: The transcript levels of genes encoding sulfate transporters (light-gray bars), ATPS (white bars), sulfate-reducing enzymes (striped bars), SATs (black bars), OAS-TLs (inclined dashed bars), proteins participating in GSH synthesis (dark gray bars), and sulfolipid biosynthesis enzymes (declined dashed bars) in sir1-1 plants are shown as percentage of wild-type levels. Asterisks indicate statistically significant (P < 0.05) differences from wild-type expression levels of the same gene.
The genes encoding two vegetative storage proteins (VSP1 and VSP2) that are supposed to serve as transient reservoirs for surplus of amino acids in vegetative tissues were ∼10-fold upregulated in expression in sir1-1 leaves. VSP2 upregulation was also confirmed by real-time PCR (Figure 5D). In agreement with the lower contents of total glucosinolates in the mutant (Figure 4E), genes encoding thioglucoside glucohydrolase (TGG1 and TGG2; myrosinases), enzymes that are potentially involved in breakdown of glucosinolates upon sulfur and the usage of respective breakdown products (NITRILASE1 [NIT1] and NIT2), were significantly upregulated (see Supplemental Table 1 online). The significant upregulation of the gene for the chlorophyll degrading enzyme CHLOROPHYLLASE1 in sir1-1 (see Supplemental Table 1 online) could be a hint toward the pale phenotype (Figure 3A). Changes in chlorophyllase, VSP, and glucosinolate breakdown-related gene expression are in agreement with the disturbed coordination of C, N, and S metabolism that became evident by analysis of anion, sugar, amino acid, and elemental composition of sir1-1 leaves (Figures 4 and 5).
DISCUSSION
Sulfite Reductase Is Essential for Development and Growth
Genetic analysis of the assimilatory sulfate reduction pathway is hampered by functional redundancy of most steps due to gene families in Arabidopsis, rice, and other species (Kopriva, 2006; Patron et al., 2008). Here, SiR as the only single copy gene in assimilatory sulfate reduction in Arabidopsis was investigated using two T-DNA insertion mutants. Mutant line sir1-2 stays pale and dies at the two cotyledon stage. The T-DNA insertion in the promoter of SiR allowed for a SiR transcription level in young sir1-2 seedlings of 14% of that in the wild type. This must have led to sufficient enzyme activity to support development to the two-leaf stage, since reduced sulfur compounds such as GSH are known to be required for complete embryo development (Cairns et al., 2006). However, an enhanced requirement for reduced sulfur during further development evidently led to the early death of sir1-2 seedlings. The sir1-2 phenotype could be completely reverted by genetic complementation with wild-type SiR, confirming the identity of the lowered SiR activity as cause for the phenotype.
Germinating seedlings of the second promoter insertion mutant sir1-1 had 44% of the wild-type SiR transcript levels. This apparently allowed for translation of sufficient SiR enzyme to complete development. During the following vegetative development toward mature leaves, the presence of 17 to 28% of mRNA, protein, and enzyme activity levels of SiR strongly limited growth, suggesting a lack of sulfide for Cys synthesis and the deregulation of nitrogen and carbon metabolism. A toxic effect from accumulated sulfite in the cells is unlikely to be responsible for the slow growth for several reasons: First, the increase of steady state levels of sulfite was low (less than twofold compared with the wild type); second, flux experiments using 35S-sulfate showed increased label in sulfate but no significant label in sulfite; and third, excess sulfite might be oxidized to sulfate by elevated SO activity (Brychkova et al., 2007; Hänsch et al., 2007). Sulfite is in equilibrium with hydrogen sulfite and sulfur dioxide (SO2), which was reported to passively diffuse through biomembranes (Furihata et al., 1997). Thus, the possibility of migration along a gradient from plastids to peroxisomes where SO resides cannot be entirely ruled out (Hänsch and Mendel, 2005). Fourth, sir1-1 growth could be significantly improved by exogenous sulfide and GSH despite the presence of elevated sulfite. The incomplete complementation was probably a consequence of deleterious side effects of high sulfide doses or limited transport and metabolization of GSH, respectively. Indeed, Arabidopsis mutants with lowered GSH contents due to mutations in the GSH biosynthestic enzymes could be only partially rescued by growth in the presence of GSH (Cobbett et al., 1998; Vernoux et al., 2000; Pasternak et al., 2008). Finally, transcripts of ATPS4 and APR2 were decreased, suggesting downregulation of flux in the pathway and, therefore, less sulfite in sir1-1.
Decreased SiR Activity Causes Reduced Flux in Assimilatory Sulfate Reduction
It has often been assumed that key enzymes catalyzing irreversible reactions in pathways are subject to fine regulation by allosteric effectors or posttranslational modification, have high flux control coefficients, and exert strong control of pathways. By contrast, enzymes with apparently little fine-tuning and mostly constitutive expression have been assumed to operate in large excess and to have low flux control coefficients. However, theoretical analyses predict that all enzymes in a pathway can contribute to the control of flux (Fell and Thomas, 1995). Indeed, detailed analysis of carbon assimilation and metabolism using transgenic plants demonstrated that rather small decreases in the activity of seemingly unregulated enzymes, such as phosphoribulokinase and transketolase, limited flux of carbon and distribution into downstream pathways. Phenotypes of such transgenic plants included reduced rates of photosynthesis and growth retardation (Stitt and Sonnewald, 1995; Henkes et al., 2001).
The results here show that, unexpectedly, SiR is such a case. A flux-limiting role for SiR had been hardly considered, probably due to its semiconstitutive expression pattern (Bork et al., 1998; Zimmermann et al., 2004). With respect to control of the pathway, considerations of Vmax activities of the enzymatic steps from sulfate to sulfide in leaves of Arabidopsis remain inconclusive. Reported maximal APR activities in Col-0 plants range from 1 to 10 nmol mg−1 protein min−1 and depend on age, light, and media conditions (Kopriva et al., 1999, 2000; Rotte and Leustek, 2000; Tsakraklides et al., 2002; Loudet et al., 2007). APR expression has been reported to be transcriptionally regulated in response to sulfur and nitrogen nutrition, heavy metal stress, addition of reduced sulfur compounds and reactive oxygen species, and the day/night cycle, strongly suggesting a regulatory function (for review, see Leustek, 2002; Kopriva, 2006). In comparison, SiR activities in crude leaf extracts from Arabidopsis were 2 to 3.5 nmol mg−1 protein min−1 (Figure 2; Hänsch et al., 2007; Heeg et al., 2008), while ATP sulfurylase activity ranged one order of magnitude higher (Rotte and Leustek, 2000). Flux control studies with Arabidopsis root cultures showed that sulfate uptake at the plasmalemma is most important for primary sulfur metabolism but confirmed that, once sulfate was inside the cell, the highly regulated reaction catalyzed by APR exerted the strongest control over the pathway (Vauclare et al., 2002). However, the moderate decrease of SiR activity and substantial flux reduction in the sir1-1 mutant resulted in strong growth limitation under normal conditions and a cadmium-sensitive phenotype that is indicative of insufficient phytochelatin synthesis from GSH and Cys (Cobbett et al., 1998). This now demonstrates that SiR activity is not in high excess as assumed earlier (Leustek, 2002; Kopriva, 2006) but can also contribute to control of the pathway.
The described major regulatory function of the first reduction step catalyzed by APR (Vauclare et al., 2002) is not in contrast with the newly defined role of SiR. Constitutive overexpression using bacterial APR from Pseudomonas aeruginosa in Arabidopsis and maize resulted in massive deregulation of primary sulfur metabolism (Tsakraklides et al., 2002; Martin et al., 2005). Cys, sulfite, and thiosulfate contents accumulated threefold to sixfold. The Arabidopsis plants were slightly stunted and chlorotic compared with the wild type, suggesting symptoms of toxicity (Tsakraklides et al., 2002). The 3- to 30-fold increases in free sulfite and 7- to 70-fold increases in thiosulfate in maize correlated with severely aberrant growth phenotypes and infertility (Martin et al., 2005). In comparison, the moderate increases in Cys and sulfite and the absence of thiosulfate, together with normal flowering and production of perfectly viable seeds, in sir1-1 further decrease the possibility of toxic side effects. APR overexpression was observed to yield no greater effect on Cys and GSH levels than overexpression of SAT in other transgenic approaches (reviewed in Sirko et al., 2004), and accumulation of reduced sulfur compounds was not proportional to transgenic APR activity. Thus, either OAS production by SAT or ATP sulfurylase or SiR activity could have been limiting under these conditions (Tsakraklides et al., 2002; Martin et al., 2005).
In this study, the block in the sir1-1 mutant resulted in 28% SiR activity and 3.6% residual flux (28-fold decreased) into Cys and other organic sulfur compounds compared with the wild type in mature leaves. This discrepancy can be explained, for one, by the difference between Vmax and in vivo activities due to substrate availability, and second, by the combined downregulation of ATPS, APR, and SULTR2.1 expression upstream of SiR. This coordination was probably achieved by feedback signals that were created by changes in metabolites in the pathway. In contrast with general expectation and the small flux rate, the steady state levels of Cys were even increased and those of GSH at least maintained. This situation differs significantly from the sulfate starvation situation in which contents of both thiols and sulfate are strongly reduced and genes of sulfate uptake and reduction are upregulated, forming a typical sulfur deficiency response (Hirai et al., 2005; Nikiforova et al., 2005; Höfgen and Nikiforova, 2008). The maintained Cys and GSH contents in sir1-1 may be linked to this difference, acting as repressors of ATPS, APR, and SULTR2.1. Maintenance of steady state levels of GSH in particular is supported by the high affinity of the first enzyme of biosynthesis, GSH1, for Cys and its allosteric feedback inhibition by GSH (Jez et al., 2004; Hothorn et al., 2006). Cys and GSH might also overrule the activating signal on sulfur-related gene expression that would be expected from elevated OAS contents like in the sulfur deficiency response. This observation is in line with results from downregulation of the major site of OAS biosynthesis in mitochondria using amiRNA against the gene encoding SAT3 (Haas et al., 2008). Arabidopsis amiSAT3 plants were strongly stunted in growth and showed severely reduced incorporation of 3H-Ser into OAS, Cys, and GSH, while the steady state levels of both thiols were not reduced, but rather were 1.5- to 2.5-fold increased. Indeed, APR2 mRNA and protein levels were reduced in such plants together with other genes related to sulfur metabolism (Haas et al., 2008). It is also very likely that the maintained steady state presence of both thiols supports the redox state and protein functions (Meyer and Hell, 2005; Meyer, 2008).
The Block in Sulfite Reduction Causes Adaptation of Carbon and Nitrogen Metabolism
Constitutive overexpression of SiR in sir1-1 and sir1-2 restored the wild-type phenotype but led to neither accelerated growth nor increased thiol contents, suggesting that SiR activity is sufficient at least under nonstressed conditions and that flux control takes place upstream of SiR. On the other hand, sir1-1 plants were clearly cadmium sensitive, arguing that even a moderate reduction of SiR activity not only retards growth but impairs stress resistance. Furthermore, the secondary sulfur compounds glucosinolates were reduced by ∼50%, an effect observed only under prolonged sulfate starvation (Blake-Kalff et al., 1998; Hirai et al., 2005, 2007). Therefore, the activity of SiR needs to be considered for regulatory processes not only in sulfate reduction but also in other assimilatory pathways. Indeed, dramatic changes in C, N, and S metabolism of leaves were observed. The most pronounced deregulation occurred at sulfate and nitrate contents that were ninefold enhanced and threefold decreased, respectively. These changes were reflected by significant shifts in elemental composition of sir1-1 compared with the wild type. The enhanced sulfate content was almost entirely responsible for the threefold increase of relative sulfur content. By contrast, both the decrease of nitrate content and the increase in free amino acids were indicative of a shift toward organically bound nitrogen compounds. The lowered total nitrogen correlated with the also significantly decreased total carbon content. Such massive effects are most unusual considering the general stability of these parameters (see Houba and Uittenbogaard, 1994). These findings point to comprehensive adaptations at the metabolic level. The lack of reduced sulfur for Cys synthesis most likely resulted in a decreased demand for reduced nitrogen and carbon. To counteract this imbalance at least partially, nitrate uptake (as indicated by contents) and carbon assimilation (as indicated by less chlorophyll and CHLOROPHYLLASE gene upregulation) were reduced. The strongly reduced levels of starch, glucose, fructose, and sucrose are contrasted by the increased content of free amino acids. Sugar levels may be reduced due to lowered photosynthetic activity as outlined before but also by an increased demand for carbon backbones for amino acid synthesis. This might be necessary to eliminate excess ammonia from nitrate reduction and/or slowed protein biosynthesis. Lowered nitrate contents point to at least partial downregulation of nitrate uptake and possibly nitrate reduction that may still be too effective, hence, the accumulation of amino acids. By contrast, sulfate accumulated strongly in sir1-1. Evidently, the severely lowered flux in the sulfate assimilation pathway caused a demand signal that led to enhanced sulfate uptake. This assumption was confirmed by apparently constitutive elevated sulfate uptake rates in roots of sir1-1 plants. Further evidence for the severe metabolic disturbance came from the strong induction of VSP1 and VSP2. VSPs are believed to serve as transient storage proteins to keep unused amino acids during plant development when excess photosynthates are produced but may have additional functions, such as plant defense. Their expression can be induced by wounding and phosphate starvation (Fujiwara et al., 2002; Liu et al., 2005). Remarkably, they carry no Met residues besides the starting Met and only two to three Cys residues. They thus contain less sulfur than average proteins in vegetative plant organs (Miseta and Csutora, 2000; Pe'er et al., 2004). Interestingly, sulfate import into leaves could not be efficiently downregulated, although expression of SULTR2.1, a major sulfate transporter in vascular tissue that is expressed in leaves (Takahashi et al., 2000; Hawkesford, 2008), was downregulated.
These adaptations in metabolism were accompanied by changes in expression of several genes related to redox homeostasis (20 genes), pathogen resistance (11 genes), glucosinolate metabolism (10 genes), and hormone synthesis and signaling (5 genes) (see Supplemental Table 1 online). The transcript levels of many of the pathogenesis related genes, including PR1, PR2, PR5, RPP1, RPP4, RPP5, EIN3, and PEN3, were significantly decreased in the leaves of sir1-1 compared with the wild-type control. The downregulation of many of the pathogen and redox-related genes in the sir1-1 leaves pointed toward the inability of Arabidopsis plants to cope with a variety of biotic and abiotic stresses under the long-term inadequate availability of reduced sulfur and underlines the importance of SiR for growth and development.
METHODS
Plant Genotypes and Growth Conditions
Arabidopsis thaliana ecotype Col-0 was used as wild-type control, since the mutants used in all experiments were in the Col-0 genetic background. Two T-DNA insertion lines for SiR, further annotated as sir1-1 (550A09) and sir1-2 (727B08), were obtained from the GABI-Kat collection center. The experiments were performed either on soil-grown plants or on hydrophonically grown plants. In all cases, including growth curves, plants were kept in the climate chamber under short-day conditions (8.5 h light). The light intensity in the growth chamber was set to 100 μE m−2 s−1, whereas the relative humidity was kept at 50%. The temperatures during the day and night were set at 22 and 18°C, respectively. Seeds were stratified on soil, and after 2 weeks, seedlings were transferred to individual pots for further growth. For hydroponic cultures, seeds were germinated in Eppendorf tubes placed in small boxes (0.25 liters) as described by Tocquin et al. (2003), containing half-strength Hoagland solution [2.5 mM Ca(NO3)2, 2.5 mM KNO3, 0.5 mM MgSO4, 0.5 mM KH2PO4, 40 μM Fe-EDTA, 25 μM H3BO3, 2.25 M MnCl2, 1.9 μM ZnSO4, 0.15 μM CuSO4, and 0.05 μM (NH4)6Mo7O24, pH 5.8 to 6.0]. For chemical complementation, the half-strength Hoagland solution was exogenously supplied either with a final concentration of 1 mM GSH or 0.1 mM Na2S. The media were exchanged every 2 d. For cadmium stress experiments, surface-sterilized seeds were germinated on solid Arabidopsis (At-medium) medium (Haughn and Somerville, 1986), containing Fe-HBED instead of Fe-EDTA. After 14 d of germination on different concentrations of CdCl2 (0, 25, 50, and 100 μM), the root lengths and phenotypes were documented.
Molecular Cloning and Genomic Characterization
Standard molecular biology technologies, such as growth of bacteria, plasmid isolation, and PCR, were performed as described by Sambrook et al. (1989) according to Good Laboratory Practice standards. Genomic DNA from a single leaf was first isolated according to Edwards et al. (1991). For selection and genomic characterization of homozygous plants, a PCR screen was set up. Primers that were used for the identification of the insertion position in sir1-1 are as follows: T-DNA_LB1 (5′-CCCATTTGGACGTGAATGTAGACAC-3′), gene specific_LP (5′-TCTTTGATTAAGCATGAAACATTG-3′), and gene specific_RP (5′-AGGCGATTCAAAAAGCATCTC-3′). For the sir1-2 line, the primers that were used are as follows: T-DNA_LB2 (5′-ATATTGACCATCATACTCATTGC-3′), gene specific_LP (5′-TCTTTGATTAAGCATGAAACATTG-3′), and gene specific_RP (5′-AGGCGATTCAAAAAGCATCTC-3′). Amplified products of the T-DNA_LB1 and gene-specific RP primer combination (for sir1-1) and T-DNA_LB2 and gene-specific LP primer combination (for sir1-2) were sequenced. Primers that were used for the identification of right border (RB) of the T-DNA (for sir1-2) are T-DNA_RB (5′-GTGGATTGATGTGATATCTCC-3′) and gene specific_RP (5′-AGGCGATTCAAAAAGCATCTC-3′).Amplified product of the T-DNA_RB and gene-specific RP primer combination was sequenced.
Cloning and Transformation of the Complementation Construct
For genetic complementation of sir1-1 and sir1-2 mutants, the SiR open reading frame along with its plastidic transit peptide was amplified from a cDNA clone using specially designed Gateway primers with attB recombination sites. These primers were as follows: attB_N (5′-AAAAAGCAGGCTATGTCATCGACGTTTCGA-3′) and attB_C (5′-AGAAAGCTGGGTTCATTGAGAAACTCCTTT-3′). A BP recombination reaction between an attB-flanked DNA fragment and an attP-containing donor vector (pDONR 201) was followed by an LR recombination reaction according to the supplier's instructions (Invitrogen) to generate an expression clone in pB2GW7 (VIB-University of Ghent). Transformation of homozygous sir1-1 and Col-0 plants were performed by floral dip according to Clough and Bent (1998). The transformants were grown directly on soil and were sprayed with commercial BASTA herbicide (Bayer Crop Science) at a final concentration of 200 mg/L glufosinate ammonium, when they were 2 to 3 weeks old.
Analysis of Transcript Levels
For qRT-PCR and microarray analysis, RNA was isolated from 200 mg leaf material of 7-week-old soil-grown homozygous sir1-1 and Col-0 plants with the RNeasy kit (Qiagen) according to manufacturer's protocol. For microarray analyses of transcript levels of 920 selected genes, the RNA was converted to cDNA, hybridized with a custom-made microarray, and evaluated as described by Haas et al. (2008). Data were normalized and examined for statistical significance as described in the statistical analysis section. Abundances of selected transcripts were independently confirmed from the same RNA preparation by qRT-PCR after cDNA conversion with the SuperScript VILO cDNA synthesis kit (Invitrogen). The qRT-PCR reaction was set up by mixture of 10 ng of freshly synthesized cDNA with 1.6 pmol of each specific primer in onefold EXPRESS Two-Step SYBR GreenER Universal mixture (Invitrogen). The reaction was performed in the LightCycler 480 (Roche Diagnostics) according to the EXPRESS Two-Step SYBR GreenER protocol and evaluated with LightCycler software 4.0 (Roche Diagnostics) using elongation factor 1α (EF1α) as reference for normalization. Each analysis consisted of three biological replicas. Each replica was tested three times, and this test was repeated once (i.e., n = 6 per replica). The following specific primers were used for analysis of APR2, Ef1α, SiR, and VSP2, respectively: APR2_for (5′-GAGGAAGATGGTGCTGCAGAC-3′), APR2_rev (5′-CCTCCTTTGCTCAATGCAACCAC-3′), Ef1α_for (5′-TGAGCACGCTCTTCTTGCTTTCA-3′), Ef1α_rev (5′-GGTGGTGGCATCCATCTTGTTACA-3′), SiR_for (5′-ACTGCAATGGCTTGCCCAGCTTT-3′), SiR_rev (5′-TCCCGCGCTCTGCCTCAGTTATT-3′), VSP2_for (5′-CTCAGTGACCGTTGGAAATTGTGG-3′), and VSP2_rev (5′-GTAGGCCACGCCAGCAGCT-3′).
Determination of Metabolites
Hydrophilic metabolites were extracted from leaves of 7-week-old soil-grown homozygous sir1-1 and Col-0 plants according to Wirtz and Hell (2003). Thiol and amino acids were quantified after derivatization with monobromobimane (Cabiochem, EMD Chemicals) and AccQ-Tag reagent (Waters), respectively. The derivatization procedure and separation of thiol derivatives were performed as described by Wirtz et al. (2004), using the same HPLC system. Anions were separated and quantified according to Wirtz and Hell (2007) using 10-fold diluted extract in water. Glucosinolates were extracted and measured as described by Brown et al. (2003). In situ staining of starch was achieved with Lugol's iodine solution (5 g I2, 10 g KI dissolved in 85 mL water, and diluted 1:10 in water for staining; leaves were kept 2 h at 80°C in 80% ethanol, rinsed with water, stained for 10 min in Lugol's iodine solution, rinsed again, and photographed). Contents of starch, sucrose, glucose, and fructose were determined according to Smith and Zeeman (2006) with the following modifications: starting material was 30 mg fresh weight and NADP was 2 mM, instead of 0.5 mM.
Analysis of Total Carbon, Nitrogen, and Sulfur Content
Total carbon, nitrogen, and sulfur contents of 9-week-old Arabidopsis rosettes were determined using a Vario MAX CNS elemental analyzer (Elementar). After complete drying of the entire rosettes at 120°C in the incubator for 16 h, the plant material was ground to fine powder. A sample of ∼20 mg dry weight was combusted entirely. The elements were detected in the forms of CO2, N2, and SO2 by means of their thermal conductivity and quantified according to a standard calibration curve prepared with sulfadiazene.
Determination of Sulfate Uptake Rates of Roots
Arabidopsis wild-type and sir1-1 plants were grown hydroponically for 5 weeks on half-strength Hoagland solution containing 50 μM sulfate under short-day conditions. Plants were transferred for 10 min to half-strength Hoagland solution supplemented with 0.1 mCi 35SO42− (37 TBq mmol−1). The uptake of 35S label was determined after washing of roots three times with water. For quantification of the incorporated label, the root tissue was dissolved in 2 mL of scintillation liquid (Ultima Gold; Perkin-Elmer) and counted for 10 min with a L600SC Scintillation counter (Beckman).
Production of At-SiR Antibody for Immunological Detection
The full-length open reading frame of SiR was amplified from a cDNA library of Arabidopsis Col-0 leaves with the primers SiR_for (5′-CATATGGCTCCGGCGGGAGCCGCT-3′) and SiR_rev (5′-CTCGAGTCATTGAGAAACTCCTTTG-3′), which contain an NdeI and XhoI restriction endonuclease site, respectively. The resulting PCR fragment was cloned via NdeI and XhoI in the pET-28a vector (Novagen) and sequenced to confirm the identity of the amplified SiR sequence. The resulting vector was termed pET-28 At-SiR and used for expression of recombinant SiR protein in fusion with a 6x His tag in Escherichia coli HMS174 (DE3). The production and purification of recombinant SiR protein was performed according to the manufacturer's protocols. Purified protein was checked by SDS-electrophoresis for impurities and sent to a commercial antibody production company (Pineda). The antibody was tested for cross-reaction with other Arabidopsis soluble proteins and was found to be specific.
Determination of Enzymatic Activities and Immunological Detection of Proteins
Aliquots from 200 mg FW leaf material were powdered in liquid nitrogen for small scale extraction of proteins as described by Heeg et al. (2008). Protein concentration was determined according to Bradford, (1976), using BSA as a standard. The enzymatic activity of SiR was measured in a total volume of 0.1 mL, containing 25 mM HEPES, pH 7.8, 1 mM Na2SO3, 5 mM OAS, 1 μg of recombinant OAS-TL C (Wirtz et al., 2004), 10 mM DTT, 30 mM NaHCO3, 15 mM Na2S2O4, and 5 mM methyl viologen along with the crude leaf extract. SAT activity was assayed by coupling to the OAS-TL reaction (Nakamura et al., 1987). To ensure high excess of OAS-TL activity during coupling of both reactions, all SAT activity determinations were supplemented with 4 units of purified recombinant OAS-TL. The activity of SO extracted from leaves of hydroponically grown sir1-1 and Col-0 plants was assayed according to Lang et al. (2007). For immunological detection of proteins, 50 μg of the total proteins of leaves were separated according to Laemmli (1970) by discontinuous SDS-PAGE in Mini-protean II cells (Bio-Rad) and subsequently blotted on nitrocellulose membrane as described by Wirtz and Hell (2007). The same samples were separated by a second SDS-PAGE and stained with Coomassie Brilliant Blue to visualize equal loading of proteins including LSU. The dilution of the primary At-SiR antibody was 1:2000, whereas the dilutions of purified primary antibodies for At-SAT3 (Heeg et al., 2008), At-OAS-TLC (Heeg et al., 2008), and At-SO (Lang et al., 2007) were 1:7500, 1:1000, and 1:500, respectively.
Determination of Incorporation Rates Using 35SO42−
Leaf pieces (30 mg) were cut out from 7-week-old hydroponically or soil-grown Arabidopsis plants. Incubation of the leaf discs on the half-strength Hoagland solution containing radioactively labeled 35SO42−, and the subsequent steps for the extraction, derivatization, and detection of metabolites were performed according to Heeg et al. (2008).
Statistical Analysis
The microarray data sets were analyzed with the M-CHiPS software (Fellenberg et al., 2001; see Supplemental Methods online). In M-CHiPS, differentially expressed genes were identified using the linear models of the Limma package (Smyth, 2004). Regression coefficients, moderated t-statistic, and P values were calculated using Empirical Bayesian statistics, accounting for a complete pairwise comparisons contrast matrix (across all samples). P values computed for the F-statistic (Smyth, 2004) were adjusted for multiple testing by Benjamini and Hochberg's algorithm (Benjamini and Hochberg, 1995) to control the false discovery rate at 5%. We selected genes that show intensity levels of >1000 in at least one of the conditions and at the same time show adjusted P values <0.05.
Means and standard errors were calculated according to standard procedures. In addition, comparisons of means from metabolite data sets were analyzed for statistical significance with the unpaired t test. Constant variance and normal distribution of data were checked with SigmaStat 3.0 prior to statistical analysis. The Mann-Whitney rank sum test was used to analyze groups of samples that did not follow normal Gaussian distribution. Significant differences between two groups (P < 0.05) are indicated by asterisks. The χ2 test was performed as described by Gardner and Osmond (1984).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank databases under the following accession numbers: SiR cDNA (Y10157), SiR gene (At5g04590), EF1-α (AT1G07920), and those listed in Supplemental Table 1 online. The microarray data reported in this publication have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the series number GSE20670.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Thiol Levels of Genetically Complemented sir1-1 Plants.
Supplemental Figure 2. Spectrum of Glucosinolates in sir1-1 Plants.
Supplemental Figure 3. Contribution of Anions to Total Sulfur and Nitrogen Content.
Supplemental Figure 4. Spectrum of Amino Acids in sir1-1 Plants.
Supplemental Figure 5. Immunological Detection of Sulfite Oxidase in sir1-1 Plants.
Supplemental Figure 6. Incorporation Rates of Sulfate in Wild-Type and sir1-1 Plants Grown on Soil.
Supplemental Figure 7. Transcript Levels of APR2 in sir1-1 Determined by qRT-PCR.
Supplemental Table 1. Significantly Regulated Transcripts in Leaves of 7-Week-Old sir1-1 Plants.
Supplemental Methods. Processing of Microarray Data.
Supplementary Material
Acknowledgments
We gratefully acknowledge funding of M.S.K. by the Higher Education Commission of Pakistan and the German Ministry for Education and Research, and of F.H.H. by the Stiftung der Deutschen Wirtschaft and the Schmeil-Stiftung Heidelberg. We thank S. Hassel (University of Heidelberg) for excellent technical assistance and S. Kopriva (John-Innes-Institute, Norwich, UK) for critical reading of the manuscript.
References
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc., B 57: 289–300 [Google Scholar]
- Benning C. (2007). Questions remaining in sulfolipid biosynthesis: A historical perspective. Photosynth. Res. 92: 199–203 [DOI] [PubMed] [Google Scholar]
- Blake-Kalff M.M., Harrison K.R., Hawkesford M.J., Zhao F.J., McGrath S.P. (1998). Distribution of sulfur within oilseed rape leaves in response to sulfur deficiency during vegetative growth. Plant Physiol. 118: 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork C., Schwenn J.D., Hell R. (1998). Isolation and characterization of a gene for assimilatory sulfite reductase from Arabidopsis thaliana. Gene 212: 147–153 [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brown P., Tokuhisa J., Reichelt M., Gershenzon J. (2003). Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62: 471–481 [DOI] [PubMed] [Google Scholar]
- Brychkova G., Xia Z., Yang G., Yesbergenova Z., Zhang Z., Davydov O., Fluhr R., Sagi M. (2007). Sulfite oxidase protects plants against sulfur dioxide toxicity. Plant J. 50: 696–709 [DOI] [PubMed] [Google Scholar]
- Cairns N.G., Pasternak M., Wachter A., Cobbett C.S., Meyer A.J. (2006). Maturation of Arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiol. 141: 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cobbett C.S., May M.J., Howden R., Rolls B. (1998). The glutathione-deficient, cadmium-sensitive mutant, cad2-1, of Arabidopsis thaliana is deficient in γ-glutamylcysteine synthetase. Plant J. 16: 73–78 [DOI] [PubMed] [Google Scholar]
- Crane B.R., Siegel L.M., Getzoff E.D. (1995). Sulfite reductase structure at 1.6 A: Evolution and catalysis for reduction of inorganic anions. Science 270: 59–67 [DOI] [PubMed] [Google Scholar]
- Edwards K., Johnstone C., Thompson C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers T., Schwarz G., Brinkmann H., Witt C., Richter T., Nieder J., Koch B., Hille R., Hansch R., Mendel R.R. (2001). Identification and biochemical characterization of Arabidopsis thaliana sulfite oxidase. A new player in plant sulfur metabolism. J. Biol. Chem. 276: 46989–46994 [DOI] [PubMed] [Google Scholar]
- Fell D., Thomas S. (1995). Physiological control of metabolic flux: The requirement for multisite modulation. Biochem. J. 311: 35–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellenberg K., Hauser N.C., Brors B., Neutzner A., Hoheisel J.D., Vingron M. (2001). Correspondence analysis applied to microarray data. Proc. Natl. Acad. Sci. USA 98: 10781–10786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Nambara E., Yamagishi K., Goto D.B., Naito S. (2002). Storage proteins. The Arabidopsis Book, Somerville C.R., Meyerowitz E.M., (Rockville, MD: American Society of Plant Biologists; ), doi/10.1199/tab.0020, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T., Pomprasirt S., Sakurai H. (1997). Characteristics of sulfite transport by Chlorella vulgaris. Plant Cell Physiol. 38: 398–403 [Google Scholar]
- Gardner M.J., Osmond C. (1984). Interpretation of time trends in disease rates in the presence of generation effects. Stat. Med. 3: 113–130 [DOI] [PubMed] [Google Scholar]
- Ha S.-B., Smith A.P., Howden R., Dietrich W.M., Bugg S., O'Connell M.J., Goldsbrough P.B., Cobbett C.S. (1999). Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe. Plant Cell 11: 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas F.H., Heeg C., Queiroz R., Bauer A., Wirtz M., Hell R. (2008). Mitochondrial serine acetyltransferase functions as a pacemaker of cysteine synthesis in plant cells. Plant Physiol. 148: 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänsch R., Lang C., Rennenberg H., Mendel R.R. (2007). Significance of plant sulfite oxidase. Plant Biol. (Stuttg.) 9: 589–595 [DOI] [PubMed] [Google Scholar]
- Hänsch R., Mendel R. (2005). Sulfite oxidation in plant peroxisomes. Photosynth. Res. 86: 337–342 [DOI] [PubMed] [Google Scholar]
- Haughn G.W., Somerville C. (1986). Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204: 430–434 [Google Scholar]
- Hawkesford M.J. (2008). Uptake, distribution and subcellular transport of sulfate. Sulfur Metabolism in Phototrophic Prganisms, Hell R., Dahl C., Leustek T., (Dordrecht, The Netherlands: Springer; ), pp. 17–32 [Google Scholar]
- Heeg C., Kruse C., Jost R., Gutensohn M., Ruppert T., Wirtz M., Hell R. (2008). Analysis of the Arabidopsis O-acetylserine(thiol)lyase gene family demonstrates compartment-specific differences in the regulation of cysteine synthesis. Plant Cell 20: 168–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell R., Wirtz M. (2008). Metabolism of cysteine in plants and phototrophic bacteria. Sulfur Metabolism in Phototrophic Organisms, Hell R., Dahl C., Leustek T., (Dordrecht, The Netherlands: Springer; ), pp. 61–94 [Google Scholar]
- Henkes S., Sonnewald U., Badur R., Flachmann R., Stitt M. (2001). A small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell 13: 535–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M.Y., et al. (2005). Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J. Biol. Chem. 280: 25590–25595 [DOI] [PubMed] [Google Scholar]
- Hirai M.Y., et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA 104: 6478–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfgen R., Nikiforova V.J. (2008). Metabolomics integrated with transcriptomics: Assessing systems response to sulfur-deficiency stress. Physiol. Plant. 132: 190–198 [DOI] [PubMed] [Google Scholar]
- Hothorn M., Wachter A., Gromes R., Stuwe T., Rausch T., Scheffzek K. (2006). Structural basis for the redox control of plant glutamate cysteine ligase. J. Biol. Chem. 281: 27557–27565 [DOI] [PubMed] [Google Scholar]
- Houba V.J.G., Uittenbogaard J. (1994). International plant-analytical exchange (IPE). Chemical Composition of Various Plant Species, Houba V.J.G., Uittenbogaard J., (Wageningen, The Netherlands: Wageningen Agricultural University; ), pp. 8–219 [Google Scholar]
- Jez J.M., Cahoon R.E., Chen S. (2004). Arabidopsis thaliana glutamate-cysteine ligase: functional properties, kinetic mechanism, and regulation of activity. J. Biol. Chem. 279: 33463–33470 [DOI] [PubMed] [Google Scholar]
- Kopriva S. (2006). Regulation of sulfate assimilation in Arabidopsis and beyond. Ann. Bot. (Lond.) 97: 479–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S., Muheim R., Koprivova A., Trachsel N., Catalano C., Suter M., Brunold C. (1999). Light regulation of assimilatory sulphate reduction in Arabidopsis thaliana. Plant J. 20: 37–44 [DOI] [PubMed] [Google Scholar]
- Koprivova A., Suter M., den Camp R.O., Brunold C., Kopriva S. (2000). Regulation of sulfate assimilation by nitrogen in Arabidopsis. Plant Physiol. 122: 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger S., Niehl A., Lopez Martin M., Steinhauser D., Donath A., Hildebrandt T., Romero L., Hoefgen R., Gotor C., Hesse H. (2009). Analysis of cytosolic and plastidic serine acetyltransferase mutants and subcellular metabolite distributions suggests interplay of the cellular compartments for cysteine biosynthesis in Arabidopsis. Plant Cell Environ. 32: 349–367 [DOI] [PubMed] [Google Scholar]
- Krüger R.J., Siegel L.M. (1982). Evidence for siroheme-Fe4S4 interaction in spinach ferredoxin-sulfite reductase. Biochemistry 21: 2905–2909 [DOI] [PubMed] [Google Scholar]
- Kruse C., Jost R., Lipschis M., Kopp B., Hartmann M., Hell R. (2007). Sulfur-enhanced defence: Effects of sulfur metabolism, nitrogen supply, and pathogen lifestyle. Plant Biol. 9: 608–619 [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lang C., Popko J., Wirtz M., Hell R., Herschbach C., Kreuzwieser J., Rennenberg H., Mendel R., Hänsch R. (2007). Sulphite oxidase as key enzyme for protecting plants against sulphur dioxide. Plant Cell Environ. 30: 447–455 [DOI] [PubMed] [Google Scholar]
- Leustek T. (2002). Sulfate metabolism. The Arabidopsis Book, Somerville C.R., Meyerowitz E.M., (Rockville, MD: American Society of Plant Biologists; ), doi/10.1199/tab.0017, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T., Martin M.N., Bick J.-A., Davies J.P. (2000). Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51: 141–165 [DOI] [PubMed] [Google Scholar]
- Liu Y., Ahn J.-E., Datta S., Salzman R.A., Moon J., Huyghues-Despointes B., Pittendrigh B., Murdock L.L., Koiwa H., Zhu-Salzman K. (2005). Arabidopsis vegetative storage protein is an anti-insect acid phosphatase. Plant Physiol. 139: 1545–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O., Saliba-Colombani V., Camilleri C., Calenge F., Gaudon V., Koprivova A., North K., Kopriva S., Daniel-Vedele F. (2007). Natural variation for sulfate content in Arabidopsis thaliana is highly controlled by APR2. Nat. Genet. 39: 896–900 [DOI] [PubMed] [Google Scholar]
- Martin M.N., Tarczynski M.C., Shen B., Leustek T. (2005). The role of 5′-adenylylsulfate reductase in controlling sulfate reduction in plants. Photosynth. Res. 86: 1–15 [DOI] [PubMed] [Google Scholar]
- Meyer A. (2008). The integration of glutathione homeostasis and redox signaling. J. Plant Physiol. 165: 1390–1403 [DOI] [PubMed] [Google Scholar]
- Meyer A.J., Hell R. (2005). Glutathione homeostasis and redox-regulation by sulfhydryl groups. Photosynth. Res. 86: 435–457 [DOI] [PubMed] [Google Scholar]
- Miseta A., Csutora P. (2000). Relationship between the occurrence of cysteine in proteins and the complexity of organisms. Mol. Biol. Evol. 17: 1232–1239 [DOI] [PubMed] [Google Scholar]
- Mugford S.G., et al. (2009). Disruption of adenosine-5′-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell 21: 910–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M., Akashi T., Hase T. (2000). Plant sulfite reductase: Molecular structure, catalytic function and interaction with ferredoxin. J. Inorg. Biochem. 82: 27–32 [DOI] [PubMed] [Google Scholar]
- Nakamura K., Hayama A., Masada M., Fukushima K., Tamura G. (1987). Measurement of serine acetyltransferase activity in crude plant extracts by a coupled assay system using cysteine synthase. Plant Cell Physiol. 28: 885–891 [Google Scholar]
- Nikiforova V.J., Kopka J., Tolstikov V., Fiehn O., Hopkins L., Hawkesford M.J., Hesse H., Hoefgen R. (2005). Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol. 138: 304–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak M., Lim B., Wirtz M., Hell R., Cobbett C.S., Meyer A.J. (2008). Restricting glutathione biosynthesis to the cytosol is sufficient for normal plant development. Plant J. 53: 999–1012 [DOI] [PubMed] [Google Scholar]
- Patron N.J., Durnford D.G., Kopriva S. (2008). Sulfate assimilation in eukaryotes: Fusions, relocations and lateral transfers. BMC Evol. Biol. 8: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe'er I., Felder C., Man O., Silman I., Sussman J., Beckmann J. (2004). Proteomic signatures: Amino acid and oligopeptide compositions differentiate among phyla. Proteins 54: 20–40 [DOI] [PubMed] [Google Scholar]
- Rotte C., Leustek T. (2000). Differential subcellular localization and expression of ATP sulfurylase and 5′-adenylylsulfate reductase during ontogenesis of Arabidopsis leaves indicates that cytosolic and plastid forms of ATP sulfurylase may have specialized functions. Plant Physiol. 124: 715–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Schmidt A., Jäger K. (1992). Open questions about sulfur metabolism in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43: 325–349 [Google Scholar]
- Sirko A., Blaszczyk A., Liszewska F. (2004). Overproduction of SAT and/or OASTL in transgenic plants: A survey of effects. J. Exp. Bot. 55: 1881–1888 [DOI] [PubMed] [Google Scholar]
- Smith A.M., Zeeman S.C. (2006). Quantification of starch in plant tissues. Nat. Protoc. 1: 1342–1345 [DOI] [PubMed] [Google Scholar]
- Smith F.W., Hawkesford M.J., Ealing P.M., Clarkson D.T., Vanden Berg P.J., Belcher A.R., Warrilow A.G. (1997). Regulation of expression of a cDNA from barley roots encoding a high affinity sulphate transporter. Plant J. 12: 875–884 [DOI] [PubMed] [Google Scholar]
- Smyth G.K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article3 [DOI] [PubMed] [Google Scholar]
- Stitt M., Sonnewald U. (1995). Regulation of metabolism in transgenic plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46: 341–368 [Google Scholar]
- Swamy U., Wang M., Tripathy J., Kim S., Hirasawa M., Knaff D., Allen J. (2005). Structure of spinach nitrite reductase: implications for multi-electron reactions by the iron-sulfur:siroheme cofactor. Biochemstry 44: 16054–16063 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Saito K. (2008). Molecular biology and functional genomics for identification of regulatory networks of plant sulfate uptake and assimilatory metabolism. Sulfur Metabolism in Phototrophic Organisms, Hell R., Dahl C., Leustek T., (Dordrecht, The Netherlands: Springer; ), pp. 151–164 [Google Scholar]
- Takahashi H., Watanabe-Takahashi A., Smith F.W., Blake-Kalff M., Hawkesford M.J., Saito K. (2000). The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J. 23: 171–182 [DOI] [PubMed] [Google Scholar]
- Tocquin P., Corbesier L., Havelange A., Pieltain A., Kurtem E., Bernier G., Perilleux C. (2003). A novel high efficiency, low maintenance, hydroponic system for synchronous growth and flowering of Arabidopsis thaliana. BMC Plant Biol. 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakraklides G., Martin M., Chalam R., Tarczynski M.C., Schmidt A., Leustek T. (2002). Sulfate reduction is increased in transgenic Arabidopsis thaliana expressing 5′-adenylylsulfate reductase from Pseudomonas aeruginosa. Plant J. 32: 879–889 [DOI] [PubMed] [Google Scholar]
- Vauclare P., Kopriva S., Fell D., Suter M., Sticher L., von Ballmoos P., Krahenbuhl U., den Camp R.O., Brunold C. (2002). Flux control of sulphate assimilation in Arabidopsis thaliana: adenosine 5′-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. Plant J. 31: 729–740 [DOI] [PubMed] [Google Scholar]
- Vernoux T., Wilson R.C., Seeley K.A., Reichheld J.P., Muroy S., Brown S., Maughan S.C., Cobbett C.S., Van Montagu M., Inze D., May M.J., Sung Z.R. (2000). The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Kusano M., Oikawa A., Fukushima A., Noji M., Saito K. (2008a). Physiological roles of the β-substituted alanine synthase gene family in Arabidopsis. Plant Physiol. 146: 310–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Mochida K., Kato T., Tabata S., Yoshimoto N., Noji M., Saito K. (2008b). Comparative genomics and reverse genetics analysis reveal indispensable functions of the serine acetyltransferase gene family in Arabidopsis. Plant Cell 20: 2484–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz M., Droux M., Hell R. (2004). O-acetylserine (thiol) lyase: An enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana. J. Exp. Bot. 55: 1785–1798 [DOI] [PubMed] [Google Scholar]
- Wirtz M., Hell R. (2003). Production of cysteine for bacterial and plant biotechnology: Application of cysteine feedback-insensitive isoforms of serine acetyltransferase. Amino Acids 24: 195–203 [DOI] [PubMed] [Google Scholar]
- Wirtz M., Hell R. (2006). Functional analysis of the cysteine synthase protein complex from plants: structural, biochemical and regulatory properties. J. Plant Physiol. 163: 273–286 [DOI] [PubMed] [Google Scholar]
- Wirtz M., Hell R. (2007). Dominant-negative modification reveals the regulatory function of the multimeric cysteine synthase protein complex in transgenic tobacco. Plant Cell 19: 625–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Onda Y., Ashikari T., Tanaka Y., Kusumi T., Hase T. (2000). Analysis of reductant supply systems for ferredoxin-dependent sulfite reductase in photosynthetic and nonphotosynthetic organs of maize. Plant Physiol. 122: 887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.