Flavonoids, including proanthocyanidins (PAs; also called condensed tannins), play a multitude of roles in plants (Winkel-Shirley, 2001). The presence of certain types of flavonoids in crops is associated with desirable and important agronomic traits; therefore, metabolic engineering of flavonoid biosynthesis has attracted considerable interest (Dixon, 2005). PAs are oligomers or polymers of flavan-3-ol units and are prominent flavonoid compounds in seed coats (where they become oxidized and confer a brownish color to the testa), leaves, fruits, flowers, and bark (Dixon et al., 2005). Characterization of a series of transparent testa (tt) and tannin-deficient seed (tds) mutants from Arabidopsis (Arabidopsis thaliana) has led to an in-depth understanding of flavonoid biosynthesis at the molecular level (Shirley et al., 1995; Winkel-Shirley, 2001; Abrahams et al., 2002; Lepiniec et al., 2006; Fig. 1). These studies clearly suggest that, in addition to structural enzymes and regulatory factors, transport proteins are also essential for flavonoid biosynthesis. Ablation of single transporter genes can result in defects in flavonoid and PA production associated with dysfunction of the central vacuole (Baxter et al., 2005; Marinova et al., 2007a, 2007b). With increasing evidence of such links between biochemistry and cell biology, transport and trafficking of plant secondary metabolites is emerging as an important, but technically challenging, research frontier (Grotewold, 2004). In this Update, we briefly review the status of our knowledge concerning PA biosynthesis and assembly, with particular emphasis on the still controversial mechanisms for transport and polymerization of the PA monomers.
Figure 1.
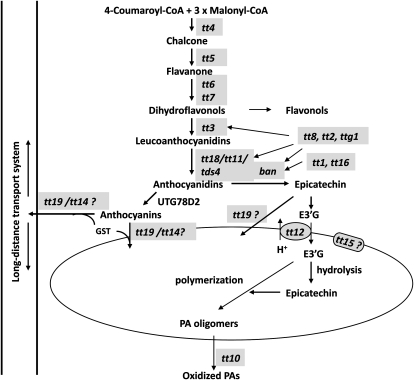
Genetically mapped TT genes involved in PA biosynthesis in Arabidopsis. tt mutants and their encoding enzymes are as follows: tt4, chalcone synthase; tt5, chalcone isomerase; tt6, flavanone 3-hydroxylase; tt7, flavonoid 3′ hydroxylase; tt3, dihydroflavonol 4-reductase; ban, ANR; tt18 (at the same locus as tt11), leucocyanidin dioxygenase; tt10, laccase-like polyphenol oxidase; tt12, MATE antiporter; tt15, UDP-Glc:sterol glycosyltransferase; tt19 (at the same locus as tt14), GST; aha10, P-type H+-ATPase. tt mutants encoding regulatory proteins are as follows: tt1, WIK-type zinc finger transcription factor; ttg1 (for transparent testa glabra1), WD40 repeat transcription factor; tt2, R2R3 Myb transcription factor; tt8, basic helix-loop-helix transcription factor; tt16, MADS domain transcription factor.
MODEL SYSTEMS AND THE BASIC PATHWAY FOR PA BIOSYNTHESIS
Arabidopsis has, until recently, been the major model system for molecular studies of PA biosynthesis (Lepiniec et al., 2006). The model legume Medicago truncatula has recently emerged as another useful system, with the ultimate goal of introducing PAs into the foliar tissues of its close relative, the major forage crop alfalfa (Medicago sativa), to protect ruminant animals from pasture bloat and enhance ruminant nutrition (Xie et al., 2003; Dixon et al., 2005; Pang et al., 2007, 2008; Peel et al., 2009; Zhao and Dixon, 2009).
PAs in Arabidopsis and M. truncatula consist of the same monomeric building unit, epicatechin, and have similar biological features (Abrahams et al., 2002; Lepiniec et al., 2006; Pang et al., 2007). Epicatechin shares a common biosynthetic pathway with anthocyanins from Phe to anthocyanidin (Dixon et al., 2005), from which epicatechin is formed by the action of anthocyanidin reductase (ANR; Xie et al., 2003; Fig. 1). The Medicago glycosyltransferase UGT72L1 catalyzes UDP-Glc-dependent glycosylation of epicatechin to form epicatechin 3′-O-glucoside (E3′G; Pang et al., 2008), which is the substrate for a vacuolar multidrug and toxic compound extrusion (MATE) transporter (Zhao and Dixon, 2009; Fig. 1). M. truncatula MATE1 is an ortholog of Arabidopsis TT12, and both proteins transport cyanidin 3-O-glucoside (Cy3G), although E3′G is transported with a higher affinity and velocity (Marinova et al., 2007b; Zhao and Dixon, 2009).
Grapevine (Vitis vinifera), with a recently sequenced genome, has become another model plant for studying PA biosynthesis. Grapevine PAs consist of two major flavan 3-ol monomers, catechin and epicatechin. Their seed and skin PAs have average degrees of polymerization of around 10 and 30, respectively. Epicatechin, and to a lesser extent epigallocatechin, is the extension unit and catechin the terminal unit in grapevine PAs (Terrier et al., 2009). Characterization of several transcription factors that regulate grapevine PA biosynthesis has revealed both similar and distinct regulatory mechanisms from those in Arabidopsis and M. truncatula (Bogs et al., 2007; Deluc et al., 2008; Terrier et al., 2009). Two TT2-like Myb transcription factors, VvMYBPA1 and VvMYBPA2, are essential for the expression of PA biosynthetic genes and PA accumulation in grapevine hairy roots (Bogs et al., 2007; Terrier et al., 2009). VvMYBPA1, which is mainly expressed during seed development, specifically regulates PA biosynthetic genes such as ANR and LEUCOANTHOCYANIDIN REDUCTASE. It can complement the seed PA-deficient phenotype of the Arabidopsis tt2 mutant, suggesting that VvMYBPA1 is an ortholog of TT2, although it is not clear whether grapevine also possesses a TT2-TT8-TTG1-like ternary transcription complex such as controls PA biosynthesis in Arabidopsis (Lepiniec et al., 2006; Bogs et al., 2007). VvMYBPA2 is mainly expressed in berries and leaves, but constitutive expression of either VvMYBPA1 or VvMYBPA2 in hairy roots induces similar sets of genes (Terrier et al., 2009). A new player in PA biosynthesis, VvMYB5 (Deluc et al., 2008), was also found to regulate the biosynthesis of PAs and other flavonoids in grapevine, and other MYB transcription factors, DkMYB4 and PtMYB134, regulate PA biosynthesis in persimmon (Diospyros kaki) fruit and poplar (Populus spp.) leaves, respectively (Akagi et al., 2009; Mellway et al., 2009). These data suggest that MYB family transcription factors are common regulators of PA biosynthesis in different tissues.
GLUCOSYLATION OF EPICATECHIN
Several studies show that PAs are stored in the vacuoles of Arabidopsis seed coat endothelial cells during the early stages of seed development (Abrahams et al., 2003; Kitamura et al., 2004), and it is likely, but not yet definitively proven, that PA oligomerization/polymerization occurs in the vacuole. As biosynthesis of PA precursors is believed to occur on the cytosolic face of the endoplasmic reticulum (ER) surface, the PA starter and extension units must first be transported into the vacuole. The tt12 mutation was mapped to a MATE family transporter (Debeaujon et al., 2001), and TT12 protein was shown to localize to the vacuolar membrane. Yeast-expressed TT12 could transport Cy3G but not catechin 3-O-glucoside, which could inhibit Cy3G uptake (Marinova et al., 2007b). This was perhaps the first comprehensive characterization of a plant secondary metabolite transporter, incorporating tissue-specific expression, subcellular localization, transport activity, and metabolite profiling of loss-of-function lines. However, Cy3G is not considered to be a PA precursor. Comparative studies of MATE1 and TT12 revealed that the kinetically preferred substrate for both transporters is E3′G rather than Cy3G, which explains the seed phenotypes of the tt12 and mate1 mutants (Marinova et al., 2007b; Zhao and Dixon, 2009). The MATE transporter (CAO69962) from grapevine may carry out similar functions that remain to be determined experimentally (Terrier et al., 2009).
If E3′G is universally the vacuolar transport form of epicatechin, its subsequent fate inside the vacuole is still not clear. E3′G can be detected during the early stages of seed development in M. truncatula (Pang et al., 2008), but its levels decrease to zero as PA levels increase, suggesting that it undergoes hydrolysis and subsequent incorporation into the PA polymer. However, other interpretations of the data are possible. For example, glucosylation by UGT72L1 could function in part as a detoxification mechanism to “mop up” excess epicatechin, consistent with the fact that this enzyme shows considerably higher sequence similarity to multifunctional UGTs, ascribed roles in xenobiotic detoxification, than to other flavonoid-specific UGTs (Pang et al., 2008). Alternatively, glucosylation of epicatechin could be an integral part of the PA polymerization mechanism as well as essential for monomer transport. In this respect, it is important to note that the exact nature of the starter and extension units of PAs remains to be determined. Gain- and loss-of-function experiments with UGT72L1 could give valuable information about its in vivo function.
THE ROLE OF TT19
TT19, a glutathione S-transferase (GST), is an essential protein for anthocyanin and PA accumulation in Arabidopsis and has orthologs in other plants such as maize (Zea mays), petunia (Petunia hybrida), and grapevine (Marrs et al., 1995; Alfenito et al., 1998; Kitamura et al., 2004). GST is believed to function in flavonoid production through its binding to flavonoids rather than by catalyzing the conjugation of glutathione to anthocyanins (Mueller et al., 2000). GST may act as an anthocyanin-binding matrix to protect anthocyanins from oxidation, or the GST-flavonoid complex may facilitate flavonoid transport mediated either by membrane transporters (Smith et al., 2003) or via vesicle trafficking. However, the exact mechanisms underlying such processes are not understood.
The tissue expression patterns of GSTs may provide additional clues to their functions in flavonoid transport. TT19 is expressed in leaves (particularly during senescence), stems, and young siliques (Kitamura et al., 2004). A TT19 promoter::GUS transgene directed high GUS activity to vascular tissues in the mid vein of Arabidopsis leaves, and a flavonoid glycosyltransferase gene exhibits a similar expression pattern (Wenzel et al., 2008). These data suggest that TT19 may be involved in long-distance transport of flavonoid-GST complexes (Fig. 2) in addition to its proposed role in anthocyanin and PA production in the seed coat, consistent with the lack of pigmentation in leaves and stems of tt19 plants (Kitamura et al., 2004). Studies on flavonoid-auxin transport interactions in vegetative axillary bud branching also support this hypothesis (Lazar and Goodman, 2006), and long-distance transport of flavonoids between cells or organs (e.g. roots and shoots) has recently been demonstrated (Buer et al., 2007).
Figure 2.
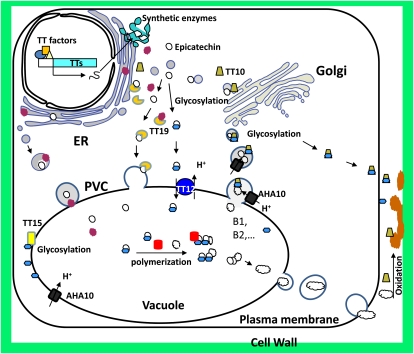
Model for PA transport and polymerization. PA regulatory transcription factors (TT factors) activate PA biosynthesis structural genes (TTs) in the nuclei of seed coat endothelial cells under appropriate conditions. PA pathway proteins are translocated to the cytosolic side of the ER for synthesis of epicatechin (white circles) and anthocyanins (red circles). These flavonoids can be compartmentalized in different ways. Epicatechin and anthocyanins are readily glycosylated, and the conjugates are transported into the vacuole by MATE (TT12) transporters. They could also be loaded into the ER membrane system or derived membrane vesicles, which are transported to the central vacuole through prevacuole compartment (PVC)-dependent vesicle trafficking, or else they could be bound to the TT19 GST, which facilitates their transport into the ER, vacuole, or other compartments. The acidic vacuolar conditions may facilitate nonenzymatic condensation of PA units, or the units may undergo enzymatic condensation catalyzed by TT10, with E3′G as a possible extension unit, or by yet unidentified proteins (red cylinders). After synthesis on the ER and modification by glycosylation in the Golgi, TT10 could be sorted and targeted within membrane vesicles that also contain epicatechin and other PA biosynthetic units. TT10 may also catalyze the condensation of PA units into oligomers (such as procyanidin B1 and B2) in the vesicles, which are transported to the vacuole, where PA chain elongation could be further catalyzed by TT10 using epicatechin glucoside and PA oligomers as substrates. PAs can also be transported through membrane vesicles or other mechanisms to the apoplastic space, where they are subjected to oxidative polymerization and further cross-linked with other cell wall components, catalyzed by apoplastic TT10-like polyphenol oxidases. Blue hexagons represent glycosylated moieties (epicatechin, membrane sterols, and TT10 protein).
A Medicago GST gene is up-regulated by ectopic expression of either the Medicago Legume Anthocyanin Production1 MYB transcription factor (which controls anthocyanin biosynthesis) or the Arabidopsis TT2 MYB transcription factor (which controls PA biosynthesis; Pang et al., 2008; Peel et al., 2009). The Medicago GST gene is expressed in vegetative buds, petioles, stems, leaves, and flower tissues. Interestingly, a Medicago anthocyanin transporter, MtMATE2, shows a similar tissue-specific expression pattern to that of the GST, particularly around vascular bundles in petals (J. Zhao and R.A. Dixon, unpublished data). Therefore, both a GST and MtMATE2 may be involved in long-distance transport of anthocyanins. It is possible that transported anthocyanins could ultimately serve as precursors of PAs through deglycosylation and subsequent conversion to epicatechin, thus providing an alternative explanation for the reduced PA phenotype of the tt19 mutation.
THE ROLE OF THE AHA10 PROTON PUMP
AHA10 is a putative P-type H+-ATPase, and Arabidopsis aha10 seeds show a typical tt phenotype with drastic reduction of PA levels (Baxter et al., 2005). It is not surprising that an H+-ATPase is involved in seed coat PA production, since one would be predicted to be necessary to support the activities of the TT12 and MATE1 H+ antiporters in the Arabidopsis and Medicago seed coats, respectively. Both tt12 and aha10 mutants show vacuolar morphological defects, consistent with the hypothesis that AHA10 may be tonoplast localized and provide the H+ gradient across the vacuolar membrane to support the MATE transporter's H+/flavonoid antiport function (Baxter et al., 2005). However, it is surprising that tt12 seeds do not accumulate epicatechin, whereas aha10 seeds contain higher levels of epicatechin than wild-type seeds.
One possible explanation for the above paradox is that transporters in addition to TT12 may exist for PA transport. If AHA10 is located on the tonoplast like its ortholog PH5 from petunia (Verweij et al., 2008) and provides an H+ gradient for antiporters like TT12, knockout of AHA10 may not completely eliminate TT12 function because other vacuolar ATPases or vacuolar pyrophosphatases could partially support the E3′G transport function of TT12. Alternatively, AHA10 may be localized on the ER or on vesicles involved in PA transport from the cytosol to the vacuole (Fig. 2). Vesicle trafficking-mediated PA transport may require AHA10 to supply energy. Therefore, definitively determining the subcellular localization of AHA10 is critical for explaining its biological functions in PA transport.
ARE PAS AND THEIR PRECURSORS TRANSPORTED BY VESICLE TRAFFICKING, MEMBRANE TRANSPORTERS, OR BOTH?
Early studies suggested that PAs are contained in cytoplasmic vesicle-like structures and that these prevacuole-like vesicles can fuse with the central vacuole (Abrahams et al., 2002). Vesicle trafficking has also been proposed to be involved in the transport of anthocyanins from the cytosol into the central vacuole (Poustka et al., 2007; Pourcel et al., 2010). Recent studies show that Arabidopsis mutants with reduced PA production exhibit morphological defects in the central vacuole of the seed coat endothelial cells (Abrahams et al., 2003; Kitamura et al., 2004; Baxter et al., 2005); for example, PAs with a patchy, filamentous distribution pattern confined to prevacuole-like small vesicles are found in the tt18/tds4 mutants (Abrahams et al., 2003; Kitamura et al., 2004; Baxter et al., 2005).
Tapetosomes, a unique type of vesicle from tapetum cells of Arabidopsis anthers, contain ER-derived flavonoids, oleosins from oil droplets, and alkanes and flavonoids that are released to the pollen surface upon tapetum cell death (Hsieh and Huang, 2007). In wild-type Arabidopsis, flavonoids are localized in the ER network and ER-derived tapetosomes but are mostly in the cytosol and not associated with the tapetosome in the tt12 and tt19 mutants (Hsieh and Huang, 2007). This suggests the possibility that TT12 and TT19 may be involved in the transport of flavonoids into the ER lumen. If so, it is not clear if this is relevant for PA transport. PAs might be carried through ER transport vesicles to the vacuole (Fig. 2), and TT12 might be on any of the endomembranes involved in such trafficking, either in a functional state or as a passenger for the membrane protein targeting pathway, or both.
It is generally believed that PAs localize to cell walls in the coats of mature seeds. How epicatechin or PA oligomers are transported out of endothelial cells across the vacuolar and plasma membranes remains unknown. It has been proposed that they are released when the vacuole bursts as a result of cell death during the seed desiccation process (Pourcel et al., 2005). Alternatively, both vesicle trafficking and membrane transporters could be involved in the release of PAs into the apoplast. According to a recent study on the release of callose and antifungal compounds in Arabidopsis, both membrane vesicle-mediated (marked with SNARE PEN1 syntaxin and SNAP33 complex) and membrane transporter-mediated (marked with PDR8/PEN3 ATP-binding cassette transporter) secretion systems are involved (Meyer et al., 2009). PA oligomers could be transported to the extracellular space by a similar mechanism.
The most recently characterized tt gene, tt15, encodes the UDP-Glc:sterol glycosyltransferase UGT8B1 (Pourcel et al., 2005; Debolt et al., 2009). In addition to reduced PA levels in seeds, tt15 has reduced cyanidin and quercetin levels (Focks et al., 1999), reduced sterol glycoside (SG) and acyl-SG levels in leaf, stem, and silique, and lower suberin and cutin-like polymer levels in the seed coat (Debolt et al., 2009). It is not clear how TT15 affects flavonoid and PA accumulation in seeds. However, it seems likely that the mechanism involves membrane properties, since SG and acyl-SG are important lipid membrane components (Debolt et al., 2009). Proteomic studies showed that UGT8B1 is localized to the vacuolar membrane (Carter et al., 2004; Jaquinod et al., 2007). Therefore, loss of function of TT15 may indirectly impair vacuolar transporter activity or vesicle trafficking associated with flavonoid sequestration. It will be of great interest to further pursue these ideas.
IS TT10 INVOLVED IN INITIAL PA OLIGOMERIZATION?
Among all characterized tt mutants, tt10 is probably the most interesting and mysterious. TT10 encodes a putative laccase-like polyphenol oxidase that was proposed to participate in the oxidation and/or polymerization of PAs. TT10 has a predicted signal peptide for secretion into the apoplastic space, where it has been proposed to oxidize colorless PA oligomers into the oxidized, insoluble, brown PAs characteristic of the mature seed coat (Pourcel et al., 2005). However, several questions remain about the tt10 mutant phenotype.
tt10 mutant seeds show a clear tt phenotype, which is generally associated with a reduction in the levels of brown, oxidized PAs (Pourcel et al., 2005); the seeds, however, do gradually brown with age. Surprisingly, metabolite profiling shows that tt10 seeds have increased levels of soluble PAs and some flavonoids but do not have lower levels of oxidized, brown PA products than wild-type seeds (Pourcel et al., 2005). So why do tt10 seeds show a tt phenotype without the reduction in PA levels found in other tt mutants? Are there additional oxidized PA products that account for the difference but that cannot be extracted or detected using current techniques?
Although TT10 has been proposed to catalyze the formation of oxidized PAs linked with cell wall substances to form an insoluble complex in the apoplastic space, studies in which epicatechin was fed to split seeds of tt10 and wild-type Arabidopsis indicated that the products of oxidation of free epicatechin by TT10 are nonconventional, “incorrectly linked” PA oligomers such as dehydrodiepicatechin A (Pourcel et al., 2005). Thus, if TT10 is indeed the initial polymerizing enzyme, either epicatechin is not the natural substrate or additional proteins (perhaps similar to the dirigent proteins involved in dimeric lignan formation; Davin et al., 1997) are necessary and do not function well in the split seed feeding assay. Perhaps the somewhat unusual glycosylation pattern of E3′G is of significance here. Most flavonoids with a free 3-hydroxyl group on the central C-ring (as found in epicatechin) are glycosylated at this position. Glycosylation at the 3′ position would prevent the formation of a B-ring orthodiquinone during oxidation, and this would likely inhibit the formation of B-ring-linked oxidation products such as dehydrodiepicatechin A. It remains to be determined whether E3′G can serve as a substrate for TT10 to produce a dimer or higher oligomer with the typical C- to A-ring linkage found in natural PAs.
Observations on the timing of seed color changes and PA accumulation in the vacuole show that epicatechin and PAs first accumulate in the central vacuole and then disappear during the later seed desiccation period (Kitamura et al., 2004). However, levels of TT10 transcripts temporally parallel those of the PA biosynthetic genes (Pourcel et al., 2005). This seems to be inconsistent with the proposed function of TT10 in oxidizing already formed PAs during the late stages of seed maturation (Pourcel et al., 2005), unless the TT10 protein has a very long half-life.
If TT10 only acts as an extracellular enzyme, catalyzing oxidative polymerization of epicatechin and PA oligomers in the apoplastic space, the tt10 mutation should not affect epicatechin and procyanidin oligomers stored intracellularly (i.e. in the vacuole; Kitamura et al., 2004). Unless epicatechin or PA oligomers accumulate in the extracellular space from the onset of seed development, the proposed functions of TT10 as currently proposed are hard to understand. Better understanding of the temporal and spatial expression and localization of TT10 is clearly required.
Does TT10 also function as a PA polymerase to catalyze PA chain elongation into higher degree PA oligomers? TT10 was excluded as being a PA polymerizing enzyme mainly on the basis of experiments in which immature tt4-8 seeds fed with epicatechin failed to accumulate PAs (Pourcel et al., 2005). However, if epicatechin were not the extension unit, or if TT10 catalyzed PA biosynthesis only under specific conditions found within the vacuole, the above feeding experiment would not necessarily be informative.
If TT10 does act as the initial PA-polymerizing enzyme, some of the controversial phenotypes of the tt10 mutant could be explained. However, other possible polymerization mechanisms, such as nonenzymatic polymerization, cannot yet be ruled out (Dixon et al., 2005). These proposed mechanisms are based on an early proposal by Haslam (1977), who showed that acid cleavage of a procyanidin dimer resulted in the formation of an active intermediate (flav-3-en-3-ol) that could then condense with epicatechin to initiate the formation of higher oligomers. Several models have been proposed whereby such an intermediate could be generated under physiological conditions (Dixon et al., 2005), but these remain theoretical.
WHY ARE THERE DIFFERENCES IN PA CONTENT IN SEEDS OF DIFFERENT TT MUTANTS?
The differences in levels of epicatechin and PAs in tt mutants compromised in expression of the various structural, regulatory, and transport-related genes are quite dramatic and diverse. Although different analytical methods may account for some of the observed differences (Marles et al., 2003), genetic and biological factors seem more likely. Dissection of such differences between transport-related tt mutants and tt10 is of particular interest, since it may provide clues for understanding the complex PA transport and polymerization processes.
One important issue concerns the stability of epicatechin or PAs in seed endothelium cells. As observed in Arabidopsis, these cells accumulate epicatechin and PAs in the central vacuole, and these do not disappear until the desiccation period of late seed development (Kitamura et al., 2004); thus, epicatechin or PA oligomers should be stable inside the vacuole, and vacuolar PA oligomers can be detected in wild-type and tt10 seeds. However, the PA transport-related mutants tt12, tt19, and aha10 have an intact biosynthetic pathway but their seeds only have trace levels of PAs, which could perhaps be transported to the vacuole through other pathways or be present elsewhere in the cell. Epicatechin is present in Arabidopsis developing siliques but is hardly detectable in mature seeds (Abrahams et al., 2002). However, among the tt19, tt12, tt10, and aha10 mutants, only tt10 has dramatically increased free epicatechin levels throughout seed development (Pourcel et al., 2005), suggesting that TT10 is directly related to the metabolic fate of epicatechin via PA stability, polymerization, or subsequent catabolism.
Determining whether 3′-O-glucosylation is essential for stabilizing epicatechin, providing a substrate for TT12, or forming E3′G as an extension unit for PA polymerization is critical for furthering our understanding of PA biosynthesis. While mature Arabidopsis seeds (except tt10) do not accumulate detectable levels of free epicatechin and E3′G, young M. truncatula seeds contain high levels of both (Marinova et al., 2007b; Pang et al., 2008). M. truncatula UGT72L1 is expressed in the seed coat and shows high specificity for E3′G biosynthesis (Pang et al., 2008), but the corresponding Arabidopsis epicatechin glycosyltransferase has yet to be identified.
PERSPECTIVES
Although TT12- and MATE1-mediated sequestration of E3′G into the central vacuole may provide the building unit(s) for PA polymerization, additional mechanisms may exist for the transport of monomers for PA biosynthesis, since tt12, aha10, and tt19 all retain low levels of PAs. Furthermore, the PAs of alfalfa, grapevine, and other species consist of both epicatechin, as extension units, and catechin, as the starter unit, suggesting the possible need for additional transporters for catechin or some derivative thereof.
Among 19 tt mutants identified from Arabidopsis, tt9, tt13, and tt17 remain to be characterized. These may represent lesions in genes encoding proteins similar to TT15 or AHA10, not directly involved in PA biosynthesis but indirectly affecting PA transport or polymerization pathways. Characterization of these mutants, coupled with biochemical analysis of polymerization mechanisms informed by the nature of the intermediates that are actually transported to the vacuole, may help reveal the remaining mysteries of PA transport and polymerization. Better understanding of these processes will ultimately help us to manipulate PA production in crop species.
In summary, in spite of the significant progress that has been made in understanding PA biosynthesis and its regulation in the past 5 years, many questions still remain unanswered, among which are the following. Is TT12 only localized to the tonoplast, and do all PA monomers have to directly cross this membrane from the cytosol? If PAs are polymerized in the vacuole, how are they then transported to the extracellular space? How is TT19 involved in flavonoid transport? Is it also involved in long-distance transport of flavonoids, and, if so, does this process impact PA biosynthesis? How does AHA10 function in PA biosynthesis, and what is its subcellular localization? Does glycosylation of epicatechin only facilitate vacuolar transport, or is it integral for PA polymerization? Is TT10 involved in the initial polymerization of PAs in addition to oxidation of existing PA oligomers/polymers? What are the fine structures of insoluble PA polymers and naturally occurring colored PA polymers? How can we better quantify them, and do our current methods give a true representation of their compositions?
References
- Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin P, Ashton AR. (2003) The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J 35: 624–636 [DOI] [PubMed] [Google Scholar]
- Abrahams S, Tanner GJ, Larkin PJ, Ashton AR. (2002) Identification and biochemical characterization of mutants in the proanthocyanidin pathway in Arabidopsis. Plant Physiol 130: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T, Ikegami A, Tsujimoto T, Kobayashi S, Sato A, Kono A, Yonemori K. (2009) DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol 151: 2028–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfenito MR, Souer E, Goodman CD, Buell R, Mol J, Koes R, Walbot V. (1998) Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell 10: 1135–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter IR, Young JC, Armstrong G, Foster N, Bogenschutz N, Cordova T, Peer WA, Hazen SP, Murphy AS, Harper JF. (2005) A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc Natl Acad Sci USA 102: 2649–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J, Jaffe FW, Takos AM, Walker AR, Robinson SP. (2007) The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143: 1347–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Muday GK, Djordjevic MA. (2007) Flavonoids are differentially taken up and transported long distances in Arabidopsis. Plant Physiol 145: 478–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV. (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16: 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin LB, Wang HB, Crowell AL, Bedgar DL, Martin DM, Sarkanen S, Lewis NG. (1997) Stereoselective bimolecular coupling by an auxiliary (dirigent) protein without an active center. Science 275: 362–366 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Peeters AJ, Leon-Kloosterziel KM, Koornneef M. (2001) The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13: 853–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debolt S, Scheible WR, Schrick K, Auer M, Beisson F, Bischoff V, Bouvier-Nave P, Carroll A, Hematy K, Li Y, et al. (2009) Mutations in UDP-glucose:sterol glucosyltransferase in Arabidopsis cause transparent testa phenotype and suberization defect in seeds. Plant Physiol 151: 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L, Bogs J, Walker AR, Ferrier T, Decendit A, Merillon JM, Robinson SP, Barrieu F. (2008) The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol 147: 2041–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA. (2005) Engineering plant natural product pathways. Curr Opin Plant Biol 8: 329–336 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Xie DY, Sharma SB. (2005) Proanthocyanidins: a final frontier in flavonoid research? New Phytol 165: 9–28 [DOI] [PubMed] [Google Scholar]
- Focks N, Sagasser M, Weisshaar B, Benning C. (1999) Characterization of tt15, a novel transparent testa mutant of Arabidopsis thaliana (L.) Heynh. Planta 208: 352–357 [DOI] [PubMed] [Google Scholar]
- Grotewold E. (2004) The challenges of moving chemicals within and out of cells: insights into the transport of plant natural products. Planta 219: 906–909 [DOI] [PubMed] [Google Scholar]
- Haslam E. (1977) Symmetry and promiscuity in procyanidin biochemistry. Phytochemistry 16: 1625–1640 [Google Scholar]
- Hsieh K, Huang AH. (2007) Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell 19: 582–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquinod M, Villiers F, Kieffer-Jaquinod S, Hugouvieux V, Bruley C, Garin J, Bourguignon J. (2007) A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol Cell Proteomics 6: 394–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura S, Shikazono N, Tanaka A. (2004) TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J 37: 104–114 [DOI] [PubMed] [Google Scholar]
- Lazar G, Goodman HM. (2006) MAX1, a regulator of the flavonoid pathway, controls vegetative axillary bud outgrowth in Arabidopsis. Proc Natl Acad Sci USA 103: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M. (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57: 405–430 [DOI] [PubMed] [Google Scholar]
- Marinova K, Kleinschmidt K, Weissenbock G, Klein M. (2007a) Flavonoid biosynthesis in barley primary leaves requires the presence of the vacuole and controls the activity of vacuolar flavonoid transport. Plant Physiol 144: 432–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinova K, Pourcel L, Weder B, Schwarz M, Barron D, Routaboul JM, Debeaujon I, Klein M. (2007b) The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 19: 2023–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marles MAS, Ray H, Gruber MY. (2003) New perspectives on proanthocyanidin biochemistry and molecular regulation. Phytochemistry 64: 367–383 [DOI] [PubMed] [Google Scholar]
- Marrs KA, Alfenito MR, Lloyd AM, Walbot V. (1995) A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375: 397–400 [DOI] [PubMed] [Google Scholar]
- Mellway RD, Tran LT, Prouse MB, Campbell MM, Constabel CP. (2009) The wound-, pathogen-, and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiol 150: 924–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, Pajonk S, Micali C, O'Connell R, Schulze-Lefert P. (2009) Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J 57: 986–999 [DOI] [PubMed] [Google Scholar]
- Mueller LA, Goodman CD, Silady RA, Walbot V. (2000) AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol 123: 1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Peel GJ, Sharma SB, Tang Y, Dixon RA. (2008) A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of Medicago truncatula. Proc Natl Acad Sci USA 105: 14210–14215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Peel GJ, Wright E, Wang Z, Dixon RA. (2007) Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol 145: 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel GJ, Modolo LV, Pang Y, Dixon RA. (2009) The LAP1 MYB transcription factor orchestrates anthocyanidin biosynthesis and glycosylation in Medicago. Plant J 59: 136–149 [DOI] [PubMed] [Google Scholar]
- Pourcel L, Irani NG, Lu Y, Riedl K, Schwartz S, Grotewold E. (2010) The formation of anthocyanic vacuolar inclusions in Arabidopsis thaliana and implications for the sequestration of anthocyanin pigments. Mol Plant 3: 78–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Kerhoas L, Caboche M, Lepiniec L, Debeaujon I. (2005) TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17: 2966–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poustka F, Irani NG, Feller A, Lu Y, Pourcel L, Frame K, Grotewold E. (2007) A trafficking pathway for anthocyanins overlaps with the endoplasmic reticulum-to-vacuole protein-sorting route in Arabidopsis and contributes to the formation of vacuolar inclusions. Plant Physiol 145: 1323–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Smith AP, Nourizadeh SD, Peer WA, Xu J, Bandyopadhyay A, Murphy AS, Goldsbrough PB. (2003) Arabidopsis AtGSTF2 is regulated by ethylene and auxin, and encodes a glutathione S-transferase that interacts with flavonoids. Plant J 36: 433–442 [DOI] [PubMed] [Google Scholar]
- Terrier N, Torregrosa L, Ageorges A, Vialet S, Verries C, Cheynier V, Romieu C. (2009) Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol 149: 1028–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij W, Spelt C, Di Sansebastiano GP, Vermeer J, Reale L, Ferranti F, Koes R, Quattrocchio F. (2008) An H+ P-ATPase on the tonoplast determines vacuolar pH and flower colour. Nat Cell Biol 10: 1456–1462 [DOI] [PubMed] [Google Scholar]
- Wenzel CL, Hester Q, Mattsson J. (2008) Identification of genes expressed in vascular tissues using NPA-induced vascular overgrowth in Arabidopsis. Plant Cell Physiol 49: 457–468 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA. (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299: 396–399 [DOI] [PubMed] [Google Scholar]
- Zhao J, Dixon RA. (2009) MATE transporters facilitate vacuolar uptake of epicatechin 3′-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 21: 2323–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]