Cellulose is intimately associated with multiple facets of human civilization: central to clothing, shelter, heat, medicine, and food, there are few moments in the average human life that are not spent in direct contact with cellulose or a by-product of its composition. It should come as no surprise then that a considerable amount of time and energy have been spent, and a couple of Nobel Prizes awarded, in research involved with the analysis of cellulose structure and metabolism from various sources. As the understanding of this important biomacromolecule has developed, numerous analytical techniques have been put to use to decipher cellulose biosynthesis, structure, and function within the plant cell wall. The content of this Update is designed to introduce the reader to current and developing tools for cellulose characterization. To begin, “a brief history” on the analysis of cellulose describes how many of the modern analytical techniques used to determine cellulose structure came into use. This leads into the various imaging techniques that interrogate cellulose biosynthesis, especially those that have arisen since the identification of the CELLULOSE SYNTHASE A (CESA) genes. We then turn our attention to recent in vitro biochemical studies of CESA, and in this context we discuss the relationship between CESA and detergent-resistant fractions of the plasma membrane (PM), which have the opportunity to shine new light on the PM-cell wall continuum.
CHARACTERIZATION OF CELLULOSE USING INTEGRATED ANALYTICAL TOOLS: A BRIEF HISTORY
Cellulose was named by the French Academy over 171 years ago (Brongniart et al., 1839), subsequent to its characterization in various plant tissues by the famous French plant scientist Anselme Payen (Payen, 1838). His use of different treatments based on sodium hydroxide, potassium hydroxide, or nitric acid to extract and partially digest cellulose from oak and beech wood revealed an element composition comparable to that of starch (Payen, 1838). Classical organic chemistry then allowed for the determination of the β-(1→4) linkage that separates Glc residues in the cellobiose unit (for review, see Hon, 1994). The remarkable nature of cellulose as a polymer of repeating Glc (cellobiose) units (Staudinger, 1926; Haworth, 1932) contributed greatly to the 1937 and 1953 Nobel Prizes in Chemistry. Today, it is understood that cellulose fibrils from many natural sources result from individual glucan chains of cellulose aggregating via hydrogen bonds and Van der Waals forces to form a long thread-like paracrystalline structure termed the microfibril. The route that led to a sophisticated model of cellulose structure began with x-ray diffraction (XRD) studies. The first XRD patterns of cellulose fibers were generated from wood, hemp, and bamboo samples, and although detailed structural data were not initially obtained, it was determined that the crystallites were of a rod-like shape (Nishikawa and Ono, 1913). However, cellulose in a majority of higher plants forms crystalline domains that are not large enough to produce high-resolution crystallographic structure determination (Lai Kee Him et al., 2002; Müller et al., 2002). Therefore, many of the early molecular models developed for the monoclinic unit cell (Meyer and Misch, 1937) and triclinic unit cell (Honjo and Watanabe, 1958) of cellulose were based on algal or tunicate (animal) model systems (Fischer and Mann, 1960). In addition, modern XRD data have been collected at even higher resolution than before using a synchrotron light source and can be paired with the separate analytical technique of neutron diffraction, which in combination with specific deuteration has greatly increased the power to locate hydrogen atoms involved in the intermolecular or intramolecular hydrogen bonding of the cellulose microfibril (Nishiyama et al., 2002, 2003).
Simultaneous to the development of XRD methods, Rowen et al. (1947) analyzed cellulose from cotton (Gossypium hirsutum) by infrared (IR) absorption spectroscopy, and later, Marrinan and Mann (1956) recognized that algae (Valonia) and bacterial cellulose yielded IR spectra that were different from those of tunicates, cotton, or ramie (Boehmeria nivea). Eventually, in the early 1980s, the spectroscopic technique of solid-state 13C cross-polarization magic-angle spinning NMR spectroscopy was able to resolve this issue by showing that native cellulose I diffraction data from many natural sources were a composite of diffraction from the two crystalline allomorphs Iα (triclinic unit cell) and Iβ (monoclinic unit cell; Atalla and VanderHart, 1984). 13C-NMR spectroscopy would not only confirm that the crystalline structure of the cellulose microfibril in most plants was a composite of Iα and Iβ crystalline forms (Viëtor et al., 2002) but, in combination with Fourier transform IR spectroscopy spectra, suggested that the more crystalline inner chains of the microfibril core are composed primarily of cellulose Iβ, while both forms of cellulose compose the chains in the surrounding paracrystalline sheath (Sturcová et al., 2004).
Collectively, these early crystallographic and spectroscopic studies laid the foundation for elucidation of the native cellulose structure in plant cell walls. Simultaneously, microscopic analyses showed that cellulose microfibrils were dimensionally different in different cell types of the plant (Roelofsen and Houwink, 1953; Balashov and Preston, 1955). Malcolm Brown and coworkers revealed, first in the green alga Oocystis (Brown and Montezinos, 1976) and then in higher plants (Mueller and Brown, 1980), that the ends of nascent cellulose microfibrils were often associated with globular structures designated as terminal complexes embedded in the PM. The freeze fracture of the PM was imaged on the P-fracture face by transmission electron microscopy (TEM), showing a structure with a 6-fold symmetry (rosette), and remains to this day a fundamental piece of evidence suggesting that this type of structure is involved in cellulose synthesis in plants (Mueller and Brown, 1980). Furthermore, in the study by Kimura and coworkers (1999), TEM was used in combination with an immunoaffinity probe to show that the catalytic subunit of cellulose synthase is associated with the rosette complex in vascular plants. Despite the disadvantages of extensive and often destructive sample preparation and the inability to study live specimens, TEM is unsurpassed in its resolution and has been the method of choice for ultrastructural analysis of the cell wall. Newer techniques in TEM sample preparation are being developed to reduce sample destruction along with the use of promising new affinity tools to be used in conjunction with TEM, such as carbohydrate-binding modules, that will most likely provide enhanced molecular resolution imaging of the cellulose microfibril network (for review, see Sarkar et al., 2009).
Integrated with the above-mentioned analytical tools of IR, NMR, XRD, and TEM, the relatively new imaging technique of atomic force microscopy (AFM) has the capacity to provide atom-level resolution of the cellulosic matrix in the cell wall of fresh tissue. Therefore, beyond the compositional structure of cellulose, AFM can offer a spatial view of cellulose microfibril orientation in the polylaminate cell wall. AFM is based on scanning probe microscopy (Binnig and Rohrer, 1986) and uses a physical tip to scan the surface of a specimen to determine its topography, physical properties, and chemical structure (Drake et al., 1989). AFM operates by driving a cantilever with a sharp tip mounted at its end to allow faster scanning across the specimen surface. AFM images are the result of convolutions of the tip and the “true” structure of the specimen at an atomic resolution. Plant cell walls were one of the first biological samples that were examined by AFM (Kirby et al., 1996; van der Wel et al., 1996). The motivation of using AFM for characterizing cell walls is obvious: plant cell walls are relatively stiff and flat, and the molecular features of the microfibril network occur at the nanometer scale. Ideally, AFM could be used to answer some of the key questions regarding the nanostructures within plant cell wall cellulose (Kirby et al., 1996; van der Wel et al., 1996; Engel et al., 1999; Morris et al., 1999; Thimm et al., 2000; Davies and Harris, 2003) beyond that defined by the previously mentioned analytical techniques.
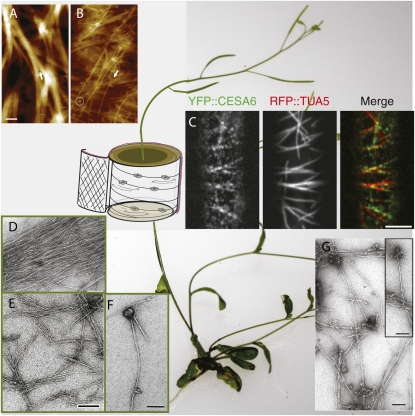
To take full advantage of AFM and reduce interference by artifacts, an approach with minimized sample preparation is ideal. Greatest success has used hand-cut sections of fresh or naturally aged dry tissue while operating the AFM device in tapping mode and imaging the inner surface of the native cell wall (Ding and Himmel, 2006; Himmel et al., 2007). Using this strategy, it was possible to image primary and secondary cell walls from maize (Zea mays). AFM imaging of dry primary cell walls documented microfibril dimensions precisely measured at 3 to 5 nm (Ding and Himmel, 2006), consistent with the 36-glucan chain model of cellulose elementary fibril (CEF) biosynthesis. In addition, cellulose macrofibrils, consisting of a bundle of CEFs that split at the end to form smaller bundles and eventually single CEFs, were also observed by AFM (Ding and Himmel, 2006). Each microfibril observed in mature primary cell walls contained only a single CEF with noncellulosic polymers associated with its surface (Ding and Himmel, 2006; Himmel et al., 2007). AFM images of maize cell walls from fresh cells further confirmed these observations (Fig. 1, A and B). AFM images of 3-d-old developing maize coleoptiles (Fig. 1A) showed macrofibrils of 50 to 100 nm in diameter with rather clean surface features, which contained multiple CEFs. By contrast, 4-week old maize stem piths containing mature parenchyma (Fig. 1B) displayed two distinguishable layers. In the upper layer, the fibrils appeared to be small macrofibrils with diameters of 7 to 10 nm and single CEFs with diameters of 3 to 5 nm. In the posterior layer, all microfibrils of 3 to 5 nm diameter were arranged in a parallel orientation (Fig. 1B). Furthermore, in the mature cell walls (Fig. 1B), particle-like features decorated the macrofibrils that may have been formed by hemicelluloses, proteins, or other cell wall polymers (Ding and Himmel, 2006) and were not observed in the macrofibrils of elongating cells (Fig. 1A). A plausible explanation for these differences among cell types is a delay between cellulose biosynthesis and the incorporation of other cell wall polymers into the cell wall. Ultimately, AFM imaging representing the native structure of cellulose provides an enormous opportunity to better understand the molecular architecture in dermal cell layers, particularly when combined with the confocal live cell imaging described below.
Figure 1.
Emerging tools for cellulose analysis in plant cell walls. The background is set with the model plant Arabidopsis; a simple schematic is overlaid on the stem to indicate various facets of cell wall cellulose synthesis. A, AFM height image of fibril structures in the elongating primary cell wall of maize contained primarily macrofibrils that split at the end (indicated by the arrow). B, The mature primary cell wall contained both small macrofibrils in the upper-most layer that can also split at the end (indicated by the arrow) and parallel-arranged single elementary fibrils. Particle-like features were clearly seen and were typically associated with the macrofibrils (indicated by the circle). Bar in A = 100 nm. C, Confocal image of upper hypocotyl cells of transgenic plants expressing YFP::CESA6, red fluorescent protein (RFP)::TUA5 (tubulin), and the overlay of the two compatible fluorophors in a merged imaged reveals the coincidence of CESA with microtubules. Bar = 5 μm. D to F, Electron micrographs of cellulose microfibrils extracted from primary walls of blackberry cells (D and E) and of cellulose synthesized in vitro (F) using detergent extracts as a source of enzyme. The cellulose from cell walls was observed before (D) and after (E) treatment with the Updegraff reagent (Lai Kee Him et al., 2002). A cellulose microfibril synthesized in vitro associated to a globular particle, which most likely corresponds to the enzyme complex (not treated with the Updegraff reagent; F). G, Electron micrograph of cellulose microfibrils synthesized in vitro by DRMs isolated from hybrid aspen cells grown as suspension cultures (negative staining with 2% uranyl acetate). The inset shows the magnification of putative DRM structures that carry the active cellulose synthase machinery and from which cellulose microfibrils are being formed. Bars in E to G = 0.1 μm.
As an analytical tool, confocal microscopy has rapidly evolved from being a challenging technique with limited accessibility to a high-throughput tool providing quantitatively precise localization data for 30% of the proteome in the model plant Arabidopsis (Arabidopsis thaliana; Chalfie et al., 1994; Cutler et al., 2000; Tian et al., 2004; Heazlewood et al., 2007). The capacity to visualize a protein relies on excitation of an autofluorescent protein (AFP), such as the GFP derived from the jellyfish Aqueora victoriae, fused to a plant protein of interest. Despite the most famous AFP being the GFP characterized 16 years ago (Chalfie et al., 1994), there are now numerous forms of AFP, including blue, cyan, and yellow (YFP) fluorescent proteins (Goodin et al., 2007). Designing an AFP expression fusion can still be a daunting task, given the numerous expression vectors available, the choice of where to fuse the AFP, what promoter to use, and whether to use stable or transient expression of the chimeric protein fusion (for review, see Goodin et al., 2007). Live cell imaging of CESA has led to a remarkable increase in our understanding of the enzyme's subcellular localization, regulation, trafficking, and guidance in growing cells during primary cell wall synthesis (Paredez et al., 2006; DeBolt et al., 2007a, 2007b; Desprez et al., 2007; Persson et al., 2007; Crowell et al., 2009; Gutierrez et al., 2009) and in vascular tissue during secondary cell wall synthesis (Wightman and Turner, 2008). Labeling of the primary wall complex with YFP::CESA6 or GFP::CESA3 in Arabidopsis hypocotyl cells revealed discrete particles at the focal plane of the PM. The observation that these particles moved along linear trajectories at constant velocity (average of 270 nm min−1; Paredez et al., 2006) suggests that they represent actively synthesizing complexes. The majority of labeled CESA, however, resides in the cytoplasm, located in Golgi stacks (confirmed by colocalization with labeled β-mannosidase, a Golgi-resident protein) and in a heterogenous population of smaller compartments (Paredez et al., 2006; Crowell et al., 2009; Gutierrez et al., 2009).
In vivo visualization of cellulose synthase was initially used to investigate the dynamic interaction between CESA complexes and the underlying cortical microtubule array (Paredez et al., 2006), first described nearly 40 years ago by Hepler and Newcomb (1964). It was long believed that the microtubule cytoskeleton guided CESA complexes and thus cellulose deposition; however, the mechanism for this guidance was not known. Time-lapse confocal microscopy showed that CESA trajectories were coincident with labeled microtubules, indicating that microtubules act as molecular rails rather than as passive barriers to CESA movement (Fig. 1C; Paredez et al., 2006). More recently, the microtubule array was found to participate in the delivery of CESA complexes to the PM (Crowell et al., 2009; Gutierrez et al., 2009). Analysis of individual delivery events indicated that complexes are preferentially delivered to sites that are coincident with cortical microtubules (Gutierrez et al., 2009). Interestingly, small CESA-containing compartments have been shown to associate with the cortical microtubule array (Crowell et al., 2009; Gutierrez et al., 2009). Taken together, these observations suggest that microtubules may position CESA delivery to the PM by interacting with secretory compartments in the cytoplasm.
Recent work has also revealed that CESA motility is coincident with microtubule arrays during secondary cell wall synthesis (Wightman and Turner, 2008). Importantly, these authors also deduced that actin filaments are instrumental in rapid intracellular trafficking of CESA, which is necessary for cell wall thickening (Wightman and Turner, 2008). The actin cytoskeleton is also required for proper CESA trafficking during primary cell wall synthesis: application of the actin-depolymerizing drug latrunculin B disrupts the global distribution of CESA at the PM (Gutierrez et al., 2009). Further details have been gleaned from careful observation of time-lapse imaging; for instance, it was observed that after arriving at the PM focal plane, the YFP::CESA6 began to move almost immediately (within 1 min) after arrival (Paredez et al., 2006). From this, a tentative deduction would be that the CESA complex is activated and begins making cellulose very soon after arrival at the PM. Particles also are observed to disappear from the PM, suggesting a further level of regulation that terminates CESA activity. However, it has not been possible to track a PM-localized CESA particle from the initiation of its lifespan to termination. One reason for this is that the inevitable curvature of the epidermal cell being imaged makes it difficult to conclude that a particle has not simply moved out of the focal plane rather than disappeared from the PM. Indeed, many questions remain unanswered for the budding microscopist and will require future improvements in confocal resolution.
Small effector molecules have been invaluable in dissecting aspects of cellulose biosynthesis by live cell imaging. For instance, the inhibitor of microtubule polymerization, oryzalin, was useful in demonstrating that the CESA insertion and directional motility were independent of microtubules (Paredez et al., 2006; DeBolt et al., 2007b). As mentioned above, inhibiting actin polymerization using latrunculin B showed the requirement for actin in CESA trafficking (Wightman and Turner, 2008; Gutierrez et al., 2009). Assessing the localization behavior of CESA after treatment with inhibitors of cellulose biosynthesis such as isoxaben (N-[3(1-ethyl-1-methylpropyl)-5-isoxazolyl]; Heim et al., 1989), 2,6,-dichlorobenzonitrile (DCB; Montezinos and Delmer, 1980), 1-cyclohexyl-5-(2,3,4,5,6-pentafluorophenoxyl)-1λ4,2,4,6-thiatriazin-3-amine (CGA; Peng et al., 2001), and thaxtomin (Loria et al., 2006) has allowed for the study of the mechanisms of action for these drugs. For instance, CGA, thaxtomin, and isoxaben cause clearance of CESA from the PM; therefore, either secretion of CESA is compromised or CESA is unable to assemble once at the PM (Paredez et al., 2006; Bischoff et al., 2009; Crowell et al., 2009). By contrast, DCB does not stop complexes from forming at the PM, but once there, CESA movement ceases and CESA hyperaccumulates (DeBolt et al., 2007b). Interestingly, DCB has been shown recently to bind to a microtubule-associated protein in hybrid aspen (Populus species), further supporting an action of the drug on the movement of CESA (Rajangam et al., 2008). New inhibitors of cellulose biosynthesis have been screened and identified using live cell imaging, a good example being morlin (DeBolt et al., 2007a), which inhibits both microtubule dynamicity and CESA, further demonstrating the intimate association between these processes. Obtaining the protein targets of all these small molecule inhibitors presents an enormous opportunity to define new players or interactions in cell wall cellulose biosynthesis.
BIOCHEMICAL ANALYSES OF CELLULOSE BIOSYNTHESIS
Despite all the previous evidence presented, the biochemical analysis of the CESA rosettes has been a major challenge in the field of plant cell wall biosynthesis. The enzyme complex is highly unstable, and this has limited the possibility of purifying it and studying its biochemical properties in vitro. Detergent extractions of proteins from plant PMs typically lead to the loss of the β-(1→4)-glucosyltransferase activity of cellulose synthase (Delmer, 1999; Bessueille and Bulone, 2008). In addition, further complication is due to the unavoidable concomitant extraction of callose synthase from the PM. This enzyme uses the same substrate as cellulose synthase (UDP-Glc) and exhibits high activity in vitro (Okuda et al., 1993; Lai Kee Him et al., 2001, 2002; Colombani et al., 2004), even though callose is normally a minor cell wall component that forms transiently at the cell plate prior to cell division or in specialized cells (e.g. pollen tubes) or cell structures (e.g. plasmodesmata; Stone and Clarke, 1992). Thus, the formation of β-(1→3) glucan by callose synthase is typically prevalent in in vitro reactions, and this has further complicated the detection of the highly unstable and low cellulose synthase activity. In addition, due to nonrigorous polysaccharide characterization, the incorporation of Glc from UDP-Glc into ethanol-insoluble β-(1→3) glucans has sometimes been wrongly associated with cellulose biosynthesis (for review, see Colombani et al., 2004; Bulone, 2007). It has been proposed that callose and cellulose formation might be catalyzed by the same enzyme (Delmer, 1999), but the isolation of different genes encoding the presumed catalytic subunits of callose and cellulose synthases is contradictory with this hypothesis (for review, see Bessueille and Bulone, 2008). However, one may still argue that experimental evidence demonstrating that the products of the CESA genes cannot catalyze the formation of callose, and vice versa, is still missing. The definitive answer to this question will be obtained when an active cellulose synthase preparation devoid of callose synthase activity becomes available.
The first successful in vitro synthesis of cellulose from plant cell-free extracts was achieved using cotton fiber and mung bean (Vigna radiata) enzymes (Kudlicka et al., 1995, 1996; Kudlicka and Brown, 1997). However, it was only several years later that the amount of cellulose synthesized in vitro was significantly improved by the careful selection of detergents that allow the extraction of enzyme complexes in an intact form (Lai Kee Him et al., 2002). The use of taurocholate and Brij 58 to extract cellulose synthase from PMs of cell suspension cultures of blackberry (Rubus fruticosus) allowed the synthesis of cellulose in milligram amounts. This made possible the complete and unequivocal characterization of the cellulose synthesized in vitro (Lai Kee Him et al., 2002). Interestingly, the microfibrillar cellulose formed in the in vitro reactions was 10% to 15% more crystalline than the cellulose extracted from the primary walls of the same cells, as measured by XRD analysis (Lai Kee Him et al., 2002). This was further supported by the fact that cellulose from primary walls was sensitive to the Updegraff reagent (Updegraff, 1969; Fig. 1, D and E) while the in vitro-synthesized microfibrils were not (Lai Kee Him et al., 2002). When the samples were not treated with this reagent, the individual microfibrils synthesized in vitro were associated with globular structures that most likely correspond to the enzyme complex responsible for their formation (Fig. 1F). Since this significant progress, the method has been optimized in other plant systems, such as hybrid aspen (Colombani et al., 2004) and tobacco (Nicotiana tabacum) BY2 cells (Cifuentes et al., 2010), for which digitonin was selected as the best detergent. It seems that the type of detergent used for enzyme extraction needs to be determined for different plant species, which perhaps reflects different lipid environments. As for the blackberry cellulose synthase (Lai Kee Him et al., 2002), higher levels of activity were obtained with the detergent-extracted cellulose synthase from hybrid aspen when the cells were harvested at their stationary phase, with up to 50% cellulose and 50% callose synthesized in vitro (Colombani et al., 2004). To date, this represents the highest proportion of cellulose to callose reported in in vitro synthesis experiments.
Despite these important advances, however, it remains that the in vitro assays need to be systematically combined with careful analyses of the in vitro product to determine the extent of β-(1→4) linkages formed. This can be performed routinely using highly specific cellulases that are not contaminated by β-(1→3)-glucanase activities, which is not always the case for commercial enzymes. Typically, the biochemical evidence of the de novo synthesis of cellulose is provided by the sensitivity of the polymer synthesized in the presence of radioactive UDP-Glc to the specific cellulases. If needed, more complete characterization can be performed, for instance by gas chromatography-mass spectrometry analysis after derivatization of the glucans (methylation analysis), electron and/or XRD analysis, TEM, and NMR (for review, see Bulone, 2007). However, for XRD and NMR analyses, the scale-up of the in vitro reactions is required to obtain enough polymer for the characterization. Solid-state NMR is particularly demanding in terms of the amount of polysaccharide (typically at least 10 mg), but a sensitive method based on the use of UDP-Glc in which the Glc is enriched in 13C has been developed and allows the analysis of in vitro glucans with as little as 100 μg of polysaccharide (Fairweather et al., 2004). The analysis of the cellulose synthesized in vitro can be further facilitated by dissolving the β-(1→3)-glucan that is cosynthesized with cellulose in NaOH solutions. Indeed, crystalline cellulose such as that synthesized in vitro by the detergent-extracted enzyme from blackberry is not soluble in NaOH (Lai Kee Him et al., 2002). However, it is important to keep in mind that some poorly crystalline β-(1→4)-glucan may be synthesized and lost by dissolution in NaOH. In summary, the tools to assay cellulose synthase in vitro and unequivocally characterize the cellulose formed are currently available. This opens a great opportunity to perform detailed biochemical analysis of the cellulose synthase complex when a purified preparation can be obtained. To date, this has been extremely challenging, but important progress has been made recently in this area with the purification of complexes from Arabidopsis using dual-epitope tagging and the specific corresponding purification steps (Atanassov et al., 2009). However, it was not possible in this work to search for enzymatic activity, because the purification procedure was not efficient enough. The enzymatic assays and sensitive tools mentioned above for the characterization of cellulose synthesized in vitro will be most useful also for the biochemical analysis of recombinant individual catalytic subunits of cellulose synthase, which may be possible in the near future with the development of more efficient expression systems for membrane-bound proteins.
Owing to the availability of these tools and of specific anti-cellulose synthase antibodies, it was recently demonstrated that callose and cellulose synthases are located in detergent-resistant structures exhibiting similar biochemical properties as lipid rafts in animal cells (Bessueille et al., 2009). The preparations were active in vitro and able to synthesize microfibrillar glucans that were identified as callose and cellulose (Bessueille et al., 2009). The glucan sample identified as cellulose, shown in Figure 1G, was not treated with any procedure prior to observation. This allowed for the preservation of the detergent-resistant membrane microdomains (DRMs), which are visible as globular structures or aggregates of globular structures (Fig. 1G). It was not determined, though, whether both callose and cellulose synthases are located in the same or different subpopulations of DRMs (Bessueille et al., 2009). The relationship between DRMs and lipid rafts is debated (Lichtenberg et al., 2005; Hancock, 2006), essentially because the experimental conditions used for DRM isolation may artificially induce the formation of such structures (Lichtenberg et al., 2005). Nonetheless, it remains that the extractions of DRMs reflect differential affinities of specific sets of membrane proteins to various lipid environments (Lichtenberg et al., 2005). Thus, the isolation of DRMs is a valuable tool for understanding the interactions of callose and cellulose synthases with the lipids that cosegregate with them and that consist of a higher relative proportion of sterols and sphingolipids than the total PMs (Bessueille et al., 2009). In addition, DRMs represent a form of isolated carbohydrate synthases that can be further fractionated using detergents or other compounds, such as chaotropic agents that disrupt interactions between specific lipids and proteins. This approach, combined with the enzymatic assays available and the detailed proteomics analysis of the subfractions recovered after treatment with chaotropic agents, represents a promising strategy to identify proteins that interact directly with the enzyme complexes.
Acknowledgments
We thank Ryan Gutierrez for helpful comments. Due to space requirements, it was simply not possible to cite all of the papers that have contributed to forward progress, and we apologize for any missed.
References
- Atalla RH, VanderHart DL. (1984) Native cellulose: a composite of two distinct crystalline forms. Science 223: 283–285 [DOI] [PubMed] [Google Scholar]
- Atanassov II, Pittman JK, Turner SR. (2009) Elucidating the mechanisms of assembly and subunit interaction of the cellulose synthase complex of Arabidopsis secondary cell walls. J Biol Chem 284: 3833–3841 [DOI] [PubMed] [Google Scholar]
- Balashov V, Preston RD. (1955) Fine structure of cellulose and other microfibrillar substances. Nature 176: 64–65 [Google Scholar]
- Bessueille L, Bulone V. (2008) A survey of cellulose biosynthesis in higher plants. Plant Biotechnol 25: 315–322 [Google Scholar]
- Bessueille L, Sindt N, Guichardant M, Djerbi S, Teeri TT, Bulone V. (2009) Plasma membrane microdomains from hybrid aspen cells are involved in cell wall polysaccharide biosynthesis. Biochem J 420: 93–103 [DOI] [PubMed] [Google Scholar]
- Binnig G, Rohrer H. (1986) Scanning tunneling microscopy. IBM J Res Develop 30: 355–369 [Google Scholar]
- Bischoff V, Cookson SJ, Wu S, Scheible WR. (2009) Thaxtomin A affects CESA-complex density, expression of cell wall genes, cell wall composition, and causes ectopic lignification in Arabidopsis thaliana seedlings. J Exp Bot 60: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brongniart A, Pelouze TJ, Dumas AB. (1839) Rapport sur un mémoire de M. Payen, relatif à la composition de la matière ligneuse. C R Hebd Seances Acad Sci 8: 51–53 [Google Scholar]
- Brown RM, Jr, Montezinos D. (1976) Cellulose microfibrils: visualization of biosynthetic and orienting complexes in association with the plasma membrane. Proc Natl Acad Sci USA 73: 143–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulone V. (2007) Analysis of (1-3)-β-d-glucans and cellulose synthesized in vitro: a key step towards the characterization of glucan synthases. Brown RM, Jr, Saxena IM, , Cellulose: Molecular and Structural Biology. Springer, London, pp 123–145 [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward W, Prasher D. (1994) Green fluorescent protein as a marker for gene expression. Science 263: 802–805 [DOI] [PubMed] [Google Scholar]
- Cifuentes C, Bulone V, Emons AMC. (2010) Biosynthesis of callose and cellulose by detergent extracts of tobacco cell membranes and quantification of the polymers synthesized in vitro. J Integr Plant Biol 52: 221–233 [DOI] [PubMed] [Google Scholar]
- Colombani A, Djerbi S, Bessueille L, Blomqvist K, Ohlsson A, Berglund T, Teeri TT, Bulone V. (2004) In vitro synthesis of (1→3)-β-d-glucan (callose) and cellulose by detergent extracts of membranes from cell suspension cultures of hybrid aspen. Cellulose 11: 313–327 [Google Scholar]
- Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Hofte H, Vernhettes S. (2009) Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville C. (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies LM, Harris PJ. (2003) Atomic microscopy of microfibrils in primary cell walls. Planta 217: 283–289 [DOI] [PubMed] [Google Scholar]
- DeBolt S, Gutierrez R, Ehrhardt DW, Melo CV, Ross L, Cutler SR, Somerville C, Bonetta D. (2007a) Morlin, an inhibitor of cortical microtubule dynamics and cellulose synthase movement. Proc Natl Acad Sci USA 104: 5854–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBolt S, Gutierrez R, Ehrhardt DW, Somerville C. (2007b) Nonmotile cellulose synthase subunits repeatedly accumulate within localized regions at the plasma membrane in Arabidopsis hypocotyl cells following 2,6-dichlorobenzonitrile treatment. Plant Physiol 145: 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer DP. (1999) Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol 50: 245–276 [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Hofte H, Gonneau M, Vernhettes S. (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SY, Himmel ME. (2006) The maize primary cell wall microfibril: a new model derived from direct visualization. J Agric Food Chem 54: 597–606 [DOI] [PubMed] [Google Scholar]
- Drake B, Prater C, Weisenhorn A, Gould S, Albrecht T, Quate C, Cannell D, Hansma H, Hansma P. (1989) Imaging crystals, polymers, and processes in water with the atomic force microscope. Science 243: 1586–1589 [DOI] [PubMed] [Google Scholar]
- Engel A, Lyubchenko Y, Müller D. (1999) Atomic force microscopy: a powerful tool to observe biomolecules at work. Trends Cell Biol 9: 77–80 [DOI] [PubMed] [Google Scholar]
- Fairweather JK, Lai Kee Him J, Heux L, Driguez H, Bulone V. (2004) Structural characterization by 13C-NMR spectroscopy of products synthesized in vitro by polysaccharide synthases using 13C-enriched glycosyl donors: application to a UDP-glucose:(1→3)-β-d-glucan synthase from blackberry (Rubus fruticosus). Glycobiology 14: 775–781 [DOI] [PubMed] [Google Scholar]
- Fischer DG, Mann J. (1960) Crystalline modifications of cellulose. Part VI. Unit cell and molecular symmetry of cellulose I. J Polym Sci 62: 189–194 [Google Scholar]
- Goodin M, Chakrabarty R, Banerjee R, Yelton S, DeBolt S. (2007) New gateways to discovery. Plant Physiol 145: 1100–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AMC, Ehrhardt DW. (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol 11: 797–806 [DOI] [PubMed] [Google Scholar]
- Hancock JF. (2006) Lipid rafts: contentious only from simplistic standpoints. Nat Rev Mol Cell Biol 7: 456–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haworth WN. (1932) Molecular structure of cellulose and of amylose. Nature 129: 365 [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. (2007) SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res 35: D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim DR, Roberts JL, Pike PD, Larrinua IM. (1989) Mutation of a locus of Arabidopsis thaliana confers resistance to the herbicide isoxaben. Plant Physiol 90: 146–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler PK, Newcomb EH. (1964) Microtubules and fibrils in the cytoplasm of coleus cells undergoing secondary wall deposition. J Cell Biol 20: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315: 804–807 [DOI] [PubMed] [Google Scholar]
- Hon DNS. (1994) Cellulose: a random walk along its historical path. Cellulose 1: 1–25 [Google Scholar]
- Honjo G, Watanabe M. (1958) Examination of cellulose fiber by the low-temperature specimen method of electron diffraction and electron microscopy. Nature 181: 326–328 [Google Scholar]
- Kimura S, Laosinchai W, Itoh T, Cui X, Linder CR, Brown RM., Jr (1999) Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell 11: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby AR, Gunning AP, Waldron KW, Morris VJ, Ng A. (1996) Visualization of plant cell walls by atomic force microscopy. Biophys J 70: 1138–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlicka K, Brown RM., Jr (1997) Cellulose and callose biosynthesis in higher plants. I. Solubilization and separation of (1→3)- and (1→4)-β-glucan synthase activities from mung bean. Plant Physiol 115: 643–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlicka K, Brown RM, Jr, Li LK, Lee JH, Shin H, Kuga S. (1995) β-Glucan synthesis in the cotton fiber. IV. In vitro assembly of the cellulose-I allomorph. Plant Physiol 107: 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlicka K, Lee JH, Brown RM., Jr (1996) A comparative analysis of in vitro cellulose synthesis from cell-free extracts of mung bean (Vigna radiata, Fabaceae) and cotton (Gossypium hirsutum, Malvaceae). Am J Bot 83: 274–284 [Google Scholar]
- Lai Kee Him J, Chanzy H, Müller M, Putaux JL, Imai T, Bulone V. (2002) In vitro versus in vivo cellulose microfibrils from plant primary wall synthases: structural differences. J Biol Chem 277: 36931–36939 [DOI] [PubMed] [Google Scholar]
- Lai Kee Him J, Pelosi L, Chanzy H, Putaux JL, Bulone V. (2001) Biosynthesis of (1→3)-β-glucan (callose) by detergent extracts of a microsomal fraction from Arabidopsis thaliana. Eur J Biochem 268: 4628–4638 [DOI] [PubMed] [Google Scholar]
- Lichtenberg D, Goñi FM, Heerklotz H. (2005) Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem Sci 30: 430–436 [DOI] [PubMed] [Google Scholar]
- Loria R, Kers J, Joshi M. (2006) Evolution of plant pathogenicity in Streptomyces. Annu Rev Phytopathol 44: 469–487 [DOI] [PubMed] [Google Scholar]
- Marrinan HJ, Mann J. (1956) Infrared spectra of the crystalline modifications of cellulose. J Polym Sci 21: 301–311 [Google Scholar]
- Meyer KH, Misch L. (1937) Position des atomes dans le nouveau modele spatial de la cellulose: sur la constitution de la partie cristallisee de la cellulose VI. Helv Chim Acta 20: 232–244 [Google Scholar]
- Montezinos D, Delmer DP. (1980) Characterization of inhibitors of cellulose synthesis in cotton fibers. Planta 148: 305–311 [DOI] [PubMed] [Google Scholar]
- Morris VJ, Kirby AR, Gunning AP. (1999) Atomic Force Microscopy for Biologists. Imperial College Press, London, p 352 [Google Scholar]
- Mueller SC, Brown RM., Jr (1980) Evidence for an intramembranous component associated with a cellulose microfibril synthesizing complex in higher plants. J Cell Biol 84: 315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Hori R, Itoh T, Sugiyama J. (2002) X-ray microbeam and electron diffraction experiments on developing xylem cell walls. Biomacromolecules 3: 182–186 [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Ono S. (1913) Transmission of X-rays through fibrous, lamellar and granular substances. Proceedings of the Tokyo Mathematico-Physical Society 7: 131–138 [Google Scholar]
- Nishiyama Y, Langan P, Chanzy H. (2002) Crystal structure and hydrogen-bonding system in cellulose Iβ from synchrotron X-ray and neutron fiber diffraction. J Am Chem Soc 124: 9074–9082 [DOI] [PubMed] [Google Scholar]
- Nishiyama Y, Sugiyama J, Chanzy H, Langan P. (2003) Crystal structure and hydrogen bonding system in cellulose Iα from synchrotron X-ray and neutron fibre diffraction. J Am Chem Soc 125: 14300–14306 [DOI] [PubMed] [Google Scholar]
- Okuda K, Li L, Kudlicka K, Kuga S, Brown MR., Jr (1993) β-Glucan synthesis in the cotton fiber. I. Identification of β-1,4- and β-1,3-glucans synthesized in vitro. Plant Physiol 101: 1131–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW. (2006) Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Payen A. (1838) Mémoire sur la composition du tissu propre des plantes et du ligneux. C R Hebd Seances Acad Sci 7: 1052–1056 [Google Scholar]
- Peng L, Xiang F, Roberts E, Kawagoe Y, Greve LC, Kreuz K, Delmer DP. (2001) The experimental herbicide CGA 325'615 inhibits synthesis of crystalline cellulose and causes accumulation of non-crystalline β-1,4-glucan associated with CesA protein. Plant Physiol 126: 981–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR. (2007) Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci USA 104: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajangam AS, Kumar M, Aspeborg H, Guerriero G, Arvestad L, Pansri P, Brown CJL, Hober S, Blomqvist K, Divne C, et al. (2008) MAP20, a microtubule-associated protein in the secondary cell walls of hybrid aspen, is a target of the cellulose synthesis inhibitor 2,6-dichlorobenzonitrile. Plant Physiol 148: 1283–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofsen PA, Houwink AL. (1953) Architecture and growth of the primary wall in some plant hairs and in the Phycomyces sporangiophore. Acta Bot Neerl 2: 218–225 [Google Scholar]
- Rowen JW, Hunt CM, Plyler EK. (1947) Absorption spectra in the detection of chemical changes in cellulose and cellulose derivatives. Text Res J 17: 504–511 [Google Scholar]
- Sarkar P, Bosneaga E, Auer M. (2009) Plant cell walls throughout evolution: towards a molecular understanding of their design principles. J Exp Bot 60: 3615–3635 [DOI] [PubMed] [Google Scholar]
- Staudinger H. (1926) Die Chemie der organischen hochmolekularen Stoffe im Sinne der Kekuléschen Strukturlehre. Ber Dtsch Chem Ges 59: 3019–3043 [Google Scholar]
- Stone BA, Clarke AE. (1992) Chemistry and Biology of (1,3)-β-Glucans. La Trobe University Press, Melbourne, Australia [Google Scholar]
- Sturcová A, His I, Apperley DC, Sugiyama J, Jarvis MC. (2004) Structural details of crystalline cellulose from higher plants. Biomacromolecules 5: 1333–1339 [DOI] [PubMed] [Google Scholar]
- Thimm JC, Burritt DJ, Ducker WA, Melton LD. (2000) Celery (Apium graveolens L) parenchyma cell walls examined by atomic force microscopy: effect of dehydration on cellulose microfibrils. Planta 212: 25–32 [DOI] [PubMed] [Google Scholar]
- Tian GW, Mohanty A, Chary SN, Li S, Paap B, Drakakaki G, Kopec CD, Li J, Ehrhardt D, Jackson D, et al. (2004) High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol 135: 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff DM. (1969) Semi-micro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- van der Wel NN, Putman CAJ, vanNoort SJT, deGrooth BG, Emons AMC. (1996) Atomic force microscopy of pollen grains, cellulose microfibrils, and protoplasts. Protoplasma 194: 29–39 [Google Scholar]
- Viëtor RJ, Newman RH, Ha MA, Apperley DC, Jarvis MC. (2002) Conformational features of crystal-surface cellulose from higher plants. Plant J 30: 721–731 [DOI] [PubMed] [Google Scholar]
- Wightman R, Turner SR. (2008) The roles of the cytoskeleton during cellulose deposition at the secondary cell wall. Plant J 54: 794–805 [DOI] [PubMed] [Google Scholar]