Abstract
To define the mechanisms involved in the molecular response to the carcinogenic metal cadmium, two novel metal-inducible genes from C. elegans were characterized: numr-1 and numr-2 (nuclear localized metal responsive). numr-1 and numr-2 sequences and cellular patterns of expression are identical, indicating that these are functionally equivalent genes. Constitutive transcription of numr-1 and numr-2 is developmentally regulated and occurs in the intestine, in head and tail neurons, and vulva muscles. Exposure to metals induces numr-1 and numr-2 transcription in pharyngeal and intestinal cells. Other environmental stressors do not affect transcription, indicating that these are metal-specific, stress-responsive genes. NUMR-1 and NUMR-2 target to nuclei and colocalize with HSF-1, suggesting that they may be components of nuclear stress granules. Nematodes overexpressing NUMR-1 and NUMR-2 are resistant to stress and live longer than control animals; likewise reducing expression increases sensitivity to metals and decreases neuromuscular functions. Upstream regulatory regions of both genes contain potential binding sites for DAF-16 and SKN-1, which are components of the insulin-IGF-like signaling pathway. This pathway regulates longevity and stress responses in C. elegans. NUMR-1 and NUMR-2 may function to promote resistance to environmental stressors and longevity, which is mediated by the insulin-IGF-like signaling pathway.
Keywords: C. elegans, Cadmium, Stress response, Lifespan, Longevity
Introduction
The transition metal cadmium is continuously introduced into the environment through anthropogenic activities. It is a persistent toxicant of occupational and environmental health concern (Waalkes et al., 1992). Numerous adverse health effects, including chronic respiratory disease, osteoporosis, impaired renal function and a variety of cancers are associated with cadmium exposure (Waalkes et al., 1992). Cadmium is also a potent animal teratogen (Holt and Webb, 1987). Molecular mechanisms of cadmium toxicity include the generation of reactive oxygen species, inactivation and denaturation of proteins, DNA damage, and the activation of intracellular signaling cascades, including mitogen-activated protein kinases, protein kinase C, tyrosine kinase, and casein kinase 2 (Huang et al., 2001; Joseph et al., 2001; Stohs et al., 2000; Watkin et al., 2003).
Although organisms are continuously exposed to cadmium, cells resist metal-induced toxicity through a variety of mechanisms. One of the primary mechanisms for cadmium detoxification is chelation and sequestration of the metal by metallothioneins (Klaassen et al., 2009). Additional cellular processes that counteract the toxic effects of cadmium include the activation of the heat shock response and DNA repair pathways. Furthermore, cells have the capacity to remove toxic by-products of cadmium exposure, such as reactive oxygen species (Liu et al., 2009).
Many of the proteins induced in response to cadmium exposure and the cognate signaling pathways are involved in cellular repair or removal of the metal. Using genomics and bioinformatics, novel proteins and signaling pathways have been indentified that are important for resistance to metal toxicity. Their exact role in cadmium detoxification, however, is unclear. In addition, many of these pathways and proteins are associated with increased lifespan.
Genetic studies in many species have identified several processes that regulate stress response and lifespan including caloric intake, mitochondrial respiration, insulin-IGF-1 (IIS), and Jun N-terminal kinase (Kaeberlein et al., 2006; Neumann-Haefelin et al., 2008; Rea and Johnson, 2003). A strong correlation between stress response and longevity has been established in the nematode Caenorhabditis elegans (Johnson et al., 2001). In C. elegans, members of the daf-2 (insulin) and the TOR signaling networks are associated with resistance to metals, oxidative stress and longevity (Barsyte et al., 2001). Although several stress-response and/or longevity pathways have been identified in C. elegans, many of the down-stream effector molecules remain unknown.
Previously, whole genome C. elegans DNA microarray and RNAi analysis were used to explore global changes in the nematode transcriptome to understand mechanisms involved in resistance to cadmium toxicity (Cui et al., 2007). Several of the genes identified in this analysis were previously associated with the C. elegans cadmium response, including mtl-1, mtl-2 and cdr-1 (Freedman et al., 1993; Liao et al., 2002). Many of the highly inducible genes, however, had no known biological or molecular function. One of these genes, originally designated F08F8.5, showed a sevenfold increase in the steady-state mRNA level following a 24-hour cadmium exposure (Cui et al., 2007). A second C. elegans gene, originally designated F08F8.1, was subsequently identified based on sequence identity. These genes are now designated numr-1 and numr-2 (nuclear localized metal responsive), respectively.
In the present report, the genomic organization, in vivo cellular patterns of expression, effects of transition metals and other stressors on transcription, and phenotypes associated with increased and decreased expression of numr-1 and numr-2 are presented. Phenotypes associated with these genes include changes in neuromuscular activities, as well as increased resistance to metal toxicity and increased lifespan.
Results
Sequence analysis of numr-1 and numr-2
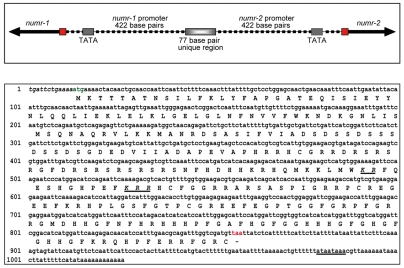
numr-1 and numr-2 were located less than 1 kb apart in a divergent orientation on chromosome III (Fig. 1). numr-1 mRNA was predicted to be encoded by a single exon. By contrast, numr-2 mRNA was predicted to be encoded by five exons, the first of which was ~99% identical to the numr-1 exon. Sequences of the full-length mRNAs and the exon arrangement for numr-1 and numr-2 were determined using 3′- and 5′-RACE. The sequence of the 3′-end of numr-1 was consistent with that reported in WormBase (version WS201). Using 3′ RACE, however, cDNAs from predicted exons 2-5 of numr-2 were not isolated. This suggested that numr-2 was annotated incorrectly and that it also was a single exon gene. Sequences for the 5′-ends of numr-1 and numr-2 were 100% identical.
Fig. 1.
Sequences of NUMR-1/-2 cDNA and protein. (Upper panel) Chromosomal organization of numr-1 and numr-2. The red boxes indicate the translation start sites. (Lower panel) The nucleotide and predicted protein sequence for numr-1/-2 cDNA are shown. The 5′-untranslated region is presented in italics; the translation start and stop codons are presented in green and red, respectively; and the polyadenylation signal is underlined. Amino acids corresponding to the predicted bipartite nuclear targeting sequence are shown in italic and underlined.
The full-length cDNA sequences of numr-1 and numr-2 were 1029 bp and contained open reading frames of 846 bp (281 amino acids). The 5′-untranslated regions were approximately 12 bp in length and were not trans spliced. The 3′-untranslated regions consisted of 142 bp that terminated with poly(A) tails and contained typical polyadenylation signals (Fig. 1).
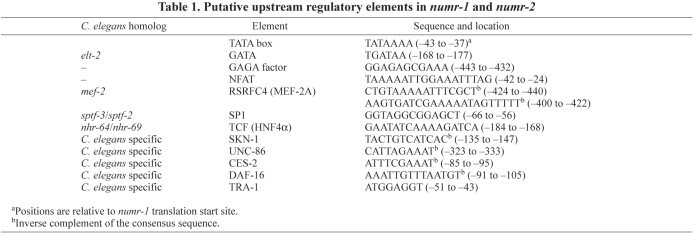
The genomic organization of both genes was similar. Both genes contained 499 bp promoters that were 99% identical. They were separated by 77 bp of unique DNA. Each gene had a single consensus TATA sequence that was ~30 bp upstream from the transcription start site. Both numr-1 and numr-2 contained a single exon that shared 100% sequence identity. (In cases where the gene, mRNA or protein can not be assigned to either numr-1 or numr-2 the material is identified as numr-1/-2 or NUMR-1/-2, respectively.) Analysis of the 499 bp upstream regions of numr-1 and numr-2 predicted several upstream regulatory elements (Table 1). Several of these elements are C. elegans specific, whereas others were conserved in higher eukaryotes.
Table 1.
Putative upstream regulatory elements in numr-1 and numr-2
BLAST analysis of the numr-1/-2 cDNA sequence identified only numr-1/-2 homologs in other Caenorhabditis species. The current assemblies of these genomes indicated that C. briggsae and C. remanei each had a single homolog of numr-1/-2, which were also single exon genes. C. brenneri contained four single exon genes with high sequence homology to numr-1/-2. C. japonica contained a single homolog of numr-1/-2.
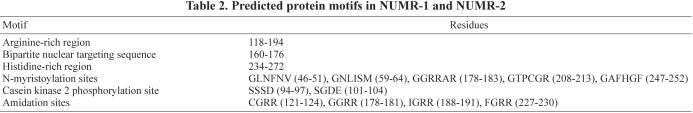
The predicted proteins encoded by numr-1 and numr-2 are 281 amino acids in length (Fig. 1). PROSITE analysis (Sigrist et al., 2002) of NUMR-1/-2 predicted several functional domains (Table 2). NUMR-1/-2 contained an arginine-rich region near a predicted nuclear localization signal. Potential casein kinase 2 phosphorylation, N-myristoylation, and amidation sites were also identified. A histidine-rich region was identified, which could function in metal binding.
Table 2.
Predicted protein motifs in NUMR-1 and NUMR-2
In vivo transcription of numr-1 and numr-2
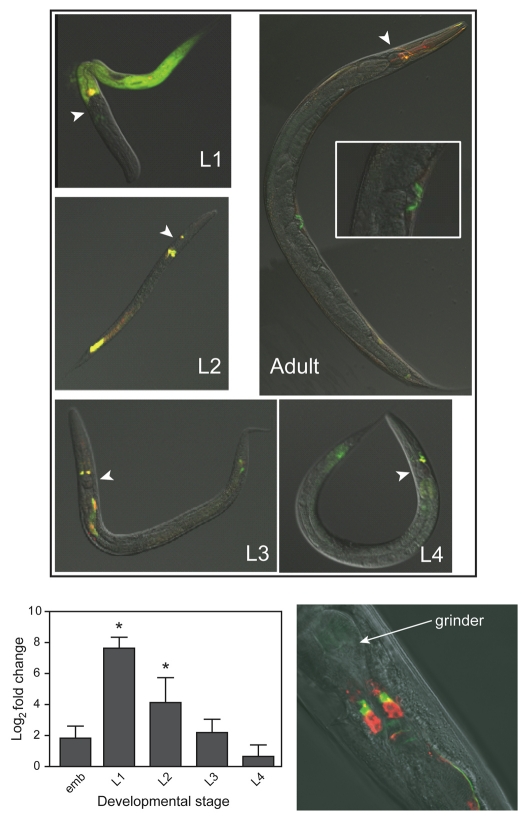
To determine cell-specific, developmental patterns of numr-1 and numr-2 transcription, JF86(mtEx61) C. elegans, which contained both Pnumr-1::mCherry and Pnumr-2::GFP transgenes, were examined. In the absence of any metal, numr-1 and numr-2 showed identical patterns of transcription (Fig. 2). Both genes showed developmental patterns of expression, where maximal levels of constitutive expression were observed throughout the intestine of L1 nematodes. This expression decreased as the nematodes developed into adults, at which point it was minimal. qRT-PCR analysis confirmed that the steady-state level of numr-1/-2 mRNA was highest in L1 C. elegans and that it gradually decreased as nematodes developed into adults.
Fig. 2.
Constitutive expression of numr-1 and numr-2. (Upper panel) Superimposed fluorescence and DIC images of JF86(mtEx61) C. elegans at four larval stages (L1, L2, L3 and L4) and adults. The insert in the adult panel shows transgene expression in vulval muscles. Coexpression of numr-1 and numr-2 is denoted by yellow fluorescence. The arrowheads indicate the location of the pharyngeal grinder in each nematode. (Lower left panel) qRT-PCR analysis of numr-1/-2 steady-state mRNA levels at different developmental stages. Total RNA was isolated from embryos (emb), L1, L2, L3, L4 larvae, and adult C. elegans. All measurements were normalized to the mRNA level of rbp-12 and the fold change was relative to the steady-state level mRNA measured in adult nematodes. Values are mean log2fold change ± s.e.m. (n=3). Asterisks indicate statistically significant difference from adult nematodes (P<0.0001). (Lower right panel) The head region of JF85(mtEx60) showing numr-1 expression and DiI-stained neurons. numr-1 expression (green) does not coincide with the ciliated amphid neurons (red). The position of the grinder is indicated.
In addition to developmentally regulated intestinal cell expression, numr-1 and numr-2 were constitutively expressed at all developmental stages in a few cells in the posterior of the nematode and in a pair of sensory neurons in the head. They were also constitutively expressed in egg-laying muscles of the adult vulva (Fig. 2). To distinguish which sensory neurons expressed numr-1, JF85(mtEx60) nematodes were treated with DiI. numr-1 expression was detected in a pair of sensory neurons with processes extending towards the external environment, but the expression did not overlap with cells stained by DiI. This suggested that numr-1 was expressed in AW sensory neurons (Fig. 2).
Inducible expression of numr-1 and numr-2
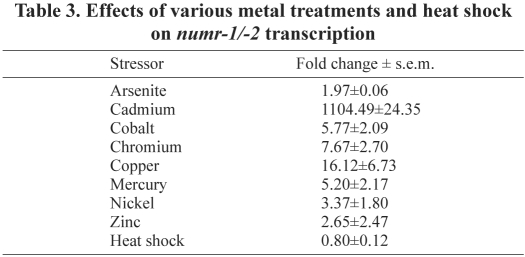
Exposure of JF86(mtEx61) C. elegans to cadmium caused an increase in numr-1 and numr-2 transcription throughout the intestine and in the posterior bulb, isthmus and corpus of the pharynx. Exposure to copper caused an increase in transcription in the pharynx, but did not induce numr-1 or numr-2 transcription in the intestine (Fig. 3).
Fig. 3.
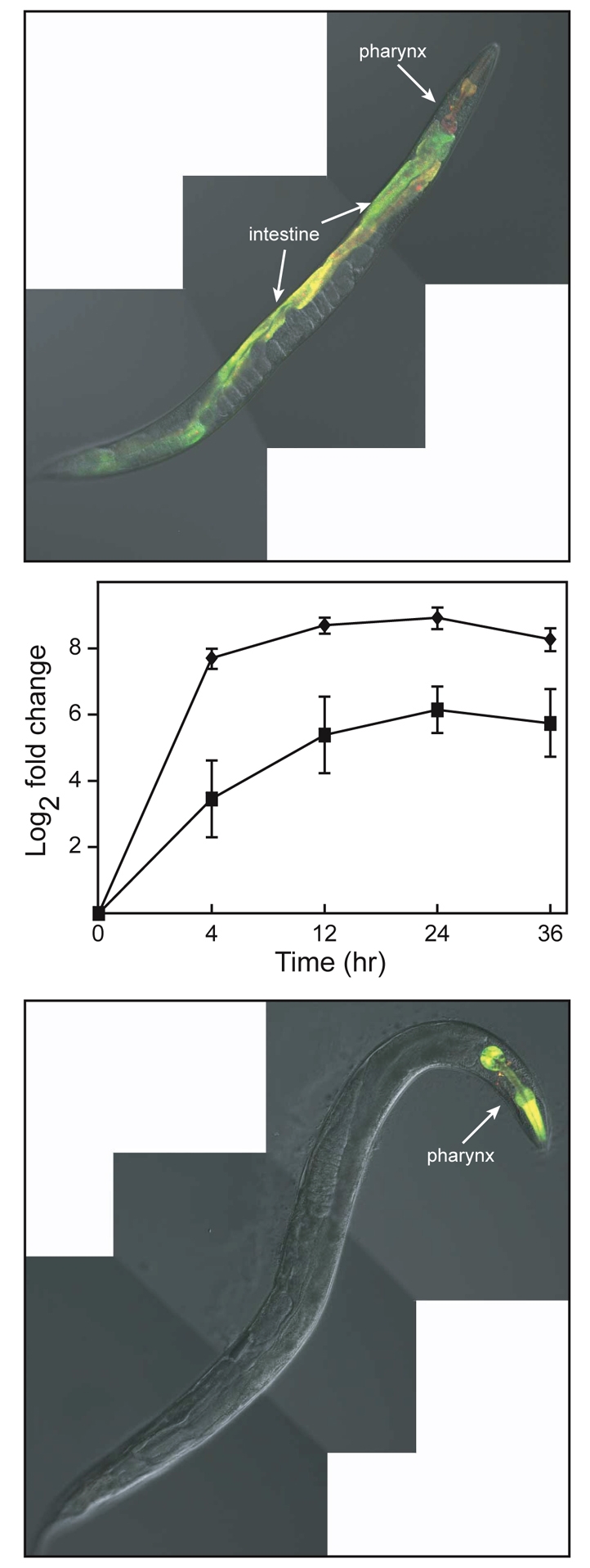
Effects of metals on numr-1 and numr-2 expression. Superimposed fluorescence and DIC images of adult JF86(mtEx61) C. elegans maintained on plates containing 100 μM cadmium (upper panel) or 100 μM copper (lower panel) for 24 hours. Coexpression of numr-1 and numr-2 is indicated by yellow fluorescence. (Middle panel) Time course for numr-1/-2 and cdr-1 mRNA accumulation following exposure to cadmium. Total RNA was isolated from wild-type C. elegans treated with 100 μM cadmium for 4, 12, 24, or 36 h, and untreated nematodes. Steady-state levels of numr-1/-2 (squares) and cdr-1 (diamonds) mRNAs were measured using qRT-PCR. All measurements were normalized to the mRNA level of mlc-2 and the fold change was relative to the mRNA levels in untreated (t=0 hours) nematodes. Values are mean log2fold change ± s.e.m. (n=4).
Changes in the steady-state level of numr-1/-2 mRNAs in response to cadmium exposure over time were determined using qRT-PCR. In a mixed stage population of wild-type C. elegans, the level of numr-1/-2 mRNA increased following a 4-hour exposure to cadmium (Fig. 3). The maximal level of numr-1/-2 mRNA was detected following a 24-hour exposure. As a control, changes in the steady-state levels of mRNA for the cadmium-inducible gene cdr-1 were also measured (Liao et al., 2002). Both cdr-1 and numr-1/-2 showed the same pattern of cadmium-responsive mRNA accumulation.
The effects of other transition metals and stressors on numr-1/-2 mRNA accumulation were determined. Mixed stage wild-type animals were treated with copper, cobalt, chromium, cadmium, nickel, arsenic, zinc and mercury, and exposed to heat shock. There was an increase in numr-1/-2 mRNA levels following exposure to all of the metals (Table 3). Cadmium was the strongest inducer followed by copper. Treatment with arsenic caused a small increase (approximately twofold) in numr-1/-2 mRNA levels.
Table 3.
Effects of various metal treatments and heat shock on numr-1/-2 transcription
The cellular patterns of expression for other transition metals were identical to that observed for either cadmium or copper (supplementary material Fig. S3). Exposure to 400 μM zinc or 2 μM silver for 24 hours induced numr-1 transcription in the pharynx and intestine, similar to that observed following cadmium exposure. Heat shock did not produce an increase in numr-1/-2 mRNA abundance. In addition, exposure of adult JF85(mtEx60) C. elegans to other stressors including oxidative stress (200 μM juglone or hydrogen peroxide), starvation, endoplasmic reticulum stress (5 μg/ml tunicamycin), heat shock (37°C 15 minutes and 35°C 1 hour), and pathogen infection (Serratia marcesans or Pseudomonas aeruginosa) did not cause an increase in NUMR-GFP expression (results not shown). These results suggested that inducible transcription of numr-1/-2 may be metal-specific and that these are not general stress response genes.
Intracellular localization of NUMR-1
The intracellular location of NUMR-1 was determined using JF88(mtEx63) C. elegans that contained the Pnumr-1::NUMR-1–GFP transgene, which expressed a NUMR-1–GFP fusion protein. In the absence of metal, the cellular pattern of NUMR-1 expression was identical to that observed in JF85(mtEx60) nematodes (Fig. 4). NUMR-1 was constitutively expressed in intestinal cells of L1 nematodes and the level of expression decreased as the nematodes developed (data not shown). NUMR-1 was also expressed in neurons in the head, egg-laying muscles of the vulva, and a few cells in the tail (Fig. 4, inset). NUMR-1 was concentrated in the nuclei of the expressing cells. This indicated that the predicted NUMR-1 nuclear localization signal was functional.
Fig. 4.

Intracellular expression of NUMR-1. (Upper panel) Constitutive expression of NUMR-1 in adult JF88(mtEx63) animals in the absence of metal. The insets show the terminal bulb (upper) and the vulva (lower) of the same nematode at higher magnification. (Lower panel) JF88(mtEx63) C. elegans exposed to 100 μM cadmium (left) or 100 μM copper (right) for 24 hours.
When exposed to cadmium, NUMR-1 expression increased in intestinal and pharyngeal nuclei. This pattern was similar to that observed for JF85(mtEx60) C. elegans. Following copper exposure, NUMR-1 expression in adult nematodes increased only in pharyngeal nuclei. Similar patterns of expression were observed in larval C. elegans in the presence and absence of the metals (results not shown).
Nuclear-accumulated NUMR-1–GFP was clearly observed as punctate structures within the intestinal nuclei of cadmium-treated nematodes (Fig. 5). Punctate nuclear structures are not commonly observed for C. elegans nuclear-targeted proteins. For example, the DAF-16 transcription factor localizes to intestinal nuclei in response to stress (Henderson and Johnson, 2001), but unlike NUMR-1–GFP, DAF-16–GFP was observed throughout the nucleus. The expression pattern of DAF-16–GFP was diffuse, occupying the entire nucleus (Fig. 5).
Fig. 5.
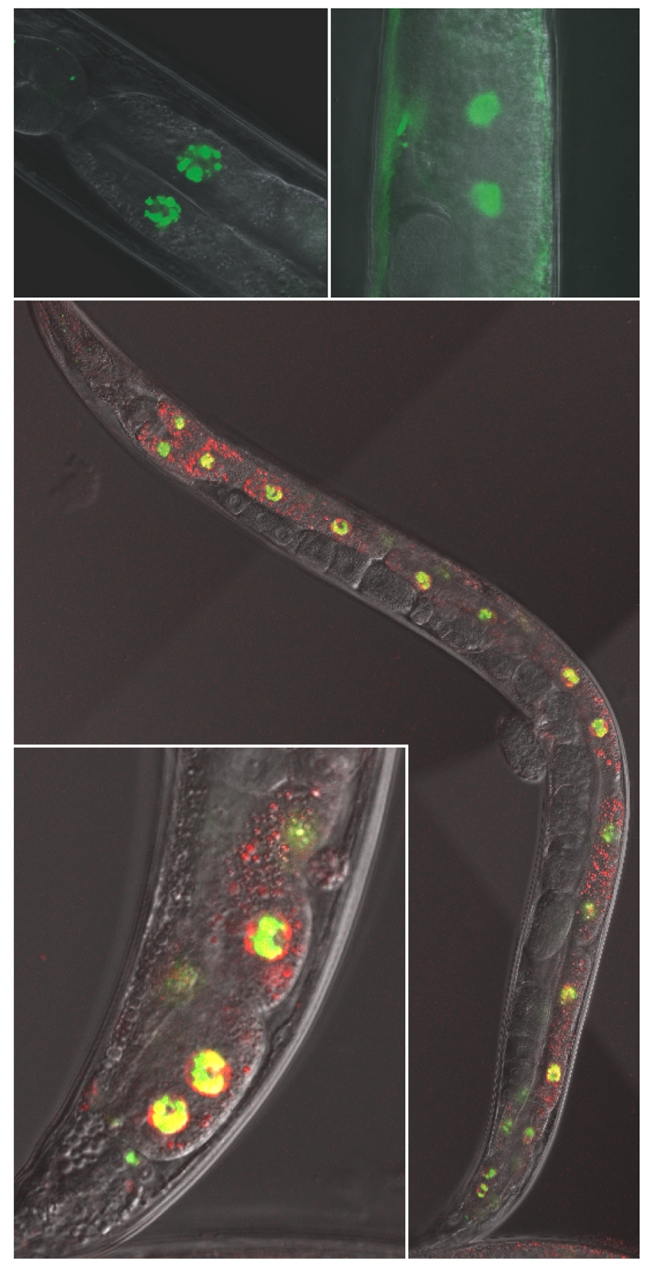
Intranuclear localization of NUMR-1. (Upper left panel) Intestinal cell nuclei of JF88(mtEx63) C. elegans following a 24-hour exposure to 100 μM cadmium. (Upper right panel) Nuclear localization of DAF-16–GFP in TJ356(zIs356) nematodes following incubation at 35°C for 1 hour; lower panel, JF87(mtEx62) nematodes coexpressing numr-1::NUMR-1–GFP and hsf-1::HSF-1–mCherry after exposure to 100 μM cadmium for 24 hours. Yellow fluorescence indicates coincident expression of NUMR-1 and HSF-1. (Inset) A higher magnification of intestinal cell nuclei from another transgenic nematode. The original magnification in the upper panels and inset was ×630 and the lower panel was ×400. Individual fluorescence images for NUMR-1–GFP, HSF-1–mCherry, and DIC for the inset can be found in supplementary material Fig. S5.
Nuclear punctate structures have been observed in mammalian cells and were defined as nuclear stress granules (Hong et al., 2001; Jolly et al., 1999). These nuclear stress granules contained the heat shock transcription factor, HSF-1. HSF-1 localizes to nuclear stress granules in eukaryotic cells following exposure to various stressors, including metal treatment (Jolly et al., 2002). To determine if NUMR-1 localized to C. elegans nuclear stress granules, colocalization of NUMR-1–GFP and HSF-1–mCherry was assessed in JF87(mtEx62) nematodes. NUMR-1 colocalized with HSF-1 after cadmium treatment (Fig. 5). This result suggests that NUMR-1 may be a component of nuclear stress granules.
Phenotypes caused by numr-1/-2
The biological function of numr-1/-2 was examined in C. elegans where NUMR-1/-2 expression was reduced using either numr-1 deletion strains or numr-1/-2 RNAi. C. elegans numr-1(ok2239) has a deletion that spans most of the coding region for numr-1. The regulatory and coding regions of numr-2 were unaffected. C. elegans numr-1(tm2775) has a deletion that removes approximately 60 bp of the numr-1 promoter, including the TATA box, and 147 bp from the numr-1 coding region; numr-2 was also unaffected. numr-1(ok2239) and numr-1(tm2775) nematodes were egg-laying defective (Egl phenotype). This was demonstrated by an increase in the number of nematodes with a ‘bagging’ phenotype, where nematodes fail to lay their eggs and embryos hatch inside the parent (Fig. 6). Microscopic examination of the vulva in L4 nematodes showed that there were structural abnormalities, which may have contributed to the Egl phenotype (Fig. 6). numr-1(ok2239) and numr-1(tm2775) nematodes often lacked organized epithelial-ring structures that line the vulva lumen at the L4 stage. Furthermore, numr-1 mutants possessed abnormal uterine seams (Hall and Altun, 2008). In addition to the Egl phenotype, numr-1(ok2239) and numr-1(tm2775) nematodes did not feed as much (i.e. Eat phenotype) as the wild-type nematodes (Fig. 6). These results suggested that numr-1/-2 may be involved in C. elegans neuromuscular activity or development.
Fig. 6.
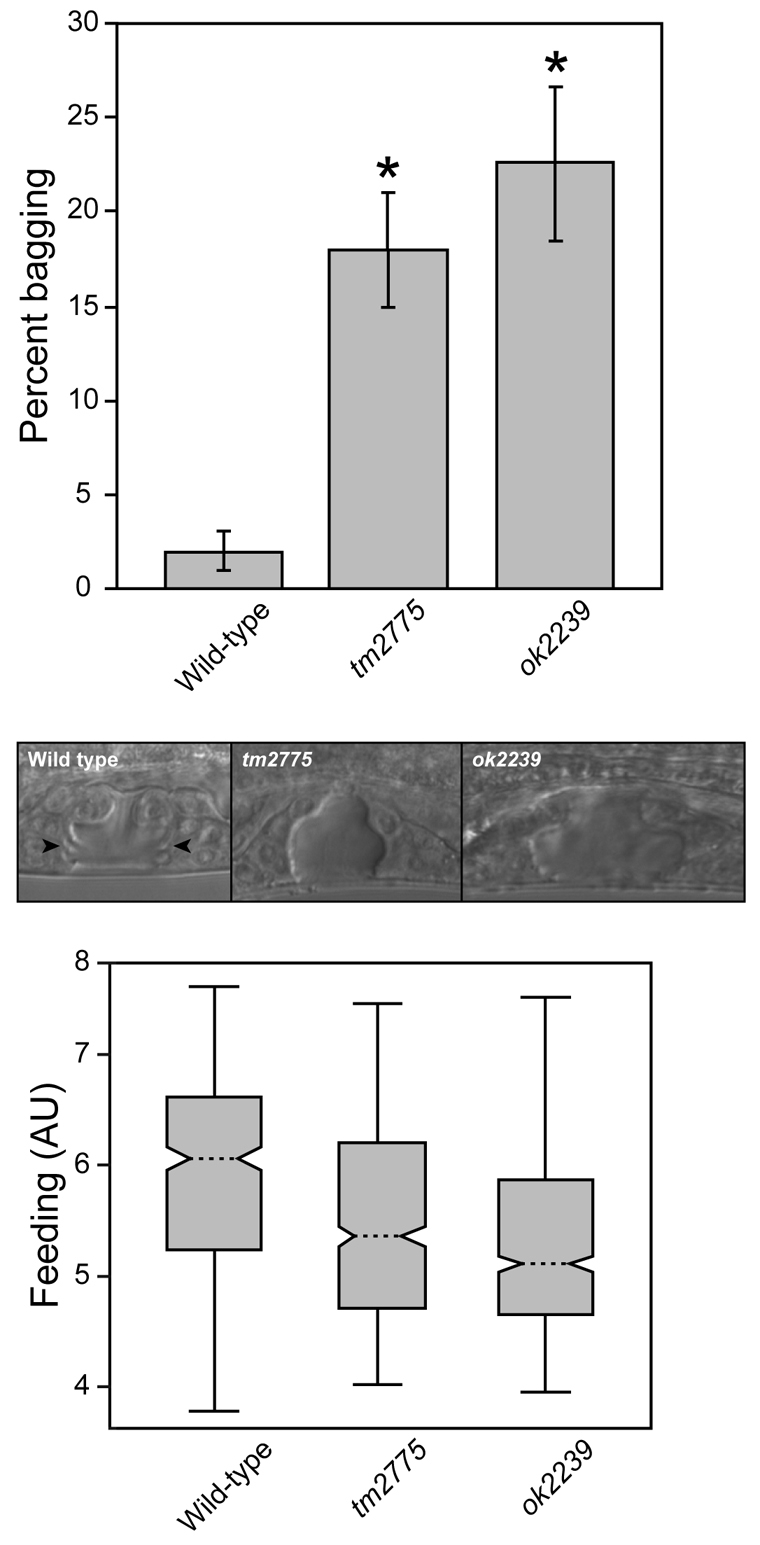
Effect of decreased numr-1 expression on egg-laying and feeding. (Upper panel) Percentage of adult wild-type, numr-1(tm2775) and numr-1(ok2239) nematodes with an egg-laying phenotype. Fisher's exact test indicated a highly significant impact of genotype on the propensity of worms to bag *P<0.001, compared with the wild-type. (Middle panels) DIC micrographs of wild-type, numr-1(tm2775) and numr-1(ok2239) L4 nematodes. Arrowheads indicate the position of the structure absent in the vulva of the numr-1 deletion strains. (Lower panel) Comparison of wild-type and numr-1 mutant C. elegans feeding. Box and whisker plots of normalized feeding for adult nematodes aggregated over three independent experiments are presented. Dotted lines indicate the median, the 25th and 75th percentiles of the data are shown by the upper and lower limits of each box, respectively. AU, arbitrary units.
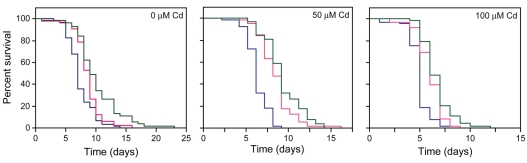
To determine the role of NUMR-1/-2 in ameliorating metal stress; the effect of cadmium exposure on longevity in transgenic C. elegans that overexpressed NUMR-1/-2 was determined. Initially, the effect of NUMR-1 overexpression on nematode longevity in the absence of metal was assessed. JF88(mtEx63) nematodes, which contained extrachromosomal copies of numr-1, lived longer than control nematodes (P<0.003; Fig. 7). JF88(mtEx63) nematodes that were treated with 50 or 100 μM cadmium had significantly longer lifespans (P<0.0001), compared with cadmium-treated wild-type nematodes (Fig. 7). This indicated that NUMR-1/-2 contributed to C. elegans resistance to cadmium toxicity. The level of resistance associated with NUMR-1 overexpression was similar to that observed in TJ356(zIs356) C. elegans, which overexpressed DAF-16–GFP (Fig. 7). Overexpression of DAF-16 is associated with increased longevity and stress resistance (Henderson and Johnson, 2001).
Fig. 7.
Effect of NUMR-1/-2 overexpression on longevity and sensitivity to cadmium toxicity. Transgenic C. elegans expressing HE1006(su1006), which resulted in a mutant form of ROL-6 (blue line); JF88(mtEx63), which overexpressed NUMR-1 and ROL-6 (red line); and TJ356(zIs356), which overexpressed DAF-16 and ROL-6 (green line) were exposed to different concentrations of cadmium. (Left panel) C. elegans not treated with cadmium. HE1006(su1006): n=60, m=7; JF88(mtEx63): n=50, m=9; TJ356(zIs356): n=54, m=9. There was a significant difference between HE1006(su1006) and JF88(mtEx63) nematodes, P=0.0028. There was no significant difference between JF88(mtEx63) and TJ356(zIs356). (Middle panel) C. elegans treated with 50 μM cadmium. HE1006(su1006): n=63, m=6; JF88(mtEx63): n=62, m=8; TJ356(zIs356): n=58, m=9. There was a significant difference between HE1006(su1006) and JF88(mtEx63) nematodes, P<0.0001. (Right panel) C. elegans treated with 100 μM cadmium: HE1006(su1006): n=64, m=5; JF88(mtEx63): n=61, m=6; TJ356(zIs356): n=65, m=6. There was a significant difference between HE1006(su1006) and JF88(mtEx63) nematodes, P<0.0001. There was no significant difference between JF88(mtEx63) and TJ356(zIs356). All experiments were repeated at least once with similar results; n, number of nematodes sampled; m, median survival in days.
In a complementary study, the effects of decreased NUMR-1/-2 expression on longevity and resistance to metal stress were determined. Treatment of numr-1(ok2239) with cadmium did not significantly affect longevity, relative to wild-type nematodes. This may have been due to the presence of the intact numr-2 and continued NUMR-2 expression. qRT-PCR showed a significant increase in the steady-state level of numr-1/-2 mRNA in numr-1 mutant nematodes following cadmium exposure (supplementary material Fig. S2). Therefore, numr-1(ok2239) nematodes were treated with numr-1/-2 RNAi to more thoroughly knock down NUMR-1/-2 expression.
RNAi-mediated knockdown of NUMR-1/-2 expression in wild-type animals did not produce obvious phenotypes or increase sensitivity to cadmium exposure (Fig. 8). To confirm that RNAi significantly reduced numr-1/-2 expression, the level of transgene expression following cadmium exposure in JF88(mtEx63) nematodes that were fed numr-1/-2 RNAi was examined. numr-1/- 2 RNAi suppressed NUMR-1–GFP expression throughout the intestine but did not appear to affect pharyngeal or neuronal expression, indicating numr-1/-2 RNAi does not abolish all NUMR-1/-2 expression (supplementary material Fig. S4).
Fig. 8.
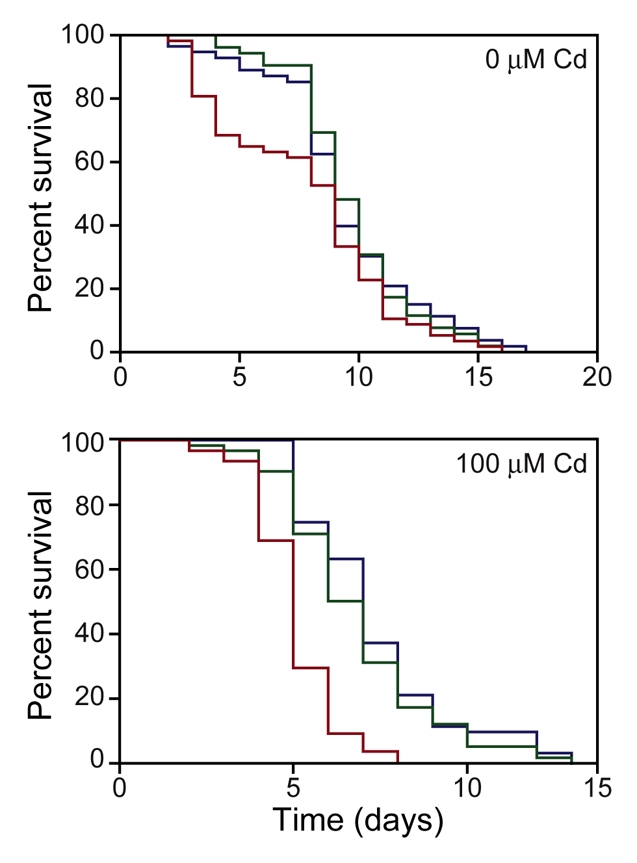
Effect of decreased NUMR-1/-2 expression on longevity and sensitivity to cadmium toxicity. Wild-type nematodes were fed RNAi control vector (blue line) or numr-1/-2 RNAi (green line) and numr-1(ok2239) nematodes were fed numr-1/-2 RNAi (red line) and exposed to 100 μM cadmium. (Upper panel) Non-exposed C. elegans numr-1/-2 knockdown does not affect life-span in the absence of metal treatment [N2: n=53, m=9. N2; numr-1/-2(RNAi): n=52, m=9. numr-1(ok2239); numr-1/-2(RNAi): n=57, m=9] but (lower panel) numr-1/-2 knockdown increases sensitivity of animals to 100 μM CdCl2 [N2 versus numr-1(ok2239) fed numr-1/-2(RNAi], [N2: n=62, m=7. numr-1(ok2239); numr-1/-2(RNAi): n=59, m=5, P<0.0001]. There was no significant difference between N2 fed RNAi control vector and N2 fed numr-1/-2(RNAi) (N2; numr-1/-2(RNAi): n=59, m=7). All experiments were repeated at least once with similar results.
In the absence of cadmium, exposure of numr-1(ok2239) C. elegans to numr-1/-2 RNAi produced an initial, rapid decrease in survival but did not significantly influence the median lifespan of animals (Fig. 8). The initial decrease in C. elegans survival could be attributed to bagging. By contrast, RNAi-treated numr-1(ok2239) C. elegans that were exposed to 100 μM cadmium had a significantly reduced lifespan, compared with both numr-1/-2 RNAi-treated and vector control-treated wild-type nematodes (P=0.0001; Fig. 8). Both NUMR-1/-2 overexpression and knockdown studies indicated that numr-1/-2 is important in cadmium detoxification and may contribute to longevity.
Discussion
Cadmium-regulated gene expression on a genome-wide scale has been documented in a number of different organisms and cell types (Jin et al., 2008; Tan et al., 2006; Yamada and Koizumi, 2002; Yepiskoposyan et al., 2006). Unfortunately, many of the differentially expressed genes identified in these studies have no known biological function. From the C. elegans cadmium-response transcriptome, a novel metal-inducible gene, numr-1 was identified that showed a sevenfold increase in its steady-state mRNA level following a 24-hour exposure to metal (Cui et al., 2007). Based on sequence homology, a second C. elegans gene was subsequently identified, numr-2. Both genes are located less than 1 kb apart in a head to head orientation. They are encoded by single exons that share 100% nucleotide sequence identity and their regulatory regions are 99% identical (Fig. 1). Protein and nucleotide sequences, genomic organization, and cellular patterns of expression for numr-1 and numr-2 are identical, indicating that these are functionally equivalent genes that arose by a gene duplication event. Genes with highly similar sequences to numr-1 and numr-2 have been found in other Caenorhabditis species. These homologs are present in single and multiple copies.
PROSITE analysis of NUMR-1/-2 identified several potential protein motifs (Table 2). NUMR-1 and NUMR-2 each contain a predicted nuclear localization signal (NLS). The NLS of NUMR-1 and NUMR-2 is most likely functional since NUMR-1–GFP is targeted to nuclei (Figs 4 and 5). NUMR-1 and NUMR-2 also contain an arginine-rich region near the predicted NLS. This domain is similar to that observed in RNA-binding proteins (Bayer et al., 2005), which suggests that they may function in RNA processing. The presence of a histidine-rich region in NUMR-1 and NUMR-2 suggests that these proteins may chelate transition metals. The ability of poly-histidine to bind transition metals has been well documented (Sovago and Osz, 2006). The presence of potential casein kinase 2 phosphorylation sites suggests that NUMR-1 and NUMR-2 activity may be regulated through this signaling cascade. Environmental stress has been shown to modulate casein kinase 2 activity and affect cognate signal transduction cascades (Filhol and Cochet, 2009).
Several biological roles can be proposed for numr-1 and numr-2, based on their cellular patterns of transcription, and phenotypes associated with over- and underexpression of these genes. NUMR-1/-2 may be involved in neuromuscular function or development. In the adult nematode, numr-1 and numr-2 are actively transcribed in the egg-laying muscles of the vulva (Figs 2 and 4). Furthermore, nematodes with numr-1 mutant alleles exhibit a more severe egg-laying defect (Fig. 6). Egg-laying defects can arise from abnormal vulva formation or alterations in the functional activity of the vulval muscles or hermaphrodite-specific (HSN) neurons (Riddle, 1997). In both numr-1 mutants, morphological changes to the vulva are observed (Fig. 6). In addition to egg-laying phenotypes, lowered levels of numr-1/-2 expression result in decreased feeding (Fig. 6). NUMR-1 and NUMR-2 are expressed in muscles and neurons in the pharynx (Figs 2 and 4; supplementary material Fig. S3). These data further support the idea that proper numr-1/-2 expression may be required for normal neuromuscular function of the vulva and pharynx.
Constitutive expression of numr-1/-2 was observed throughout the intestinal cells during early larval stages of development and decreased as animals developed into adults (Fig. 2). During early larval stages of development, C. elegans intestinal cells experience endogenous endoplasmic reticulum (ER) stress (Shen et al., 2001). Cadmium has been reported to induce ER stress in mouse renal tubule and hepatoma cells, 3T3 cells, and C. elegans (Biagioli et al., 2008; Hiramatsu et al., 2007; Urano et al., 2002). numr-1 and numr-2 are actively transcribed in the intestine in the presence of cadmium, suggesting that ER stress may act as a signal to regulate numr-1/-2 intestinal cell transcription. The precise mechanisms by which development- and cadmium-induced ER stress affects numr-1/-2 transcription remain to be determined. It is unlikely that misfolded proteins are activating numr-1/-2 since heat shock and tunicamycin, which induce ER stress through the accumulation of misfolded proteins, do not affect numr-1/-2 transcription. It has been proposed that cadmium can increase intracellular calcium levels by inducing release of calcium from ER stores via stimulation of inositol-trisphosphate-gated channels and inhibition of sarco(endo)plasmic reticulum calcium ATPase pumps (Biagioli et al., 2008). This raises the possibility that the expression of numr-1/-2 may be regulated by alterations in intracellular calcium. In accordance with the idea that variations in calcium levels might act as a signal to regulate numr-1/-2, calcium signaling plays an important role in egg-laying and feeding in C. elegans (Robatzek and Thomas, 2000; Steger and Avery, 2004). Both of these activities are abnormal in the two numr-1 mutant alleles when compared with wild-type nematodes (Fig. 6).
NUMR-1 and NUMR-2 are constitutively expressed in chemosensory neurons in the nematode head (Fig. 2). This raises the possibility that these proteins may be part of a peripheral signaling pathway that is involved in sensing environmental stress and mediating a specific stress response in other cells in the nematodes. A direct link between sensory neuron function and the ability to respond at the transcriptional level to environmental stress has been demonstrated (Prahlad et al., 2008). The constitutive neuronal expression of numr-1 and numr-2 in head and tail neurons may be regulated by the POU transcription factor UNC-86. The upstream regulatory regions of both genes contain consensus binding sites for UNC-86. UNC-86 is required for the transcription of several C. elegans neuronally expressed genes, including ric-4, mec-3 and tph-1 (Hwang and Lee, 2003; Sze et al., 2002; Xue et al., 1992). Although unc-86 null mutants are available, these nematodes exhibit a basal state of elevated stress, as indicated by constitutive DAF-16 nuclear accumulation (Liang et al., 2006). Since numr-1/-2 responds to stress, the elevated level of continuous stress confounds the interpretation of the effect of unc-86 loss of function on numr-1/-2 expression. Thus, the exact molecular mechanism by which UNC-86 may affect numr-1 and numr-2 neuronal transcription remains to be confirmed.
The C. elegans intestine is the primary site of metal detoxification in the nematode. Many of the genes associated with metal detoxification are actively transcribed in the nematode intestine including mtl-1, mtl-2, cdr-1, gsto-1 and hmt-1 (Burmeister et al., 2008; Freedman et al., 1993; Liao et al., 2002; Vatamaniuk et al., 2005). Exposure to cadmium and other transition metals produce significant increases in numr-1/-2 transcription in the nematode intestine. Intestinal numr-1/-2 expression is consistent with the finding of a highly conserved GATA motif within the numr-1/-2 promoter (Table 1). Like numr-1/-2, three other well-characterized cadmium-responsive C. elegans genes, mtl-1, mtl-2 and cdr-1, contain GATA elements (Freedman et al., 1993; Liao et al., 2002). The GATA motif is highly over-represented in the promoters of intestinally expressed genes and the ELT-2 GATA-type transcription factor regulates the expression of the majority of these genes (McGhee et al., 2009). Metal inducible transcription of other C. elegans genes that contribute to resistance to metal toxicity, mtl-1, mtl-2 and gsto-1 is also limited to the intestine (Burmeister et al., 2008; Freedman et al., 1993; Liao et al., 2002). Furthermore, the ELT-2–GATA interaction controls mtl-1 and mtl-2 intestinal cell transcription (Moilanen et al., 1999). Additional studies are still needed to confirm the role of ELT-2 in regulating numr-1 and numr-2 transcription.
Transgenic nematodes and qRT-PCR results indicate that numr-1 and numr-2 transcription is influenced by metal exposure, as well as developmental factors. The response of numr-1/-2 to metals is atypical, compared with other known metal-responsive genes. The transcription of many metal-inducible genes, such as the metallothioneins and glutathione S-transferases, is generally affected by other stressors, such as heat shock, oxidative stress, damaged proteins or radiation. By contrast, the only environmental stressors that affect numr-1/-2 transcription are transition metals. This is similar to the response of cdr-1 (Liao et al., 2002). cdr-1 transcription, however, is only inducible following cadmium exposure, whereas, numr-1/-2 transcription increases in response to a variety of transition metals.
numr-1 and numr-2 metal-inducible transcription is also regulated by cell-specific factors. Exposure to cadmium induces transcription in intestinal and pharyngeal cells. A similar pattern of expression is observed following zinc and mercury treatment (results not shown). Copper, however, induces transcription only in pharyngeal cells (Fig. 3). This pattern of transcription is similar to that observed for hsp16, which encodes the C. elegans small heat shock protein (Stringham and Candido, 1994). This phenomenon is probably not the result of the bioavailability of copper to intestinal cells, since these cells express a copper transporter and copper activates mtl-2 transcription in intestinal cells (Wakabayashi et al., 1998) (J.H.F., unpublished observation). The variation in expression pattern could be a result of a difference in the toxicity of the two metals. Exposure to low-level cadmium (10 μM) for 24 hours increases numr-1/-2 expression only in the pharynx but longer exposures to cadmium increase numr-1/-2 transcription throughout the intestine. The mechanism limiting numr-1/-2 inducible transcription to the pharynx following copper exposure remains to be elucidated.
The expression of NUMR-1/-2 is associated with resistance to metal toxicity. This conclusion is based on the observations that (1) knockdown of numr-1/-2 expression increases sensitivity to cadmium exposure and (2) overexpression of numr-1/-2 increases resistance (Figs 7 and 8). In addition to increased resistance to metals, overexpression of numr-1/-2 results in an increase in lifespan in both the presence and absence of metal. It is possible that overexpression of numr-1/-2 increases the overall well-being of the nematode and in turn extends the lifespan of C. elegans. However, decrease in numr-1/-2 expression does not significantly shorten wild-type lifespan under control conditions but does decrease lifespan in the presence of metals, suggesting that the contribution of numr-1/-2 to longevity is complemented by other genes.
There is a strong association between longevity and resistance to environmental stress (Neumann-Haefelin et al., 2008). C. elegans genes have been identified that both influence nematode lifespan, as well as sensitivity to environmental stressors, including cadmium (Barsyte et al., 2001; Neumann-Haefelin et al., 2008). DAF-2 (insulin receptor-like protein), AGE-1 (phosphatidylinositol 3-kinase), and DAF-16 (FOXO transcription factor) are members of the evolutionarily conserved insulin-IGF-like signaling pathway (Vanfleteren and Braeckman, 1999). Activation of this pathway ultimately inhibits the nuclear localization of transcription factors such as: DAF-16, SKN-1 and HSF-1 (Inoue et al., 2005; Lin et al., 1997; Tullet et al., 2008). These transcription factors regulate the expression of numerous stress-responsive genes and when overexpressed, they increase stress resistance and extend lifespan (Johnson et al., 2000; Lin et al., 1997; Morley and Morimoto, 2004; Sykiotis and Bohmann, 2008; Tullet et al., 2008). It has been suggested that ELT-2 also plays a critical role in regulating components of the daf-2-daf-16 pathway. ELT-2 may interact with SKN-1 to control expression of some SKN-1 downstream target genes (McGhee et al., 2009). The regulatory regions of numr-1 and numr-2 contain consensus binding sites for both SKN-1 and DAF-16 (Table 1). The regulatory pathways controlling longevity and stress resistance have been characterized in a variety of species. The downstream targets that ultimately affect C. elegans physiology to modulate lifespan and stress resistance, however, have not been identified. NUMR-1 and NUMR-2 may perform the biological activities necessary to increase resistance and longevity that are mediated by the insulin-IGF-like signaling pathway.
The response to cadmium exposure involves the integration of a number of cadmium-responsive signaling pathways and genes. numr-1 and numr-2 represent a new class of metal-responsive genes that are required for neuromuscular functions such as egg-laying and feeding. In addition, they contribute to longevity and mediate resistance to metal toxicity in C. elegans. Characterization of cadmium-responsive genes can provide insights into the mechanism by which metal-activated signal-transduction pathways elicit cellular responses and define the biological consequences associated with those responses. Many of the regulatory pathways affecting longevity have been defined. However, the downstream genes that they regulate, and how the products of the downstream targets affect longevity have not been identified. NUMR-1/-2 may be the products that contribute to the longevity phenotype.
Materials and Methods
Strains
The following C. elegans strains were obtained from the C. elegans Genetic Center (Minneapolis, MN): wild-type N2 Bristol, TJ356(zIs356[daf-16::daf-16–GFP; rol-6(su1006)]), HE1006 rol-6(su1006), and numr-1(ok2239) mutant. The numr-1 mutant allele, ok2239, was backcrossed to N2 seven times prior to use. A second numr-1 mutant allele, tm2775, was obtained from the National Bioresource Project for the Nematode (Tokyo, Japan) and backcrossed ten times. Several transgenic C. elegans strains were generated to examine the in vivo expression of numr-1 and numr-2 (see below). These strains are JF85(mtEx60[numr-1::GFP + rol-6(su1006)]), JF86(mtEx61[numr-1::mCherry, numr-2::GFP + rol-6(su1006)]), JF87(mtEx62[numr-1::NUMR-1–GFP, hsf-1::HSF-1–mCherry + rol-6 (su1006)]) and JF88(mtEx63[numr-1::NUMR-1–GFP + rol-6(su1006)]). Nematodes were maintained at 20°C, as previously described (Sulston and Hodgkin, 1988).
Growth and treatment of C. elegans
Developmentally staged C. elegans were prepared by hypochlorite treatment, as previously described (Khanna et al., 1997). C. elegans at six developmental stages: embryo, L1, L2, L3, L4, and adult, were collected by sucrose flotation, washed, rapidly frozen in liquid nitrogen, and stored at −80°C, as previously described (Sulston and Hodgkin, 1988). Developmental stages were confirmed by visual examination.
For cadmium exposures, mixed populations of nematodes were cultured in S-medium [100 mM sodium chloride, 50 mM potassium phosphate, 10 mM potassium citrate pH 6.0, 3 mM calcium chloride, 3 mM magnesium sulfate, 50 μM EDTA, 25 μM iron sulfate, 10 μM manganese chloride, 10 μM zinc sulfate, 1 μM copper sulfate, 13 μM cholesterol, K-medium (51 mM sodium chloride, 32 mM potassium chloride)] at 20°C for 5 days using E. coli OP50 as a food source (Sulston and Hodgkin, 1988). Additional E. coli was added after 3 days to maintain an adequate food supply. Cadmium chloride (100 μM, final concentration) was then added to the culture and incubation was continued for an additional 4, 12, 24, or 36 hours. Nematodes were collected and stored, as described above.
The effect of various metals and heat shock on numr-1/-2 mRNA levels was determined. Mixed populations of C. elegans were cultured in K-Plus medium (Anderson et al., 2004; Williams and Dusenbery, 1988) at 20°C for 4 days. Metal salts were then added singly to the nematode cultures at the following final concentrations: 100 μM sodium arsenite, 100 μM cadmium chloride, 100 μM cobalt chloride hexahydrate, 100 μM chromium oxide, 100 μM copper sulfate, 25 μM mercury chloride, 100 μM nickel sulfate hexahydrate and 100 μM zinc sulfate heptahydrate. C. elegans were incubated in the presence of the metal for an additional 24 hours. Heat shock was performed by incubating 5-day-old C. elegans cultures at 37°C for 30 minutes. Treated nematodes were collected and stored as described above.
Isolation of total RNA
Frozen nematode pellets were ground into a fine powder using a liquid-nitrogen-cooled mortar and pestle before being homogenized in RNeasy Lysis buffer and total RNA was then isolated using RNeasy Midi kits (Qiagen Inc., Valencia, CA). To eliminate potential DNA contamination, RNA was subsequently purified using RNase-Free DNase Sets (Qiagen). The quality of the purified RNA was assessed with an Agilent 2100 Bioanalyzer using RNA 6000 Nano LabChip kits (Agilent Technologies, Santa Clara, CA).
Rapid amplification of cDNA ends
To obtain full-length cDNAs corresponding to numr-1 and numr-2,3′- and 5′-RACE were performed following the manufacturer's protocol (Invitrogen Corp., Carlsbad, CA). For 3′-RACE, a 3′-RACE oligonucleotide primer, which has an adapter region annealed to oligo(dT), was used to reverse transcribe 2 μg of total RNA isolated from mixed populations of untreated C. elegans. Gene-specific primers were then combined with 3′-RACE primers UAP and AUAP to amplify 3′ ends of the cDNA. For 5′-RACE, 2 μg of total RNA was reverse transcribed using gene-specific primers located near the 5′ end of numr-1 and numr-2. PCR was performed to amplify dC-tailed cDNA using the 5′-RACE Abridged Anchor Primer in combination with a GSP (gene specific primer) and a nested GSP, respectively. All 3′- and 5′-RACE PCR fragments were cloned into a pCR2.1 TA vector (Invitrogen) and sequenced. DNAStar 5.05 sequence analysis software (DNAStar, Madison, WI) was used to assemble continuous cDNA sequences. Gene-specific primers for 3′- and 5′-RACE were designed according to the predicted mRNA sequence of numr-1 and numr-2 published in WormBase, version WS201 (Harris et al., 2004) and are presented in supplementary material Table S1.
Quantitative RT-PCR
To quantify changes in the steady-state levels of numr-1/-2 mRNA during development and in response to metal exposure, qRT-PCR was used. qRT-PCR was performed using QuantiTect SYBR Green RT-PCR kits (Qiagen) following the manufacturer's instructions in an ABI Prism 7900HT system (Applied Biosystems, Foster City, CA, USA). A minimum of three biological replicates for each treatment were prepared and each biological replicate was measured in triplicate. Primer sequences for qRT-PCR experiment are presented in supplementary material Table S2.
Changes in the steady-state mRNA level of numr-1, -2 during C. elegans development were first normalized to the control gene, rpb-12 (RNA polymerase II (B) subunit) (Link et al., 2003). To determine relative levels of expression, the normalized data at each developmental stage was then compared to that observed in adult nematodes.
Changes in numr-1/-2 mRNA following metal exposure and heat shock were normalized to mlc-2 (myosin light chain) (Cummins and Anderson, 1988) and compared with their level of expression in untreated C. elegans. Results are presented as mean log2(fold change) ± standard error (s.e.m.; n=3) or fold change ± s.e.m. Statistical significance was determined using one-way ANOVA followed by Dunnett's multiple comparison post-hoc test with the level of significance set at P<0.05.
Construction of transgenic C. elegans
Transgenic strains of C. elegans containing numr-1 or numr-2 promoters regulating the expression of GFP or mCherry reporter genes were generated to determine the developmental, cell-specific pattern of expression of these genes. Two additional transgenic strains were generated to examine the intracellular location of NUMR-1. The first expressed a NUMR-1–GFP fusion protein under the transcriptional regulation of the numr-1 promoter. The second expressed an HSF-1-mCherry fusion protein under the control of the hsf-1 promoter.
Reporter transgenes were generated using overlap extension PCR (Hobert, 2002). numr-1 and numr-2 transgenes were made by amplifying upstream regulatory regions from C. elegans genomic DNA and fusing them to PCR fragments containing mCherry- or GFP-coding regions, and the unc-54 3′ untranslated region (Fire et al., 1990). Because numr-1 and numr-2 upstream regulatory regions are >99% identical, 5′-end primers were designed to bind to the unique 77 bp intergenic region between the genes. The numr-1 and numr-2 promoter transgenes each contain 499 bp of upstream regulatory sequence, including the 77 bp intergenic region, and the respective start codons (supplementary material Fig. S1).
A NUMR-1–GFP fusion protein under the transcriptional control of the numr-1 regulatory region was generated by amplifying the genomic region containing 499 bp of the numr-1 upstream regulatory region and the entire numr-1 coding region, without the translation termination codon (supplementary material Fig. S1). Both numr-1 and numr-2 are single exon genes (see below). This product was fused to the GFP coding region using overlap extension PCR.
The HSF-1–mCherry fusion protein under the control of the hsf-1 regulatory region was generated in two steps using overlap extension PCR. First, HSF-1 cDNA was amplified and fused to 2 kb of the hsf-1 regulatory region to generate a transgene that expresses HSF-1 under the control of the hsf-1 promoter, Phsf-1::HSF-1. Next, the mCherry coding region was fused in-frame with HSF-1 in Phsf-1::HSF-1 to generate the Phsf-1::HSF-1–mCherry transgene. Sequences for all reporter transgenes were verified by DNA sequencing. Primer sequences used in transgene construction are presented in supplementary material Table S3.
Transgenic animals were generated as previously described (Fire, 1986). To generate strains containing single reporter transgenes, JF85(mtEx60) and JF88(mtEx63), Pnumr-1::GFP and Pnumr-1::NUMR-1–GFP were injected at 50 and 10 ng/μl, respectively. Transgenes were co-injected with 50 ng/μl of the pRF4 rol-6(su1006) selectable marker plasmid (Mello et al., 1991). Three independent C. elegans strains were obtained for each transgene. To generate transgenic strains that express multiple transgenes, Pnumr-1::mCherry (100 ng/μl) was co-injected with Pnumr-2::GFP (100 ng/μl) for strain JF86(mtEx61) and Pnumr-1::NUMR-1–GFP (5 ng/μl) was co-injected with Phsf-1::HSF-1–mCherry (5 ng/μl) for strain JF87(mtEx62). To determine cellular and intracellular patterns of numr-1, numr-2, or fusion protein expression, transgenic C. elegans were mounted on agar pads and then anesthetized with 150 mM sodium azide (Sulston and Hodgkin, 1988). Images were then acquired using a Zeiss LSM 510 confocal microscope (Carl Zeiss, Inc., Thornwood, NY).
Dye-filling assay
To determine which head neurons of C. elegans expressed numr-1, JF85(mtEx60) nematodes were stained by dye filling using DiI. DiI preferentially fills six amphid head neurons (ASI, ADL, ASK, AWS, ASH and ASJ) and the two phasmid tail neurons (PHA and PHB) (Herman, 1984; Perkins et al., 1986). Nematodes were grown on K-agar plates and then collected into K-medium. DiI solution (Molecular Probes, Invitrogen) was then added to the medium and the mixture incubated for 2-3 hours at room temperature in the dark. After incubation, nematodes were washed twice with K-medium and placed on to K-agar plates, without food for 1 hour prior to microscopic examination. Assignment of numr-1-transcribing cells were determined by comparing patterns of GFP and DiI fluorescence to the published C. elegans neuronal anatomy (Hall and Altun, 2008).
Phenotype characterization
Lifespan assay
To determine the role of numr-1/-2 in resistance to cadmium toxicity, lifespan assays were performed using wild-type and numr-1(ok2239) nematodes. Lifespans of nematodes overexpressing numr-1/-2 were compared with nematodes expressing only the selectable marker. Assays were carried out at 20°C on NGM plates containing 0, 50 or 100 μM cadmium. Embryos were prepared as described above and allowed to develop for 2 days. L4 stage or young adult nematodes were then transferred to fresh NGM plates containing cadmium and scored daily. Nematodes were scored as dead when they no longer responded to gentle prodding with a platinum wire. The lifespan of an individual was defined as the time elapsed from when it was placed onto the fresh NGM plate (t=0) to when it was scored as dead. Nematodes that crawled off plates during the assay were excluded from calculations. The data was analyzed using GraphPad Prism 5.0. Survival curves were compared using the log-rank (Mantel-Cox) test, and a Bonferroni post-hoc test (P<0.05) was used to adjust for multiple comparisons.
Quantitative RT-PCR indicated that numr-2 mRNA levels increased in response to cadmium exposure in the numr-1 null background (supplementary material Fig. S2). To reduce numr-1/-2 mRNA levels, RNAi was used in wild-type and numr-1(ok2239) C. elegans. The effects of reduced numr-1/-2 expression levels on lifespan and cadmium resistance were then measured. RNAi was performed as previously described (Kamath and Ahringer, 2003). First, synchronized L1 nematodes were maintained on E. coli that express numr-1/-2 dsRNA and allowed to develop into gravid adults. The progeny of these nematodes were maintained on dsRNA-expressing bacteria until they were L4 or young adults. They were then transferred to fresh RNAi plates and lifespan scored. Nematodes were also scored for the bagging phenotype (Trent et al., 1983). All lifespan assays were repeated at least once.
Feeding assay
Feeding assays were modified from Boyd et al. (Boyd et al., 2007). Briefly, 25 adult C. elegans were dispensed into each well of a 96-well plate, containing K-medium and OP50 E. coli, using a COPAS Biosort (Union Biometrica Inc., Somerville, MA, USA). After 4 hours, Fluoresbrite polychromatic red microspheres (Polysciences, Inc., Warrington, PA) were added into each well and mixed for 5 minutes. Nematodes were allowed to ingest the microspheres for 10 additional minutes and then anesthetized by adding sodium azide (10 mM final concentration) to prevent additional bead ingestion. The size and level of fluorescence of individual C. elegans were measured. The feeding data were analyzed as previously described (Boyd et al., 2007).
Supplementary Material
Acknowledgments
This work was supported (in part) by the Intramural Research Program of the NIH, and NIEHS (Z01ES102045) and the National Toxicology Program (Z01ES102046). Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). We thank Daniel Snyder (Biomolecular Screening Branch, NTP) for generating transgenic strains and Leping Li (Biostatistics Branch, NIEHS) for numr-1 and numr-2 promoter analyses. We also thank Roger Tsien (UCSD) for the mCherry construct and Scott Alper for his comments on this manuscript and his helpful discussions. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/12/2124/DC1
References
- Anderson G. L., Cole R. D., Williams P. L. (2004). Assessing behavioral toxicity with Caenorhabditis elegans. Environ. Toxicol. Chem. 23, 1235-1240 [DOI] [PubMed] [Google Scholar]
- Barsyte D., Lovejoy D. A., Lithgow G. J. (2001). Longevity and heavy metal resistance in daf-2 and age-1 long-lived mutants of Caenorhabditis elegans. FASEB J. 15, 627-634 [DOI] [PubMed] [Google Scholar]
- Bayer T. S., Booth L. N., Knudsen S. M., Ellington A. D. (2005). Arginine-rich motifs present multiple interfaces for specific binding by RNA. RNA 11, 1848-1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagioli M., Pifferi S., Ragghianti M., Bucci S., Rizzuto R., Pinton P. (2008). Endoplasmic reticulum stress and alteration in calcium homeostasis are involved in cadmium-induced apoptosis. Cell Calcium 43, 184-195 [DOI] [PubMed] [Google Scholar]
- Boyd W. A., McBride S. J., Freedman J. H. (2007). Effects of genetic mutations and chemical exposures on Caenorhabditis elegans feeding: evaluation of a novel, high-throughput screening assay. PLoS ONE 2, e1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister C., Luersen K., Heinick A., Hussein A., Domagalski M., Walter R. D., Liebau E. (2008). Oxidative stress in Caenorhabditis elegans: protective effects of the Omega class glutathione transferase (GSTO-1). FASEB J. 22, 343-354 [DOI] [PubMed] [Google Scholar]
- Cui Y., McBride S. J., Boyd W. A., Alper S., Freedman J. H. (2007). Toxicogenomic analysis of Caenorhabditis elegans reveals novel genes and pathways involved in the resistance to cadmium toxicity. Genome Biol. 8, R122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins C., Anderson P. (1988). Regulatory myosin light-chain genes of Caenorhabditis elegans. Mol. Cell. Biol. 8, 5339-5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filhol O., Cochet C. (2009). Protein kinase CK2 in health and disease: Cellular functions of protein kinase CK2: a dynamic affair. Cell. Mol. Life Sci. 66, 1830-1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A. (1986). Integrative transformation of Caenorhabditis elegans. EMBO J. 5, 2673-2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Harrison S. W., Dixon D. (1990). A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene 93, 189-198 [DOI] [PubMed] [Google Scholar]
- Freedman J. H., Slice L. W., Dixon D., Fire A., Rubin C. S. (1993). The novel metallothionein genes of Caenorhabditis elegans. Structural organization and inducible, cell-specific expression. J. Biol. Chem. 268, 2554-2564 [PubMed] [Google Scholar]
- Hall D. H., Altun Z. F. (2008). C. elegans Atlas Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Harris T. W., Chen N., Cunningham F., Tello-Ruiz M., Antoshechkin I., Bastiani C., Bieri T., Blasiar D., Bradnam K., Chan J., et al. (2004). WormBase: a multi-species resource for nematode biology and genomics. Nucleic Acids Res. 32, D411-D417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S. T., Johnson T. E. (2001). daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 11, 1975-1980 [DOI] [PubMed] [Google Scholar]
- Herman R. K. (1984). Analysis of genetic mosaics of the nematode Caenorhabditis elegans. Genetics 108, 165-180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu N., Kasai A., Du S., Takeda M., Hayakawa K., Okamura M., Yao J., Kitamura M. (2007). Rapid, transient induction of ER stress in the liver and kidney after acute exposure to heavy metal: evidence from transgenic sensor mice. FEBS Lett. 581, 2055-2059 [DOI] [PubMed] [Google Scholar]
- Hobert O. (2002). PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32, 728-730 [DOI] [PubMed] [Google Scholar]
- Holt D., Webb M. (1987). Teratogenicity of ionic cadmium in the Wistar rat. Arch. Toxicol. 59, 443-447 [DOI] [PubMed] [Google Scholar]
- Hong Y., Rogers R., Matunis M. J., Mayhew C. N., Goodson M. L., Park-Sarge O. K., Sarge K. D. (2001). Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 276, 40263-40267 [DOI] [PubMed] [Google Scholar]
- Huang C., Zhang Q., Li J., Shi X., Castranova V., Ju G., Costa M., Dong Z. (2001). Involvement of Erks activation in cadmium-induced AP-1 transactivation in vitro and in vivo. Mol. Cell Biochem. 222, 141-147 [DOI] [PubMed] [Google Scholar]
- Hwang S. B., Lee J. (2003). Neuron cell type-specific SNAP-25 expression driven by multiple regulatory elements in the nematode Caenorhabditis elegans. J. Mol. Biol. 333, 237-247 [DOI] [PubMed] [Google Scholar]
- Inoue H., Hisamoto N., An J. H., Oliveira R. P., Nishida E., Blackwell T. K., Matsumoto K. (2005). The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 19, 2278-2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y. H., Dunlap P. E., McBride S. J., Al-Refai H., Bushel P. R., Freedman J. H. (2008). Global transcriptome and deletome profiles of yeast exposed to transition metals. PLoS Genet. 4, e1000053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. E., Cypser J., de Castro E., de Castro S., Henderson S., Murakami S., Rikke B., Tedesco P., Link C. (2000). Gerontogenes mediate health and longevity in nematodes through increasing resistance to environmental toxins and stressors. Exp. Gerontol. 35, 687-694 [DOI] [PubMed] [Google Scholar]
- Johnson T. E., de Castro E., Hegi de Castro S., Cypser J., Henderson S., Tedesco P. (2001). Relationship between increased longevity and stress resistance as assessed through gerontogene mutations in Caenorhabditis elegans. Exp. Gerontol. 36, 1609-1617 [DOI] [PubMed] [Google Scholar]
- Jolly C., Usson Y., Morimoto R. I. (1999). Rapid and reversible relocalization of heat shock factor 1 within seconds to nuclear stress granules. Proc. Natl. Acad. Sci. USA 96, 6769-6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C., Konecny L., Grady D. L., Kutskova Y. A., Cotto J. J., Morimoto R. I., Vourc'h C. (2002). In vivo binding of active heat shock transcription factor 1 to human chromosome 9 heterochromatin during stress. J. Cell Biol. 156, 775-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph P., Muchnok T. K., Klishis M. L., Roberts J. R., Antonini J. M., Whong W. Z., Ong T. (2001). Cadmium-induced cell transformation and tumorigenesis are associated with transcriptional activation of c-fos, c-jun, and c-myc proto-oncogenes: role of cellular calcium and reactive oxygen species. Toxicol. Sci. 61, 295-303 [DOI] [PubMed] [Google Scholar]
- Kaeberlein T. L., Smith E. D., Tsuchiya M., Welton K. L., Thomas J. H., Fields S., Kennedy B. K., Kaeberlein M. (2006). Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell 5, 487-494 [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J. (2003). Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313-321 [DOI] [PubMed] [Google Scholar]
- Khanna N., Cressman C. P., 3rd, Tatara C. P., Williams P. L. (1997). Tolerance of the nematode Caenorhabditis elegans to pH, salinity, and hardness in aquatic media. Arch. Environ. Contam. Toxicol. 32, 110-114 [DOI] [PubMed] [Google Scholar]
- Klaassen C. D., Liu J., Diwan B. A. (2009). Metallothionein protection of cadmium toxicity. Toxicol. Appl. Pharmacol. 238, 215-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B., Moussaif M., Kuan C. J., Gargus J. J., Sze J. Y. (2006). Serotonin targets the DAF-16/FOXO signaling pathway to modulate stress responses. Cell Metab. 4, 429-440 [DOI] [PubMed] [Google Scholar]
- Liao V. H., Dong J., Freedman J. H. (2002). Molecular characterization of a novel, cadmium-inducible gene from the nematode Caenorhabditis elegans. A new gene that contributes to the resistance to cadmium toxicity. J. Biol. Chem. 277, 42049-42059 [DOI] [PubMed] [Google Scholar]
- Lin K., Dorman J. B., Rodan A., Kenyon C. (1997). daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278, 1319-1322 [DOI] [PubMed] [Google Scholar]
- Link C. D., Taft A., Kapulkin V., Duke K., Kim S., Fei Q., Wood D. E., Sahagan B. G. (2003). Gene expression analysis in a transgenic Caenorhabditis elegans Alzheimer's disease model. Neurobiol. Aging 24, 397-413 [DOI] [PubMed] [Google Scholar]
- Liu J., Qu W., Kadiiska M. B. (2009). Role of oxidative stress in cadmium toxicity and carcinogenesis. Toxicol. Appl. Pharmacol. 238, 209-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Fukushige T., Krause M. W., Minnema S. E., Goszczynski B., Gaudet J., Kohara Y., Bossinger O., Zhao Y., Khattra J., et al. (2009). ELT-2 is the predominant transcription factor controlling differentiation and function of the C. elegans intestine, from embryo to adult. Dev. Biol. 327, 551-565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V. (1991). Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959-3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moilanen L. H., Fukushige T., Freedman J. H. (1999). Regulation of metallothionein gene transcription. Identification of upstream regulatory elements and transcription factors responsible for cell-specific expression of the metallothionein genes from Caenorhabditis elegans. J. Biol. Chem. 274, 29655-29665 [DOI] [PubMed] [Google Scholar]
- Morley J. F., Morimoto R. I. (2004). Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell 15, 657-664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann-Haefelin E., Qi W., Finkbeiner E., Walz G., Baumeister R., Hertweck M. (2008). SHC-1/p52Shc targets the insulin/IGF-1 and JNK signaling pathways to modulate life span and stress response in C. elegans. Genes Dev. 22, 2721-2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L. A., Hedgecock E. M., Thomson J. N., Culotti J. G. (1986). Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117, 456-487 [DOI] [PubMed] [Google Scholar]
- Prahlad V., Cornelius T., Morimoto R. I. (2008). Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science 320, 811-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S., Johnson T. E. (2003). A metabolic model for life span determination in Caenorhabditis elegans. Dev. Cell 5, 197-203 [DOI] [PubMed] [Google Scholar]
- Riddle D. L. (1997). C. elegans II Plainview, NY: Cold Spring Harbor Laboratory Press; [PubMed] [Google Scholar]
- Robatzek M., Thomas J. H. (2000). Calcium/calmodulin-dependent protein kinase II regulates Caenorhabditis elegans locomotion in concert with a G(o)/G(q) signaling network. Genetics 156, 1069-1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Ellis R. E., Lee K., Liu C. Y., Yang K., Solomon A., Yoshida H., Morimoto R., Kurnit D. M., Mori K., et al. (2001). Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107, 893-903 [DOI] [PubMed] [Google Scholar]
- Sigrist C. J., Cerutti L., Hulo N., Gattiker A., Falquet L., Pagni M., Bairoch A., Bucher P. (2002). PROSITE: a documented database using patterns and profiles as motif descriptors. Brief. Bioinform. 3, 265-274 [DOI] [PubMed] [Google Scholar]
- Sovago I., Osz K. (2006). Metal ion selectivity of oligopeptides. Dalton Trans. 32, 3841-3854 [DOI] [PubMed] [Google Scholar]
- Steger K. A., Avery L. (2004). The GAR-3 muscarinic receptor cooperates with calcium signals to regulate muscle contraction in the Caenorhabditis elegans pharynx. Genetics 167, 633-643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohs S. J., Bagchi D., Hassoun E., Bagchi M. (2000). Oxidative mechanisms in the toxicity of chromium and cadmium ions. J. Environ. Pathol. Toxicol. Oncol. 19, 201-213 [PubMed] [Google Scholar]
- Stringham E. G., Candido E. P. M. (1994). Transgenic hsp16-lacZ strains of the soil nematode Caenorhabditis elegans as biological monitors of environmental stress. Environ. Toxicol. Chem. 13, 1211-1220 [Google Scholar]
- Sulston J., Hodgkin J. (1988). Methods. In The Nematode Caenorhabditis elegans (ed. Wood W. B.), pp. 587-606 Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Sykiotis G. P., Bohmann D. (2008). Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell 14, 76-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze J. Y., Zhang S., Li J., Ruvkun G. (2002). The C. elegans POU-domain transcription factor UNC-86 regulates the tph-1 tryptophan hydroxylase gene and neurite outgrowth in specific serotonergic neurons. Development 129, 3901-3911 [DOI] [PubMed] [Google Scholar]
- Tan Y., Shi L., Hussain S. M., Xu J., Tong W., Frazier J. M., Wang C. (2006). Integrating time-course microarray gene expression profiles with cytotoxicity for identification of biomarkers in primary rat hepatocytes exposed to cadmium. Bioinformatics 22, 77-87 [DOI] [PubMed] [Google Scholar]
- Trent C., Tsuing N., Horvitz H. R. (1983). Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics 104, 619-647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet J. M., Hertweck M., An J. H., Baker J., Hwang J. Y., Liu S., Oliveira R. P., Baumeister R., Blackwell T. K. (2008). Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132, 1025-1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F., Calfon M., Yoneda T., Yun C., Kiraly M., Clark S. G., Ron D. (2002). A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J. Cell Biol. 158, 639-646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanfleteren J. R., Braeckman B. P. (1999). Mechanisms of life span determination in Caenorhabditis elegans. Neurobiol. Aging 20, 487-502 [DOI] [PubMed] [Google Scholar]
- Vatamaniuk O. K., Bucher E. A., Sundaram M. V., Rea P. A. (2005). CeHMT-1, a putative phytochelatin transporter, is required for cadmium tolerance in Caenorhabditis elegans. J. Biol. Chem. 280, 23684-23690 [DOI] [PubMed] [Google Scholar]
- Waalkes M. P., Coogan T. P., Barter R. A. (1992). Toxicological principles of metal carcinogenesis with special emphasis on cadmium. Crit. Rev. Toxicol. 22, 175-201 [DOI] [PubMed] [Google Scholar]
- Wakabayashi T., Nakamura N., Sambongi Y., Wada Y., Oka T., Futai M. (1998). Identification of the copper chaperone, CUC-1, in Caenorhabditis elegans: tissue specific co-expression with the copper transporting ATPase, CUA-1. FEBS Lett. 440, 141-146 [DOI] [PubMed] [Google Scholar]
- Watkin R. D., Nawrot T., Potts R. J., Hart B. A. (2003). Mechanisms regulating the cadmium-mediated suppression of Sp1 transcription factor activity in alveolar epithelial cells. Toxicology 184, 157-178 [DOI] [PubMed] [Google Scholar]
- Williams P. L., Dusenbery D. B. (1988). Using the nematode Caenorhabditis elegans to predict mammalian acute lethality to metallic salts. Toxicol. Ind. Health 4, 469-478 [DOI] [PubMed] [Google Scholar]
- Xue D., Finney M., Ruvkun G., Chalfie M. (1992). Regulation of the mec-3 gene by the C. elegans homeoproteins UNC-86 and MEC-3. EMBO J. 11, 4969-4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H., Koizumi S. (2002). DNA microarray analysis of human gene expression induced by a non-lethal dose of cadmium. Ind. Health 40, 159-166 [DOI] [PubMed] [Google Scholar]
- Yepiskoposyan H., Egli D., Fergestad T., Selvaraj A., Treiber C., Multhaup G., Georgiev O., Schaffner W. (2006). Transcriptome response to heavy metal stress in Drosophila reveals a new zinc transporter that confers resistance to zinc. Nucleic Acids Res. 34, 4866-4877 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.