Abstract
This study reports DNA barcodes for more than 1300 Lepidoptera species from the eastern half of North America, establishing that 99.3 per cent of these species possess diagnostic barcode sequences. Intraspecific divergences averaged just 0.43 per cent among this assemblage, but most values were lower. The mean was elevated by deep barcode divergences (greater than 2%) in 5.1 per cent of the species, often involving the sympatric occurrence of two barcode clusters. A few of these cases have been analysed in detail, revealing species overlooked by the current taxonomic system. This study also provided a large-scale test of the extent of regional divergence in barcode sequences, indicating that geographical differentiation in the Lepidoptera of eastern North America is small, even when comparisons involve populations as much as 2800 km apart. The present results affirm that a highly effective system for the identification of Lepidoptera in this region can be built with few records per species because of the limited intra-specific variation. As most terrestrial and marine taxa are likely to possess a similar pattern of population structure, an effective DNA-based identification system can be developed with modest effort.
Keywords: DNA barcoding, cytochrome c oxidase 1, species identification, cryptic species, Lepidoptera
1. Introduction
The need for an advance in our ability to identify and discriminate species is widely acknowledged (Sutherland of Houndswood 2008). Most eukaryotes remain undescribed and the entry of a species into the Linnaean system does little to ensure its subsequent recognition because the subtle morphological characters that separate closely allied species often demand expert interpretation. Sequence diversity, in short, standardized gene regions (DNA barcodes), provides an alternative approach for both the identification of known species and the discovery of new ones (Hebert et al. 2003; Savolainen et al. 2005; Mitchell 2008). However, questions persist concerning the efficacy of DNA barcoding (Hickerson et al. 2006) and the level of effort that is required to parameterize an effective identification system (Zhang et al. in press).
2. Material and methods
We tested the ability of DNA barcodes to both identify known species and reveal overlooked taxa within the Lepidoptera of North America. With nearly 13 000 species, this assemblage includes 1 per cent of all described animal species. We acquired DNA barcodes from 11 289 individuals representing 1327 species (electronic supplementary material, figure S1) collected from the eastern half of this continent (38–59°N, 70–90°W). These taxa included representatives of 62 different families of micro- and macro-Lepidoptera (electronic supplementary material, figure S2), but with stronger representation for macros because of the greater maturity of their taxonomy. PCR amplification using a single pair of primers consistently recovered the 648 bp region near the 5′ terminus of the mitochondrial cytochrome c oxidase I (COI) gene that serves as the barcode region for the animal kingdom (Hebert et al. 2003). DNA isolation, PCR amplification and DNA sequencing followed standard protocols (deWaard et al. 2008). COI pseudogenes (NUMTS) have been encountered in some invertebrate lineages (e.g. Buhay 2009), but we detected none in our work, a result that coincides with their rarity in taxa with small genome sizes. Sequences were deposited in GenBank with accession codes GU087155–GU097197. Complete specimen data are available from the Barcode of Life Data System (www.barcodinglife.org) in the project ‘Lepidoptera of Eastern North America Phase I’.
3. Results and discussion
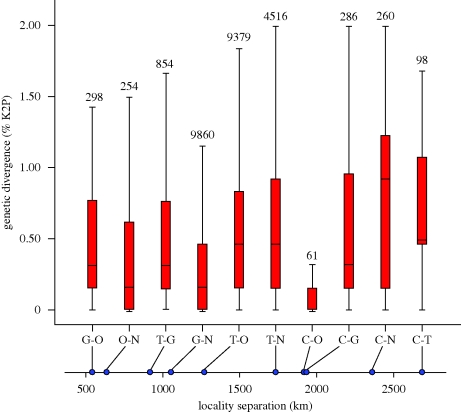
Sequence analysis of the COI amplicon (electronic supplementary material, figure S2) established that members of a species usually showed low sequence variation, averaging 0.43 per cent (s.e. = 0.017%) while congeneric species possessed 18-fold higher mean divergences (7.70%, s.e. = 0.033%). The present study provides the first comprehensive analysis of barcode divergences in populations of single species separated by large geographical distances. Comparison of intraspecific divergences for populations collected from 500–2800 km apart revealed no significant increase in genetic distances with geographical separation (figure 1). This lack of substantial regional variation in barcode sequences indicates that an effective identification system can be constructed for the Lepidoptera fauna of eastern North America without extensive geographical surveys of each species. We anticipate that similarly muted levels of intraspecific variation will be shared by most taxa in other insect orders such as Coleoptera, Diptera and Hymenoptera from this region. We expect more differentiation in groups with low vagility and in other areas, such as western North America, where higher topographic roughness provides more opportunities for population isolation and differentiation. It will also be intriguing to probe the patterns of regional divergence in areas such as Australia where Pleistocene glaciations had a much less dramatic impact on species distributions.
Figure 1.
Box plots of intraspecific divergence observed for populations of species that were collected from two or more localities. The median distances of separation are given for the general localities of Churchill, MB (C), Guelph, ON (G), Ottawa, ON (O), St Andrew's, NB (N) and Great Smoky Mountains National Park, TN/NC (T). The number of comparisons used to calculate genetic divergence is denoted above each box plot.
We detected only nine cases of barcode sharing in the 1327 species included in our study, all involving situations in which a pair of species shared the same barcode. These cases always involved congeneric species with close morphological similarity. Because 99.3 per cent of the 1327 species had barcode sequences distinct from those of other taxa, a COI reference library can generate identifications very effectively.
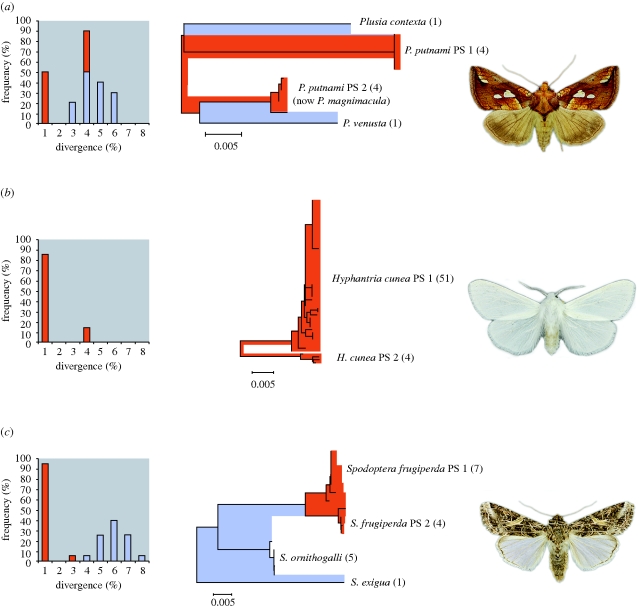
Although most species possessed low intra-specific divergence, 67 taxa included two or three barcode groups with more than 2 per cent sequence divergence. Many of these cases probably reflect overlooked species pairs or triads. As evidence, we note that individuals of Plusia putnami separated into two barcode groups with 3.8 per cent COI divergence (figure 2a). Subsequent investigation revealed differences in genitalia, host plant use and habitats, leading to the description of a new species (Handfield & Handfield 2006). Other cases of deep barcode divergence involved species where there is independent evidence for unrecognized taxa. For example, two barcode lineages with 2.8 per cent sequence divergence were detected in the fall webworm, Hyphantria cunea (figure 2b), which has long been thought to include two species with differing larval morphologies (Itô & Warren 1973). Young species pairs will be overlooked by a 2 per cent screening threshold, but they can still show barcode differentiation. For example, the fall armyworm, Spodoptera frugiperda, includes two barcode lineages with 1.3 per cent divergence (figure 2c). This species consists of two ‘host races’ that not only have different primary hosts (rice versus corn), but show allozyme and mitochondrial DNA divergence as well as reproductive isolation (Levy et al. 2002), justifying their recognition as distinct species. As this last example reveals, barcodes can highlight young species pairs, but studies of biological covariates are critical to confirm their status.
Figure 2.
Histograms and neighbour-joining trees showing deep sequence divergences at COI among species in three genera of Lepidoptera from eastern North America: (a) Plusia, (b) Hyphantria and (c) Spodoptera. For the species displaying deep divergences, intraspecific divergences are shaded red and divergences among congeneric taxa are shaded blue. Individuals showing deep ‘intraspecific’ barcode divergence occur in sympatry for all three taxa.
Our work affirms the validity of most Lepidoptera species recognized though prior taxonomy and suggests that relatively few species have been overlooked as just 5.1 per cent of the 1327 taxa included deeply divergent barcode lineages. However, there are two provisos. Young species pairs, such as those comprising S. frugiperda, will often be morphologically cryptic, and will also show low barcode divergence. Such taxa can be revealed, but only through a search for covariation between barcode splits and ecological or morphological traits. Secondly, the constrained species discovery in this study probably reflects both the intensity of prior taxonomic work on Lepidoptera and their flamboyant phenotypes. Interestingly, the incidence of overlooked species encountered in the present study shows close congruence to the value reported for a well-studied fauna of tropical Lepidoptera (Hajibabaei et al. 2006). In contrast, barcode analyses on insect groups with cryptic morphologies have encountered much higher rates of species discovery (Smith et al. 2006, 2008).
In summary, this study has assembled DNA barcodes for 0.1 per cent of the animal species described over the past 250 years. Our results confirm the effectiveness of a DNA barcode reference library in the identification of a continental fauna of Lepidoptera, reinforcing conclusions from studies that examined fewer species and that probed diversity on smaller geographical scales. Our work has also provided further examples of deep barcode divergences, illuminating probable overlooked species, and setting the stage for their detailed taxonomic investigation. There is no reason to expect that Lepidoptera are a particularly compliant target for barcode-based identification systems. Instead, it is likely that the key findings of this investigation apply to most other taxonomic groups occupying continental or oceanic habitats. In such situations, the barcode analysis of very few individuals of each species will provide the basis for a highly effective identification system. More effort will be required to gain a good understanding of sequence diversity in taxa from insular or freshwater habitats where local population differentiation is more pronounced, but such taxa form a minor component of global biodiversity.
We conclude that DNA barcoding can deliver—in its promise both to enable the automated identification of known species and to aid the detection of overlooked taxa. Further, as this study indicates, a comprehensive barcode library for animal life can be assembled rapidly, promising massive improvement in our knowledge of biodiversity.
Acknowledgements
We thank James Adams, John Brown, Don Davis, Don Lafontaine, Michael Pogue, Brian Scholtens, Bo Sullivan and David Wagner for aid with collections and identifications. Natalia Ivanova and Janet Topan played key roles in the oversight of sequencing facilities, Suz Bateson and Andrea Brauner assisted with the figures, while Sujeevan Ratnasingham aided on the informatics front. This research was supported by NSERC, by Genome Canada through the Ontario Genomics Institute and by the Gordon and Betty Moore Foundation.
References
- Buhay J. E.2009‘COI-like’ sequences are becoming problematic in molecular systematic and DNA barcoding studies. J. Crust. Biol. 29, 96–110 (doi:10.1651/08-3020.1) [Google Scholar]
- deWaard J. R., Ivanova N. V., Hajibabaei M., Hebert P. D. N.2008Assembling DNA barcodes: analytical protocols. In Methods in molecular biology: environmental genetics (ed. Martin C.), pp. 275–293 Totowa, NJ: Humana Press; [DOI] [PubMed] [Google Scholar]
- Hajibabaei M., Janzen D. H., Burns J. M., Hallwachs W., Hebert P. D. N.2006DNA barcodes distinguish species of tropical Lepidoptera. Proc. Natl Acad. Sci. USA 103, 968–971 (doi:10.1073/pnas.0510466103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handfield D., Handfield L.2006A new species of Plusia (Lepidoptera: Noctuidae) from North America. Can. Entomol. 138, 853–859 (doi:10.4039/N06-041) [Google Scholar]
- Hebert P. D. N., Cywinska A., Ball S. L., deWaard J. R.2003Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B 270, 313–321 (doi:10.1098/rspb.2002.2218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickerson M. J., Meyer C. P., Moritz C.2006DNA barcoding will often fail to discover new animal species over broad parameter space. Syst. Biol. 55, 729–739 (doi:10.1080/10635150600969898) [DOI] [PubMed] [Google Scholar]
- Itô Y., Warren L. O.1973Status of black-headed and red-headed types of Hyphantria cunea (Drury) (Lepidoptera: Arctiidae): biology of two types and results of crossing experiment. Appl. Entomol. Zool. 8, 157–171 [Google Scholar]
- Levy H. C., Garcia-Maruniak A., Maruniak J. E.2002Strain Identification of Spodoptera frugiperda (Lepidoptera: Noctuidae) insects and cell line: PCR-RFLP of cytochrome oxidase subunit I gene. Fla Entomol. 85, 186–190 [Google Scholar]
- Mitchell A.2008DNA barcoding demystified. Aust. J. Entomol. 47, 169–173 (doi:10.1111/j.1440-6055.2008.00645.x) [Google Scholar]
- Savolainen V., Cowan R. S., Vogler A. P., Roderick G. K., Lane R.2005Towards writing the encyclopedia of life: an introduction to DNA barcoding. Phil. Trans. R. Soc. B 360, 1805–1811 (doi:10.1098/rstb.2005.1730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. A., Woodley N. E., Janzen D. H., Hallwachs W., Hebert P. D. N.2006DNA barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae). Proc. Natl Acad. Sci. USA 103, 3657–3662 (doi:10.1073/pnas.0511318103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. A., Rodriguez J. J., Whitfield J. B., Deans A. R., Janzen D. H., Hallwachs W., Hebert P. D. N.2008Extraordinary diversity of parasitoid wasps exposed by iterative integration of natural history, DNA barcoding, morphology and collections. Proc. Natl Acad. Sci. USA 105, 12 359–12 364 (doi:10.1073/pnas.0805319105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland of Houndswood. Systematics and taxonomy: follow-up. House of Lords; 2008. Science and Technology Committee. [Google Scholar]
- Zhang A. B., He L. J., Crozier R. H., Muster C., Zhu D.In press Estimating sample sizes for DNA barcoding. Mol. Phylogen. Evol. (doi:10.1016/j.ympev.2009.09.014) [DOI] [PubMed] [Google Scholar]