Abstract
Phototransduction in retinal rods is one of the most extensively studied G-protein signaling systems. In recent years, our understanding of the biochemical steps that regulate the deactivation of the rod’s response to light has greatly improved. Here, we summarize recent advances and highlight some of the remaining puzzles in this model signaling system.
Major Features of Rod Phototransduction
The absorption of photons by the G-protein-coupled receptor (GPCR), rhodopsin, in the outer segments of retinal rod photoreceptors activates a series of biochemical reactions, called the phototransduction cascade, which generates the electrical response to light and thus mediates the first steps in vision. The understanding of the molecular basis of phototransduction and its regulation has greatly expanded with the application of mouse gene-targeting techniques, which made it possible to knock out or otherwise perturb the phototransduction machinery of intact photoreceptors. With the adaptation of suction electrode recording to mouse rods in the mid-1990s in Denis Baylor’s laboratory, it became feasible to assess how the photoresponse is affected by deletions, mutations, and transgenic overexpression of proteins thought to regulate the cascade. Many lines of mice in which phototransduction proteins have been knocked out or mutated have been characterized physiologically; some of these are given in Table 1. The goal of this review is to summarize recent findings in the deactivation of this prototypical G-protein cascade and to articulate several remaining questions. We begin by a brief introduction to the activation and deactivation steps in phototransduction and then focus on the quantitative physiological measures that provide kinetic insights into the biochemical events underlying response recovery in living photoreceptors, as exemplified by mouse rods.
Table 1.
Mouse genetic manipulations affecting rod phototransduction deactivation and response kinetics
| Gene | Refs. | Protein | Perturbation | Phenotype |
|---|---|---|---|---|
| Sag | 129 | Arrestin 1 | Knockout | Delayed final recovery |
| 10 | Transgenic | Restored knockout | ||
| 110 | Mutated transgenic | Restored knockout | ||
| Arr3/Arr4 | 14 | Cone arrestin | Transgenic in Arrl ko rods | Incomplete rescue |
| Grk1 | 10, 17 | Rhodopsin kinase | Knockout | Long-lasting step-like responses |
| 59 | Transgenic overexpressor | None | ||
| 15 | Chimera | Slower recovery | ||
| Rho | 13, 64 | Rhodopsin | Underexpression | Faster kinetics - diffusion and volume effects |
| 19 | COOH-terminal deletion | Long-lasting step-like responses | ||
| 121 | Palmitoylation deletion | Shorter integration time | ||
| Rcvn | 15, 68 | Recoverin | Knockout | Shorter integration time |
| Rgs9 | 16 | RGS9-1 | Knockout | Slow, graded recovery |
| 72 | DEPless transgenic | Slow recovery like RGS9 ko | ||
| 71 | RGS9-2 | Transgenic | Faster recovery to very bright flashes | |
| Gnb5 | 58 | Gβ5-L | Knockout | Slow recovery like RGS9 ko |
| Rgs9bp | 54 | R9AP | Knockout | Slow recovery like RGS9 ko |
| 15, 59 | Transgenic overexpressor | Much faster τrec and τD | ||
| Gucy2e | 3, 131 | GC1 | Knockout | Increased sensitivity |
| Gucy2f | 3 | GC2 | Knockout | Normal dark-adapted rod responses |
| Double KO (GC1 and GC2) | No photoresponses; abnormal trafficking | |||
| Guca1a | 11, 76 | GCAP1 | Double KO (GCAP1 and GCAP2) | Large, slow responses |
| 46 | Transgenic | Restores rod recovery in double ko | ||
| Guca1b | 69 | GCAP2 | Knockout | Slow recovery; saturation in dim light |
| 76 | Transgenic in double GCAP | Like GC1 ko | ||
| knockout | ||||
| Pde6g | 115 | PDEγ | Knockout | Photoreceptor degeneration |
| 116 | Overexpression | Reduced gain; faster recovery in RGS9 ko background | ||
| 117, 127 | Phosphorylation site mutants | Modulated recovery by background light | ||
| 114 | W70A mutation | Reduced sensitivity and slower recovery |
Activation reactions are highly amplifying
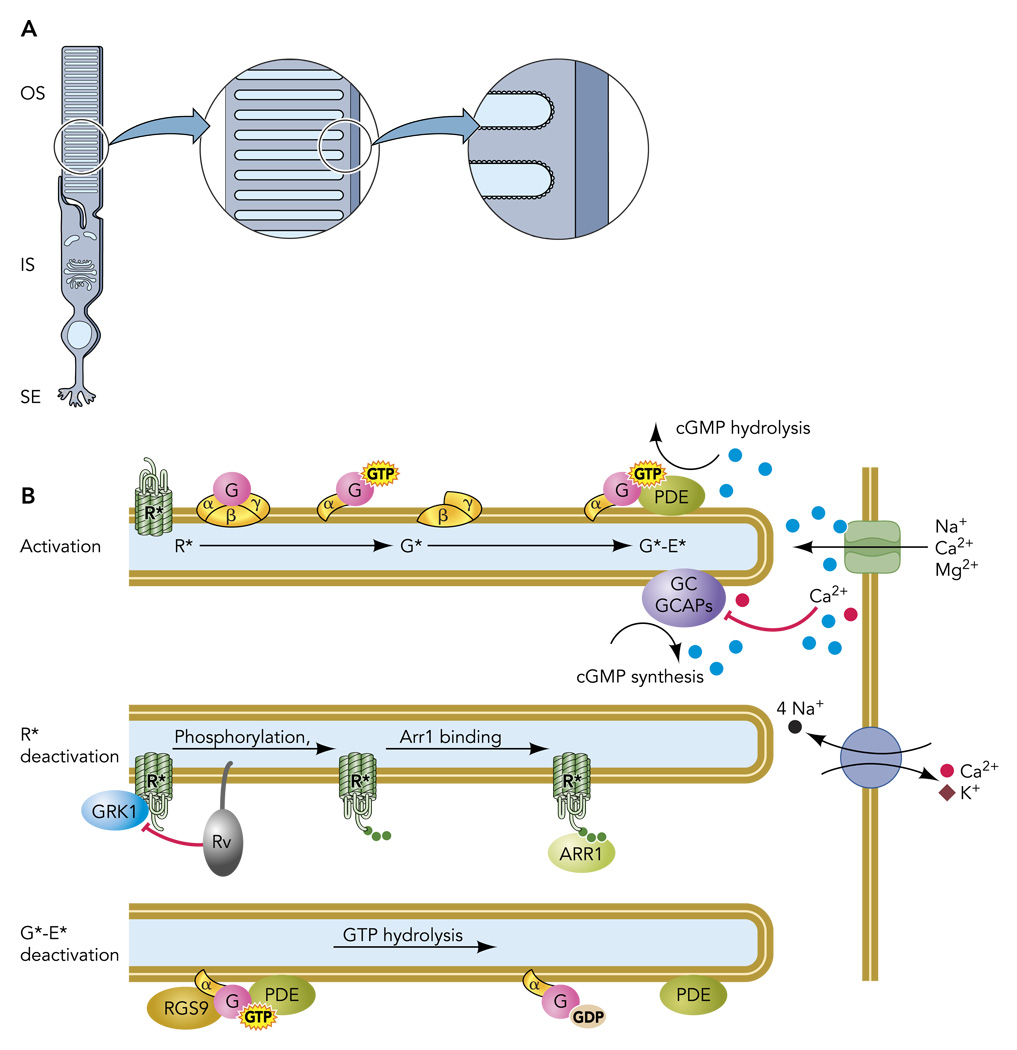
In rods, the molecular machinery of the cascade is concentrated in the outer segment, a subcellular reaction chamber containing several thousand square microns of highly structured, protein-laden membranes, the disc stack (FIGURE 1A). The biochemical events that initiate signaling are known in detail (FIGURE 1B): the absorption of a photon isomerizes the 11-cis retinal chromophore of rhodopsin (120), triggering a conformational change to a catalytically active state [metarhodopsin II (R*)] within milliseconds. R* catalyzes GDP/GTP exchange on multiple copies of the heterotrimeric G-protein transducin. Each activated transducin alpha subunit (Gtα-GTP; hereafter G*), binds to the gamma subunit of the cGMP phosphodiesterase (PDEγ), relieving its inhibition of the PDE6 catalytic subunits (50, 122); the activated transducin-PDE6 complex (G*-E*) hydrolyzes cGMP. The resulting fall in the cGMP concentration causes the cGMP-gated channels in the plasma membrane to close (21, 51), leading to membrane hyperpolarization that reduces the rate of glutamate release onto second-order retinal neurons. For a more detailed, historically developed perspective on the activation reactions, see other reviews (e.g., Refs. 1, 99).
FIGURE 1. The rod photoreceptor and the disc-associated proteins that mediate the response to light.
A: rod photoreceptors are highly compartmentalized cells, with most organelles retained in the inner segment (IS) and synaptic ending (SE). The specialized outer segment (OS) is filled with a stack of intracellular membranous discs that house the transduction machinery. B: schematic of a portion of three disc membranes and plasma membrane shown in A, illustrating activation and deactivation reactions.
The ability of rods to signal the absorption of single photons arises from amplification contributed by three primary molecular or biophysical mechanisms: 1) G-protein activation by R*; 2) hydrolysis of cGMP by G*-E*; 3) cooperative gating of the cGMP-gated channels. After photon capture, G-protein activation proceeds at a rate of ~150 s−1 per R* in amphibian rods and more than twice that in mammalian rods (1, 42). PDE activation per R* is thought to proceed at very nearly the same rate (63). These high rates of G-protein and effector activation result in part from the rate constants of the reactions but also from the high density of G proteins (~3,000 µm−2) and PDE molecules (~80 µm−2) in the disc membrane. At the next stage of amplification, each G*-E* hydrolyzes cGMP at a rate near the diffusion limit (kcat/Km ~ 108 M−1·s−1) (63), causing the local cGMP concentration to fall quickly. An important biophysical contribution to amplification arises from the cytoplasmic reaction volume: the same number of G*-E* in a smaller outer segment more rapidly changes the cGMP concentration than it would in a larger volume. This reaction-volume effect provides a basis for the > 100-fold higher amplification constant of slender mammalian vs. hefty amphibian rods (99). Finally, the cooperative activation of the cGMP-gated channels (Hill coefficient of ~3; Ref. 105) causes a given fractional change in Cgmp concentration to produce a threefold larger fractional change in the inward current. In summary, the amplification needed for high sensitivity is determined primarily by the two enzymatic amplifiers, the activated GPCR, R*, and the activated phosphodiesterase complex, G*-E*, with a further boost from the cGMP channel cooperativity.
Many deactivation reactions are required for normal recovery of the photoresponse
Because of the high amplification of phototransduction, relatively modest light intensities could easily drive rods into saturation, i.e., close all of the cGMP-gated channels were it not for timely deactivation. If the active lifetimes of R* and G*-E* are prolonged, this saturation occurs at lower light intensities, and when not in saturation a rod with a prolonged response is less able to signal rapid changes in illumination. Therefore, timely recovery of the photoresponse is critical for maintaining sensitivity in steady light and for signaling increments and decrements of light intensity. In rods, the timing of response recovery is determined by regulatory proteins that act to 1) speed deactivation of R*, 2) speed de-activation of G*-E*, and 3) accelerate the rate of cGMP synthesis. In humans, mutations that cause defects in each of these three categories of biochemical reactions cause 1) nightblindness (2, 20, 30, 40, 130), 2) bradyopsia (39, 77, 86), and 3) early onset Leber’s congenital amaurosis and cone-rod dystrophy (48), respectively.
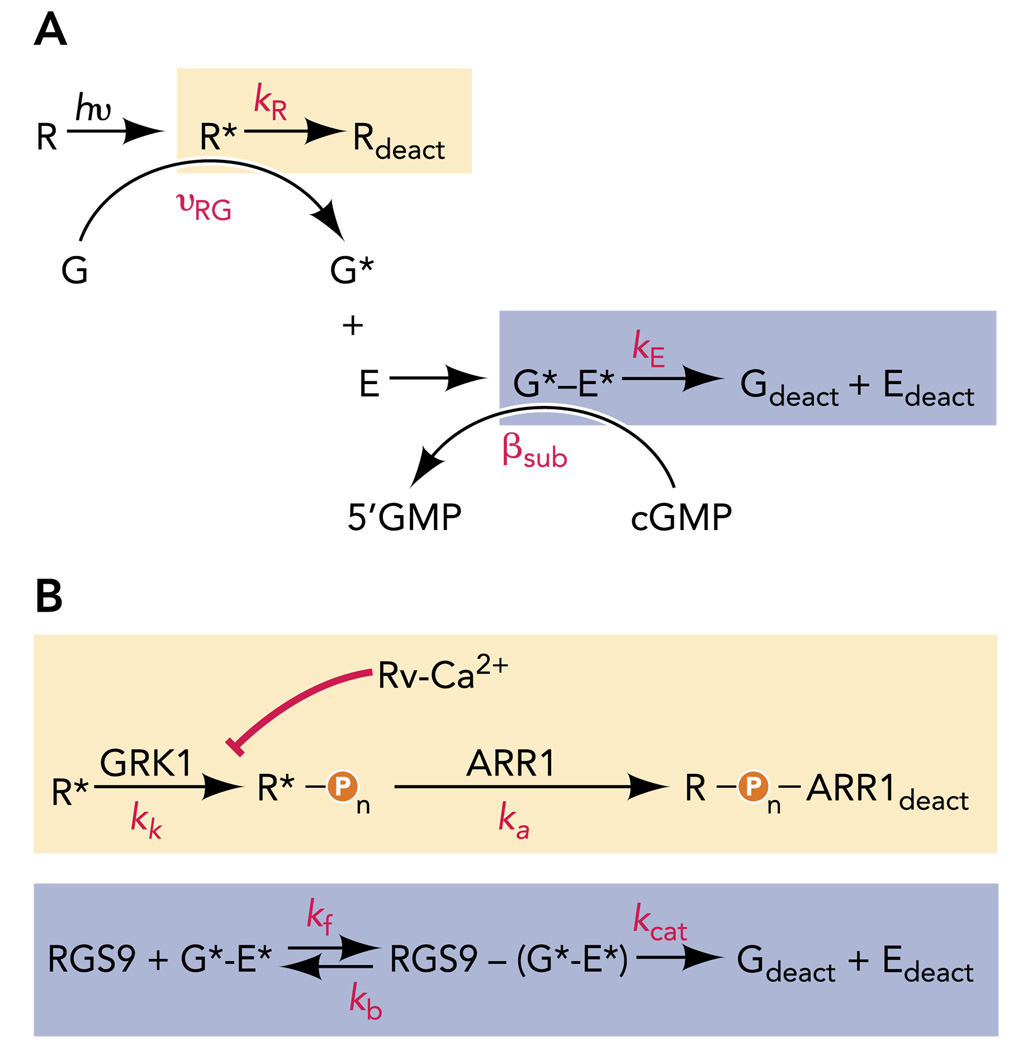
A vast literature has revealed rich details of the biochemical reactions that govern R* and G*-E* deactivation and cGMP synthesis, and parallel physiology experiments have sought to understand the relative rates of R* (kR) and G*-E* (kE) deactivation in living rods in this context (FIGURE 2). R* activity persists until it is quenched by two successive reactions: phosphorylation by rhodopsin kinase (GRK1) and the binding of the protein arrestin (Arr1; Refs. 61, 126). Likewise, G*-E* activity persists until the GTP bound to Gtα is hydrolyzed to GDP, a reaction that is speeded by RGS9-1, a retina-specific regulator of G-protein signaling (41), in a complex with Gβ5-L (70) and the membrane anchoring protein R9AP (47). Finally, guanylate cyclase activating proteins, or GCAPs (27, 36, 95), activate guanylate cyclases (GC-1 and GC-2) when intracellular calcium levels fall during the light-induced decline in inward current. Understanding the contributions of these biochemical steps to normal physiology requires manipulating the reactions in intact rods and quantitatively evaluating the kinetic consequences for the light response.
FIGURE 2. Kinetic schemes of phototransduction deactivation.
A: standard schemes for R* and G*-E* deactivation, each assuming a single rate constant (kR or kE, respectively) for the shut-off reactions. B: expanded schemes for the deactivation events boxed in A, which include rate constants for kinase (kk) and arrestin (ka) actions known to be essential for R* deactivation and the forward (kf), reverse (kb), and catalytic (kcat) rate constants needed to describe the RGS9-catalyzed GTP hydrolysis needed for G*-E* deactivation.
The Recovery of the Rod Photoresponse is Rate-Limited by a Single Process
Normal response kinetics need to be defined in native cells
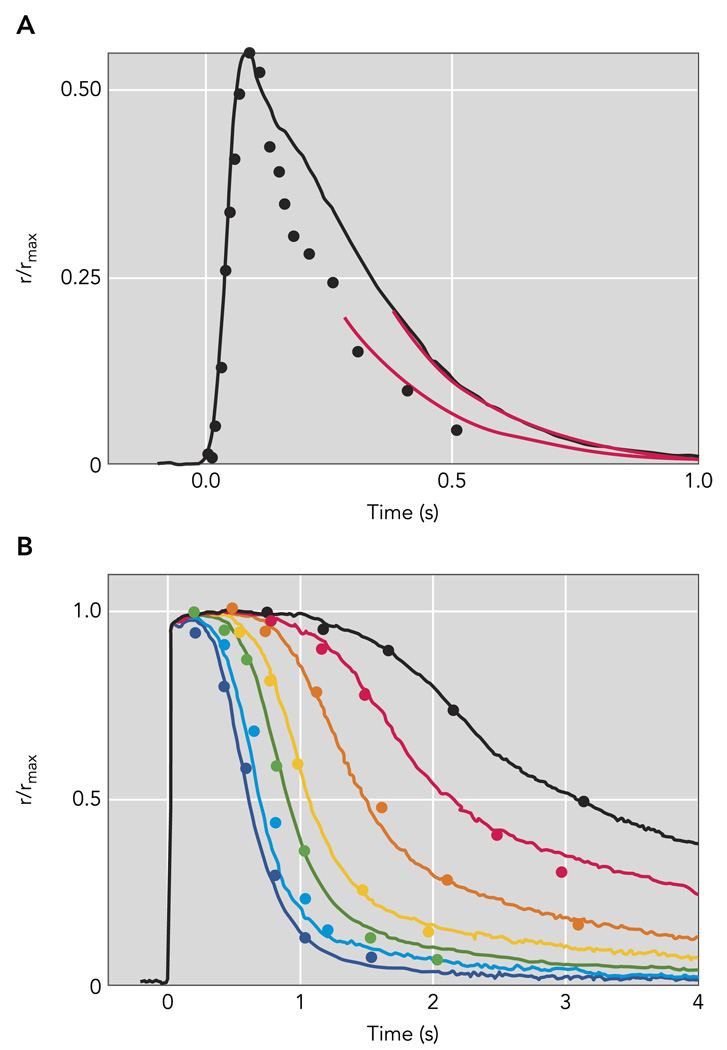
The compartmentalization of phototransduction in the outer segment permits the light-induced changes in cGMP-gated current to be readily recorded by drawing an individual outer segment of an intact rod into a highly polished suction pipette electrode. The primary advantage of suction electrode recordings is that the integrity of the cell membrane and consequently the natural protein concentrations and their subcellular localizations are maintained. In contrast, in vitro assays that rely on cellular homogenates or reconstituted enzymes do not reproduce the more rapid kinetics observed in intact cells. The validity of using suction electrode recordings to determine deactivation kinetics from acutely isolated retinal slices is demonstrated by the remarkable agreement of the sensitivity and kinetics of rod responses recorded in Locke’s solution at 37°C to those recorded in vivo with electroretinographic methods (FIGURE 3).
FIGURE 3. Kinetics of responses recorded from mouse rods using suction electrodes closely match the kinetics of in vivo electroretinographic (ERG) recordings from mice.
A: rod photoresponse extracted from paired-flash ERG experiments of Hetling and Pepperberg (symbols; Ref. 43; Figure 5B); flash produced a response of ~55% the saturating a-wave amplitude corneal ERG a-waves (symbol). The black trace is a population average half-maximal response from 15 c57Bl/6 mouse rods recorded with suction electrodes; the flash strength was 48 photons/µm2. Red curves are exponential functions, both with a 202-ms time constant. B: paired-flash mouse a-wave data of Lyubarsky and Pugh (1996) for a series of flashes of increasing strength that completely suppressed the a-wave (symbols) are compared with population average suction electrode recordings (smooth traces) from 15 mouse rods made in bicarbonate-buffered Locke’s solution at 37°C to flashes that ranged from 580 to 25,200 pho-tons/µm2 by factors of 2. The average maximal response amplitude for the suction electrode recordings in A and B was 16.5 pA; the average effective collecting area was 0.50 µm2.
The membrane currents recorded with suction electrodes provide a real-time readout of the cGMP concentration, owing to the rapid gating and voltage independence of the cGMP-gated channels at physiological membrane potentials. The time course of the cGMP concentration in turn can be further analyzed to derive the time course of PDE activity (44, 99). Conversely, theoretical modeling of the underlying biochemical reactions can be used to predict changes in cGMP concentration and thereby the time course of the photoresponse (e.g., Ref. 38).
Recovery time constant of dim and bright flash responses
Since the seminal investigation of Hodgkin and Nunn (44), it has been known that rod responses to both dim and moderately bright flashes recover to baseline along a time course that is exponential at adequately late times (83). For mouse rods, this exponential “tail phase” has a time constant of recovery (τrec) of ~200 ms.
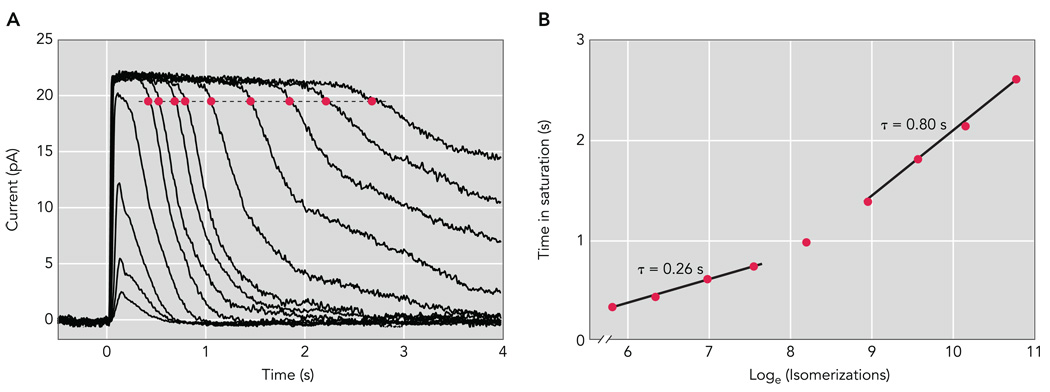
The exponential recovery can also be revealed through the analysis of saturating responses (FIGURE 4). The time that a response to a bright flash remains in saturation (Tsat; red circles in FIGURE 4A) is directly proportional to the logarithm of the flash strength, a graphical analysis colloquially termed a “Pepperberg plot” (FIGURE 4B; Ref. 83). The slope of the Tsat vs. flash strength relationship in this semilog plot measures the time constant τD of the slowest kinetic step in photoresponse recovery, the so-called “dominant recovery time constant.” For the mouse, τD is ~250 ms for flashes producing up to ~1 R* per disc face (loge isomerizations = 7.5 in FIGURE 4B). This value for τD is close to τrec determined from the tail-phase recovery of dim flash responses and the time constant of recovery measured in vivo (FIGURE 3). The similarity of τrec and τD in normal rods and in many genetic lines in which deactivation mechanisms have been perturbed suggests that the same biochemical step rate-limits recovery over an ~1,000-fold range, from a single photon up to ~1 R* per disc face.
FIGURE 4. Dark-adapted responses of a wild-type mouse rod.
A: population average response family from a Sv129 mouse rod. Brief (10 ms) flashes were delivered at t = 0 s and ranged in strength from 7.78 to 85,600 photons/µm2 by factors of 2–4. B: Pepperberg plot of wild-type mouse rods. The time that a bright flash response remained in saturation (Tsat) is plotted as a function of the natural logarithm of the flash strength in photoisomerizations per flash (R*). The slopes of the fitted lines, 0.26 and 0.80 s, give the dominant time constants of recovery over the respective range of flash strengths.
Hypotheses regarding the mechanism underlying τD
The molecular mechanism rate-limiting response recovery has been intensely investigated and debated since 1992. Theoretical analysis of the cascade reactions revealed that the exponential recovery could arise from either of two mechanisms, the deactivation of the G*-E* complex via GTP hydrolysis or the decay of R* activity (Ref. 83; FIGURE 2A). In the first case, R* would deactivate relatively rapidly, so that at the “long times” of the response tail phase the brief pulse of R* activity would have decayed completely, leaving only the slower deactivation of the G*-E* complexes to determine the recovery time course. In the second case, R* would continue to activate G* during the response tail phase, but the lifetimes of the resultant G*-E* complexes would be relatively short, so that the response recovery would track R* deactivation.
Early experiments aimed at identifying the biochemical step underlying τD characterized its calcium dependence. Experiments of Lyubarsky et al. (67) showed that τD was identical whether a rod’s internal calcium was clamped to its dark level or free to change during the light response. This result favored the identification of τD with deactivation of G*-E*, which has no known calcium dependence. It also complemented earlier work by Matthews (73, 74), which had uncovered a short-lived calcium dependence of deactivation well before recovery of the current began; this latter dependence was hypothesized to be calcium-dependent regulation of R* deactivation and to be mediated by the calcium-binding protein recoverin (52). In contrast, other experiments led authors to suggest R* lifetime to be the rate-limiting step in recovery (98, 104), whereas still others favored the contrary conclusion that hydrolysis of GTP to GDP by G*-E* is rate limiting (57, 83, 106). These disagreements propagated through the analysis of many features of photoreceptors, including the molecular basis of reproducible single photon responses (38) and the molecular basis of the functional differences between rod and cone photoreceptors (24, 132). For many years, the principal difficulty in settling the controversy was that most experimental manipulations abnormally prolonged deactivation, either by diluting endogenous regulators in biochemical experiments or by slowing the time course of physiological responses through pharmacological or genetic loss-of-function manipulations. What was needed was a targeted molecular manipulation that dramatically sped the rate-limiting step in deactivation and thus shortened the dominant time constant of recovery. Thus, if the targeted molecular mechanism were rate-limiting, speeding it up would shorten τD, whereas were it not rate-limiting, further speeding it would not alter τD.
RGS9 Rate Limits the Recovery of the Normal Photoresponse
The shortening of the dominant recovery time constant was finally achieved in experiments with rods that transgenically overexpressed R9AP, which in turn led to overexpression of the entire RGS9 complex (59). As mentioned above, the RGS9 complex (RGS9-1/Gβ5-L/R9AP) resides in the disc membrane (FIGURE 1), where it stimulates the GTPase activity of the G*-E* complex. Comparison of recovery kinetics for both dim and saturating responses (measurements of τrec and τD, respectively) showed that overexpression of the RGS9 complex speeds response recovery in a dose-dependent manner (59). In rods in which RGS9-expression was approximately fourfold higher than wild-type levels, the recovery time constant for both dim and bright flash responses was sped roughly threefold, from 250 to 80 ms (59). In a more recent study, roughly sixfold over-expression of the RGS9 complex further reduced both time constants to 54 ms (15). These results establish that, for flashes activating from 1 to 4,000 R* in mouse rods, the rate-limiting step in recovery is RGS9-catalyzed GTP hydrolysis by the G*-E* complex.
To understand how the rate of G*-E* deactivation can be controlled by the RGS9 expression level, the RGS9-catalyzed GTP hydrolysis reaction needs to be considered in a framework of enzyme kinetics (FIGURE 2B). Generally, when an enzyme with Michaelis constant Km eliminates a substrate S, the terminal phase of substrate removal (S < < Km) is an exponential decay with time constant 1/(Vmax/Km), where Vmax = kcat[Enzyme], i.e., the product of enzyme concentration and the turnover number of the enzyme. Remarkably, these simple ideas apply directly to RGS9-catalysis of G*-E* deactivation, with RGS9 identified as the enzyme and G*-E*as its substrate.
Incorporating a Michaelis scheme for RGS9 into a standard model of phototransduction (gray boxes in FIGURE 2) revealed that the dominant recovery time constant obeys τD = 1/(Vmax/Km) = 1/{(kcat [RGS9])/Km} over most of the 20-fold range of RGS9 expression levels in the mice investigated by Krispel et al. (12). Thus the concentration dependence of τD on RGS9 expression level is a consequence of classic enzyme kinetics and reveals how the dominant time constant of the GPCR cascade can be dramatically tuned by changing the expression level of a single gene, in this case R9AP driving expression of the RGS complex.
Mechanism of slowed recovery for flashes producing more than 1 R* per disc face
A mouse rod outer segment comprises ~800 membranous discs with 1,600 disc faces. Recovery slows dramatically when flashes produce in excess of 4,000 R* or ~2R* per disc face, as shown by a sharp break in the slope of the Pepperberg plot (FIGURE 4B). Recent work suggests that, for flashes producing 4,000–160,000 R*, the rate-limiting step is RGS9-catalyzed GTP hydrolysis of G* that is uncomplexed with E*, i.e., G* produced in excess of total PDE (71). Evidence for this came from experiments on mouse rods that expressed a brain-specific isoform of RGS9, RGS9-2, that does not require PDEγ for stimulating GTP hydrolysis of G*.
Unresolved questions about the RGS9 reaction in rods and cones
The shortest recovery time constant observed thus far with RGS9 complex overexpression is ~50 ms, corresponding to an overall GTPase rate constant of 1/τD =20 s−1 (15). Could τD be reduced further in rods with still higher expression levels? Theoretical analysis of the data of Krispel et al. (59) suggests that indeed should be the case: RGS9-catalyzed hydrolysis of G*-E* likely has a kcat in situ exceeding 50 s−1 (12), a turnover generally consistent with biochemical data (4, 65). Thus the τD of rods could in theory become as short as 1/50 s−1 or 20 ms, assuming other processes, like R* deactivation, are sufficiently fast. Likewise, could the higher expression level of the RGS9 complex in cones underlie their more rapid deactivation (132)? To date, the rates of cone opsin deactivation and G*-E* deactivation in cones remain undetermined.
Questions remain about the molecular steps that immediately follow RGS9-catalyzed GTP hydrolysis and, in particular, about the role of PDEγ in the G*-E* complex. For example, what are the structural changes that drive the dissociation of the G*-E*-RGS9 complex on GTP hydrolysis and thus permit the high turnover number of the RGS9 reaction? Furthermore, how rapidly does PDEγ re-inhibit the PDEαβ catalytic subunits? It is unknown whether the binding of G* to PDEγ causes PDEγ to completely dissociate during activation or whether it merely opens up the catalytic site while remaining bound. Experiments in truncated toad rods showed no change in cellular dark noise associated with dialysis of exogenous PDEγ into the rod, leading to the conclusion that PDEγ does not completely dissociate (103). However, in mice, overexpression of PDEγ can decrease the amplitude and speed recovery of rods lacking RGS9, suggesting that the exogenous PDEγ can indeed compete with the endogenous protein (116). The extent of PDEγ dissociation may also depend on the adaptational state of the cell: biochemical studies have suggested that cGMP-dependent changes in the affinity of the GAF domain of PDEγ may indeed cause complete dissociation under some conditions, such as during light adaptation (23). Additional evidence for this change in affinity with light adaptation stems from experiments showing steady light can alter the phosphorylation state of PDEγ, which in turn alters the kinetics of the flash response (117, 127). Thus many questions remain about the role of PDEγ in the regulation of G*-E* activity in both dark and light adapted conditions.
Evidence for Rapid Deactivation of the GPCR Rhodopsin
Steps in R* deactivation necessary for normal recovery in intact rods
Following activation by light, R* becomes phosphorylated by rhodopsin kinase (GRK1; Refs. 17, 66, 87, 91, 108, 109, 112, 126). The sites of phosphorylation are clustered at the COOH-terminus of rhodopsin, and either truncating the COOH-terminus entirely (19) or mutating these COOH-terminal serine and threonine residues to alanines (28, 75) leads to abnormally slow response recovery. Indeed, responses of rods expressing either the COOH-terminal rhodopsin truncation mutant or those expressing a rhodopsin mutant lacking all six of the potential phosphorylation sites are indistinguishable from those of rods in which GRK1 has been knocked out (17). Furthermore, biochemical studies suggest that at least three phosphoryl groups on rhodopsin’s COOH-terminus are required to trigger high-affinity binding of arrestin (Arr1; Refs. 31, 33, 34, 118, 125, 129), the subsequent step that fully quenches R*’s activity and prevents further activation of G-protein subunits (129). Multiple phosphorylation and Arr1 binding are likewise required for normal recovery, since rhodopsin mutants with fewer than three phosphorylation sites give responses that are abnormally slowed (28, 75).
R* deactivation is normally rapid
One of the major consequences of the identification of G*-E* deactivation as the rate-limiting step of recovery is that it necessarily follows that R* deactivation is still faster. However, the question remains of exactly how much faster. The dominant time constant of recovery for both dim and saturating responses (τrec and τD) provide strong constraints on the lifetime of R*: in the highest RGS9 overexpression attained thus far (approximately sixfold), the recovery time constant, and thus the upper limit for R* lifetime in dark-adapted rods, is 54 ms (15). Such a short R* lifetime is consistent with manipulations of R* deactivation that cause little or no change in τD or τrec, including overexpression of GRK1 (59, 123), knocking out the calcium-dependent inhibitor of GRK1, recoverin (68), and underexpression of Arr1 (10, 32, 129). Nonetheless, it is puzzling that these perturbations have little effect on the dim flash response, because changes in R* lifetime should produce corresponding changes in the total number of G*-E* and thus would be expected to alter the response amplitude. One possible resolution to this puzzle is that calcium feedback to guanylate cyclase dampens amplitude variation that would otherwise arise from different R* lifetimes.
Measuring R* lifetime
Given that R* deactivation is much more rapid than G*-E* deactivation, and the apparent insensitivity of the early phases of the response to changes in R* lifetime, how can one determine R* lifetime in the living rod? In a recent study, Gross and Burns (32) observed that a 50% decrease in Arr1 expression increased the absolute times that responses to strong flashes remained in saturation but did not change τD. This result is consistent with a lengthening of R* lifetime, without a change in τD, which remained determined by G*-E* deactivation. When Arr1 underexpression was accompanied by an increase in RGS9-overexpression (Arr1+/−RGS9-ox rods), an identical increase in the Tsat values was observed with no change in the 80-ms τD, indicating that even the slowed R* deactivation remained faster than 80 ms.
Gross and Burns also noted that the prolonged R* lifetime lengthened the apparent τrec in Arr1+/−RGS9-ox rods. As noted previously, normally τrec ≈ τD, indicating that one and the same process rate limits recovery from both dim and bright flashes. The reason τrec is longer than τD in Arr1+/−RGS9-ox rods appears to be straight-forward: τrec only reflects the slower of τR and τE if the two time constants are sufficiently different from each other and τrec is extracted from the tail phase at sufficiently late times (31). When the two time constants become similar, as in Arr1+/−RGS9-ox rods, the “sufficiently late time” for dominance would only be reached well after the dim flash response has completely recovered. In contrast, τD in Arr1+/−RGS9-ox rods is extracted from responses at later times, after dominance has occurred (32).
Gross and Burns used the increase in the Tsat values for bright flash responses and the increase in τrec for dim flash responses to extract the rate of phosphorylation and Arr1 binding under normal conditions and conditions of reduced Arr1 expression. Using a scheme of photoresponse deactivation consistent with established biochemistry, they found that R* deactivation normally follows a single exponential decay with a time constant of ~40 ms, suggesting that phosphorylation and Arr1 binding proceed at effectively the same rate in wild-type rods. However, in rods expressing 50% of normal levels of Arr1, a single exponential time course for R* deactivation could not explain the experimental results. Rather, reduced Arr1 expression resulted in data best described by an R* decay with two time constants in which the first time constant was the same as for wild-type and represented phosphorylation by GRK1 (τK = 40 ms), whereas the second time constant was slower and represented slowed Arr1 binding (τa+/− = 67 ms). Given the association rate constant for Arr1 binding of ~1 µM−1·s−1 (101), the calculated binding rate for Arr1 in normal rods is consistent with an effective Arr1 concentration of 28 µM, which is similar to previous biochemical estimates of Arr1 monomer in bovine rods (35).
R* deactivation is speeded during light adaptation
Rhodopsin phosphorylation is inhibited by the protein recoverin in a calcium-dependent manner in vitro (18, 52, 55), providing a mechanism by which rhodopsin lifetime can be regulated by intracellular calcium levels. As discussed above, calcium clamp experiments in salamander rods show that τD is not calcium sensitive (83), whereas another faster mechanism, presumably R* deactivation, is dependent on the calcium concentration at the time of the flash (73, 74, 81). Consistent with the idea that R* deactivation is normally very rapid, mouse rods lacking recoverin have the same τD as wild-type rods. However, the time that a bright flash response remains in saturation is dramatically reduced in rods lacking recoverin, consistent with a shorter R* lifetime. Recoverin knockout rods also fail to exhibit the “step-flash” effect, the shortening of response duration in the presence of steady light (68), supporting the notion that recoverin prolongs R* lifetime and that this prolongation is relieved during adaptation. Very strong evidence for the role of recoverin in directly controlling R* lifetime was provided by a recent study by Chen et al. (15). These authors made use of the transgenic overexpression of RGS9 to speed G*-E* deactivation and, in addition, slowed R* lifetime by expressing a GRK1 chimera with reduced enzymatic activity. Together, these manipulations made the R* lifetime rate-limiting for recovery. In these rods in which R* was rate limiting, background light shortened τD, but only in the presence of recoverin. The remarkable conclusion to be drawn from the cumulative results of all of these studies is that, in dark-adapted rods, recoverin prolongs R* lifetime by inhibiting GRK1, resulting in the “long” 40-ms R* lifetime. When calcium falls during light adaptation, this inhibition is relieved, and the lifetime of R* is shorter still. Precisely how much shorter remains to be determined.
Unresolved issues in R* deactivation
Given the importance of R* deactivation for maintaining the balance between signal amplification and rapid, reliable recovery, understanding the molecular details of R* deactivation in intact rods remains one of the most fundamental unresolved issues in phototransduction. How can R* deactivation, which requires phosphorylation by GRK1 and the binding of Arr1, occur within ~40 ms and be faster still when recoverin’s inhibition is relieved during light adaptation?
How many phosphoryl transfers occur during deactivation of R* in the intact rod?
Biochemical studies of light-dependent rhodopsin phosphorylation have reported widely varying stoichiometries. Although many in vitro studies have reported stoichiometries in excess of six phosphates per R* (e.g., Refs. 80, 124, 126), in vivo studies have reported that only one or a few phosphates get incorporated (e.g., Refs. 49, 88), at least on a time scale comparable to the time course of the flash response (53). The individual contributions of phosphorylation and Arr1 binding to R* deactivation in intact, normal rods have been difficult to determine because phosphorylation not only increases rhodopsin’s binding affinity for Arr1 (31, 33, 60, 118) but also itself reduces R* activity (94, 129).
Control of GRK1 activity
Following photoactivation of rhodopsin to R*, GRK1 itself can become “activated” by R*, leading to the phosphorylation of non-photoexcited rhodopsins, a process referred to as “high-gain phosphorylation” (6, 8, 25, 26, 78, 79, 92, 109). In addition, autophosphorylation of GRK1 (62) lowers its affinity for phosphorylated R* (9, 102), and mutagenesis of GRK1’s autophosphorylation sites increases phosphorylation of rhodopsin in vitro (93), supporting the idea that autophosphorylation normally limits the extent or rate of rhodopsin phosphorylation by triggering dissociation. Thus it may be the dissociation of GRK1 from R* that determines the timing of arrestin binding. The activity of GRK1, as well as its counterpart in many mammalian cones, GRK7, is reduced by phosphorylation by cAMP-dependent kinase (45), and in cones dephosphorylation of GRK7 accompanies light exposure, suggesting that GRK7 activity is regulated during light adaptation (90). As yet, there have been no physiological investigations of GRK1 modulation by phosphorylation.
Despite the rapid rate of R* deactivation in rods, there are suggestions that the rate of deactivation may be higher still for cone opsins. GRK7 has higher specific activity for rhodopsin than GRK1 in vitro (119). Furthermore, the cone visual pigments have higher rates of spontaneous decay (107) and seem to rely on phosphorylation (85) and arrestin (Arr4; Ref. 84) binding to lesser extents than rhodopsin. Whether cone pigment deactivation is likewise subject to calcium-dependent modulation by recoverin during light adaptation remains unknown, but recoverin is expressed in cones and does bind to GRK7 (113).
Does Arr1 movement into the outer segment alter R* deactivation?
Prolonged steady illumination that cumulatively bleaches only a few percent of all of the rhodopsin molecules is sufficient to cause light-dependent movement of Arr1 into the outer segment, an intriguing phenomenon that has been intensely studied in recent years (e.g., Refs. 82, 89, 96, 111). A drastic increase in total Arr1 concentration could reduce the rate at which R* activates the G-protein transducin by competition, thereby decreasing phototransduction gain and thus decreasing response amplitude. In addition, a higher level of Arr1 could shorten R* lifetime by decreasing the time needed for Arr1 to bind following phosphorylation. Theoretically, this could also affect response amplitude by decreasing the number of G-proteins that any given R* could activate during its (even briefer) lifetime. However, because high-affinity Arr1 binding to R* first requires multiple phosphorylation, increased Arr1 levels presumably could not shorten R* lifetime beyond that determined by the rate of phosphorylation.
Role of Calcium Regulation of cGMP Synthesis in Shaping the Photoresponse
Molecular components of calcium-dependent guanylyl cyclase regulation
The deactivation reactions discussed above are only part of the story: for the photoresponse to recover, cGMP concentration must be restored to its pre-flash levels. In rods, cGMP synthesis is maintained by two guanylyl cyclases, GC1 and GC2 (3), whose activities are regulated by calcium-sensitive, GC-activating proteins or GCAPs (in mouse rods, only GCAP1 and GCAP2). In normal rods, the light-induced decline of cGMP that leads to closure of some of the cGMP-sensitive channels causes a reduction in calcium influx. Because the Na+/Ca2+, K+(NCKX) exchanger continues to extrude calcium, the reduced calcium influx leads to a reduction in intracellular [Ca2+]. This decline in [Ca2+] promotes dissociation of calcium from GCAPs, leading to activation of GC and an increased rate of cGMP synthesis. The increased rate of cGMP synthesis acts to more rapidly restore cGMP levels, opposing the light-induced decline triggered by the G*-E* activity. Loss of either GC1 or GC2 causes little change in rods, indicating functional redundancy at maintaining the physiological level of cGMP (3, 131). However, GCAP1 activates GC1 more efficiently than GC2, and GCAP1 and GCAP2 have slightly different calcium sensitivities, causing them to have different time courses and potencies in regulating cyclase activity in rods (69, 76). As a result, in normal mouse rods, the physiological consequences for calcium feedback activation of cGMP synthesis affect both the time course of the flash response (dynamic feedback) as well as the steady-state level of current in the presence of continuous illumination (static feedback).
Dynamic and static feedback
Calcium-dependent activation of cGMP synthesis during the dim flash response produces a sharply sculpted recovery phase shortly after the peak of the response. Evidence that this sharp initial downstroke just after the peak (the “chop”) in mouse rod responses arises from GC activation comes from the pronounced absence of this feature in rods loaded with the calcium buffer BAPTA (114) and in rods in which both GCAP1 and GCAP2 were knocked out (GCAPs−/−; Refs. 11, 76). The comparison of flash responses of wild-type and GCAPs−/− rods clearly illustrates this sculpting effect: the single photon responses (SPRs) of GCAPs−/− rods begin to diverge from those of wild-type by 40 ms after a flash, peak later, and recover more slowly (11). When only GCAP1 is expressed in rods, photoresponses do show the “chop” (46, 69), whereas rods that express only GCAP2 do not (76). These results indicate that rapid calcium feedback through GCAP1/GC1 sculpts the amplitude and time course of the dim flash response near the peak. However, GCAP2 also shapes the photoresponse dynamically, although at slightly later times, so that the integration time and recovery are affected in mouse rods lacking only GCAP2 (69). These results indicate that both GCAP1 and GCAP2 regulate GC activity, even during the presumably small changes in intracellular [Ca2+] that occur during the SPR. Since the apparent K1/2 of calcium for GCAP2 is ~50 nM (69), the involvement of GCAP2 during the single photon response suggests that the local changes in calcium that are produced during the single photon response are larger and/or faster than what is predicted by calcium imaging of the bulk cytoplasm in response to a bright flash (128). Larger and more rapid changes in calcium might also arise due to the close proximity of the cGMP-sensitive channels and the NCKX exchanger (5). This remains an important unresolved issue in photoreceptor physiology, since the time course of calcium feedback is important for regulating sensitivity, light adaptation, and perhaps even reproducibility of the SPR.
It is precisely the dynamic, regulated feedback to cGMP synthesis that enables photoresponse recovery to track G*-E* deactivation. The power and speed of the GC-GCAP-calcium feedback is such that during the tail phase of the flash response the decaying G*-E* activity is effectively in equilibrium with GC activity so that the photoresponse recovery of normal rods closely tracks the decay of the light-evoked PDE activity (11, 44, 83). In addition, steady-state GC-GCAP feedback plays a crucial role in allowing the rod to operate in higher steady illumination than would be possible in the absence of such feedback (56, 100). In mouse, this effect is ~13-fold; thus steady illumination suppresses a given fraction of the cGMP current of GCAPs−/− rods at 13-fold lower levels than in wild-type rods (11). The loss of GCAP2 causes rods to be more sensitive than normal to steady light and to lose roughly half of this static feedback control that normally extends their operating range in steady light (69). Thus both GCAP1/GC1 and GCAP2/GC2 serve distinct but overlapping roles resulting from their differences in Ca2+ sensitivity.
Efficient Signal Integration: The Single Photon Response
Reproducibility in the framework of fast R* deactivation
The SPR of vertebrate rods has been investigated for more than 30 years and generally found to be “unexpectedly” reproducible. The expectation of high variability arose in part from the realization that deactivation of a single R* would necessarily be stochastic. Historically, much of the research on the reproducibility of the rod SPR has taken place in a framework in which the deactivation of R* was considered rate limiting for recovery. In such a framework, accounting for the observed reproducibility has been argued to require many stochastic steps sequentially diminishing the activity of R*, which would include multiple phosphorylation events and subsequent Arr1 binding (28, 29, 38, 104).
SPR reproducibility remains poorly understood, especially in the context of a short R* lifetime. In an investigation by Bisgena and colleagues (7), it was found that longitudinal diffusion of cGMP contributes spatio-temporal filtering that materially improves predicted reproducibility. In addition, reproducibility similar to that observed experimentally could be achieved with three graded steps in R* deactivation, two phosphoryl transfers by GRK1, followed by Arr1 binding. Thus explaining SPR reproducibility does not require the postulation of a large number of R* deactivation steps, nor that the overall R* lifetime dominates recovery.
Signaling is efficient when R* deactivation is fast
Given that the active lifetime of R* in situ in mouse rods is ~40 ms (32) and that G*-E* deactivation in wild-type rods is normally ~250 ms, the ratio of the average lifetimes of the two enzymatic amplifiers, R* and G*-E*, stands as ~6:1. An interesting feature of this ratio is that it implies that the GPCR signaling is highly efficient (12): at the time to peak of the light response (125 ms), >70% of the E* that have been produced are simultaneously active. In contrast, in a situation in which the two lifetimes are reversed, the E* activity is <20% efficient–thus most of the E* produced would not contribute to signaling but would be “lost” by rapid hydrolysis before the response reaches its maximum.
Overall Summary
The identification of deactivation of the G-protein-effector complex as the slowest step in rod response recovery has resolved a long-standing controversy about the phototransduction cascade in living rods. This resolution has sharpened the focus on other unresolved problems in photoreceptor physiology, including the kinetics and molecular details of the mechanisms underlying the brief (<50 ms) lifetime of photoactivated rhodopsin R* and the molecular and biophysical processes that contribute to the reliability of the single photon response. The resolution has also revealed how the concentration of an RGS, acting through classic enzyme kinetics, can precisely control the principal time constant of a GPCR cascade.
Acknowledgments
The authors are supported by National Eye Institute Grants EY-14047 (M. E. Burns) and EY-02660 (E. N. Pugh, Jr.).
References
- 1.Arshavsky VY, Lamb TD, Pugh EN., Jr G proteins and phototransduction. Annu Rev Physiol. 2002;64:153–187. doi: 10.1146/annurev.physiol.64.082701.102229. [DOI] [PubMed] [Google Scholar]
- 2.Azam M, Collin RW, Khan MI, Shah ST, Qureshi N, Ajmal M, den Hollander AI, Qamar R, Cremers FP. A novel mutation in GRK1 causes Oguchi disease in a consanguineous Pakistani family. Mol Vis. 2009;15:1788–1793. [PMC free article] [PubMed] [Google Scholar]
- 3.Baehr W, Karan S, Maeda T, Luo DG, Li S, Bronson JD, Watt CB, Yau KW, Frederick JM, Palczewski K. The function of guanylate cyclase 1 and guanylate cyclase 2 in rod and cone photoreceptors. J Biol Chem. 2007;282:8837–8847. doi: 10.1074/jbc.M610369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker SA, Martemyanov KA, Shavkunov AS, Arshavsky VY. Kinetic mechanism of RGS9-1 potentiation by R9AP. Biochemistry. 2006;45:10690–10697. doi: 10.1021/bi060376a. [DOI] [PubMed] [Google Scholar]
- 5.Bauer PJ. Binding of the retinal rod Na+/Ca2+-K+ exchanger to the cGMP-gated channel indicates local Ca2+-signaling in vertebrate photoreceptors. Ann NY Acad Sci. 2002;976:325–334. doi: 10.1111/j.1749-6632.2002.tb04755.x. [DOI] [PubMed] [Google Scholar]
- 6.Binder BM, Biernbaum MS, Bownds MD. Light activation of one rhodopsin molecule causes the phosphorylation of hundreds of others. A reaction observed in electropermeabilized frog rod outer segments exposed to dim illumination. J Biol Chem. 1990;265:15333–15340. [PubMed] [Google Scholar]
- 7.Bisegna P, Caruso G, Andreucci D, Shen L, Gurevich VV, Hamm HE, DiBenedetto E. Diffusion of the second messengers in the cytoplasm acts as a variability suppressor of the single photon response in vertebrate phototransduction. Biophys J. 2008;94:3363–3383. doi: 10.1529/biophysj.107.114058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown NG, Fowles C, Sharma R, Akhtar M. Mechanistic studies on rhodopsin kinase. Light-dependent phosphorylation of C-terminal peptides of rhodopsin. Eur J Biochem. 1992;208:659–667. doi: 10.1111/j.1432-1033.1992.tb17232.x. [DOI] [PubMed] [Google Scholar]
- 9.Buczylko J, Gutmann C, Palczewski K. Regulation of rhodopsin kinase by autophosphorylation. Proc Natl Acad Sci USA. 1991;88:2568–2572. doi: 10.1073/pnas.88.6.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burns ME, Mendez A, Chen CK, Almuete A, Quillinan N, Simon MI, Baylor DA, Chen J. Deactivation of phosphorylated and nonphosphorylated rhodopsin by arrestin splice variants. J Neurosci. 2006;26:1036–1044. doi: 10.1523/JNEUROSCI.3301-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burns ME, Mendez A, Chen J, Baylor DA. Dynamics of cyclic GMP synthesis in retinal rods. Neuron. 2002;36:81–91. doi: 10.1016/s0896-6273(02)00911-x. [DOI] [PubMed] [Google Scholar]
- 12.Burns ME, Pugh EN., Jr RGS9 concentration matters in rod phototransduction. Biophys J. 2009;97:1538–1547. doi: 10.1016/j.bpj.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calvert PD, Govardovskii VI, Krasnoperova N, Anderson RE, Lem J, Makino CL. Membrane protein diffusion sets the speed of rod phototransduction. Nature. 2001;411:90–94. doi: 10.1038/35075083. [DOI] [PubMed] [Google Scholar]
- 14.Chan S, Rubin WW, Mendez A, Liu X, Song X, Hanson SM, Craft CM, Gurevich VV, Burns ME, Chen J. Functional comparisons of visual arrestins in rod photoreceptors of transgenic mice. Invest Ophthalmol Vis Sci. 2007;48:1968–1975. doi: 10.1167/iovs.06-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CK, Woodruff ML, Chen S, Chen D, Fain GL. Background light produces a recoverin-dependent modulation of activated-rhodopsin lifetime in mouse rods. J Neurosci. doi: 10.1523/JNEUROSCI.4353-09.2010. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen CK, Burns ME, He W, Wensel TG, Baylor DA, Simon MI. Slowed recovery of rod photoresponse in mice lacking the GTPase accelerating protein RGS9-1. Nature. 2000;403:557–560. doi: 10.1038/35000601. [DOI] [PubMed] [Google Scholar]
- 17.Chen CK, Burns ME, Spencer M, Niemi GA, Chen J, Hurley JB, Baylor DA, Simon MI. Abnormal photoresponses and light-induced apoptosis in rods lacking rhodopsin kinase. Proc Natl Acad Sci USA. 1999;96:3718–3722. doi: 10.1073/pnas.96.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen CK, Inglese J, Lefkowitz RJ, Hurley JB. Ca2+-dependent interaction of recoverin with rhodopsin kinase. J Biol Chem. 1995;270:18060–18066. doi: 10.1074/jbc.270.30.18060. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Makino CL, Peachey NS, Baylor DA, Simon MI. Mechanisms of rhodopsin inactivation in vivo as revealed by a COOH-terminal truncation mutant. Science. 1995;267:374–377. doi: 10.1126/science.7824934. [DOI] [PubMed] [Google Scholar]
- 20.Cideciyan AV, Zhao X, Nielsen L, Khani SC, Jacobson SG, Palczewski K. Null mutation in the rhodopsin kinase gene slows recovery kinetics of rod and cone phototransduction in man. Proc Natl Acad Sci USA. 1998;95:328–333. doi: 10.1073/pnas.95.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cobbs WH, Pugh EN., Jr Kinetics and components of the flash photocurrent of isolated retinal rods of the larval salamander, Ambystoma tigrinum. J Physiol. 1987;394:529–572. doi: 10.1113/jphysiol.1987.sp016884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cote RH. Photoreceptor Phosphodiesterase (PDE6): a G-Protein-Activated PDE Regulating Visual Excitation in Rod and Cone Photoreceptor Cells. Boca Raton, FL: CRC Press; 2006. [Google Scholar]
- 24.Cowan CW, Fariss RN, Sokal I, Palczewski K, Wensel TG. High expression levels in cones of RGS9, the predominant GTPase accelerating protein of rods. Proc Natl Acad Sci USA. 1998;95:5351–5356. doi: 10.1073/pnas.95.9.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dean KR, Akhtar M. Novel mechanism for the activation of rhodopsin kinase: implications for other G protein-coupled receptor kinases (GRK’s) Biochemistry. 1996;35:6164–6172. doi: 10.1021/bi952480q. [DOI] [PubMed] [Google Scholar]
- 26.Dean KR, Akhtar M. Phosphorylation of solubilised dark-adapted rhodopsin. Insights into the activation of rhodopsin kinase. Eur J Biochem. 1993;213:881–890. doi: 10.1111/j.1432-1033.1993.tb17832.x. [DOI] [PubMed] [Google Scholar]
- 27.Dizhoor AM, Olshevskaya EV, Henzel WJ, Wong SC, Stults JT, Ankoudinova I, Hurley JB. Cloning, sequencing, and expression of a 24-kDa Ca2+-binding protein activating photoreceptor guanylyl cyclase. J Biol Chem. 1995;270:25200–25206. doi: 10.1074/jbc.270.42.25200. [DOI] [PubMed] [Google Scholar]
- 28.Doan T, Mendez A, Detwiler PB, Chen J, Rieke F. Multiple phosphorylation sites confer reproducibility of the rod’s single-photon responses. Science. 2006;313:530–533. doi: 10.1126/science.1126612. [DOI] [PubMed] [Google Scholar]
- 29.Field GD, Rieke F. Mechanisms regulating variability of the single photon responses of mammalian rod photoreceptors. Neuron. 2002;35:733–747. doi: 10.1016/s0896-6273(02)00822-x. [DOI] [PubMed] [Google Scholar]
- 30.Fuchs S, Nakazawa M, Maw M, Tamai M, Oguchi Y, Gal A. A homozygous 1-base pair deletion in the arrestin gene is a frequent cause of Oguchi disease in Japanese. Nat Genet. 1995;10:360–362. doi: 10.1038/ng0795-360. [DOI] [PubMed] [Google Scholar]
- 31.Gibson SK, Parkes JH, Liebman PA. Phosphorylation modulates the affinity of light-activated rhodopsin for G protein and arrestin. Biochemistry. 2000;39:5738–5749. doi: 10.1021/bi991857f. [DOI] [PubMed] [Google Scholar]
- 32.Gross OP, Burns ME. Control of rhodopsin’s active lifetime by arrestin-1 expression in mammalian rods. J Neurosci. doi: 10.1523/JNEUROSCI.5391-09.2010. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurevich VV. The selectivity of visual arrestin for light-activated phosphorhodopsin is controlled by multiple nonredundant mechanisms. J Biol Chem. 1998;273:15501–15506. doi: 10.1074/jbc.273.25.15501. [DOI] [PubMed] [Google Scholar]
- 34.Gurevich VV, Benovic JL. Visual arrestin interaction with rhodopsin. Sequential multisite binding ensures strict selectivity toward light-activated phosphorylated rhodopsin. J Biol Chem. 1993;268:11628–11638. [PubMed] [Google Scholar]
- 35.Gurevich VV, Hanson SM, Gurevich EV, Vishnivetskiy SA. How rod arrestin achieved perfection: regulation of its availability and binding selectivity. In: Fliesler SJ, Kisselev O, editors. Signal Transduction in the Retina. Boca Raton, FL: CRC Press; 2007. pp. 55–88. [Google Scholar]
- 36.Haeseleer F, Sokal I, Li N, Pettenati M, Rao N, Bronson D, Wechter R, Baehr W, Palczewski K. Molecular characterization of a third member of the guanylyl cyclase-activating protein subfamily. J Biol Chem. 1999;274:6526–6535. doi: 10.1074/jbc.274.10.6526. [DOI] [PubMed] [Google Scholar]
- 38.Hamer RD, Nicholas SC, Tranchina D, Liebman PA, Lamb TD. Multiple steps of phosphorylation of activated rhodopsin can account for the reproducibility of vertebrate rod single-photon responses. J Gen Physiol. 2003;122:419–444. doi: 10.1085/jgp.200308832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartong DT, Pott JW, Kooijman AC. Six patients with bradyopsia (slow vision): clinical features and course of the disease. Ophthalmology. 2007;114:2323–2331. doi: 10.1016/j.ophtha.2007.04.057. [DOI] [PubMed] [Google Scholar]
- 40.Hayashi T, Gekka T, Takeuchi T, Goto-Omoto S, Kitahara K. A novel homozygous GRK1 mutation (P391H) in 2 siblings with Oguchi disease with markedly reduced cone responses. Ophthalmology. 2007;114:134–141. doi: 10.1016/j.ophtha.2006.05.069. [DOI] [PubMed] [Google Scholar]
- 41.He W, Cowan CW, Wensel TG. RGS9, a GTPase accelerator for phototransduction. Neuron. 1998;20:95–102. doi: 10.1016/s0896-6273(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 42.Heck M, Hofmann KP. Maximal rate and nucleotide dependence of rhodopsin-catalyzed transducin activation: initial rate analysis based on a double displacement mechanism. J Biol Chem. 2001;276:10000–10009. doi: 10.1074/jbc.M009475200. [DOI] [PubMed] [Google Scholar]
- 43.Hetling JR, Pepperberg DR. Sensitivity and kinetics of mouse rod flash responses determined in vivo from paired-flash electroretinograms. J Physiol. 1999;516:593–609. doi: 10.1111/j.1469-7793.1999.0593v.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hodgkin AL, Nunn BJ. Control of light-sensitive current in salamander rods. J Physiol. 1988;403:439–471. doi: 10.1113/jphysiol.1988.sp017258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horner TJ, Osawa S, Schaller MD, Weiss ER. Phosphorylation of GRK1 and GRK7 by cAMP-dependent protein kinase attenuates their enzymatic activities. J Biol Chem. 2005;280:28241–28250. doi: 10.1074/jbc.M505117200. [DOI] [PubMed] [Google Scholar]
- 46.Howes KA, Pennesi ME, Sokal I, Church-Kopish J, Schmidt B, Margolis D, Frederick JM, Rieke F, Palczewski K, Wu SM, Detwiler PB, Baehr W. GCAP1 rescues rod photoreceptor response in GCAP1/GCAP2 knockout mice. EMBO J. 2002;21:1545–1554. doi: 10.1093/emboj/21.7.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu G, Wensel TG. R9AP, a membrane anchor for the photoreceptor GTPase accelerating protein, RGS9-1. Proc Natl Acad Sci USA. 2002;99:9755–9760. doi: 10.1073/pnas.152094799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hunt DM, Buch P, Michaelides M. Guanylate cyclases and associated activator proteins in retinal disease. Mol Cell Biochem. 2010;334:157–168. doi: 10.1007/s11010-009-0331-y. [DOI] [PubMed] [Google Scholar]
- 49.Hurley JB, Spencer M, Niemi GA. Rhodopsin phosphorylation and its role in photoreceptor function. Vision Res. 1998;38:1341–1352. doi: 10.1016/s0042-6989(97)00459-8. [DOI] [PubMed] [Google Scholar]
- 50.Hurley JB, Stryer L. Purification and characterization of the gamma regulatory subunit of the cyclic GMP phosphodiesterase from retinal rod outer segments. J Biol Chem. 1982;257:11094–11099. [PubMed] [Google Scholar]
- 51.Karpen JW, Zimmerman AL, Stryer L, Baylor DA. Gating kinetics of the cyclic-GMP-activated channel of retinal rods: flash photolysis and voltage-jump studies. Proc Natl Acad Sci USA. 1988;85:1287–1291. doi: 10.1073/pnas.85.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawamura S. Rhodopsin phosphorylation as a mechanism of cyclic GMP phosphodiesterase regulation by S-modulin. Nature. 1993;362:855–857. doi: 10.1038/362855a0. [DOI] [PubMed] [Google Scholar]
- 53.Kennedy MJ, Lee KA, Niemi GA, Craven KB, Garwin GG, Saari JC, Hurley JB. Multiple phosphorylation of rhodopsin and the in vivo chemistry underlying rod photoreceptor dark adaptation. Neuron. 2001;31:87–101. doi: 10.1016/s0896-6273(01)00340-3. [DOI] [PubMed] [Google Scholar]
- 54.Keresztes G, Martemyanov KA, Krispel CM, Mutai H, Yoo PJ, Maison SF, Burns ME, Arshavsky VY, Heller S. Absence of the RGS9. Gbeta5 GTPase-activating complex in photoreceptors of the R9AP knockout mouse. J Biol Chem. 2004;279:1581–1584. doi: 10.1074/jbc.C300456200. [DOI] [PubMed] [Google Scholar]
- 55.Klenchin VA, Calvert PD, Bownds MD. Inhibition of rhodopsin kinase by recoverin. Further evidence for a negative feedback system in phototransduction. J Biol Chem. 1995;270:16147–16152. doi: 10.1074/jbc.270.27.16147. [DOI] [PubMed] [Google Scholar]
- 56.Koutalos Y, Nakatani K, Tamura T, Yau KW. Characterization of guanylate cyclase activity in single retinal rod outer segments. J Gen Physiol. 1995;106:863–890. doi: 10.1085/jgp.106.5.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krispel CM, Chen CK, Simon MI, Burns ME. Novel form of adaptation in mouse retinal rods speeds recovery of phototransduction. J Gen Physiol. 2003;122:703–712. doi: 10.1085/jgp.200308938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krispel CM, Chen CK, Simon MI, Burns ME. Prolonged photoresponses and defective adaptation in rods of Gbeta5−/− mice. J Neurosci. 2003;23:6965–6971. doi: 10.1523/JNEUROSCI.23-18-06965.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krispel CM, Chen D, Melling N, Chen YJ, Martemyanov KA, Quillinan N, Arshavsky VY, Wensel TG, Chen CK, Burns ME. RGS expression rate-limits recovery of rod photoresponses. Neuron. 2006;51:409–416. doi: 10.1016/j.neuron.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 60.Krupnick JG, Gurevich VV, Benovic JL. Mechanism of quenching of phototransduction. Binding competition between arrestin and transducin for phosphorhodopsin. J Biol Chem. 1997;272:18125–18131. doi: 10.1074/jbc.272.29.18125. [DOI] [PubMed] [Google Scholar]
- 61.Kuhn H, Hall SW, Wilden U. Light-induced binding of 48-kDa protein to photoreceptor membranes is highly enhanced by phosphorylation of rhodopsin. FEBS Lett. 1984;176:473–478. doi: 10.1016/0014-5793(84)81221-1. [DOI] [PubMed] [Google Scholar]
- 62.Lee RH, Brown BM, Lolley RN. Autophosphorylation of rhodopsin kinase from retinal rod outer segments. Biochemistry. 1982;21:3303–3307. doi: 10.1021/bi00257a009. [DOI] [PubMed] [Google Scholar]
- 63.Leskov IB, Klenchin VA, Handy JW, Whitlock GG, Govardovskii VI, Bownds MD, Lamb TD, Pugh EN, Jr, Arshavsky VY. The gain of rod phototransduction: reconciliation of biochemical and electrophysiological measurements. Neuron. 2000;27:525–537. doi: 10.1016/s0896-6273(00)00063-5. [DOI] [PubMed] [Google Scholar]
- 64.Liang Y, Fotiadis D, Maeda T, Maeda A, Modzelewska A, Filipek S, Saperstein DA, Engel A, Palczewski K. Rhodopsin signaling and organization in heterozygote rhodopsin knockout mice. J Biol Chem. 2004;279:48189–48196. doi: 10.1074/jbc.M408362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lishko PV, Martemyanov KA, Hopp JA, Arshavsky VY. Specific binding of RGS9-Gbeta 5L to protein anchor in photoreceptor membranes greatly enhances its catalytic activity. J Biol Chem. 2002;277:24376–24381. doi: 10.1074/jbc.M203237200. [DOI] [PubMed] [Google Scholar]
- 66.Lorenz W, Inglese J, Palczewski K, Onorato JJ, Caron MG, Lefkowitz RJ. The receptor kinase family: primary structure of rhodopsin kinase reveals similarities to the beta-adrenergic receptor kinase. Proc Natl Acad Sci USA. 1991;88:8715–8719. doi: 10.1073/pnas.88.19.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lyubarsky A, Nikonov S, Pugh EN., Jr The kinetics of inactivation of the rod phototransduction cascade with constant Ca2+i. J Gen Physiol. 1996;107:19–34. doi: 10.1085/jgp.107.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Makino CL, Dodd RL, Chen J, Burns ME, Roca A, Simon MI, Baylor DA. Recoverin regulates light-dependent phosphodiesterase activity in retinal rods. J Gen Physiol. 2004;123:729–741. doi: 10.1085/jgp.200308994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Makino CL, Peshenko IV, Wen XH, Olshevskaya EV, Barrett R, Dizhoor AM. A role for GCAP2 in regulating the photoresponse. Guanylyl cyclase activation and rod electrophysiology in GUCA1B knock-out mice. J Biol Chem. 2008;283:29135–29143. doi: 10.1074/jbc.M804445200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Makino ER, Handy JW, Li T, Arshavsky VY. The GTPase activating factor for transducin in rod photoreceptors is the complex between RGS9 and type 5 G protein beta subunit. Proc Natl Acad Sci USA. 1999;96:1947–1952. doi: 10.1073/pnas.96.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martemyanov KA, Krispel CM, Lishko PV, Burns ME, Arshavsky VY. Functional comparison of RGS9 splice isoforms in a living cell. Proc Natl Acad Sci USA. 2008;105:20988–20993. doi: 10.1073/pnas.0808941106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martemyanov KA, Lishko PV, Calero N, Keresztes G, Sokolov M, Strissel KJ, Leskov IB, Hopp JA, Kolesnikov AV, Chen CK, Lem J, Heller S, Burns ME, Arshavsky VY. The DEP domain determines subcellular targeting of the GTPase activating protein RGS9 in vivo. J Neurosci. 2003;23:10175–10181. doi: 10.1523/JNEUROSCI.23-32-10175.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matthews HR. Actions of Ca2+ on an early stage in phototransduction revealed by the dynamic fall in Ca2+ concentration during the bright flash response. J Gen Physiol. 1997;109:141–146. doi: 10.1085/jgp.109.2.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matthews HR. Effects of lowered cytoplasmic calcium concentration and light on the responses of salamander rod photoreceptors. J Physiol. 1995;484:267–286. doi: 10.1113/jphysiol.1995.sp020664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mendez A, Burns ME, Roca A, Lem J, Wu LW, Simon MI, Baylor DA, Chen J. Rapid and reproducible deactivation of rhodopsin requires multiple phosphorylation sites. Neuron. 2000;28:153–164. doi: 10.1016/s0896-6273(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 76.Mendez A, Burns ME, Sokal I, Dizhoor AM, Baehr W, Palczewski K, Baylor DA, Chen J. Role of guanylate cyclase-activating proteins (GCAPs) in setting the flash sensitivity of rod photoreceptors. Proc Natl Acad Sci USA. 2001;98:9948–9953. doi: 10.1073/pnas.171308998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Michaelides M, Li Z, Rana NA, Richardson EC, Hykin PG, Moore AT, Holder GE, Webster AR. Novel mutations and electrophysiologic findings in RGS9- andR9AP-associated retinal dysfunction (Bradyopsia) Ophthalmology. 2009;117:120–127. doi: 10.1016/j.ophtha.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 78.Miller JA, Paulsen R. Phosphorylation and dephosphorylation of frog rod outer segment membranes as part of the visual process. J Biol Chem. 1975;250:4427–4432. [PubMed] [Google Scholar]
- 79.Miller JA, Paulsen R, Bownds MD. Control of light-activated phosphorylation in frog photoreceptor membranes. Biochemistry. 1977;16:2633–2639. doi: 10.1021/bi00631a009. [DOI] [PubMed] [Google Scholar]
- 80.Miller JL, Dratz EA. Phosphorylation at sites near rhodopsin’s carboxyl-terminus regulates light initiated cGMP hydrolysis. Vision Res. 1984;24:1509–1521. doi: 10.1016/0042-6989(84)90313-4. [DOI] [PubMed] [Google Scholar]
- 81.Murnick JG, Lamb TD. Kinetics of desensitization induced by saturating flashes in toad and salamander rods. J Physiol. 1996;495:1–13. doi: 10.1113/jphysiol.1996.sp021569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nair KS, Hanson SM, Mendez A, Gurevich EV, Kennedy MJ, Shestopalov VI, Vishnivetskiy SA, Chen J, Hurley JB, Gurevich VV, Slepak VZ. Light-dependent redistribution of arrestin in vertebrate rods is an energy-independent process governed by protein-protein interactions. Neuron. 2005;46:555–567. doi: 10.1016/j.neuron.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nikonov S, Engheta N, Pugh EN., Jr Kinetics of recovery of the dark-adapted salamander rod photoresponse. J Gen Physiol. 1998;111:7–37. doi: 10.1085/jgp.111.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nikonov SS, Brown BM, Davis JA, Zuniga FI, Bragin A, Pugh EN, Jr, Craft CM. Mouse cones require an arrestin for normal inactivation of phototransduction. Neuron. 2008;59:462–474. doi: 10.1016/j.neuron.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nikonov SS, Daniele LL, Zhu X, Craft CM, Swaroop A, Pugh EN., Jr Photoreceptors of Nrl−/− mice coexpress functional S- and M-cone opsins having distinct inactivation mechanisms. J Gen Physiol. 2005;125:287–304. doi: 10.1085/jgp.200409208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nishiguchi KM, Sandberg MA, Kooijman AC, Martemyanov KA, Pott JW, Hagstrom SA, Arshavsky VY, Berson EL, Dryja TP. Defects in RGS9 or its anchor protein R9AP in patients with slow photoreceptor deactivation. Nature. 2004;427:75–78. doi: 10.1038/nature02170. [DOI] [PubMed] [Google Scholar]
- 87.Ohguro H, Palczewski K, Ericsson LH, Walsh KA, Johnson RS. Sequential phosphorylation of rhodopsin at multiple sites. Biochemistry. 1993;32:5718–5724. doi: 10.1021/bi00072a030. [DOI] [PubMed] [Google Scholar]
- 88.Ohguro H, Van Hooser JP, Milam AH, Palczewski K. Rhodopsin phosphorylation and dephosphorylation in vivo. J Biol Chem. 1995;270:14259–14262. doi: 10.1074/jbc.270.24.14259. [DOI] [PubMed] [Google Scholar]
- 89.Orisme W, Li J, Goldmann T, Bolch S, Wolfrum U, Smith WC. Light-dependent translocation of arrestin in rod photoreceptors is signaled through a phospholipase C cascade and requires ATP. Cell Signal. 2010;22:447–456. doi: 10.1016/j.cellsig.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Osawa S, Jo R, Weiss ER. Phosphorylation of GRK7 by PKA in cone photoreceptor cells is regulated by light. J Neurochem. 2008;107:1314–1324. doi: 10.1111/j.1471-4159.2008.05691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Palczewski K, Benovic JL. G-protein-coupled receptor kinases. Trends Biochem Sci. 1991;16:387–391. doi: 10.1016/0968-0004(91)90157-q. [DOI] [PubMed] [Google Scholar]
- 92.Palczewski K, Buczylko J, Kaplan MW, Polans AS, Crabb JW. Mechanism of rhodopsin kinase activation. J Biol Chem. 1991;266:12949–12955. [PubMed] [Google Scholar]
- 93.Palczewski K, Ohguro H, Premont RT, Inglese J. Rhodopsin kinase autophosphorylation. Characterization of site-specific mutations. J Biol Chem. 1995;270:15294–15298. doi: 10.1074/jbc.270.25.15294. [DOI] [PubMed] [Google Scholar]
- 94.Palczewski K, Rispoli G, Detwiler PB. The influence of arrestin (48K protein) and rhodopsin kinase on visual transduction. Neuron. 1992;8:117–126. doi: 10.1016/0896-6273(92)90113-r. [DOI] [PubMed] [Google Scholar]
- 95.Palczewski K, Subbaraya I, Gorczyca WA, Helekar BS, Ruiz CC, Ohguro H, Huang J, Zhao X, Crabb JW, Johnson RS, et al. Molecular cloning and characterization of retinal photoreceptor guanylyl cyclase-activating protein. Neuron. 1994;13:395–404. doi: 10.1016/0896-6273(94)90355-7. [DOI] [PubMed] [Google Scholar]
- 96.Peet JA, Bragin A, Calvert PD, Nikonov SS, Mani S, Zhao X, Besharse JC, Pierce EA, Knox BE, Pugh EN., Jr Quantification of the cytoplasmic spaces of living cells with EGFP reveals arrestin-EGFP to be in disequilibrium in dark adapted rod photoreceptors. J Cell Sci. 2004;117:3049–3059. doi: 10.1242/jcs.01167. [DOI] [PubMed] [Google Scholar]
- 98.Pepperberg DR, Cornwall MC, Kahlert M, Hofmann KP, Jin J, Jones GJ, Ripps H. Light-dependent delay in the falling phase of the retinal rod photoresponse. Vis Neurosci. 1992;8:9–18. doi: 10.1017/s0952523800006441. [DOI] [PubMed] [Google Scholar]
- 99.Pugh EN, Jr, Lamb TD. Amplification and kinetics of the activation steps in phototransduction. Biochim Biophys Acta. 1993;1141:111–149. doi: 10.1016/0005-2728(93)90038-h. [DOI] [PubMed] [Google Scholar]
- 100.Pugh EN, Jr, Nikonov S, Lamb TD. Molecular mechanisms of vertebrate photoreceptor light adaptation. Curr Opin Neurobiol. 1999;9:410–418. doi: 10.1016/S0959-4388(99)80062-2. [DOI] [PubMed] [Google Scholar]
- 101.Pulvermuller A, Maretzki D, Rudnicka-Nawrot M, Smith WC, Palczewski K, Hofmann KP. Functional differences in the interaction of arrestin and its splice variant, p44, with rhodopsin. Biochemistry. 1997;36:9253–9260. doi: 10.1021/bi970772g. [DOI] [PubMed] [Google Scholar]
- 102.Pulvermuller A, Palczewski K, Hofmann KP. Interaction between photoactivated rhodopsin and its kinase: stability and kinetics of complex formation. Biochemistry. 1993;32:14082–14088. doi: 10.1021/bi00214a002. [DOI] [PubMed] [Google Scholar]
- 103.Rieke F, Baylor DA. Molecular origin of continuous dark noise in rod photoreceptors. Biophys J. 1996;71:2553–2572. doi: 10.1016/S0006-3495(96)79448-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rieke F, Baylor DA. Origin of reproducibility in the responses of retinal rods to single photons. Biophys J. 1998;75:1836–1857. doi: 10.1016/S0006-3495(98)77625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ruiz M, Brown RL, He Y, Haley TL, Karpen JW. The single-channel dose-response relation is consistently steep for rod cyclic nucleotide-gated channels: implications for the interpretation of macroscopic dose-response relations. Biochemistry. 1999;38:10642–10648. doi: 10.1021/bi990532w. [DOI] [PubMed] [Google Scholar]
- 106.Sagoo MS, Lagnado L. G-protein deactivation is rate-limiting for shut-off of the phototransduction cascade. Nature. 1997;389:392–395. doi: 10.1038/38750. [DOI] [PubMed] [Google Scholar]
- 107.Sakurai K, Onishi A, Imai H, Chisaka O, Ueda Y, Usukura J, Nakatani K, Shichida Y. Physiological properties of rod photoreceptor cells in green-sensitive cone pigment knock-in mice. J Gen Physiol. 2007;130:21–40. doi: 10.1085/jgp.200609729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sitaramayya A, Liebman PA. Mechanism of ATP quench of phosphodiesterase activation in rod disc membranes. J Biol Chem. 1983;258:1205–1209. [PubMed] [Google Scholar]
- 109.Sitaramayya A, Liebman PA. Phosphorylation of rhodopsin and quenching of cyclic GMP phosphodiesterase activation by ATP at weak bleaches. J Biol Chem. 1983;258:12106–12109. [PubMed] [Google Scholar]
- 110.Song X, Vishnivetskiy SA, Gross OP, Emelianoff K, Mendez A, Chen J, Gurevich EV, Burns ME, Gurevich VV. Enhanced arrestin facilitates recovery and protects rods lacking rhodopsin phosphorylation. Curr Biol. 2009;19:700–705. doi: 10.1016/j.cub.2009.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Strissel KJ, Sokolov M, Trieu LH, Arshavsky VY. Arrestin translocation is induced at a critical threshold of visual signaling and is superstoichiometric to bleached rhodopsin. J Neurosci. 2006;26:1146–1153. doi: 10.1523/JNEUROSCI.4289-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thompson P, Findlay JB. Phosphorylation of ovine rhodopsin. Identification of the phosphorylated sites. Biochem J. 1984;220:773–780. doi: 10.1042/bj2200773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Torisawa A, Arinobu D, Tachibanaki S, Kawamura S. Amino acid residues in GRK1/GRK7 responsible for interaction with S-modulin/recoverin. Photochem Photobiol. 2008;84:823–830. doi: 10.1111/j.1751-1097.2007.00292.x. [DOI] [PubMed] [Google Scholar]
- 114.Tsang SH, Burns ME, Calvert PD, Gouras P, Baylor DA, Goff SP, Arshavsky VY. Role for the target enzyme in deactivation of photoreceptor G protein in vivo. Science. 1998;282:117–121. doi: 10.1126/science.282.5386.117. [DOI] [PubMed] [Google Scholar]
- 115.Tsang SH, Gouras P, Yamashita CK, Kjeldbye H, Fisher J, Farber DB, Goff SP. Retinal degeneration in mice lacking the gamma subunit of the rod cGMP phosphodiesterase. Science. 1996;272:1026–1029. doi: 10.1126/science.272.5264.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tsang SH, Woodruff ML, Chen CK, Yamashita CY, Cilluffo MC, Rao AL, Farber DB, Fain GL. GAP-independent termination of photoreceptor light response by excess gamma subunit of the cGMP-phosphodiesterase. J Neurosci. 2006;26:4472–4480. doi: 10.1523/JNEUROSCI.4775-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tsang SH, Woodruff ML, Janisch KM, Cilluffo MC, Farber DB, Fain GL. Removal of phosphorylation sites of gamma subunit of phosphodiesterase 6 alters rod light response. J Physiol. 2007;579:303–312. doi: 10.1113/jphysiol.2006.121772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vishnivetskiy SA, Raman D, Wei J, Kennedy MJ, Hurley JB, Gurevich VV. Regulation of arrestin binding by rhodopsin phosphorylation level. J Biol Chem. 2007;282:32075–32083. doi: 10.1074/jbc.M706057200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wada Y, Sugiyama J, Okano T, Fukada Y. GRK1 and GRK7: unique cellular distribution and widely different activities of opsin phosphorylation in the zebrafish rods and cones. J Neurochem. 2006;98:824–837. doi: 10.1111/j.1471-4159.2006.03920.x. [DOI] [PubMed] [Google Scholar]
- 120.Wald G, Durell J, St. George CC. The light reaction in the bleaching of rhodopsin. Science. 1950;111:179–181. doi: 10.1126/science.111.2877.179. [DOI] [PubMed] [Google Scholar]
- 121.Wang Z, Wen XH, Ablonczy Z, Crouch RK, Makino CL, Lem J. Enhanced shutoff of phototransduction in transgenic mice expressing palmitoylation-deficient rhodopsin. J Biol Chem. 2005;280:24293–24300. doi: 10.1074/jbc.M502588200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wensel TG, Stryer L. Reciprocal control of retinal rod cyclic GMP phosphodiesterase by its gamma subunit and transducin. Proteins. 1986;1:90–99. doi: 10.1002/prot.340010114. [DOI] [PubMed] [Google Scholar]
- 123.Whitcomb T, Sakurai K, Brown BM, Young JE, Sheflin L, Dlugos C, Craft CM, Kefalov VJ, Khani SC. Effect of G protein-coupled receptor kinase 1 (Grk1) overexpression on rod photoreceptor cell viability. Invest Ophthalmol Vis Sci. 2009;51:1728–1737. doi: 10.1167/iovs.09-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wilden U. Duration and amplitude of the light-induced cGMP hydrolysis in vertebrate photoreceptors are regulated by multiple phosphorylation of rhodopsin and by arrestin binding. Biochemistry. 1995;34:1446–1454. doi: 10.1021/bi00004a040. [DOI] [PubMed] [Google Scholar]
- 125.Wilden U, Hall SW, Kuhn H. Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48-kDa protein of rod outer segments. Proc Natl Acad Sci USA. 1986;83:1174–1178. doi: 10.1073/pnas.83.5.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wilden U, Kuhn H. Light-dependent phosphorylation of rhodopsin: number of phosphorylation sites. Biochemistry. 1982;21:3014–3022. doi: 10.1021/bi00541a032. [DOI] [PubMed] [Google Scholar]
- 127.Woodruff ML, Janisch KM, Peshenko IV, Dizhoor AM, Tsang SH, Fain GL. Modulation of phosphodiesterase6 turnoff during background illumination in mouse rod photoreceptors. J Neurosci. 2008;28:2064–2074. doi: 10.1523/JNEUROSCI.2973-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Woodruff ML, Sampath AP, Matthews HR, Krasnoperova NV, Lem J, Fain GL. Measurement of cytoplasmic calcium concentration in the rods of wild-type and transducin knock-out mice. J Physiol. 2002;542:843–854. doi: 10.1113/jphysiol.2001.013987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xu J, Dodd RL, Makino CL, Simon MI, Baylor DA, Chen J. Prolonged photoresponses in transgenic mouse rods lacking arrestin. Nature. 1997;389:505–509. doi: 10.1038/39068. [DOI] [PubMed] [Google Scholar]
- 130.Yamamoto S, Sippel KC, Berson EL, Dryja TP. Defects in the rhodopsin kinase gene in the Oguchi form of stationary night blindness. Nat Genet. 1997;15:175–178. doi: 10.1038/ng0297-175. [DOI] [PubMed] [Google Scholar]
- 131.Yang RB, Robinson SW, Xiong WH, Yau KW, Birch DG, Garbers DL. Disruption of a retinal guanylyl cyclase gene leads to cone-specific dystrophy and paradoxical rod behavior. J Neurosci. 1999;19:5889–5897. doi: 10.1523/JNEUROSCI.19-14-05889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang X, Wensel TG, Kraft TW. GTPase regulators and photoresponses in cones of the eastern chipmunk. J Neurosci. 2003;23:1287–1297. doi: 10.1523/JNEUROSCI.23-04-01287.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]