Abstract
Background
The completion of 19 insect genome sequencing projects spanning six insect orders provides the opportunity to investigate the evolution of important gene families, here tubulins. Tubulins are a family of eukaryotic structural genes that form microtubules, fundamental components of the cytoskeleton that mediate cell division, shape, motility, and intracellular trafficking. Previous in vivo studies in Drosophila find a stringent relationship between tubulin structure and function; small, biochemically similar changes in the major alpha 1 or testis-specific beta 2 tubulin protein render each unable to generate a motile spermtail axoneme. This has evolutionary implications, not a single non-synonymous substitution is found in beta 2 among 17 species of Drosophila and Hirtodrosophila flies spanning 60 Myr of evolution. This raises an important question, How do tubulins evolve while maintaining their function? To answer, we use molecular evolutionary analyses to characterize the evolution of insect tubulins.
Results
Sixty-six alpha tubulins and eighty-six beta tubulin gene copies were retrieved and subjected to molecular evolutionary analyses. Four ancient clades of alpha and beta tubulins are found in insects, a major isoform clade (alpha 1, beta 1) and three minor, tissue-specific clades (alpha 2-4, beta 2-4). Based on a Homarus americanus (lobster) outgroup, these were generated through gene duplication events on major beta and alpha tubulin ancestors, followed by subfunctionalization in expression domain. Strong purifying selection acts on all tubulins, yet maximum pairwise amino acid distances between tubulin paralogs are large (0.464 substitutions/site beta tubulins, 0.707 alpha tubulins). Conversely orthologs, with the exception of reproductive tissue isoforms, show little sequence variation except in the last 15 carboxy terminus tail (CTT) residues, which serve as sites for post-translational modifications (PTMs) and interactions with microtubule-associated proteins. CTT residues overwhelming comprise the co-evolving residues between Drosophila alpha 2 and beta 3 tubulin proteins, indicating CTT specializations can be mediated at the level of the tubulin dimer. Gene duplications post-dating separation of the insect orders are unevenly distributed, most often appearing in major alpha 1 and minor beta 2 clades. More than 40 introns are found in tubulins. Their distribution among tubulins reveals that insertion and deletion events are common, surprising given their potential for disrupting tubulin coding sequence. Compensatory evolution is found in Drosophila beta 2 tubulin cis-regulation, and reveals selective pressures acting to maintain testis expression without the use of previously identified testis cis-regulatory elements.
Conclusion
Tubulins have stringent structure/function relationships, indicated by strong purifying selection, the loss of many gene duplication products, alpha-beta co-evolution in the tubulin dimer, and compensatory evolution in beta 2 tubulin cis-regulation. They evolve through gene duplication, subfunctionalization in expression domain and divergence of duplication products, largely in CTT residues that mediate interactions with other proteins. This has resulted in the tissue-specific minor insect isoforms, and in particular the highly diverse α3, α4, and β2 reproductive tissue-specific tubulin isoforms, illustrating that even a highly conserved protein family can participate in the adaptive process and respond to sexual selection.
Background
Proteins vary in the stringency of their structure/function relationships, which may affect their ability to participate in the adaptive process [1]. Nature has ready opportunity to shape the phenotype through selection on proteins that show non-synonymous allelic variation, for example esterases [2] and glycolytic enzymes [3]. Other proteins, for example actins, show little amino acid variation (~5-7% across metazoans, [4]), and tend to loose function entirely rather than provide altered function in response to change in their amino acid sequence [5]. Such proteins may not typically admit allelic variation, which raises an old, but important question: is selection a shaper of diversity, or merely an executioner [6]?
One of the best-studied proteins with respect to its structure/function relationship is tubulin. In vivo studies of alpha and beta tubulin in the Drosophila melanogaster spermtail axoneme find that small changes in the amino acid sequence of the major alpha 1 or testis-specific beta 2 tubulin protein render each unable to generate a motile axoneme [7-10]. This stringency has evolutionary implications; comparisons of beta 2 sequences among different species of Drosophila find not a single non-synonymous substitution, indicating the protein has not changed in sequence for more than 60 million years [11]. Together these results indicate that only rarely does beta 2 participate in the adaptive process.
For proteins with stringent structure/function relationships, evolving while maintaining function is problematic. Gene duplication is a fundamental mechanism in answer to this problem [12], yet without additional changes, in expression domain and/or in the proteins with which it co-functions, a duplicate copy will have the same function as the original, will experience the same selective regime as the original and so will not evolve.
Here we characterize insect tubulin evolution, to identify events that release tubulins to evolve, and to more generally serve as a model for the evolution of functionally constrained proteins. Tubulins are a family of eukaryotic structural proteins that comprise microtubules, fundamental components of the spindle in cell division, the axoneme in cilia and flagella, mediators of cell shape, and dynein/kinesin-based cell trafficking [13-15]. Two members of the tubulin family, alpha and beta tubulin, form a dimer that is the building block of the microtubule. All eukaryotes contain at least one major alpha (α1) and beta (β1) tubulin. In addition, Drosophila melanogaster express minor, tissue-specific isoforms in the motile spermtail axoneme (β2), pre-adult tissues (β3, β4, and α2), and the ovary (α4) [16,17].
We studied tubulin evolution in two hemimetabolous insect orders, Phthiraptera (Pediculus humanus corporis, body louse) and Hemiptera (Acyrthosiphon pisum, pea aphid), and four holometabolous orders, Hymenoptera (Apis millifera honeybee, Nasonia vitripennis jewel wasp), Coleoptera (Tribolium castenatum flour beetle), Lepidoptera (Bombyx mori silkmoth), and Diptera (Aedes aegypti, Anopheles gambiae, and Drosophila melanogaster, D. sechellia, D. yakuba, D. erecta, D. simulans, D. mojavensis, D. grimshawi, D. ananassae, D. persimilis, D. psuedoobscura, D. virilis, D. willistoni). These orders represent well over 80% of the diversity in all insect species [18]. Their evolutionary relationships are not controversial, and each of these orders is considered to be monophyletic [19]. They are known to be quite ancient, the origin of these insect orders has been dated to be >300 Mya using a molecular clock [20], with the oldest beetle (Coleopteran) fossils from the Lower Permian (about 265 million years ago [21]) and the earliest fly (Diptera) fossil from the Upper Triassic of the Mesozoic geological period, some 225 million years ago [22].
We find four clades of alpha and beta tubulins in insects that, for the most part, do not evolve without a gene duplication event. Yet gene duplication is not sufficient to release tubulin evolution, most duplication products are lost, and major tubulin duplication products do not evolve unless followed by subfunctionalization in expression domain. Subfunctionalization has resulted in a number of reproductive tissue-specific tubulins that are diverse in sequence, particularly in CTT residues that mediate integrations with other proteins. Together these results indicate that tubulin evolution is constrained, yet tubulins can in fact participate in the adaptive process.
Methods
Sequence retrieval
Insect tubulins were obtained through BLAST [23] searches of the sequenced insect genome databases http://flybase.bio.indiana.edu/ and NCBI http://www.ncbi.nlm.nih.gov/ databases using Drosophila melanogaster tubulin cDNAs as query sequences. Tubulin exon/intron structure was determined by aligning retrieved genomic sequences to their Drosophila cDNA orthologs using Sequencher 4.1 (Gene Codes Corporation [24]).
Genealogical reconstruction
DNA sequence alignments were made using ClustalW [25] in the MEGA v. 4.0 software package [26]. Translated sequences were aligned using BLOSUM [27] (gap opening penalty 100, extension penalty 0.1), refined by hand, and untranslated for genealogical analysis using the Bayesian method as implemented in Mr. Bayes v. 3.1.2 [28]). For both alpha and beta tubulins a GTR model was used with 4 rate categories, gamma corrections were estimated by the program, and gaps were coded. For beta tubulins, the analysis was done using first and second codon positions (88 sequences, 947 sites, gamma correction α = 0.542); for alpha tubulins zero-fold degenerate codon positions (69 sequences, 811 sites, gamma correction α = 0.486). Analyses were run until the standard deviation of split frequencies was below 0.01, for the alpha tubulins 4,000,000 generations and beta tubulins 1,500,000 generations. A 25% burnin was performed [28], and the majority-rule consensus tree is reported.
Pairwise amino acid distances
The average and maximum pairwise amino acid distances (number of amino acid differences/site) between paralogs and orthologs are presented, these were generated using the Poisson correction method in MEGA v. 4 [26]; standard error estimates were obtained by a bootstrap procedure (2000 replications).
Test of selection
Since tubulins do not appear to be evolving at appreciable rates, they are probably under severe purifying selection. On the other hand, the carboxy terminus tails, which mediate tubulin functional specializations via interactions with other proteins, and are released from protein folding constraints because they lie on the surface of the MT [29], are the most rapidly evolving, and possibly under positive selection. These two hypotheses were tested, first by means of estimating dN (the number of nonsynonymous substitutions per nonsynonymous site), dS (the number of synonymous substitutions per synonymous site), and the ratio dN/dS (ω) by the ML method as implemented in PAML v4.3b http://abacus.gene.ucl.ac.uk/software/paml.html[30], and then by means of a likelihood ratio test comparing the null model H0 with ω = 1 fixed and the alternative model H1 with ω estimated from the data.
Rate tests
The tubulins were tested for the molecular clock, to determine: 1) if paralogous tubulins resulting from ancient duplication events (preceding separation of insect orders) evolve at the same rate, 2) if orthologous tubulins evolve at the same rate, and 3) if paralogous tubulins resulting from recent duplication events (post-dating separation of the insect orders) diverge following duplication. All rate tests were performed using Tajima's relative rates test in MEGA v. 3.0 [26,31].
Test of co-evolution
Stringency in tubulin structure/function relationships may result from the need to maintain proper lateral and longitudinal contacts between alpha and beta tubulin in the microtubule, such that the alpha and beta tubulins must co-evolve for either tubulin component to evolve. Sato's mirror-tree method [32] compares partial correlation coefficients between candidate co-evolving proteins' distance matrices to identify co-evolution. This test requires 1) multiple sequences for comparison, 2) sequence variation among those sequences, and 3) knowledge that the alpha and beta isoforms are co-expressed in a cell. These conditions are met only by the Drosophila α2 and β3 tubulins, which co-function in visceral mesoderm, the testis cyst cells, and pre-adult sensory neurons [7,8].
Evolution of Drosophila Beta 2 tubulin cis-regulation
An opportunity to study cis-regulatory aspects of tubulin evolution is provided by the D. melanogaster β2 gene. Three aspects of Dmβ2 regulation have been identified, the β2UE1 element, required for testis-specific gene expression, and the β2UE2 and β2DE1 elements for proper expression levels [33]; these elements are identifiable in most Drosophila species. The core promoter does not contain a TATA box, but uses an Inr sequence found in many TATA-less promoters. We identified these sequences in Drosophila species by scanning the 1000 base pairs 5' to transcription start for identical and degenerate sequence matches to Dmβ2 regulation, using Sequencher.
To determine if Drosophila β2 is expressed in the testis in species in which these sequences were not found, a PCR approach was used. RNA was extracted from 20 pairs of dissected testes using a guanidine hydrochloride, phenol/chloroform method [34], and DNAase treated to remove DNA contaminants (New England Biolabs). Beta 2-specific primers were used in a reverse transcription reaction, followed by PCR amplification, to identify β2 mRNA in testes. For a negative control, PCR was performed on the same RNA template, minus the reverse transcription reaction.
Results
Sequence retrieval and genealogical reconstruction
A total of 86 beta tubulins and 66 alpha tubulins were obtained through blast searches of the completed insect genome projects and NCBI databases (Tables 1, 2; Additional File 1). Homarus americana (American lobster; Crustacea, Decapoda) tubulins were used as an outgroup in the genealogical reconstruction, because Homarus is the closest relation to insects for which the full complement of tubulins has been identified. Homarus has two major beta tubulin isoforms and three major alpha tubulins. Based on this outgroup, duplication products of major alpha and beta tubulin isoforms gave rise to the insect major and minor tubulins.
Table 1.
Insect beta tubulin sequence features.
| Isoform | Function | Average and maximum pairwise distances | -COOH terminus sequence | Post-translational modification |
||||
|---|---|---|---|---|---|---|---|---|
| sites/Conserved | ||||||||
| Drosophila | Mosquito | All Insects | sequence features | |||||
|
β1 |
Major isoform |
0.000 +/- 0.000 (n = 12) |
0.002 +/- 0.002 (n = 2) |
Average 0.011 +/- 0.003 (n = 9) Maximum 0.027 (n = 10) |
Haβ1a EATADDEAE FEEEGEVEGE YA Haβ1b EATADDEAE FEEEGEVEGE YD Dmβ1 EATADEDAE FEEEQEAEVD EN Aeβ1 EATADEDAE FDEEQEAEVD EN Agβ1 EATADEDAE FDEEQEAEVD EN Bmβ1a EATADEDAE FDEEQEQEIE DN Bmβ1b EATADEDAE FDEEQEQEIE EH Tcβ1 EATADEDAE FDEEQEAEVD EN Nvβ1a - Nvβ1b TMNGPRDAP DEDVEVVEEE LRD Amβ1 EATADEDAE FDEEQEAEVD EN Apβ1 EATADEEAE FDEEQEQEVD EN Phβ1 EATADEDAE FDEEQEEVVD EN |
PTM sites Polyglutamylation - yes Polyglycylation - yes Phosphorylation - yes |
||
|
β2 |
Testis- specific isoform |
0.000 +/- 0.000 (n = 12) |
0.060 +/- 0.011 (n = 2) |
Average 0.085 +/- 0.009 (n = 9) Maximum 0.464 (n = 17) |
Dmβ2 EATADEEGE FDEDEEGGGD E Aeβ2 EATADEEGE FDEEEEGGEE Agβ2 EATADDEGE MDEEEEGGED Bmβ2 DATADDEGE FDEEAEEGLE E Tcβ2a DATAEEEGE FDEEEEGDNE GEN Tcβ2b DAEVDEEYG DEDETEEDKF EEET Nvβ2a EATAEEDTE FDEDEGENEG N Nvβ2b EATADEFAD YEEDEEEEED YA Nvβ2c EATTEE--D FETEDAGDD FETCDQE Amβ2a EATAEEEGE FDEEEEGEGE HP Amβ2b EATAEDEGE FDEEEETEK Apβ2a DATVDEDGE GDDDEEDADA Apβ2b EATIDETGE-EDEDEDADA Apβ2c DATVDEEGE GDDDDEDAEA Apβ2d EATVDAPGG VNEE Phβ2a EATADEEGE DEEDEGGED Phβ2b EATSYEYDE DEGEENEVEE EEEKEMTNWL PA |
PTM sites Polyglutamylation - yes, except Apβ2d Polyglycylation -- yes, except Apβ2d Phosphorylation -- yes Conserved sequence features Axoneme motif - yes, except Agβ2, Tcβ2b, Nvβ2a-c, Apβ2a-d, Phβ2a, b Gly56-- yes, except Phβ2b |
||
|
β3 |
Minor isoform expressed in variety of pre-adult mesodermal and neural domains |
0.009 +/- 0.002 (n = 10) |
0.014 +/- 0.005 (n = 2) |
Average 0.092 +/- 0.010 (n = 9) Maximum 0.148 (n = 9) |
Dmβ3 EATADDEFD PEVNQEEVEG DCI Aeβ3 EATADDEFE QEECADEMEG ECV Agβ3 EATADDEFE QEDCQDEMEG ECV Bmβ3 EATAEDDTE FDQEDLEELA QDEHHD Tcβ3 EATADEEYE AEEEAAADDF NC Nvβ3 EATTEEDFE TEDAGDDFET CDQE Amβ3 EATAEEDFE AEECADDFET CDQE Apβ3 EASVDEEYI EEEETEETDM CD Phβ3 LYISTIIKI |
PTM sites Polyglutamylation -- yes Polyglycylation -- yes, except Tcβ3, Nvβ3, Amβ3, Nvβ3, Apβ3 Phosphorylation - yes Conserved sequence features Nucleotide-binding domain amino acid insert (aa56) - yes |
||
|
β4 |
Minor isoform, pre-adult tissues in Dm, absent in Bm) |
0.032 +/- 0.005 (n = 11) |
0.104 +/- 0.012 (n = 3) |
Average 0.136 +/- 0.015 (n = 5) Maximum 0.186 (n = 6) |
Dmβ4 EATADDEVE FDDEQAEQEG YESEVLQNGN GE Aeβ4a EASADDYVE GEHDFDDEEE IQQ Aeβ4b DASVEDYED GEEMIEEEGE QHVE Agβ4 DAEVEDYDE MEEIPEEEQQ QQQE Apβ4 EATAEEVEF DDEEVVEEVD DNKDY Phβ4 VRSSLHLSN AANIEIQKNE ILNRNT |
PTM sites Polyglutamylation - ? Polyglycylation - ? Phosphorylation - yes |
||
Features of the four beta tubulin isoforms identified in insects are presented. The function and/or expression domain of each sequence in D. melanogaster [16,17] and B. mori [35], the two insects in which tubulin expression and function have been studied, are presented in Column 2. Average and maximum pairwise distance calculations in Column 3 refer to the average # amino acid differences/site among conserved isoforms, and the maximum pairwise distance between any two orthologs, including divergent duplication products, respectively. For "all insects", the only Drosophila species included is Dm, to avoid a Dipteran skew in the results. CTT sequences are presented in Column 4, for purposes of inspection as they constitute ~50% of the differences among tubulins. Tubulin post-translational modifications (PTMs) occur on sequence motifs whose presence and absence are presented in the Column 5. Polyglutamylation and polyglycylation sequence motifs are degenerate, the "?" indicates that a potentially modifiable, but experimentally uncharacterized residue(s) is present for these PTMs. Unusual sequence features or motifs known to mediate specific tubulin functional specializations are also noted. The full-length sequences for Nvβ1, Phβ3, Dpβ3, Dsβ3 were not available. Key: Pediculus humanus corporisPh, Acyrthosiphon pisum Ap, Apis millifera Am, Nasonia vitripennis Nv, Tribolium castenatum Tc, Bombyx mori Bm, Aedes aegypti Ae, Anopheles gambiae Ag, Drosophila melanogaster Dm, D. sechellia Dc, D. yakuba Dy, D. erecta De, D. simulans Ds, D. mojavensis Do, D. grimshawi Dg, D. ananassae Da, D. persimilis Dp, D. psuedoobscura Du, D. virilis Dv, D. willistoni Dw).
Table 2.
Insect alpha tubulin sequence features.
| Isoform | Function | Average and maximum pairwise distances | -COOH terminus sequence | Post-translational modification sites/Conserved sequence features | ||||
|---|---|---|---|---|---|---|---|---|
| Drosophila | Mosquito | All Insects | ||||||
|
α1 |
Major isoform α1a: Somatic and Testis function in Dm, somatic only in Bm α1b: low level somatic in Dm |
0.000 +/- 0.000 (n = 17) |
0.004 +/- 0.002 (n = 4) |
Average 0.010 +/- 0.003 (n = 9) Maximum 0.086 (n = 18) |
Haα1a DLAALEKDY EEVGVDSADA EGEEEGEEY Haα1b DLAALEKDY EEVGMDSADG EDIEGGDEY Haα1c DLATLEKDY EEVGIDTADG EDDEEANDY Dmα1a DLAALEKDY EEVGMDSGDG EGEGAEEY Dmα1b DLAALEKDY EEVGMDSGDG EGEGAEEY Aeα1a DLAALEKDY EEVGMDSGEG EGEGAEEY Aeα1b DLAALEKDY EEVGMDSGEG EGEGAEEY Agα1a DLAALEKDY EEVGMDSGEG EGEGAEEY Agα1b DLAALEKDY EEVGMDSGDG EGEGAEEY Bmα1 DLAALEKDY EEVGMDSAEG EGEGAEEY Tcc1 DLAALEKDY EEVGMDSGEG EGEGGEEY Amα1a DLAALEKDY EEVGMDSAEG EGEGAEEY Amα1b DLAALEKDY EEVGMDSAEG EGEGAEEY Nvα1a DLAALEKDY EEVGMDSVEG EGEGAEEY Nvα1b DLAALEKDY EEVGMDSTEG EGEGAEEY Nvα1c DLAALEKDY EEVGMDSTEG EGEGAEEY Apα1a DLAALEKDY EEVGMDSVEG EGEGGEEY Apα1b DLAALEKDY EEVGLDSVEG QFDEGVEDF Apα1c DLAALEKDY EEVGMDSTEG DGEAGEEEI Phα1a DLAALEKDY EEVGMDSVEG EGEGGDEY |
PTM sites Polyglutamylation -- yes, except Apα1a Polyglycylation - yes, except Apα1a Acetylation -- yes, except Tcα1 Detyrosination -- yes, except Apα1a, Apα1c, Phα1a Phosphorylation - no Palmitoylation - yes |
||
|
α2 |
Minor isoform often co- expressed with β3 in variety of pre-adult mesodermal and neural domains in Dm and Bm |
0.014 +/- 0.004 (n = 10) Absent in Dp and Du |
- |
Average 0.069 +/- 0.008 (n = 6) Maximum 0.170 (n = 6) |
Dmα2 DLAALEKDY EEVGIDSTTE LGEDEEY Aeα2 DLAALEKDY EEVGVDSTEE VGEGDEY Bmα2 DLAALEKDY EEVGVDSTEG ELDEENEY Tcα2 DLAALEKDY EEVAVDSIEG EGDEGDEY Amα2 DLAALELDY REVQEDATNT DDEEEY Phα2 DLAALEKDY EEVGIDSVEE VGEGDEY |
PTM sites Polyglutamylation - ? Polyglycylation - ? Acetylation - yes Detyrosination - yes Phosphorylation - no Palmitoylation - yes |
||
|
α3 |
Testis- specific isoform in Bm, absent in Dm |
- |
- |
Average 0.231 +/- 0.003 (n = 2) Maximum 0.231 (n = 2) |
Bmα3 DLAALERDY DEVAIETSDM QPGADDEL Tcα3 DLAMLEKDY EEVSIDDIE |
PTM sites Polyglutamylation - ? Polyglycylation - ? Acetylation - no Detyrosination - no Phosphorylation - no Palmitoylation -- yes Tcα3, no Bmα3 |
||
|
α4 |
Ovary- specific isoform in Dm, absent in Bm |
0.087 +/- 0.009 (n = 12) |
0.650 +/- 0.043 (n = 2) |
Average 0.560 +/- 0.028 (n = 4) Maximum 0.707 (n = 5) |
Dmα4 NIAVLERDF EEVGLDNAEE GGDEDFDEF Agα4a DLACLERDY EEVAGDTVAS GEEYYDDDEY Agα4b NIRTLIKDY EEI Tcα4 DLTALVLDY KEVDSD Amα4 DMMTLINDY KEIEK Phα4 DLAALEKDY EEVGMDSVEG EGEGGEEM |
PTM sites Polyglutamylation -? Polyglycylation -? Acetylation - no Detyrosination -- yes, except Agα4a, Agα4b, Dyα4, Dvα4, Dsα4, Doα4, Dmα4, Dgα4, Deα4, Daα4, Tcα4, Am c4, Ph c4 Phosphorylation -- yes except Tcα4 Palmitoylation -- yes except Tcα4, Amα4 |
||
Features of the four alpha tubulin isoforms identified in insects are presented. The function and/or expression domain of each sequence in D. melanogaster [16,17] and B. mori [35], the two insects in which tubulin expression and function have been studied, are presented in Column 2. Average and maximum pairwise ("All Insects" only) distance calculations in Column 3 refer to the average # amino acid differences/site among conserved isoforms, and the maximum pairwise distance between any two orthologs, including divergent duplication products, respectively. For "all insects", the only Drosophila species included is Dm, to avoid a Dipteran skew in the results. CTT sequences are presented in Column 4, for purposes of inspection as they constitute ~50% of the differences among tubulins. Tubulin post-translational modifications (PTMs) occur on sequence motifs whose presence and absence are presented in the Column 5. Polyglutamylation and polyglycylation sequence motifs are degenerate, the "?" indicates that a potentially modifiable, but experimentally uncharacterized residue(s) is present for these PTMs. Unusual sequence features or motifs known to mediate specific tubulin functional specializations are also noted. The full-length sequences for Nvβ1, Phβ3, Dpβ3, Dsβ3 were not available. Key: Pediculus humanus corporisPh, Acyrthosiphon pisum Ap, Apis millifera Am, Nasonia vitripennis Nv, Tribolium castenatum Tc, Bombyx mori Bm, Aedes aegypti Ae, Anopheles gambiae Ag, Drosophila melanogaster Dm, D. sechellia Dc, D. yakuba Dy, D. erecta De, D. simulans Ds, D. mojavensis Do, D. grimshawi Dg, D. ananassae Da, D. persimilis Dp, D. psuedoobscura Du, D. virilis Dv, D. willistoni Dw).
Beta tubulins
There is posterior probability support (provided in parentheses for the remainder of this section) for four monophyletic beta tubulin clades in insects (Figs. 1, 2). Orthologs of the Dmβ1 isoform (0.74) form a clade of major somatic isoforms, the most conserved in sequence of the insect beta tubulins (Table 1). This gene duplicated in a Bombyx ancestor, giving rise to Bmβ1a and Bmβ1b (1.0).
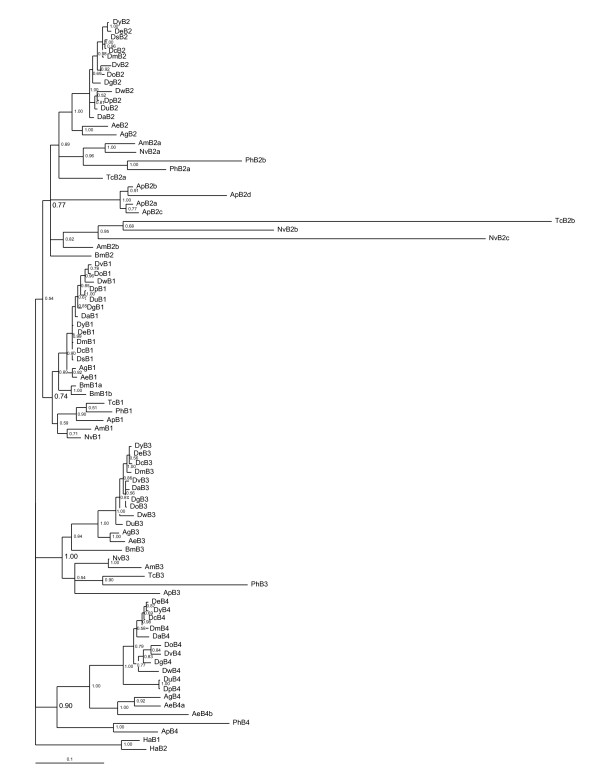
Figure 1.
Beta tubulin genealogy. Bayesian reconstruction of insect beta tubulin evolutionary relationships. Eighty-six tubulins were analyzed with Homarus americanus (Crustacea, Decapoda) beta tubulins as the outgroup. There are four beta tubulin clades ancestral to insects, the posterior probability scores in support of these clades are in larger font in the figure: β1 (0.74), β2 (0.77), β3 (1.00) and β4 (0.90). Removal of the 5 most divergent tubulins (Tcβ2b, Nvβ2b, c, Phβ4, and Apβ4) results in support >0.97 for each clade.
Figure 2.
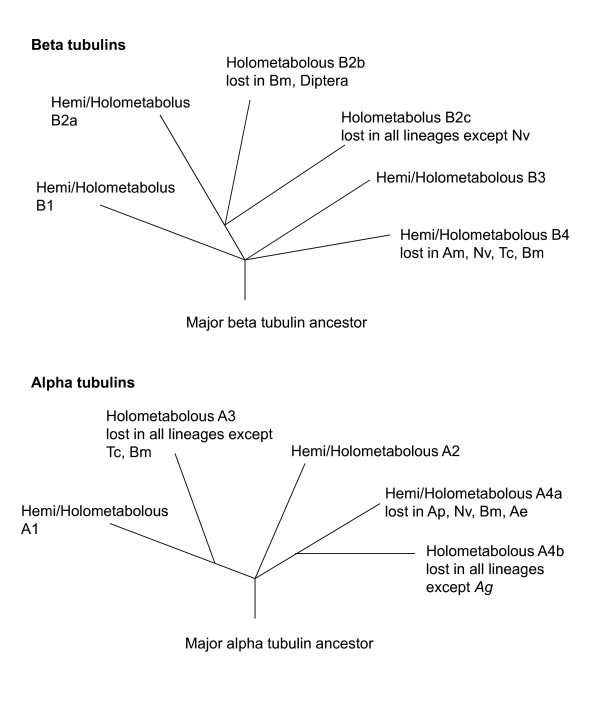
Summary of tubulin isoform relationships. Each of the β1, β2, β3, and β4 isoforms is represented in both hemimetabolous and holometabolous insect taxa, indicating they evolved prior to the separation of these taxa. The β2 isoform duplicated in holometabolous insects following their separation from hemimetabolous insects, based on the clade containing Amβ2b, Nvβ2b, Nvβ2c, Tcβ2b. The β2b isoform was lost in the Lepidoptera/Diptera ancestor, and the β2c isoform was lost in every holometabolous taxa except Nv. The β4 isoform is represented in hemimetabolous insects and Diptera, indicating independent losses in Hymenoptera, Coleoptera, and Lepidoptera. Each of the α1, α2, and α4 isoforms are represented in both hemimetabolous and holometabolous insect taxa, indicating they evolved prior to the separation of these taxa. The α3 isoforms, present in Tc and Bm, fall within the α1 isoforms, suggesting its origin in a duplication event in the common ancestor of Coleoptera, Lepidoptera, and Diptera that was lost in Dipterans.
A second clade consists of orthologs to the Dmβ2 isoform (0.77), which is testis-specific in both Bm and Dm and supports the motile axoneme [16,17,35]. Insect β2 tubulins share a Gly62 which mediates doublet microtubule interactions [36], and a carboxy terminus axoneme motif "EGEFXXX" (X = Asp or Glu, [37,38]), which serves as a substrate for polyglutamylation and polyglycylation [39,40], post-translational modifications characteristic of motile axonemes. Conversely, the Thr61 Gly62 Ala63 motif, which contributes to the extreme length of the D. melanogaster spermtail [10], is a unique feature of the Drosophila β2 protein. Beta 2 duplicated a number of times, in a Pediculus ancestor (Phβ2a, b: 1.0), an Acyrthosiphon ancestor (Apβ2a-d: 1.0), and in a holometabolous ancestor that is not resolved but inferred by the presence of multiple β2 genes in these taxa (Amβ2b; Nvβ2b, c; Tcβ2b: (0.82)) and was followed by losses of the β2b and β2c products in most taxa (Figs. 1, 2).
A third clade consists in orthologs of the Dmβ3 isoform (1.00), expressed in pre-adult visceral mesoderm, the testis cyst cells, and sensory neurons [16,17,35]. Beta 3 orthologs contain a 6 codon insertion (5 in Phβ3) in the internal variable region of the gene, and are the only beta tubulins that have not duplicated in insects. A fourth clade consists in orthologs of the Dmβ4 isoform (0.90), which is expressed in pre-adult tissues [16,17]. Beta 4 orthologs are the most variable beta tubulins in sequence and in representation among insects, having been lost in the Tc, Am, and Bm lineages. One Beta 4 duplication event is found in insects, in an Aedes ancestor (1.0).
Alpha tubulins
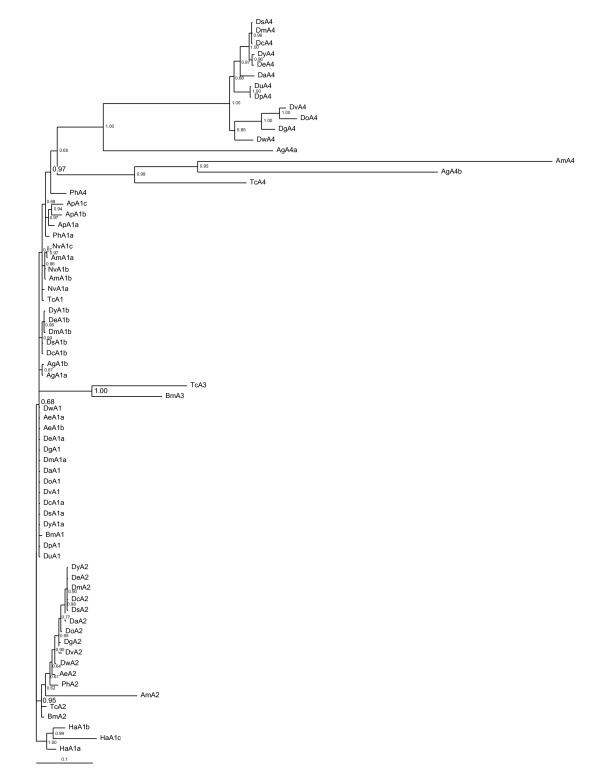
There is support for four alpha tubulin insect clades, though their relationships are less resolved than the beta tubulins (Figs. 2, 3). There is a major α1 clade with numerous polytomies (0.68). These are orthologs of the major Drosophila α1 isoform, expressed in somatic cells and the testis [16,17], and are most conserved alpha tubulins (Table 2). The second clade consists in the minor α2 isoforms (0.95) expressed in Drosophila visceral mesoderm and testis cyst cells [8,16,17]. The α2 isoform is absent in Acyrthosiphon, Nasonia, Anopheles, and D. persimilis and D. psuedoobscura. The α3 clade (1.0) are orthologs of the Bmα3 testis-specific isoforms [35]. The alpha 3 clade falls within the α1 tubulins, indicating its origin in an α1 duplication event in a Coleopteran/Lepidopteran/Dipteran ancestor that was lost in Dipterans. The fourth clade consists in the α4 isoforms (0.97), which is ovary-specific in Dm [16,17], with losses in Ap, Nv, Bm, and Ae.
Figure 3.
Alpha tubulin genealogy. Bayesian reconstruction of insect alpha tubulin evolutionary relationships. Eighty-four tubulins were analyzed with Homarus americanus (Crustacea, Decapoda) alpha tubulins as the outgroup. There are four alpha tubulin clades ancestral to insects, the posterior probability scores in support of these clades are in larger font in the figure: α1 (0.68) which contains numerous polytomies; α2 (0.95); α3 (1.0) present in Bombyx and Tribolium; and α4 (0.97).
Duplications of the major α1 isoform occurred in a number of insect lineages Acyrthosiphon (Apα1a-c: (0.97, 0.94), Hymenoptera [Apis (Amα1a, b) and Nasonia (Nvα1a-c): (0.96)]. While not supported in the zero-fold degeneracy codon tree, a three codon NJ tree (not shown) provides strong support for independent α1 duplication events in Aedes α1a, b:(99), Anopheles Agα1a, b:(98) and melanogaster subgroup Drosophila Ds, Dc, Dy, De and Dmα1a, b: (99) ancestors respectively.
Sequence distances and carboxy terminus tail sequences
The greatest pairwise distances between any two beta and alpha tubulin protein sequences are 0.464 (Tcβ4 vs. Nvβ1) and 0.700 (Agα2 vs. Amα4) respectively, which reveals that a wide range of amino acid sequence identities are capable of supporting microtubule assembly per se (Tables 1, 2). Within this overall diversity, orthologs sequence identities are highly conserved, with average pairwise distances among orthologs less than 0.011 for the major α1 and β1 isoforms, and less than 0.140 for the α2, β2 (excluding divergent duplication products), β3, and β4 minor isoforms. This suggests that tubulin evolution is not constrained by microtubule assembly, but by cell-type specific functions. The α3 testis and α4 ovary-specific isoforms, and testis β2 tubulin duplication products are exceptions, with average pairwise distances >0.23, >0.55 and >0.46 respectively. These reproductive isoforms are the most variable tubulins in sequence. The major isoforms are the most conserved, and there is not a single non-synonymous substitution among the Drosophila β1 and α1 tubulins.
Over 50% of the residues that distinguish tubulin paralogs from each other are found in the carboxy terminus tails (CTT). The CTT lies on the surface of the tubulin protein, being free from protein folding permits its greater variability [29]. Because it serves as a site for many tubulin post-translational modifications (PTMs) and for binding tubulin interacting proteins, it is an important mediator of tubulin function.
PTM motifs
Tubulins can undergo numerous post-translational modifications, which occur on specific tubulin sequence motifs. Polyglutamylation, polyglycylation, detyrosination, and acetylation are modifications associated with stable MT arrays and motile axonemes [40,41], phosphorylated MTs are excluded from mitotic arrays [42], palmitoylation may contribute to the membrane localization of tubulin [43], and interactions with +TIPS (plus-end tracking proteins) have been shown to be inhibited by detyrosination [44].
PTMs that occur on the beta tubulins are polyglutamylation and polyglycylation on a subset of glutamic acid residues on the CTTs [38-40], and phosphorylation of a conserved Ser178 [42]; these sites are found on most beta tubulins (Table 1). The same PTMs that occur on beta tubulins also occur on alpha tubulins, in addition, alpha tubulins can undergo acetylation of Lys40, detyrosination of the 3' terminal Tyr, and palmitoylation of Cys387 [[43-45]; Table 2]. Alpha tubulins show much more variation in PTM motifs than beta tubulins, both between paralogs and among orthologs, indicating alpha tubulins more typically underlie PTM-based microtubule specializations. Note that presence of a PTM sequence motif is necessary, but not sufficient for a PTM to occur; regulation of tubulin modifying proteins will play an important role in PTM-based cell type specializations.
Selection Tests
Selection tests performed between Drosophila tubulin orthologs and recent gene duplication products (Bmβ1a, b; Tcβ2a, b; Nvβ2a-c; Apβ2a-d; Aeβ4a, b; Aeα1a, b; Agα1a, b; Amα1a, b; Nvα1a-c; Apα1a-c) find strong purifying selection; dN/dS ranges from 0.070 to 0.000 in all pairwise comparisons with a high degree of statistical significance (p = 0.00) (Additional File 2). The last 60 nucleotides that comprise the CTT were also tested for mode of selection based on their different role in tubulin folding and function [29]. There is evidence of positive selection acting on the CTT of insect β2 duplication products, and on some D. spp. α4 tubulins (p < 0.01), however, the small alignment length after gap removal requires this result be taken with caution.
Rate Tests
The branch lengths in the genealogy indicate clear differences in tubulin evolutionary dynamics not captured by tests of selection, such that we used Tajima's rate test to identify substitution rate difference among tubulin proteins.
Rates tests between ancient tubulin paralogs
The major and minor insect isoforms have their origin in duplication events on major tubulin ancestors. The lack of overlap in major and minor isoform expression domains in Bombyx and Drosophila, the two insects in which expression data is available, indicates subfunctionalization followed these duplication events, resulting in the minor isoforms. Amino acid rate tests find that minor tubulins evolve more rapidly than major α1 and β1 tubulins (Table 3); divergence in sequence followed duplication and subfunctionalization.
Table 3.
Rate tests between major and minor tubulin paralogs.
| Major isoform | Minor isoform | Rate test (mean Chi-Sq. +/- Sdv) |
|
|---|---|---|---|
| Insect β1 | Insect β2 | 5.12 +/- 3.81 | n = 9 |
| Insect β1 | Insect β3 | 11.85 +/- 4.33 | n = 9 |
| Insect β1 | Insect β4 | 21.34 +/- 21.15 | n = 6 |
| Insect α1 | Insect α2 | 10.66 +/- 17.34 | n = 6 |
| Insect α1 | Insect α3 | 71.19 +/- 9.91 | n = 2 |
| Insect α1 | Insect α4 | 97.87 +/- 62.34 | n = 4 |
Protein evolutionary rates were compared between major and minor tubulin paralogs resulting from ancient duplication events predating separation of insect orders, using Ha major alpha and beta tubulins as outgroups. The average and standard deviation of chi-square values for pairwise rate tests are presented (eg. Insect β1 vs. Insect β2 = (Chi-sq. Dmβ1 vs. Dmβ2 + Chi-sq. Agβ1 vs. Agβ2 + Chi-sq. Aeβ1 vs. Agβ2...)/9). In taxa with multiple copies of an isoform, rate tests are performed using the conserved isoform. The only Drosophila species included is Dm, to avoid a Dipteran skew in the results. Chi-Sq. values > 3.8 have a probability of p < 0.05; Chi-Sq. values > 5.2, p < 0.01.
Rate tests between tubulin orthologs
Tubulin major isoform orthologs vary in the amount of pleiotropy in their function, which may have rate effects. All insects have a β2 and β3 isoforms, but some do not have β4, α2, α3 and/or α4 isoforms. In these taxa the major isoform takes on this minor isoform function, resulting in different amounts of pleiotropy for the major isoforms. For example, the major Nvα1 isoform supports both somatic, testis, and ovary alpha tubulin function, vs. the Tcα1 isoform that only supports somatic function. These differences in pleiotropy do not affect their rates of evolution, all of the insect major α1 and β1 proteins evolve at the same rate respectively (Table 4).
Table 4.
Rate tests on tubulin orthologs.
| Beta 1 | Mean Chi-Sq. Vs. Orthologs (n = 8) |
Beta 2 | Mean Chi-Sq. Vs. Orthologs (n = 8) |
Beta 3 | Mean Chi-Sq. Vs. Orthologs (n = 8) |
Beta 4 | Mean Chi-Sq. Vs. Orthologs (n = 4) |
|---|---|---|---|---|---|---|---|
| Dmβ1 | 1.08 +/- 0.91 | Dmβ2 | 1.02 +/- 2.09 | Dmβ3 | 0.71 +/- 0.70 | Dmβ4 | 1.62 +/- 1.72 |
| Aeβ1 | 1.49 +/- 1.51 | Aeβ2 | 1.47 +/- 3.28 | Aeβ3 | 1.61 +/- 1.46 | Aeβ4a | 1.29 +/- 0.44 |
| Agβ1 | 0.96 +/- 0.98 | Agβ2 | 5.54 +/- 2.11 | Agβ3 | 1.19 +/- 1.20 | Agβ4 | 1.68 +/- 1.65 |
| Bmβ1a | 1.65 +/- 1.05 | Bmβ2 | 1.01 +/- 1.42 | Bmβ3 | 2.31 +/- 1.89 | Apβ4 | 2.53 +/- 1.49 |
| Tcβ1 | 1.19 +/- 1.58 | Tcβ2a | 1.56 +/- 2.47 | Tcβ3 | 2.82 +/- 1.75 | Phβ4 | 29.84 +/-24.42 |
| Amβ1 | 0.97 +/- 0.98 | Amβ2a | 1.50 +/- 1.94 | Amβ3 | 1.44 +/- 1.22 | ||
| Nvβ1 | 0.74 +/- 1.09 | Nvβ2a | 1.25 +/- 2.06 | Nvβ3 | 1.48 +/- 1.35 | ||
| Apβ1 | 2.18 +/- 2.17 | Apβ2a | 0.57 +/- 0.93 | Apβ3 | 2.69 +/- 1.81 | ||
| Phβ1 | 0.51 +/- 0.44 | Phβ2a | 0.18 +/- 0.16 | Phβ3 | 5.91 +/- 3.03 | ||
| Alpha 1 |
Mean Chi-Sq. Vs. Orthologs (n = 8) |
Alpha 2 |
Mean Chi-Sq. Vs. Orthologs (n = 4*) |
Alpha 3 |
Mean Chi-Sq. Vs. Orthologs (n = 1) |
Alpha 4 |
Mean Chi-Sq. Vs. Orthologs (n = 5) |
| Dmα1a | 0.29 +/- 0.23 | Dmα2 | 2.37 +/- 3.34 | Bmα3 | 0.56 | Dmα4 | 7.29 +/- 10.41 |
| Aeα1a | 1.42 +/- 0.98 | Aeα2 | 4.76 +/- 0.60 | Tcα3 | 0.56 | Agα4a | 5.46 +/- 5.83 |
| Agα1a | 0.48 +/- 0.42 | Bmα2 | 1.69 +/- 2.91 | Tcα4 | 13.31 +/- 18.91 | ||
| Bmα1 | 0.74 +/- 1.05 | Tcα2 | 1.51 +/- 2.31 | Amα4 | 22.17 +/- 11.79 | ||
| Tcα1 | 0.43 +/- 0.71 | Amα2 | 56.30 +/- 6.55 | ||||
| Amα1a | 0.45 +/- 0.69 | Phα2 | 1.63 +/- 1.72 | ||||
| Nvα1a | 0.29 +/- 0.25 | ||||||
| Apα1a | 0.44 +/- 0.44 | ||||||
| Phα1a | 0.83 +/- 1.05 | ||||||
Protein evolutionary rates were compared among tubulin orthologs. The average +/- Sdv of chi-square values between each tubulin isoform and its insect orthologs is presented (eg. Dmβ1 = (Chi-sq. Dmβ1 vs. Agβ1 + Chi-sq. Dmβ1 vs. Aeβ1 +...)/8), using Pediculus humanus corporis orthologs as outgroups; Pediculus rates tested with Acyrthosiphon pisum outgroups. In taxa with multiple copies of an isoform, the conserved isoform is used in the rate test. *The highly divergent Amα2 isoform is an outlier, and was removed from the analysis. Chi-Sq. values > 3.8 have a probability of p < 0.05; Chi-Sq. values > 5.2, p < 0.01.
The tubulin minor orthologs in general evolve at the same rate (Table 4), with the exception of the α4 ovary-specific proteins, and divergent testis β2 duplication products (Table 5).
Table 5.
Rate tests on tubulin gene duplication products.
| Product 1 | Product 2 | Rate test (Chi-Sq., p) |
|
|---|---|---|---|
| Bmβ1a | Bmβ1b | 1.80 | p = 0.179 |
| Tcβ2a | Tcβ2b | 79.45 | p = 0.000 |
| Amβ2a | Amβ2b | 5.40 | p = 0.020 |
| Nvβ2a | Nvβ2b | 61.81 | p = 0.000 |
| Nvβ2a | Nvβ2c | 76.17 | p = 0.000 |
| Apβ2a | Apβ2b | 0.14 | p = 0.705 |
| Apβ2a | Apβ2c | 0.00 | p = 1.00 |
| Apβ2a | Apβ2d | 24.00 | p = 0.000 |
| Phβ2a | Phβ2b | 18.96 | p = 0.000 |
| Aeβ4a | Aeβ4b | 11.31 | p = 0.001 |
| Dmα1a | Dmα1b | 2.00 | p = 0.157 |
| Aeα1a | Aeα1b | 1.00 | p = 0.317 |
| Agα1a | Agα1b | 0.33 | p = 0.563 |
| Amα1a | Amα1b | 0.00 | p = 1.000 |
| Nvα1a | Nvα1b | 0.20 | p = 0.654 |
| Nvα1a | Nvα1c | 1.29 | p = 0.256 |
| Apα1a | Apα1b | 7.36 | p = 0.007 |
| Apα1a | Apα1c | 4.00 | p = 0.046 |
| Agα4a | Agα4b | 10.64 | p = 0.001 |
Protein evolutionary rates are compared between tubulin duplication products that postdate separation of insect orders, using Pediculus humanus corporis outgroups. Apβ2a is used as the outgroup in Pediculus rate tests.
Recent duplication events
With respect to more recent duplication events, those post-dating separation of the insect orders, gene duplication did not necessarily result in divergent duplication products. Most recent gene duplications appear in the major alpha 1 and minor beta 2 clades. Recent duplication events always generated at least one conserved product (evolving at the same rate as its orthologs in taxa that did not experience gene duplication). The second product was also conserved in two of eight minor β2 isoform duplication events, and in six of the eight major α1 isoform duplications (Table 5).
The Bmβ1a and Bmβ1b duplication products are undergoing subfunctionalization. Both have wide, but only partially overlapping expression domains; of 16 expression domains tested, they overlap in five, nine are unique to Bmβ1a, and two are unique to Bmβ1b [35]. Both Bmβ1a and Bmβ1b proteins have the same substitution rates as other β1 isoforms (Table 5). The Dmα1a, b products of the melanogaster subgroup α1 duplication are also undergoing subfunctionalization, but of a different kind. Dmα1b it is expressed in the same domain as the Dmα1a duplication product, but at much reduced levels [16,17]. These products are evolving at the same, slow rate as the other α1 proteins (Table 5).
Co-evolution between alpha and beta tubulin
Co-functional links between alpha and beta tubulin must be relatively strong to be detected in co-evolution between proteins. Nonetheless, Sato's Mirror-Tree test of co-evolution finds that the alpha-beta tubulins in the Drosophila α2-β3 dimer co-evolve (Correlation 0.4258, p < 0.01). As there is nothing unusual about the α2-β3 dimer to indicate it is not representative of other tubulins, co-function in the dimer may attenuate rates of tubulin evolution, as change in one tubulin to some extent requires change in the other. Contacts between alpha and beta tubulin along the protofilament and between filaments (inter- and intradimer contacts) are known [46], based on these, none of the α2-β3 co-evolving amino acids are in contact with each other. On the other hand, the CTT residues are sites of PTMs, and co-function in this regard could underlie α2-β3 co-evolution. Evolving amino acids in α2: (first, nucleotide-binding domain: 42, 50, 68, 70, 128; second, taxol-binding domain: 236, 289; third, carboxy-terminus domain: 430, 448, CTT 451-57). Evolving amino acids in β3: (nucleotide-binding domain: 5, 23, 32, 33, 47, 58, 60, 98; taxol-domain: 291; CTT: 442-444.
Intron evolution
Forty-five different introns are present in alpha and beta tubulins (Fig. 4, Additional Files 3, 4), and their distribution indicates they are very mobile. Given the potential for intron insertion and removal to alter tubulin coding sequence, the abundance and dynamic movement of introns among tubulins is surprising.
Figure 4.
Beta and alpha tubulin introns. Beta and alpha tubulin introns are plotted on an insect phylogeny [19], and on Dmα1 and Dmβ1 protein sequences. Introns are labeled A-W and A-V, from the most 5' to most 3' intron found in beta and alpha tubulins. Introns for taxa with multiple copies of an isoform are presented in order, ie. Nvα1a/Nvα1b/Nvα1c.
One way to assess intron evolution is to plot their presence on an insect phylogeny [19], and assume that introns present in more than one isoform were present in the major isoform ancestor to all insect tubulins (Fig. 4). Twenty-three beta tubulin introns were identified. The DGIPRST and U introns each are unique gains in the beta tubulins in which they reside. The CJMP and Q introns are common to insect β4, with losses of MP and Q in Dipteran β4. The remaining 9 introns, ABEFHKLNO, are present in >1 beta tubulin isoform, suggesting they were present in the major beta ancestor to the insect tubulins. However, except for the A intron, the number of independent losses required for this explanation seems sufficiently large to argue against it. Rather, a combination of independent gains, losses, and lateral transfer via recombination between paralogs, for example in EN and P, likely explains their representation.
Twenty-two alpha tubulin introns were identified. Eleven introns, BFHIKLPQRTU, are unique gains in the alpha tubulins in which they reside. Three introns, DM and S, are found only in α2 isoforms. The remaining seven introns, ACEGJN and O, are present in >1 isoform. Again, except for the A intron, their presence/absence patterns require too many independent losses to assume they were present in the major alpha ancestor to the insect tubulins.
An important mechanism of intron loss is through recombination with reverse transcribed tubulin mRNA sequences [47,48]. The most 5' introns are in general the most conserved in both alpha and beta tubulins, consistent with this mechanism. Mechanisms of intron insertion remain largely a mystery [49,50]. Introns found in two paralogs in the same species, such as the beta tubulin EN and P introns and the alpha tubulin E intron, indicate horizontal transfer of the intron through gene conversion or double recombination between paralogs [50].
The majority of introns are found at sites that are highly conserved across all tubulins (Additional Files 3, 4), suggesting intron insertion must accommodate sequence requirements of the protein, rather than visa versa. There are preferences for certain amino acids bracketing insertion sites, for example, glycine resides bracket 16% of intron splice sites, more than twice their frequency in insect tubulins. There are a few observations indicating intron insertion either altered coding sequence or unusual coding sequence facilitated intron insertion. Five of the 20 unique introns (found only in a single tubulin) are correlated with unusual amino acid identities at the insertion sites, the Amα4 F and I introns, the Amα2 H and G introns, and the Phβ2b D intron.
Tubulin introns are typically large and could therefore be prone to splicing mistakes [51,52]. While ortholog-specific introns are likely promoted by selection, by virtue of the intron insertion event coinciding with the duplication event that led to the isoform, the evolutionary benefit of unique introns in established isoforms requires an explanation. Alternate splicing is not known in tubulin, and thus does not provide a utility to introns. However, regulatory sequences are known to reside in tubulin introns [7,53,54], and if present provide a plausible benefit for intron insertion into established isoforms. Large introns could benefit tubulins, as they reduce Hill-Robertson interference within genes [55].
There is evidence that movement from phase 0 to 1 or 2 accompanies the evolution of old introns as splice signals move from the exon to the intron, while disrupting coding sequence might bias recent introns to phase 0 insertion [56]. There is no association between intron phase and age, 6/11 conserved (old) introns and 7/20 unique (new) introns are phase zero, and except in the few previously mentioned cases, intron insertion regardless of phase does not affect tubulin coding sequence. Intron splice sites tend to remain conserved over time; only the beta tubulin Q and R introns and alpha J and K introns are possibly the same intron undergoing splice site movement.
Evolution of Drosophila beta 2 tubulin cis-regulation
D. melanogaster β2 regulatory elements are conserved in some Diptera, in others they are not found (Table 6). In a subset of these species (D. willistoni, D. ananassae, D. persimilis, D. pseudoobscura) testis expression of the β2 gene was tested, and confirmed through RT-PCR (Fig. 5). The maintenance of testis expression in view of the loss of previously identified testis regulatory elements indicates that compensatory evolution has occurred in their β2 cis-regulation. While the basis of this compensation is not known, it may be more complex than simple re-positioning of regulatory elements, as this would have been identified through our analysis.
Table 6.
Drosophila β2 cis-regulatory sequences.
| Species | B2UE1 | B2UE2 | Inr | B2DE1 | ATG |
|---|---|---|---|---|---|
| D. melanogaster |
(-51) ATCGTAGTAGCCTA |
(-32) GAACAT |
(-3) TTCAGTT |
(+51) AAAATTATACGTTTAAAT |
+172 |
| D. ananassae |
(-103) ACCCGAGTATCGTT |
(-57) GAACAG |
(-3) TCCACCT |
(+47) AAAATTGTACGTTAAAAA |
+212 |
| D. erecta |
(-51) ATCGTAGTAGCCCA |
(-32) GAACAT |
(-3) TTCAGTC |
(+51) AAAATTATACGTTTAAAT |
+211 |
| D. grimshawi |
(-328) ATCAGAATTGTTCG |
(-256) GAATAT |
(-3) CTCATTC |
(+49) AAAATTAAACGTGAAAAA |
+155 |
| D. mojavensis |
(-51) ATCCCAGTAGTTCC |
(-32) GTACAT |
(-3) CTCATTC |
(+48) AAAATTATACGTTAAAAT |
+187 |
| D. persimilis |
(-304) CATGTAGAGACCCA |
(-55) GAACAA |
(-3) CTCATTC |
(+43) TAACTTAAAAAATTCATT |
+196 |
| D. pseudoobscura |
(-304) CATGTAGAGACCCA |
(-55) GAACAA |
(-3) CTCATTC |
(+43) TAACTTAAAAAATTCATT |
+195 |
| D. sechellia |
(-51) ATCGCAGTAGCCTA |
(-32) GAACAT |
(-3) TTCAGTT |
(+51) AAAATTATACGTTTAAAT |
+169 |
| D. simulans |
(-51) ATCGCAGTAGCCTA |
(-32) GAACAT |
(-3) TTCAGTT |
(+51) AAAATTATACGTTTAAAT |
+165 |
| D. virilis |
(-51) ATCGAAGTAGTCTA |
(-32) GGACAT |
(-3) CTCATTC |
(+48) AAAATTATACGTAAAAAT |
+169 |
| D. willistoni |
(-189) ATCGAAGAATATTA |
(-165) GAACAT |
-(3) TCCAGCT |
(+46) AAAATTATTCGTACAAAA |
+205 |
| D. yakuba |
(-51) ATCGTAGTAGCCCA |
(-32) GAATAT |
(-3) CTCAGTC |
(+51) AAAATTATACGTTTAAAT |
+155 |
| Anopheles gambiae |
(-79) GCCGTACGTGCCGG |
(-52) GAACCT |
(-3) TCCATTC |
(+45) AAACTAGAAATTTGTGTA |
+188 |
The cis-regulatory elements required for β2 tubulin expression in the testis at appropriate levels have been identified in D. melanogaster; these sequences (when identifiable) are presented for other Drosophila species. Numerical positions indicating sequence position relative to transcription start site (= +1). The B2UE1 element is required for testis-specific gene expression, the B2UE2 and B2DE1 elements for proper expression levels, the Inr element is part of the β2 core promotor, and ATG is the start of β2 coding sequence.
Figure 5.
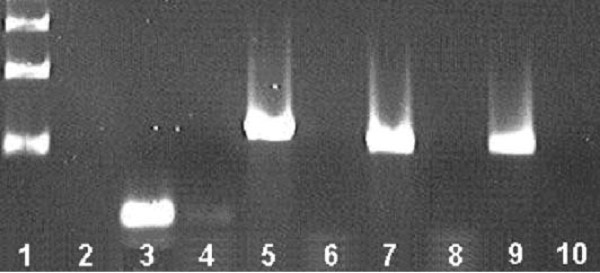
β2 mRNA expression in Drosophila testis tissue. Testis expression of Beta 2 message was tested in D. ananassae, D. willistoni, D. persimilis, and D. psuedoobscura, species in which the B2UE1 testis cis-regulatory sequence was not identifiable (Table 6). All were found to express Beta 2 in the testis, indicating compensatory mutation in testis cis-regulation has occurred to maintain their testis expression. Lane Numbers (L to R) 1. Ladder (bottom most band = 500 bp). 2. blank 3. D. willistoni RT-PCR 4. D. willistoni Taq PCR 5. D. ananassae RT-PCR 6. D. ananassae Taq PCR 7. D. persimilis RT-PCR 8. D. persimilis Taq PCR 9. D. pseudoobscura RT-PCR 10. D. pseudoobscura Taq PCR.
Discussion
Tubulins have stringent structure/function relationships, indicated by strong purifying selection, the loss of many gene duplication products, alpha-beta co-evolution in the tubulin dimer, and compensatory evolution in beta 2 tubulin cis-regulation. Gene duplication, subfunctionalization in expression domain, and divergence, particularly in CTT sequences has resulted in the specialized, minor tissue-specific insect isoforms. Conservation of ortholog sequence identities and expression patterns in Bm and Dm suggests ortholog function might be ancient and largely shared among insects, having been established in their common ancestor. The exception to conservation is in the α3, α4, and β2b, c isoforms. The great sequence variability in these reproductive tissue-specific tubulins indicates species-specific function, and illustrates that even a highly conserved protein family can participate in the adaptive process and respond to sexual selection [57,58].
Pairwise distances between tubulin paralogs reveals a wide variety of tubulin sequences are able to generate an MT array, such that the slow rate of ortholog evolution does not result from a lack of sequences able to generate microtubule arrays. Furthermore, testis-specific isoforms support a wider diversity of microtubule arrays than do major somatic tubulins, yet show more sequence diversity than major isoforms, moreover, somatic tubulins with reproductive function, like Nvα1, do not evolve more slowly than those without. These observations indicate that pleiotropy in microtubule array support does not constrain tubulin evolution; more generally, that support of MT arrays per se is not main source of purifying selection on tubulin sequence.
Path-dependence in the order of amino acid change has been proposed as an important constraint in the evolution of beta 2 tubulin residues that participate in an amino acid synergism [1], and may be a general constraint in residues involved in protein folding. This local constraint would result in purifying selection, yet allow for variation among paralogs to build over time as viable evolutionary pathways are found.
In addition, ortholog conservation may result from support of more subtle, cell-type specific aspects of tubulin function that involve sorting among different MT arrays and the timing of MT array generation. These aspects are mediated by CTT sequences. CTTs do not participate in protein folding, but mediate the tubulin code by providing sequence motifs for PTMs, and by mediating interactions with tubulin associated proteins. CTT sequences can influence subcellular localization of different MT arrays, interactions with plus-end tracking proteins (+TIPS) that influence dynamic instability, and sites for motor proteins to preferentially bind [41]. CTT variation can provide MT specializations, for example, insects with unusual axonemes show reduced levels of both polyglutamylation and polyglycylation [39,59]. Conversely, avoiding unusual MT arrays may contribute to the conservation of major somatic isoform CTTs, which need to function in "normal" MT arrays across a diversity of cell types.
Role of gene duplication in tubulin evolution
Insight into the ancient duplication events that generated the major and minor insect tubulins can be found in more recent duplication events. Duplication events that post-date separation of the insect orders are unevenly distributed among tubulins, with most occurring on alpha 1 and beta 2 templates. Many duplication products are lost, likely because they are deleterious; tubulins are incorporated into MT arrays as a function of cellular concentration, thus diverging duplication products have the potential to poison existing MT arrays, resulting in selection against them. This argues against the classical model of duplication and divergence [60], as without positive selection, in most cases a duplicate gene would be lost before finding novel function. The duplication-degeneration-complementation model [61] proposes that degenerative mutations may accumulate in each duplication product, resulting in subfunctionalization. This alleviates the need for positive selection to operate in order to maintain duplicated genes. Subfunctionalization may explain why major and minor duplication products differ in their fate: 8/10 minor isoform duplications result in a rapidly-evolving and a conserved product, while 7/9 major isoform duplications result in two equally conserved products. Minor isoform expression is already confined to narrow expression domain, removing cis-evolution (and subfunctionalization) as a potential prerequisite for their retention and diversification.
Roles of beta and alpha tubulin in specialized MT arrays
Beta tubulins vary little in PTM sites, such that functional variation among them resides in the tubulin modifying protein composition of a cell, not the beta tubulin. Conversely, the alpha tubulins both experience a wider range of PTMs, and show more variation in PTM sequence motifs, and therefore might be more fundamental in mediating the tubulin code.
In addition to this role in PTMs, alpha tubulin may also have the greater potential to specialize in function, thereby playing a role in adaptation, because it seems more dispensable. Only one alpha tubulin, α1, is present in every insect order, as compared to three beta tubulins, β1, β2, β3. Loss of the α2 gene in D. persimilis and D. pseudoobscura correlates with short sperm and oval testis morphology unique in their genus. Alpha tubulins also show a great amount of standing variation in "unevolved" α1 duplication products that have the potential to participate in the adaptive process.
On the other hand, Tuszynski in his review of vertebrate tubulins [15] suggests the beta tubulin component may be more associated with MT array specializations, the number of beta minor isoforms is greater in most vertebrates than alpha minor isoforms, and more beta minor isoforms co-function with a major, "vanilla" alpha major isoform than visa versa. This seems to also hold largely true for the insects, as many insect species express only the major alpha isoform, but multiple beta isoforms, while all but Bombyx have 4 distinct beta isoforms.
Co-evolution in the alpha/beta tubulin dimer reveals that both the alpha and the beta component are associated with MT array specializations. Although the particular co-evolving residues do not suggest a clear structural basis for co-evolution in α2 and β3, a basis for co-evolution in mediating the tubulin code is quite possible in co-evolving CTT residues. It has been shown that either the alpha or beta tubulin CTT can serve as the donor of a PTM site [62], an interrelationship that provides a mechanism for PTM-based alpha/beta co-evolution. More generally, specialized MT arrays can be mediated by either dimer component: the specialized Dmβ2 functions with the major Dmα1 tubulin in Drosophila axoneme, the major Dmβ1 functions with a specialized Dmα4 in the ovary, the minor α3/β2 isoforms support the Bombyx motile axoneme, and the minor α2/β3 isoforms support the Drosophila testis cyst cell in which the gigantic spermtails distinctive of Drosophila are generated. Given this variety of relationships between alpha and beta tubulin and MT specializations, which component provides specialization may well be due to evolutionary chance.
Conclusion
Gene duplication alone is not sufficient for tubulin evolution. Most gene duplication products do not survive, whether through direct elimination or pseudogenization. Those that do survive evolve only when followed by a narrowing in the expression domain. Given the number of rare events that must occur (viable gene duplication, subfunctionalization, path-dependent evolution of coding sequence under purifying selection) to result in a novel tubulin, their slow rate of evolution seems best explained by limitations on such opportunities, especially given the great variation in tubulin proteins found in distance comparisons. Such opportunities do not seem exhausted given vertebrates express 7-8 alpha and beta tubulins; the rapid evolution of the reproductive tubulins also reveals a use for divergent tubulins. It also provides chance a fundamental role in shaping tubulin evolution it terms of when these events occur, providing an allele for selection to choose from. "Evidence" of this role being real is seen in the odd distribution of isoforms, duplication events, and divergent duplication products, and which component of the dimer, alpha, beta, or both, underlies a microtubule specialization.
One important exception is in reproductive tissue-specific isoforms, which show a large amount of variation potentially capable of responding to sexual selection, a fundamental force in insect evolution. Reproductive isoforms have the fewest PTMs, and the most unusual spermtail axonemes are accompanied by reduction of PTM modifications [59]. Relaxation of the informational aspect of tubulin function might release tubulins to contribute to specialized testis phenotypes typical of insect evolution. Continued study might show more such relaxations, a form of co-evolution fostering the evolvability of an important gene family.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MN conceived of the study, carried out the sequence retrieval, determination of intron/exon structure, the Bayesian genealogy, rate tests, and drafted the manuscript. SG carried out ML selection test, contributed to the data analysis, and editing the manuscript. LG performed the testes RNA preparations and RT-PCR for the different Drosophila species. All authors read and approved the final manuscript.
Supplementary Material
Insect tubulin sequence accessions. Insect species, tubulin isoform, and accessions are presented. Accessions beginning with gi derive from Flybase blast searches, other accessions from Genbank.
Likelihood tests of mode of selection acting on tubulin. Maximum likelihood tests of selection are performed on Drosophila tubulins, and genes resulting from recent duplication events (those restricted to a single insect order) show that duplicated genes evolve under purifying selection. The entire coding sequence (beta tubulin nt 1-1422, alpha tubulin nt 1-1422) was analyzed. Analyses were conducted using the Nei-Gojobori method in PAML [30]. Alignment gaps were eliminated by complete deletion.
Beta Tubulin Intron Features. Intron representation in insect tubulins, intron phase, length, and splice donor/acceptor sites are presented. The amino acids bracketing the splice site, whether the spice site is within unique 5' and 3' codon sequence, and whether tubulins with the intron share the same sequence is presented, to indicate associations between intron presence/absence and tubulin coding sequence. Key: M = A or C, K = G or T, R = A or G, Y = C or T, W = A or T, S = C or G, V = not T, H = not G.
Alpha Tubulin Intron Features. Intron representation in insect tubulins, intron phase, length, and splice donor/acceptor sites are presented. The amino acids bracketing the splice site, whether the spice site is within unique 5' and 3' codon sequence, and whether tubulins with the intron share the same sequence is presented, to indicate associations between intron presence/absence and tubulin coding sequence. Key: M = A or C, K = G or T, R = A or G, Y = C or T, W = A or T, S = C or G, V = not T, H = not G.
Contributor Information
Mark G Nielsen, Email: mark.nielsen@notes.udayton.edu.
Sudhindra R Gadagkar, Email: sgadagkar@centralstate.edu.
Lisa Gutzwiller, Email: Lisa.Gutzwiller@cchmc.org.
Acknowledgements
The authors thank the National Science Foundation (MCB-0316359 to MN) for their support of this research, and four anonymous reviewers for very helpful comments.
References
- Nielsen MG, Raff EC. The best of all worlds or the best possible world? Developmental constraint in the evolution of beta tubulin and the sperm tail axoneme. Evolution & Development. 2002;4:303–315. doi: 10.1046/j.1525-142X.2002.02015.x. [DOI] [PubMed] [Google Scholar]
- Lewontin RC, Hubby JL. A molecular approach to the study of genic heterozygosity in natural populations. II. of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics. 1966;54:595–609. doi: 10.1093/genetics/54.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheat CW, Watt WB, Pollock DD, Schulte PM. From DNA to fitness differences: sequences and structures of adaptive variants of Colias phosphoglucose isomerase (PGI) Mol Biol Evol. 2006;3:499–512. doi: 10.1093/molbev/msj062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher RB, Hightower RC. The Molecular Evolution of Actin. Genetics. 1986;114:315–332. doi: 10.1093/genetics/114.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrberg EA, Fyrberg CC, Biggs JR, Saville D, Beall CJ, Ketchum A. Functional nonequivalence of Drosophila actin isoforms. Biochem Genet. 1998;36:271–287. doi: 10.1023/A:1018785127079. [DOI] [PubMed] [Google Scholar]
- St. Mivart. On the Genesis of Species (London 1871)
- Hoyle HD, Raff EC. Two Drosophila beta tubulin isoforms are not functionally equivalent. J Cell Biol. 1990;111:1009–26. doi: 10.1083/jcb.111.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchens JA, Hoyle HD, Turner FR, Raff EC. Structurally similar Drosophila alpha-tubulins are functionally distinct in vivo. Mol Biol Cell. 1997;8:481–500. doi: 10.1091/mbc.8.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff EC, Hutchens JA, Hoyle HD, Nielsen MG, Turner FR. Conserved axoneme symmetry altered by a component beta-tubulin. Curr Biol. 2000;10:1391–4. doi: 10.1016/S0960-9822(00)00784-3. [DOI] [PubMed] [Google Scholar]
- Nielsen MG, Turner FR, Hutchens JA, Raff EC. Axoneme-specific β-tubulin specialization a conserved C-terminal motif specifies the central pair. Curr Biol. 2001;11:529–533. doi: 10.1016/S0960-9822(01)00150-6. [DOI] [PubMed] [Google Scholar]
- Nielsen MG, Caserta JM, Kidd SJ, Phillips CM. Functional constraint underlies 60 million year stasis of Dipteran testis-specific β-tubulin. Evol Dev. 2006;8:23–29. doi: 10.1111/j.1525-142X.2006.05072.x. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Gene duplication and the origin of novel proteins. Proc Natl Acad Sci USA. 2005;102(25):8933–8. doi: 10.1073/pnas.0502350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK. The tubulin fraternity: alpha to eta. Curr Opin Cell Biol. 2001;13:49–54. doi: 10.1016/S0955-0674(00)00173-3. [DOI] [PubMed] [Google Scholar]
- McKean PG, Vaughan S, Gull K. The extended tubulin superfamily. J Cell Sci. 2001;114:2723–2733. doi: 10.1242/jcs.114.15.2723. [DOI] [PubMed] [Google Scholar]
- Tuszynski JA, Carpenter EJ, Huzil JT, Malinski W, Luchko T, Luduena RF. The evolution of the structure of tubulin and its potential consequences for the role and function of microtubules in cells and embryos. Int J Dev Biol. 2006;50:341–358. doi: 10.1387/ijdb.052063jt. [DOI] [PubMed] [Google Scholar]
- Matthews KA, Miller DF, Kaufman TC. Developmental distribution of RNA and protein products of the Drosophila alpha-tubulin gene family. Dev Biol. 1989;132:45–61. doi: 10.1016/0012-1606(89)90203-0. [DOI] [PubMed] [Google Scholar]
- Natzel JE, McCarthy BJ. Regulation of Drosophila alpha- and beta-tubulin genes during development. Dev Biol. 1984;104:187–98. doi: 10.1016/0012-1606(84)90047-2. [DOI] [PubMed] [Google Scholar]
- Whiting M. Phylogeny of holometabolus insect orders: molelcular evidence. Zoologica Scripta. 2002;31:3–17. doi: 10.1046/j.0300-3256.2001.00093.x. [DOI] [Google Scholar]
- Savard J, Tautz D, Richards S, Weinstock GM, Gibbs RA, Werren JH, Tettelin H, Lercher MJ. Phylogenomic analysis reveals bees and wasps (Hymenoptera) at the base of the radiation of Holometabolous insects. Genome Res. 2006;16(11):1334–8. doi: 10.1101/gr.5204306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarenko AG. In: Biology, phylogeny, and classification of Coleoptera: papers celebrating the 80th birthday of Roy A. Crowson. Pakaluk, Slipinski, editor. Museum and Institute of Zoology Warsaw, Poland; 1995. The geological history of beetles; pp. 155–171. [Google Scholar]
- Evenhuis NL. Catalogue of the fossil flies of the world (Insecta: Diptera) Leiden: Backhuys Publishers; 1994. [Google Scholar]
- Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Sequencher. Gene Codes, Ann Arbor, MI.
- Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1998;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings in Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, John P, Huelsenbeck JP, Mark P van der. Mr. Bayes v. 3.1. 2005.
- Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: a program package for phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Yamanishi Y, Horimoto K, Kanehisa M, Hiroyuki Toh. Partial correlation coefficient between distance matrices as a new indicator of protein-protein interactions. Bioinformatics. 2006;22(20):2488–2492. doi: 10.1093/bioinformatics/btl419. [DOI] [PubMed] [Google Scholar]
- Santel A, Kaufmann J, Hyland R, Renkawitz-Pohl R. The initiator element of the Drosophila beta2 tubulin gene core promoter contributes to gene expression in vivo but is not required for male germ-cell specific expression. Nucleic Acids Res. 2000;28:1439–46. doi: 10.1093/nar/28.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. Drosophila. A laboratory handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY; 1998. [Google Scholar]
- Kawasaki H, Sugaya K, Quan GX, Nohata J, Mita K. Analysis of alpha- and beta-tubulin genes of Bombyx mori using an EST database. Insect Biochem Mol Biol. 2003;33:131–7. doi: 10.1016/S0965-1748(02)00184-4. [DOI] [PubMed] [Google Scholar]
- Raff EC, Hoyle HD, Popodi EM, Turner FR. Axoneme β-tubulin sequence determines attachment of outer dynein arms. Current Biology. 2008;18:911–914. doi: 10.1016/j.cub.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff EC, Fackenthal JD, Hutchens JA, Hoyle HD, Turner FR. Microtubule architecture specified by a β-tubulin isoform. Science. 1997;275:70–73. doi: 10.1126/science.275.5296.70. [DOI] [PubMed] [Google Scholar]
- Vent J, Wyatt TA, Smith DD, Banerjee A, Ludueña RF, Sisson JH, Hallworth R. Direct involvement of the isotype-specific C-terminus of beta tubulin in ciliary beating. J Cell Sci. 2005;118(Pt 19):4333–41. doi: 10.1242/jcs.02550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle HD, Turner FR, Raff EC. Axoneme-dependent tubulin modifications in singlet microtubules of the Drosophila sperm tail. Cell Motil Cytoskeleton. 2008;65(4):295–313. doi: 10.1002/cm.20261. [DOI] [PubMed] [Google Scholar]
- Xia L, Hai B, Gao Y, Burnette D, Thazhath R, Duan J, Bré M-H, Levilliers N, Gorovsky MA, Gaertig J. Polyglycylation of tubulin is essential and affects cell motility and division in Tetrahymena thermophila. J Cell Biol. 2000;149:1097–1106. doi: 10.1083/jcb.149.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey K, Gaertig J. The Tubulin Code. Cell Cycle. 2007;6:2152–2160. doi: 10.4161/cc.6.17.4633. [DOI] [PubMed] [Google Scholar]
- Fourest-Lieuvin A, Peris L, Gache V, Garcia-Saez I, Juillan-Binard C, Lantez V, Job D. Microtubule regulation in mitosis: tubulin phosphorylation by the cyclin-dependent kinase Cdk1. Mol Biol Cell. 2006;17(3):1041–50. doi: 10.1091/mbc.E05-07-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambito AM, Wolff J. Palmitoylation of tubulin. J Biochem Biophys Res Commun. 1997;239(3):650–4. doi: 10.1006/bbrc.1997.7525. [DOI] [PubMed] [Google Scholar]
- Peris L, Thery M, Fauré J, Saoudi Y, Lafanechère L, Chilton JK, Gordon-Weeks P, Galjart N, Bornens M, Wordeman L, Wehland J, Andrieux A, Job D. Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. J Cell Biol. 2006;174(6):839–49. doi: 10.1083/jcb.200512058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo1 A, Ackerman B, Gundersen GG. Tubulin acetylation and cell motility. Nature. 2003;421:230. doi: 10.1038/421230a. [DOI] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;96(1):79–88. doi: 10.1016/S0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Trelles F, Tarrío R, Ayala FJ. Origins and evolution of spliceosomal introns. Annu Rev Genet. 2006;40:47–76. doi: 10.1146/annurev.genet.40.110405.090625. [DOI] [PubMed] [Google Scholar]
- Sverdlov AV, Babenko VN, Rogozin IB, Koonin EV. Preferential loss and gain of introns in 3' portions of genes suggests a reverse-transcription mechanism of intron insertion. Gene. 2004;338(1):85–91. doi: 10.1016/j.gene.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Koonin EV. The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biology Direct. 2006;1:22. doi: 10.1186/1745-6150-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW. The origin of recent introns: transposons? Genome Biology. 2004;5:251. doi: 10.1186/gb-2004-5-12-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox-Walsh KL, Dou Y, Lam BJ, Hung SP, Baldi PF, Hertel KJ. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc Natl Acad Sci USA. 2005;102(45):16176–81. doi: 10.1073/pnas.0508489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel KJ. Combinatorial control of exon recognition. JBC. 2008;283(3):1211–1215. doi: 10.1074/jbc.R700035200. [DOI] [PubMed] [Google Scholar]
- Gasch A, Hinz U, Renkawitz-Pohl R. Intron and upstream sequences regulate expression of the Drosophila f83-tubulin gene in the visceral and somatic musculature, respectively. Proc Natl Acad Sci USA. 1989;86:3215–3218. doi: 10.1073/pnas.86.9.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianıà S, Morelloa L, Bardinib M, Breviarioa D. Tubulin intron sequences: multi-functional tools. Cell Biology International. 2003;27:203–205. doi: 10.1016/S1065-6995(02)00301-3. [DOI] [PubMed] [Google Scholar]
- Marais G, Nouvellet P, Keightley PD, Charlesworth B. Intron size and exon evolution in Drosophila. Genetics. 2005;170(1):481–5. doi: 10.1534/genetics.104.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sverdlov AV, Rogozin IB, Babenko VN, Koonin EV. Evidence of splice signal migration from exon to intron during intron evolution. Curr Biol. 2003;13(24):2170–4. doi: 10.1016/j.cub.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Civetta A, Singh RS. High divergence of reproductive tract proteins and their association with postzygotic reproductive isolation in Drosophila melanogaster and Drosophila virilis group species. J Mol Evol. 1995;41:1085–1095. doi: 10.1007/BF00173190. [DOI] [PubMed] [Google Scholar]
- Torgerson DG, Singh RS. Rapid Evolution Through Gene Duplication and Subfunctionalization of the Testes-Specific α4 Proteasome Subunits in Drosophila. Genetics. 2004;168:1421–1432. doi: 10.1534/genetics.104.027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencarelli C, Caroti D, Bré MH, Levilliers N, Dallai R. Tubulin glycylation and glutamylation deficiencies in unconventional insect axonemes. Cell Motil Cytoskeleton. 2005;61(4):226–36. doi: 10.1002/cm.20081. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by Gene Duplication. Springer Verlag, New York; 1970. [Google Scholar]
- Force A, Lynch M, F Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative and speciation: a new paradigm. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Gorovsky MA. Both carboxy-terminal tails of alpha- and beta-tubulin are essential, but either one will suffice. Curr Biol. 2002;12(4):313–6. doi: 10.1016/S0960-9822(02)00651-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Insect tubulin sequence accessions. Insect species, tubulin isoform, and accessions are presented. Accessions beginning with gi derive from Flybase blast searches, other accessions from Genbank.
Likelihood tests of mode of selection acting on tubulin. Maximum likelihood tests of selection are performed on Drosophila tubulins, and genes resulting from recent duplication events (those restricted to a single insect order) show that duplicated genes evolve under purifying selection. The entire coding sequence (beta tubulin nt 1-1422, alpha tubulin nt 1-1422) was analyzed. Analyses were conducted using the Nei-Gojobori method in PAML [30]. Alignment gaps were eliminated by complete deletion.
Beta Tubulin Intron Features. Intron representation in insect tubulins, intron phase, length, and splice donor/acceptor sites are presented. The amino acids bracketing the splice site, whether the spice site is within unique 5' and 3' codon sequence, and whether tubulins with the intron share the same sequence is presented, to indicate associations between intron presence/absence and tubulin coding sequence. Key: M = A or C, K = G or T, R = A or G, Y = C or T, W = A or T, S = C or G, V = not T, H = not G.
Alpha Tubulin Intron Features. Intron representation in insect tubulins, intron phase, length, and splice donor/acceptor sites are presented. The amino acids bracketing the splice site, whether the spice site is within unique 5' and 3' codon sequence, and whether tubulins with the intron share the same sequence is presented, to indicate associations between intron presence/absence and tubulin coding sequence. Key: M = A or C, K = G or T, R = A or G, Y = C or T, W = A or T, S = C or G, V = not T, H = not G.