Abstract
Molecular dissociation rates have long been known to be sensitive to applied force. We use a laser trap to provide evidence that rates of association may also be force-dependent. We use the thermal fluctuation assay to study single bonds between E-selectin and sialyl Lewisa (sLea), the sugar on PSGL-1 to which the three selectins bind. Briefly, an E-selectin-coated bead is held in a laser trap and pressed with various compressive loads against the vertical surface of a bead coated with sLea. The time it takes for a bond to form is used to calculate a specific two-dimensional on-rate, . We observe an increase in with increasing compressive force, providing single molecule evidence that on-rate, in addition to off-rate, is influenced by load. By measuring bond lifetimes at known tensile loads, we show that E-selectin, like its family members L- and P-selectin, is capable of forming catch bonds. Our data support a reverse Bell model, in which compressive forces lower the activation energy for binding. Load-dependent on-rates may be a general feature of all intermolecular bonds.
Keywords: Catch bonds, On-rate, Single molecule, Dynamic force spectroscopy
Introduction
The selectins are a family of transmembrane adhesion molecules responsible for the initial capture of flowing leukocytes by the vessel wall, as well as the subsequent rolling of leukocytes along the endothelial surface. The selectin family is comprised of L-selectin, found on most leukocytes, P-selectin, expressed primarily on activated endothelial cells as well as activated platelets, and E-selectin, found on activated endothelium. The most common selectin ligand is P-selectin glycoprotein ligand-1 (PSGL-1), a dimeric glycoprotein found on the surface of both leukocytes and the endothelium. E-selectin/PSGL-1 binding has been shown to be crucial for leukocyte tethering under flow.60 Selectin/ligand bonds must possess on-rates high enough to form bonds between a stationary endothelial cell and a rapidly moving leukocyte, and off-rates high enough for the rapid bond dissociation necessary for leukocyte rolling. Rolling rates on the three types of selectins vary greatly, suggesting the molecules do not share the same kinetic properties.39,44 The underlying molecular kinetics responsible for leukocyte rolling have been investigated primarily in L- and P-selectin,10,20,26,29,34,42,43,48,58,61 while single molecule studies of E-selectin kinetics remain few.20,48,51
L- and P-selectin have been shown to form catch-slip bonds with PSGL-1.29,43 Catch–slip bonds are bonds that initially increase in lifetime with increasing force, and subsequently decrease in lifetime when a critical force is reached. The existence of catch bonds was first proposed by Dembo9 in 1988, but they were not observed experimentally until recent years.17,29,54 Though catch bonds have been found in such diverse molecules as the actin–myosin system,17 integrin-mediated adhesion,24 and bacterial fimbria,54 their properties have been most widely investigated at the single molecule level using bonds between selectins and their corresponding ligands. Though bonds between P-selectin and PSGL-129 and L-selectin and PSGL-143 behave as catch–slip bonds, E-selectin has not been shown to form such bonds with any of its ligands. This is perhaps due to the fewer56 and less well defined ligands for E-selectin, causing it to be the least studied member of its family.
A recent flow chamber study34 suggested that like the rates of bond rupture, the rate of L-selectin/PSGL-1 bond formation may be a function of force. This complements an earlier study showing that increasing contact stress, while controlling for contact area, results in higher intrinsic rates of bond formation for bonds between β2-integrins and intercellular adhesion molecule-1 (ICAM-1).46 The authors attributed this to microvillus compression, but a direct increase in molecular binding rate is certainly a possibility.
Here, we use an optical trapping system to study single bonds between E-selectin and sialyl Lewisa (sLea), a sugar on PSGL-1 to which the three selectins bind. The sialyl Lewis sugars (sLea and sLex) of PSGL-1 are known to be recognized by E-selectin23,27,37 and evidence suggests that their affinity for E-selectin is stronger than that for either L- or P-selectin.55 We observe for the first time clear evidence that E-selectin is capable of forming catch bonds with one of its ligands. We also find that the intrinsic rate of bond formation between E-selectin and sLea is enhanced by compressive load. Our laser trap method allows us to observe multiple bonds rupturing sequentially, enabling us to directly determine which of these binding events are in fact single bonds. We present simple physical models to explain how on-rate can be influenced by compressive force, and relate these to the catch bond phenomenon.
Methods
Bead Preparation
Chimeric recombinant human E-selectin (R&D Systems, Minneapolis, MN, USA) was covalently coupled to 0.92 μm carboxylated polystyrene beads (Bangs Laboratories, Fishers, IN, USA) using carbodiimide crosslinking.5 First, 12.5 μL (10% solids) beads were suspended in 1 mL of 50 mM MES, pH 6.1 (MES activation buffer). Beads were washed by centrifugation 3× with 1 mL MES activation buffer and resuspended in 50 μL MES activation buffer following the final wash. Next, 500 μL of 200 mM NHS, followed by 500 μL of 4% (w/v) EDAC, was added while mixing. This was allowed to react for 30 min at room temperature on a circular rotator. Beads were then washed twice with 1 mL of 50 mM MES, pH 7.4 (MES coupling buffer) and were resuspended in 100 μL MES coupling buffer following the final wash. The desired amount of E-selectin was added to the beads and they were rotated overnight at 4 °C. Beads were then washed twice with 1 mL of MES coupling buffer and resuspended in 1 mL of 0.1 M ethanolamine, followed by rotation for 30 min at room temperature. Beads were then washed twice with 1 mL of “flow cell buffer” (150 mM NaCl, 25 mM Imidazole, 1 mM CaCl2, 1 mM NaN3, pH 7.4), resuspended in 1 mL of flow cell buffer with 0.5% Tween-20 for blocking, and rotated at room temperature for 1 h. Beads are stored at 4 °C.
Trapping Protocol
Selectin-ligand association kinetics have traditionally been studied in three-dimensional (solution) experiments, where at least one species of the receptor– ligand pair is in solution.21 Leukocyte binding to the vessel wall is approximately two-dimensional, with a receptor confined to the plane of the leukocyte membrane and a ligand confined to the plane of the endothelial cell membrane. Thus 2-D kinetic constants are perhaps more appropriate than the 3-D kinetic constants. While previous studies have examined the load-dependent 2-D off-rates of selectin/PSGL-1 binding,10,29 only a few studies to date have attempted to directly measure 2-D on-rates for any selectin interaction.7,8,48 We used a variation on one of these assays7 to measure the time to bond formation, from which 2-D on-rates are directly calculated; our variation used a laser trap transducer rather than a biomembrane force probe.
The laser trap used in these studies is described elsewhere,16 except that a 1090 nm fiber laser was used (SP25C, SPI, Santa Clara, CA, USA). Trap stiffness and sensitivity were determined by step response and power spectral density methods.50
Coverslips were prepared as described15,17 by spraying a suspension of ∼3 μm glass beads (Polysciences, Warrington, PA, USA) onto a 22 × 22 mm2 glass coverslip and then coating with a thin layer of nitrocellulose. An 18 × 18 mm2 glass coverslip was then used to assemble a 25 μL flow cell. Streptavidin (MP Biomedicals, Solon, OH, USA) was incubated at the desired concentration in the flow cell for 15 min at room temperature. The flow cells were then blocked with 1% Tween-20 overnight at 4 °C to reduce nonspecific interactions. The desired concentration of sialyl Lewisa–PAA–biotin (GlycoTech, Gaithersburg, MD, USA) was then incubated in the flow cells for 15 min at room temperature. E-selectin-coated beads were sonicated to minimize aggregation, diluted in flow cell buffer, and added to the flow cell.
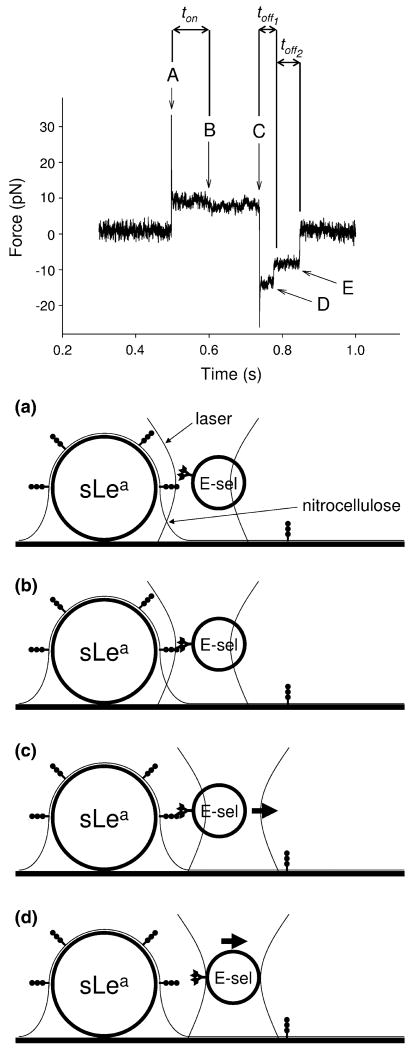
An E-selectin-coated bead was then trapped and brought into contact with a sLea-coated bead on the surface of the flow cell (Fig. 1a). Note that the laser is typically moved closer to the sLea-coated bead than necessary to bring the receptor and ligand molecules on opposing bead surfaces within binding range, thus generating a compressive load between the beads. The beads were held in contact for a sufficiently long time to form a bond (Fig. 1b). The trapped bead was then stepped away from the stationary bead (Fig. 1c), applying a load to any bond or bonds that had formed until bond rupture (Fig. 1d). The trap data were analyzed as follows. When the trapped bead was initially brought into contact with the stationary bead (Fig. 1a), there was a characteristic mean and variance of the signal determined by noise and overlap of the laser with the stationary bead. When a bond formed between receptor and ligand on opposing bead surfaces (Fig. 1b), there was a small shift of the signal mean and a change in variance. The shift of the signal mean is presumably due to interacting receptor/ligand pairs not being centered within the contact area prior to binding. Bond formation thus results in the trapped bead moving slightly out of trap center. The change in variance is a result of the trapped bead being tethered to the stationary bead once a bond has formed. These measurable changes allowed the time to bond formation (ton) to be measured. When the trapped bead was stepped away (Fig. 1c), the signal shifted to a new baseline and the laser exerted a near-instantaneous force on the bead, while the receptor–ligand bond effectively prevented the bead from moving to the new trap position. When the bond broke (Fig. 1d), the bead moved toward the trap center and the signal returned to baseline. This allowed us to definitively say if a bond indeed formed between molecules on the opposing bead surfaces. The load applied to the bond and the lifetime of the bond (toff) under the applied load could be measured. If multiple bonds formed, it was possible to observe the rupture of each individual bond separately (Fig. 1d, e). This allowed us to state with some confidence whether an individual event was due to a single bond or multiple bonds. Upon stepping the trapped bead toward the stationary bead, we compared the known distance the laser was displaced to the measured displacement of the trapped bead. This allowed us to measure how much compressive force was being exerted when the beads were brought together.
FIGURE 1.
Experimental setup and representative laser trap data of a bond event. Letters in the data trace correspond with similarly lettered panels in the experimental setup diagrams. The trapped bead is stepped toward the stationary bead (a), a bond forms (b), the trapped bead is stepped away from the stationary bead (c), a bond ruptures (d), and if there was a second bond present, a second bond ruptures (e). Measurements of ton and toff are used to determine on-rate and off-rates, respectively. In the case of multiple bonds, the lifetime of the nth bond is designated toffn.
Protein Site Density Determination
The site density of E-selectin on microspheres was determined by time-resolved fluorescence detection of Eu-labeled streptavidin (PerkinElmer Wallac, Turku, Finland).18,49 Beads were prepared as described previously. They were resuspended in biotinylated anti-E-selectin antibody (HAE-1f, Ancell Corporation, Bayport, MN, USA) at 10 μg/mL in flow cell buffer and incubated on ice for 1 h. After three washes in flow cell buffer, beads were resuspended in Eu-streptavidin at 100 ng/mL in PBS and incubated on ice for 1 h. Beads were then washed five times with flow cell buffer, resuspended in DELFIA enhancer solution (PerkinElmer), and aliquoted into a 384-well plate. After 5 min of incubation at room temperature, the time resolved fluorescence was measured at 355 nm excitation and 615 nm emission with a detection interval of 50–1450 μs on a BMG spectrofluorometer.
The total streptavidin (active and inactive) site density on the flow cell surface was measured by incubation of known concentrations of Eu-streptavidin for 15 min at room temperature in flow cells made with a nitrocellulose-coated coverslip. After washing with five volumes of PBS, 100 μL of DELFIA enhancer solution was simultaneously injected and removed from each flow cell. The solution was transferred to a 384-well plate and the time resolved fluorescence was measured as described previously. The site density of active streptavidin and sLea on the flow cell surface was determined by the rate of enzymatic production of a chemiluminescent signal by biotinylated alkaline phosphatase (biotin-AP, New England BioLabs, Ipswich, MA, USA). Briefly, streptavidin at various concentrations was incubated for 15 min at room temperature in flow cells made with a nitrocellulose-coated coverslip. After washing with five volumes of flow cell buffer, flow cells were blocked by overnight incubation of 1% 40 kDa polyvinylpyrrolidone (PVP) at 4 °C. sLea was then incubated for 15 min at room temperature in one flow cell at each streptavidin concentration. Flow cells were washed with 5 vol of PBS and biotinylated alkaline phosphatase at 10 μg/mL was then incubated in each flow cell for 1 h at room temperature. After washing with 5 vol of PBS, chemiluminescent substrate with 5% enhancer by volume was added (Invitrogen, Calsbad, CA, USA). Flow cells were immediately imaged for 30 min with 1 min exposures on an Alpha Innotech FluorChem 8900 imager. The density of biotin binding sites was found for flow cells with and without the addition of sLea, and the difference between the two values indicated the site density occupied by sLea–PAA molecules.
Results
Sequential Bond Ruptures Can Be Observed in Real-Time
A problem that plagues many experimental systems built to study single intermolecular interactions is ascertaining whether one is actually observing single molecules or bonds. The presence of multiple bonds is a concern as most force spectroscopy methods are incapable of definitively distinguishing them from bonds that are truly between a single receptor–ligand pair. A common method of inferring single bonds based on Poisson's law of small numbers is to argue that when a low frequency of test cycles results in a binding event, a very low percentage of these binding events should be multiple bonds.62 Other methods employed to argue that single bonds are being measured include looking at how adhesion frequency varies with protein site density,8 fitting tether lifetime data from flow chamber studies to the equations of single-bond kinetics,1,2,6,40,41,44 looking for quantal behavior in measurements of low number molecular interactions,2,12 and looking for binding events that have the kinetic and mechanical properties on the order of magnitude of those that can be predicted or inferred.8 None of these methods are able to discern the presence of single bonds on datum-by-datum basis.
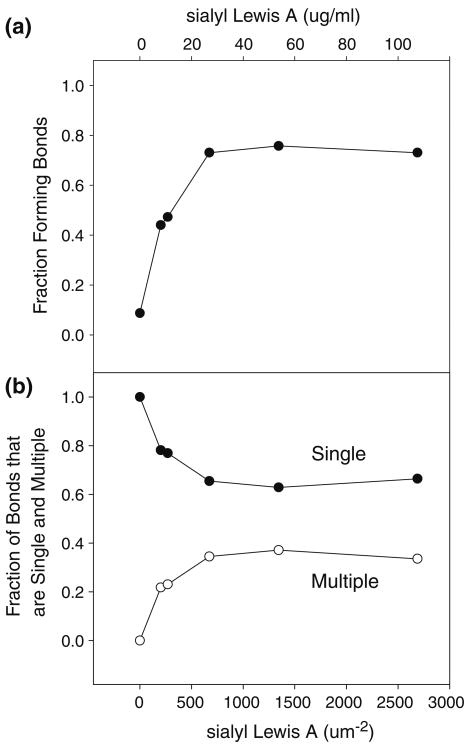
The method utilized here allowed us to accurately determine the presence and frequency of single bond events, and, in the event of multiple bonds, the number of bonds comprising each multiple bond. We saw that the fraction of bonds that are multiple bonds increased with increasing sLea concentration (Fig. 2b), suggesting the bonds were specific receptor–ligand bonds. The fraction of total contacts that form a bond (Fig. 2a) did not reach 1, suggesting that the E-selectin-coated surface was not saturated with protein. If a receptor– ligand interaction is comprised of multiple bonds, we were able to observe the dissociation of each individual bond (Fig. 1).
FIGURE 2.
(a) The fraction of bead contacts forming bonds (●) as a function of sLea concentration (top x-axis) and sLea site density (bottom x-axis). (b) The fraction of bonds that are single bonds (●) and multiple bonds (○) as a function of sLea concentration and sLea site density.
Bonds Between E-Selectin and sLea Exhibit Catch–Slip Behavior
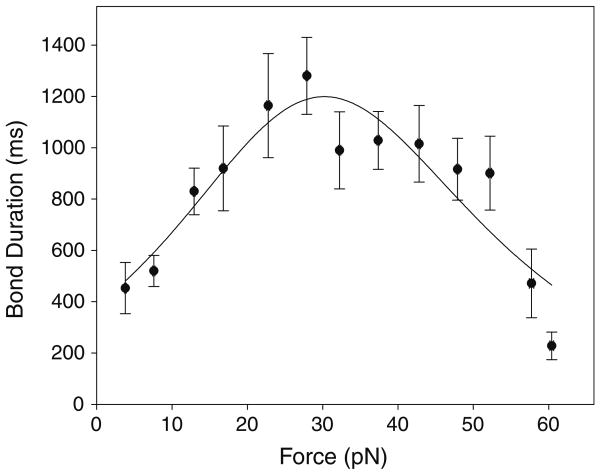
The lifetime of E-selectin/sLea bonds was measured over a range of applied loads using a laser trap (Fig. 1). An E-selectin-coated bead was held in the laser trap and brought into contact with a stationary target bead that was coated with sLea. The trapped bead was then stepped away from the target, applying a load to any bond (or bonds) that had formed. Bond lifetime and applied load were measured for a total of ∼1200 bonds. As can be seen in Fig. 3, single bond lifetimes increased with increasing load, characteristic of a catch bond, until reaching a maximum at a force of 30 ± 14 pN (propagated from the individual fitted parameter errors). Above 30 pN, bond lifetimes decrease with increasing load, characteristic of a slip bond. The critical force was obtained by fitting the data with a two-pathway model for catch–slip adhesion,35
FIGURE 3.
Bond lifetime as a function of applied load for single bonds. Data is fit by the two-pathway model for catch–slip adhesion. Error bars indicate standard error of the mean.
| (1) |
where t(f) is the mean bond lifetime at force f, and are the dissociation constants for unbinding through the catch and slip pathways, respectively, xc and xs are the transition state distances for unbinding through the catch and slip pathways, respectively, f is the applied load, T the absolute temperature, and kB the Boltzmann constant. The critical force of 30 pN is nearly identical to the catch–slip transition force reported for P-selectin/PSGL-1 bonds in both AFM and flow chamber experiments,29 but less than the ∼50 pN transition force observed for L-selectin/PSGL-1 bonds in AFM and biomembrane force probe (BFP) experiments, and L-selectin/sLex bonds in BFP experiments.26,43 The measured E-selectin/sLea bond lifetime at the transition force is 1.2 s, similar to that seen by others for P-selectin/PSGL-1 bonds,29 and significantly longer than the lifetimes seen for L-selectin/PSGL-1 and L-selectin/sLex bonds.26,43
2-D On-Rates Are Enhanced by Compressive Force
We studied directly the effects of compressive force on the rate of bond formation between E-selectin and sLea. An E-selectin-coated bead was brought into contact with a stationary target bead that was coated with sLea. When a bond formed between receptor and ligand on opposing bead surfaces, there was an appreciable shift of the signal mean and a change in variance (Fig. 1) that allowed the time to bond formation (ton) to be measured, from which an apparent 2-D on-rate (kon) was calculated. Upon stepping the trapped bead away from the stationary bead, comparing the known distance the laser was stepped to the measured displacement of the bead allowed us to measure how much compressive force was being exerted on opposing bead surfaces. A similar method of detecting bond formation has been reported previously in conjunction with a biomembrane force probe7 rather than a laser trap, though no attempt was reported to measure compressive force.
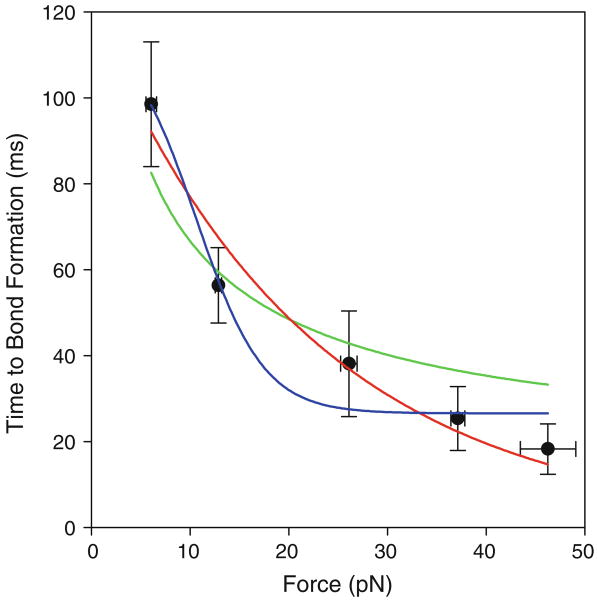
As seen in Fig. 4, ton decreases with increasing compressive force, showing that force does indeed increase the rate of bond formation. In fact, ton decreases approximately fivefold as the inward load increases from 6 to 46 pN. This corresponds to a similarly large change in the specific 2-D on-rate, , calculated as , where mr and ml are the site densities of receptor and ligand, respectively, on opposing bead surfaces, and Ac is the bead interaction area. Using our measured E-selectin site density of 1641 μm−2, the sLea–PAA site density of 2687 μm−2, and interaction area of 0.325 μm2, ranges from 7.1 × 10−6 μm2/s at a compressive force of 6 pN to 3.8 × 10−5 μm2/s at 46 pN.
FIGURE 4.
Time to bond formation (ton) as a function of compressive force. Data fit by two-state conformational change model (blue line), tilting energy landscape model (red), and surface layer deformation model (green). Error bars indicate standard error of the mean.
Discussion
We show that E-selectin, like L- and P-selectin, forms catch bonds with at least one of its ligands. We also present the first direct evidence that compressive load increases the association rates of single receptor– ligand pairs. These combined effects—increasing rates of association with compressive load, and decreasing rates of dissociation with tensile load—suggest a new aspect of force-enhanced adhesion.
Catch–slip behavior has been established for bonds of both P- and L-selectin29,43 with PSGL-1, but has not previously been observed for any E-selectin bonds. The single molecule studies that have previously been conducted with E-selectin20,51 find no evidence of a catch pathway, and instead analyze data in the context of the traditional Bell model3 for force-induced dissociation, which predicts that bond lifetimes decrease exponentially with applied load. The fact that our values for critical force and maximum bond lifetime are similar to those seen for P-selectin catch bonds, but different from those seen for L-selectin, is not surprising as P- and E-selectin serve similar roles on the endothelial surface. Catch bonds and the catch–slip transition are thought to be important in mediating leukocyte adhesion and subsequent rolling. It has been shown that a minimum shear stress is often necessary for leukocytes to tether and roll on selectins,1,11,25 a concept known as the shear threshold phenomenon. This shear threshold effect may be important in preventing undesired leukocyte adhesion and aggregation in slow flow vessels.29,61 It has been speculated that catch bonds may be the underlying cause of the shear threshold phenomenon, whereby bond lifetimes are too short to support rolling at low shear stresses, while high shear stresses slow bond dissociation at the trailing edge to allow rolling. Indeed, evidence for this was recently shown for L- and P-selectin using adhesive dynamics simulations.4 Our data suggest that the lack of a consistent display of a shear threshold for rolling on E-selectin is not the result of an inability for E-selectin to form catch bonds.
Our adaptation of the recently published thermal fluctuation assay7 allowed it to be used with a laser trap to measure the time to bond formation, from which 2-D on-rates were directly calculated. Our specific 2-D on-rate of 7.1 × 10−6 μm2/s for the lowest value of compressive load is equivalent to an effective on-rate, , of 2.3 × 10−6μm4/s, which is somewhat lower than the effective on-rate of 5.1 × 10−5 μm4/s found by others for L-selectin/PSGL-1 bonds.7 This lower on-rate may be due to differences in ligand, as the initial tethering rate of sLex microspheres to E-selectin has been shown to be lower that that of PSGL-1 microspheres,63 though an even lower effective on-rate was recently found for E-selectin/PSGL-1 bonds using an E-selectin lacking consensus repeats.48
The apparent sensitivity of this on-rate to compressive force is intriguing. While it is accepted that molecular rates of dissociation are impacted by force, the question of whether the rate of association may be influenced by force has largely been ignored. It has been shown that attachment of platelets to the endothelium occurs in orientations that are predicted to result in compression along the bond being formed.30 It has also been observed that increasing contact stress, while controlling for contact area, results in higher intrinsic rates of bond formation for bonds between β2-integrins and intercellular adhesion molecule-1 (ICAM-1).46 The authors attributed this to microvillus compression, but a more fundamental increase in binding rate is certainly a possibility. Evidence for a selectin/ligand on-rate that is enhanced by load was recently seen in a flow chamber study in which the authors observe an amplification of L-selectin binding rates to PSGL-1 by shear-induced compressive forces.34 Our data lends further support to this being an intrinsic property of at least some selectins.
Physical Mechanisms
We considered four possible physical mechanisms to explain load-dependent on-rates. One possibility is that there are two E-selectin conformations whose distribution is determined by force, with each conformation having a distinct association rate. Another potential mechanism is that force is causing the molecules to bind faster by tilting the energy landscape and thus reducing the activation energy for binding. Alternatively, force may be increasing the interaction area by deforming the molecular layers on the bead and/or flow cell surface. Finally, one might postulate that force is causing a reduction in Brownian motion such that the trapped bead is spending a greater fraction of time with its surface molecules within binding range of the molecules on the opposing bead surface.
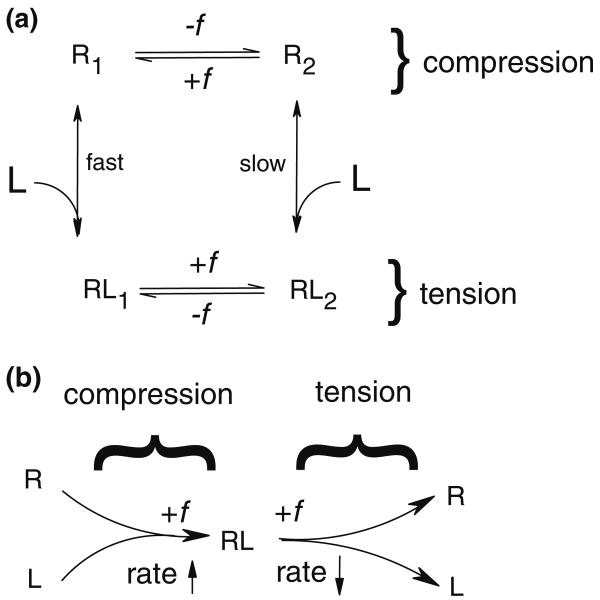
First we consider the conformational change model. This model assumes the existence of two E-selectin conformations with different association rates, such that compressive load generates a conformational change in E-selectin, biasing it toward the state with a faster binding rate. P-selectin has been shown to have two conformations, closed- and open-angle, each with a different angle between the lectin and EGF domains.45 The structures suggest that the lectin domain can pivot about the EGF domain in a hingelike fashion. The EGF domains of L- and E-selectin have a similar hinge region but with an Asn at residue 138 rather than a Gly, allowing for an additional hydrogen bond between Tyr37 and Asn138.14,45 It has been shown how force-induced conformational changes between the two hinge states can regulate the kinetics of L-selectin association and dissociation with PSGL-1 and 6-sulfo-sLex.26 Tensile force shifts the molecular equilibrium toward the open-angle state, which is postulated to lower off-rate by allowing the ligand to slide across the lectin domain as preexisting interactions dissociate, leading to the catch bond phenomenon. We envision compressive force between the two surfaces generating a conformational change in the hinge region of E-selectin, converting the molecule to a state with a faster rate of association. This can be viewed as an extension of a two-state catch bond model to include different binding pathways for each state. As can be seen in Fig. 5a, we have modified the two-state allosteric model53 to include two unbound states, R1 and R2. The application of a compressive load favors unbound state R1, which has a faster rate of association. We describe the association pathways of our model by fitting ton as a function of compressive load, f, using the parallel sum of two different association rates, as follows. Let the overall kon = kfastzfast + kslowzslow where kfast and kslow are the specific 2-D on-rates for the fast and slow association pathways, respectively, and zfast and zslow are the fractions of receptors in the fast- and slow-binding states. zfast + zslow = 1. The equilibrium distribution between the two states is given by a Boltzmann distribution modified by force, as in Bell3:
FIGURE 5.
(a) A schematic for the load-dependent association model integrated into the two-state model. Compressive force shifts the equilibrium of unbound receptors to conformational state R2, which has a faster association rate than state R1. Tensile force shifts the equilibrium of bound complex to state RL2, which has a slower dissociation rate than state RL1. In the compressive regime (top half of figure), +f indicates compressive force, while in the tensile regime, +f indicates tensile force. (b) A schematic for the reverse Bell model. Compressive force increases the rate at which bonds form, while tensile force decreases the rate at which bonds rupture.
| (2) |
where ΔE is the energy difference between the two states and xδ represents the transition state distance between them. If z is zslow at zero load, algebraic rearrangement gives
| (3) |
where Keq = z/(1 − z). Our model assumes rapid equilibrium between the two unbound states relative to force application, the rate of binding, and the rate of unbinding. This two-pathway association model fits our data only when z > 0.98, and with large parameter errors. We therefore set z = 0.99, which minimizes the parameter errors, and fitted the data (Fig. 4) with a kslow of 6.0 ± 0.9 × 10−6 μm2/s, a kfast of 2.6 ± 0.6 × 10−5 μm2/s, and an xδ of 1.2 ± 0.2 nm. The ratio of kfast to kslow is 4.3, meaning that molecules in the conformation favored by force bind 4.3-fold faster than molecules in the low-force conformation. Both association rate values are reasonable when compared to that measured directly in this study (7.1 × 10−6 μm2/s) and when converted to an effective on-rate and compared to that determined by others for L-selectin/PSGL-1 bonds (5.9 × 10−5 μm2/s). The transition state distance of 1.2 nm is reasonable given the 0.4 nm movement in the direction of tensile force found for the transition between the open and closed conformation in molecular dynamics simulations.47 The high fraction of receptors in the slow-binding state at zero force that is necessary for this model to fit our data is a point of concern, as molecular dynamics simulations have found L-selectin to exhibit a stable closed angle conformation 70% of the time.26 Additionally, the extended conformation state seen at equilibrium in this model is thought to be a high affinity state,36 thus one would expect selectin-ligand bonds to form readily at low or no force; this is not the case.25
We next consider the possibility that force is causing a direct enhancement of binding rate by forcing the receptor and ligand together once they have aligned stereospecifically with one another (Fig. 5b). This would require the opposing binding sites to be approximately opposite one another prior to force application. Force will then compress the binding sites together, effectively tilting the energy landscape to reduce the activation energy for binding, and thus allowing the receptor and ligand to bind more rapidly. This is analogous to the Bell model for force-induced dissociation,3 but with the energy landscape tilting in the opposite direction.
| (4) |
where kon(f) is the load-dependent on-rate, is the unloaded on-rate, f the applied load, xγa the characteristic bond length of association, kB the Boltzmann constant, and T the absolute temperature. This model fits our data well (Fig. 4), and yields a of 5.7 ± 0.6 μm2/s and an xγ of 0.19 ± 0.03 nm. The value of is similar to but slightly larger than that found for bonds between E-selectin and E-selectin ligand-1 (ESL-1) by surface plasmon resonance.59 The value of xγa is comparable to values previously reported for the dissociation characteristic bond length for L- and P-selectin,13,19,41,42,44 though this need not be the case because our measurement approaches the transition state from the unbound, rather than the bound state. A limitation of this model is that the receptor and ligand must already be stereospecifically aligned as the compressive force is applied. At any given instant in time the compressive force may not fully manifest at the binding interface; this is the inevitable result of thermal fluctuations in the system, and finite rigidities of the receptors and ligands. Nonetheless, the time-averaged energy landscape for bond formation should be “tilted inward” under compressive force.
A third possible physical mechanism is that compressive force causes an increase in interaction area, and thus the number of potential binding sites, by deforming the surface-adsorbed layer of molecules on the bead and/or flow cell. For example, the likelihood of bond formation between β2-integrins on neutrophils and ICAM-1 has been shown to increase linearly with contact area,46 though the present experiments use beads that are much less compressible than cells. To determine whether a similar mechanism was feasible to explain our data, we fit a molecular compression model to the ton vs. f data. This allowed us to determine the bulk moduli and/or molecular stiffnesses that would be required for the increase in interaction area necessary to generate the observed decrease in ton with increasing inward load. Briefly, using expressions for the compression of two spheres in contact,22,57 the contact radius, a, was calculated as
| (5) |
where f is the applied force, and R* and E* are the equivalent contact curvature and equivalent elastic modulus, respectively. The parameters R* and E* are calculated as
| (6) |
and
| (7) |
where R1 and R2 are the radii of the contacting spheres, E1 and E2 are the Young's moduli of the two spheres, and the effect of the Poisson's ratio is minimal. From this, the inward displacement, d, of opposing points on the two spheres was calculated as
| (8) |
The contact area of each sphere42 was found as
| (9) |
where hx is the height of the cap on sphere x, and rx is the radius of the sphere. Given a bond that is assumed to be parallel to the x-axis, the cap height for one sphere42 is found as
| (10) |
and the second cap height is given by h1 = l − h2, where the bond length l = lo − d, where lo is the original bond length. The total contact area, Ac = A1 + A2, is then used to model ton as
| (11) |
where E* and kon are fitted parameters. Fitting this model to our ton vs. f data yields a kon of 3.1 ± 0.3 × 10−6 μm2/s and an E* of 152 ± 752 Pa. This model, with large parameter errors, does not fit the data nearly as well as the two pathway association model (Fig. 4). Additionally, the value of E* does not seem reasonable; assuming the surface layer undergoing compression on each bead is of similar stiffness, E1 and E2 are both equal to 304 Pa. This is outside the range of any possible modulus for a continuous surface layer of Tween-20, nitrocellulose, or polystyrene, which are more likely to be of GPa magnitude.28,32,38 Considering the possibility that the modulus is due to individual E-selectin and sLea–PAA molecules being compressed, we used the equation E = kn(L/As) to calculate the molecular stiffness, k, that would be necessary to generate the modulus value determined by the model, where n is the number of molecules in the interaction area, L is the molecule length, and As is the surface area taken up by each molecule based on the measured site densities. For E-selectin and sLea–PAA this yields a k of 1.6e−5 and 1.3e−5 pN/nm, respectively. These values are far below experimental measurements of single molecule stiffness. In fact, P-selectin/PSGL-1 complexes have been shown to have a molecular stiffness of 5.3 pN/nm,13 many fold larger than that calculated using the compression model. Furthermore, we find that the fraction of bonds that are multiple bonds does not increase with increasing compressive force, suggesting that the number of available receptor/ligand molecules and therefore the contact area is not increasing.
A final possibility is that compressive force is reducing Brownian motion of the trapped bead by simply pushing it against the stationary bead, such that it spends a greater fraction of time within a range that allows its surface molecules to bind to those on the opposing bead surface. This seems unlikely as the standard deviation of bead fluctuation, σ, calculated as
| (12) |
where α is the trap stiffness, is only 6.5 nm, many fold smaller than the ∼55 nm combined length of the receptor and ligand. The compressive force is likely also reducing rotational Brownian motion; however, this is unlikely to lead to a large increase in association rate, and may in fact decrease association rate as the receptor and ligand are able to cover a smaller volume per unit time in their search for a binding partner.
Significance
The one model with which our data are fully consistent is the “reverse Bell model,” where compressive force causes a direct enhancement of binding rate by lowering the activation energy of bond formation. If this model is indeed correct, all bonds should exhibit load-dependent on-rates regardless of whether they are also catch bonds. This physical mechanism is likely to depend on the relative orientation of the molecular binding sites, a complex situation that may depend on the orientation of the molecules with respect to the surfaces to which they are bound. The molecules used in our system are not especially rigid and thus it is difficult for us to say how we would expect binding sites to be oriented.
Though the particular two-state conformational change model presented here seems unlikely, a mechanism involving a conformational change of the hinge region should not be ruled out. Some have pointed out that many of the molecules found to exhibit catch–slip behavior contain similar hinge regions,26,36,52 lending further credence to this possibility. It would be exciting to perform similar tests with selectin mutants that have a modified hinge region to test how the molecules' ability to change lectin-EGF hinge angle affects its binding rate at the single molecule level. Further investigations should examine the relationship between association and dissociation to determine how binding and unbinding pathways are related and how these pathways respond to force.
The localization of PSGL-1 on the tips of neutrophil microvilli31 suggests that the effects of these structures on the tensile and compressive forces of adhesion33 must be considered. The implications of having both a load-dependent association and dissociation rate are intriguing, particularly potential effects on the kinetics of cell capture and rolling. Bonds at the leading edge of a rolling cell may be more likely to form prior to rupture of the initial load-bearing bond at the trailing edge, promoting continued rolling and shorter skips along the surface. Load-dependent association may even play a role in the shear threshold effect, as higher shear increases compressive load, resulting in increased tethering. A detailed knowledge of load dependent kinetics is vital to understand this and other phenomena, and are crucial for the further development of leukocyte adhesion cascade simulations.
Acknowledgments
The authors thank Sasha Klibanov, Michael Lawrence, and Brian Schmidt of the University of Virginia for helpful discussions. The authors also acknowledge the support of the Department of Biomedical Engineering at the University of Virginia, and the National Institutes of Health (EB002185).
References
- 1.Alon R, Chen S, Puri KD, Finger EB, Springer TA. The kinetics of L-selectin tethers and the mechanics of selectin-mediated rolling. J Cell Biol. 1997;138:1169–1180. doi: 10.1083/jcb.138.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alon R, Hammer DA, Springer TA. Lifetime of the P-selectin-carbohydrate bond and its response to tensile force in hydrodynamic flow. Nature. 1995;374:539–542. doi: 10.1038/374539a0. [DOI] [PubMed] [Google Scholar]
- 3.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 4.Beste MT, Hammer DA. Selectin catch-slip kinetics encode shear threshold adhesive behavior of rolling leukocytes. Proc Natl Acad Sci USA. 2008;105:20716–20721. doi: 10.1073/pnas.0808213105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunk DK, Hammer DA. Quantifying rolling adhesion with a cell-free assay: E-selectin and its carbohydrate ligands. Biophys J. 1997;72:2820–2833. doi: 10.1016/S0006-3495(97)78924-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen S, Springer TA. Selectin receptor-ligand bonds: formation limited by shear rate and dissociation governed by the Bell model. Proc Natl Acad Sci USA. 2001;98:950–955. doi: 10.1073/pnas.98.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W, Evans EA, McEver RP, Zhu C. Monitoring receptor-ligand interactions between surfaces by thermal fluctuations. Biophys J. 2008;94:694–701. doi: 10.1529/biophysj.107.117895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesla SE, Selvaraj P, Zhu C. Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys J. 1998;75:1553–1572. doi: 10.1016/S0006-3495(98)74074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dembo M, Torney DC, Saxman K, Hammer D. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc R Soc Lond B Biol Sci. 1988;234:55–83. doi: 10.1098/rspb.1988.0038. [DOI] [PubMed] [Google Scholar]
- 10.Evans E, Leung A, Heinrich V, Zhu C. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc Natl Acad Sci USA. 2004;101:11281–11286. doi: 10.1073/pnas.0401870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finger EB, Puri KD, Alon R, Lawrence MB, von Andrian UH, Springer TA. Adhesion through L-selectin requires a threshold hydrodynamic shear. Nature. 1996;379:266–269. doi: 10.1038/379266a0. [DOI] [PubMed] [Google Scholar]
- 12.Florin EL, Moy VT, Gaub HE. Adhesion forces between individual ligand-receptor pairs. Science. 1994;264:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 13.Fritz J, Katopodis AG, Kolbinger F, Anselmetti D. Force-mediated kinetics of single P-selectin/ligand complexes observed by atomic force microscopy. Proc Natl Acad Sci USA. 1998;95:12283–12288. doi: 10.1073/pnas.95.21.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graves BJ, Crowther RL, Chandran C, Rumberger JM, Li S, Huang KS, Presky DH, Familletti PC, Wolitzky BA, Burns DK. Insight into E-selectin/ligand interaction from the crystal structure and mutagenesis of the lec/EGF domains. Nature. 1994;367:532–538. doi: 10.1038/367532a0. [DOI] [PubMed] [Google Scholar]
- 15.Guilford WH, Dupuis DE, Kennedy G, Wu J, Patlak JB, Warshaw DM. Smooth muscle and skeletal muscle myosins produce similar unitary forces and displacements in the laser trap. Biophys J. 1997;72:1006–1021. doi: 10.1016/S0006-3495(97)78753-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guilford WH, Tournas JA, Dascalu D, Watson DS. Creating multiple time-shared laser traps with simultaneous displacement detection using digital signal processing hardware. Anal Biochem. 2004;326:153–166. doi: 10.1016/j.ab.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 17.Guo B, Guilford WH. Mechanics of actomyosin bonds in different nucleotide states are tuned to muscle contraction. Proc Natl Acad Sci USA. 2006;103:9844–9849. doi: 10.1073/pnas.0601255103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ham AS, Goetz DJ, Klibanov AL, Lawrence MB. Microparticle adhesive dynamics and rolling mediated by selectin-specific antibodies under flow. Biotechnol Bioeng. 2007;96:596–607. doi: 10.1002/bit.21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanley W, McCarty O, Jadhav S, Tseng Y, Wirtz D, Konstantopoulos K. Single molecule characterization of P-selectin/ligand binding. J Biol Chem. 2003;278:10556–10561. doi: 10.1074/jbc.M213233200. [DOI] [PubMed] [Google Scholar]
- 20.Hanley WD, Wirtz D, Konstantopoulos K. Distinct kinetic and mechanical properties govern selectin-leukocyte interactions. J Cell Sci. 2004;117:2503–2511. doi: 10.1242/jcs.01088. [DOI] [PubMed] [Google Scholar]
- 21.Hulme EC, Birdsall NJM. Strategy and Tactics in Receptor-Binding Studies. Oxford, UK: IRL Press; 1992. pp. 63–176. [Google Scholar]
- 22.Johnson KL. Contact Mechanics. 2nd. Cambridge, UK: Cambridge University Press; 1985. [Google Scholar]
- 23.Kimura N, Mitsuoka C, Kanamori A, Hiraiwa N, Uchimura K, Muramatsu T, Tamatani T, Kansas GS, Kannagi R. Reconstitution of functional L-selectin ligands on a cultured human endothelial cell line by cotransfection of alpha1 → 3 fucosyltransferase VII and newly cloned GlcNAcbeta:6-sulfotransferase cDNA. Proc Natl Acad Sci USA. 1999;96:4530–4535. doi: 10.1073/pnas.96.8.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong F, Garcia AJ, Mould AP, Humphries MJ, Zhu C. Demonstration of catch bonds between an integrin and its ligand. J Cell Biol. 2009;185:1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence MB, Kansas GS, Kunkel EJ, Ley K. Threshold levels of fluid shear promote leukocyte adhesion through selectins (CD62L, P, E) J Cell Biol. 1997;136:717–727. doi: 10.1083/jcb.136.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lou J, Yago T, Klopocki AG, Mehta P, Chen W, Zarnitsyna VI, Bovin NV, Zhu C, McEver RP. Flow-enhanced adhesion regulated by a selectin interdomain hinge. J Cell Biol. 2006;174:1107–1117. doi: 10.1083/jcb.200606056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowe JB, Stoolman LM, Nair RP, Larsen RD, Berhend TL, Marks RM. ELAM-1-dependent cell adhesion to vascular endothelium determined by a transfected human fucosyltransferase cDNA. Cell. 1990;63:475–484. doi: 10.1016/0092-8674(90)90444-j. [DOI] [PubMed] [Google Scholar]
- 28.Lubarsky GV, Davidson MR, Bradley RH. Elastic modulus, oxidation depth and adhesion force of surface modified polystyrene studied by AFM and XPS. Surf Sci. 2004;558:135–144. [Google Scholar]
- 29.Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 30.Mody NA, Lomakin O, Doggett TA, Diacovo TG, King MR. Mechanics of transient platelet adhesion to von Willebrand factor under flow. Biophys J. 2005;88:1432–1443. doi: 10.1529/biophysj.104.047001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore KL, Patel KD, Bruehl RE, Li F, Johnson DA, Lichenstein HS, Cummings RD, Bainton DF, McEver RP. P-selectin glycoprotein ligand-1 mediates rolling of human neutrophils on P-selectin. J Cell Biol. 1995;128:661–671. doi: 10.1083/jcb.128.4.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park SE, Ryoo R, Ahn WS, Lee CW, Chang JS. Nanotechnology in mesostructured materials. Proceedings of the 3rd International Mesostructured Materials Symposium. 2003;146:393. [Google Scholar]
- 33.Park EY, Smith MJ, Stropp ES, Snapp KR, DiVietro JA, Walker WF, Schmidtke DW, Diamond SL, Lawrence MB. Comparison of PSGL-1 microbead and neutrophil rolling: microvillus elongation stabilizes P-selectin bond clusters. Biophys J. 2002;82:1835–1847. doi: 10.1016/S0006-3495(02)75534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paschall CD, Guilford WH, Lawrence MB. Enhancement of L-selectin, but not P-selectin, bond formation frequency by convective flow. Biophys J. 2008;94:1034–1045. doi: 10.1529/biophysj.106.098707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pereverzev YV, Prezhdo OV, Forero M, Sokurenko EV, Thomas WE. The two-pathway model for the catch-slip transition in biological adhesion. Biophys J. 2005;89:1446–1454. doi: 10.1529/biophysj.105.062158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phan UT, Waldron TT, Springer TA. Remodeling of the lectin-EGF-like domain interface in P- and L-selectin increases adhesiveness and shear resistance under hydrodynamic force. Nat Immunol. 2006;7:883–889. doi: 10.1038/ni1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phillips ML, Nudelman E, Gaeta FC, Perez M, Singhal AK, Hakomori S, Paulson JC. ELAM-1 mediates cell adhesion by recognition of a carbohydrate ligand, sialyl-Lex. Science. 1990;250:1130–1132. doi: 10.1126/science.1701274. [DOI] [PubMed] [Google Scholar]
- 38.Pierce GW. The Songs of Insects. Cambridge, MA: Harvard University Press; 1948. [Google Scholar]
- 39.Puri KD, Finger EB, Springer TA. The faster kinetics of L-selectin than of E-selectin and P-selectin rolling at comparable binding strength. J Immunol. 1997;158:405–413. [PubMed] [Google Scholar]
- 40.Ramachandran V, Nollert MU, Qiu H, Liu WJ, Cummings RD, Zhu C, McEver RP. Tyrosine replacement in P-selectin glycoprotein ligand-1 affects distinct kinetic and mechanical properties of bonds with P- and L-selectin. Proc Natl Acad Sci USA. 1999;96:13771–13776. doi: 10.1073/pnas.96.24.13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramachandran V, Yago T, Epperson TK, Kobzdej MM, Nollert MU, Cummings RD, Zhu C, McEver RP. Dimerization of a selectin and its ligand stabilizes cell rolling and enhances tether strength in shear flow. Proc Natl Acad Sci USA. 2001;98:10166–10171. doi: 10.1073/pnas.171248098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rinko LJ, Lawrence MB, Guilford WH. The molecular mechanics of P- and L-selectin lectin domains binding to PSGL-1. Biophys J. 2004;86:544–554. doi: 10.1016/S0006-3495(04)74133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarangapani KK, Yago T, Klopocki AG, Lawrence MB, Fieger CB, Rosen SD, McEver RP, Zhu C. Low force decelerates L-selectin dissociation from P-selectin glycoprotein ligand-1 and endoglycan. J Biol Chem. 2004;279:2291–2298. doi: 10.1074/jbc.M310396200. [DOI] [PubMed] [Google Scholar]
- 44.Smith MJ, Berg EL, Lawrence MB. A direct comparison of selectin-mediated transient, adhesive events using high temporal resolution. Biophys J. 1999;77:3371–3383. doi: 10.1016/S0006-3495(99)77169-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Somers WS, Tang J, Shaw GD, Camphausen RT. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1. Cell. 2000;103:467–479. doi: 10.1016/s0092-8674(00)00138-0. [DOI] [PubMed] [Google Scholar]
- 46.Spillmann CM, Lomakina E, Waugh RE. Neutrophil adhesive contact dependence on impingement force. Biophys J. 2004;87:4237–4245. doi: 10.1529/biophysj.103.031773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Springer TA. Structural basis for selectin mechanochemistry. Proc Natl Acad Sci USA. 2009;106:91–96. doi: 10.1073/pnas.0810784105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun G, Zhang Y, Huo B, Long M. Surface-bound selectin-ligand binding is regulated by carrier diffusion. Eur Biophys J. 2009;38:701–711. doi: 10.1007/s00249-009-0428-y. [DOI] [PubMed] [Google Scholar]
- 49.Suonpaa M, Markela E, Stahlberg T, Hemmila I. Europium-labelled streptavidin as a highly sensitive universal label indirect time-resolved immunofluorometry of FSH and TSH. J Immunol Methods. 1992;149:247–253. doi: 10.1016/0022-1759(92)90256-s. [DOI] [PubMed] [Google Scholar]
- 50.Svoboda K, Block SM. Biological applications of optical forces. Annu Rev Biophys Biomol Struct. 1994;23:247–285. doi: 10.1146/annurev.bb.23.060194.001335. [DOI] [PubMed] [Google Scholar]
- 51.Tees DF, Waugh RE, Hammer DA. A micro-cantilever device to assess the effect of force on the lifetime of selectin-carbohydrate bonds. Biophys J. 2001;80:668–682. doi: 10.1016/S0006-3495(01)76047-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas W. For catch bonds, it all hinges on the inter-domain region. J Cell Biol. 2006;174(7):911–913. doi: 10.1083/jcb.200609029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas WE, Forero M, Yakovenko O, Nilsson LM, Vicini P, Sokurenko EV, Vogel V. Catch bond model derived from allostery explains force-activated bacterial adhesion. Biophys J. 2006;90:753–764. doi: 10.1529/biophysj.105.066548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas WE, Nilsson LM, Forero M, Sokurenko EV, Vogel V. Shear-dependent ‘stick-and-roll’ adhesion of type 1 fimbriated. Escherichia coli Mol Microbiol. 2004;53:1545–1557. doi: 10.1111/j.1365-2958.2004.04226.x. [DOI] [PubMed] [Google Scholar]
- 55.Ushakova NA, Preobrazhenskaya ME, Bird MI, Priest R, Semenov AV, Mazurov AV, Nifantiev NE, Pochechueva TV, Galanina OE, Bovin NV. Monomeric and multimeric blockers of selectins: comparison of in vitro and in vivo activity. Biochemistry (Moscow) 2005;70:432–439. doi: 10.1007/s10541-005-0133-0. [DOI] [PubMed] [Google Scholar]
- 56.Varki A. Selectin ligands. Proc Natl Acad Sci USA. 1994;91:7390–7397. doi: 10.1073/pnas.91.16.7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vu-Quoc L, Zhang X, Lesburg L. A normal force-displacement model for contacting spheres accounting for plastic deformation: force-driven formulation. J Appl Mech. 2000;67:363–371. [Google Scholar]
- 58.Waldron TT, Springer TA. Transmission of allostery through the lectin domain in selectin-mediated cell adhesion. Proc Natl Acad Sci USA. 2009;106:85–90. doi: 10.1073/pnas.0810620105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wild MK, Huang MC, Schulze-Horsel U, van der Merwe PA, Vestweber D. Affinity, kinetics, and thermodynamics of E-selectin binding to E-selectin ligand-1. J Biol Chem. 2001;276:31602–31612. doi: 10.1074/jbc.M104844200. [DOI] [PubMed] [Google Scholar]
- 60.Xia L, Sperandio M, Yago T, McDaniel JM, Cummings RD, Pearson-White S, Ley K, McEver RP. P-selectin glycoprotein ligand-1-deficient mice have impaired leukocyte tethering to E-selectin under flow. J Clin Invest. 2002;109:939–950. doi: 10.1172/JCI14151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yago T, Wu J, Wey CD, Klopocki AG, Zhu C, McEver RP. Catch bonds govern adhesion through L-selectin at threshold shear. J Cell Biol. 2004;166:913–923. doi: 10.1083/jcb.200403144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu C, Long M, Chesla SE, Bongrand P. Measuring receptor/ligand interaction at the single-bond level: experimental and interpretative issues. Ann Biomed Eng. 2002;30:305–314. doi: 10.1114/1.1467923. [DOI] [PubMed] [Google Scholar]
- 63.Zou X, Shinde VR, Patil, Dagia NM, Smith LA, Wargo MJ, Interliggi KA, Lloyd CM, Tees DF, Walcheck B, Lawrence MB, Goetz DJ. PSGL-1 derived from human neutrophils is a high-efficiency ligand for endothelium-expressed E-selectin under flow. Am J Physiol Cell Physiol. 2005;289:C415–C424. doi: 10.1152/ajpcell.00289.2004. [DOI] [PubMed] [Google Scholar]