Abstract
The control of mRNA decay is emerging as an important control point and a major contributor to gene expression in both immune and non-immune cells. The identification of protein factors and cis-acting elements responsible for transcript degradation has illuminated a comprehensive picture of precisely orchestrated events required to both regulate and establish the decay process. One gene that is highly regulated at the posttranscriptional level is CD40 ligand (CD154 or CD40L). CD154 on CD4+ T cells is tightly controlled by an interacting network of transcriptional and posttranscriptional processes that result in precise surface levels of protein throughout an extended time course of antigen stimulation. The activation-induced stabilization of the CD154 transcript by a polypyrimidine tract-binding protein (PTB)-complex is a key event that corresponds to the temporal expression of CD154. In this review, we discuss known and potential roles of major mRNA decay pathways in lymphocytes and focus on the unique posttranscriptional mechanisms leading to CD154 expression by activated CD4+ T cells.
Progression of an immune response requires the coordinated expression of ligands, receptors and cytokines. The expression of these regulatory molecules is tightly controlled at multiple levels upon cell activation. At the epigenetic level these events are orchestrated by changes in chromatin conformation allowing binding of an array of transcription factors and rapid induction of gene expression.1 Further control is modulated at the posttranscriptional level by regulating the decay rate and localization of transcripts such that the required level of translation is achieved. During the last two decades the study of posttranscriptional regulation of immune and non-immune related genes has advanced significantly and is now recognized as being central in regulating gene expression. 2–7
Eukaryotic mRNAs transcribed by RNA polymerase II are made resistant from exonuclease degradation by incorporation of the 5´ 7-methylguanosine cap and the 3´ poly(A) tail.8, 9 These two essential elements are added during transcription and their interaction with cytoplasmic eIF4E and poly(A)-binding protein (PABP) enhances both translation and protection from exonuclease activity.10 Although these factors are common to every mRNA, a subset of transcripts are also susceptible to regulation mediated by RNA binding proteins interacting with cis-acting elements encoded within the 3´- or 5´-untranslated regions (UTRs). Binding of specific trans-acting factors to these regulatory elements has a wide range of consequences resulting in enhanced or diminished transcript stability and/or changes in the overall translational capacity of a specific mRNA.
Upon activation, lymphocytes must undergo a series of rapid phenotypic and functional changes that allow them to proliferate and express specific effector functions in order to neutralize pathogen. A balanced pattern of gene expression during such changes is an essential prerequisite to establish an appropriate immune response. Short-lived mRNA transcripts allow for a much more rapid response to changing environmental or developmental cues and can limit transcript availability at times when expression of a specific set of proteins would otherwise be detrimental.11 Conversely, long-lived transcripts are a more energy efficient alternative when sustained expression of a protein is required.12 Since lymphocyte activation is characterized by transitions between different checkpoints during which the fate of an immune response is decided, diversification at the level of mRNA stability of different transcripts provide a valuable tool to regulate the magnitude of a response. These changes in mRNA stability are controlled by the regulatory activity of many different RNA binding proteins whose function is dictated by signaling pathways during distinct stages of cell maturation and inflammation.5, 13 This review will explore these mechanisms of posttranscriptional regulation of immune related genes with a focus on factors controlling CD154 mRNA stability.
AU-rich elements
The most studied transcript instability signature in lymphocytes is the AU-rich element (ARE).14 This element is present in the 3´UTRs of many rapidly induced mRNAs and is often characterized by different length repeats of the cis-acting AUUUA pentamers organized in stem loop structures.15 The number, length and surrounding sequences of ARE repeats are decisive factors for the recruitment of the family of ARE binding proteins (AUBP), which includes the AU-rich binding factor-1 (AUF1), tristetraprolin (TTP), KH splicing regulatory protein (KSRP), Human-antigen R (HuR) as well as others.14, 16, 17 To date, the stability of several immune related-transcripts has been linked to AUBP binding including TNFα,18 IL-2,19 IL-3,20 IL-8,21 IL-10,21, 22 VEGF,23 COX224 and MMP13.25 In addition to experimentally identified transcripts, microarray technology has established the cohort of ARE-containing genes that constitute the ARE-mRNA Database (ARED)26 and provided a grouping of ARE containing mRNAs based on their role during immune cell activation.27
The mechanisms by which AUBPs regulate mRNA decay is under extensive investigation and is known to include mRNA targeting to the 3´–5´ cleavage activity of the cytoplasmic multiprotein component known as the exosome28, or by formation of processing bodies (P-bodies); cytoplasmic messenger ribonucleoproteins foci characterized by the presence of decapping enzymes and 5´-3´ RNA processing proteins.29 Differential AUBPs binding has also being shown to either increase or decrease endo-ribonuclease activity or translation of target mRNAs.3, 30, 31 Specifically, AUBPs binding to AREs can have opposite outcomes on the half-life (t1/2) of transcripts either by recruitment of the deadenylation machinery and subsequent rapid degradation by the exosome32, 33 or by masking the ARE to increase transcript stability and prevent endonuclease activity.34 In some instances AUBPs with different properties compete for binding to the same cis-acting element. This is the case with TTP binding to an ARE within the TNFα 3´UTR that results in enhanced degradation by recruitment of the exosome.35 However, TTP binding to the TNFα ARE can be out competed by HuR resulting in the increased stabilization of the transcript.18 Also, the IL-2 mRNA, which has a 3´UTR rich in AREs, is stabilized by the NF90 AUBP following CD28 co-stimulation of CD4+ T cells suggesting that NF90 binding prevents the interaction of other decay promoting AUBPs.36 Finally, the same AUBP can function to stabilize or decay an mRNA which is the case with AUF1 which exits as four distinct isoforms and functions as a destabilizing factor in K562 cells37 and a stabilizing factor in NIH 3T3 cells.38
GU-rich elements
Global analysis of mRNA degradation patterns in resting and activated T lymphocytes identified a sizeable number of transcripts exhibiting activation dependent changes in posttranscriptional regulation.39 Only a small minority of these transcripts was found to contain ARE sequences within their 3´UTRs. In contrast, the vast majority of mRNA regulated at the posttranscriptional level in T lymphocytes contained a GU-rich element (GRE). The presence of GU repeats in the 3´UTR corresponded to enhanced transcript instability40 and the CUG-binding protein 1 (CUGBP1) was identified as a factor that bound GREs41 and stabilized transcripts.40 While specific studies have shown that CUGBP1 can recruit the deadenylation machinery42 others report that this factor functions to enhance translation of target genes.43 Therefore, it would appear that a subset of GREcontaining transcripts, like their ARE-containing counterparts, are targeted for stability or degradation through a GRE-specific pathway in response to varying cellular conditions.
CA-rich elements
The most common di-nucleotide motif in mammals is the CA repeat.44 This element is widely used in genetic linkage analyses and a correspondence between microsatelite CA polymorphisms within 3´UTRs and incidence of disease has been demonstrated in Type I diabetes, rheumatoid arthritis and systemic lupus erythematousus.45–48 The ubiquitously expressed heterogeneous ribonuclear protein (hnRNP)-L maintains a high affinity for ribo-CA-repeats in introns, exons and regulatory regions through the interaction of three distinct RNA binding domains (RBDs).49 This factor is implicated in multiple steps of RNA processing including splice-site selectivity,50, 51 nuclear-cytoplasmic transport,52, 53 IRESmediated translation54 and RNA stability.51 In T cells, the expression of different spliced isoforms of CD45 is mediated through an activation-induced pathway of exon exclusion through selective binding of both hnRNP-L and a homologue hnRNP-LL.55–57 Also, hnRNP-L binding to 3’UTR CA-repeats has been shown to impact the stability of transcripts encoding endothelial isoform of nitric oxide synthase (eNOS),58 inducible nitric oxide synthase (iNOS) mRNA59, human vascular endothelial growth factor (VEGF) 60 and CD154.61
CU-rich elements
Two distinct CU-rich elements have been implicated in posttranscriptional regulation of multiple eukaryotic mRNA transcripts. These are the Differentiation Control Element (DICE) and the CU-Rich Element (CURE).62, 63 Although the distinction between these two elements is debatable,63 DICE is characterized by the consensus architecture (C/U)CCANx CCC(U/A) (C/U)y UC(C/U)CC.64 This sequence motif is bound by the heterogeneous ribonuclear protein (hnRNP) K (E2/E1, αCP, PCBP) resulting in the stabilization of the target molecule65 with a corresponding induction66 or repression67 of translation. Examples of DICE-directed post-transcriptional regulation include the modulation of 15-lipoxygenase mRNA stability by hnRNP-K and -E1.66 Also, DICE sequences within the 3´UTR of human p21-activated kinase 1 (Pak1) were shown to bind hnRNP-E1.68
A CURE binding protein that has been extensively characterized is the polypyrimidine tract-binding protein (PTB). PTB has been implicated in the post-transcriptional regulation of inducible nitric oxide synthetase (iNOS),69 insulin,70 VEGF,71 and CD154.72, 73 In lymphocytes, this protein is present in two ubiquitously expressed isoforms of 50 and 55 kDa (PTB-1 and PTB-4), which contain four RNA Binding Domains (RBD) separated by unstructured linker sequences.74 Both PTB-1 and PTB-4 are shuttling proteins with a novel bipartite N-terminal nuclear localization sequence (NLS) that is bound by importin-α.75 This protein functions as a cytoplasmic receptor to promote nuclear localization of bipartite NLS-containing proteins.76 Translocation of PTB between the nuclear and cytoplasmic compartments is controlled by phosphorylation at Ser-16 by 3´–5´ cAMP dependant protein kinase A (PKA)77 and this process per se appears to be uncoupled to RNA export.78 The activity of PTB in post-transcriptional regulation requires the formation of protein complexes that may include nucleolin,79 hnRNP-L,80 Cold Shock Domain (CSD)71 as well as additional heterologous RNA binding proteins (reviewed in ref. 81).
Posttranscriptional regulation of CD154 (CD40 ligand) mRNA
A critical immune regulatory protein that is controlled at the level of posttranscriptional control is CD40 ligand (CD40L or CD154). The interaction of transiently expressed CD154 on CD4+ T cells with the constitutively expressed CD40 on antigen-presenting cells (APCs) generates critical thymus dependent (TD) responses and enhances a subset of innate responses to bacterial and viral pathogens.82–84
Several studies have shown that CD154 mRNA expression is regulated at both the transcriptional and posttranscriptional levels in response to T cell receptor (TCR) activation. Within 10 min of activation intracellular stores of CD154 protein are translocated to the extracellular surface and CD154 expression is quickly enhanced by increased gene transcription, which results in maximal mRNA levels occurring after 6 h of continuous stimulation. At approximately 12 h of stimulation the CD154 mRNA levels drop to a basal level that remains constant throughout the subsequent activation period.85–87 Early expression analysis revealed that CD154 transcription is dependent on the activation of the Ca2+/calmodulin pathway88 whereas transcript stabilization is increased in response to treatment with cAMP analogues in ionomycin-stimulated peripheral blood mononuclear cells (PBMC).89 Other reports indicated that co-culture of PHA-activated CD4+ T cells with human endothelial cells (EC) causes direct changes in CD154 mRNA stability through an LFA-3-dependent process.90 However, unlike its effect on ARE-mediated decay,91, 92 co-stimulation through CD28 was shown to induce only a modest increase in CD154 transcript stability.93
The posttranscriptional mechanisms underlying CD154 expression have been extensively investigated.73, 93, 94 In human CD4+ T cells the CD154 transcript was found to decay with a t1/2 of less than 40 min during the first 12 h of TCR activation and with a t1/2 of approximately 2.2 h following 24 h of continuous stimulation.93 Biphasic posttranscriptional regulation of this mRNA was found to depend on the activation-induced binding of ribonucleoprotein (RNP) complexes containing PTB to a region of the CD154 3’UTR at extended times following activation.94 A similar mechanism of posttranscriptional regulation was demonstrated in response to TLR9 stimulation in primary B cells where multiple transcripts were stabilized by the binding of a PTB-containing complex (B-cpx I) to CU-rich elements within their 3´UTRs.95 Thus, both T- and B-lymphocytes maintain a similar non-ARE pathway of mRNA stability that is directly linked to antigen activation.
cis-elements and trans-factors involved in CD154 mRNA regulation
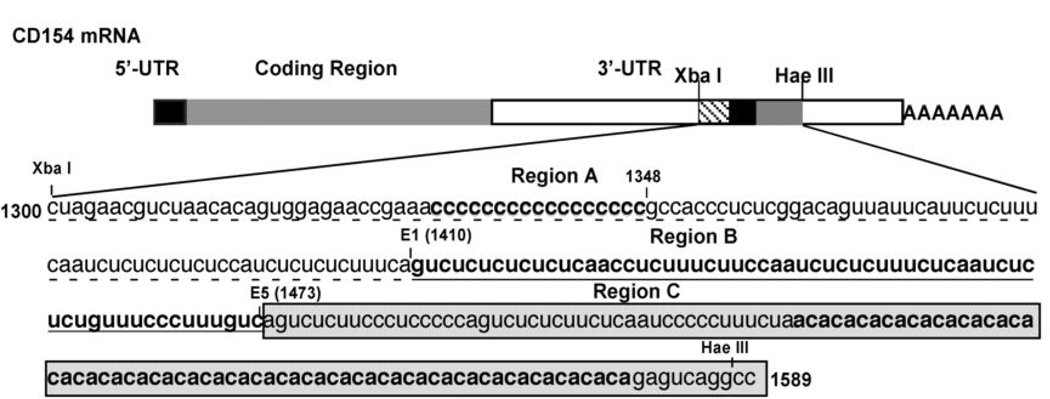
A thorough analysis of cis- and trans-acting factors involved in CD154 mRNA decay revealed that two PTB containing complexes (Complex I and Complex II) bind to three distinct sequences spanning nucleotides 1300 to 1589 of CD154 mRNA (XbaI-E1, E1-E5 and E5-HaeIII), within the 3´UTR region defined by the Xba I and Hae III restriction sites (X-H) (Fig 1).94 Further analysis revealed that PTB is the major RNA binding component of the RNP complexes72, 73 and that nucleolin and hnRNP-L are additional components of Complex I and II, respectively.61, 79, 80 Formation of these RNP complexes is only seen at extended times of activation corresponding to the increased stability of the transcript, suggesting their direct involvement in modulating activation-induced CD154 mRNA stability.72, 94
Figure 1.
Schematic diagram of the CD154 mRNA showing the three distinct Complex I/II binding sites in the 3’UTR. The CD154 stability element, defined by the restriction sites Xba I to Hae III (nt 1300 to 1589), is divided into three sub-regions: A (Xba I-E1, dashed underlined), B (E1–E5, underlined) and C (E5-Hae III, boxed). 5’ A (1300–1348) is destabilizing and 1348-E1 is stabilizing and contains a Complex I binding site (see Table I) (see Laughlin, et. al. 2008).
The minimal sequences required for each complex binding were identified in in vitro studies.80 The contribution of each minimal sequence to stabilizing a heterologous transcript (Renilla luciferase) and modulating reporter activity was tested in Jurkat/D1.1 cells which constitutively express Complex I and II (Table I). The centrally located E1-E5 region (Fig 1, Site B), which is one of the two minimum Complex I binding sequences94 was found to provide the highest level of transcript stability and activity.80 A second unit of Complex I was found to bind to a region defined by Xba I and E1 (Fig. 1, Site A) and insertion of this sequence at the 3’UTR of Renilla luciferase was also shown to increase expression, although at a much lower level than that seen with Site B. Further analysis of Site A revealed that the region between Xba I and nucleotide 1351 was responsible for the reduced luciferase activity and that this reduction was caused by enhanced luciferase mRNA instability (Table I). Together these findings support a model where binding of Complex I to Sites A and B results in enhanced protection of the transcript from rapid degradation mediated by adjacent sequences between the Xba I site and nucleotide 1348 (Fig. 2). This model also explains the high level of mRNA decay at early times following T cell activation when there is an absence of Complex I binding to the CD154 transcript.94
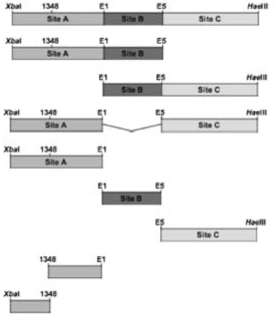
TABLE I.
Activity of the Different Regions of the CD154 Stability Element1
| Region Tested | Luciferase Activity (Fold over pRLSV40) |
|---|---|
 |
2.93+/−0.32 |
| 3.1+/−0.29 | |
| 3.88+/−0.8 | |
| 0.26+/−0.05 | |
| 1.08+/−0.05 | |
| 4.52+/−0.55 | |
| 1.33+/−0.12 | |
| 2.25+/−0.11 | |
| 0.22+/−0.2 | |
The different regions of the CD154 stability regions were inserted into the 3′ UTR of the Renilla luciferase operon contained within the pRLSV40 vector. Jurkat D1.1 cells were transfected with the various constructs and luciferase activity measured 48 h later. Shown are the mean values +/− SEMs calculated for each construct over pRLSV40 vector alone (see Laughlin, et. al. 2008).
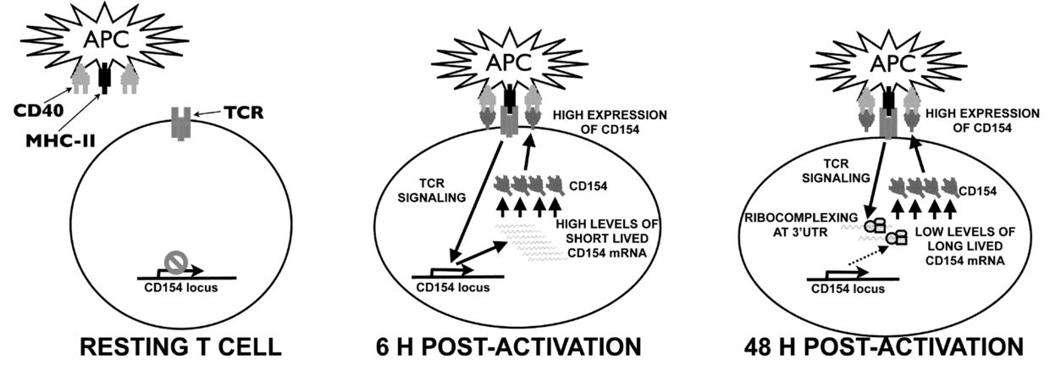
Figure 2.
Model of posttranscriptional regulation of CD154. Resting CD4+ T cells retain low CD154-specific transcription, minimal amounts of short-lived CD154 mRNA and virtually little to no detectable surface expression of the protein. Within 6 h following antigen presentation by APCs a peak in transcription drives high levels of surface expression. At this time the CD154 message is being rapidly degraded. At extended times of activation, low levels of CD154 mRNA are stabilized by ribocomplexing on the 3’UTR which drives a second peak of surface expression.
Analysis of the 3´ sequence of the X-H region downstream of the E5 site identified the binding site for Complex II (Fig. 1, Site C) This region contains a contracted CU-rich stretch and a polymorphic CA repeat72 and is the weakest of the three individual regions with respect to its ability to enhance luciferase expression (Table I). Interestingly, Complex II binding was shown to require the whole length of this region. In addition to PTB, hnRNP-L was identified as a protein that bound to this sequence through interactions with the CArepeats.80 Hamilton and colleagues recently reported that the binding of hnRNP-L to the CD154 CA-repeat in HeLa cells correlated both with transcripts containing shortened poly(A) tails and increased translation of a heterologous transcript.61 The allelic and genotypic distribution of the polymorphic CD154 repeat has been studied as a genetic marker in specific autoimmune diseases and the higher number of CA repeats correlate with disease incidence.46, 47 Notably, deregulating CD154 expression is associated with an increase in autoantibodies in both mouse and humans and this may be achieved in part through posttranscriptional and translational processes that result in enhanced expression during an immune response.96–100
Analysis of CD154 transcript upon in vivo activation
Similar to the regulation of human, the mouse CD154 transcript maintains a biphasic pattern of stability that is activation-dependent, however, the t1/2 values in both the early and late stages are considerably shortened (∼23 min and ∼45min) (Vavassori, et. al., submitted). The question of whether the activation induced program of mRNA stability functions in vivo following antigen challenge has been addressed by priming animals with antigen plus adjuvant or adjuvant alone and challenging the T cells ex vivo with the same antigen. Analysis of RNA t1/2 profiles of the unprimed and primed lymphocyte populations demonstrated that several days following injection, CD154 mRNA decayed in unprimed T cells with a t1/2 of approximately 30 min which increased two-fold to approximately 50 min in in vivo primed cells. These findings revealed that restimulating CD4+ T cells with antigen resulted in an increase in CD154 transcript stability relative to cells that were exposed to antigen for the first time and strongly suggested a role for regulated CD154 mRNA stability in vivo (Vavassori et. al., submitted).
A comparison of CD154 mRNA decay with the overall pattern of CD154 regulation led us to propose that mRNA stability is largely responsible for appropriate protein levels at late times of activation. This is based on the unexpected finding that overall steady state levels of CD154 mRNA are inversely linked to transcript stability where at 6 h post-activation the message level is highest and the stability lowest and vice versa. Previous data suggested that purified CD4+ T cells express CD154 for extended times of activation in the presence or absence of co-stimulation101–105 and that expression is biphasic with an early peak detected at 6 h and a second peak at 24 h.106–109 Since this pattern of expression closely corresponds to our biphasic pattern of mRNA stability it suggests that there is as yet an undefined role for enhanced CD154 expression at late times of T cell activation. Future work will focus on identifying the signaling pathways of CD154 mRNA stability and defining the functional consequences of this pattern of expression in both B cell and T cell activation.
ACKNOWLEDGEMENTS
We thank Mike Kiledjian for insightful and helpful comments and acknowledge the contributions of past and present members of the Covey lab in the overall development of this story. This work was supported by grants from The National Institutes of Health (PO1 AI-57596) and The American Heart Association to L. R. C.
REFERENCES
- 1.Sawalha AH. Epigenetics and T-cell immunity. Autoimmunity. 2008;41:245–252. doi: 10.1080/08916930802024145. [DOI] [PubMed] [Google Scholar]
- 2.Anderson CJ, Hoare SF, Ashcroft M, Bilsland AE, Keith WN. Hypoxic regulation of telomerase gene expression by transcriptional and post-transcriptional mechanisms. Oncogene. 2006;25:61–69. doi: 10.1038/sj.onc.1209011. [DOI] [PubMed] [Google Scholar]
- 3.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 4.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 5.Khabar KS. Rapid transit in the immune cells: the role of mRNA turnover regulation. J Leukoc Biol. 2007;81:1335–1344. doi: 10.1189/jlb.0207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoecklin G, Anderson P. Posttranscriptional mechanisms regulating the inflammatory response. Adv Immunol. 2006;89:1–37. doi: 10.1016/S0065-2776(05)89001-7. [DOI] [PubMed] [Google Scholar]
- 7.Vlasova MA, Moshkovskii SA. Molecular interactions of acute phase serum amyloid A: possible involvement in carcinogenesis. Biochemistry (Mosc) 2006;71:1051–1059. doi: 10.1134/s0006297906100014. [DOI] [PubMed] [Google Scholar]
- 8.Proudfoot N. New perspectives on connecting messenger RNA 3’ end formation to transcription. Curr Opin Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic Acid Res Mol Biol. 2001;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- 10.Bentley D. The mRNA assembly line: transcription and processing machines in the same factory. Curr Opin Cell Biol. 2002;14:336–342. doi: 10.1016/s0955-0674(02)00333-2. [DOI] [PubMed] [Google Scholar]
- 11.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 12.Russell JE, Morales J, Liebhaber SA. The role of mRNA stability in the control of globin gene expression. Prog Nucleic Acid Res Mol Biol. 1997;57:249–287. doi: 10.1016/s0079-6603(08)60283-4. [DOI] [PubMed] [Google Scholar]
- 13.Frevel MA, Bakheet T, Silva AM, Hissong JG, Khabar KS, Williams BR. p38 Mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol Cell Biol. 2003;23:425–436. doi: 10.1128/MCB.23.2.425-436.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez de Silanes I, Galban S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, et al. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, et al. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Lai WS, Parker JS, Grissom SF, Stumpo DJ, Blackshear PJ. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol Cell Biol. 2006;26:9196–9208. doi: 10.1128/MCB.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dean JL, Wait R, Mahtani KR, Sully G, Clark AR, Saklatvala J. The 3’ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Mol Cell Biol. 2001;21:721–730. doi: 10.1128/MCB.21.3.721-730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogilvie RL, Abelson M, Hau HH, Vlasova I, Blackshear PJ, Bohjanen PR. Tristetraprolin down-regulates IL-2 gene expression through AU-rich elementmediated mRNA decay. J Immunol. 2005;174:953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- 20.Ming XF, Stoecklin G, Lu M, Looser R, Moroni C. Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol Cell Biol. 2001;21:5778–5789. doi: 10.1128/MCB.21.17.5778-5789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma P, Cui X, Wang S, Zhang J, Nishanian EV, Wang W, et al. Nitric oxide post-transcriptionally up-regulates LPS-induced IL-8 expression through p38 MAPK activation. J Leukoc Biol. 2004;76:278–287. doi: 10.1189/jlb.1203653. [DOI] [PubMed] [Google Scholar]
- 22.Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, et al. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J Biol Chem. 2008;283:11689–11699. doi: 10.1074/jbc.M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pages G, Berra E, Milanini J, Levy AP, Pouyssegur J. Stress-activated protein kinases (JNK and p38/HOG) are essential for vascular endothelial growth factor mRNA stability. J Biol Chem. 2000;275:26484–26491. doi: 10.1074/jbc.M002104200. [DOI] [PubMed] [Google Scholar]
- 24.Lasa M, Mahtani KR, Finch A, Brewer G, Saklatvala J, Clark AR. Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol Cell Biol. 2000;20:4265–4274. doi: 10.1128/mcb.20.12.4265-4274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rydziel S, Varghese S, Canalis E. Transforming growth factor beta1 inhibits collagenase 3 expression by transcriptional and post-transcriptional mechanisms in osteoblast cultures. J Cell Physiol. 1997;170:145–152. doi: 10.1002/(SICI)1097-4652(199702)170:2<145::AID-JCP6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 26.Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raghavan A, Dhalla M, Bakheet T, Ogilvie RL, Vlasova IA, Khabar KS, et al. Patterns of coordinate down-regulation of ARE-containing transcripts following immune cell activation. Genomics. 2004;84:1002–1013. doi: 10.1016/j.ygeno.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Shen V, Kiledjian M. A view to a kill: structure of the RNA exosome. Cell. 2006;127:1093–1095. doi: 10.1016/j.cell.2006.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Espel E. The role of the AU-rich elements of mRNAs in controlling translation. Semin Cell Dev Biol. 2005;16:59–67. doi: 10.1016/j.semcdb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 31.Lu JY, Bergman N, Sadri N, Schneider RJ. Assembly of AUF1 with eIF4G–poly(A) binding protein complex suggests a translation function in AU-rich mRNA decay. Rna. 2006;12:883–893. doi: 10.1261/rna.2308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Kiledjian M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- 34.Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. Embo J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 36.Shim J, Lim H, J RY, Karin M. Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol Cell. 2002;10:1331–1344. doi: 10.1016/s1097-2765(02)00730-x. [DOI] [PubMed] [Google Scholar]
- 37.Loflin P, Chen CY, Shyu AB. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu N, Chen CY, Shyu AB. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol Cell Biol. 2001;21:6960–6971. doi: 10.1128/MCB.21.20.6960-6971.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raghavan A, Ogilvie RL, Reilly C, Abelson ML, Raghavan S, Vasdewani J, et al. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002;30:5529–5538. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vlasova IA, Bohjanen PR. Posttranscriptional regulation of gene networks by GU-rich elements and CELF proteins. RNA Biol. 2008;5:201–207. doi: 10.4161/rna.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le Bec C, et al. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem J. 2006;400:291–301. doi: 10.1042/BJ20060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. Rna. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barreau C, Paillard L, Mereau A, Osborne HB. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 44.Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, et al. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 45.Jahromi M, Millward A, Demaine A. A CA repeat polymorphism of the IFN-gamma gene is associated with susceptibility to type 1 diabetes. J Interferon Cytokine Res. 2000;20:187–190. doi: 10.1089/107999000312595. [DOI] [PubMed] [Google Scholar]
- 46.Citores MJ, Rua-Figueroa I, Rodriguez-Gallego C, Durantez A, Garcia-Laorden MI, Rodriguez-Lozano C, et al. The dinucleotide repeat polymorphism in the 3’UTR of the CD154 gene has a functional role on protein expression and is associated with systemic lupus erythematosus. Ann Rheum Dis. 2004;63:310–317. doi: 10.1136/ard.2003.006148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin-Donaire T, Losada-Fernandez I, Perez-Chacon G, Rua-Figueroa I, Erausquin C, Naranjo-Hernandez A, et al. Association of the microsatellite in the 3’ untranslated region of the CD154 gene with rheumatoid arthritis in females from a Spanish cohort: a case-control study. Arthritis Res Ther. 2007;9:R89. doi: 10.1186/ar2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Citores MJ, Perez-Aciego P, Rodriguez-Gallego C, Contreras-Martin B, Garcia-Laorden I, Durantez A. CD154 polymorphism in Spanish populations. Differences in the allelic distribution between Canary islanders and Peninsulars. Eur J Immunogenet. 2000;27:141–144. doi: 10.1046/j.1365-2370.2000.00220.x. [DOI] [PubMed] [Google Scholar]
- 49.Pinol-Roma S, Swanson MS, Gall JG, Dreyfuss G. A novel heterogeneous nuclear RNP protein with a unique distribution on nascent transcripts. J Cell Biol. 1989;109:2575–2587. doi: 10.1083/jcb.109.6.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hui J, Bindereif A. Alternative pre-mRNA splicing in the human system: unexpected role of repetitive sequences as regulatory elements. Biol Chem. 2005;386:1265–1271. doi: 10.1515/BC.2005.143. [DOI] [PubMed] [Google Scholar]
- 51.Hung LH, Heiner M, Hui J, Schreiner S, Benes V, Bindereif A. Diverse roles of hnRNP L in mammalian mRNA processing: a combined microarray and RNAi analysis. Rna. 2008;14:284–296. doi: 10.1261/rna.725208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guang S, Felthauser AM, Mertz JE. Binding of hnRNP L to the pre-mRNA processing enhancer of the herpes simplex virus thymidine kinase gene enhances both polyadenylation and nucleocytoplasmic export of intronless mRNAs. Mol Cell Biol. 2005;25:6303–6313. doi: 10.1128/MCB.25.15.6303-6313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu X, Mertz JE. HnRNP L binds a cis-acting RNA sequence element that enables intron-dependent gene expression. Genes Dev. 1995;9:1766–1780. doi: 10.1101/gad.9.14.1766. [DOI] [PubMed] [Google Scholar]
- 54.Hahm B, Kim YK, Kim JH, Kim TY, Jang SK. Heterogeneous nuclear ribonucleoprotein L interacts with the 3’ border of the internal ribosomal entry site of hepatitis C virus. J Virol. 1998;72:8782–8788. doi: 10.1128/jvi.72.11.8782-8788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melton AA, Jackson J, Wang J, Lynch KW. Combinatorial control of signal-induced exon repression by hnRNP L and PSF. Mol Cell Biol. 2007;27:6972–6984. doi: 10.1128/MCB.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oberdoerffer S, Moita LF, Neems D, Freitas RP, Hacohen N, Rao A. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321:686–691. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Topp JD, Jackson J, Melton AA, Lynch KW. A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. Rna. 2008;14:2038–2049. doi: 10.1261/rna.1212008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hui J, Reither G, Bindereif A. Novel functional role of CA repeats and hnRNP L in RNA stability. Rna. 2003;9:931–936. doi: 10.1261/rna.5660803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soderberg M, Raffalli-Mathieu F, Lang MA. Inflammation modulates the interaction of heterogeneous nuclear ribonucleoprotein (hnRNP) I/polypyrimidine tract binding protein and hnRNP L with the 3’untranslated region of the murine inducible nitric-oxide synthase mRNA. Mol Pharmacol. 2002;62:423–431. doi: 10.1124/mol.62.2.423. [DOI] [PubMed] [Google Scholar]
- 60.Shih SC, Claffey KP. Regulation of human vascular endothelial growth factor mRNA stability in hypoxia by heterogeneous nuclear ribonucleoprotein L. J Biol Chem. 1999;274:1359–1365. doi: 10.1074/jbc.274.3.1359. [DOI] [PubMed] [Google Scholar]
- 61.Hamilton BJ, Wang XW, Collins J, Bloch D, Bergeron A, Henry B, et al. Separate cis-trans pathways post-transcriptionally regulate murine CD154 (CD40 ligand) expression: a novel function for CA repeats in the 3’-untranslated region. J Biol Chem. 2008;283:25606–25616. doi: 10.1074/jbc.M802492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Messias AC, Harnisch C, Ostareck-Lederer A, Sattler M, Ostareck DH. The DICE-binding activity of KH domain 3 of hnRNP K is affected by c-Src-mediated tyrosine phosphorylation. J Mol Biol. 2006;361:470–481. doi: 10.1016/j.jmb.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 63.Wang S, Zhang J, Theel S, Barb JJ, Munson PJ, Danner RL. Nitric oxide activation of Erk1/2 regulates the stability and translation of mRNA transcripts containing CU-rich elements. Nucleic Acids Res. 2006;34:3044–3056. doi: 10.1093/nar/gkl386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Holcik M, Liebhaber SA. Four highly stable eukaryotic mRNAs assemble 3’ untranslated region RNA- protein complexes sharing cis and trans components. Proc Natl Acad Sci U S A. 1997;94:2410–2414. doi: 10.1073/pnas.94.6.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skalweit A, Doller A, Huth A, Kahne T, Persson PB, Thiele BJ. Posttranscriptional control of renin synthesis: identification of proteins interacting with renin mRNA 3’-untranslated region. Circ Res. 2003;92:419–427. doi: 10.1161/01.RES.0000059300.67152.4E. [DOI] [PubMed] [Google Scholar]
- 66.Ostareck-Lederer A, Ostareck DH, Cans C, Neubauer G, Bomsztyk K, Superti-Furga G, et al. c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol Cell Biol. 2002;22:4535–4543. doi: 10.1128/MCB.22.13.4535-4543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ostareck DH, Ostareck-Lederer A, Shatsky IN, Hentze MW. Lipoxygenase mRNA silencing in erythroid differentiation: The 3’UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 2001;104:281–290. doi: 10.1016/s0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
- 68.Meng Q, Rayala SK, Gururaj AE, Talukder AH, O’Malley BW, Kumar R. Signaling-dependent and coordinated regulation of transcription, splicing, and translation resides in a single coregulator, PCBP1. Proc Natl Acad Sci U S A. 2007;104:5866–5871. doi: 10.1073/pnas.0701065104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pautz A, Linker K, Hubrich T, Korhonen R, Altenhofer S, Kleinert H. The polypyrimidine tract-binding protein (PTB) is involved in the post-transcriptional regulation of human inducible nitric oxide synthase expression. J Biol Chem. 2006;281:32294–32302. doi: 10.1074/jbc.M603915200. [DOI] [PubMed] [Google Scholar]
- 70.Tillmar L, Carlsson C, Welsh N. Control of insulin mRNA stability in rat pancreatic islets. Regulatory role of a 3’-untranslated region pyrimidine-rich sequence. J Biol Chem. 2002;277:1099–1106. doi: 10.1074/jbc.M108340200. [DOI] [PubMed] [Google Scholar]
- 71.Coles LS, Bartley MA, Bert A, Hunter J, Polyak S, Diamond P, et al. A multi-protein complex containing cold shock domain (Y-box) and polypyrimidine tract binding proteins forms on the vascular endothelial growth factor mRNA. Potential role in mRNA stabilization. Eur J Biochem. 2004;271:648–660. doi: 10.1111/j.1432-1033.2003.03968.x. [DOI] [PubMed] [Google Scholar]
- 72.Kosinski PA, Laughlin J, Singh K, Covey LR. A Complex Containing Polypyrimidine Tract-Binding Protein Is Involved in Regulating the Stability of CD40 Ligand (CD154) mRNA. J Immunol. 2003;170:979–988. doi: 10.4049/jimmunol.170.2.979. [DOI] [PubMed] [Google Scholar]
- 73.Hamilton BJ, Genin A, Cron RQ, Rigby WF. Delineation of a novel pathway that regulates CD154 (CD40 ligand) expression. Mol Cell Biol. 2003;23:510–525. doi: 10.1128/MCB.23.2.510-525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 75.Li B, Yen TS. Characterization of the nuclear export signal of polypyrimidine tract- binding protein. J Biol Chem. 2002;277:10306–10314. doi: 10.1074/jbc.M109686200. [DOI] [PubMed] [Google Scholar]
- 76.Romanelli MG, Morandi C. Importin alpha binds to an unusual bipartite nuclear localization signal in the heterogeneous ribonucleoprotein type I. Eur J Biochem. 2002;269:2727–2734. doi: 10.1046/j.1432-1033.2002.02942.x. [DOI] [PubMed] [Google Scholar]
- 77.Xie J, Lee JA, Kress TL, Mowry KL, Black DL. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc Natl Acad Sci U S A. 2003;100:8776–8781. doi: 10.1073/pnas.1432696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kamath RV, Leary DJ, Huang S. Nucleocytoplasmic shuttling of polypyrimidine tract-binding protein is uncoupled from RNA export. Mol Biol Cell. 2001;12:3808–3820. doi: 10.1091/mbc.12.12.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Singh K, Laughlin J, Kosinski PA, Covey LR. Nucleolin is a second component of the CD154 mRNA stability complex that regulates mRNA turnover in activated T cells. J Immunol. 2004;173:976–985. doi: 10.4049/jimmunol.173.2.976. [DOI] [PubMed] [Google Scholar]
- 80.Laughlin J, Oghlidos S, Porter JF, Matus-Nicodemos R, Sinquett FL, Marcelli V, et al. Functional analysis of a tripartite stability element within the CD40 ligand 3’ untranslated region. Immunology. 2008;124:368–379. doi: 10.1111/j.1365-2567.2007.02783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fred RG, Welsh N. The importance of RNA binding proteins in preproinsulin mRNA stability. Mol Cell Endocrinol. 2009;297:28–33. doi: 10.1016/j.mce.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 82.Banchereau J, Bazan F, Blanchard D, Briere F, Galizzi JP, Kooten Cv, et al. The CD40 antigen and its ligand. Annu Rev Immunol. 1994;12:881–922. doi: 10.1146/annurev.iy.12.040194.004313. [DOI] [PubMed] [Google Scholar]
- 83.Foy TM, Aruffo A, Bajorath J, Buhlmann JE, Noelle RJ. Immune regulation by CD40 and its ligand GP39. Annu Rev Immunol. 1996;14:591–617. doi: 10.1146/annurev.immunol.14.1.591. [DOI] [PubMed] [Google Scholar]
- 84.Noelle RJ. CD40 and its ligand in host defense. Immunity. 1996;4:415–419. doi: 10.1016/s1074-7613(00)80408-2. [DOI] [PubMed] [Google Scholar]
- 85.Armitage RJ, Fanslow WC, Strockbine L, Sato TA, Clifford KN, Macduff BM, et al. Molecular and biological characterization of a murine ligand for CD40. Nature. 1992;357:80–82. doi: 10.1038/357080a0. [DOI] [PubMed] [Google Scholar]
- 86.Castle BE, Kishimoto K, Stearns C, Brown ML, Kehry MR. Regulation of expression of the ligand for CD40 on T helper lymphocytes. J Immunol. 1993;151:1777–1788. [PubMed] [Google Scholar]
- 87.Roy M, Waldschmidt T, Aruffo A, Ledbetter JA, Noelle RJ. The regulation of the expression of gp39, the CD40 ligand, on normal and cloned CD4+ T cells. J Immunol. 1993;151:2497–2510. [PubMed] [Google Scholar]
- 88.Nusslein HG, Frosch K-H, Woith W, Lane P, Kalden JR, Manger B. Increase of intracellular calcium is the essential signal for the expression of cd40 ligand. Eur J Immunol. 1996;26:846–850. doi: 10.1002/eji.1830260418. [DOI] [PubMed] [Google Scholar]
- 89.Suarez A, Mozo L, AbelGayo, Zamorano J, Gutierrez C. Requirement of a second signal via protein kinase C or protein kinase A for maximal expression of CD40 ligand Involvement of transcriptional and posttranscriptional mechanisms. Eur J Immunol. 1997;27:2822–2829. doi: 10.1002/eji.1830271112. [DOI] [PubMed] [Google Scholar]
- 90.Murakami K, Ma W, Fuleihan R, Pober JS. Human endothelial cells augment early CD40 ligand expression in activated CD4+ T cells through LFA-3-mediated stabilization of mRNA. J Immunol. 1999;163:2667–2673. [PubMed] [Google Scholar]
- 91.Ledbetter JA, Imboden JB, Schieven GL, Grosmaire LS, Rabinovitch PS, Lindsten T, et al. CD28 ligation in T-cell activation: evidence for two signal transduction pathways. Blood. 1990;75:1531–1539. [PubMed] [Google Scholar]
- 92.Powell JD, Ragheb JA, Kitagawa-Sakakida S, Schwartz RH. Molecular regulation of interleukin-2 expression by CD28 co-stimulation and anergy. Immunol Rev. 1998;165:287–300. doi: 10.1111/j.1600-065x.1998.tb01246.x. [DOI] [PubMed] [Google Scholar]
- 93.Ford GS, Barnhart B, Shone S, Covey LR. Regulation of CD154 (CD40 ligand) mRNA stability during T cell activation. J Immunol. 1999;162:4037–4044. [PubMed] [Google Scholar]
- 94.Barnhart B, Kosinski PA, Wang Z, Ford GS, Kiledjian M, Covey LR. Identification of a complex that binds to the CD154 3’ untranslated region: implications for a role in message stability during T cell activation. J Immunol. 2000;165:4478–4486. doi: 10.4049/jimmunol.165.8.4478. [DOI] [PubMed] [Google Scholar]
- 95.Porter JF, Vavassori S, Covey LR. A polypyrimidine tract-binding protein-dependent pathway of mRNA stability initiates with CpG activation of primary B cells. J Immunol. 2008;181:3336–3345. doi: 10.4049/jimmunol.181.5.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Santos-Argumedo L, Alvarez-Maya I, Romero-Ramirez H, Flores-Romo L. Enforced and prolonged CD40 ligand expression triggers autoantibody production in vivo. Eur J Immunol. 2001;31:3484–3492. doi: 10.1002/1521-4141(200112)31:12<3484::aid-immu3484>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 97.Blossom S, Gilbert KM. Antibody production in autoimmune BXSB mice. I. CD40L-expressing B cells need fewer signals for polyclonal antibody synthesis. Clin Exp Immunol. 1999;118:147–153. doi: 10.1046/j.1365-2249.1999.01032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Desai-Mehta A, Lu L, Ramsey-Goldman R, Datta SK. Hyperexpression of CD40 ligand by B and T cells in human lupus and its role in pathogenic autoantibody production. J Clin Invest. 1996;97:2063–2073. doi: 10.1172/JCI118643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Devi BS, Van Noordin S, Krausz T, Davies KA. Peripheral blood lymphocytes in SLE--hyperexpression of CD154 on T and B lymphocytes and increased number of double negative T cells. J Autoimmun. 1998;11:471–475. doi: 10.1006/jaut.1998.0213. [DOI] [PubMed] [Google Scholar]
- 100.Crow MK, Kirou KA. Regulation of CD40 ligand expression in systemic lupus erythematosus. Curr Opin Rheumatol. 2001;13:361–369. doi: 10.1097/00002281-200109000-00004. [DOI] [PubMed] [Google Scholar]
- 101.de Boer M, Dasran A, Kwekkeboom J, Walter H, Vandenberghe P, Ceupens JL. Ligation of B7 with CD28/CTLA-4 on T cells results in CD40 ligand expression, interkeukin-4 secretion and efficient help for antibody production by B cells. Eur J Immunol. 1993;23:3120. doi: 10.1002/eji.1830231212. [DOI] [PubMed] [Google Scholar]
- 102.Ding L, Green JM, Thompson CB, Shevach EM. B7/CD28-dependent and -independent induction of CD40 ligand expression. J Immunol. 1995;155:5124–5132. [PubMed] [Google Scholar]
- 103.Jaiswal AI, Dubey C, Swain SL, Croft M. Regulation of CD40 ligand expression on naive CD4 T cells: a role for TCR but not co-stimulatory signals. Int Immunol. 1996;8:275–285. doi: 10.1093/intimm/8.2.275. [DOI] [PubMed] [Google Scholar]
- 104.Lane P, Traunecker A, Inui S, Lanzavecchia A, Gray D. Activated human T cells express a ligand for the human B cell-associated antigen CD40 which participates in T cell-dependent activation of B lymphocytes. Eur J Immunol. 1992;22:2573–2578. doi: 10.1002/eji.1830221016. [DOI] [PubMed] [Google Scholar]
- 105.Johnson-Leger C, Christensen J, Klaus GG. CD28 co-stimulation stabilizes the expression of the CD40 ligand on T cells. Int Immunol. 1998;10:1083–1091. doi: 10.1093/intimm/10.8.1083. [DOI] [PubMed] [Google Scholar]
- 106.Karmann K, Hughes CC, Fanslow WC, Pober JS. Endothelial cells augment the expression of CD40 ligand on newly activated human CD4+ T cells through a CD2/LFA-3 signaling pathway. Eur J Immunol. 1996;26:610–617. doi: 10.1002/eji.1830260316. [DOI] [PubMed] [Google Scholar]
- 107.Peng X, Remacle JE, Kasran A, Huylebroeck D, Ceuppens JL. IL-12 up-regulates CD40 ligand (CD154) expression on human T cells. J Immunol. 1998;160:1166–1172. [PubMed] [Google Scholar]
- 108.Skov S, Bonyhadi M, Odum N, Ledbetter JA. IL-2 and IL-15 regulate CD154 expression on activated CD4 T cells. J Immunol. 2000;164:3500–3505. doi: 10.4049/jimmunol.164.7.3500. [DOI] [PubMed] [Google Scholar]
- 109.Lee BO, Haynes L, Eaton SM, Swain SL, Randall TD. The biological outcome of CD40 signaling is dependent on the duration of CD40 ligand expression: reciprocal regulation by interleukin (IL)-4 and IL-12. J Exp Med. 2002;196:693–704. doi: 10.1084/jem.20020845. [DOI] [PMC free article] [PubMed] [Google Scholar]