Abstract
Synthesis of oligoribonucleotide primers for lagging-strand DNA synthesis in the DNA replication system of bacteriophage T7 is catalyzed by the primase domain of the gene 4 helicase-primase. The primase consists of a zinc-binding domain (ZBD) and an RNA polymerase (RPD) domain. The ZBD is responsible for recognition of a specific sequence in the ssDNA template whereas catalytic activity resides in the RPD. The ZBD contains a zinc ion coordinated with four cysteine residues. We have examined the ligation state of the zinc ion by X-ray absorption spectroscopy and biochemical analysis of genetically altered primases. The ZBD of primase engaged in catalysis exhibits considerable asymmetry in coordination to zinc, as evidenced by a gradual increase in electron density of the zinc together with elongation of the zinc–sulfur bonds. Both wild-type primase and primase reconstituted from purified ZBD and RPD have a similar electronic change in the level of the zinc ion as well as the configuration of the ZBD. Single amino acid replacements in the ZBD (H33A and C36S) result in the loss of both zinc binding and its structural integrity. Thus the zinc in the ZBD may act as a charge modulation indicator for the surrounding sulfur atoms necessary for recognition of specific DNA sequences.
DNA primases are RNA polymerases that synthesize oligoribonucleotides for use as primers by DNA polymerases (1). The synthesis of oligoribonucleotides by DNA primase occurs continuously on the lagging strand to provide primers for the lagging-strand DNA polymerase to initiate the synthesis of Okazaki fragments. The DNA primase encoded by bacteriophage T7 is located in the N-terminal half of the multifunctional gene 4 helicase-primase. The primase domain is composed of an RNA polymerase (RPD)1 and a zinc-binding (ZBD) domain (Figure 1). The ZBD is essential for the recognition of a specific sequence, 5′-GTC-3′, where primer synthesis is initiated by the synthesis of the dinucleotide pppAC (2). The “cryptic” 3′-cytosine in the recognition sequence is essential for recognition but is not copied into the product. The functional tetranucleotide primer arises by extension of the dinucleotide by the primase providing the proper bases are present in the template.
Figure 1.
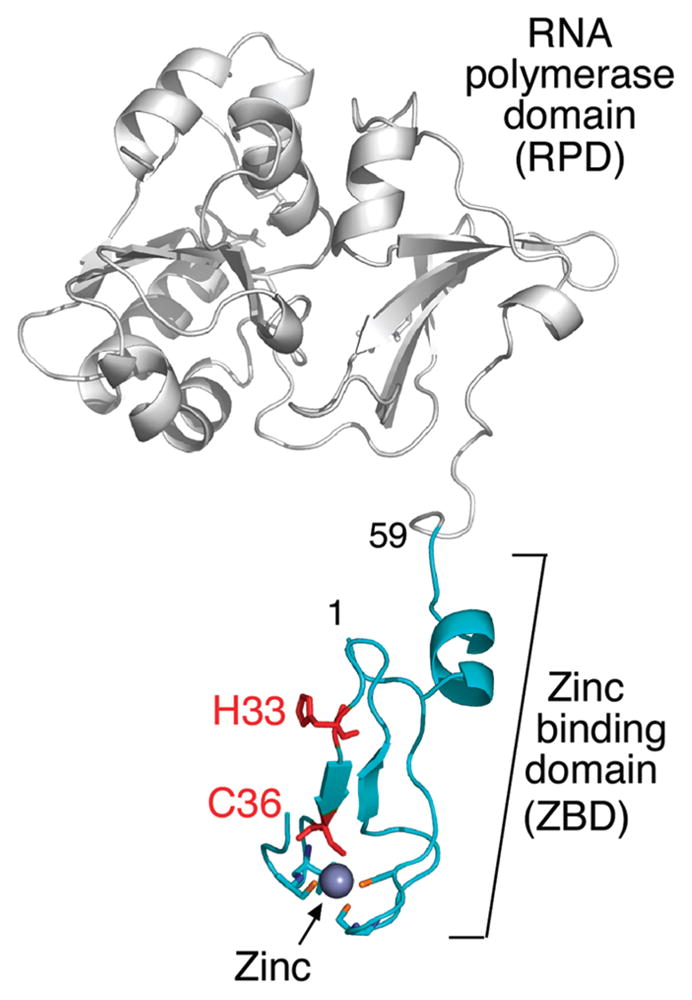
Crystal structure of the phage T7 primase fragment (PDB entry 1nui). The ZBD is green, and the genetically altered residues are indicated in red. The RNA polymerase domain is colored in gray. The figure was created using PyMOL (http://www.pymol.org). The residue numbers are indicated.
The zinc-binding motif in the ZBD contains a zinc ion coordinated to four cysteine residues. Alteration of any of the cysteines decreases the zinc content drastically and impairs template-directed synthesis of primers (3). Using crystallography, NMR spectroscopy, and biochemical assays, Kato et al. showed that the ZBD and the RPD were connected by a flexible linker and were in an “open” conformation. Upon the addition of nucleotides and DNA template, the enzyme adopts a “closed” conformation (4). Consequently, it was suggested that the RPD and the ZBD physically contact one another during the synthesis of RNA primers. The flexible linker between the domains enables the ZBD to recognize the template and to transfer the primer to T7 DNA polymerase (4). In the hexameric gene 4 protein, the ZBD of one primase domain can contact the RPD of an adjacent primase domain. This interaction between two adjacent primases is made possible by the linker (5). Furthermore, this “trans” synthesis of primers may be important in the coordination of leading and lagging-strand synthesis.
Interestingly, the ZBD alone can stimulate primer extension by the DNA polymerase upon primer delivery (4).
Zinc ions that are important in structural determinants are often buried and surrounded by an elaborate network of hydrogen bonds provided by a second coordination layer (6). Moreover, cysteine residues in structural zinc sites can also serve as nucleophiles. For example, in the Zn–Cys4 system of the Ada protein, one of the Zn-bound thiolates acts as a nucleophile to react with the methyl group of a DNA methyl phosphotriester (7). This observation was the first example of a structural Zn–Cys4 site that has a reactive sulfur ligand and challenged the concept that a thiolate cannot be reactive in a structural site.
The chemical properties of zinc and its electronic configuration among the other transition metal ions have important consequences that make zinc-binding domains very selective (8). In biological systems zinc ions are always in the Zn(II) state and are not themselves redox active (8). However, binding of a zinc ion changes the properties of the thiol group in cysteine (9). Oxidation of the sulfur ligands mobilizes zinc, while reduction of the oxidized ligands enhances zinc binding. Some proteins have been shown to be redox sensors in which zinc release is coupled to conformational changes that control various functions such as enzymatic activity, binding interactions, and molecular chaperone activity (9).
Removal or disruption of the Zn–Cys4 motif in T7 primase destroys recognition of the specific sequence 5′-GTC-3′ (2, 3) and suggests contact between the Zn–Cys4 motif and its trinucleotide recognition sequence. Substitution of His33 for alanine has a striking effect on primase activity (10). This altered protein cannot synthesize primers on a template that contain the canonical T7 primase-recognition sequence 5′-GTC-3′. Surprisingly, this altered primase does have activity, albeit low, on templates containing 5′-GTA/G-3′. Apparently, this altered protein can now recognize sites in which the cryptic nucleotide is a purine rather than a cytosine (3). Therefore, His33 may play a role in the recognition of the cryptic cytosine.
Simonson and Calimet used ab initio calculations to examine the possibility of thiolate protonation in zinc-binding motifs in proteins (11). They found, as expected, that protonating thiolates increases the Zn–S (thiol) distance. Thus, the Zn–S(H) bond length can serve as a structural marker for the protonation state. Thus, Zn–S4 and ZnSH4 structures are predicted to be symmetrical with Zn–S(H) bond lengths differing by no more than 0.1 Å, while a mixed distribution of thiols versus thiolates in the Zn–Cys4 motif is moderately asymmetrical. They predicted several structures with different combinations: one structure with four thiolates and others with a combination of one or more thiols.
Structural studies of Zn sites in metalloenzymes are limited to X-ray crystallography, X-ray absorption spectroscopy, and other methods that require the substitution of zinc with a “visible” metal ion (12), inasmuch as the Zn ion is silent to most spectroscopic methods. X-ray absorption spectroscopy (XAS) allows detection of changes in the metal sites of metalloenzymes during the course of an enzymatic reaction (13). XAS provides high resolution structural and electronic information about a metal site by measuring the transition from core electronic states of the metal to the excited electronic or continuum states. X-ray absorption near-edge structure (XANES) spectral analysis near the electronic transition provides information about the charge and the geometry of the metal ion (14). The edge energy corresponds to the amount of energy needed to extract core electron to the continuum. The extraction of the electron occurs when an incident X-ray beam has energy equal to the binding energy of the core level. When the atom is relatively reduced, the X-ray energy needed for extraction is lower; when the atom is relatively oxidized, higher energy is needed to promote the core electron. The spectral region of this electronic promotion is called the absorption edge. The edge energy of many elements shows significant edge shifts (binding-energy shifts) with an oxidation state of about 4 eV per change of one electron. Therefore, with a higher resolution, the analysis of XANES enables the identity of even subelectronic changes that can be associated with ultramolecular aspects that one wishes to describe. Thus, fingerprints of binding of metalostructure (as for example a metal-binding site in a protein) to its ligand can be easily identified. Spectral analysis above the absorption edge, in the extended X-ray absorption fine structure (EXAFS) region, allows the determination of complementary structural information. Such information includes coordination number, type of scattering ligands, thermal disorder, Debye–Waller factors, and distances from neighboring atoms to the absorbing metal (14). Therefore, EXAFS can be used to construct a model based on spectroscopic data by fitting analysis of the EXAFS equation to the measured spectra. Using XAS, Powers and Griep found that a change in the ligation state of the zinc site in the E. coli primase occurs upon binding of various substrates and cofactors (15). This was the first attempt to study a primase-bounded zinc ion at the atomic level, among the other studies that were aimed at identification of the need of ZBD to recognize initiation sites. Specifically, XAS has established that the E. coli primase zinc will adopt a different ligation state when the enzyme is bound to poly dT and ATP or has an inhibitory condition, such as a high concentration of magnesium acetate. Using these different binding states of the primase, they demonstrated the zinc-binding region of the primase plays a critical role during primer synthesis, and the zinc is at or near the site of action. T7 primase is presented here as a model for studies on structural zinc sites. We address several biological questions that are difficult to answer by conventional methods: What is the effect of specific DNA interactions on the charge of the zinc ion in the ZBD of the primase? What is the specific role of the His-33 residue in sequence recognition? Is the Zn ion affected by the nature of the binding of DNA to the primase? What is the relationship between the charge of the zinc ion, the binding state of the primase, and its activity?
To answer these questions, we carried out XANES analysis on the intact primase domain of the gene 4 protein, the isolated ZBD, and a mixture of RPD and ZBD. We show that the effective charge on the Zn(II) ion in T7 primase can vary with concomitant changes in DNA-binding affinity and primase activity. The measured charge transfer is fragmental since it is not a traditional oxidation/reduction that conventionally requires full electron changes in the oxidation state. Therefore, the zinc ion serves as a sensitive indicator within the ZBD for DNA binding. EXAFS analysis was used to probe Zn–S(H) bond lengths to provide complementary information on the protonation state of the cysteine sulfurs.
Using mutation analysis, we show that altering the conserved Cys36 or His33 mobilizes zinc from the ZBD, changes the local structure of the ZBD, and impairs the primase activity. These findings emphasize the importance of direct binding of cysteines to zinc as well as the importance of a nearby residue (His33). The latter plays a crucial role in the structural maintenance of the zinc-binding site and/or regulates the effective protonation state of the Zn–S(H) ligands.
EXPERIMENTAL PROCEDURES
Proteins and Reagents
All chemical reagents were of molecular biology grade (Sigma). ATP and CTP, were purchased from Roche Molecular Biochemicals. Molecular weight markers used in the SDS-PAGE studies, Precision Plus Protein prestained standard, and Ready gel 10–20% linear gradient were purchased from BioRad (Hercules, CA). T7 primase fragment (residues: 1–271), or ZBD (residues: 1–59) and RPD (residues: 64–271) were overproduced and purified using metal-free buffers as previously described (4, 16).
Oligoribonucleotide Synthesis Assay
Synthesis of oligoribonucleotides by DNA primase was measured as described (17, 18) in reactions containing various concentrations (1.1, 3.3, and 10 μM) of gene 4 primase fragment. Standard 10 μL reactions contained 5 μM DNA templates (5′-GGGT-CAA-3′ or 5′-CACACAA-3′ as a negative control), 200 μM ATP, 200 μM [α-32P]-CTP, and primase fragment in a buffer containing 40 mM Tris–HCl pH 7.5, 10 mM MnCl2, 10 mM DTT, and 50 mM potassium glutamate. After incubation at room temperature for 10 min, the reaction was terminated by adding an equal volume of sequencing buffer containing 98% formamide, 0.1% bromophenol blue, and 20 mM EDTA. The samples were loaded onto 25% polyacrylamide sequencing gel containing 3 M urea and visualized using autoradiography. For reconstitution of primase by ZBD and RPD, ZBD (10 μM) was preincubated with an increasing concentration of RPD (4, 8, and 16 μM) for 10 min at room temperature. Oligoribonucleotide synthesis was measured under the same conditions described above for the standard reaction.
Inductively Coupled Plasma-Mass Spectrometry (ICP-MS)
Samples were prepared by adding 0.5 mL of concentrated HNO3 acid for 24 h and then diluted to 5 mL with deionized water. The zinc content of proteins was analyzed using a dynamic reaction cell-inductively coupled plasma-mass spectrometer (DRC-ICP-MS, Elan 6100, Perkin-Elmer, Norwark, CT) using indium as the internal standard. The ICP-MS was calibrated using a zinc standard from SPEX Certi Prep (Metuchen, NJ). Quality assurance and quality control procedures were performed to ensure the accuracy of the zinc analysis. Method blanks were analyzed to monitor contamination. NIST SRM 1643e, trace elements in water, was used as the ICP-MS calibration verification standard. The limit of detection (LOD) was determined as 3 times the standard deviation of 10 replicate measurements of the method blank. None of the sample concentrations were below the LOD (primase, primase-H33A, primase-C36S, ZBD, ZBD-H33A, and ZBD-C36S).
Sample Preparation for X-ray Absorption Spectroscopy (XAS)
The proteins were concentrated by centrifugation using a Millipore Centricon (Bedford, MA) device to make a final millimolar concentration range. T7 primase fragment or ZBD and RPD were mixed with DNA (primase-recognition sequence: 5′-GGGTCAA-3′ and the nonspecific sequence: 5′-CACACAA-3′), ATP, CTP, and magnesium at 0 °C and immediately frozen in copper sample holders (10 × 5 × 0.5 mm) covered with Mylar using liquid nitrogen. The sample cells were mounted in a Displex closed-cycle cryostat. In order to minimize thermal disorder and enhance signal-to-noise, the temperature was kept at 20 K.
XAS Data Collection
XAS data collection was performed at the National Synchrotron Light Source at Brookhaven National Laboratory, beam line X3B. The spectra were recorded as previously described (19). Specifically the Zn(II) K-edge was measured in a fluorescence mode at a cryogenic temperature (20 K). The beam energy was selected using a flat Si(III) crystal monochromator with an energy resolution (dE/E) of ~2 × 10−4. The incident beam intensity was recorded using a transmission ion chamber and the florescence signal from the sample was measured using a 13-element germanium detector. For the calibration of the X-ray energy, the transmission signal of a Zn(II) foil reference was measured simultaneously with the sample fluorescence. For each sample, several scans were collected to obtain a total of 1 × 106 counts. The beam position was varied for each scan to minimize radiation damage, and samples were checked for visual signs of photoreduction (burn marks) after each scan (30 min). A further protection of the sample from radiation damage was obtained by using buffer containing 1 mM DTT to serve as a free-radical scavenger. In addition, the protein sample was exposed to X-ray under cryogenic temperature. Examination of the samples on SDS-PAGE after the exposure to the X-ray beam revealed no evidence for protein degradation caused by radiation damage (not shown).
XAS Data Processing and Analysis
The average Zn(II) K-edge adsorption coefficient μ(E) was obtained after four to six independent XAS measurements for each sample. To calibrate the X-ray energy, all spectra in every data set (sample) were aligned using the first inflection point of a reference Zn(II) foil X-ray absorption spectrum (9659 eV). The data measured intensities were converted to μ(E), and the smooth pre-edge function was subtracted from the spectra to get rid of any instrumental background and absorption from other atomic edges. The data (μ(E)) were normalized and a smooth postedge background function was removed to approximate μ(E) in order to isolate the EXAFS signal. The threshold E0 was identified and was converted from energy to wavenumber space. The EXAFS signal was weighted and Fourier transformed to real space.
The XAS data processing and the followed fitting analysis were performed using an iFEFFIT package (20, 21) to the EXAFS equation
| (1) |
where f(k) and δj(k) are scattering amplitude and the phase shift of the neighboring atom, respectively. Knowing these values, the distance to the neighboring atom (R), the coordination numbers of the neighboring atom (N), and the mean-square disorder of neighbor distance (σ) were determined. Furthermore, based on scattering properties of the neighboring atoms, their atomic number (Z) value was also determined. This data analysis strategy, while employing the chemical and physically reasonable constraints between the fitting parameters, provides general trends in the dynamic changes in coordination number and metal–ligand bond distances.
The same theoretical photoelectron path corresponding to the first-shell Zn–S distances were used to fit all of the data set systematically to examine combinations of various Zn–S bond lengths for the first-shell model. The input model was based on the crystallographic coordinates of T7 primase fragment (PDB entry 1nui). EXFAS analysis depends on the data points (K-range and R-range) to determine the limited number of parameters. The number of the variables to be determined in the process of EXAFS fitting must be smaller than the relevant independent points that are presented in the EXAFS spectrum (22). Therefore, to reduce the degrees of freedom to the fit, the following parameters were fixed for all data sets: amplitude factor (SO2 = 0.9), the number of first-shell ligands (N = 4), and the data range for fitting (Δr and Δk). The following parameters were varied: the correction to the photoelectron energy origin (ΔE0), the relative movement of the S atom relatively to the Zn (dr), and the mean-square disorder of the corrections (ΔR) to the Zn–S (σ2). Therefore, the total number of variables was smaller than the number of independent data points in the experimental XAS spectra. The best fit for each value of the starting-phase fraction was found by locating the minimum of the statistical 〈χ〉2 values obtained in each fit.
Circular Dichroism (CD) Spectroscopy
CD measurement was performed by employing an Aviv 202 spectropolarimeter using a 1 mm path-length quartz cuvette at 25 °C. Wavelength scans were done in the 200–300 nm far-ultraviolet range. The CD spectra of primase fragments (including the mutants H33A and C36A) and ZBD (including the mutants H33A and C36S) were recorded at the concentration 0.16 mg/mL in a buffer (50 mM HEPES pH 7.5, 1 mM DTT, and 30% glycerol). The data sets were averaged and then normalized to the baseline at 300 nm.
Computer Modeling
The pKa of cysteines in the zinc-binding motif was predicted using PROPKA2.0 (23, 24) from the crystal structure of the T7 primase fragment (PDB entry 1nui). PROPKA data were obtained using the publicly available web interface (http://propka.ki.ku.dk). Several other methods for the prediction of pKa values are available, e.g., MCCE (25, 26), MEAD (27), and PKAcal (28). However, PROPKA performed well in several benchmarking studies (e.g., refs (23), (24), and (29)) and was therefore our method of choice. The two monomers present in the crystal asymmetric unit were aligned using PyMOL (www.pymol.org).
RESULTS
Precisely how the T7 DNA primase recognizes the sequence 5′-GTC-3′ in single-stranded DNA is not known. However, several lines of evidence suggest that the ZBD plays a role in recognition and in delivering the site to the catalytic RPD domain for oligoribonucleotide synthesis (1). We have examined the association of the ZBD with the RPD using X-ray absorption spectroscopy to obtain structural information on the events that occur during primer synthesis. We have monitored the various steps using biochemical assays to measure primase activity. Purification of the ZBD and the RPD allows for an examination of their interaction and a comparison with the isolated primase fragment that contains both the ZBD and the RPD. The primase fragment (residues 1–271) used in these studies is expressed from a plasmid harboring the sequence for the major portion of the primase domain of gene 4 of bacteriophage T7. The ZBD (residues 1–59) and the RPD (residues 64–271) were likewise expressed from plasmids harboring the respective sequences for each (Experimental Procedures).
Template-Directed Oligoribonucleotide Synthesis
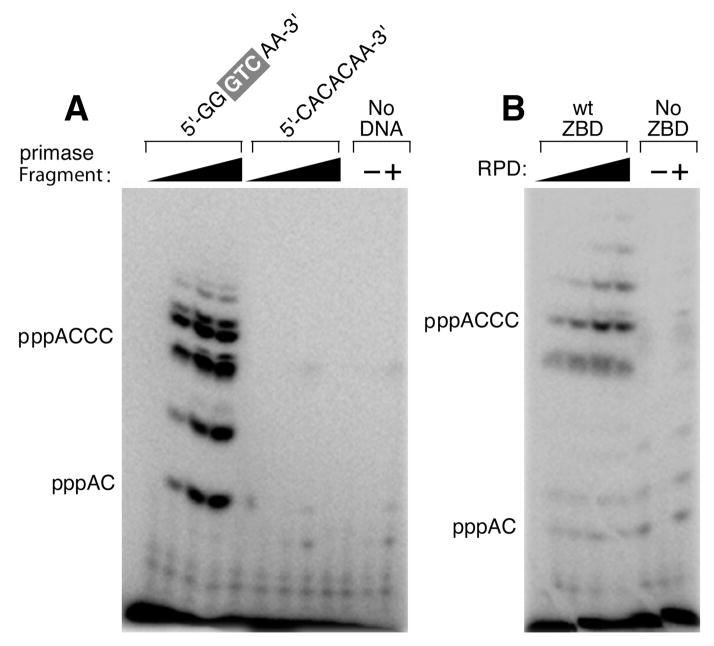
On a DNA template containing the T7 primase-recognition site 5′-GGGTC-3′, the T7 DNA primase catalyzes the synthesis of the di-, tri-, and tetraribonucleotides pppAC, pppACC, and pppACCC (1). We examined the ability of the primase fragment as well as primase reconstituted from the ZBD and the RPD to synthesize oligoribonucleotides under the same conditions as those used for the spectroscopic measurements (Figure 2). The reaction conditions involve incubating the primase with an oligonucleotide containing a primase-recognition sequence, [α-32P]-CTP, and ATP. The radioactively labeled oligoribonucleotide products are separated on a denaturing polyacrylamide gel, and the radioactivity is measured on an autoradiogram. The quantity of product was measured in order to compare the biochemical results to those of the structural studies. The primase fragment catalyzes the synthesis of oligoribonucleotides in the presence of the synthetic DNA template containing the primase-recognition sequence 5′-GGGTCAA-3′ (700 fmol of tetraribonucleotide primer/pmol of primase/min) but not in the presence of a control oligonucleotide (5′-CACACAA-3′) that lacks the recognition sequence (Figure 2A). Neither the ZBD nor the RPD alone catalyzes the synthesis of oligonucleotides (30), but activity can be restored in a reconstitution assay (Figure 2B) where increasing amounts of the ZBD are added to a fixed amount of the RPD. The reconstitution assay requires relatively high concentrations of the proteins presumably due to the absence of the linker that covalently connects the two in the primase fragment. These results ensure that the proteins used are fully active and the conditions are optimal for structural studies.
Figure 2.
Template-directed oligoribonucleotide synthesis catalyzed by T7 DNA primase. (A) Oligonucleotide synthesis by primase fragment. The reaction contained the oligonucleotide 5′-GGGT-CAA-3′ containing the primase-recognition sequence and a control oligonucleotide 5′-CACACAA-3′ lacking the primase-recognition sequence and [α-32P]-CTP and ATP in the standard reaction mixture (Experimental Procedures). (B) Oligoribonucleotide synthesis by reconstituted DNA primase. The reaction contained the oligonucleotide 5′-GGGTCA-3′, 100 μM ZBD, and increasing amounts of the RPD (0, 25, 50, and 100 μM) using the standard reaction conditions. In both (A) and (B), the reaction contained [α-32P]-CTP and ATP. After incubation, the radioactive products were analyzed by electrophoresis through a 25% polyacrylamide gel containing 3 M urea and visualized using autoradiography. Exposure time in (B) was longer than in (A) to increase the bands intensity in the reconstitution assay.
X-ray Absorption Spectroscopy of ZBD
The Cys4-zinc motif of T7 DNA primase binds a zinc ion in a molar ratio of 1:1 (3). We used X-ray absorption spectroscopy (XAS) to probe the local structure of the zinc-binding motif when it interacts with ATP and CTP, its recognition site in DNA, or with the RPD. XAS is a method that uses the metal ion itself as a sensitive probe to report changes in local structure and intrinsic chemical properties that occur in metallocenters when the protein interacts with other relevant biological molecules (14). For example, XAS has shown that the local structure around the zinc in the E. coli DNA primase, which has a zinc-binding domain similar to that of T7 primase (31), undergoes changes on substrate binding (15). A typical X-ray absorption spectrum is divided into two regimes: the near edge spectrum (XANES) and the extended fine structure spectrum (EXAFS). XANES provides information on the formal valency (oxidation state) of the metal ion probe and its geometry. EXAFS provides information on (1) the distances between central and neighboring atoms, (2) the number of neighboring atoms, (3) the nature of neighboring atoms that are embedded in the scattering factors that are the structural models in the analysis, and (4) the changes in central-atom coordination with changes in experimental conditions. A significant advantage of EXAFS analysis over X-ray crystallography is that structures can be studied in noncrystalline forms, including liquid and frozen solutions.
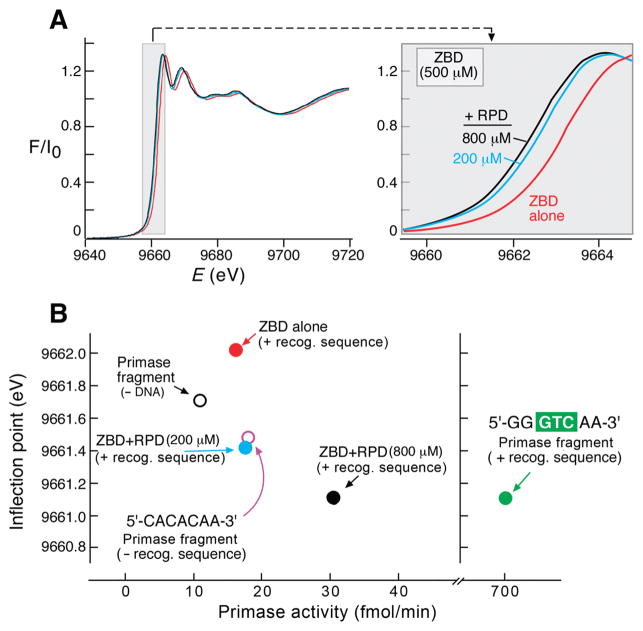
Using reaction conditions similar to those used for the primase assays described above, frozen mixtures of DNA template-protein complexes were prepared in HEPES buffer containing potassium glutamate, MgCl2, ATP, and CTP. The components were mixed in an ice bath and immediately frozen in liquid nitrogen to avoid the start of the reaction prior to EXAFS data collection at the synchrotron light source. The relative formal charge on the zinc atom is reflected by the X-ray energy that is required to promote a core-level electron in the zinc absorption edge region of the spectrum (inflection point). Thus, the charge state of the zinc ion in the primase and in the reconstituted primase (ZBD and RPD) was extracted from the XANES part of the spectrum. With the energy resolution (dE/E) of ~2 × 10−4, the energy difference of one electron (as of Fe(II) versus Fe(III)) is ~4 eV. In the presence of an oligonucleotide (5′-GGGTCAA-3′) containing the primase-recognition site and nucleotides, the addition of the RPD to the ZBD resulted in a gradual and fragmental increase in electron density of the zinc (i.e., decrease in formal charge) as the concentration of the RPD increased (Figure 3A). The primase fragment presents an X-ray energy value for the inflection point that is similar to the free ZBD when no DNA is present (Figure 3B). When DNA lacking a primase-recognition site (5′-CACACAA-3′) replaces the one containing a recognition site, the inflection point energy value is lower, indicating that the formal charge on zinc is fractionally reduced. However, when the primase-recognition sequence is present, this energy value is decreased further and corresponds to the value of the highest ratio of RPD to ZBD (Figure 3B). The change in edge energy as a function of primase activity (Figure 3B) indicates that the zinc is partially reduced and that the local zinc environment is modified during DNA binding, the first step in the primase reaction. Previous studies have shown that significant shifts are correlated directly with structural rearrangements of the metal ion coordination sphere (13, 32).
Figure 3.
Near edge X-ray absorption spectroscopy. (A) X-ray absorption near edge spectra of the titration of the RPD to a constant concentration of the ZBD. RPD was titrated (0, 200, and 800 μM) to a constant concentration of ZBD (500 μM) in the presence of ATP, CTP, and DNA template containing a primase-recognition sequence (5′-GGGTCAA-3′). The inflection point values were derived from the near edge X-ray absorption spectra (9662.01, 9661.41, and 9661.11 eV, respectively). (B) The primase activity is presented as a function of the change in the inflection point value of each sample. Titration of RPD to ZBD is labeled as in (A). Key: ZBD (500 μM, red circle), ZBD:RPD (500:200 μM, blue circle), ZBD:RPD (500:800 μM, black circle), primase fragment (500 μM, open black circle), primase fragment:DNA template 5′-GGGTCAA-3′ (500:495 μM, green circle), primase fragment:DNA template 5′-CACACAA-3′ (500:495 μM, open purple circle). The activity was calculated from densitometry analysis following separation of the reaction products on sequencing gels and visualized using autoradiography as described in the Experimental Procedures. The same mixing ratios of protein components were used as in Figure 2.
The ZBD and primase fragment have slightly different energies in the absence of DNA, indicating that Zn can sense the RPD domain. In the presence of rNTPs and DNA containing the recognition sequence, Zn in the primase and the reconstituted primase (ZBD + 800 μM RPD) has a transition at the same absorption energy. This value is distinctly lower than in the absence of DNA and rNTPs. This finding indicates that the formal charge on Zn is reduced further when the productive complex is made. An intermediate situation is found when DNA lacking the primase-recognition sequence replaces the DNA containing the recognition sequence. Thus, the data indicate that more or less subtle structural changes occur close to the zinc atom in the ZBD on interaction with the RPD whereas larger changes occur on formation of a productive priming complex. These changes were further probed by extensive analysis of the EXAFS apparent in these spectra.
EXAFS analysis was used to examine zinc ligation to the four cysteines using amplitude and phase functions based on FEFF calculations embedded in the iFFEFIT package (20). The input model was based on the crystallographic coordinates of T7 primase fragment (PDB entry 1nui). The model corresponds to four sulfur atoms coordinated to the zinc and is referred to as the four-atom model. The two-atom model refers to two Zn–S distances used in the fitting analysis (Supporting Information, Table S1). Starting models were generated on the basis of 26 possible combinations of the four different Zn–S bond lengths observed in the crystal structure. Initial fits were performed for all models on the four protein samples (the results for three representative fits are presented in Table S1 (Supporting Information). Best-fitting results revealed that a two-atom model had a better fit than a four-atom model to the EXAFS spectrum of the ZBD and the primase fragment in the absence of DNA template (Table 1, Figure 4A). However, a four-atom model is best fit to the EXAFS spectra of ZBD-RPD and primase fragment, both in the presence of the DNA template. The spectral evolution that corresponds to the RPD titration to ZBD demonstrates that the spectra are a linear combination of the two principal factors of the complex mixture. Factor A corresponds to the spectrum of ZBD, whereas factor B resembles a mixture of primase and ZBD-RPD. Thus, each XAS spectrum along the binding cycle of primase with DNA is presumably a result of a linear combination of these two components.
Table 1.
Analysis of EXAFS Dataa
| 1x Zn–S I |
1x Zn– S II |
1x Zn–S III |
1x Zn–S IV |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sample | χ2 | R (Å) | ΔE0 (eV) | σ2 (Å2) | R (Å) | ΔE0 (eV) | σ2 (Å2) | R (Å) | ΔE0 (eV) | σ2 (Å2) | R (Å) | ΔE0 (eV) | σ2 (Å2) |
| ZBD | 59.65 | 2.31 | 11.99 | −3.22E–04 | 2.31 | 11.99 | −3.22E–04 | 2.31 | 11.99 | −2.67E–03 | 2.42 | 11.99 | −2.67E–03 |
| ZBD + RPD | 11.62 | 2.23 | 10.18 | −1.00E–02 | 2.36 | 10.18 | −1.17E–02 | 2.72 | 10.18 | −3.12E–03 | 2.76 | 10.18 | −7.10E–04 |
| primase | 8.06 | 2.19 | 6.45 | −1.48E–02 | 2.34 | 6.45 | −1.48E–02 | 2.76 | 2.68 | −1.66E–02 | 2.80 | −7.16 | −4.02E–03 |
| primase (§) | 38.40 | 2.23 | 1.31 | −2.12E–03 | 2.23 | 1.31 | −2.12E–03 | 2.23 | 1.31 | 4.81E–04 | 2.38 | 15.68 | 4.81E–04 |
EXAFS curve-fitting results for the T7 DNA primase, ZBD, and ZBD and RPD. The ratios of components and their concentrations were used as in Figure 2. Zn–S I, Zn–S II, Zn–S III, and Zn–S IV represent the different first-shell bond distance in the model. The best fit for ZBD and primase (§) was obtained using a two-atom model, and the best fit was obtained for the other samples using a four-atom model. The local structure around zinc was taken from the crystal structure of T7 primase fragment (4) and used as the model for the curve fitting. χ2 stands for the goodness of fit; R, the distance of sulfurs from Zn in angstroms; ΔE0, the correction to the energy origin; and σ, the Debye–Waller factor. The Zn–S distances are accurate to within 0.06 Å.
without DNA, with nucleotides. All of the other samples contain DNA with a primase-recognition sequence and nucleotides (see Experimental Procedures).
Figure 4.
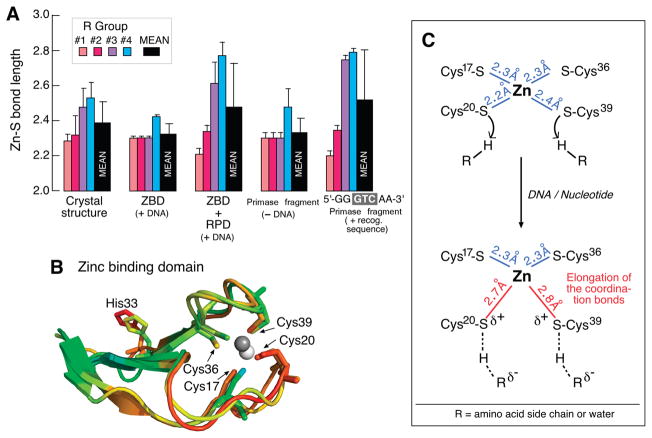
Distance distribution of cysteine ligands around the zinc in the ZBD, ZBD:RPD, and the primase fragment. (A) Columns representing the Zn–S coordination bond distances based on EXAFS-fitting results. The Zn–S distances are presented in the various states of ZBD: free in solution, mixed with RPD, and as a part of the primase, all in the presence of ATP and CTP. With the exception of the primase fragment without DNA, all assays are performed in the presence of DNA containing the primase-recognition sequence 5′GGGTCAA-3′. The averaged distance value and the corresponding rmsd were obtained from four best-fitting results for every sample. (B) The crystallographic B-factor supports the two labile coordinated cysteines observed by EXAFS. The artificial color scheme red to blue represents high to low B-factor. This figure was created using PyMOL (http://www.pymol.org). (C) Schematic representation of the ZBD in the absence (upper) and the presence (lower) of DNA. Of note is the elongation of the Zn–S distance caused by protonation of the cysteine sulfurs at Cys20 and Cys39 upon DNA binding. Bond distances were obtained from EXAFS results.
A Fourier transformation of the XAS spectra was performed to resolve the contribution of coordination shell atoms to the spectrum. The Fourier transform (right panel in Figure S1, Supporting Information) describes the radial distribution of the atoms around the zinc ion in three states of the ZBD:ZBD alone, ZBD and RPD, and ZBD in the primase fragment. The first and the most pronounced peak intensity describes the ligating cysteine sulfurs in the first coordination shell (Supporting Information, Figure S1). The ZBD in the primase without DNA behaves like the ZBD alone (Figure 4A and Table 1); it demonstrates shorter Zn–S distances (Figure 4A, Table 1). We have observed an increase in the Zn–S distances upon addition of RPD to the ZBD. Specifically, two of the four Zn–S distances increased. Thus the zinc-binding motif became asymmetrical (Figure 4A). Additional ligands beyond the first coordination shell that could have led to a higher coordination number were not considered in the EXAFS-fitting analysis. The conformational change in the ZBD upon DNA binding is supported by previous NMR studies (4, 33). The data show once again that when the productive priming complex is formed, lengthening of two of the Zn–S bonds occurs, which could be due to protonation of or hydrogen bond donation to the sulfur atoms.
Structural Properties of T7 Primase
Conformational changes of structures within the zinc-binding motif of the T7 primase almost certainly play a critical role in the recognition of specific DNA sequences. Analysis of the EXAFS results show that the rmsd of the averaged Zn–S bond length is smaller in the ZBD than in the intact primase or in the reconstituted primase consisting of the ZBD and the RPD (Figure 4A). This difference in the distance of ligating sulfurs corresponds to a symmetrical zinc-binding motif in the truncated ZBD and to a break of symmetry when the RPD is present along with DNA and nucleotides. Superposition of the two chains in the asymmetric unit of the crystal structure of the T7 primase fragment (PDB entry 1nui) reveals that the cysteines differ in their position between the two molecules of the asymmetric unit (Figure 4B). The rmsd for the alignment of the ZBD (residues 1–59) of the two chains is 1.65 Å, and the biggest difference in the spatial position of the two protein chains was found to be in the zinc-binding ligands. The crystallographic B-factor, representing the distribution of electron density in the raw data, supports the notion that two of the cysteine residues in the ZBD are more mobile than the other two (Figure 4B). In this context, the dissociation constant (pKa) of thiols provides an indication of their functional state. Normally, the thiolate form of free cysteine has a pKa of 8.3. We have used PROPKA (23) to predict the pKa values of the Zn-bound cysteine residues based on the crystal structure where the Zn is included in the calculations. The coordination to Zn normally lowers the pKa’s of the cysteine residues by about 4–7 units, so the high pKa predicted residues can fit into a model where these sulfurs are protonated when bound to the zinc as demonstrated previously (11). These protonated cysteines would have long Zn–S bonds and could potentially act as H-bond donors in recognizing the DNA sequence. The pKa predication of each individual cysteine residue comes along with bond length obtained by XAS. Hence we have assigned bond length to particular cysteines to produce a plausible model (Figure 4C) that shows the distortion of the Zn coordination sphere upon binding of the RPD and recognition sequences (assuming tetrahedral geometry is maintained). Importantly, if His33 in its protonated state is sufficiently close to the H-bond to one of these sulfurs or to participate in a water-mediated H-bond, thus raising the pKa of its conjugate acid (–SH) as predicted from the bond length, then it might explain how His33 contributes to the integrity of the Zn site and how its protonation state varies on interaction with DNA.
Cys36 and His33 Stabilize Zinc
In order to examine the importance of the first and higher shell residues around the zinc to the structure and function of the ZBD, we have used mutation analysis of the zinc-binding motif. Specifically, we examined the effect of substitution of serine and alanine for Cys36 and His33, respectively, in the ZBD on both structure and function. Both residues are important in primase function; primase in which Cys36 is replaced by serine (primase-C36S) or His33 is replaced by alanine (primase-H33A) cannot support the growth of phage T7Δ4 lacking gene 4 (3, 10). Cys36 directly coordinates the zinc and primase-C36S has been shown previously to have one-half the zinc content of wild-type primase (3). His33 has an exceptionally high crystallographic B-factor among other residues in the ZBD (4) and has been implicated previously in sequence recognition (10).
We analyzed the zinc content in the two genetically altered primases as well as in the ZBD using XAS. The zinc content of the primase fragments derived from primase-C36S and primase-H33A was very low compared with that found for the wild-type primase fragment (Supporting Information, Figure S2B). We also purified the ZBD of primase-C36S and primase-H33A and compared their zinc content with that of the wild-type ZBD using ICP-MS. Zinc was essentially undetectable in the ZBD of both mutants, whereas the Zn content of the wild-type ZBD was one gram of atom per mole of protein. This is the first demonstration that the ZBD alone has a full content of zinc (Supporting Information, Figure S2C). Similar results were found for the zinc content of the full-length primase domain containing these alterations (Supporting Information, Figure S2C).
Oligoribonucleotide Synthesis by Primase-C36S and Primase-H33A
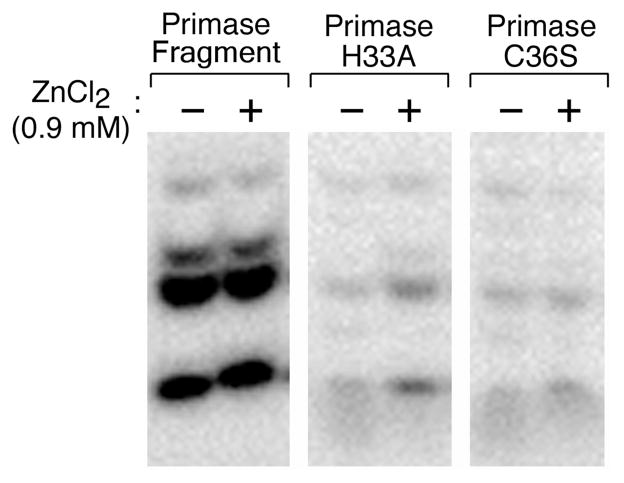
The wild-type primase catalyzes the synthesis of oligoribonucleotides on templates containing the primase-recognition site 5′-GGGTC-3′ whereas synthesis by primase-H33A and primase-C36S is diminished greatly (Supporting Information, Figure S2D). The small amount of synthesis observed most likely arises from the DNA-independent synthesis catalyzed by the RPD (3, 31). This interpretation is supported by the results obtained with templates lacking the cryptic cytosine necessary for sequence recognition. Primase-H33A and primase-C36S catalyze the same low level of oligonucleotide synthesis on these templates as they do on the template containing the recognition sequence (Supporting Information, Figure S3). The addition of zinc chloride to reactions containing primase-H33A slightly increase primase activity (Figure 5). However, the addition of zinc to primase-C36S had no detectable effect on primase activity, most likely due to the absence of the cysteine required to coordinate the zinc ion. Above the 0.9 mM zinc concentration used in these experiments, inhibition of oligoribonucleotide synthesis occurred, presumably due to competition of zinc with magnesium ions for the metal-binding site in the catalytic RPD.
Figure 5.
Effect of ZnCl2 on the template-directed synthesis of oligonucleotides by genetically altered and wild-type primases. Primase (0.5 μM), primase-H33A (40 μM), and primase-C36S (40 μM) were incubated in the standard reaction conditions as described in Figure 2 with additional ZnCl2 (0.9 mM). The radioactive oligoribonucleotides synthesized in the reaction were analyzed on a polyacrylamide gel and visualized using autoradiography.
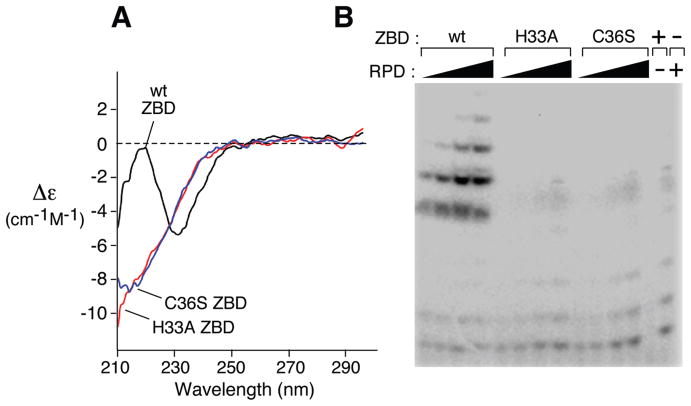
His33 and Cys36 Are Critical for Structural Integrity of the ZBD
The far UV CD spectra of wild-type ZBD and the ZBD of primase-C36S and primase-H33A are shown in Figure 6A. The loss in secondary structure in primase-C36S and primase-H33A is apparent in the structural changes observed relative to the wild-type ZBD. The change observed in the 220–240 nm region is typical for alterations in the protein helical content. However, the CD spectrum of the primase fragment (not shown) is more stable than is that of the ZBD alone. The spectra of the primase fragment, primase C36S, and primase H33A are similar suggesting that the RPD makes the major contribution to the spectrum. The stability of the primase secondary structure indicates that amino acid substitution in the ZBD affects only the local structure of the zinc-binding motif while leaving the global structure of the primase intact. In a reconstitution assay, the genetically modified ZBDs (ZBD-C36S and ZBD-H33A) did not function with the RPD to catalyze the synthesis of oligoribonucleotides as did the wild-type ZBD (Figure 6B).
Figure 6.
Effect of alteration in the ZBD of primase. (A) CD spectra of the ZBD of DNA primases. The effect of substitution of serine and alanine for Cys36 and His33, respectively, on the structure of the ZBD. Far-ultraviolet circular dichroism spectra of wild-type ZBD and the ZBD of primase-C36S and primase-H33A. Measurements were carried out at 25 °C. Spectra were background subtracted and normalized to the signal at 300 nm. (B) Reconstituted primase activity from a mixture of RPD and ZBD containing amino acid substitution. Reactions were carried out as described in Figure 2B.
DISCUSSION
In this study, we show that the zinc itself in the zinc-binding site of the DNA primase of bacteriophage T7 senses the other components required for template-directed oligoribonucleotide synthesis. We find that interactions of the ZBD with the catalytic RPD decrease the effective charge on the zinc. These results complement the arrangement of four cysteines around the Zn as reflected by the subsequent EXAFS analysis. One of the pronounced observations is the lower efficiency of oligoribonucleotide synthesis catalyzed by a primase reconstituted from the ZBD and RPD relative to the intact primase. This decreased efficiency most likely arises from the difficulties in precisely orienting the ZBD with the RPD in the absence of the polypeptide linking them. Hence, adding the RPD to the ZBD is not sufficient to increase the probability of productive matching. In chemical catalysis, two molecules must collide in the correct orientation with enough kinetic energy to pass the energetic barrier. Likewise, the flexible linker between the ZBD and RPD helps in positioning the two domains relative to each other for effective catalysis. In addition, the primase-recognition site (5′-GGGTCAA-3′) is sensed by the zinc whereas without DNA or with DNA lacking this sequence (5′-CACACAA-3′) is not. The inflection point value of ZBD + RPD (200 μM) (Figure 3, blue) nearly saturates the XANES signal. The deviation from linearity of the titration curve with concentration of RPD could be explained by the following: (1) There is sample heterogeneity of the RPD + ZBD mixture; free ZBD may be partially unfolded. However, in this case if the ZBD was fully folded, the inflection point value for free ZBD was slightly closer to the ZBD + RPD (200 μM). (2) The RPD was in excess (800 μM), and if the primase reconstitution effect was nonlinear, the RPD was almost fully active at a concentration lower than 800 μM. In other words, a lower concentration of RPD could be sufficient for the “saturating” effect on the ZBD. (3) At 800 μM, the RPD did not behave well and is partially unfolded/inactive. Therefore, the effective activity of it is closer to the effect of 200 μM rather than the effect of higher concentration. As in many other zinc finger proteins that bind DNA, the structural zinc is buried in a hydrophobic cleft rather than exposed on the surface of the protein. The free energy of exchange (ΔGex) between a metal bound to waters and to protein side chains is sensitive to the dielectric constants (34). Thus, a low dielectric constant as in the zinc-binding cleft in the T7 primase would be associated with high ΔGex. Such a buried cleft favors the inner sphere binding of cysteine to the zinc. In zinc sites that serve a catalytic role, there is a high dielectric constant where the outer-sphere binding dominates. The cysteines in the zinc-binding motif are partially exposed to the medium; the zinc is buried in the cavity (Supporting Information, Figure S4). Normally, binding of zinc (Lewis acid) causes the pKa of the cysteines to drop from 8 to 9 resulting in deprotonation. There is a large variation in the predicted pKa values of the ligating cysteines in the ZBD from the pKa value of free cysteine in solution (Table 2). This variation may indicate an interplay between protonation–deprotonation states of the sulfhydryl group coordinated to the zinc ion (explained elsewhere (35)). The crystal structure of the primase fragment shows that the Zn–S distance for the four first-shell cysteine ligands is distributed between values of 2.25 and 2.6 Å. Using theoretical calculations, Simenson and Calimet have shown that the single thiol in the Zn–Cys4 system could achieve a distance of 2.81 Å from zinc (11). Furthermore, protonating one (two) thiolate increases the Zn–S (thiol) distance by 0.4 Å (0.3 Å) which can be used as a structural marker for protonation. Thus, the higher pKa of Cys20 and Cys39 may reflect their protonated state. The knowledge on the identity of the protonated cysteines was used for assigning the Zn–S distances obtained by EXAFS to their corresponded cysteines (see Figure 4C).
Table 2.
Predicted pKa of Cysteines Ligated to Zinca
| pKaI | pKaII | |
|---|---|---|
| Cys17 | 1.9 | 1.5 |
| Cys20 | 5.6 | 9 |
| Cys36 | 2 | 5 |
| Cys39 | 6.5 | 7 |
The values were obtained by the PROPKA web server (http://pdb2pqr.sourceforge.net/) using the coordinates of the T7 primase fragment (PDB entry 1nui) as an input file. pKaI stands for chain 1 and pKaII stands for chain 2 in the asymmetric unit.
The detailed mechanism of metal-cysteine binding is poorly understood. The change in the protonation state of cysteines is a zinc-assisted process and occurs normally during protein folding. The protonation state of cysteines in zinc-binding systems remains controversial. There is a consensus that the first two cysteines bind to zinc as thiolate, but the protonation states of the third and fourth cysteines remain contentious (36). Dudev and Lim have elucidated the factors that govern the cysteine deprotonation in a Zn–Cys4 system using a combined ab initio and continuum dielectric approach (37). Their calculation predicts that the zinc ion could assist deprotonation of cysteines during the folding of the zinc-binding motif. They provide a possible explanation for the loss of zinc binding as in the case of primase-H33A. Although second or higher shell amino acid residues do not contribute significantly to the protonation of first-shell cysteines, they can contribute to the stability of the Zn–Cys4 binding site. Our XAS results support their theoretical approach regarding the ZBD and DNA binding, where cysteines can be protonated upon change in the solvent exposed surface of ZBD as a result of DNA binding. In addition, all cysteines in Cys4 structural binding sites may become deprotonated on a solvent exposed surface.
Amino acid substitutions of H33 and C36 in the ZBD lead to loss of zinc, remodeling of the ZBD, and poor recognition of primase-recognition sites. The template-directed primase activity is increased slightly by the addition of zinc to primase-H33A. There are two possible explanations as to how the zinc couples structure and function of the ZBD. Zinc is involved in direct sensing of DNA as supported by XAS results, or it maintains the ZBD structure that is required for recognition of the cryptic cytosine in the recognition sequence. In the latter case, XAS results reflect the side effect of the phenomenon. To achieve a higher level of zinc, binding zinc was supplied to the reaction mixture. Thus, with a higher concentration of zinc ions primase-H33A regains primase activity, and the CD spectrum changes slightly. We conclude, therefore, that notwithstanding the low affinity to the binding site, zinc can be used as an external cofactor. Primase with amino acid substitutions of H33 or C36 failed to produce RNA primers on a template containing the primase-recognition sequence (5′-GGGTC-3′). However, low primase activity was observed on DNA lacking the cryptic cytosine. This nonspecific activity together with the loss in sequence-dependent primase activity of the altered proteins may indicate that zinc has a primary role in the recognition of the cryptic cytosine. Hydrogen bonding between cysteine sulfurs and more distant side chains (NH–S) drastically influences the reactivity of the cysteine sulfurs. This reactivity may explain the decrease of binding affinity of zinc to the ZBD of primase-H33A, an alteration that affects primase activity.
Zinc binding changes the properties of thiol group in cysteine (9). One aspect is lowering the pKa value of the thiol, as was shown here by the predicted pKa values of the cysteine residues in the ZBD. It is possible that mutations in close proximity to zinc (e.g., H33A) affect the protonation state of the cysteine-sulfur ligands. As a result, the zinc is mobilized, and the ZBD becomes unstructured. Protonation of sulfur in the Zn–Cys4 system is still a source of debate. Our approach shows that the thiolate–zinc bond length can provide a marker for protonation. In comparison with the effect caused by the first coordination sphere residues, the second coordination sphere residues, the electrostatic environment of the enzyme, and the presence of solvent molecules all play a small role in determining Zn–ligand bond lengths (38).
Overall, the zinc in the ZBD functions as a redox indicator for changes in the Zn–S oxidation state that are coupled to (i) structural changes, (ii) binding properties, and (iii) enzymatic activity. We have shown that zinc is the crucial element for sequence recognition by T7 primase, and that the fragmental reduction of the zinc ion is accompanied by remodeling of the Zn–S bond. Mutations altering the conserved Cys-36 and His-33 mobilize zinc and lead to a change in the local structure resulting in a decrease in primase activity. This finding emphasizes the importance of direct binding of cysteines to zinc and also the importance of the second coordination sphere that plays a crucial role in the structural maintenance of zinc-binding sites and/or regulation of the oxidation balance of the Zn–S. The second coordination sphere, the enzyme backbone, the electrostatic environment of the enzyme, and the presence of solvent molecules, all play small roles in Zn-ligand bond lengths (38). Our findings shed light on the role of the zinc in such zinc-binding biosystems and provide a sensitive indicator for the binding state of the enzyme. Zinc in the Zn–Cys4 motif aligns the thiol groups and establishes their proximity. Therefore, high resolution structural studies of the metal-binding site are crucial for elucidating the manner in which zinc ions mediate DNA binding.
Supplementary Material
Acknowledgments
We thank Michael Sullivan (Beamline X-3B, National Synchrotron Light Source, Upton, NY) for the technical support to Robert O. Wright and Chitra J. Amarasiriwardena (Harvard School of Public Health, and Channing Laboratory, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA) for the ICP-MS measurements, and to Stephen C. Blacklow and Kelly Arnett (Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA) for the use of the circular dichroism facility. We are also grateful to Nicholas J. Dixon and Antoine van-Oijen for critical reading of the manuscript.
Footnotes
Abbreviations: RPD, RNA polymerase domain; ZBD, zinc-binding domain; ssDNA, single-stranded DNA; Zn/Zn–S, zinc/zinc–sulfur; Cys, cysteine; XAS, X-ray absorption spectroscopy; XANES, X-ray near edge spectroscopy; EXAFS, extended X-ray absorption spectroscopy; ICP-MS, inductively coupled plasma mass spectrometry.
This research was supported by the United States Public Health Services Grant GM 54397.
SUPPORTING INFORMATION AVAILABLE
XAS results (plots and table), primase activity, ICP-MS results of the modified primase, and figure of solvent accessibility to the cysteine ligands. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Frick DN, Richardson CC. DNA primases. Annu Rev Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein JA, Richardson CC. A 7-kDa region of the bacteriophage T7 gene 4 protein is required for primase but not for helicase activity. Proc Natl Acad Sci USA. 1988;85:396–400. doi: 10.1073/pnas.85.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mendelman LV, Beauchamp BB, Richardson CC. Requirement for a zinc motif for template recognition by the bacteriophage T7 primase. EMBO J. 1994;13:3909–3916. doi: 10.1002/j.1460-2075.1994.tb06702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato M, Ito T, Wagner G, Richardson CC, Ellenberger T. Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol Cell. 2003;11:1349–1360. doi: 10.1016/s1097-2765(03)00195-3. [DOI] [PubMed] [Google Scholar]
- 5.Lee SJ, Richardson CC. Interaction of adjacent primase domains within the hexameric gene 4 helicase-primase of bacteriophage T7. Proc Natl Acad Sci USA. 2002;99:12703–12708. doi: 10.1073/pnas.202471499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dudev T, Lim C. Principles governing Mg, Ca, and Zn binding and selectivity in proteins. Chem Rev. 2003;103:773–788. doi: 10.1021/cr020467n. [DOI] [PubMed] [Google Scholar]
- 7.Myers LC, Cushing TD, Wagner G, Verdine GL. Metal-coordination sphere in the methylated Ada protein-DNA co-complex. Chem Biol. 1994;1:91–97. doi: 10.1016/1074-5521(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 8.Berg JM, Shi Y. The galvanization of biology: a growing appreciation for the roles of zinc. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 9.Maret W. Zinc coordination environments in proteins as redox sensors and signal transducers. Antioxid Redox Signaling. 2006;8:1419–1441. doi: 10.1089/ars.2006.8.1419. [DOI] [PubMed] [Google Scholar]
- 10.Kusakabe T, Hine AV, Hyberts SG, Richardson CC. The Cys4 zinc finger of bacteriophage T7 primase in sequence-specific single-stranded DNA recognition. Proc Natl Acad Sci USA. 1999;96:4295–4300. doi: 10.1073/pnas.96.8.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simonson T, Calimet N. Cys(x)His(y)-Zn2+interactions: thiol vs. thiolate coordination. Proteins. 2002;49:37–48. doi: 10.1002/prot.10200. [DOI] [PubMed] [Google Scholar]
- 12.Penner-Hahn JE. Characterization of “spectroscopically quiet” metals in biology. Coord Chem Rev. 2005;249:161–177. [Google Scholar]
- 13.Kleifeld O, Frenkel A, Martin JM, Sagi I. Active site electronic structure and dynamics during metalloenzyme catalysis. Nat Struct Biol. 2003;10:98–103. doi: 10.1038/nsb889. [DOI] [PubMed] [Google Scholar]
- 14.Scott RA, Schwartz JR, Cramer SP. Effect of cyanide binding on the copper sites of cytochrome c oxidase: an X-ray absorption spectroscopic study. J Inorg Biochem. 1985;23:199–205. doi: 10.1016/0162-0134(85)85026-1. [DOI] [PubMed] [Google Scholar]
- 15.Powers L, Griep MA. Escherichia coli primase zinc is sensitive to substrate and cofactor binding. Biochemistry. 1999;38:7413–7420. doi: 10.1021/bi983059f. [DOI] [PubMed] [Google Scholar]
- 16.Frick DN, Baradaran K, Richardson CC. An N-terminal fragment of the gene 4 helicase/primase of bacteriophage T7 retains primase activity in the absence of helicase activity. Proc Natl Acad Sci USA. 1998;95:7957–7962. doi: 10.1073/pnas.95.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusakabe T, Richardson CC. Template recognition and ribonucleotide specificity of the DNA primase of bacteriophage T7. J Biol Chem. 1997;272:5943–5951. doi: 10.1074/jbc.272.9.5943. [DOI] [PubMed] [Google Scholar]
- 18.Mendelman LV, Richardson CC. Requirements for primer synthesis by bacteriophage T7 63-kDa gene 4 protein. Roles of template sequence and T7 56-kDa gene 4 protein. J Biol Chem. 1991;266:23240–23250. [PubMed] [Google Scholar]
- 19.Kleifeld O, Kotra LP, Gervasi DC, Brown S, Bernardo MM, Fridman R, Mobashery S, Sagi I. X-ray absorption studies of human matrix metalloproteinase-2 (MMP-2) bound to a highly selective mechanism-based inhibitor. comparison with the latent and active forms of the enzyme. J Biol Chem. 2001;276:17125–17131. doi: 10.1074/jbc.M011604200. [DOI] [PubMed] [Google Scholar]
- 20.Newville M. IFEFFIT: interactive XAFS analysis and FEFF fitting. J Synchrotron Radiat. 2001;8:322–324. doi: 10.1107/s0909049500016964. [DOI] [PubMed] [Google Scholar]
- 21.Ravel B, Newville M. ATHENA, ARTEMIS, HEPHAESTUS: data analysis for X-ray absorption spectroscopy using IFEFFIT. J Synchrotron Radiat. 2005;12:537–541. doi: 10.1107/S0909049505012719. [DOI] [PubMed] [Google Scholar]
- 22.Stern EA. Number of relevant independent points in x-ray-absorption fine-structure spectra. Phys Rev B Condens Matter Mater Phys. 1993;48:9825–9827. doi: 10.1103/physrevb.48.9825. [DOI] [PubMed] [Google Scholar]
- 23.Bas DC, Rogers DM, Jensen JH. Very fast prediction and rationalization of pK(a) values for protein-ligand complexes. Proteins. 2008;73:765–783. doi: 10.1002/prot.22102. [DOI] [PubMed] [Google Scholar]
- 24.Davies MN, Toseland CP, Moss DS, Flower DR. Benchmarking pK(a) prediction. BMC Biochem. 2006;7:18. doi: 10.1186/1471-2091-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexov EG, Gunner MR. Incorporating protein conformational flexibility into the calculation of pH-dependent protein properties. Biophys J. 1997;72:2075–2093. doi: 10.1016/S0006-3495(97)78851-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Georgescu RE, Alexov EG, Gunner MR. Combining conformational flexibility and continuum electrostatics for calculating pK(a)s in proteins. Biophys J. 2002;83:1731–1748. doi: 10.1016/S0006-3495(02)73940-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bashford D. Scientific Computing in Object-Oriented Parallel Environments. Vol. 1343. Springer; Berlin: 1997. An object-oriented programming suite for electrostatic effects in biological molecules; pp. 233–240. Lecture Notes in Computer Science. [Google Scholar]
- 28.He Y, Xu J, Pan XM. A statistical approach to the prediction of pK(a) values in proteins. Proteins. 2007;69:75–82. doi: 10.1002/prot.21478. [DOI] [PubMed] [Google Scholar]
- 29.Stanton CL, Houk KN. Benchmarking pK(a) prediction methods for residues in proteins. J Chem Theory Comput. 2008;4:951–966. doi: 10.1021/ct8000014. [DOI] [PubMed] [Google Scholar]
- 30.Qimron U, Lee SJ, Hamdan SM, Richardson CC. Primer initiation and extension by T7 DNA primase. EMBO J. 2006;25:2199–2208. doi: 10.1038/sj.emboj.7601112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kusakabe T, Richardson CC. The role of the zinc motif in sequence recognition by DNA primases. J Biol Chem. 1996;271:19563–19570. doi: 10.1074/jbc.271.32.19563. [DOI] [PubMed] [Google Scholar]
- 32.Pagel K, Vagt T, Kohajda T, Koksch B. From α helix to beta-sheet--a reversible metal ion induced peptide secondary structure switch. Org Biomol Chem. 2005;3:2500–2502. doi: 10.1039/b505979h. [DOI] [PubMed] [Google Scholar]
- 33.Kato M, Ito T, Wagner G, Ellenberger T. A molecular handoff between bacteriophage T7 DNA primase and T7 DNA polymerase initiates DNA synthesis. J Biol Chem. 2004;279:30554–30562. doi: 10.1074/jbc.M403485200. [DOI] [PubMed] [Google Scholar]
- 34.Dudev M, Wang J, Dudev T, Lim C. Factors governing the metal coordination number in metal complexes from Cambridge Structural Database analyses. J Phys Chem B. 2006;110:1889–1895. doi: 10.1021/jp054975n. [DOI] [PubMed] [Google Scholar]
- 35.Van Faassen E, Vanin AF. Radicals for Life: The Various Forms of Nitric Oxide. Elsevier; Amsterdam: 2007. [Google Scholar]
- 36.Maret W. Zinc and sulfur: a critical biological partnership. Biochemistry. 2004;43:3301–3309. doi: 10.1021/bi036340p. [DOI] [PubMed] [Google Scholar]
- 37.Dudev T, Lim C. Factors governing the protonation state of cysteines in proteins: an Ab initio/CDM study. J Am Chem Soc. 2002;124:6759–6766. doi: 10.1021/ja012620l. [DOI] [PubMed] [Google Scholar]
- 38.Tamames B, Sousa SF, Tamames J, Fernandes PA, Ramos MJ. Analysis of zinc-ligand bond lengths in metalloproteins: trends and patterns. Proteins. 2007;69:466–475. doi: 10.1002/prot.21536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.