SUMMARY
The retinoblastoma protein (pRB) tumor suppressor blocks cell proliferation by repressing the E2F transcription factors. This inhibition is relieved through mitogen-induced phosphorylation of pRB, triggering E2F release and activation of cell cycle genes. E2F1 can also activate pro-apoptotic genes in response to genotoxic or oncogenic stress. However, pRB’s role in this context has not been established. Here we show that DNA damage and E1A-induced oncogenic stress promotes formation of a pRB-E2F1 complex even in proliferating cells. Moreover, pRB is bound to pro-apoptotic promoters that are transcriptional active and pRB is required for maximal apoptotic response in vitro and in vivo. Together, these data reveal a direct role for pRB in the induction of apoptosis in response to genotoxic or oncogenic stress.
SIGNIFICANCE
pRB function is disrupted in many human tumors through either inactivation of the Rb gene or alterations in its upstream regulators. pRB’s tumor suppressive activity is at least partially dependent upon its ability to arrest cells through E2F inhibition. Our data now establish a second role for pRB as a stress-induced activator of apoptosis. Notably, pRB’s ability to promote either arrest versus apoptosis seems to be context dependent, with apoptosis being favored in proliferating cells. This finding has the potential to explain why cells are typically more resistant to apoptosis when in the arrested state. Most importantly, our observations suggest that Rb status will influence tumor response to chemotherapy by impairing both the arrest and apoptotic checkpoint responses.
INTRODUCTION
The retinoblastoma gene (RB1), a member of the pocket protein family with p130 and p107, was the first known tumor suppressor. The retinoblastoma protein (pRB) is targeted by the transforming proteins of the DNA tumor viral proteins (e.g. Adenoviral E1A), and it is functionally inactivated in a large proportion of human tumor cells, due to mutations of either the RB1 gene itself or its upstream regulators (Trimarchi and Lees, 2002). pRB’s tumor suppressive activity is thought to be largely dependent upon its ability to directly bind members of the E2F transcription factors family and prevent them from promoting transcription of genes required for cell proliferation (Trimarchi and Lees, 2002). This inhibition can occur via two distinct mechanisms: pRB binds to sequences within E2F’s transactivation domain and inhibits its function, and the resulting pRB/E2F complex recruits a number of transcriptional co-repressors, including HDACs, methyltrasferases and polycomb group proteins to actively repress the promoters of E2F target genes.
In normal cells, pRB’s repressive activity is controlled by its cell cycle dependent phosphorylation (Trimarchi and Lees, 2002). In response to mitogenic signaling, pRB is sequentially phosphorylated by the cdk complexes, cyclinD-cdk4/6 and cyclinE-cdk2. This phosphorylation is sufficient to induce pRB to release E2F, thereby allowing activation of E2F-responsive genes in late G1. However, phosphorylated pRB (ppRB) persists in the nucleus through the remainder of the cell cycle until it is dephosphorylated by protein phosphatase 1 at the end of mitosis (Ludlow et al., 1993). It is widely assumed that ppRB is functionally inactive and that dephosphorylation restores pRB to the active state. The majority of human tumors carry mutations that disable pRB-mediated repression of E2F (Sherr and McCormick, 2002). These mutations either inactivate the Rb gene itself or they promote pRB phosphorylation in the absence of normal mitogenic signals, through activation of the cyclinD-cdk4/6 kinases or inactivation the cdk inhibitor p16. These changes result in the inappropriate release of E2F, thereby inducing transcriptional activation of E2F target genes and consequently cell proliferation.
It is well established that E2F1, among other E2F family members, also contributes to the induction of apoptosis in response either to DNA damage or to oncogenic stress (Iaquinta and Lees, 2007). This is thought to be a critical event in suppressing the formation of tumors. Work from many labs has shown that genotoxic stress induces E2F1 recruitment to the promoters of pro-apoptotic genes including p73 and caspase 7, coincident with their transcriptional activation, even in cells that retain wildtype pRB (Pediconi et al., 2003). This led us to consider how pRB influences DNA damage-induced apoptosis. The prevailing view is that pRB is an anti-apoptotic regulator. Early support for this model came from the finding that several tissues in the Rb mutant mice display both ectopic proliferation and apoptosis (Jacks et al., 1992). However, it is now clear that much of this apoptosis is non-cell autonomous, resulting from a proliferation defect in the extra-embryonic tissues (de Bruin et al., 2003; Wenzel et al., 2007; Wu et al., 2003). The analysis of tissue-specific Rb mutant models reinforces the notion that pRB plays a much more nuanced role in apoptosis. Loss of pRB in neuronal tissue (MacPherson et al., 2003), lung (Mason-Richie et al., 2008; Wikenheiser-Brokamp, 2004), skin (Ruiz et al., 2004), and intestine (Haigis et al., 2006; Wang et al., 2007) drives ectopic proliferation but has no effect on apoptosis. In contrast, pRB inactivation in the lens (de Bruin et al., 2003) and myoblasts (Huh et al., 2004) does induce apoptosis but this is specifically observed in the differentiating cells. Thus, taken together, these mouse studies support two general conclusions. First, in many different settings, Rb-inactivation can induce inappropriate proliferation without triggering apoptosis. Second, when apoptosis is observed, it seems to result from an inability to cease proliferation and undergo terminal differentiation.
pRB’s apoptotic role has also been analyzed in established tissue culture cell lines. However, these studies yield conflicting results: some conclude that pRB suppresses apoptosis (Almasan et al., 1995; Bosco et al., 2004; Knudsen et al., 2000), whereas others suggest that it is pro-apoptotic (Araki et al., 2008; Bowen et al., 2002; Bowen et al., 1998; Knudsen et al., 1999). None of these studies address the molecular basis for the observed role of pRB. In this study, we have investigated how pRB influences the ability of E2F to induce apoptosis in response to genotoxic stress.
RESULTS
Stabilization of the pRB-E2F1 complex in response to DNA damage
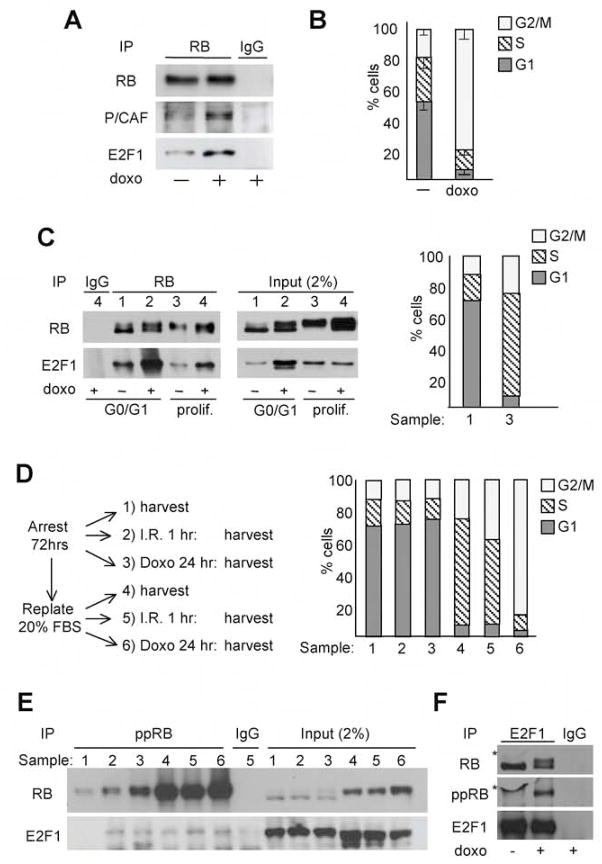
In cells committed to die by apoptosis in response to either DNA damage or oncogenic stress, E2F1 transcriptional activity is directed toward promoters of a subset of apoptotic genes that include caspase7, p73 and Apaf1 (Iaquinta and Lees, 2007). Thus, we hypothesized that DNA damage must somehow inactivate pRB’s repressive function to allow the release of transcriptionally active E2F1. To test this hypothesis, we assessed the binding of pRB to E2F1 either before or after doxorubicin treatment of human T98G cells. However, contrary to our expectations, we found that the pRB-E2F1 complex was stabilized by this genotoxic stress (Fig. 1A). Notably, T98G cells are p53-defective and consequently treatment with doxorubicin causes them to accumulate in G2/M (Fig. 1B). Thus, we can conclude that the observed DNA damage induced formation of the pRB-E2F1 complex is neither dependent on p53 nor simply an indirect consequence of a G1 arrest.
Figure 1. DNA damage promotes formation of a pRB-E2F1 complex in proliferating cells.
(A, B) Asynchronous T98G cells untreated (−) or treated (+) with 2μM doxorubicin (doxo) for 24 hours were (A) screened by immunoprecipitation (IP) using an antibody against pRB followed by western blotting (WB) to assess levels of pRB and associated E2F1 and P/CAF or (B) assayed for cell cycle distribution by FACS analysis. Bars represent the mean of three independent experiments (± SD). (C) T98G cells were highly enriched for G0/G1 or S/G2/M (prolif.) cells, as determined by FACS (right panel), by culturing in 0.1% FBS for 72 hours and then maintaining in 0.1% or replating in 20% FBS for 16hrs. The cells were collected (samples 1 and 3) or treated with 2μM doxo for additional 48 hours (samples 2 and 4) and then assayed in parallel for pRB-E2F1 complexes by IP-WB (left panel). (D, E) Enriched populations of G0/G1 or proliferating T98G cells were untreated (samples 1 and 4) or treated with I.R. (10 Gy) for 1 hour (samples 2 and 5) or 2μM doxo for 24 hours (samples 3 and 6) as indicated (D, left panel). These samples were analyzed for: (D, right panel) cell cycle phasing by FACS; (E, left panel) the presence of ppRB-E2F1 complexes by IP with either IgG control or ppRB antibodies and then WB for pRB or associated E2F1; or (E, right panel) the levels of total pRB and E2F1 by WB of the input lysates. (F) Asynchronous T98G cells, untreated (−) or treated (+) with 2μM doxo for 24 hours, were screened for the presence of ppRB-E2F1 complexes by IP with either IgG control or E2F1 antibodies and then WB, first for pRB and subsequently (after stripping the blot) for ppRB. A bubble in the blot yielded the non-specific signal (*).
pRB’s ability to bind to E2F is normally limited to the early stages of the cell cycle when pRB exists in the hypo-phosphorylated form. Therefore, it was surprising to observe pRB-E2F1 complexes in a population that was highly enriched for G2/M phase cells. To more directly address the influence of cell cycle phasing, we used serum deprivation and re-addition to generate two populations of T98G cells that were greatly enriched for either G0/G1 (70%) or proliferating (95% S or G2/M phase) cells (Fig. 1C) and then treated these with doxorubicin. Consistent with the known cell cycle-dependent phosphorylation of pRB, the pRB protein was present in its slower mobility form in untreated proliferating cells and it bound little E2F1 (Fig. 1C). Notably, doxorubicin treatment was still able to induce formation of the pRB-E2F1 complex in this proliferating population (Fig. 1C). This occurred independent of any change in total levels of E2F1 protein (Fig. 1C) and it correlated with full activation of the apoptotic program, as judged by Parp-p85 induction (data not shown). Importantly, the G0/G1 cells within the proliferating population cannot fully account for this DNA damage induced pRB-E2F1 complex because the treated proliferating cells and untreated G0/G1 cells had comparable levels of pRB-associated E2F1 (Fig. 1C) but the fraction of G0/G1 cells in the enriched populations differed by 14 fold (5% versus 70%). Thus, these data show that pRB-E2F1 complexes can form in S/G2/M phase cells in response to DNA damage.
pRB is sequentially phosphorylated by the cyclinD-cdk4/6 and cyclinE-cdk2 complexes and this is thought to disrupt the interaction between pRB and E2F. Since DNA damage causes pRB to bind to E2F1 irrespective of cell cycle phase, we wished to determine whether ppRB could participate in this complex. To this end, we generated enriched populations of G0/G1 and proliferating T98G cells, exposed them to either ionizing radiation (IR) or doxorubicin (see schema Fig. 1D, left panel) and then assessed both the levels and E2F1-binding properties of ppRB, by using antibodies that specifically recognize known cdk phosphorylation sites within pRB, pSer780, pSer795 and pSer807-811. FACS analysis confirmed the high degree of enrichment of the G0/G1 and proliferating populations both before, and after, IR or doxorubicin treatment (Fig. 1D, right panel). As expected, ppRB was present at much higher levels in the proliferating versus the G0/G1 cells, as judged by both the immunoprecipitation of ppRB and the mobility shift of the total pRB (Fig. 1E). We found that a small subset of the E2F1 co-immunoprecipitated with ppRB before treatment, and IR and doxorubicin both increased this level (Fig. 1E, left panel). This increased binding occurred independently of any change in the total levels of E2F1 (Fig. 1E, right panel). Interestingly, IR and doxorubicin actually increased the levels of ppRB in the G0/G1 population (Fig. 1E, compare lane 1 with lanes 2 and 3), even though neither treatment altered the cell cycle distribution of these cells (Fig. 1D, right panel). The increased level of ppRB in the treated G0/G1 cells clearly contributes to, but seems insufficient to fully account for, the increased levels of E2F1 in the ppRB immunoprecipitates. Indeed, for all of the cell cycle fractions, we clearly recover a smaller fraction of total E2F1 in ppRB-immunoprecipitates (Fig. 1E) versus total pRB-immunoprecipitates (Fig. 1C). It was unclear whether the phospho-specific pRB antibody cocktail disrupts the ppRB/E2F1 complex, leading to an underestimation of its levels, or whether hypo-phosphorylated or other phosphorylated pRB species are the main constituent of this complex. To distinguish between these possibilities, we conducted a reciprocal IP. Specifically, we immunoprecipitated E2F1 from T98G cells before or after doxorubicin treatment, and then screened for associated pRB by western blotting with an antibody that recognizes all forms of pRB (Fig. 1F). In the untreated cells, the E2F1 immunoprecipitate contained a single pRB species. In contrast, doxorubicin caused E2F1 to bind two distinct pRB bands that were present at approximately equal levels. One of these co-migrated with the single pRB species seen in the untreated cells, while the other had a slower mobility characteristic of ppRB. To verify this, we stripped, and re-probed, the blot with the anti-ppRB antibody cocktail. This recognized only the slower migrating pRB species specific to the doxorubicin-treated cells, confirming that this is ppRB. Based on the relative levels of the two bands in the anti-pRB blot, we conclude that ppRB accounts from at least half of the E2F1-associated pRB activity in the doxorubicin treated T98G cells. Taken together, these experiments clearly show that DNA damage induces formation of pRB-E2F1 complexes in both arrested and proliferating cells and that ppRB is able to participate in this complex.
pRB participates in transcriptional activation of proapoptotic genes in response to stress
Histone acetyltransferases (HATs) and deacetylases (HDACs) are known to play key roles in mediating the transcriptional properties of pRB and the E2F proteins (Frolov and Dyson, 2004). The pRB-E2F complex is thought to act as a repressor of classic E2F target genes through recruitment of HDACs, while HATs acetylate E2F1 and promote its transcriptional activity. Importantly, DNA damage triggers the HAT P/CAF to bind and acetylate E2F1 (Ianari et al., 2004), and this modification is required for E2F1 association with pro-apoptotic promoters (Pediconi et al., 2003). Given these observations, we screened for the presence of P/CAF in pRB immunoprecipitates in untreated versus doxorubicin-treated cells (Fig. 1A). We found that DNA damage promotes P/CAF-pRB complex formation (Fig. 1A). This raised the possibility that pRB might participate in a transcriptionally active complex under proapoptotic conditions.
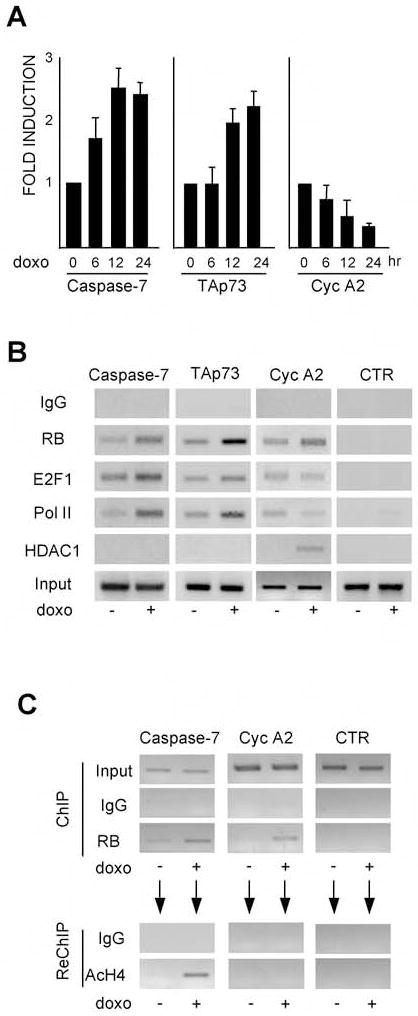
To understand the transcriptional relevance of the DNA damage-induced pRB-E2F1 complex, we examined the regulation of representative cell cycle control and proapoptotic E2F1-responsive genes in DNA damaged T98G cells. This analysis revealed that doxorubicin caused a differential response of these two target gene classes: activation of the proapoptotic genes Caspase 7 and p73 and repression of the cell cycle-regulator CyclinA2 (Fig. 2A). To further understand this differential response, we conducted chromatin immunoprecipitation (ChIP) assays. Notably, doxorubicin treatment induced pRB recruitment to both the cell cycle and proapoptotic promoters (Fig. 2B). A quantitative analysis of these results (Supplementary Fig. 1) showed that the increase of pRB levels was slightly higher at Caspase 7 (2 fold) and p73 (2.2 fold), than at CyclinA2 (1.5 fold) gene promoters. For all of the other proteins that we assayed, doxorubicin treatment caused differential changes at cell cycle versus proapoptotic promoters (Fig. 2B). At the CyclinA2 gene promoter, we found a reduction in the binding of both E2F1 and RNA polymerase II. In addition, the transcriptional co-repressor HDAC1 was specifically recruited to the CyclinA2 promoter in treated, but not untreated, cells (Fig. 2B). These changes are entirely consistent with the observed down-regulation of cyclinA2 mRNA and the prevailing view that pRB mediates the transcriptional repression of cell cycle promoters. At the same time, doxorubicin treatment induced the recruitment of both E2F1 (1.4 and 2.7 fold) and RNA polymerase II (2.3 and 2 fold) to the Caspase 7 and p73 promoters. Importantly, we did not detect any recruitment of HDAC1 to the pro-apoptotic promoters either in the damaged cells or undamaged cells (Fig. 2B).
Figure 2. DNA damage-induces pRB-dependent activation of proapoptotic gene promoters.
(A) Untreated and doxo treated asynchronously growing T98G cells were assessed for the levels of Caspase 7, p73 and CyclinA2 mRNAs by real time RT-PCR analysis. Results are normalized with GAPDH and shown relative to the levels seen in untreated cells (set to 1). Bars represent the mean of three independent experiments (± SD). (B) T98G cells were untreated or treated with 2μM doxo for 16 hours and the binding of pRB, E2F1, RNA Pol II and HDAC1 to the Caspase7, p73 and Cyclin A2 gene promoters determined by ChIP. (C) AcH4 re-ChIP analysis of the pRB ChIP shows that pRB is bound to the transcriptionally active Caspase7 gene promoter following doxo treatment.
The coordinated enrichment of pRB, E2F1 and RNA polymerase II at the Caspase 7 and p73 promoters fits with the hypothesis that pRB contributes to activation of pro-apoptotic genes. However, we could not rule out that there were two distinct populations of cells in which these promoters are either bound by pRB and repressed, or associated with RNA polymerase II and activated. To address this possibility, we performed ChIP-reChIP experiments in which immunoprecipitated pRB-chromatin complexes were eluted and then subjected to a second round of immunoprecipitation with either control IgG or an antibody against acetylH4, a marker of transcriptional activation (Fig. 2C). As with our previous experiment, the primary ChIP showed that doxorubicin promoted recruitment of pRB to both the caspase 7 and cyclinA2 promoters (Fig. 2C). However, when we analyzed the eluate from the pRB-immunoprecipitates, we found that acetylH4 was specifically detected at the Caspase 7, but not the CyclinA2, gene promoter (Fig. 2C). This analysis showed unequivocally that pRB was bound to the transcriptionally active Caspase 7 gene promoter, presumably via its participation in the pRB-E2F1-P/CAF complex that is promoted by DNA damage. At the same time, pRB binds to cell cycle promoters and recruits HDAC1 to mediate their repression.
pRB is required for maximal induction of the apoptotic response in vitro and in vivo
These data show that pRB is associated with proapoptotic promoters that are transcriptionally active in DNA damaged cells. However, they do not establish whether pRB contributes to their transcriptional activation, or to the apoptotic response. To address these questions, we took advantage of the lentivirus pPRIME-GFP-shRB and its control pPRIME-GFP, producing respectively either a short hairpin against human pRB (shRB), or a hairpin targeting the luciferase gene, in the context of Mir30 (Stegmeier et al., 2005; elledgelab.bwh.harvard.edu/protocols/pPRIME/pPRIME_vectors.doc). We used these viruses to infect T98G cells and selected parallel populations of GFP-positive cells. The shRb reduced pRB levels to less than 50% of that seen in the control cells (Fig. 3A). Importantly, this partial knockdown had no effect on cell cycle phasing (Fig. 3A). This allowed us to assess pRB’s contribution to apoptosis independent of its role in cell cycle. We found that this partial pRB knockdown significantly reduced the fraction of cells undergoing apoptosis in response to either doxorubicin or to another Topoisomerase II inhibitor, etoposide (respectively 50% and 30% reduction; Fig. 3C). This correlated with a reduction in the levels of p73 (>50%) and caspase 7 (approximately 20%) mRNA in the shRb-expressing cells (Fig. 3D). Thus, we conclude that pRB-loss can impair the apoptotic response in the absence of any cell cycle defects.
Figure 3. pRB loss impairs the apoptotic response to DNA damage.
(A–D) T98G cells were infected with pPRIME-GFP (GFP) or pPRIME-GFP-shRB (GFPshRB) lentivirus and sorted for >20% GFP positive cells. GFP and GFPshRB sorted cells were screened for (A) the level of pRB by WB using actin as a loading control, (B) cell cycle phasing by FACS analysis or (C) the percentage of early apoptotic cells by FACS (AnnexinV+, AAD−) after culturing for 48 hours in either the absence (−) or presence of 2μM doxo or 25μM etoposide (et). Bars represent the mean of three independent experiments (± SD). (D) Caspase 7 and p73 mRNAs levels measured by real time RT-PCR analysis. Results are normalized with GAPDH and expressed relative to levels seen in GFP infected cells (set as 1). Bars represent the mean of three independent experiments (± SD). (E) Wild-type (WT) or Rb2lox/2lox (cRb) MSCs were infected with GFP or GFP-Cre expressing adenoviruses. The percentage of GFP+ apoptotic cells was measured by FACS analysis (AnnexinV+, 7AAD−). The mean of three independent experiments (± SD) is shown.
All of the previous experiments have been conducted in the p53-deficient tumor cell line T98G. To determine whether our findings are more broadly relevant, we repeated this analysis in a second tumor cell line, U2OS (Supplementary Fig. 2). These cells express wildtype p53 and pRB is constitutively hyperphosphorylated due to hypermethylation and silencing of the p16INK4a gene promoter (Park et al., 2002). In accordance with our prior results, we found that numerous genotoxic agents (including doxorubicin, etoposide and campthotecin) triggered pRB to bind to E2F1, and promoted both activation of pro-apoptotic and repression of cell cycle-related E2F target genes (Supplementary Fig. 2A–C). Moreover, pRB knockdown using either the pPRIME-GFP-shRB lentivirus, or a tTA-inducible RB hairpin, impaired the apoptotic response and the transcriptional activation of the pro-apoptotic gene p73 (Supplementary Fig. 2D–G). Thus, pRB can play a positive role in DNA damage-induced apoptosis in both the absence, and presence, of p53.
It has previously been reported that Rb-inactivation renders mouse embryonic fibroblasts (MEFs) more, not less, sensitive to DNA damage-induced apoptosis (Almasan et al., 1995; Knudsen et al., 2000). We have repeated these experiments in both germline and conditional Rb mutant MEFs and obtained similar results (AI and JAL, unpublished observations). Thus, in different settings, pRB can either promote or inhibit apoptosis. It seemed possible that the differential consequences of pRB-loss reflect fundamental differences between tumor versus normal, or mouse versus human, cells. To address these possibilities, we examined a second source of primary murine cells, mesenchymal stem cells (MSCs). We generated these MSCs from mice carrying either wildtype (WT) or conditional Rb (cRb) alleles, infected them with adenoviruses expressing either the Cre recombinase gene (+Cre) or a GFP control (−Cre) and confirmed recombination of the cRb alleles by PCR (data not shown). The four cell populations were then treated with ionizing radiation (Fig. 3E) or doxorubicin (data not shown), and the fraction of apoptotic cells quantified by FACS analysis. Cre expression had no significant effect on the level of apoptosis in the wildtype cells, but it reduced apoptosis in the cRb cells by more than 30% (Fig. 3E). Thus, pRB loss also impairs the apoptotic response of these primary murine cells.
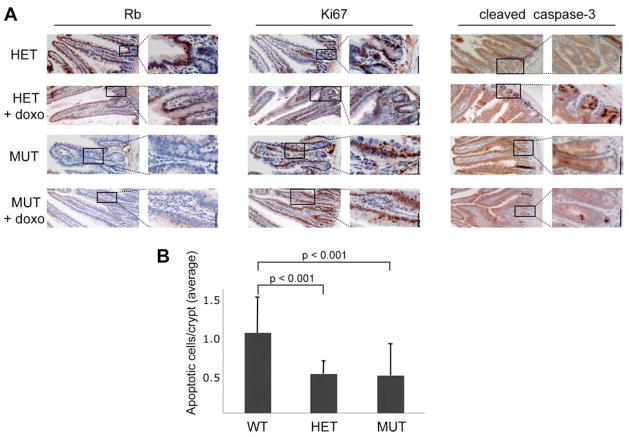
To further extend this analysis, we next examined pRB’s role in the DNA damage response in vivo. For this, we used mice carrying conditional Rb alleles and a VillinCre transgene, which is expressed in the adult intestinal epithelium. To eliminate any potential contribution of Cre-mediated deletion to the DNA damage response, we selected Rb+/2lox;Villin-Cre+ mice (effectively Rb heterozygous) as controls for our analysis of Rb2lox/2lox;Villin-Cre+ (Rb mutant) mice. Rb2lox/2lox;Villin-Cre+ mice are known to have histologically normal intestinal crypts but these contain proliferating cells at ectopic locations (Kucherlapati et al., 2006). Immunohistochemical staining confirmed that pRB was expressed in the intestinal epithelium of the Rb+/2lox;Villin-Cre+ controls but not in the Rb2lox/2lox;Villin-Cre+ animals (Fig. 4A, left panel). We then assessed the level of proliferating cells by screening for the cell cycle marker Ki67 (Fig. 4A, middle panel). Consistent with prior studies (Haigis et al., 2006; Kucherlapati et al., 2006), the proliferating cells are restricted to the intestinal crypts of the Rb+/2lox;Villin-Cre+ controls, but they exist in both the crypts and throughout the height of the villi of Rb-deficient intestinal epithelium. We then subjected these Rb2lox/2lox;Villin-Cre+ mice and their Rb+/2lox;Villin-Cre+ littermate controls to intraperitoneal injections of doxorubicin and examined the levels of apoptosis of the proximal small intestines by staining for cleaved caspase 3 (Fig. 4A, right panel). As expected, apoptosis was essentially absent in the untreated intestinal epithelia of both genotypes (Fig. 4A, right panel). In response to treatment, we observed high levels of cleaved caspase 3 in the Rb+/2lox;Villin-Cre+ control tissues. Interestingly, these apoptotic cells were localized exclusively within the intestinal crypts, clearly correlating with the proliferative region of the intestinal epithelium. This is consistent with the prevailing view that the cycling cells are more predisposed to undergo apoptosis than their arrested counterparts. Notably, quantification of 60 villi from each genotype showed that the fraction of proliferating cells undergoing apoptosis was reduced in the Rb mutant (1.9±0.88%) versus the control (3.8±0.72%) tissue. This effect was most striking in the height of the villi, where we observed few, if any, apoptotic cells in the Rb2lox/2lox;Villin-Cre+ mice even though this zone was highly proliferative (Fig. 4A). However, we did not observe a significant difference in the level of apoptotic cells in the intestinal crypts of Rb2lox/2lox;Villin-Cre+ mice versus Rb+/2lox;Villin-Cre+ controls (data not shown). Since our cell studies had shown that a partial knockdown of pRB was sufficient to impair the apoptotic response, we wondered whether there might be a heterozygous mutant phenotype in the Rb+/2lox;Villin-Cre+ intestinal crypts. To address this possibility, we subjected a second cohort of Rb+/+;Villin-Cre+ (n=5), Rb+/2lox;Villin-Cre+ (n=5) and Rb2lox/2lox;Villin-Cre+ (n=6) mice to intraperitoneal injections of doxorubicin, stained for cleaved caspase 3 and quantified the level of apoptosis in the intestinal crypts. Consistent with our prior analysis, doxorubicin treatment induced a similar apoptotic response in the intestinal crypts of Rb+/2lox;Villin-Cre+ (HET) and Rb2lox/2lox;Villin-Cre+ (MUT) mice (Fig 4B). However, these two genotypes had a twofold lower level of apoptosis than the Rb+/+;Villin-Cre+ (WT) controls (Fig. 4B). In both cases, this difference was statistically significant (p<0.001). Thus, in this tissue, pRB-loss is promoting inappropriate proliferation while reducing the ability of these cells to undergo DNA damage induced apoptosis. Moreover, mutation of a single Rb allele is sufficient to impair the apoptotic response, without altering proliferation. These observations show that Rb influences the apoptotic response to DNA damage in vivo. They also raise the possibility that this impaired response could occur in patients carrying germline Rb mutations.
Figure 4. Conditional pRB knockout mice are less sensitive to genotoxic stress.
(A) Analysis of the proximal small intestines of Rb+/2lox;VillinCre+ (HET) and Rb2lox/2lox;VillinCre+ (MUT) mice treated with vehicle control or doxo. Left panel: Immunostaining confirmed Cre-mediated loss of pRb in the proximal small intestine of MUT animals. Middle panel: Analysis of Ki67, a marker of proliferating cells, shows that pRb-loss caused proliferation throughout the villi. Ki67 levels were similar in the absence and presence of doxorubicin treatment. Right panel: Staining for cleaved caspase 3 showed high levels of apoptotic cells specifically in the base of the crypts in doxo treated, by not untreated, WT and MUT mice. Scale bar, 50μm for all panels. (B) The average number of apoptotic cells/intestinal crypt (± SE) in the proximal small intestines of doxo treated Rb+/+;VillinCre+ (WT; n=5), Rb+/2lox;VillinCre+ (HET; n=5), and Rb2lox/2lox;VillinCre+ (MUT; n=6) mice was determined by counting cleaved caspase 3-positive cells in 21 crypts for each animal.
E1A promotes the formation of transcriptionally active pRB-E2F1 complexes
Given our findings, we decided to extend our analysis to look at the mechanism of action of adenoviral E1A. E1A is a potent oncogene that induces uncontrolled proliferation and also sensitizes cells to apoptosis. It binds to pRB with high affinity and this requires the LXCXE motif that is essential for E1A’s transforming activity (Helt and Galloway, 2003). The prevailing view is that E1A acts to sequester pRB allowing release of transcriptionally active E2Fs. This model can explain the increased proliferation rate seen for E1A-infected cells. However, prior studies have shown that the interaction between E1A and pRB is insufficient to promote apoptosis in response to doxorubicin (Samuelson et al., 2005). Indeed, their mutant analysis suggests that this requires E1A binding to both pRB and p400, a component of the TRAAP/Tip60 HAT chromatin remodeling complex. Given our observations, we hypothesized that E1A’s proapoptotic function might reflect its ability to promote formation of transcriptionally active pRB-E2F1 complexes, in an analogous manner to genotoxic stress. To test this notion, we investigated the interplay between E1A and the pRB-E2F1 complex using primary human diploid fibroblasts IMR90 that express the murine ecotropic receptor. We selected these cells because they will apoptose in response to genotoxic agents or E1A, but they are more resistant to apoptosis than many other cell lines, and thus are better able to tolerate E1A expression. Importantly, in a similar manner to T98G and U2OS cells, doxorubicin promotes formation of pRB-E2F1 complexes in IMR90 cells in the absence of E1A (Fig. 5A). To study the role of E1A, IMR90 cells were retrovirally transduced with either the pBabe-puro vector or pBabe-puro expressing the 12S form of E1A. We found that E1A and E2F1 both coimmunoprecipitated with pRB (Fig. 5B). Doxorubicin treatment caused a modest additional increase (1.6 fold) in the level of E2F1 co-immunoprecipitating with pRB in the E1A expressing IMR90 cells (Fig. 5B). These observations show that E1A potently induces formation of pRB-E2F1-E1A complexes and exogenous DNA damage reinforces this response.
Figure 5. pRB facilitates E1A’s ability to promote apoptosis.
(A) In the absence of E1A, a pRB-E2F1 complex was induced by treatment of IMR90 cells with doxo as judged by IP for pRB and WB for pRB and E2F1. (B–I) IMR90 cells were retrovirally transduced with either pBabe-hygro vector (−E1A) or pBabe-hygro-E1A12S (+E1A), selected with hygromycin 75μg/ml for four days and then assayed as follows. (B) The cells were treated with 2μM doxo (16 hours) and WB was used to determine the levels of pRB, E2F1, E2F3A, E2F4 and E1A in (left panel) pRB IPs and (right panel) the total lysates. The far right panel shows a longer exposure of the key input lanes. The E1A expression strongly increased both the total levels of pRB, E2F1 and E2F3A and the level of pRB-E2F1/E2F3A complexes. At the exposure shown, the pRB-E2F1 interaction is not visible in the −EIA;+doxo cells. (C) The binding of E1A to pRB and E2F1 is also detected via IP of E1A. (D) IPs with antibodies against ppRB and WB for pRB and E2F1. (E) The levels of pRB, E1A, E2F1 and AcH4 associated with the p73 and the CyclinA2 gene promoters were determined by ChIP. Densitometric quantification of each signal, relative to the input, is shown. (F) Real time RT-PCR analysis of Caspase 7, p73 and CyclinA2 mRNA levels. Results are expressed as arbitrary units normalized with GAPDH and they show the mean of three independent experiments (± SD). (G–I) Control or E1A-transduced cells were infected with either GFP or GFPshRB lentiviruses. (G) WB confirmed a reduction in pRB levels after shRB infection, using actin as the loading control. (H) Caspase 7, p73 and CyclinA2 mRNA levels were determined by real time RT-PCR analysis in cells cultured in the absence (−) or presence of 1μM etoposide (et) for 12 hours. Results are normalized with GAPDH and expressed relative to levels seen in the untreated cells (set as 1). Bars represent the mean of three independent experiments (± SD). (I) FACS analysis of early apoptotic cells (AnnexinV+, 7AAD−) after culture in the absence (−) or presence of 1μM et for 16 hours. Values are the percentage of apoptotic cells over the GFP+ cells (set at 1) and they represent the mean of three independent experiments (±SD).
In the presence of E1A, pRB also associated with E2F3A, another activating E2F that is known to participate in the oncogenic stress response, but not with the repressive E2F, E2F4 (Fig. 5B). Importantly, the reciprocal immunoprecipitation using antibodies against E1A also recovered both pRB and E2F1 (Fig. 5C). Since it is well established that a pocket protein, such as pRB, is required to bridge the interaction between E1A and E2F (Fattaey et al., 1993), we can infer that these three proteins must be part of the same complex. Similar to our analysis of the DNA damage response, E1A also enhanced the formation of ppRB species and their binding to E2F1 (Fig. 5D). Additionally, E1A yielded a dramatic increase in the intracellular levels of pRB, E2F1 and E2F3A, but not E2F4 (Fig. 5B). DNA binding is known to protect E2F-pocket protein complexes from degradation (Hofmann et al., 1996). Given the observed stabilization of both pRB and the activating E2Fs in E1A-expressing cells, we hypothesized that E1A induces the formation of pRB-E2F1-E1A complexes that bind to DNA and activate transcription. Thus, we used ChIP assays to assess binding to the p73 and cyclin A2 promoters (Fig. 5E). In the absence of E1A, we observed a significant pRB ChIP signal at both promoters, but little or no binding of either E2F1 or acetylH4 (Fig. 5E). Since the uninfected IMR90 cells are predominantly in G0/G1 cells, we speculate that pRB contributes to the repression of both p73 and cyclin A in this setting. Accordingly, these genes appeared to be transcriptionally silent, as judged by the lack of p73 and cyclin A2 mRNA (Fig. 5F). In contrast, in the E1A infected cells, pRB, E2F1, E1A and the acetylated form of H4 all associated with both the cyclin A and p73 promoters, coincident with the dramatic induction of both of these mRNAs (Fig. 5E, F). Given the existence of a pRB-E2F1-E1A complex and the positive ChIP signals for E1A, pRB and E2F1 we speculate that E1A’s ability to activate apoptosis and cell cycle genes reflects, at least in part, its direct action at their promoters.
As noted above, the current view of E1A action is that is acts to sequester pRB and thereby release transcriptionally active E2F. If this model fully explains the relationship between E1A and pRB, E1A’s ability to induce apoptosis should not be impaired by pRB-loss. To test this, we performed a pRB knockdown in E1A infected IMR90 cells. The shRb yielded significant, but not complete, pRB knockdown in both uninfected and in E1A-infected IMR90 cells (Fig. 5G). Strikingly, the shRb caused a twofold reduction in the levels of caspase 7 and p73 mRNAs in either the absence, or presence, of DNA damage (Fig. 5H). Consistent with previous studies (Samuelson et al., 2005), expression of E1A alone induced only low level of apoptosis in these cells and this was unaffected by pRB levels (Fig. 5I). However, when combined with genotoxic stress, E1A induced programmed cell death at two fold higher levels in control versus shRb-expressing cells (Fig. 5I). Thus, pRB plays a positive role in E1A-induced apoptosis. Taken together our findings suggest that E1A associates with both pRB and E2F1, stabilizing these proteins, and the resulting complex associates with both proapoptotic promoters to promote their transcription.
DISCUSSION
The E2F transcription factors play a key role in promoting cellular proliferation under the control of the pRB tumor suppressor. It is well established that E2F1, among other E2F family members, also contributes to the induction of apoptosis in response either to DNA damage or to oncogenic stress (Iaquinta and Lees, 2007). However, the role of pRB in this process is poorly understood. We anticipated that DNA damage would have to release E2F1 from pRB to allow it to activate pro-apoptotic genes. Instead, our data suggest an unexpected mechanism of pRB action - that DNA damage induces pRB to participate in a transcriptionally active complex that drives expression of pro-apoptotic genes. This model is supported by three central observations: pRB is induced to bind both E2F1 and the histone acetylase P/CAF in DNA damaged cells; ChIP-reChIP assays show unequivocally that pRB is bound to the promoters of pro-apoptotic genes that are transcriptionally active; and knockdown and genetic ablation experiments confirm that pRB-loss typically reduced the apoptotic response to DNA damage by 34–50%. Since the apoptotic threshold is determined by many different factors, the degree of impairment is striking. Moreover, this is seen in many different settings including both primary and tumor cells derived from either mice or humans, and it is independent of p53. Thus, our finding that pRB has pro-apoptotic activity has broad relevance.
The concept that pRB can participate in transcriptional active complexes is not without precedent, since pRB has been shown to cooperate with differentiation-specific transcription factors in the activation of key target genes (Charles et al., 2001; Gery et al., 2004; Thomas et al., 2001). However, this pro-apoptotic function of pRB is not observed in all situations. As we outlined in the introduction, the analysis of Rb mutant embryos lead to the prevailing view that pRB is anti-apoptotic. Although much of this apoptosis in non-cell autonomous (de Bruin et al., 2003; Wenzel et al., 2007; Wu et al., 2003), pRB-loss does promote apoptosis in a cell autonomous manner in some tissues of the developing embryo and this seems to reflect an inability to undergo terminal differentiation (de Bruin et al., 2003; Haigis et al., 2006; Huh et al., 2004; MacPherson et al., 2003; Mason-Richie et al., 2008; Ruiz et al., 2004; Wang et al., 2007; Wikenheiser-Brokamp, 2004). Moreover, some cell types, such as MEFs, have a heightened sensitivity to DNA damage in the absence of pRB. Thus, taken together, the published literature and this current study indicate that pRB can either suppress or promote apoptosis depending on the cellular context. We note that there is a strong correlation between the proliferative properties of the cell and the observed role of pRB in apoptosis. Thus, we propose the following model of pRB action (Fig. 6). In G0/G1 cells, genotoxic stress induces pRB recruitment into the classic repressive pRB-E2F-HDAC complex. This prevents cell cycle entry and thus acts indirectly to protect cells from apoptosis. Thus, in G0/G1 cells, pRB loss would impair arrest and thereby promote apoptosis. In contrast, in proliferating cells, genotoxic stress favors formation of the transcriptionally active pRB-E2F1-P/CAF complexes because the hyperphosphorylation of pRB inhibits its participation in the repressive complexes. Consequently, in this setting, pRB is pro-apoptotic. Interestingly, this context-dependent model of pRB function has the potential to explain the well-established phenomenon that proliferating cells have a higher predisposition to undergo apoptosis compared to their quiescent counterparts. Importantly, both our cell line and in vivo studies show that a reduction in Rb levels is sufficient to impair its pro-apoptotic function without any disruption of proliferation control. This is particularly striking in our animal experiments where Rb haploinsufficiency reduces the apoptotic response to doxorubicin just as efficiently as the complete inactivation of Rb. This dose-dependent effect raises the possibility that mutation of a single Rb allele might increase the probability of cellular transformation by impairing the apoptotic response to DNA damage and thereby enabling the acquisition of mutations within other genes. If this is true, individuals who carry germline Rb mutations would be particularly at risk.
Figure 6.
Model of pRB-E2F1 complexes involved in the regulation of proliferation and pro-apoptotic genes in response to DNA damage and E1A-induced oncogenic stress
Clearly, additional questions remain about how genotoxic stress triggers formation of the pRB-E2F1-P/CAF complex and directs it specifically to pro-apoptotic promoters. Our data show that ppRB can participate in the DNA damage-induced pRB-E2F1 complexes. However, it is still unclear whether pRB phosphorylation actively promotes, or is just permissive for, formation of the propapoptotic pRB-E2F1-P/CAF complex. Moreover, although we have conducted these studies using antibodies against known cdk phosphorylation sites (Ser 780, Ser 795 and Ser 807-811), we cannot be sure whether cyclin/cdk complexes, or other kinases, are responsible for this modification in the DNA damaged cells. Moreover, it is entirely possible that additional post-translational modifications of pRB and/or E2F1 may facilitate formation of the pro-apoptotic complex. We note that pRB has been shown to contain a second E2F binding site that does not interfere with E2F1’s transactivation domain (Dick and Dyson, 2003). Thus, it is intriguing to speculate that DNA damage somehow induces pRB and E2F1 to adopt this alternate structure. If this model is true, this conformation must also enable recruitment of P/CAF, which is known to be critical for E2F1-dependent activation of pro-apoptotic target genes in response to DNA damage (Ianari et al., 2004; Pediconi et al., 2003).
Our data also cause us to revise our view of E1A’s mechanism of action. The prevailing view proposes that E1A acts to disrupt pRB-E2F complexes, releasing free E2F1 to induce transcription of its target genes. However, our data clearly show that E1A forms a stable complex with both pRB and the activating E2Fs. Moreover, pRB, E2F1 and E1A are all recruited to the promoters of both apoptosis and cell cycle genes, coincident with their transcriptional activation. Interestingly, mutant analysis has shown that E1A must interact with both pRB and p400 in order to promote apoptosis in response to doxorubicin (Samuelson et al., 2005). Thus, we now propose that E1A’s ability to promote both apoptosis and proliferation reflects, at least in part, its direct action at pro-apoptotic and cell cycle gene promoters through its association with the pRB-E2F1 complex and the concomitant recruitment of p400 and other transcriptional coactivators (Fig. 6). Given the recent finding that oncogenic stress activates the DNA damage response (Bartkova et al., 2006; Di Micco et al., 2006), formation of the transcriptionally active pRB-E2F1 complexes may be further reinforced through E1A’s activation of the DNA damage-dependent process. Essentially, E1A would trigger the DNA damage response, and then piggyback onto the resultant pRB-E2F1 complex to create a stable super-activator. Notably, in contrast to the DNA damage-induced pRB-E2F1 complex, which specifically activates only pro-apoptotic genes, the E1A-containing species has an expanded target specificity that now includes both apoptosis and cell cycle targets. Importantly, in agreement with this super-activator model, pRB knockdown impairs E1A’s ability to promote the transcription of both apoptosis and cell cycle-related genes and induce apoptosis in response to DNA damaging agents.
The elucidation of the mechanism by which pRB acts as a tumor suppressor has been complicated by various factors. In addition to its role in cell-cycle control, pRB has been implicated in regulating a wide variety of cellular processes, including DNA replication, differentiation and apoptosis (Classon and Harlow, 2002). Whereas the decreased differentiation potential and the increase in proliferative rate seen in pRB-deficient cells could contribute to tumorigenesis, it is more difficult to reconcile pRB’s role as a tumor suppressor with the notion that loss of pRB may lead to increased apoptosis. Our finding that pRB plays a positive role in DNA damage-induced apoptosis widens our understanding of pRB’s functions. Notably, the behavior of pRB in the DNA damage response bears strong parallels with that of p53: both of these tumor suppressors appear capable of triggering either cell arrest or apoptosis depending on the cellular context. Given this model, we propose that Rb-inactivation in tumor cells promotes tumorigenicity by yielding both a proliferative advantage and resistance to apoptotic stimuli such as chemotherapeutic treatments.
EXPERIMENTAL PROCEDURES
Cell culture and infections
T98G, U2OS and IMR90 cell lines were cultured in DMEM with 10% heat inactivated FBS. The IMR90 cells overexpressed the murine ecotropic receptor. Lentiviral and retroviral preparations and infections were performed as described (Samuelson et al., 2005; Stegmeier et al., 2005). MSCs were generated by mechanically crushing the femurs and tibias of 6–8 weeks old mice and culturing in α-MEM with 10% heat inactivated FBS. These cells were infected either with Ad5CMVCre-eGFP or Ad5CMVeGFP at about 100pfu/cell for 4h (U. of Iowa Gene Transfer Vector Core) and three days later treated with 2μM doxo or irradiated for 15 minutes for a total dose of 10Gy and analyzed by FACS after 24h. The hairpins used in this study are shown in Supplementary Table 1.
FACS analysis
Suspensions of T98G or U2OS cells were processed for DNA content as described (Pozarowski and Darzynkiewicz, 2004). For apoptosis assays, cell suspensions were stained with AnnexinV-APC and 7AAD (Becton Dickinson). The cells were analyzed using a FACScan machine (Becton-Dickinson) and the data was analyzed using ModFit LT software (Verity Software).
Immunoprecipitations and western blotting
Proteins were extracted with RIPA buffer (Pediconi et al., 2003) and quantified by the BCA protein assay reagent (Pierce, Inc). Extracts were immunoprecipitated with the indicated antibodies and either protein A or protein G plus (Santa Cruz Biotechnology, Inc). Antibodies were obtained from Santa Cruz Biotechnology, Inc. [E2F1 (C-20), RNAPol-II (N-20), E1A (M73-HRP), actin (I-19), E2F3 (C-18) and E2F4 (C-20)], Cell Signaling Technologies [RB (4-H1), 780-795-807-811 ppRB and cleaved caspase 3], BD Pharmingen [pRB and Ki67], Upstate Biotechnology, Inc. (AcH4 and HDAC1) and P. Nakatani (P/CAF).
ChIP assay
ChIP was performed as previously described (Pediconi et al., 2003). For re-ChIP experiments, RB immunoprecipitates were eluted with DTT and then subjected to a second round of IP with the AcH4 antibody or with IgG. Densitometric quantification of ChIP results has been performed using the ImageJ 1.4 program. Primers sequences are described in Supplementary Table 2.
Real time RT PCR
Total RNA was extracted with Qiagen RNeasy Kit and reverse-transcribed with oligo (dT) primers and Superscript II reverse transcriptase (Invitrogen). Quantitative analysis of caspase 7, TAp73 and cyclinA2 mRNA expression was performed employing ABI PRISM 7900 Sequence Detection System (Applied Biosystems). Gene expression values were normalized with β-actin. Results are expressed as mean±SD. Statistical differences were analyzed with the Mann-Whitney non-parametric test. Values of p<0.05 were considered statistically significant.
Animal maintenance and tissue analyses
The Rblox;Villin-Cre mice were maintained and genotyped as described (Haigis et al., 2006). The relevant genotypes were injected intraperitoneally with either saline vehicle (0.9% NaCl) or doxorubicin (10 mg/Kg) and 3 hours later mice were sacrificed and intestines collected for histology. All animal procedures followed protocols approved by MIT’s Committee on Animal Care. pRB immunostaining was conducted using the UltraVision LP Detection System (Lab Vision Corporation) with the primary antibody (G3-245, BD) at a concentration of 1:100. IHC was performed as previously described for cleaved caspase 3 (Haigis et al., 2006) and Ki67 (Danielian et al., 2007). All samples were counterstained with hematoxylin.
Supplementary Material
Acknowledgments
Vectors and cell lines were kindly provided by F. Stegmeier (pPRIME-CMV-GFP and pPRIME-CMV-GFP-shRB) and S. Lowe (pBabe-Hygro, pBabe-E1A-Hygro and U2OS rtTA RB.670). We thank Keren Hilgendorf and Ann Cheung for technical help and Massimo Levrero, Koch Institute colleagues and members of the Gulino and Lees labs for helpful discussion throughout this work. This project was supported by grants to A.G. (Associazione Italiana per la Ricerca sul Cancro, Telethon Grant GGP04168, Istituto Pasteur Cenci Bolognetti, MiUR, Ministry of Health and the Rome Oncogenomic Center) and J.A.L. (NIH-2-P01-CA42063). A.I. was supported by postdoctoral fellowships from the American Italian Cancer Foundation and from the Marie Curie OIF. J.A.L. is a Ludwig Scholar at MIT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almasan A, Yin Y, Kelly RE, Lee EY, Bradley A, Li W, Bertino JR, Wahl GM. Deficiency of retinoblastoma protein leads to inappropriate S-phase entry, activation of E2F-responsive genes, and apoptosis. Proc Natl Acad Sci U S A. 1995;92:5436–5440. doi: 10.1073/pnas.92.12.5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Ahmad SM, Li G, Bray DA, Jr, Saito K, Wang D, Wirtz U, Sreedharan S, O’Malley BW, Jr, Li D. Retinoblastoma RB94 enhances radiation treatment of head and neck squamous cell carcinoma. Clin Cancer Res. 2008;14:3514–3519. doi: 10.1158/1078-0432.CCR-07-4538. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Bosco EE, Mayhew CN, Hennigan RF, Sage J, Jacks T, Knudsen ES. RB signaling prevents replication-dependent DNA double-strand breaks following genotoxic insult. Nucleic Acids Res. 2004;32:25–34. doi: 10.1093/nar/gkg919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen C, Birrer M, Gelmann EP. Retinoblastoma protein-mediated apoptosis after gamma-irradiation. J Biol Chem. 2002;277:44969–44979. doi: 10.1074/jbc.M202000200. [DOI] [PubMed] [Google Scholar]
- Bowen C, Spiegel S, Gelmann EP. Radiation-induced apoptosis mediated by retinoblastoma protein. Cancer Res. 1998;58:3275–3281. [PubMed] [Google Scholar]
- Charles A, Tang X, Crouch E, Brody JS, Xiao ZX. Retinoblastoma protein complexes with C/EBP proteins and activates C/EBP-mediated transcription. J Cell Biochem. 2001;83:414–425. doi: 10.1002/jcb.1239. [DOI] [PubMed] [Google Scholar]
- Classon M, Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Bender Kim CF, Caron AM, Vasile E, Bronson RT, Lees JA. E2f4 is required for normal development of the airway epithelium. Dev Biol. 2007;305:564–576. doi: 10.1016/j.ydbio.2007.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin A, Wu L, Saavedra HI, Wilson P, Yang Y, Rosol TJ, Weinstein M, Robinson ML, Leone G. Rb function in extraembryonic lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proc Natl Acad Sci U S A. 2003;100:6546–6551. doi: 10.1073/pnas.1031853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Dick FA, Dyson N. pRB contains an E2F1-specific binding domain that allows E2F1-induced apoptosis to be regulated separately from other E2F activities. Mol Cell. 2003;12:639–649. doi: 10.1016/s1097-2765(03)00344-7. [DOI] [PubMed] [Google Scholar]
- Fattaey AR, Harlow E, Helin K. Independent regions of adenovirus E1A are required for binding to and dissociation of E2F-protein complexes. Mol Cell Biol. 1993;13:7267–7277. doi: 10.1128/mcb.13.12.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov MV, Dyson NJ. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J Cell Sci. 2004;117:2173–2181. doi: 10.1242/jcs.01227. [DOI] [PubMed] [Google Scholar]
- Gery S, Gombart AF, Fung YK, Koeffler HP. C/EBPepsilon interacts with retinoblastoma and E2F1 during granulopoiesis. Blood. 2004;103:828–835. doi: 10.1182/blood-2003-01-0159. [DOI] [PubMed] [Google Scholar]
- Haigis K, Sage J, Glickman J, Shafer S, Jacks T. The related retinoblastoma (pRb) and p130 proteins cooperate to regulate homeostasis in the intestinal epithelium. J Biol Chem. 2006;281:638–647. doi: 10.1074/jbc.M509053200. [DOI] [PubMed] [Google Scholar]
- Helt AM, Galloway DA. Mechanisms by which DNA tumor virus oncoproteins target the Rb family of pocket proteins. Carcinogenesis. 2003;24:159–169. doi: 10.1093/carcin/24.2.159. [DOI] [PubMed] [Google Scholar]
- Hofmann F, Martelli F, Livingston DM, Wang Z. The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes Dev. 1996;10:2949–2959. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]
- Huh MS, Parker MH, Scime A, Parks R, Rudnicki MA. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J Cell Biol. 2004;166:865–876. doi: 10.1083/jcb.200403004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianari A, Gallo R, Palma M, Alesse E, Gulino A. Specific role for p300/CREB-binding protein-associated factor activity in E2F1 stabilization in response to DNA damage. J Biol Chem. 2004;279:30830–30835. doi: 10.1074/jbc.M402403200. [DOI] [PubMed] [Google Scholar]
- Iaquinta PJ, Lees JA. Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol. 2007;19:649–657. doi: 10.1016/j.ceb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- Knudsen KE, Booth D, Naderi S, Sever-Chroneos Z, Fribourg AF, Hunton IC, Feramisco JR, Wang JY, Knudsen ES. RB-dependent S-phase response to DNA damage. Mol Cell Biol. 2000;20:7751–7763. doi: 10.1128/mcb.20.20.7751-7763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen KE, Weber E, Arden KC, Cavenee WK, Feramisco JR, Knudsen ES. The retinoblastoma tumor suppressor inhibits cellular proliferation through two distinct mechanisms: inhibition of cell cycle progression and induction of cell death. Oncogene. 1999;18:5239–5245. doi: 10.1038/sj.onc.1202910. [DOI] [PubMed] [Google Scholar]
- Kucherlapati MH, Nguyen AA, Bronson RT, Kucherlapati RS. Inactivation of conditional Rb by Villin-Cre leads to aggressive tumors outside the gastrointestinal tract. Cancer Res. 2006;66:3576–3583. doi: 10.1158/0008-5472.CAN-05-2699. [DOI] [PubMed] [Google Scholar]
- Ludlow JW, Glendening CL, Livingston DM, DeCarprio JA. Specific enzymatic dephosphorylation of the retinoblastoma protein. Mol Cell Biol. 1993;13:367–372. doi: 10.1128/mcb.13.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson D, Sage J, Crowley D, Trumpp A, Bronson RT, Jacks T. Conditional mutation of Rb causes cell cycle defects without apoptosis in the central nervous system. Mol Cell Biol. 2003;23:1044–1053. doi: 10.1128/MCB.23.3.1044-1053.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason-Richie NA, Mistry MJ, Gettler CA, Elayyadi A, Wikenheiser-Brokamp KA. Retinoblastoma function is essential for establishing lung epithelial quiescence after injury. Cancer Res. 2008;68:4068–4076. doi: 10.1158/0008-5472.CAN-07-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Park MJ, Kimura K, Shimizu K, Lee SH, Yokota J. Alterations in the INK4a/ARF locus and their effects on the growth of human osteosarcoma cell lines. Cancer Genet Cytogenet. 2002;133:105–111. doi: 10.1016/s0165-4608(01)00575-1. [DOI] [PubMed] [Google Scholar]
- Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L, Porcellini A, Screpanti I, Balsano C, Alesse E, et al. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol. 2003;5:552–558. doi: 10.1038/ncb998. [DOI] [PubMed] [Google Scholar]
- Pozarowski P, Darzynkiewicz Z. Analysis of cell cycle by flow cytometry. Methods Mol Biol. 2004;281:301–311. doi: 10.1385/1-59259-811-0:301. [DOI] [PubMed] [Google Scholar]
- Ruiz S, Santos M, Segrelles C, Leis H, Jorcano JL, Berns A, Paramio JM, Vooijs M. Unique and overlapping functions of pRb and p107 in the control of proliferation and differentiation in epidermis. Development. 2004;131:2737–2748. doi: 10.1242/dev.01148. [DOI] [PubMed] [Google Scholar]
- Samuelson AV, Narita M, Chan HM, Jin J, de Stanchina E, McCurrach ME, Fuchs M, Livingston DM, Lowe SW. p400 Is Required for E1A to Promote Apoptosis. J Biol Chem. 2005;280:21915–21923. doi: 10.1074/jbc.M414564200. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci U S A. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, Hinds PW. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell. 2001;8:303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ray SK, Hinds PW, Leiter AB. The retinoblastoma protein, RB, is required for gastrointestinal endocrine cells to exit the cell cycle, but not for hormone expression. Dev Biol. 2007;311:478–486. doi: 10.1016/j.ydbio.2007.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel PL, Wu L, de Bruin A, Chong JL, Chen WY, Dureska G, Sites E, Pan T, Sharma A, Huang K, et al. Rb is critical in a mammalian tissue stem cell population. Genes Dev. 2007;21:85–97. doi: 10.1101/gad.1485307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikenheiser-Brokamp KA. Rb family proteins differentially regulate distinct cell lineages during epithelial development. Development. 2004;131:4299–4310. doi: 10.1242/dev.01232. [DOI] [PubMed] [Google Scholar]
- Wu L, de Bruin A, Saavedra HI, Starovic M, Trimboli A, Yang Y, Opavska J, Wilson P, Thompson JC, Ostrowski MC, et al. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature. 2003;421:942–947. doi: 10.1038/nature01417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.