Abstract
Altered levels of cerebrospinal fluid (CSF) peptides related to Alzheimer’s disease (AD) are associated with pathologic AD diagnosis, although cognitively normal subjects can also have abnormal levels of these AD biomarkers. To identify novel CSF biomarkers that distinguish pathologically confirmed AD from cognitively normal subjects and patients with other neurodegenerative disorders, we collected antemortem CSF samples from 66 AD patients and 25 patients with other neurodegenerative dementias followed longitudinally to neuropathologic confirmation, plus CSF from 33 cognitively normal subjects. We measured levels of 151 novel analytes via a targeted multiplex panel enriched in cytokines, chemokines and growth factors, as well as established AD CSF biomarkers (levels of Aβ42, tau and p-tau181). Two categories of biomarkers were identified: (1) analytes that specifically distinguished AD (especially CSF Aβ42 levels) from cognitively normal subjects and other disorders; and (2) analytes altered in multiple diseases (NrCAM, PDGF, C3, IL-1α), but not in cognitively normal subjects. A multiprong analytical approach showed AD patients were best distinguished from non-AD cases (including cognitively normal subjects and patients with other neurodegenerative disorders) by a combination of traditional AD biomarkers and novel multiplex biomarkers. Six novel biomarkers (C3, CgA, IL-1α, I-309, NrCAM and VEGF) were correlated with the severity of cognitive impairment at CSF collection, and altered levels of IL-1α and TECK associated with subsequent cognitive decline in 38 longitudinally followed subjects with mild cognitive impairment. In summary, our targeted proteomic screen revealed novel CSF biomarkers that can improve the distinction between AD and non-AD cases by established biomarkers alone.
Keywords: Amyloid beta, Abeta42, Diagnosis, IL-1α, MCI, NrCAM, PDGF, Resistin, TECK, TDP-43, Tau
Introduction
Alzheimer’s disease (AD), frontotemporal lobar degenerations (FTLD) and dementia with Lewy bodies (DLB) are major neurodegenerative disorders pathologically characterized by lesions composed of disease-specific misfolded proteins. Their clinical syndromes often have overlapping features, making antemortem prediction of pathology challenging. Yet, as specific disease-modifying therapies become available, it is increasingly important that such diagnoses be made. Analytes in cerebrospinal fluid (CSF) associated with AD pathology, such as total tau, tau phosphorylated at threonine 181 (p-tau181) and Aβ1-42 (or Aβ42), offer the potential for more accurate diagnosis, although cognitively normal elderly subjects could have altered levels of these traditional AD biomarkers [20, 24, 26]. Peptides in common inflammatory and apoptotic pathways, growth factors and other analytes have also been proposed as novel CSF biomarkers for AD [23] and the use of novel biomarkers on their own or in conjunction with traditional AD biomarkers may improve the specificity of CSF-based AD diagnosis. Recently, a proteomic approach targeting specific inflammatory and growth factors in the plasma identified novel biomarkers for the clinical diagnosis of AD [21], but the absence of pathological confirmation makes these results difficult to interpret since 10–20% of patients clinically diagnosed with AD are found on autopsy to have a cause for dementia other than AD. The determination of diagnostic accuracy for novel AD biomarkers thus requires studies of biofluids obtained during life from well-characterized AD patients longitudinally followed to autopsy confirmation [1, 3, 7]. Also, novel CSF biomarkers for AD can potentially facilitate disease staging and predict rates of clinical decline, as these biomarkers could represent factors that modulate AD pathogenesis associated with various stages of the disease. Characterization of a select panel of CSF biomarkers can, therefore, be critical in diagnosis and prognosis, and alterations in their levels can be further considered as secondary endpoints in future therapeutic trials.
Here, we tested the hypothesis that distinct sets of CSF peptides and proteins are associated with AD in contrast to cognitively normal subjects and other common neurodegenerative disorders, including FTLD with TDP-43-immunoreactive lesions (FTLD-TDP), FTLD with tau-immunoreactive inclusions (FTLD-Tau), and DLB. We collected and analyzed CSF samples antemortem from a total of 162 subjects, and the concentrations of 151 analytes in the Rules Based Medicine Human DiscoveryMAP™ panel (referred to below here as MAP) were measured by a Luminex-based multiplex platform. As the choice of analytical strategy in this type of high-dimensional data may significantly alter the composition of the identified biomarker panel, we analyzed the same body of data through three independent algorithms. Alterations in the levels of these candidate biomarkers were first analyzed according to the traditional statistical modeling, including Mann–Whitney U test and logistic regression. To avoid bias associated with feature pre-selection by univariate analysis and instability of linear models associated with high-dimensional data, we searched for novel AD biomarkers by two additional methods: a tree-based classification algorithm (random forest) and a nearest shrunken centroid algorithm (predictive analysis of microarrays, or PAM) [21, 29]. The diagnostic accuracy of novel analyte combinations predicted by each algorithm was then assessed. Lastly, novel AD biomarkers were then evaluated for their relationship to the severity of cognitive impairment in AD, and their potential role in predicting rates of cognitive decline in patients with mild cognitive impairment (MCI).
Materials and methods
Participants
Patients and control subjects were recruited and longitudinally followed at Penn in specialty services dedicated to the evaluation and management of neurodegenerative diseases (Supplementary Table 1). All protocols were approved by the Penn Institutional Review Board. Each patient in the autopsy cohort had undergone detailed cognitive, neurological, neuroimaging and laboratory examinations to ensure the accuracy of clinical diagnosis according to established criteria for AD [6], frontotemporal dementia (FTD) [16], amyotrophilc lateral sclerosis (ALS) [22] and DLB [13]. Autopsy-confirmed cases of AD (n = 66), FTLD (n = 16) and DLB (n = 2) were characterized neuropathologically with detailed immunohistochemical analysis for pathology associated with each major neurodegenerative disorder, including Aβ42, hyperphosphorylated tau, hyperphosphorylated TDP-43 and alpha-synuclein as described by Neumann et al. [18]. Seven patients with clinical FTD-ALS, but no autopsy was added to the FTLD-TDP group, as these cases nearly always have TDP-43 pathology. Thirty-eight patients with MCI were also recruited to assess predictors of cognitive decline. Each MCI patient was diagnosed by modified Petersen criteria [30], and followed longitudinally with serial cognitive and neurological examination. Cognitively normal subjects were evaluated at the time of CSF collection, and continued to undergo annual testing to confirm their cognitive status. ApoE genotyping was performed for all subjects (Supplementary Material).
Procedures
Baseline CSF samples were obtained during routine diagnostic lumbar puncture as previously described [3, 24]. Briefly, lumbar puncture was performed with a 20- or 24- gauge spinal needle, and CSF was transferred into polypropylene tubes. At the time of CSF collection, aliquots (0.5 mL) were prepared, bar-coded and then stored in polypropylene vials at −80°C until analysis (mean 8.7 years, SD = 3.6 years). Samples were then grouped altogether and simultaneously interrogated by Rules-Based Medicine, Inc. (Austin, TX) for levels of 151 analytes using the Human DiscoveryMAP™ panel and a Luminex 100 platform (Supplementary Material). The 151 MAP analytes were assembled by RBM into pre-formatted assays that RBM designed for studies of a number of different diseases including cancer, autoimmune disorders and AD based on the previous associations with AD of many, but by no means all of these analytes in peer-reviewed literature. Measures of CSF Aβ42, total tau and p-tau181, were performed using the multiplex xMAP Luminex platform (Luminex Corp, Austin, TX) with Innogenetics (INNO-BIA AlzBio3, Ghent, Belgium) immunoassay kit-based reagents as described [24].
Statistical analysis
Statistical analysis was performed in SPSS 12.0, Random Forests (http://www.stat.berkeley.edu/~breiman/RandomForests/) and SAM/PAM. For testing of stability associated with each analyte, Pearson’s correlation analysis was performed between analyte levels and time in −80°C storage. For each analytical strategy, diagnostic performance (sensitivity, specificity, accuracy) was determined using traditional AD biomarkers alone (tau, p-tau181, Aβ42), MAP biomarkers alone or both traditional and MAP biomarkers. For each model, performance characteristics reported were based on the cross-validation. In the first model (Model 1), analytes that differed significantly between cognitively normal and AD by Mann–Whitney U test (nominal P < 0.01) were entered into logistic regression models for AD identification, adjusting for age and gender. Sensitivity and specificity of Model 1 were obtained by leave-one-out approach in discriminant analysis. In random forest analysis, analytes were entered into the analysis with nodes optimized for best classification of AD versus cognitively normal (Model 2). Out-of-box error rate was used to derive diagnostic accuracy, with sensitivity and specificity derived from the confusion matrix. In PAM, analytes that significantly differentiated AD from cognitively normal were identified, and diagnostic accuracy was derived through internal cross-validation (Model 3). Given the number of analytes relative to the number of subjects, interaction terms were not entered in the logistic regression model (Model 1). Random forest analysis (Model 2) and PAM (Model 3) each relies less on the assumption of normal distribution and takes into account possible correlations between analytes, although each algorithm can derive different analytes to account for variations in the respective classification model. Thus, to expand our analysis beyond the strengths and constraints of any one algorithm, we sought to identify biomarkers determined by at least two of these three well-established analytical strategies as key novel biomarkers. A similar three-approach strategy was employed to determine biomarkers that distinguished between AD and non-AD neurodegenerative disorders.
For cross-sectional association between novel AD biomarker levels and severity of cognitive impairment at the time of CSF collection, Pearson’s correlation coefficient was used to relate levels of newly identified CSF AD biomarkers with cognitive performance characterized by Mini-Mental Status Examination (MMSE) in autopsyconfirmed AD cases. For correlation of CSF biomarker levels and rates of cognitive decline following CSF collection in MCI, rates of cognitive decline were first estimated by the slope of MMSE score linear regression over time. Pearson’s correlation coefficient was then determined for CSF biomarker levels and rates of cognitive decline. Effects from age and gender were adjusted for all diagnostic and progression models.
Results
All CSF was obtained from patients with informed consent as described [2, 12, 24]. Levels of 151 analytes in the MAP were measured in the CSF, with 106 analytes having measurable levels for analysis (Supplementary Table 2). Four analytes (angiotensinogen, BMP-6, endothelin-1, SGOT) demonstrated level changes that corresponded to time stored in −80°C freezer and were excluded from the analysis because of their apparent instability with increasing length of storage. To determine the best biomarkers of AD, we used three independent analytical strategies to identify MAP analytes associated with AD, and combined traditional AD biomarkers and MAP analytes to identify complementary AD biomarkers.
AD versus cognitively normal
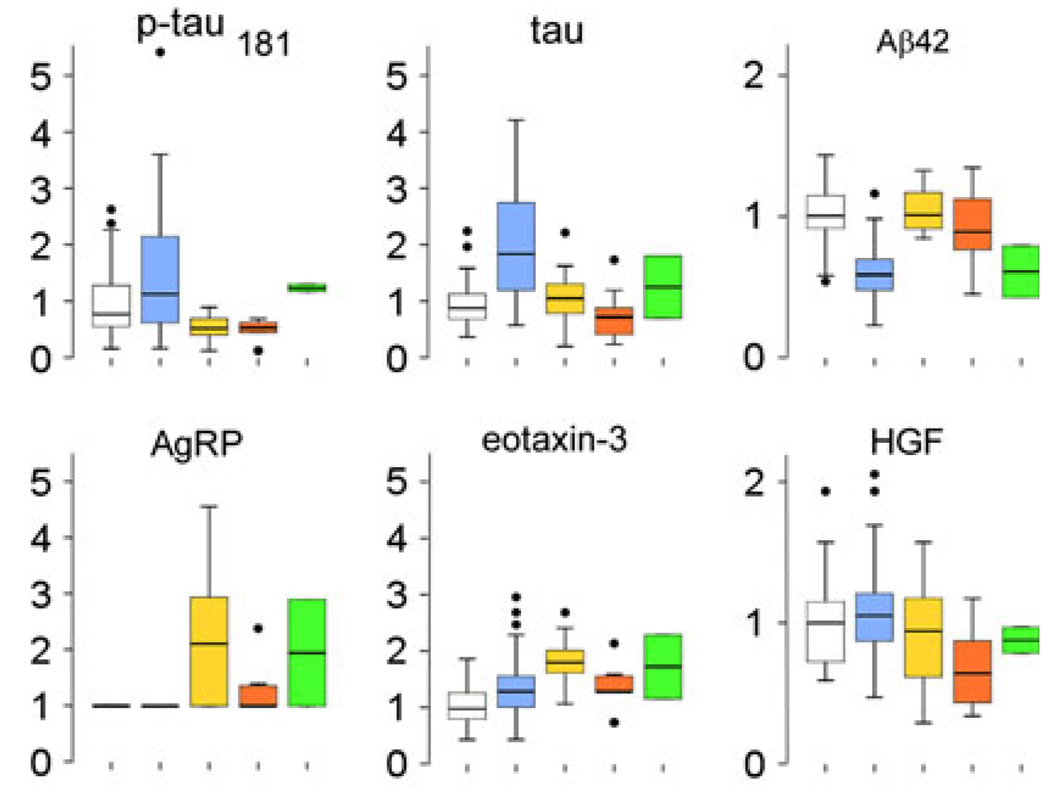
In Model 1, 21 MAP analytes were found to differ between cognitively normal subjects and AD (Fig. 1) by Mann–Whitney U test at P < 0.01, and only a minority of these were specifically changed in AD, including resistin and thrombospondin-1. MAP analytes alone, but not traditional AD biomarkers, were entered into a forward stepwise logistic regression model (Table 1). Leave-one-out discriminant analysis using the five resultant MAP analytes achieved 84.8% sensitivity and 87.9% specificity, with overall 85.9% accuracy. By comparison, traditional AD biomarkers Aβ42 and total tau yielded greater sensitivity (92.4%), but less specificity (81.8%) for overall accuracy of 88.9%. Combining MAP analytes and traditional AD biomarkers resulted in a model differentiating AD from cognitively normal subjects by the following biomarkers: levels of tau, Aβ42, complement 3 (C3), neuron-glia-CAM-related cell adhesion molecule (NrCAM) and platelet-derived growth factor (PDGF, Table 1). This combined model has high sensitivity (97.0%) and specificity (93.9%) with 96.0% accuracy, and improved upon the traditional AD model by correctly reclassifying up to four cognitively normal subjects with pathologic CSF levels of tau and Aβ42, and three AD subjects with non-pathologic levels of CSF tau and Aβ42.
Fig. 1.
Boxplots showing median values, quartiles, and outliers (circles) of traditional (i.e. tau and Aβ42) and other candidate CSF biomarkers that differed in levels between subjects with normal cognition and AD. Values shown are normalized to mean values of cognitively normal subjects. a Analytes elevated in AD as compared to cognitively normal subjects. b Analytes decreased in AD as compared to cognitively normal subjects. Levels in patients with autopsy-confirmed non-AD neurodegeneration were also shown for comparison. White box cognitively normal subjects, blue box autopsy-confirmed cases of AD, red box autopsy-confirmed cases of non-AD neurodegenerative disorders. *I-309 was found to differ between AD and cognitively normal subjects by random forest and PAM, but not Mann–Whitney U test
Table 1.
Factors predictive of AD compared with cognitively normal subjects according to logistic regression
| AD versus cognitively normal | B | P |
|---|---|---|
| MAP model | ||
| Age | −0.073 | 0.141 |
| Male gender | 1.845 | 0.092 |
| C3 | 0.932 | 0.017 |
| Fabp | 0.809 | 0.002 |
| IL-23 | 17.24 | 0.031 |
| NrCAM | −0.051 | <0.001 |
| PDGF | 0.004 | 0.064 |
| Traditional AD model | ||
| Age | −0.024 | 0.558 |
| Male gender | 0.001 | 0.999 |
| Aβ42 | −0.035 | <0.001 |
| Tau | 0.019 | 0.051 |
| Combined model | ||
| Age | −0.217 | 0.088 |
| Male gender | 2.038 | 0.309 |
| C3 | 2.376 | 0.025 |
| NrCAM | −0.063 | 0.041 |
| PDGF | 0.013 | 0.061 |
| Tau | 0.08 | 0.042 |
| Aβ42 | −0.039 | 0.01 |
Traditional AD model incorporated Aβ42 and tau levels
Coefficient (B) and P value for each factor as part of the overall model are shown
Age and gender were entered into first block of LR, while analytes identified to be different between AD and cognitively normal subjects were then entered in a forward step-wise fashion, with P < 0.05 for entry and P > 0.10 for removal
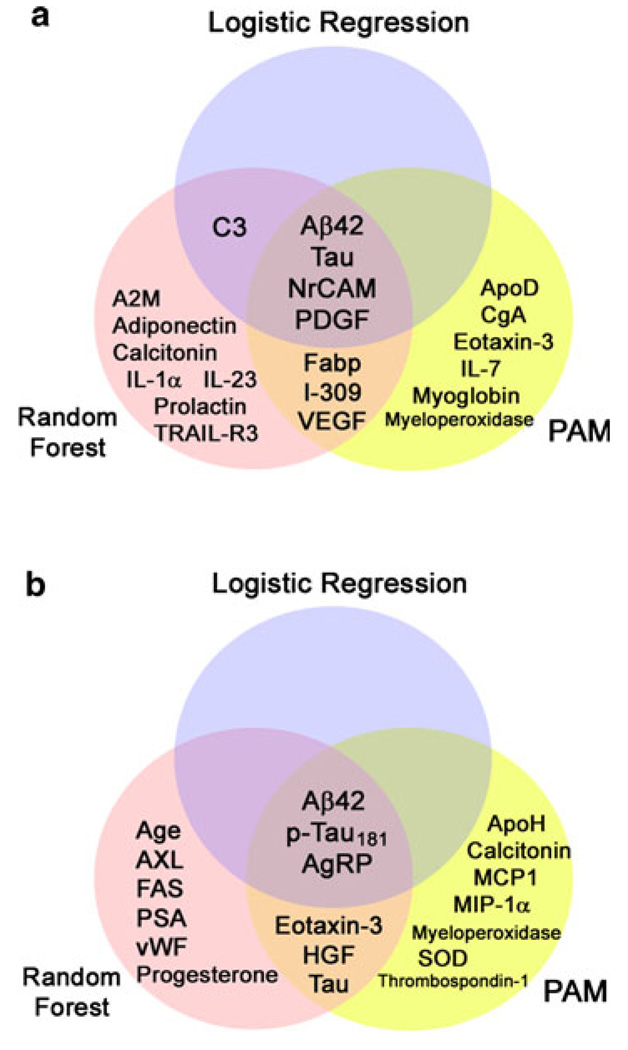
Feature pre-selection and the lack of an independent validation set may bias our classification results. Hence, we performed a similar analysis of AD versus cognitively normal through random forest (Model 2) and PAM (Model 3) using age, gender and levels of 3 traditional biomarkers and 106 MAP analytes, as each analysis incorporates internal cross-validation that is more objective than leave-one-out analysis. Model 2 using MAP analytes alone identified some analytes from Model 1, including C3, fatty acid-binding protein (Fabp), IL-23, NrCAM and PDGF, among others (Table 2; Fig. 2a). The out-of-box error rate of traditional AD biomarkers was 12.1%, which reduced to 6.1% when MAP analytes were introduced with 93.9% accuracy. Model 3 also identified NrCAM and PDGF as important biomarkers useful in distinguishing between AD and cognitively normal subjects (Tables 3, 4; Fig. 2a). Diagnostic accuracy obtained through cross-validation was 93.9% in Model 3. A summary of analytes important in distinguishing between AD and cognitive normal subjects is shown in Fig. 2a, including Aβ42, tau, NrCAM and PDGF identified by all three algorithms.
Table 2.
Analytes differentiating AD from cognitively normal according to random forest analysis
| AD versus cognitively normal |
Z score |
|---|---|
| MAP model | |
| PDGF | 27.585 |
| IL-1α | 26.656 |
| IL-23 | 20.686 |
| C3 | 18.343 |
| Fabp | 17.888 |
| NrCAM | 15.937 |
| VEGF | 13.894 |
| TRAIL-R3 | 12.064 |
| IL-17 | 11.209 |
| Eotaxin-3 | 11.037 |
| IL-7 | 10.48 |
| A2M | 9.603 |
| Prolactin | 9.549 |
| Ferritin | 8.463 |
| ThBG | 8.371 |
| I-309 | 7.743 |
| HCC-4 | 3.502 |
| Traditional AD model | |
| Aβ42 | 68.401 |
| Tau | 21.826 |
| Male gender | 10.052 |
| Combined model | |
| Aβ42 | 47.935 |
| Tau | 26.232 |
| PDGF | 22.506 |
| IL-1α | 19.806 |
| NrCAM | 17.474 |
| C3 | 16.702 |
| IL-23 | 14.472 |
| VEGF | 13.362 |
| Fabp | 11.583 |
| Prolactin | 11.533 |
| TRAIL-R3 | 9.779 |
| A2M | 9.425 |
| I-309 | 7.981 |
| Calcitonin | 5.824 |
| Adiponectin | 4.64 |
Fig. 2.
AD biomarkers identified by each of the three analytical strategies (logistic regression, random forest, and PAM). a Biomarkers useful in distinguishing between subjects with AD and normal cognition. b Biomarkers useful in distinguishing between subjects with AD and other non-AD neurodegenerative disorders. Analytes in overlapping regions were identified by multiple strategies as important biomarkers
Table 3.
Factors differentiating AD from cognitively normal according to PAM
| MAP model | Traditional AD model |
Combined model |
|---|---|---|
| AD versus cognitively normal | ||
| PDGF | Aβ42 | Aβ42 |
| VEGF | Tau | Tau |
| NrCAM | p-tau181 | PDGF |
| CgA | Age | VEGF |
| ApoD | NrCAM | |
| Fabp | CgA | |
| I-309 | ApoD | |
| Eotaxin-3 | Fabp | |
| IL-7 | Eotaxin-3 | |
| Myoglobin | I-309 | |
| Myeloperoxidase | IL-7 | |
| GRO-α | Myoglobin | |
| EN-RAGE | Myeloperoxidase | |
| TGF α | ||
| Thrombospondin-1 | ||
| Age | ||
| Stem cell factor | ||
| Tissue factor | ||
| Pancreatic polypeptide | ||
| MDC | ||
| TECK | ||
| SOD | ||
| Ferritin | ||
| EGF-R | ||
| IL-11 | ||
| FAS | ||
| IL-1ra | ||
| Prolactin | ||
| AXL | ||
| IL-17 | ||
| TRAIL-R3 | ||
| FAS-ligand | ||
| IL-16 |
Threshold was set for each model through internal cross-validation
Table 4.
Comparative diagnostic performance of biomarker panels according to random forest versus PAM analysis for AD versus cognitively normal
| Random forest | PAM | |||||
|---|---|---|---|---|---|---|
| Traditional | MAP | Combined | Traditional | MAP | Combined | |
| Sensitivity (%) | 88.6 | 86.4 | 92.4 | 97.0 | 89.4 | 97.0 |
| Specificity (%) | 86.2 | 81.8 | 97.0 | 66.7 | 75.8 | 87.9 |
| Accuracy (%) | 87.9 | 84.8 | 93.9 | 86.9 | 84.8 | 93.9 |
Traditional models included established AD biomarkers (Aβ42, tau and/or p-tau181). MAP models included novel biomarkers from the MAP 151 analyte panel. Combined models included traditional and MAP biomarkers
AD versus other neurodegenerative disorders
With the emergence of substrate-specific therapeutic interventions, it is critically important to identify biomarkers that reliably differentiate the major neurodegenerative disorders from one another. To this end, we assessed which CSF biomarkers best differentiated AD from other neurodegenerative disorders using a similar series of analytical strategies.
Among traditional AD biomarkers, Aβ42 and p-tau181 levels discriminated between AD and non-AD neurodegenerative disorders in all models. Among novel MAP analytes, agouti-related peptide (AgRP) was identified by all algorithms to distinguish between AD and non-AD disorders (Fig. 2b). Post hoc analysis showed AgRP as most altered in FTLD-TDP (Fig. 3) and its classification power may rest in identifying FTLD-TDP cases. Tau, eotaxin-3 and hepatocyte growth factor (HGF) were additionally identified by both RF and PAM to be important in distinguishing between AD and non-AD disorders (Fig. 2b). Similar to the classification role of AgRP, eotaxin-3 was most different between AD and FTLD-TDP (P = 0.001), and HGF was most different between AD and FTLD-Tau (P = 0.002, both comparisons by Mann–Whitney U test; Fig. 3). Thus, biomarkers more specifically associated with other neurodegenerative disorders can also aid in the diagnosis of AD.
Fig. 3.
Boxplots showing median values, quartiles, and outliers (circles) of traditional and candidate biomarkers that differed in levels between AD and other non-AD neurodegenerative disorders. Values shown are normalized to mean values of cognitively normal subjects. White box cognitively normal subjects, blue box AD, yellow box FTLD-TDP, orange box FTLD-Tau, green box dementia with Lewy bodies
Biomarker associations with cognitive function and decline
Some diagnostic biomarkers may reflect severity of cognitive impairment and thus be useful in disease staging. To assess this, we correlated CSF biomarker levels with MMSE scores at the time of CSF collection as a general measure of cognitive impairment. Among CSF biomarkers for AD identified by at least one approach, six (C3, CgA, IL-1α, I-309, NrCAM and VEGF) were correlated with MMSE score, and levels of these analytes did not correlate with MMSE scores in the other neurodegenerative disorders. A multivariate linear regression analysis adjusting for age, gender and education showed C3, IL-1α and I-309 levels were independently associated with MMSE scores in autopsy-confirmed cases of AD.
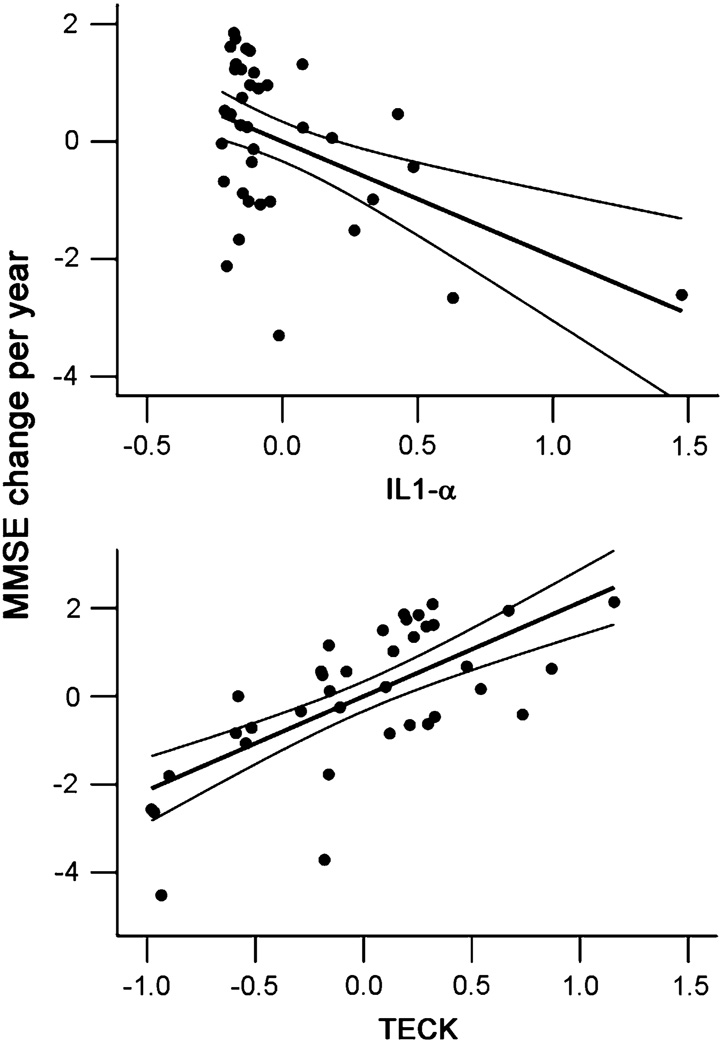
To further test the value of these CSF biomarkers in predicting cognitive decline, we determined the correlation between levels of these six biomarkers and rates of subsequent MMSE decline in MCI subjects following CSF collection. The 38 living MCI patients were similar to AD patients in age (71.39 vs. 70.79 yo, P = 0.674), education (15.66 vs. 14.64 yo, P = 0.143) and gender (42.1 vs 53.0% women), but MCI patients had higher MMSE scores (mean 26.16, SD = 2.00) as compared to AD (mean 17.55, SD = 8.57, P < 0.01). The MCI patients had a median follow-up of 52 months (range 30–129 months) and a median rate of MMSE decline of 1.2 points per year (mean 2.0, SD = 2.0). Among analytes associated with cognitive performance at the time of CSF collection in AD, IL-1α levels correlated with the rates of MMSE decline (P = 0.003), although with modest effect on decline rates (R = 0.498 for model). A search across 4 traditional and 106 MAP analytes additionally identified thymus-expressed chemokine (TECK) as significantly associated with rates of cognitive decline in MCI (P < 0.001 adjusting for age, gender and education) and had a stronger effect on the rate of decline (R = 0.745 for model, Fig. 4).
Fig. 4.
Partial residual plots of MAP analytes versus rates of subsequent cognitive decline in MCI. Linear fit and 95% confidence interval for fit are shown for each graph. The overall model includes age, gender, education, IL-1α level and TECK level
Discussion
The search for accurate CSF and plasma biomarkers in neurodegenerative diseases has intensified with the increasing need for informative biomarkers in clinical trials of disease-modifying therapies for AD, and has been facilitated by high-throughput multiplex platforms [21, 24]. Using clinically and pathologically well-characterized cases of AD and FTLD, we identified novel biomarkers useful in improving the distinction between AD and cognitively normal subjects, such as NrCAM and PDGF and biomarkers associated with other disorders that improved the classification between AD and non-AD dementia, including AgRP, eotaxin-3 and HGF. C3, IL-1α and I-309 were helpful in the staging of AD, and IL-1α and TECK levels correlated with rates of subsequent cognitive decline in MCI. We discuss these findings below.
In none of our analytical models did MAP biomarkers alone out-perform traditional AD biomarkers in identifying AD from non-AD cases, but they complemented traditional biomarkers in two ways. First, while decreased Aβ42 and increased total/phosphorylated-tau levels are strongly linked to AD, altered levels of some MAP biomarkers improved the classification of cognitively normal subjects with decreased levels of CSF Aβ, but no dementia. Alterations in MAP biomarkers (NrCAM, PDGF) were seen in multiple neurodegenerative disorders, and likely represent neuronal loss rather than AD-specific processes (Fig. 1). NrCAM is a member of the L1 family of cell adhesion molecules, and may be involved in ion channel clustering at the axon initial segment and nodes of Ranvier [4, 5]. A decrease in NrCAM levels in AD and other neurodegenerative disorders likely follows axonal degeneration, although accumulation of ankryn (which interacts with NrCAM intracellularly) in the insoluble AD proteome relative to normal and FTLD-TDP brains raises additional possibilities [10]. PDGF was previously identified as a plasma AD biomarker by Ray et al. [21]. PDGF-receptor activation can promote Aβ precursor protein processing in vitro [9], and inhibition of PDGF-receptor activation with imatinib mesylate can decrease Aβ40 and Aβ42 secretion [17]. In our cohort, PDGF was also found to be elevated in multiple disorders, and its constitutive expression by neurons [8] suggests elevated PDGF levels to also reflect more general neuronal loss. C3 and Fabp were identified as AD biomarkers by two algorithms, and CSF Fabp was elevated in AD and DLB cases in one other study [25]. Despite alterations in multiple forms of dementia, however, changes in these biomarkers associated with neuronal loss improved the distinction between AD and cognitively normal with age-associated amyloidosis by traditional AD biomarkers alone, and they can further serve as secondary endpoints in therapies aimed at Aβ42 or tau clearance.
In addition to traditional AD CSF biomarkers (i.e. tau and Aβ42), altered levels of resistin and thrombospondin-1 specifically associated with AD despite little classification value beyond analytes in Fig. 2a. Central resistin modulates leptin action and oral intake [19], and resistin as a marker of macrophage may mean preferential microglial activation. Thrombospondin-1 is a key molecule in astrocyte-induced neurogenesis [14], and can promote recovery after brain ischemia. Elevated thrombospondin-1 levels in a subgroup of AD patients may identify a unique subgroup with vascular and degenerative etiologies for their dementia. Further stratification of AD patients by their CSF resistin and/or thrombospondin-1 levels in a larger cohort should clarify their role in AD lesions.
Novel MAP biomarkers also represent candidate biomarkers of disease staging and prediction of progression. Cross-sectionally, six diagnostic AD biomarkers correlated with cognitive deficits at the time of CSF collection. Because these analytes likely mirror severity of neuro-degeneration, correlations between their levels and clinical status should be expected. Furthermore, IL-1α levels were modestly associated with rates of decline in MCI after the CSF was collected. IL-1α immunoreactive microglia in AD neuritic plaques have been implicated in plaque evolution [11], and the difference in IL-1α levels between fast and slow MCI decliners may signal cognitive deficits insensitive to MMSE alone. We also identified TECK to predict the rate of cognitive decline among MCI patients, even though TECK itself was not a robust diagnostic biomarker for AD. This can be due to the potential pathologic heterogeneity of MCI, or represent a biomarker change that is transient in nature and specifically linked to the MCI or pre-AD stage. TECK (CCL25) is best understood as a strong chemoattractant for thymocytes and intestinal T cells [15]. TECK is a ligand to CCR9 which is predominantly expressed in epithelial tissues, but also a ligand to CCX-CKR found in the human brain [27, 31]. The role of TECK in AD has never been investigated, and its role as a robust predictor of cognitive decline in MCI should prompt further examination of its role in AD.
Some analytes were identified by only one analytical strategy as a potential AD biomarker due to the non-uniqueness of multiple analytical strategies, begging the question of whether such analytes are “true” biomarkers. Notably, the number of ApoE4 alleles was only identified by one analytical strategy (logistic regression, data not shown) to be a significant predictor of AD versus cognitively normal, despite its known association with AD[23]. The ordinal nature of allele dosage (such as number of ApoE4 alleles) may be more suited for models using linear scaling and less preferred by random forest and PAM. Consequently, we elected to seek analytes of wider ranging levels as novel biomarkers for more uniformity among the algorithms. Among other analytes identified only by one algorithm, IL-1α appears to be important in disease staging, and HGF was previously found to differentiate between AD and PSP [28]. Several explanations are possible. First, some analytes may correlate strongly with others, and each strategy may select different proxy analytes to represent a group of correlated analytes from the same biological process. Second, different analytical strategies may have various strengths and weaknesses for detecting particular effects. This was the reason we chose three analytical strategies to identify putative AD biomarkers, and analytes identified by multiple strategies may be most reliable. Third, some analytes identified by only one analytical strategy may be associated with chance difference at the population level not directly associated with dementia or AD. These speculations notwithstanding, each putative novel biomarker’s value in diagnosis and prognosis needs independent validation in another single-or a multicenter study, and their biological significance should be assessed independently. Indeed, we have studies underway now to do exactly this.
In summary, we identified novel biomarkers associated with pathologically confirmed AD. Some analytes were specifically associated with AD including Aβ42, resistin, and thrombospondin-1, while others were associated with multiple neurodegenerative disorders. Some diagnostic biomarkers mirrored the severity of cognitive impairment at time of CSF collection, while TECK and IL-1α reflected the rate of cognitive decline among clinically diagnosed MCI subjects. Accordingly, we propose the inclusion of diagnostic and prognostic biomarkers in a composite AD biomarker panel. Given the variability of each candidate biomarker across individuals, their collective classifying power should be determined in a large multicenter cohort, such as the Alzheimer Disease Neuroimaging Initiative. The biological relevance of each individual and set of biomarkers should be investigated for potential targets of therapeutic developments.
Supplementary Material
Acknowledgments
This work has been supported by the Penn-Pfizer Alliance as well as AG-10124 and AG-17586. WTH is supported by the American Academy of Neurology Clinical Translational Research Fellowship. ACP is supported by a Burroughs Wellcome Fund Career Award for Medical Scientists and NIH K08 AG033101. EP, MK, YC, and HDS are employees of Pfizer Global Research and Development.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00401-010-0667-0) contains supplementary material, which is available to authorized users.
Contributor Information
William T. Hu, Department of Neurology, University of Pennsylvania School of Medicine, Philadelphia, PA, USA Center for Neurodegenerative Disease Research, University of Pennsylvania School of Medicine, HUP, Maloney 3rd Floor, 36th and Spruce Streets, Philadelphia, PA 19104-4283, USA, trojanow@mail.med.upenn.edu.
Alice Chen-Plotkin, Department of Neurology, University of Pennsylvania School of Medicine, Philadelphia, PA, USA; Center for Neurodegenerative Disease Research, University of Pennsylvania School of Medicine, HUP, Maloney 3rd Floor, 36th and Spruce Streets, Philadelphia, PA 19104-4283, USA.
Steven E. Arnold, Department of Psychiatry, University of Pennsylvania School of Medicine, Philadelphia, PA, USA
Murray Grossman, Department of Neurology, University of Pennsylvania School of Medicine, Philadelphia, PA, USA.
Christopher M. Clark, Department of Neurology, University of Pennsylvania School of Medicine, Philadelphia, PA, USA
Leslie M. Shaw, Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, HUP, Maloney 3rd Floor, 36th and Spruce Streets, Philadelphia, PA 19104-4283, USA
Eve Pickering, Pfizer Global Research and Development, Groton, CT, USA.
Max Kuhn, Pfizer Global Research and Development, Groton, CT, USA.
Yu Chen, Pfizer Global Research and Development, Groton, CT, USA.
Leo McCluskey, Department of Neurology, University of Pennsylvania School of Medicine, Philadelphia, PA, USA.
Lauren Elman, Department of Neurology, University of Pennsylvania School of Medicine, Philadelphia, PA, USA.
Jason Karlawish, Department of Medicine, University of Pennsylvania School of Medicine, Philadelphia, PA, USA.
Howard I. Hurtig, Department of Neurology, University of Pennsylvania School of Medicine, Philadelphia, PA, USA
Andrew Siderowf, Department of Neurology, University of Pennsylvania School of Medicine, Philadelphia, PA, USA.
Virginia M.-Y. Lee, Center for Neurodegenerative Disease Research, University of Pennsylvania School of Medicine, HUP, Maloney 3rd Floor, 36th and Spruce Streets, Philadelphia, PA 19104-4283, USA Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, HUP, Maloney 3rd Floor, 36th and Spruce Streets, Philadelphia, PA 19104-4283, USA; Institute on Aging, University of Pennsylvania School of Medicine, Philadelphia, PA, USA.
Holly Soares, Pfizer Global Research and Development, Groton, CT, USA.
John Q. Trojanowski, Center for Neurodegenerative Disease Research, University of Pennsylvania School of Medicine, HUP, Maloney 3rd Floor, 36th and Spruce Streets, Philadelphia, PA 19104-4283, USA Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, HUP, Maloney 3rd Floor, 36th and Spruce Streets, Philadelphia, PA 19104-4283, USA; Institute on Aging, University of Pennsylvania School of Medicine, Philadelphia, PA, USA.
References
- 1.Castano EM, Roher AE, Esh CL, Kokjohn TA, Beach T. Comparative proteomics of cerebrospinal fluid in neuropathologically-confirmed Alzheimer’s disease and non-demented elderly subjects. Neurol Res. 2006;28:155–163. doi: 10.1179/016164106X98035. [DOI] [PubMed] [Google Scholar]
- 2.Clark CM, Davatzikos C, Borthakur A, et al. Biomarkers for early detection of Alzheimer pathology. Neurosignals. 2008;16:11–18. doi: 10.1159/000109754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark CM, Xie S, Chittams J, et al. Cerebrospinal fluid tau and beta-amyloid: how well do these biomarkers reflect autopsy-confirmed dementia diagnoses? Arch Neurol. 2003;60:1696–1702. doi: 10.1001/archneur.60.12.1696. [DOI] [PubMed] [Google Scholar]
- 4.Custer AW, Kazarinova-Noyes K, Sakurai T, et al. The role of the ankyrin-binding protein Nrcam in node of Ranvier formation. J Neurosci. 2003;23:10032–10039. doi: 10.1523/JNEUROSCI.23-31-10032.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis JQ, Bennett V. Ankyrin binding activity shared by the Neurofascin/L1/Nrcam family of nervous system cell adhesion molecules. J Biol Chem. 1994;269:27163–27166. [PubMed] [Google Scholar]
- 6.Dubois B, Feldman HH, Jacova C, et al. Research criteria for the diagnosis of Alzheimer’s disease: revising the Nincds–Adrda criteria. Lancet Neurol. 2007;6:734–746. doi: 10.1016/S1474-4422(07)70178-3. [DOI] [PubMed] [Google Scholar]
- 7.Finehout EJ, Franck Z, Choe LH, Relkin N, Lee KH. Cerebrospinal fluid proteomic biomarkers for Alzheimer’s disease. Ann Neurol. 2007;61:120–129. doi: 10.1002/ana.21038. [DOI] [PubMed] [Google Scholar]
- 8.Fruttiger M, Calver AR, Richardson WD. Platelet-derived growth factor is constitutively secreted from neuronal cell bodies but not from axons. Curr Biol. 2000;10:1283–1286. doi: 10.1016/s0960-9822(00)00757-0. [DOI] [PubMed] [Google Scholar]
- 9.Gianni D, Zambrano N, Bimonte M, et al. Platelet-derived growth factor induces the beta-gamma-secretase-mediated cleavage of Alzheimer’s amyloid precursor protein through a Src-Rac-dependent pathway. J Biol Chem. 2003;278:9290–9297. doi: 10.1074/jbc.m211899200. [DOI] [PubMed] [Google Scholar]
- 10.Gozal YM, Duong DM, Gearing M, et al. Proteomics analysis reveals novel components in the detergent-insoluble subproteome in Alzheimer’s disease. J Proteome Res. 2009;8:5069–5079. doi: 10.1021/pr900474t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin WS, Sheng JG, Roberts GW, Mrak RE. Interleukin-1 expression in different plaque types in Alzheimer’s disease: significance in plaque evolution. J Neuropathol Exp Neurol. 1995;54:276–281. doi: 10.1097/00005072-199503000-00014. [DOI] [PubMed] [Google Scholar]
- 12.Grossman M, Farmer J, Leight S, et al. Cerebrospinal fluid profile in frontotemporal dementia and Alzheimer’s disease. Ann Neurol. 2005;57:721–729. doi: 10.1002/ana.20477. [DOI] [PubMed] [Google Scholar]
- 13.Lippa CF, Duda JE, Grossman M, et al. Dlb and Pdd boundary issues: diagnosis, treatment, molecular pathology, and biomarkers. Neurology. 2007;68:812–819. doi: 10.1212/01.wnl.0000256715.13907.d3. [DOI] [PubMed] [Google Scholar]
- 14.Lu Z, Kipnis J. Thrombospondin 1—a key astrocyte-derived neurogenic factor. Faseb J. doi: 10.1096/fj.09-150573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moser B, Wolf M, Walz A, Loetscher P. Chemokines: multiple levels of leukocyte migration control. Trends Immunol. 2004;25:75–84. doi: 10.1016/j.it.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Neary D, Snowden JS, Gustafson L, et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–1554. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- 17.Netzer WJ, Dou F, Cai D, et al. Gleevec inhibits beta-amyloid production but not notch cleavage. Proc Natl Acad Sci USA. 2003;100:12444–12449. doi: 10.1073/pnas.1534745100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neumann M, Kwong LK, Lee EB, et al. Phosphorylation of S409/410 of Tdp-43 is a consistent feature in all sporadic and familial forms of Tdp-43 proteinopathies. Acta Neuropathol. 2009;117:137–149. doi: 10.1007/s00401-008-0477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park S, Hong SM, Sung SR, Jung HK. Long-term effects of central leptin and resistin on body weight, insulin resistance, and beta-cell function and mass by the modulation of hypothalamic leptin and insulin signaling. Endocrinology. 2008;149:445–454. doi: 10.1210/en.2007-0754. [DOI] [PubMed] [Google Scholar]
- 20.Peskind ER, Li G, Shofer J, et al. Age and apolipoprotein E*4 allele effects on cerebrospinal fluid beta-amyloid 42 in adults with normal cognition. Arch Neurol. 2006;63:936–939. doi: 10.1001/archneur.63.7.936. [DOI] [PubMed] [Google Scholar]
- 21.Ray S, Britschgi M, Herbert C, et al. Classification and prediction of clinical Alzheimer’s diagnosis based on plasma signaling proteins. Nat Med. 2007;13:1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- 22.Ross MA, Miller RG, Berchert L, et al. Toward earlier diagnosis of amyotrophic lateral sclerosis: revised criteria: Rhcntf Als Study Group. Neurology. 1998;50:768, 772. doi: 10.1212/wnl.50.3.768. [DOI] [PubMed] [Google Scholar]
- 23.Shaw LM, Korecka M, Clark CM, Lee VM, Trojanowski JQ. Biomarkers of neurodegeneration for diagnosis and monitoring therapeutics. Nat Rev Drug Discov. 2007;6:295–303. doi: 10.1038/nrd2176. [DOI] [PubMed] [Google Scholar]
- 24.Shaw LM, Vanderstichele H, Knapik-Czajka M, et al. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinacker P, Mollenhauer B, Bibl M, et al. Heart fatty acid binding protein as a potential diagnostic marker for neurodegenerative diseases. Neurosci Lett. 2004;370:36–39. doi: 10.1016/j.neulet.2004.07.061. [DOI] [PubMed] [Google Scholar]
- 26.Tapiola T, Alafuzoff I, Herukka SK, et al. Cerebrospinal fluid {beta}-amyloid 42 and tau proteins as biomarkers of alzheimer-type pathologic changes in the brain. Arch Neurol. 2009;66:382–389. doi: 10.1001/archneurol.2008.596. [DOI] [PubMed] [Google Scholar]
- 27.Townson JR, Nibbs RJ. Characterization of mouse Ccx-Ckr, a receptor for the lymphocyte-attracting chemokines Teck/Mccl25, Slc/Mccl21 and Mip-3beta/Mccl19: comparison to human CCX-Ckr. Eur J Immunol. 2002;32:1230–1241. doi: 10.1002/1521-4141(200205)32:5<1230::AID-IMMU1230>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 28.Tsuboi Y, Kakimoto K, Nakajima M, et al. Increased hepatocyte growth factor level in cerebrospinal fluid in Alzheimer’s disease. Acta Neurol Scand. 2003;107:81–86. doi: 10.1034/j.1600-0404.2003.02089.x. [DOI] [PubMed] [Google Scholar]
- 29.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied o the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winblad B, Palmer K, Kivipelto M, et al. Mild cognitive impairment-beyond controversies, towards a consensus: report of the international working group on mild cognitive impairment. J Intern Med. 2004;256:240–246. doi: 10.1111/j.1365-2796.2004.01380.x. [DOI] [PubMed] [Google Scholar]
- 31.Youn BS, Yu KY, Oh J, Lee J, Lee TH, Broxmeyer HE. Role of the Cc chemokine receptor 9/Teck interaction in apoptosis. Apoptosis. 2002;7:271–276. doi: 10.1023/a:1015320321511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.