Abstract
The temporal and spatial regulation of histone post-translational modifications is essential for proper chromatin structure and function. The Saccharomyces cerevisiae NuA3 histone acetyltransferase complex modifies the amino-terminal tail of histone H3, but how NuA3 is targeted to specific regions of the genome is not fully understood. Yng1, a subunit of NuA3 and a member of the Inhibitor of Growth (ING) protein family, is required for the interaction of NuA3 with chromatin. This protein contains a C-terminal plant homeodomain (PHD) finger that specifically interacts with lysine 4-trimethylated histone H3 (H3K4me3) in vitro. This initially suggested that NuA3 is targeted to regions bearing the H3K4me3 mark; however, deletion of the Yng1 PHD finger does not disrupt the interaction of NuA3 with chromatin or result in a phenotype consistent with loss of NuA3 function in vivo. In this study, we uncovered the molecular basis for the discrepancies in these data. We present both genetic and biochemical evidence that full-length Yng1 has two independent histone-binding motifs: an amino-terminal motif that binds unmodified H3 tails and a carboxyl-terminal PHD finger that specifically recognizes H3K4me3. Although these motifs can bind histones independently, together they increase the apparent association of Yng1 for the H3 tail.
CHROMATIN is a nucleoprotein structure consisting of DNA, histones, and nonhistone proteins. It has been found to regulate numerous processes including transcription, DNA replication, and repair. Histones are subjected to numerous post-translational modifications (PTMs), including acetylation and methylation, that are proposed to act as docking sites for various chromatin-remodeling complexes (Peterson and Laniel 2004). As such, histone post-translational modification is one mechanism for modifying chromatin structure.
One family of proteins that specifically bind histone PTMs is the Inhibitor of Growth (ING) group of proteins. These proteins share highly conserved carboxyl-terminal plant homeodomain (PHD) fingers and less conserved amino-terminal regions (He et al. 2005). The PHD fingers of ING proteins specifically interact with the amino-terminal tail of histone H3 that has been trimethylated on lysine 4 (H3K4me3) (Martin et al. 2006a; Shi et al. 2006). The first ING protein discovered was ING1 (p33), which was later shown to act as a tumor suppressor (Garkavtsev et al. 1996). The ING family is now thought to include five proteins in humans: hING1, hING2, hING3, hING4, and hING5 (He et al. 2005). ING1 and ING2 are found in related histone deacetylase complexes, whereas ING3–5 are associated with MYST family histone acetyltransferases (HATs) (Skowyra et al. 2001; Kuzmichev et al. 2002; Doyon et al. 2006). The ING proteins interact with p53, regulate transcription, and promote cell senescence and death when overexpressed (Garkavtsev et al. 1998; Nourani et al. 2003).
There are three ING proteins in the yeast Saccharomyces cerevisiae, including Yng1, Yng2, and Pho23 (Loewith et al. 2000). Each of these proteins is found associated with a different multiprotein complex that post-translationally modifies histones. Yng1 and Yng2 are associated with the NuA3 and NuA4 HAT complexes, respectively (Loewith et al. 2000, 2001; Choy et al. 2001; Howe et al. 2002). In contrast, Pho23 is found associated with the Rpd3L HDAC complex (Loewith et al. 2001; Carrozza et al. 2005). Although the ING proteins are not required for the structural integrity of their respective complexes, both Yng1 and Yng2 are required for the ability of their complexes to bind and acetylate nucleosome substrates (Loewith et al. 2000; Choy et al. 2001; Howe et al. 2002). Additionally, loss of PHO23 causes a reduction in the HDAC activity of the Rpd3L complex (Loewith et al. 2001). These data suggest that members of the ING protein family serve general roles in permitting various enzymatic activities to interact with nucleosomes and modify histone tails.
Similar to their mammalian counterparts, the yeast ING proteins share a conserved PHD finger that preferentially interacts with H3K4me3-modified histones (Martin et al. 2006a; Shi et al. 2006, 2007; Taverna et al. 2006). However, while the ING proteins are important for the function of their cognate complexes, the PHD fingers are surprisingly dispensable (Nourani et al. 2001; Howe et al. 2002). As a result, whether these highly conserved methyl–histone binding domains actually have a role in targeting complexes to specific regions of the genome is in question. In this study we have used both biochemical and genetic approaches to show that Yng1 sequences outside of the PHD finger can directly interact with the histone H3 tail and this interaction is not dependent on the presence of H3K4me3. These results demonstrate that Yng1 possesses bivalent histone-binding motifs that function together to increase the apparent association of Yng1 for the histone H3 tail.
MATERIALS AND METHODS
Yeast strains, plasmids, and genetic methods:
All strains used in this study are isogenic to S288C. Yeast culture and genetic manipulations were carried out using standard protocols (Ausubel 1987). Genomic deletions were verified by PCR analysis and whole-cell extracts were generated as previously described (Ausubel 1987; Kushnirov 2000). The plasmids pGALYNG1, pGALYNG1ΔPHD, pGALYNG1W180A, pGALYNG1Δ2-28, pGALYNG2, and pGALPHO23 were generated by cloning sequences encoding YNG1, YNG2, and PHO23 into the BamHI sites of pGAL.415, pGAL.425, or pGAL.416 (Mumberg et al. 1994). pGALYNG1HA3, pGALYNG2HA3, and pGALPHO23HA3 were generated by replacing the CYC1 terminator of the above plasmids with an HA3-CYC1 terminator cassette (Howe et al. 2002). To construct the PHD finger swap plasmids we introduced silent mutations into codons 145 and 146 of YNG1, introducing an AatII restriction site. We then PCR amplified codons 147–219 of Yng1, codons 214–282 of Yng2, and codons 272–330 of Pho23 as AatII/NotI fragments and cloned them into the AatII/NotI-digested plasmid. To construct pHDA1 the HDA1 open reading frame, along with 230 bp of promoter and 651 bp of terminator sequences, was amplified from genomic DNA and cloned into pRS415 using XhoI and SacI restriction sites.
Synthetic dosage resistance genomewide screen:
The synthetic genetic array (SGA) starting strain Y7092 (MATα can1Δ∷STE2pr-Sp-his5 lypΔ1 his3Δ1 leu2Δ0 met15Δ ura3Δ0) was transformed with pGALYNG1. The resulting query strain was mated to the MATa deletion mutant array. SGA methodology, previously described for a plasmid-based synthetic dosage resistance screen (Martin et al. 2006a), was used with the following modifications: (i) medium lacking uracil was used to maintain the plasmid, and (ii) hits were scored against strains containing pGALYNG1 grown on dextrose. The screen was performed in triplicate and all hits were confirmed using traditional transformation and dilution plating. Confirmed hits were followed up with Western blots with anti-HA antibodies (Santa Cruz, catalog no. sc-57529) to determine the levels of Yng1HA3 expression and anti-H3K4me3 (Active Motif, catalog no. 39159) antibodies to check for changes in H3K4 methylation.
Peptide pull-down assays:
Biotinylated histone peptides were synthesized by Anaspec. For pull-down assays, ∼1 μg of GST-tagged recombinant protein was incubated with 1 μg of biotinylated histone peptides in binding buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.05% NP-40) overnight at 4° with rotation. After a 1-hr incubation with streptavidin sepharose beads (Amersham Biosciences) and extensive washing, bound proteins were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and Western blotting with anti-GST antibodies (Santa Cruz, catalog no. sc-459).
RESULTS
Sequences outside of the Yng1 PHD finger are required for growth inhibition by YNG1 overexpression:
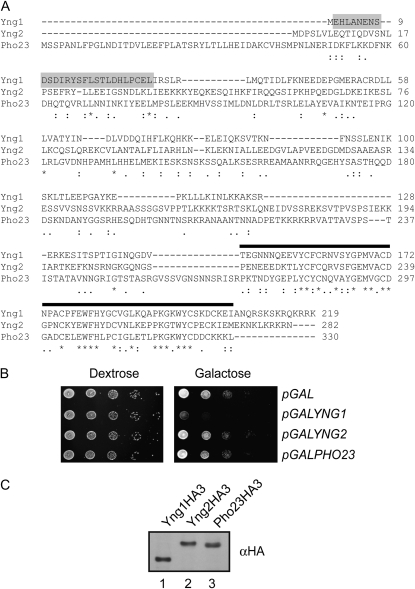
We have previously reported that full-length YNG1 inhibits yeast growth when overexpressed and this effect requires both the Yng1 PHD finger and histone H3K4 methylation (Martin et al. 2006a). We hypothesized that growth inhibition is due to excess levels of Yng1 sterically hindering the access of other factors to the H3 tail. If this is true, then other proteins that show similar methyl–histone binding properties should be toxic when expressed at similar levels. The PHD fingers of the three yeast ING proteins, Yng1, Yng2, and Pho23, share significant sequence similarity and H3K4me3 binding properties (Figure 1A) (Shi et al. 2006, 2007). It is therefore expected that YNG2 and PHO23 would inhibit growth when overexpressed as well. To address this question we fused the YNG2 and PHO23 open reading frames to the galactose inducible promoter of the GAL1 gene on a URA3-based centromeric plasmid (pGALYNG2, pGALPHO23). The resulting plasmids were transformed into wild-type cells and plated in 10-fold dilution series on media lacking uracil and containing dextrose or galactose as carbon sources (Figure 1B). Vectors carrying only the GAL1 promoter alone (pGAL) and the GALYNG1 construct were used as negative and positive controls for growth inhibition, respectively. Consistent with previously published results, cells carrying pGALYNG1 grew on dextrose, but failed to grow on galactose, confirming that overexpression of YNG1 inhibits cell growth. In contrast, yeast carrying empty vector (pGAL), pGALYNG2, or pGALPHO23 grew well on both dextrose and galactose, indicating that neither overexpression of YNG2 nor that of PHO23 is inhibitory to cell growth (Figure 1B). To verify that YNG2 and PHO23 are expressed at similar levels to YNG1, we introduced sequences encoding HA tags into each of our pGAL plasmids such that each ING protein would be expressed with a C-terminal triple-HA tag. Fortuitously, HA-tagged Yng1 is not toxic when overexpressed, allowing us to compare the levels of all of the ING protein in cells grown on galactose. Figure 1C demonstrates that Yng1HA3, Yng2HA3, and Pho23HA3 expressed from a GAL1 promoter are present in similar levels.
Figure 1.—
Overexpression of YNG1, but not YNG2 or PHO23, inhibits yeast cell growth. (A) CLUSTAL 2.0.12 multiple-sequence alignment of the ING proteins in S. cerevisiae. “*” indicates that the residues in that column are identical in all sequences in the alignment. “:” and “.” mean that conserved and semiconserved substitutions are observed, respectively. Sequences corresponding to the PHD finger are indicated by a solid bar, and amino-terminal residues important for unmodified tail binding are shaded. (B) Tenfold serial dilutions of a wild-type yeast strain containing the indicated low-copy plasmids were plated on synthetic leucine drop-out medium with either dextrose or galactose as a carbon source and incubated at 30° for 3 days. (C) Yeast YNG1, YNG2, and PHO23 are equivalently expressed in cells. Shown is an αHA Western blot analysis of whole-cell extracts from strains expressing HA-tagged versions of Yng1, Yng2, and Pho23 from a GAL1 promoter during growth in galactose.
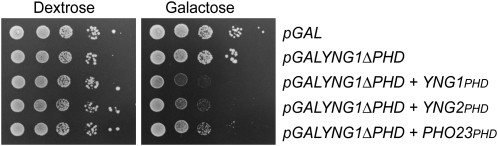
The fact that overexpression of YNG1 inhibits growth, while YNG2 and PHO23 do not, was surprising as we have previously shown that the level of growth inhibition by YNG1 correlates with the methyl–histone binding ability of the Yng1 PHD finger and that the PHD fingers of all ING proteins share similar methyl–histone binding specificities (Martin et al. 2006a; Shi et al. 2006, 2007). There are two possible explanations as to why this is the case. First, it is feasible that there is something distinct about the Yng1 PHD finger that makes it toxic when overexpressed. Indeed, it has been previously shown that the Yng1 PHD finger binds methylated peptides with a slightly higher affinity than the Yng2 or Pho23 fingers (Shi et al. 2007). A second possibility is that while the PHD finger is required for the growth-inhibiting properties of Yng1, additional sequences outside of the PHD finger may also be required. These sequences are significantly divergent from those in Yng2 and Pho23 (Figure 1A). To test these possibilities we replaced the PHD finger of Yng1 with the PHD finger of Yng2 or Pho23 and tested whether these chimeric proteins inhibit growth of yeast cells when overexpressed from a GAL1 promoter. While Yng1 lacking the PHD finger does not inhibit growth when overexpressed, addition of either the Yng2 or the Pho23 PHD finger restores the growth-inhibiting properties to Yng1 (Figure 2). The fact that the Yng2 and Pho23 PHD fingers do not inhibit growth when fused to their native amino termini, but inhibit growth when fused to Yng1, strongly suggests that the growth inhibition by YNG1 overexpression is dependent on the amino terminus of Yng1, in addition to the PHD finger.
Figure 2.—
The Yng2 and Pho23 PHD fingers confer inhibition of growth when fused to the amino-terminal domain of Yng1. Tenfold serial dilutions of a wild-type yeast strain transformed with the indicated high-copy plasmids were plated on synthetic uracil drop-out medium containing either dextrose or galactose as a carbon source and incubated at 30° for 3 days.
Full-length Yng1 binds unmodified chromatin:
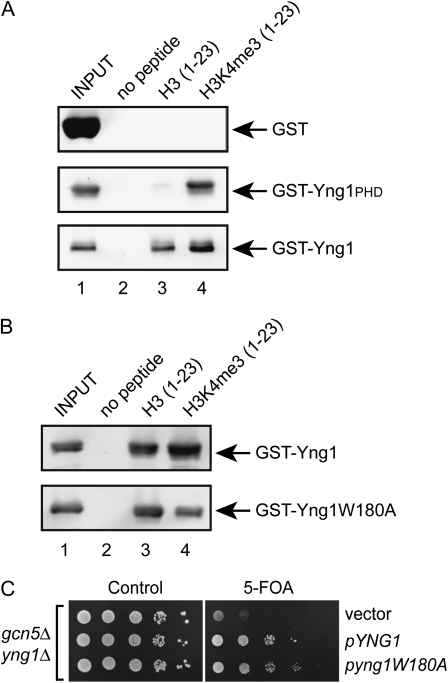
The results of this and previously published work indicate that the PHD finger of Yng1 is necessary, but not sufficient to confer growth inhibition when overexpressed. Instead, residues within the amino-terminal region of Yng1 are equally important. This result is reminiscent of other work from our lab demonstrating that while full-length Yng1 is important for NuA3 function in vivo, the PHD finger is dispensable (Howe et al. 2002). Additionally, we have shown that deletion of YNG1 or the H3 tail disrupts the interaction of NuA3 with chromatin, but we have been unable to show a similar effect due to loss of the Yng1 PHD finger alone (Martin et al. 2006a,b). Collectively, these data support the idea that Yng1 may contain bivalent chromatin-binding abilities: a PHD finger that specifically interacts with H3K4me3 and an amino-terminal motif that interacts with some other part of the H3 tail. To test this hypothesis we bacterially expressed and purified full-length Yng1 and the Yng1 PHD finger with amino-terminal GST tags. The purified proteins were incubated with biotinylated peptides corresponding to the H3 tail (residues 1–23) that were either unmodified or trimethylated at K4. The peptides were immobilized on streptavidin sepharose and the bound proteins detected by Western blotting for GST. Figure 3A shows that both full-length Yng1 and the Yng1 PHD finger alone are able to interact with trimethylated peptides (lane 4). More importantly, however, full-length Yng1, but not the PHD finger alone, is able to interact with unmodified peptides (lane 3). The fact that full-length Yng1 can bind unmodified tails, while the PHD finger cannot, suggests that residues within the amino terminus of Yng1 are important for recognition of an unmodified H3 tail. These results confirm our hypothesis that Yng1 has two histone-binding modules. The fact that full-length Yng1 binds H3K4me3 tails better than unmodified tails (Figure 3A, compare lanes 3 and 4) suggests that these modules work together to increase the strength of interaction of Yng1 for the histone H3 tail.
Figure 3.—
Full-length Yng1 binds unmodified histone H3 tails independently of the PHD finger. (A and B) Histone tail peptide-binding assays were performed with the indicated immobilized peptides and purified GST fusion proteins. Shown are αGST Western blots of precipitated material. Input lanes contain 10% of the proteins used for the pulldown. (C) A gcn5Δ yng1Δ strain expressing GCN5 from a URA3-based plasmid was transformed with the indicated plasmids, plated on synthetic complete medium with and without 5-FOA, and incubated at 30° for 3 days.
As a final confirmation of the presence of an additional histone-binding activity in Yng1, we mutated a residue within the Yng1 PHD finger that is critical for methyl–histone recognition and tested for binding to both unmodified and methylated H3 tail peptides. Yng1 W180 forms part of an aromatic cage that specifically interacts with H3K4me3 and substitution of this amino acid with an alanine disrupts H3K4me3 binding and alleviates Yng1 growth inhibition (Martin et al. 2006a). We expressed full-length Yng1 with an alanine substitution of W180 and tested the binding of this protein to both unmodified and methylated H3 tail peptides. Figure 3B shows that the mutant form of Yng1 can still interact with unmodified H3 tails, but, as expected, lacks the preferential binding to the peptide bearing H3K4me3. Finally, we tested whether this form of Yng1 could rescue a phenotype associated with loss of Yng1. Deletion of YNG1 results in no significant phenotypes in a wild-type strain, but when deleted in a gcn5Δ strain results in synthetic sickness. This phenotype is due to a redundancy of histone H3 acetylation and acetylation of Rsc4, a subunit of the RSC chromatin remodeling complex, in maintaining cell viability. Introduction of wild-type Yng1 into gcn5Δ yng1Δ restores growth, as does Yng1W180A, reiterating the fact that the PHD finger is not essential for NuA3 function in vivo (Figure 3C). These data explain why loss of the PHD finger alone does not disrupt NuA3 function since the interaction of NuA3 with chromatin is presumably still maintained by the interaction of the Yng1 amino terminus with the H3 tail.
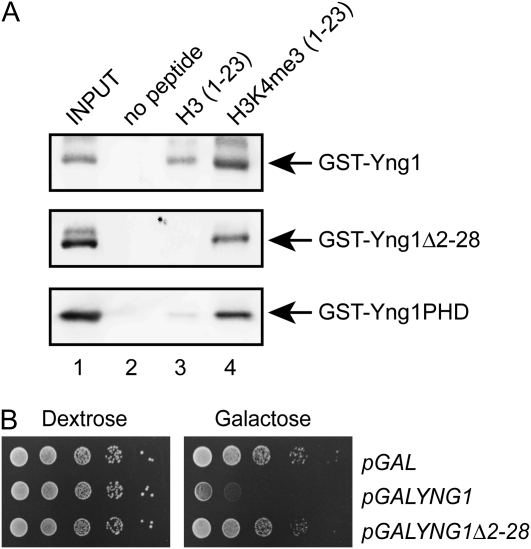
We next attempted to map the region of the amino terminus of Yng1 that binds unmodified peptides. To this end we generated deletions of both the amino and the carboxyl regions. Unfortunately, deletion of the PHD finger rendered the protein unstable and we were unable to recover any protein following bacterial expression. In contrast, deletion of residues 2–28 in the amino terminus in Yng1 did not affect protein stability and completely abolished binding to unmodified tails (Figure 4A). Concomitant with this, Yng1Δ2–28 is not toxic when overexpressed (Figure 4B). These data indicate that the first 28 residues of Yng1 are important for recognition of unmodified histone tails. Moreover, we have been able to separate the unmodified-tail and methylated-tail binding activities of Yng1 and have shown that each can bind histones independently of the other (Figures 3B and 4A).
Figure 4.—
The amino terminus of Yng1 is required for binding of unmodified histone H3 tails. (A) Histone tail peptide binding assays were performed with the indicated immobilized peptides and purified GST fusion proteins. Shown are αGST Western blots of precipitated material. Input lanes contain 10% of the proteins used for the pulldown. (B) Tenfold serial dilutions of a wild-type yeast strain containing the indicated low-copy plasmids were plated on synthetic uracil drop-out medium containing either dextrose or galactose as a carbon source and incubated at 30° for 3 days.
A synthetic dosage resistance screen identifies genes that regulate the binding of Yng1 to chromatin:
Previously we identified the PHD finger of Yng1 as a methyl–histone binding domain, using a synthetic dosage resistance screen (Martin et al. 2006a). In this screen, overexpression of YNG1 in the yeast haploid deletion collection identified a deletion in LGE1 as conferring resistance to YNG1 overexpression. LGE1 is required for methylation of H3K4 (Hwang et al. 2003); however, the screen failed to identify numerous other genes known to be required for H3K4me3. Subsequent testing confirmed that all mutants with defects in H3K4me3 are resistant to YNG1 overexpression and thus our initial synthetic dosage resistance screen had a very high false negative rate (Martin et al. 2006a). With the advent of new instrumentation for SGA analysis, we repeated the screen in an attempt to uncover additional information on the regulation of Yng1 binding to unmodified tails. A pGALYNG1 vector expressing the URA3 marker was introduced to the query strain, which was then crossed with the set of ∼5000 nonessential gene deletion mutants. The screen was performed in a 1536-plate format to reduce the plate border effects. The synthetic dosage phenotype was scored by comparing the growth of cells carrying pGALYNG1 on media lacking uracil with either glucose or galactose as the carbon source. Using this approach we identified multiple genes that conferred resistance to YNG1 overexpression (Table 1). Expectedly, we found a large group of genes that encode components of the Set1/COMPASS histone methyltransferase complex, including BRE2, SDC1, SPP1, SWD1, and SWD3 (Miller et al. 2001; Krogan et al. 2002). We also identified deletions of BRE1 and RAD6, genes required for ubiquitination of H2B, and RTF1, CDC73, and LEO1, which encode three of the five components of the PAF complex (Robzyk et al. 2000; Miller et al. 2001; Krogan et al. 2002). Both H2B ubiquitination and the PAF complex are required for H3K4 methylation (Dover et al. 2002; Krogan et al. 2003; Morillon et al. 2005). Finally, we identified CTK2 and CTK3, which encode two of the three components of the Ctk1 protein kinase complex that phosphorylates the C-terminal domain (CTD) of RNA Polymerase II (Sterner et al. 1995). Loss of Ctk1 results in spreading of H3K4 trimethylation across the coding regions of genes, presumably acting as a sink for binding of excess Yng1 (Wood et al. 2007; Xiao et al. 2007). Given these results we were confident that this synthetic dosage resistance screen was useful for identifying genes required for growth inhibition due to YNG1 overexpression.
TABLE 1.
Genes required for growth inhibition due to YNG1 overexpression
| Complex | Required for H3K4 trimethylation? | Gene identified |
|---|---|---|
| COMPASS complex | Yes | BRE2 |
| SDC1 | ||
| SPP1 | ||
| SWD1 | ||
| SWD3 | ||
| Rad6-Bre1 complex | Yes | BRE1 |
| RAD6 | ||
| PAF complex | Yes | RTF1 |
| CDC73 | ||
| LEO1 | ||
| Ctk1 protein kinase complex | Yesa | CTK2CTK3 |
| HDA histone deacetylase complex | No | HDA1HDA2 |
| HDA3 |
The Ctk1 protein kinase complex restricts the spread of H3K4 trimethylation into the 3′ ends of genes.
Deletion of HDA1 confers resistance to YNG1 overexpression:
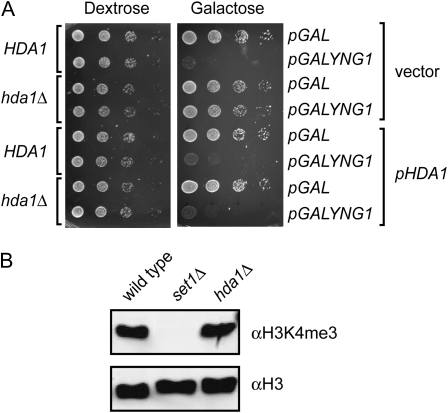
In addition to genes required for histone H3K4 methylation, we also identified deletions of HDA1, HDA2, and HDA3 as conferring resistance to YNG1 overexpression (Table 1). Hda1, Hda2, and Hda3 compose the tetrameric HDA histone deacetylase complex, which is composed of Hda1 as a catalytic subunit, Hda3, and two molecules of Hda2 (Carmen et al. 1996; Wu et al. 2001). The HDA complex has been shown to exhibit activity toward histones H2B, H3, and H4, and deletion of HDA1 results in increased acetylation of these histones. To confirm that deletion of HDA1 confers resistance to YNG1 overexpression, we reintroduced HDA1 with its native promoter on a LEU2-based plasmid (pHDA1) into an hda1Δ mutant. Wild-type (HDA1) and hda1Δ strains transformed with vector alone were used as controls. The resulting strains were tested for resistance to YNG1 overexpression by transforming with either pGAL or pGALYNG1 and spotting in 10-fold dilution series onto plates lacking leucine and uracil and containing dextrose or galactose as the carbon source. Yeast bearing a HDA1 deletion and pGALYNG1 grew on both dextrose and galactose, indicating that deletion of HDA1 confers resistance to YNG1 overexpression. When HDA1 was provided on a plasmid, hda1Δ cells grew in the presence of pGAL on both dextrose and galactose; however, they failed to grow when carrying the pGALYNG1 plasmid, indicating that a functional HDA complex is required for YNG1-mediated inhibition of growth (Figure 5A). The two obvious modes of resistance to YNG1 overexpression are a GAL1-transcriptional activation defect, which reduces the levels of Yng1 in the cell, or alternatively disruption of the histone H3K4 methylation pathway. To test whether hda1Δ strains have a GAL1 transcriptional defect, we overexpressed the HA-tagged version of Yng1 in wild-type and hda1Δ cells. The hda1Δ cells showed no decrease in the levels of Yng1HA, when normalized to levels of H3, indicating that a GAL1 transcription defect is not a likely cause of resistance (data not shown). To test the possibility that deletion of HDA1 causes a decrease in methylation we subjected yeast whole-cell extracts to Western blotting with anti-H3K4me3 antibodies. We found that deletion of HDA1 did not have an effect on histone methylation levels in cells, therefore eliminating it as a mechanism of resistance (Figure 5B).
Figure 5.—
Deletion of HDA1 confers resistance to inhibition of growth by YNG1 overexpression. (A) Tenfold serial dilutions of HDA1 and hda1Δ yeast strains containing the indicated plasmids were plated on synthetic uracil/leucine drop-out medium containing either dextrose or galactose as a carbon source and incubated at 30° for 3 days. (B) Western blot of the whole-cell extracts from wild type, set1Δ, and hda1Δ strains. The blot was probed with αH3K4me3 and αH3 antibodies.
Histone H3 acetylation does not affect Yng1 binding to chromatin:
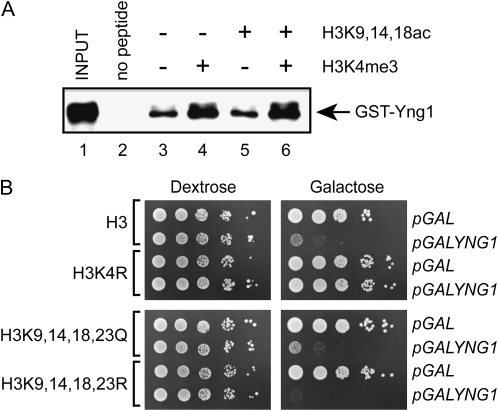
Deletion of HDA1 results in increased acetylation of histone H3 (Carmen et al. 1996; Wu et al. 2001). This led us to ask whether Yng1 binding to chromatin might be negatively regulated by acetylation of the histone H3 tail. To test this hypothesis we assayed binding of full-length Yng1 to peptides with and without H3K4me3 and/or acetylation of lysines 9, 14, and 18. In Figure 6A we show that full-length Yng1 binds to both unmodified and acetylated peptides (lanes 3 and 5). Although the apparent association of Yng1 increases for methylated peptides as compared to unmodified (compare lanes 3 to 4), we did not observe any change in binding to methylated vs. methylated acetylated peptides (compare lanes 4–6).
Figure 6.—
Acetylation does not alter the association of Yng1 with the histone H3 tail. (A) Histone peptide-binding assays were performed with the indicated biotinylated peptides and purified GST-Yng1. Shown is an αGST Western blot of precipitated material. Input lane contains 10% of the protein amount used for the pulldowns. (B) Tenfold serial dilutions of yeast strains, expressing wild-type and mutant versions of histone H3, containing the indicated plasmids, were plated on synthetic uracil drop-out medium containing either dextrose or galactose as a carbon source and incubated at 30° for 3 days.
To further confirm that the histone acetylation does not block Yng1 binding, we mutated four of the acetylated lysines within the H3 tail to glutamine and tested whether this conferred resistance to Yng1 growth inhibition. Figure 6B shows that cells expressing H3 K9, 14, 18, and 23Q showed similar growth to wild type when Yng1 is overexpressed. A strain expressing H3 with an arginine substitution of lysine 4 does rescue growth, consistent with growth inhibition being dependent on methylation of this residue. Collectively these results suggest that the HDA complex does not directly regulate the binding of Yng1 to chromatin.
If histone H3 acetylation does not prevent Yng1 binding, then why does disruption of the HDA complex result in resistance to Yng1 overexpression? The fact that Yng1 contains two independent histone-binding motifs suggests the intriguing possibility that Yng1 could simultaneously bind two H3 tails, resulting in chromatin compaction. If this is the case, then the hyperacetylation associated with loss of the HDA complex could result in decompaction of chromatin, alleviating the toxic effects of Yng1 overexpression. Consistent with this, mutation of H3 K9, 14, 18, and 23 to arginine, which would be expected to enhance chromatin compaction, renders Yng1 more toxic (Figure 6B).
DISCUSSION
NuA3 is a histone H3-specific HAT complex consisting of Sas3, Nto1, Eaf6, Taf14, and Yng1 (John et al. 2000; Howe et al. 2001, 2002; Taverna et al. 2006). Genomewide localization studies have shown that NuA3 is enriched throughout coding sequences, with a modest increase within the 5′ half of genes (Taverna et al. 2006). How this complex is targeted to these regions is unclear. We have previously shown that Yng1 is required for NuA3 association with chromatin in vitro and were interested in determining which part of this protein is required for this function (Howe et al. 2002). To this end, we developed two independent assays for Yng1 function in vivo: rescue of an yng1Δ phenotype and inhibition of growth due to overexpression of YNG1 from a GAL1 promoter. Surprisingly, conflicting results were generated when using these two assays. Yng1 lacking the PHD finger rescues a yng1Δ phenotype, suggesting that the PHD finger is dispensable for NuA3 function in vivo (Howe et al. 2002). In contrast, the Yng1 PHD finger is essential for the growth-inhibiting properties of this protein when overexpressed (Martin et al. 2006a). In this study we have been able to reconcile these conflicting observations and generate insight into how this complex is localized to transcribed regions. We show that Yng1 contains two histone-binding motifs: an amino-terminal motif that binds unmodified histone H3 tails and a carboxyl-terminal PHD finger that specifically recognizes H3K4me3. Moreover, our results suggest that the presence of both regions enhances binding of Yng1, and presumably NuA3, to the histone H3 tail.
One phenotype associated with yng1Δ is a synthetic growth defect with loss of GCN5, which encodes an alternate histone H3-specific HAT (Howe et al. 2002). We know that this phenotype is due specifically to loss of NuA3 acetytransferase activity because mutation of the acetyl-CoA binding site of Sas3, the catalytic subunit of NuA3, is lethal in a gcn5Δ background (Howe et al. 2001). Full-length Yng1, Yng1ΔPHD, and Yng1 lacking residues important for methyl–histone recognition fully rescue growth of a gcn5Δ yng1Δ mutant, indicating that targeting of NuA3 to H3K4me3 is not essential for NuA3 HAT activity in vivo (Howe et al. 2002). In this study we demonstrated that amino acids in the amino-terminal region of Yng1 can bind unmodified histone tails and thus Yng1 may be simply required to dock NuA3 on its substrate regardless of the methylation status. We cannot, however, exclude the possibility that the Yng1 PHD finger–H3K4me3 interaction strengthens the binding of the NuA3 complex to chromatin. Indeed the enhanced binding of full-length Yng1 to H3K4me3 vs. unmodified peptide may explain the slight enrichment of NuA3 at the 5′ ends of genes (Taverna et al. 2006).
Overexpression of YNG1 inhibits growth of both wild-type and sas3Δ strains, indicating that this phenotype is independent of NuA3 HAT activity (Martin et al. 2006a). This synthetic dosage lethality is dependent on the presence of both the Yng1 amino terminus and the PHD finger. While we do not fully understand why YNG1 overexpression inhibits growth, we can conclusively state that toxicity is due to the binding of excess Yng1 to the H3 tail. On the basis of the results of this study, we hypothesize that while both the PHD finger and the amino terminus of Yng1 can bind the H3 tail independently, loss of either region weakens the interaction to the extent that the protein is no longer toxic when overexpressed. Whether the enhanced binding activity of the two motifs in combination has a relevant function when expressed at normal levels is unknown, but, as mentioned above, it may explain the modest accumulation of Yng1 observed at the 5′ end of genes (Taverna et al. 2006).
This study raises a number of interesting questions. First, if Yng1 can bind to unmodified tails, why did genome-localization studies show depletion of Yng1 in intergenic regions (Taverna et al. 2006)? We feel that the most likely explanation is that the intergenic regions are depleted for histones and thus have fewer sites for NuA3 to bind (Lee et al. 2004, 2007; Yuan et al. 2005; Albert et al. 2007; Whitehouse et al. 2007). Second, why does loss of the HDA complex confer resistance to YNG1 overexpression? We have eliminated the obvious explanation that acetylation negatively regulates Yng1 binding to chromatin. Interestingly, mutation of the acetylated lysines within the H3 tail to constitutively charged residues enhances growth inhibition by Yng1, which is consistent with the idea that Yng1 may be compacting chromatin. The exact mechanism behind the regulation of Yng1 binding to chromatin, as well as the targeting of NuA3, will be a subject of future studies.
Acknowledgments
We gratefully acknowledge Or Gozani and Fred Winston for providing yeast strains and plasmids. Support for this work was provided by grants to L.J.H. and C.J.R.L. from the Canadian Institutes of Health Research. L.J.H. and C.J.R.L. are Canadian Institutes of Health Research New Investigators and Scholars of the Michael Smith Foundation for Health Research. C.J.R.L. is a Tula Foundation Investigator.
References
- Albert, I., T. N. Mavrich, L. P. Tomsho, J. Qi, S. J. Zanton et al., 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446 572–576. [DOI] [PubMed] [Google Scholar]
- Ausubel, F. M., 1987. Current Protocols in Molecular Biology. Greene Publishing Associates and Wiley-Interscience/John Wiley & Sons, New York.
- Carmen, A. A., S. E. Rundlett and M. Grunstein, 1996. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J. Biol. Chem. 271 15837–15844. [DOI] [PubMed] [Google Scholar]
- Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson et al., 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123 581–592. [DOI] [PubMed] [Google Scholar]
- Choy, J. S., B. T. Tobe, J. H. Huh and S. J. Kron, 2001. Yng2p-dependent NuA4 histone H4 acetylation activity is required for mitotic and meiotic progression. J. Biol. Chem. 276 43653–43662. [DOI] [PubMed] [Google Scholar]
- Dover, J., J. Schneider, M. A. Tawiah-Boateng, A. Wood, K. Dean et al., 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277 28368–28371. [DOI] [PubMed] [Google Scholar]
- Doyon, Y., C. Cayrou, M. Ullah, A. J. Landry, V. Cote et al., 2006. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 21 51–64. [DOI] [PubMed] [Google Scholar]
- Garkavtsev, I., A. Kazarov, A. Gudkov and K. Riabowol, 1996. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat. Genet. 14 415–420. [DOI] [PubMed] [Google Scholar]
- Garkavtsev, I., I. A. Grigorian, V. S. Ossovskaya, M. V. Chernov, P. M. Chumakov et al., 1998. The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control. Nature 391 295–298. [DOI] [PubMed] [Google Scholar]
- He, G. H., C. C. Helbing, M. J. Wagner, C. W. Sensen and K. Riabowol, 2005. Phylogenetic analysis of the ING family of PHD finger proteins. Mol. Biol. Evol. 22 104–116. [DOI] [PubMed] [Google Scholar]
- Howe, L., D. Auston, P. Grant, S. John, R. G. Cook et al., 2001. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 15 3144–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, L., T. Kusch, N. Muster, R. Chaterji, J. R. Yates, 3rd et al., 2002. Yng1p modulates the activity of Sas3p as a component of the yeast NuA3 histone acetyltransferase complex. Mol. Cell. Biol. 22 5047–5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, W. W., S. Venkatasubrahmanyam, A. G. Ianculescu, A. Tong, C. Boone et al., 2003. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 11 261–266. [DOI] [PubMed] [Google Scholar]
- John, S., L. Howe, S. T. Tafrov, P. A. Grant, R. Sternglanz et al., 2000. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 14 1196–1208. [PMC free article] [PubMed] [Google Scholar]
- Krogan, N. J., J. Dover, S. Khorrami, J. F. Greenblatt, J. Schneider et al., 2002. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 277 10753–10755. [DOI] [PubMed] [Google Scholar]
- Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt et al., 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11 721–729. [DOI] [PubMed] [Google Scholar]
- Kushnirov, V. V., 2000. Rapid and reliable protein extraction from yeast. Yeast 16 857–860. [DOI] [PubMed] [Google Scholar]
- Kuzmichev, A., Y. Zhang, H. Erdjument-Bromage, P. Tempst and D. Reinberg, 2002. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol. Cell. Biol. 22 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl and J. D. Lieb, 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36 900–905. [DOI] [PubMed] [Google Scholar]
- Lee, W., D. Tillo, N. Bray, R. H. Morse, R. W. Davis et al., 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 39 1235–1244. [DOI] [PubMed] [Google Scholar]
- Loewith, R., M. Meijer, S. P. Lees-Miller, K. Riabowol and D. Young, 2000. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol. Cell. Biol. 20 3807–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith, R., J. S. Smith, M. Meijer, T. J. Williams, N. Bachman et al., 2001. Pho23 is associated with the Rpd3 histone deacetylase and is required for its normal function in regulation of gene expression and silencing in Saccharomyces cerevisiae. J. Biol. Chem. 276 24068–24074. [DOI] [PubMed] [Google Scholar]
- Martin, D. G., K. Baetz, X. Shi, K. L. Walter, V. E. MacDonald et al., 2006. a The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol. Cell. Biol. 26 7871–7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, D. G., D. E. Grimes, K. Baetz and L. Howe, 2006. b Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin. Mol. Cell. Biol. 26 3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, T., N. J. Krogan, J. Dover, H. Erdjument-Bromage, P. Tempst et al., 2001. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. USA 98 12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon, A., N. Karabetsou, A. Nair and J. Mellor, 2005. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell 18 723–734. [DOI] [PubMed] [Google Scholar]
- Mumberg, D., R. Muller and M. Funk, 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourani, A., Y. Doyon, R. T. Utley, S. Allard, W. S. Lane et al., 2001. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell. Biol. 21 7629–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourani, A., L. Howe, M. G. Pray-Grant, J. L. Workman, P. A. Grant et al., 2003. Opposite role of yeast ING family members in p53-dependent transcriptional activation. J. Biol. Chem. 278 19171–19175. [DOI] [PubMed] [Google Scholar]
- Peterson, C. L., and M. A. Laniel, 2004. Histones and histone modifications. Curr. Biol. 14 R546–R551. [DOI] [PubMed] [Google Scholar]
- Robzyk, K., J. Recht and M. A. Osley, 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287 501–504. [DOI] [PubMed] [Google Scholar]
- Shi, X., T. Hong, K. L. Walter, M. Ewalt, E. Michishita et al., 2006. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X., I. Kachirskaia, K. L. Walter, J. H. Kuo, A. Lake et al., 2007. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 282 2450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra, D., M. Zeremski, N. Neznanov, M. Li, Y. Choi et al., 2001. Differential association of products of alternative transcripts of the candidate tumor suppressor ING1 with the mSin3/HDAC1 transcriptional corepressor complex. J. Biol. Chem. 276 8734–8739. [DOI] [PubMed] [Google Scholar]
- Sterner, D. E., J. M. Lee, S. E. Hardin and A. L. Greenleaf, 1995. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol. Cell. Biol. 15 5716–5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna, S. D., S. Ilin, R. S. Rogers, J. C. Tanny, H. Lavender et al., 2006. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell 24 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse, I., O. J. Rando, J. Delrow and T. Tsukiyama, 2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450 1031–1035. [DOI] [PubMed] [Google Scholar]
- Wood, A., A. Shukla, J. Schneider, J. S. Lee, J. D. Stanton et al., 2007. Ctk complex-mediated regulation of histone methylation by COMPASS. Mol. Cell. Biol. 27 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., A. A. Carmen, R. Kobayashi, N. Suka and M. Grunstein, 2001. HDA2 and HDA3 are related proteins that interact with and are essential for the activity of the yeast histone deacetylase HDA1. Proc. Natl. Acad. Sci. USA 98 4391–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, T., Y. Shibata, B. Rao, R. N. Laribee, R. O'Rourke et al., 2007. The RNA polymerase II kinase Ctk1 regulates positioning of a 5′ histone methylation boundary along genes. Mol. Cell. Biol. 27 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, G. C., Y. J. Liu, M. F. Dion, M. D. Slack, L. F. Wu et al., 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309 626–630. [DOI] [PubMed] [Google Scholar]