Abstract
APC/CCdh1 plays a key role in mitotic exit and has essential targets in the G1 phase; however, these mechanisms are poorly understood. In this report, we provide evidence that damaged DNA-binding protein 1 (DDB1) is capable of binding the WD40 domains of Cdh1, but not of Cdc20, through its BPA and BPC domains. Moreover, cells lacking DDB1 exhibit markedly elevated levels of the protein substrates of APC/CCdh1. Depletion of DDB1 in mitotic cells significantly delays mitotic exit, which demonstrates that the interaction between DDB1 and Cdh1 plays a critical role in regulating APC/CCdh1 activity. However, cells depleted of Cdh1 demonstrated no change in the UV-induced degradation of Cdt1, the main function of DDB1 as an E3 ligase. Strikingly, the APC/CCdh1 substrate levels are normal in cell knockdowns of Cul4A and Cul4B, which, along with DDB1, form an E3 ligase complex. This finding indicates that DDB1 modulates the function of APC/CCdh1 in a manner independent on the Cul4-DDB1 complex. Our results suggest that DDB1 may functionally regulate mitotic exit by modulating APC/CCdh1 activity. This study reveals that there may be cross-talk among DDB1, Cdh1, and Skp2 in the control of cell cycle division.
Keywords: Cell Cycle, Checkpoint Control, DNA Damage, E3 Ubiquitin Ligase, Protein-Protein Interactions, APC, Cdh1, DDB1, Mitotic Exit
Introduction
The anaphase-promoting complex/cyclosome (APC/C)3 (1, 2), a ubiquitin E3 ligase complex, plays crucial roles in late mitosis and during G1 phase by degrading cell cycle-related proteins such as cyclins, Plk1, and Skp2. The temporal cell cycle specificity of APC/C is achieved by activation of two closely related activators, Cdc20 and Cdh1, which facilitate the recruitment of substrates and ubiquitination by the core complex (3–5). Cdc20 and Cdh1 are members of the Cdc20 protein family and contain seven WD40 repeats in their C-terminal domains (6). These repeats form a seven-blade propeller structure that mediates protein-protein interactions. In early mitosis, APC/C binding to Cdc20 leads to the initiation of anaphase. In contrast, association with Cdh1 in late mitosis maintains APC/C activity throughout the subsequent G1 phase (3–5, 7–11). Both activators are essential and confer substrate specificity to APC/C activity, which in turn determines the timing of degradation for each target. Therefore, one important way that APC/C activity is governed is through the regulation of these activators. However, such regulation has not been thoroughly investigated in mitotic exit and during the G1 phase of the cell cycle.
Damaged DNA-binding protein 1 (DDB1) was initially identified as part of a heterodimer, which includes damaged DNA-binding protein 2 (DDB2). This heterodimer tightly associates with DNA following UV damage (12) and functions in nucleotide excision repair. DDB1 has since been shown to function outside of nucleotide excision repair as an adaptor molecule for the Cul4 ubiquitin E3 ligase complex. DDB1 facilitates the binding of substrates to the Cul4-Roc1 complex and the ubiquitination of selected proteins (13). Thus, as an E3 ligase capable of degrading cyclin E, p27, and Cdt1, DDB1 plays a key role in the normal cell cycle and in response to DNA damage (14–19). Recently, numerous proteins containing WD40 domains have been identified that interact with Cul4-DDB1 (20–23). However, the function and physiological significance of the interaction between DDB1 and most of the WD40-containing proteins remain unknown. Herein, we report that DDB1 may have a novel role, independent of the Cul4-DDB1 complex, in normal mitotic exit, at least partially by regulating APC/CCdh1 activity.
EXPERIMENTAL PROCEDURES
Cell Lines
Cells (HeLa, U2OS, HepG2, and CNE2) were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone), 1 mm glutamine, and 100 units/ml each of penicillin and streptomycin.
Plasmids
Human Myc-Cdh1 and Myc-Cdc20 (24) were gifts from Dr. James Hsieh (Washington University in St. Louis). V5-tagged DDB1 (25) was a gift from Dr. Helen Piwnica-Worms (Washington University in St. Louis). Mutations were introduced using the QuikChange site-directed mutagenesis kit (Stratagene), and all mutations were verified by DNA sequencing.
Antibodies
Human anti-Cdh1, anti-Cul4A, anti-Plk-1, anti-Aurora A, and anti-Cul4B anti-DDB1 antibodies were obtained from Abcam. Other primary antibodies used for Western blotting, which included anti-Cdt1, anti-hemagglutinin, anti-glyceraldehyde-3-phosphate dehydrogenase, anti-Cdc20, anti-Skp2, anti-geminin, anti-anillin, and anti-Cdc27, were obtained from Santa Cruz Biotechnology. Anti-Myc (A-14 or M10E, Santa Cruz Biotechnology) and anti-V5 (Sigma) were used for tagged proteins. Bound primary antibodies were detected with either horseradish peroxidase-conjugated goat anti-mouse antibody or horseradish peroxidase goat anti-rabbit antibody (Sigma), and proteins were visualized by chemiluminescence.
Transfection Experiments
Transfection was performed as described previously (26). Briefly, asynchronously growing cells were seeded at 2.5 × 105 cells per well of a 6-well tissue culture plate or at 1 × 106 cells/100 mm in a tissue culture dish and were transfected with 2 μg or 10 μg plasmid DNA, respectively, using LipofectamineTM 2000 (Invitrogen).
RNA Interference Treatment
Knockdown of Cdh1, DDB1, or Cdc27 was accomplished using SMARTpool siRNA (Dharmacon). Cul4A, Cul4B, and FBW5 (22), DDB2 (27), and β-TrCP (28) siRNA were designed according to previously validated oligonucleotides and synthesized by Invitrogen. Approximately 2 × 105 HeLa cells per well were seeded in a 6-well tissue culture dish the day before transfection. Transfection was performed according to the manufacturer's instructions using LipofectamineTM RNAiMAX transfection reagent (Invitrogen) and 100 nm siRNA. Forty-eight hours post-transfection, cells were harvested in MCLB (50 mm Tris-HCl, pH 8.0, 2 mm dithiothreitol, 5 mm EDTA, 0.5% Nonidet P-40, 100 mm NaCl, 1 mm microcystin, 1 mm sodium orthovanadate, 2 mm phenylmethylsulfonyl fluoride, protease (Sigma) supplemented with a phosphatase inhibitor mixture (Calbiochem)). For rescue experiments, a single oligonucleotide of Cdh1 (29), synthesized by Dharmacon, was used and transfected as above for 24 h; then, vectors Myc-Cdh1 and Myc-Cdh1-dCB-dIR were transfected using LipofectamineTM 2000 (Invitrogen) for 24 h.
Western Blotting and Immunoprecipitations
These procedures were performed as described previously (26). Briefly, cells were lysed in MCLB, and the clarified lysates were resolved by SDS-PAGE and transferred to nitrocellulose membranes for Western blotting using ECL detection reagents (Beyotime Co.). Alternatively, the clarified supernatants were first incubated with anti-Myc-agarose (Santa Cruz Biotechnology) or anti-V5-agarose (Sigma) for 1–2 h at 4 °C, and precipitates were washed four times with MCLB. To investigate the interaction between DDB1 and Cdh1 at the endogenous level, the clarified supernatants were first incubated with anti-Cdh1 or anti-DDB1 for 1–2 h at 4 °C. Then, protein A/G-agaroses were added for another 1 to 2 h, and precipitates were washed four times with MCLB and analyzed by Western blotting.
Synchronization of HeLa Cells
This method was described previously (26). Briefly, HeLa cells were cultured in the presence of 60 ng/ml nocodazole for 12–15 h, and mitotic cells were isolated by mitotic shake off. Cells were washed twice with phosphate-buffered saline and cultured for 2, 3, 4, or 5 h. Cells were collected and analyzed by flow cytometry and Western blotting.
Flow Cytometry
The procedure was detailed previously (26). Briefly, cells were harvested by trypsinization and collected by centrifugation. Cells were washed once with phosphate-buffered saline and fixed in 5 ml of 70% ethanol at 4 °C. Cells were washed once with phosphate-buffered saline/1% bovine serum albumin and then incubated with 1 ml of phosphate-buffered saline/1% bovine serum albumin containing 30 μg/ml propidium iodide and 0.25 mg/ml RNase A for 1 h at room temperature. Cells were analyzed for DNA content by flow cytometry using a FACS (fluorescene-activated cell sorter) Cytomics FC 500 (Beckman). The data were analyzed using Multicycle AV for Windows (Beckman).
RNA Extraction and Reverse Transcription PCR
The cells transfected with DDB1, DDB2, FBW5, β-TrCP, or scrambled siRNA for 48 h were collected, and RNAs were extracted using TRIzol (Invitrogen) according to the manufacturer's instructions. The primers used for amplifying FBW5 and β-TrCP were as follows: FBW5-F, 5′-CGCTGGACCACGTCATAGACA-3′; FBW5-R, 5′-CTGGGGACTGAAGACCACTGAG-3′; β-TrCP-F, 5′-AATCCTCATCTGGGACTTCCT-3′; and β-TrCP-R, 5′-CTGTTGTTGCTCATCCTGGTA-3′.
RESULTS
DDB1 Binds with WD40 Domains of Cdh1 through Its PBA and PBC Domains
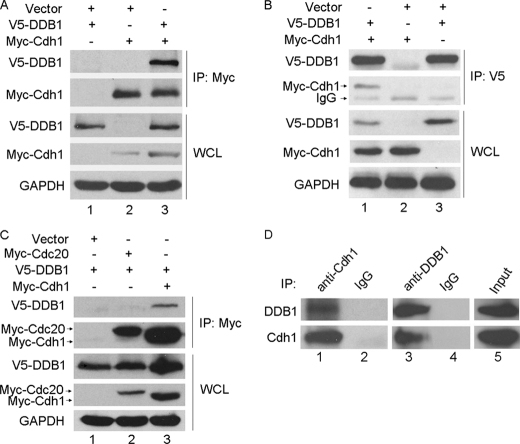
DDB1 is a key component of G2/M checkpoints and was recently found to associate with multiple WD40-containing proteins (23). Cdh1, a WD40-containing protein, is an activator of APC/C. APC/CCdh1 was recently shown to be important in response to DNA damage in G2 cells (28). Therefore, we speculated that DDB1 may bind Cdh1. To test this hypothesis, Myc-tagged Cdh1 was cotransfected with V5-tagged DDB1 into HeLa cells, and reciprocal coimmunoprecipitation using anti-Myc and anti-V5 was performed. As shown in Fig. 1, A and B, the complex containing these two proteins was clearly detected in the cell lysates. Interestingly, Cdc20, a WD40-containing protein, which is another activator of APC/C, did not form a complex with DDB1 (Fig. 1C), which is consistent with the previous report by He et al. (23). To further confirm the association between DDB1 and Cdh1, we examined this interaction at the endogenous level using coimmunoprecipitation. As shown in Fig. 1D, the endogenous complex between DDB1 and Cdh1 was clearly detected using either anti-Cdh1 or anti-DDB1 in HeLa cells. These results indicate that DDB1 specifically and physically interacts with Cdh1 in cells.
FIGURE 1.
DDB1 interacts with Cdh1 but not Cdc20. A, B, and C, HeLa cells transfected with the indicated plasmids for 24 h were lysed with MCLB, and immunoprecipitation (IP) using anti-Myc (A and C) or anti-V5 (B) was performed. Analysis by Western blotting was conducted with the indicated antibody. D, HeLa cells were lysed with MCLB, and lysates were subjected to immunoprecipitation using anti-Cdh1 or anti-DDB1 (n = 3). WCL, whole cell lysate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
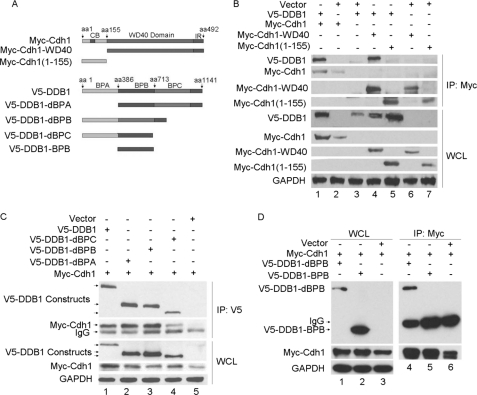
As expected, Cdh1-WD40, the WD40 domains of Cdh1, demonstrate a similar association with DDB1 as wild type Cdh1. In contrast, the N-terminal domain of Cdh1, Cdh1-(1–155) fragment, which contains a destruction box, A-box, C-box, and phosphorylation sites (2, 30–32), lacks the capacity to bind DDB1, as shown in Fig. 2B.
FIGURE 2.
Mapping of the binding domains for DDB1 and Cdh1. A, schematic description of the domains of Cdh1 and DDB1. B, the indicated plasmids of Cdh1 were cotransfected with V5-DDB1 (n = 3). C and D, the indicated constructs of DDB1 were cotransfected with Myc-Cdh1; and the formation of immunoprecipitation (IP) complex (B, C, and D) was detected as described in Fig. 1 (n = 3). WCL, whole cell lysate; aa, amino acid; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; CB, c-box; IR, IR-tail.
DDB1 is a large multidomain protein featuring an independent β-propeller domain (BPB), which is flexibly connected to a clam-shaped double propeller fold (BPA-BPC) (20, 33). To evaluate the domain responsible for DDB1 binding with Cdh1, we generated various DDB1 mutants, including deletions of BPA (V5-DDB1-dBPA), BPB (V5-DDB1-dBPB), BPC (V5-DDB1-dBPC), or BPA plus BPC (V5-DDB1-BPB), as shown in Fig. 2A. The binding of DDB1 with Cdh1 is only partially reduced in the BPC deletion mutant but is completely abolished when both BPA and BPC are absent, as shown in Fig. 2, C and D. This observation indicates that the clam-shaped double propeller fold formed by BPC and BPA is a crucial domain for DDB1 binding with Cdh1.
Knockdown of DDB1 Stabilizes the Substrates of APC/CCdh1
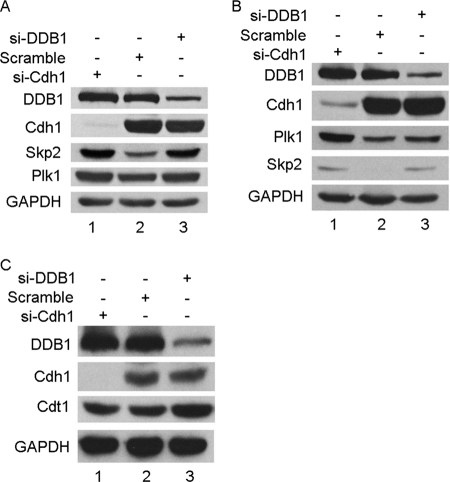
Cdh1 is one of the activators of APC/C ubiquitin E3 ligase and recruits the substrates of APC/CCdh1 for ubiquitination, and DDB1 along with Cul4 forms an E3 complex. We attempted to test whether the interaction between DDB1 and Cdh1 affects their E3 ligase activities. As shown in Fig. 3A, two representative substrates of APC/CCdh1, Skp2 and Plk1, were stabilized in HeLa cell siRNA knockdowns of either DDB1 or Cdh1. This observation was also confirmed in multiple cell lines, including U2OS, HepG2, and CNE2 (Fig. 3B, data not shown). Predominantly degraded by the Cul4-DDB1 complex in response to UV-induced DNA damage, Cdt1 was utilized to monitor the E3 ligase activity of DDB1. Interestingly, Cdt1 was only stabilized by knocking down DDB1 but not by Cdh1 siRNA when cells were exposed to UV (Fig. 3C). Taken together, the interaction between DDB1 and Cdh1 may be important for the functions of APC/CCdh1 but not of Cul4-DDB1.
FIGURE 3.
Depletion of DDB1 stabilizes Skp2 and Plk1, but knockdown of Cdh1 has no effect on the E3 ligase activity of DDB1. HeLa (A) or U2OS (B) cells were transfected with Cdh1 or DDB1 siRNA for 48 h, and extracts were separated by SDS-PAGE and analyzed by Western blotting with the indicated antibodies (n = 3). C, HeLa cells were transfected with Cdh1 (si-Cdh1) or DDB1 siRNA (si-DDB1) for 48 h and then were exposed with 20 J/m2 UV and incubated for 3 h prior to analysis by Western blotting with the indicated antibodies (n = 3). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
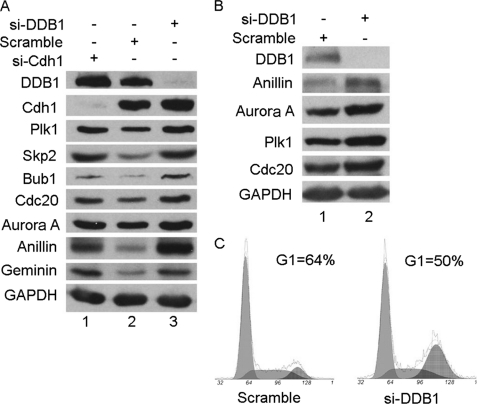
To further extend this observation, more substrates of APC/CCdh1, including Cdc20, Aurora A, anillin, geminin, and Bub1, were tested in HeLa cells depleted of DDB1 by siRNA. As shown in Fig. 4A, the depletion of DDB1 resulted in accumulation of all substrates tested, indicating that DDB1 may regulate the activity of APC/CCdh1 by binding with Cdh1. Given that DDB1 depletion can induce DNA damage and G2/M arrest (34) and that APC/CCdh1 substrates periodically oscillate during the cell cycle, such effects of DDB1 on APC/CCdh1 substrates may result from the difference of the cell cycle profile treated by DDB1 siRNA. To exclude this possibility, the well characterized Chk1 inhibitor Gö6976, which is capable of abrogating the G2/M checkpoint induced by DNA damage (35, 36), was used to diminish the difference in cell cycle profiles under the treatments of DDB1 or scrambled siRNA. The stabilization of APC/CCdh1 substrates by depleting DDB1 was still clearly observed when most of the cells were in G1 phase, which is when APC/CCdh1 is the most active, as shown in Fig. 4, B and C. Collectively, these results strongly indicate that DDB1 may functionally regulate the activity of APC/CCdh1.
FIGURE 4.
DDB1 positively regulates the activity of APC/CCdh1. A, HeLa cells were transfected with Cdh1, DDB1, or scrambled siRNA for 48 h, and extracts were separated by SDS-PAGE and analyzed by Western blotting with the indicated antibodies (n = 2). B and C, HeLa cells transfected with DDB1 or scramble siRNA for 48 h were incubated with 100 nmol/liter of Gö6976 for 24 h. Some of the cells were collected for analysis by flow cytometry (C), and the rest of the cells were analyzed by Western blotting with the indicated antibodies (B). Note: the percentages of G1 phase cells are indicated (n = 2). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; si-Cdh1, Cdh1 siRNA; si-DDB1, DDB1 siRNA.
DDB1-Cdh1 Regulation of APC/CCdh1 Activity Is Dependent on the APC/C Complex but Independent of Cul4
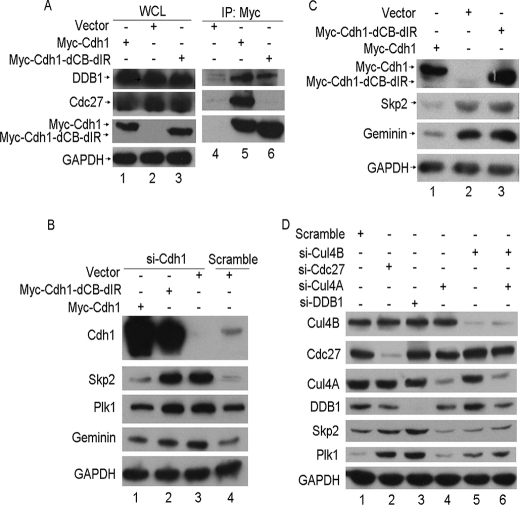
It has been reported that DDB1 interacts with multiple WD40-containing proteins, or DCAFs (DDB1 Cul4-associated factors), and forms the Cul4-DDB1-DCAF ubiquitin E3 ligase with Cul4 (23). Thus, it is possible that the DDB1-Cdh1 complex may function with Cul4 as a new E3 ligase, Cul4-DDB1-Cdh1, to target the tested substrates of APC/CCdh1. To examine this idea, we constructed a Cdh1 mutant depleted of the C box and IR tail of Cdh1 (Myc-Cdh1-dCB-IR). Consistent with another report (30), this mutant failed to bind with Cdc27, the core subunit of APC/C, as shown in Fig. 5A. However, this mutation did not alter its association with DDB1 (Fig. 5A), suggesting that the APC/C complex is not required for the interaction of DDB1 and Cdh1. Notably, wild-type Cdh1, but not the Cdh1-dCB-dIR mutant, could rescue the function of Cdh1 when endogenous Cdh1 was depleted (Fig. 5B). In addition, ectopic overexpression of wild-type Cdh1, but not Cdh1-dCB-dIR, enhanced the activity of APC/CCdh1, as judged by the protein levels of APC/CCdh1 substrates (Fig. 5C). These results indicated that Cul4-DDB1-Cdh1 is unlikely to function as an independent E3 ligase to degrade the substrates of APC/CCdh1.
FIGURE 5.
DDB1-Cdh1 regulation of APC/CCdh1 activity is dependent on the APC/C complex but independent of Cul4. A, HeLa cells transfected with the indicated plasmids were lysed and analyzed as described in Fig. 1 (n = 3). B, Cdh1 or scrambled siRNA were transfected into HeLa cells for 24 h, and then the indicated plasmids were transfected for another 24 h. The cells were subjected to Western blotting as described in Fig. 3A (n = 2). C, HeLa cells transfected with the indicated plasmids were lysed and subjected to Western blotting with the indicated antibodies (n = 2). D, HeLa cells were transfected with scramble, DDB1, Cdc27, Cul4A, Cul4B, or both Cul4A and Cul4B siRNA for 48 h, and extracts were separated by SDS-PAGE and detected by Western blotting with the indicated antibodies (n = 3). WCL, whole cell lysate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; si-Cdh1, Cdh1 siRNA; si-DDB1, DDB1 siRNA; IP, immunoprecipitation.
Because it is well documented that most characterized DDB1 functions are connected with Cul4, it is possible that the effect of DDB1 on the regulation of APC/CCdh1 activity may also require Cul4. If this is the case, we speculated that the knockdown of Cul4A, Cul4B, or both would stabilize the substrates of APC/CCdh1. However, as shown in Fig. 5D, the substrates of APC/CCdh1, such as Skp2 and Plk1, were only stabilized in the cell knockdowns of DDB1 or Cdc27 and not of Cul4A, Cul4B, or both, suggesting that DDB1 regulates the activity of APC/CCdh1 independent on the Cul4-DDB1 complex. Therefore, we conclude that DDB1-Cdh1 regulation of APC/CCdh1 activity is dependent on the APC/C complex but independent of Cul4.
Depletion of DDB1 Delays Mitotic Exit
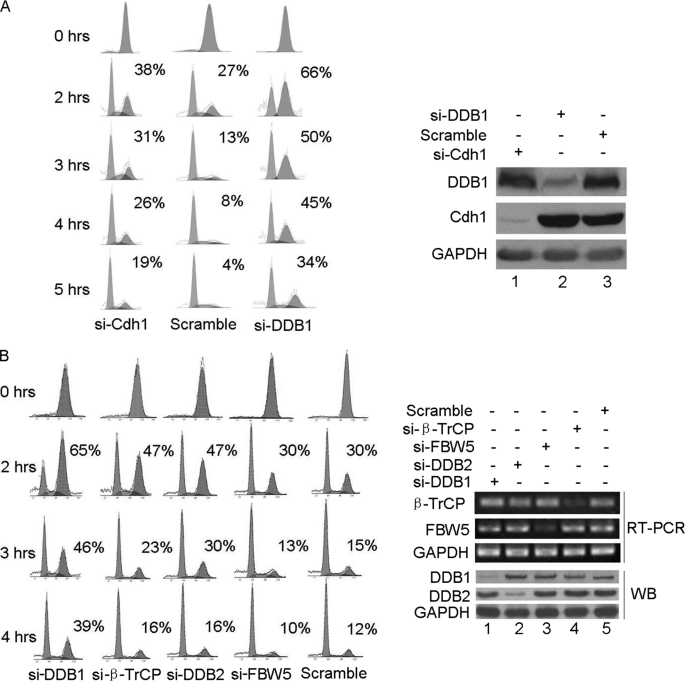
APC/CCdh1 plays a key role in mitotic exit and during G1 phase. If the above conclusion that DDB1-Cdh1 regulates APC/CCdh1 activity is true, DDB1 should play an important role in mitotic exit. To assess this speculation, HeLa cells treated with siRNAs against DDB1, Cdh1, or scrambled RNA were synchronized in M phase using nocodazole and were released. The mitotic exit progression in these cells was monitored, as shown in Fig. 6A. Mitotic exit was delayed in cells depleted of either DDB1 or Cdh1. In contrast, ectopic expression of DDB1 did not alter mitotic exit of the cells (supplemental Fig. S1), indicating that adequate endogenous DDB1 protein may be present to execute its functions during mitotic exit. Unexpectedly, cells depleted of DDB1 were much slower to exit from M phase than those depleted of Cdh1, indicating that modulation of APC/CCdh1 activity by Cdh1 binding only partially accounts for the delayed mitotic exit in cells depleted of DDB1. However, this conclusion must be made cautiously due to the likely incomplete knockdown of each protein by siRNA treatment.
FIGURE 6.
Depletion of DDB1 delays mitotic exit in HeLa cells. HeLa cells transfected with Cdh1, DDB1, or scrambled siRNA (A), or DDB1, DDB2, FBW5, β-TrCP, or control siRNA (B) were incubated with 100 nmol/liter Gö6976 and synchronized in M phase by incubation with 60 ng/liter nocodazole for 12 h. M phase cells selected by shake-off were released for the indicated time. Some of the cells were collected for analysis by flow cytometry (A and B, left panel), and the rest of the cells were analyzed by Western blotting (WB) or reverse transcription PCR (RT-PCR) as indicated (A and B, right panel). Note: the percentages of M phase cells are indicated (n = 2). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
The DDB1-Cul4-Roc1 E3 complex, when coupled with other adapter proteins, such as DCAFs and FWB5, could have a much broader target range than Cdh1, which may include mitotic regulators. Therefore, FBW5 (22) and DCAFs, such as DDB2 (12) and β-TrCP (37), were tested for their potential roles in mitotic exit. Interestingly, as shown in Fig. 6B, mitotic exit was delayed in cells depleted of DDB2 or β-TrCP but not of FBW5. Notably, mitotic exit was much slower in the cells depleted of DDB1 compared with those depleted of DDB2 or β-TrCP, indicating that the DDB1-Cul4-Roc1 E3 complex, when coupled with DCAFs, including DDB2 and β-TrCP, could also potentially target mitotic regulators. Given that APC/CCdh1 is critical for mitotic exit, DCAFs, including DDB2 and β-TrCP, may have important functions. It is possible that DDB2 or β-TrCP may also regulate APC/CCdh1 activity. However, as shown in supplemental Fig. S2, the knockdown of DDB2, FBW5, or β-TrCP had no effect on the stabilities of the substrates of APC/CCdh1, such as Skp2 and Plk1; therefore, this possibility was excluded. We conclude that DDB1 has a novel role in mitotic exit and that this function of DDB1 depends on Cdh1 and/or some DCAFs.
DISCUSSION
In this report, we have uncovered a novel function of DDB1: the regulation of mitotic exit, partially through the modulation of APC/CCdh1 activity by forming a complex with Cdh1. This new function of DDB1 does not depend on the Cul4-DDB1 complex, revealing that there may be cross-talk among DDB1, Cdh1, and Skp2 in the control of cell cycle division.
The APC/C ubiquitin E3 ligase complex plays crucial roles in mitotic exit by degrading mitotic cyclins and proteins, such as cyclin B1, Plk1, and Skp2. It has three statuses in terms of APC/C activity. It is inactive from S- to M phase and binds with Cdc20 to become active as APC/CCdc20 from metaphase to anaphase. The active form switches into APC/CCdh1 from anaphase to cytokinesis until G1 phase (3–5, 7–11). Thus, the precise regulation of APC/CCdh1 activity is essential to ensure normal cell cycle progression and genome integrity (38). Indeed, there are multiple levels of governance for APC/CCdh1 activity during the cell cycle. mRNA expression of Cdh1 is cell cycle-dependent. Cdh1 is degraded by APC/CCdh1, which is down-regulated by proteolysis of its E2 ligase, UbcH10 in G1 phase (39). Emi1/Rca1 (early mitotic inhibitor/regulator of cyclin A1) inhibits activity of APC/CCdh1 during the transition from G1 to S phase (40, 41). From S to M phase, the APC/C complex is inactive due to phosphorylation by Cdks, such as Cdk1 and Cdk2. In the metaphase-to-anaphase transition, APC/CCdh1 is inhibited by its inhibitors, such as Mad2l2 (mitotic arrest deficient-like 2) (42). Recently, it was reported that CREB-binding protein may act as an E4 ligase to promote efficient substrate ubiquitination by APC/C (43, 44). Both retinoblastoma protein and MDC1 (mediator of DNA damage checkpoint 1) physically bind to multiple APC/C subunits and functionally regulate mitotic exit (43, 45, 47). In this report, we demonstrated that DDB1 physically binds with Cdh1 but not Cdc20, and this interaction seems to be physiologically important because Cdh1 may partially account the significant delay in mitotic exit in cells depleted of DDB1. However, how DDB1 affects the function of APC/CCdh1 needs to be further investigated. Given that DDB1 is a scaffold protein that facilitates substrate recruitment for the DDB1-Cul4-Roc1 complex and that Cdh1 is also capable of recruiting substrates for APC/CCdh1, we speculate that the DDB1-Cdh1 complex may promote more efficient substrate recruitment for APC/CCdh1. Alternatively, DDB1 may act as an E4 ubiquitin ligase for APC/CCdh1, similar to CREB-binding protein. It is worth noting that, in response to DNA damage, APC/CCdh1 is reactivated to allow the elimination of the promitotic kinase Plk1, which is essential for the establishment and maintenance of an efficient G2 checkpoint (28). Considering that both MDC1 and DDB1 are important molecules in the G2/M checkpoint (34, 43), it would be very interesting to determine whether MDC1, DDB1, or both also may play a role in the reactivation of APC/CCdh1 in response to DNA damage.
Strikingly, we also demonstrated that adapter proteins for DDB1-Cul4-Roc1 E3 ligase, such as DDB2 and β-TrCP, but not FWB5, may have important roles in mitotic exit. Cells depleted of DDB2 or β-TrCP, but not of FWB5, display slower exit from M phase compared with control cells but are much faster than cells depleted of DDB1. Furthermore, DDB2 or β-TrCP does not affect APC/CCdh1 activity. Therefore, DDB1 plays a critical role in mitotic exit in at least two ways: by binding Cdh1 to modulate the APC/CCdh1 activity and by coupling with DCAFs, such as DDB2 and β-TrCP, to target unidentified substrates that regulate mitotic exit or spindle checkpoints. However, neither the cell cycle profile nor mitotic exit is altered by DDB1 overexpression, indicating that endogenous DDB1 levels are adequate to execute its functions.
The cell cycle transitions are driven by waves of ubiquitin-dependent degradation of key cell cycle regulators. APC/C, SCF (Skp1/Cullin/F-box protein), and DDB1-Cul4-Roc1 complexes are three major classes of ubiquitination ligases to fulfill the transitions, such as G1/S, G2/M, and M/G1 (48, 49). Skp2, an F-box protein, forms an SCF complex and is degraded by APC/CCdh1 during G1 phase (10). The phosphorylated Cdh1 is a substrate of the SCF complex (50), indicating that there is cross-talk between APC/C and SCF complexes. The SCF complex cooperates with the Cul4-DDB1 complex to degrade p27, cyclin E, and Cdt1 during the cell cycle and in response to DNA damage (15, 16), indicating that there is cross-talk between SCF and Cul4-DDB1 complexes. Here, we provide evidence that DDB1 forms a complex with Cdh1 and regulates mitotic exit by modulating the activity of APC/CCdh1, which demonstrates that there is a cross-talk between APC/C and Cul4-DDB1 complexes. However, this novel function of DDB1 is not dependent on the Cul4-DDB1 complex. Notably, p21 is targeted by Skp2, DDB1, and APC/CCdc20 in G1, S, and M phase, respectively (46). Therefore, these three ubiquitination E3 ligases, Cul4-DDB1, APC/C, and SCF-Skp2, may actually be cross-talk in precisely governing cell cycle division.
Supplementary Material
Acknowledgments
We thank Professor Xin-Yuan Guan (University of Hong Kong) for critical comments on this manuscript. We thank Dr. Helen Piwnica-Worms (Washington University in St. Louis) for providing V5-DDB1 and Dr. James Hsu (Washington University in St. Louis) for providing Myc-Cdh1 and Myc-Cdc20. Members of the laboratory are thanked for helpful comments on the manuscript.
This work was supported by grants from National Natural Science Foundation of China (30840005, 30930045), China's 973 project (2010CB912201) (to T. K.), China's 863 program (2006AA02A404), and the National Natural Science Foundation of China (u0732005) (to Y.-X. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- APC/C
- anaphase-promoting complex/cyclosome
- DDB1
- damaged DNA-binding protein 1
- BPB
- β-propeller domain
- siRNA
- small interfering RNA
- CREB
- cAMP-response element-binding protein
- BPA
- β-propeller domain A
- BPB
- β-propeller domain B
- BPC
- β-propeller domain C
- MCLB
- mammalian cell lysis buffer.
REFERENCES
- 1.Clarke D. J., Díaz-Martínez L. A., Giménez-Abián J. F. (2005) Cell Cycle 4, 1585–1592 [DOI] [PubMed] [Google Scholar]
- 2.Acquaviva C., Pines J. (2006) J. Cell Sci. 119, 2401–2404 [DOI] [PubMed] [Google Scholar]
- 3.Schwab M., Lutum A. S., Seufert W. (1997) Cell 90, 683–693 [DOI] [PubMed] [Google Scholar]
- 4.Sigrist S. J., Lehner C. F. (1997) Cell 90, 671–681 [DOI] [PubMed] [Google Scholar]
- 5.Visintin R., Prinz S., Amon A. (1997) Science 278, 460–463 [DOI] [PubMed] [Google Scholar]
- 6.Smith K. N., Iwanejko L., Loeillet S., Fabre F., Nicolas A. (1999) Yeast 15, 1255–1267 [DOI] [PubMed] [Google Scholar]
- 7.Kitamura K., Maekawa H., Shimoda C. (1998) Mol. Biol. Cell 9, 1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanco M. A., Sánchez-Díaz A., de Prada J. M., Moreno S. (2000) EMBO J. 19, 3945–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bashir T., Dorrello N. V., Amador V., Guardavaccaro D., Pagano M. (2004) Nature 428, 190–193 [DOI] [PubMed] [Google Scholar]
- 10.Wei W., Ayad N. G., Wan Y., Zhang G. J., Kirschner M. W., Kaelin W. G., Jr. (2004) Nature 428, 194–198 [DOI] [PubMed] [Google Scholar]
- 11.Buschhorn B. A., Peters J. M. (2006) Nat. Cell Biol. 8, 209–211 [DOI] [PubMed] [Google Scholar]
- 12.Chu G., Chang E. (1988) Science 242, 564–567 [DOI] [PubMed] [Google Scholar]
- 13.Groisman R., Polanowska J., Kuraoka I., Sawada J., Saijo M., Drapkin R., Kisselev A. F., Tanaka K., Nakatani Y. (2003) Cell 113, 357–367 [DOI] [PubMed] [Google Scholar]
- 14.Senga T., Sivaprasad U., Zhu W., Park J. H., Arias E. E., Walter J. C., Dutta A. (2006) J. Biol. Chem. 281, 6246–6252 [DOI] [PubMed] [Google Scholar]
- 15.Nishitani H., Sugimoto N., Roukos V., Nakanishi Y., Saijo M., Obuse C., Tsurimoto T., Nakayama K. I., Nakayama K., Fujita M., Lygerou Z., Nishimoto T. (2006) EMBO J. 25, 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bondar T., Kalinina A., Khair L., Kopanja D., Nag A., Bagchi S., Raychaudhuri P. (2006) Mol. Cell. Biol. 26, 2531–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cang Y., Zhang J., Nicholas S. A., Kim A. L., Zhou P., Goff S. P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2733–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishitani H., Shiomi Y., Iida H., Michishita M., Takami T., Tsurimoto T. (2008) J. Biol. Chem. 283, 29045–29052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abbas T., Sivaprasad U., Terai K., Amador V., Pagano M., Dutta A. (2008) Genes Dev. 22, 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Angers S., Li T., Yi X., MacCoss M. J., Moon R. T., Zheng N. (2006) Nature 443, 590–593 [DOI] [PubMed] [Google Scholar]
- 21.Higa L. A., Wu M., Ye T., Kobayashi R., Sun H., Zhang H. (2006) Nat. Cell Biol. 8, 1277–1283 [DOI] [PubMed] [Google Scholar]
- 22.Hu J., Zacharek S., He Y. J., Lee H., Shumway S., Duronio R. J., Xiong Y. (2008) Genes Dev. 22, 866–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y. J., McCall C. M., Hu J., Zeng Y., Xiong Y. (2006) Genes Dev. 20, 2949–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H., Cheng E. H., Hsieh J. J. (2007) Genes Dev. 21, 2385–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leung-Pineda V., Huh J., Piwnica-Worms H. (2009) Cancer Res. 69, 2630–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang T., Wei Y., Honaker Y., Yamaguchi H., Appella E., Hung M. C., Piwnica-Worms H. (2008) Cancer Cell 13, 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kattan Z., Marchal S., Brunner E., Ramacci C., Leroux A., Merlin J. L., Domenjoud L., Dauça M., Becuwe P. (2008) PloS One 3, e2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bassermann F., Frescas D., Guardavaccaro D., Busino L., Peschiaroli A., Pagano M. (2008) Cell 134, 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brummelkamp T. R., Bernards R., Agami R. (2002) Science 296, 550–553 [DOI] [PubMed] [Google Scholar]
- 30.Vodermaier H. C., Gieffers C., Maurer-Stroh S., Eisenhaber F., Peters J. M. (2003) Curr. Biol. 13, 1459–1468 [DOI] [PubMed] [Google Scholar]
- 31.Pfleger C. M., Kirschner M. W. (2000) Genes Dev. 14, 655–665 [PMC free article] [PubMed] [Google Scholar]
- 32.Liu W., Li W., Fujita T., Yang Q., Wan Y. (2008) Carcinogenesis 29, 263–272 [DOI] [PubMed] [Google Scholar]
- 33.Li T., Chen X., Garbutt K. C., Zhou P., Zheng N. (2006) Cell 124, 105–117 [DOI] [PubMed] [Google Scholar]
- 34.Lovejoy C. A., Lock K., Yenamandra A., Cortez D. (2006) Mol. Cell. Biol. 26, 7977–7990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohn E. A., Yoo C. J., Eastman A. (2003) Cancer Res. 63, 31–35 [PubMed] [Google Scholar]
- 36.Jia X. Z., Yang S. Y., Zhou J., Li S. Y., Ni J. H., An G. S., Jia H. T. (2009) Biochem. Pharmacol. 77, 770–780 [DOI] [PubMed] [Google Scholar]
- 37.Katiyar S., Liu E., Knutzen C. A., Lang E. S., Lombardo C. R., Sankar S., Toth J. I., Petroski M. D., Ronai Z., Chiang G. G. (2009) EMBO Reports 10, 866–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.García-Higuera I., Manchado E., Dubus P., Cañamero M., Méndez J., Moreno S., Malumbres M. (2008) Nat. Cell Biol. 10, 802–811 [DOI] [PubMed] [Google Scholar]
- 39.Rape M., Kirschner M. W. (2004) Nature 432, 588–595 [DOI] [PubMed] [Google Scholar]
- 40.Grosskortenhaus R., Sprenger F. (2002) Dev. Cell 2, 29–40 [DOI] [PubMed] [Google Scholar]
- 41.Hsu J. Y., Reimann J. D., Sørensen C. S., Lukas J., Jackson P. K. (2002) Nat. Cell Biol. 4, 358–366 [DOI] [PubMed] [Google Scholar]
- 42.Pfleger C. M., Salic A., Lee E., Kirschner M. W. (2001) Genes Dev. 15, 1759–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Townsend K., Mason H., Blackford A. N., Miller E. S., Chapman J. R., Sedgwick G. G., Barone G., Turnell A. S., Stewart G. S. (2009) J. Biol. Chem. 284, 33939–33948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turnell A. S., Stewart G. S., Grand R. J., Rookes S. M., Martin A., Yamano H., Elledge S. J., Gallimore P. H. (2005) Nature 438, 690–695 [DOI] [PubMed] [Google Scholar]
- 45.Binné U. K., Classon M. K., Dick F. A., Wei W., Rape M., Kaelin W. G., Jr., Näär A. M., Dyson N. J. (2007) Nat. Cell Biol. 9, 225–232 [DOI] [PubMed] [Google Scholar]
- 46.Abbas T., Dutta A. (2009) Nat. Rev. 9, 400–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coster G., Hayouka Z., Argaman L., Strauss C., Friedler A., Brandeis M., Goldberg M. (2007) J. Biol. Chem. 282, 32053–32064 [DOI] [PubMed] [Google Scholar]
- 48.Nakayama K. I., Hatakeyama S., Nakayama K. (2001) Biochem. Biophys. Res. Commun. 282, 853–860 [DOI] [PubMed] [Google Scholar]
- 49.Peters J. M. (2002) Mol. Cell 9, 931–943 [DOI] [PubMed] [Google Scholar]
- 50.Benmaamar R., Pagano M. (2005) Cell Cycle 4, 1230–1232 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.