Abstract
Considering the wide variety of effects that have been reported to occur in the developmental neurotoxicity of chlorpyrifos (CP) and the lack of consensus on their dependence of brain acethylcholinesterase (AChE) activity inhibition, we applied microarray technology to explore dose-dependent alterations in transcriptional response in the fetal and maternal C57BL/6 mouse brain after daily gestational exposure (days 6 to 17) to CP (2, 4, 10, 12 or 15 mg/kg, sc). We identified significantly altered genes across doses and assessed for overrepresentation of Gene Ontology (GO) biological processes and KEGG pathways. We further clustered genes based on their expression profiles across doses and repeated the GO/pathways analysis for each cluster. The dose-effect relationship of CP on gene expression, both at the gene and pathway levels was non-monotonic and not necessarily related to brain AChE inhibition. The largest impact was observed in the 10 mg/kg dose group which was also the LOAEL for brain AChE inhibition. In the maternal brain, lower doses (4 mg/kg) influenced GO categories and pathways such as cell adhesion, behavior, lipid metabolism, long-term potentiation, nervous system development, neurogenesis, synaptic transmission. In the fetal brain, lower doses (2 and/or 4 mg/kg) significantly altered cell division, translation, transmission of nerve impulse, chromatin modification, long-term potentiation. In addition, some genes involved in nervous system development and signaling were shown to be specifically influenced by these lower CP doses. Our approach was sensitive and reflected the diversity of responses known to be disrupted by CP and highlighted possible additional consequences of CP neurotoxicity, such as disturbance of the ubiquitin proteasome system.
Keywords: chlorpyrifos, gestational exposure, microarray, gene expression, brain
INTRODUCTION
Chlorpyrifos (CP) is a broad-spectrum organophosphorus (OP) insecticide applied worldwide. In the US, household use of this pesticide was restricted in 2000 based on its potential as a developmental neurotoxicant (USEPA, 2002). However, the continued use of CP in agriculture still poses the potential for childhood exposure through the take-home or dietary pathways in both agricultural and non-agricultural communities.
The classical mechanism of toxicity of all OPs is through the inhibition of the acetylcholinesterase (AChE) enzymatic activity, increasing the availability of the neurotransmitter acetylcholine. In many countries, including the USA, regulatory standards for human exposure of OPs are determined based on their inhibitory effects on cholinesterase activity. Recent studies raise concern regarding these standards based solely on the inhibitory effects of AChE due to observed CP-induced developmental neurotoxicity at exposure levels once considered subtoxic, i.e., exposures that neither induce overt signs of systemic intoxication nor inhibit cholinesterase activity (Eaton et al., 2008). A multitude of mechanisms have been proposed to underlie CP developmental neurotoxicity. Oxidative stress and alteration of cell signaling cascades, nuclear transcription factors, and neuronal-glial cell interactions have been suggested to occur at doses below AChE inhibition (Song et al., 1997; Pope, 1999; Crumpton et al., 2000b; Crumpton et al., 2000a). Even though these results would suggest a lack of AChE participation in the low-dose CP-induced effects, this may not necessarily be always the case. Evidences suggest that AChE plays a role in promoting axonal growth in developing neurons (Brimijoin and Koenigsberger, 1999; Bigbee et al., 2000). Comparative analyses of the effects of CP and its oxon metabolite on axonal growth in dorsal root ganglia (DRG) neurons cultured from AChE nullizygous (AChE−/−) versus wild type (AChE+/+) mice indicated that while these OPs inhibited axonal growth in AChE+/+ DRG neurons, they had no effect on axonal growth in AChE−/− DRG neurons. However, transfection of AChE−/− DRG neurons with cDNA encoding full-length AChE restored the wild type response to the axon inhibitory effects of OPs (Yang et al., 2008). These data seems to suggest that inhibition of axonal growth by OPs requires the presence of AChE, but the mechanism involves inhibition of the morphogenic rather than enzymatic activity of AChE. To add more complexity to the developmental neurotoxicity of the OPs, it should be pointed out that differently from the common anticholinesterasic activity that all OPs share, the developmental neurotoxicity of OPs may result from mechanisms that are not necessarily the same for all the compounds belonging to this class of pesticides (Slotkin and Seidler, 2007; Slotkin et al., 2007; Slotkin and Seidler, 2008), i.e., for this endpoint, these pesticides may not behave as a class and each one may induce a different repertoire of effects.
Considering the wide variety of effects that have been reported to occur in the developmental neurotoxicity of CP and the lack of consensus on their dependence of brain AChE activity inhibition (Eaton et al., 2008), in this study we applied microarray technology using a genome wide systems-based approach to explore in depth the dose-dependent alterations in transcriptional response both in the maternal and fetal brains after gestational exposure to CP (gestational days 6 to 17). Toxicogenomic analysis across doses of CP at and sub brain AChE inhibition provided evidence of biological processes/pathways impacted by CP which may be independent of AChE activity inhibition both in the adult and developing brain.
METHODS AND MATERIALS
Animals and CP Exposure
Mice (C57BL/6) were supplied from JAX Mice and Services (Bar Harbor, ME). They were maintained at the animal care facility in the University of Washington, in filter covered transparent plastic cages and under controlled temperature and 12-h light/dark cycle. Water and food were available ad libitum. All procedures involving animals were performed in accordance with the approved protocol and guidelines of the University of Washington’s Institutional Animal Care and Use Committee (IACUC) in compliance with guidelines established by the National Institutes of Health.
Timed matings were produced by placing individual male mice into cages containing two females. Copulatory plugs were identified in the early morning (8:00 A.M. ± 0.5h) the following day and designated as gestational day (GD) 0. Pregnant mice received a daily subcutaneous (sc) injection of CP (99.5% purity, ChemService, West Chester, PA) from GD 6 to 17. Control animals received dimethyl sulfoxide (DMSO, Sigma, St Louis, MO) at the volume of 1 ml/kg. Dams were weighed daily and CP doses were adjusted for recorded body weight changes.
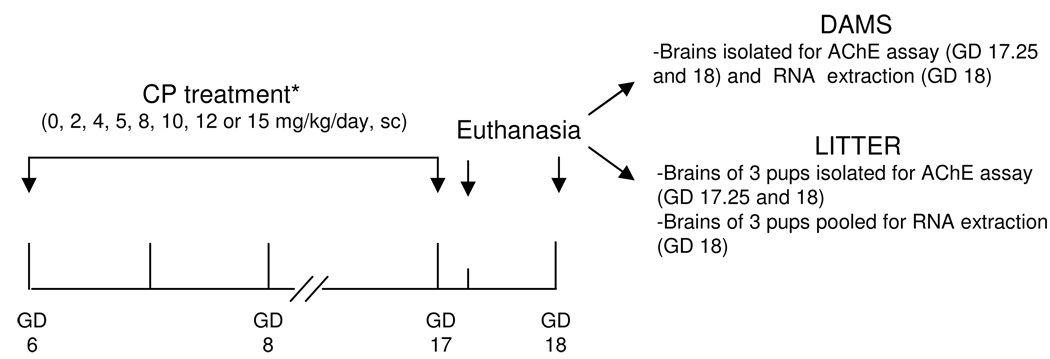
As illustrated in Fig. 1, for the microarray study, two sets of experiments were conducted with different doses of CP. Experiment 1 (Exp 1) was conducted with 0 (DMSO), 10 and 15 mg/kg. Since both doses significantly decreased AChE activity in the maternal and fetal brain, experiment 2 (Exp 2) was conducted with lower doses of CP (2 and 4 mg/kg), in addition with another set of control animals and an intermediate dose (12 mg/kg) to the previously tested in Exp 1. Three pregnant mice were assigned to each dose group. On GD 18 (24h after the last CP injection), dams were euthanized via inhalation of isofluorane followed by cervical dislocation. The uterus was removed and fetuses were euthanized by overexposure to isofluorane. Maternal and fetal brains were removed for AChE activity assay and RNA isolation. For AChE activity assessments, two additional doses of CP were evaluated (5 and 8 mg/kg) as well as an additional time point: GD 17.25 (6h after the last CP injection). CP from the same lot was used throughtout the study.
Fig 1.
Diagram of the experimental design. The day the copulatory plug was observed was considered gestational day (GD) 0. CP was dissolved in DMSO. *For the microarray study, the doses of 5 and 8 mg/kg were not evaluated.
Brain sample preparation
After removal, each maternal brain was homogenized in 2 ml 0.1 M sodium phosphate buffer containing 1% triton X-100 (PBS-triton) in a Polytron PT 10–35 homogenizer. Homogenized brains were used for the AChE activity assay, protein quantification and RNA isolation. Fetal brains were homogenized in the same manner as the maternal brain. For each litter, AChE activity was assayed in the brain of 4 different littermates, in average. Each calculated value of AChE activity at each dose group represents the mean value among those biological replicates. For RNA isolation, brains of three littermates were pooled together in order to provide enough RNA amount for the microarray analysis.
AChE assay
AChE activity was assessed using 50 or 300 µl of the maternal or fetal brain homogenate, respectively, added to 450 µl PBS-triton. Adaptated from Ellman et al. (Ellman et al., 1961) using the automated microplate reader (Bio-Tek, EL3401), briefly, AChE activity was measured spectrophotometrically using acetylthiocholine iodide (Sigma, St Louis, MO) as the substrate, 5,5’-dithiobis-2-nitrobenzoic acid (DTNB, Sigma, St Louis, MO) as the chromagen and tetra-isopropylpyrophosphoaramide (iso-OPMA, Sigma, St Louis, MO) as the butirylcholinesterase inhibitor. Protein concentration of the homogenates was quantified (Bradford, 1976) and activity of AChE was calculated and expressed as µmoles product produced per min per mg protein. The AChE assay had 2 to 4 replicates for each animal.
RNA isolation and microarrays hybridization
RNA was extracted from the brain of dams and fetuses (3 fetuses per litter, undefined gender) using the RNAeasy Mini Kit (Qiagen, Valencia, CA). RNA quality was assessed using the 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Genome wide gene expression analysis was conducted within the Functional Genomics Laboratory at the NIEHS Center for Ecogenetic and Environmental Health (CEEH), using the Affymetrix Mouse Whole Genome 430 2.0 platform following the manufacturer' protocol (Yu et al., 2009). Three arrays were conducted for each dose level except for the 2 mg/kg. At this dose level, only two arrays were conducted since one of the dams delivered only one pup. Scanned images of the arrays were analyzed to obtain an intensity value for each probe.
Statistical analysis
The results being reported here are part of a study conducted in our laboratory to evaluate gestational impacts of CP exposure on brain development. For the weight gain and AChE analysis, we are reporting data from all the dams/litters used in the complete CP study, and, because of this, these analysis have a higher sample size than the microarray analysis.
Mixed effects analysis of variance was used for the evaluation of weights and AChE. In these analyses dose was a fixed effect that had a factor at each of the individual dose levels of CP (0, 2, 4, 5, 8, 10, 12, and 15 mg/kg) and as random effects we had the cohort in which the animal was exposed, the litter to which the animal belonged, and the individual animal used in the AChE analysis to incorporate replicate assays of AChE. AChE measured values were normalized to concurrent controls measured on the same date of analysis before being statistically analyzed with a mixed effects model.
Trend analysis was also performed on the AChE data and changes with dose were treated as a linear function of doses in the fixed effects in the mixed effects model. The trend analysis was an analysis of covariance to determine the differences and similarities between the data at 6 h and 24 h after the last exposure. In order to perform this analysis both the 6 h and 24 h data was normalized to concurrent controls at 24 h. The analysis of covariance allowed us to test whether there was a change in AChE controls between 6 h and 24 h, and whether the slope was different between these timepoints.
For the statistical analysis of weights by the mixed effects statistical analysis the fixed effect was dose and the random effect for the dams was the cohort and for the fetuses was the cohort and litter.
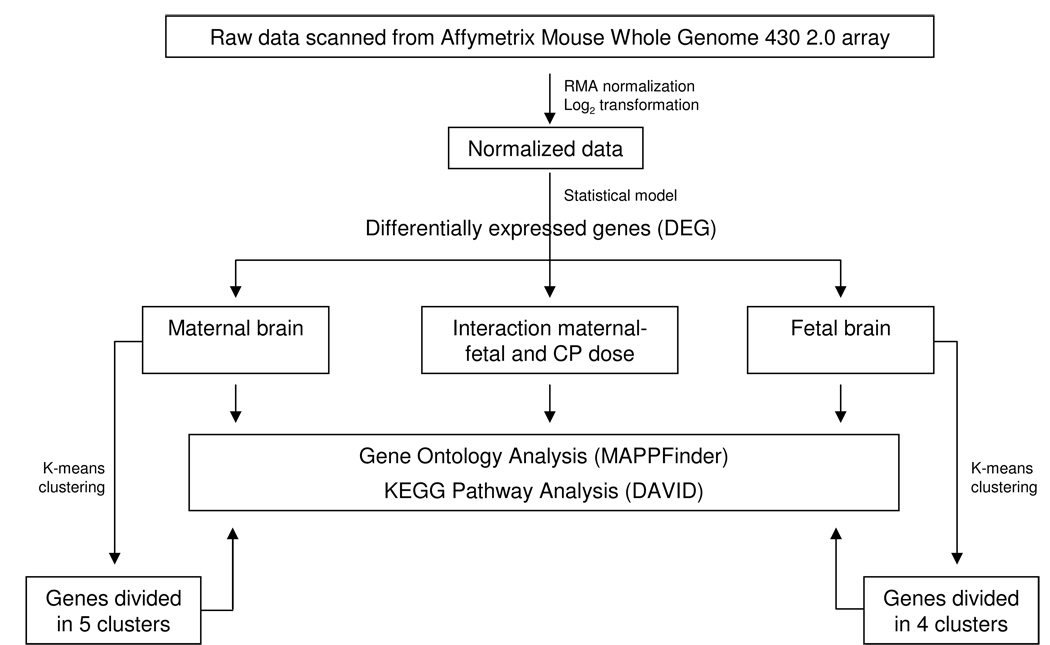
As observed in Fig 2, microarray data analysis was conducted using a systems-based approach to identify significant CP-induced gene expression alterations and their respective Gene Ontology (GO)–based classifications in both maternal and fetal brains. Briefly, raw microarray data was processed and analyzed with Bioconductor (Gentleman et al., 2004) and normalized using the RMA (Robust Multi-Array) method as implemented in the Bioconductor Affy package (Bolstad et al., 2003). From the normalized data, genes with significant evidence for differential expression were identified using the limma package (Smyth, 2004) in Biocunductor. We employed a mixed effects linear model to detect genes that were differentially expressed based on the fixed effects of dose, age (either dam or fetus), experiment, and interaction of maternal-fetal with CP doses, and a random effect for maternal/fetus pairs. P-values were calculated with modified t or F-tests in conjunction with an empirical Bayes method to moderate the standard errors of the estimated log-fold changes.
Fig 2.
Toxicogenomic approach to analyze and identify CP-induced differentially expressed genes in the maternal and fetal brain. For details on the statistical model used please refer to the text.
To establish the association between CP treatment and the altered gene ontology (GO) terms, we applied MAPPFinder to identify enriched biological processes/pathways (Dahlquist et al., 2002) among genes found to be differentially expressed (P≤0.005 ) in each one of the 3 different models (fetal across doses, maternal across doses, interaction of maternal/fetal with CP doses). Z score (Z) and permutation P value (P) were used to rank the terms of biological significance (Dahlquist et al., 2002). Significant GO categories (GOIDs) were determined based on the following cut-off values: P ≤ 0.05, Z ≥ 2.0 and a minimum of 3 genes altered within each specific GOID. Due to the higher number of differentially expressed genes (DEG) in the fetal brain, a minimum of 5 genes was ultimately used as a cut-off for GOIDs displayed. Using the GO-based application, GO-Quant, enriched GOIDs were examined quantitavely across CP doses by calculating the absolute average fold change of genes within each GOID (Yu et al., 2006). Additionally, a KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis was conducted using DAVID (Database for Annotation, Visualization and Integrated Discovery) Bioinformatics Resources (Huang da et al., 2009).
To explore the observed non-monotonic relationship of CP on gene expression, we further applied K-means clustering using MultiExperiment Viewer (MeV) software (Saeed et al., 2003) to cluster genes based on the pattern of their expression profiles across doses. The number of clusters was determined based on the amount of unique relationships observed across doses. We conducted GO analysis with MAPPFinder as well as pathway analysis using DAVID for each separate cluster to investigate differential biological processes/pathways enriched in each cluster. Enriched pathways were selected with at least 3 DEG and a P ≤ 0.05.
RESULTS
The number of litters and dams varied for each dose and group of animals and are shown in Table 1 for the 24 h animals. For the 6 h animals there was a minimum of 2 litters at 10 mg/kg and an average of 5 litters for the other doses.
Table 1.
General toxicity parameters from dams/litters exposed to CP from GD 6 to GD 17.
| CP dose (mg/kg, sc) |
||||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 4 | 5 | 8 | 10 | 12 | 15 | |
| Number of litters per dose | 13 | 7 | 5 | 6 | 5 | 3 | 6 | 3 |
| Number of live fetuses per litter | 7.0 ± 1.7 | 6.1 ± 2.4 | 7.2 ± 1.3 | 6.8 ± 1.0 | 6.8 ± 3.6 | 7.3 ± 0.6 | 6.0 ± 1.1 | 7.3 ± 0.6 |
| Number of resorptions per litter | 1.1 ± 1.0 | 0.7 ± 0.8 | 0.8 ± 1.3 | 0.8 ± 0.8 | 0.4 ± 0.5 | 0.3 ± 0.6 | 1.0 ± 0.9 | 1.0 ± 0.0 |
| Probability of live fetuses | 0.87 (0.79, 0.92) |
0.90 (0.77, 0.96) |
0.90 (0.76, 0.96) |
0.89 (0.76, 0.95) |
0.94 (0.80, 0.99) |
0.96 (0.75, 0.99) |
0.86 (0.72, 0.93) |
0.88 (0.69, 0.96) |
| Fetus weight on GD18 (g) | 1.08 (1.00, 1.16) |
1.09 (0.99, 1.19) |
1.08 (0.97, 1.19) |
1.12 (1.02, 1.23) |
1.03 (0.92, 1.15) |
1.20 (1.07, 1.34) |
1.14 (1.03, 1.24) |
1.11 (0.98, 1.25) |
| Dam weight gain from GD6-17 (g) | 11.4 (9.9, 12.9) |
10.4 (8.4, 12.5) |
12.6 (10.2, 15.0) |
11.5 (9.3, 13.7) |
11.5 (9.1, 13.8) |
13.2 (10.1, 16.3) |
9.8 (7.5, 12.9) |
12.5 (9.4, 15.6) |
Data are means and SD (number of live fetuses and resorptions) or 95% confidence interval (fetus weight, dam weight gain and probability of live fetuses). A mixed effects analysis of variance was used to analyze the data (P > 0.05). The 0 mg/kg group refers to the control group that was treated with DMSO.
General Toxicity
The analyses of fetus body weights on GD18 and weight changes for dams from GD6 to 17 are shown in Table 1. There were no significant changes in the weight gain of dams (P = 0.5) nor fetuses (P = 0.3) across doses. The number of litters and dams is the same as the 24 h numbers in the AChE analysis (refer to the next subsection). We had from 1 to 12 fetuses per litter with a median of 7 fetuses and 90% of the litters having 5 to 9 fetuses.
The average number of live fetuses and resorptions per litter at GD 18 for each dose are shown in Table 1. To statistically analyze whether dose had an effect on survival of the fetuses we used a generalized linear model for binomial data to estimate the probability of a live fetus at each dose which is shown in Table 1 along with 95% confidence limits. Dose had no significant effect on the probability of a live fetus (P = 0.8).
AChE activity
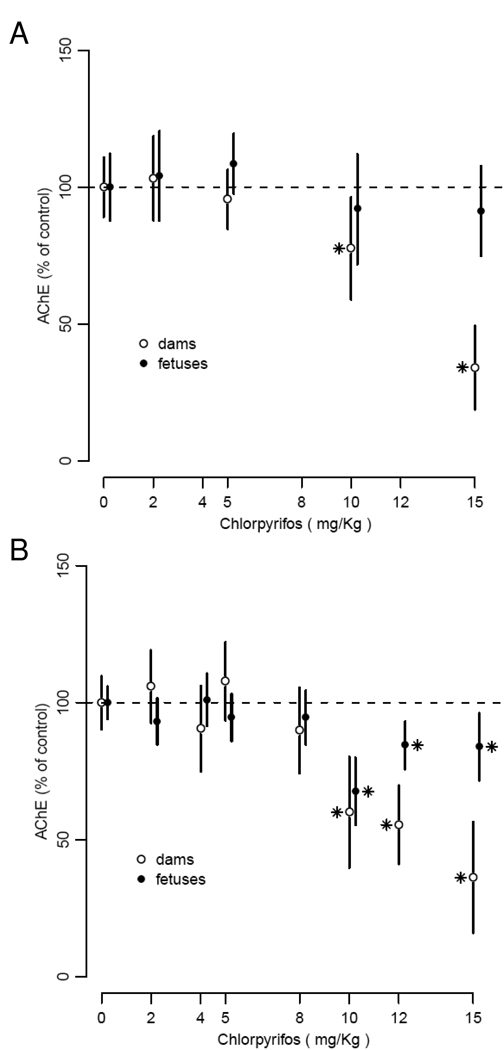
The analysis of AChE in the mixed effects model with individual dose levels is shown in Fig. 3 with 95% confidence levels from the mixed effects model about the estimated value of AChE inhibition for each dose. The overall analysis of variance for dose indicated that at 6 h after the last exposure (Fig. 3A) the changes in AChE with dose were not significant for fetuses, P = 0.35, but were significant for dams, P < 0.001 (10 and 15 mg/kg). At 24 h (Fig. 3B) the changes in AChE were statistically significant (P < 0.001) for both fetuses and dams (10, 12 and 15 mg/kg). There was an average of 4 fetuses per litter analyzed for AChE in both the 6 h and 24 h groups. Mean control fetuses AChE values were 21% (standard error 2.1%) of dam values with a range of 7.4% to 32% in 15 litters.
Fig 3.
AChE activity inhibition (percentage to control value) in the maternal and fetal brain evaluated 6 h (A) and 24 h (B) after the last CP treatment (GD 17). Data are means and 95% confidence interval of 2 to 13 litters in each group . * P≤0.05 compared to the control group (0 mg/kg, DMSO) as indicated by a mixed effects analysis of variance with dose as a fixed covariate and experiment and litter as random.
The trend analysis of AChE showed that the slope with dose was statistically significant for both dams and fetuses with values of −3.3% per mg/kg (P < 0.001) for dams and −1.2% per mg/kg (P < 0.001) for fetuses. The change in AChE in the dam controls was a 0.5% increase from 6 to 24 h which was not significant (P = 0.9) and in the fetuses controls was a 11% increase per mg protein (P = 0.03). There were no significant differences in AChE slopes with dose between 6 and 24 h for dams (P = 0.7) and fetuses (P = 0.17). For the 24 h animals significant AChE inhibition (P < 0.05) in maternal and fetal brains occurred at doses of 10mg/kg and above.
CP-induced differential gene expression in maternal and fetal brain
The Affymetrix Mouse Whole Genome 430 2.0 platform is composed by 45,101 probes that analyzes the expression level from over 34,000 mouse genes. We established our cut-off value (P ≤ 0.005) to identify a maximum of 4% of the probes to be differentially expressed across doses. Approximately 5–15% of all significantly altered genes were represented by more than one probe. In this manuscript, the term “genes” is used instead of probes due to the relatively small proportion of genes represented by more than one probe and to increase the ease of readership. Redundant probes were assessed separately and corresponding fold change values were not averaged.
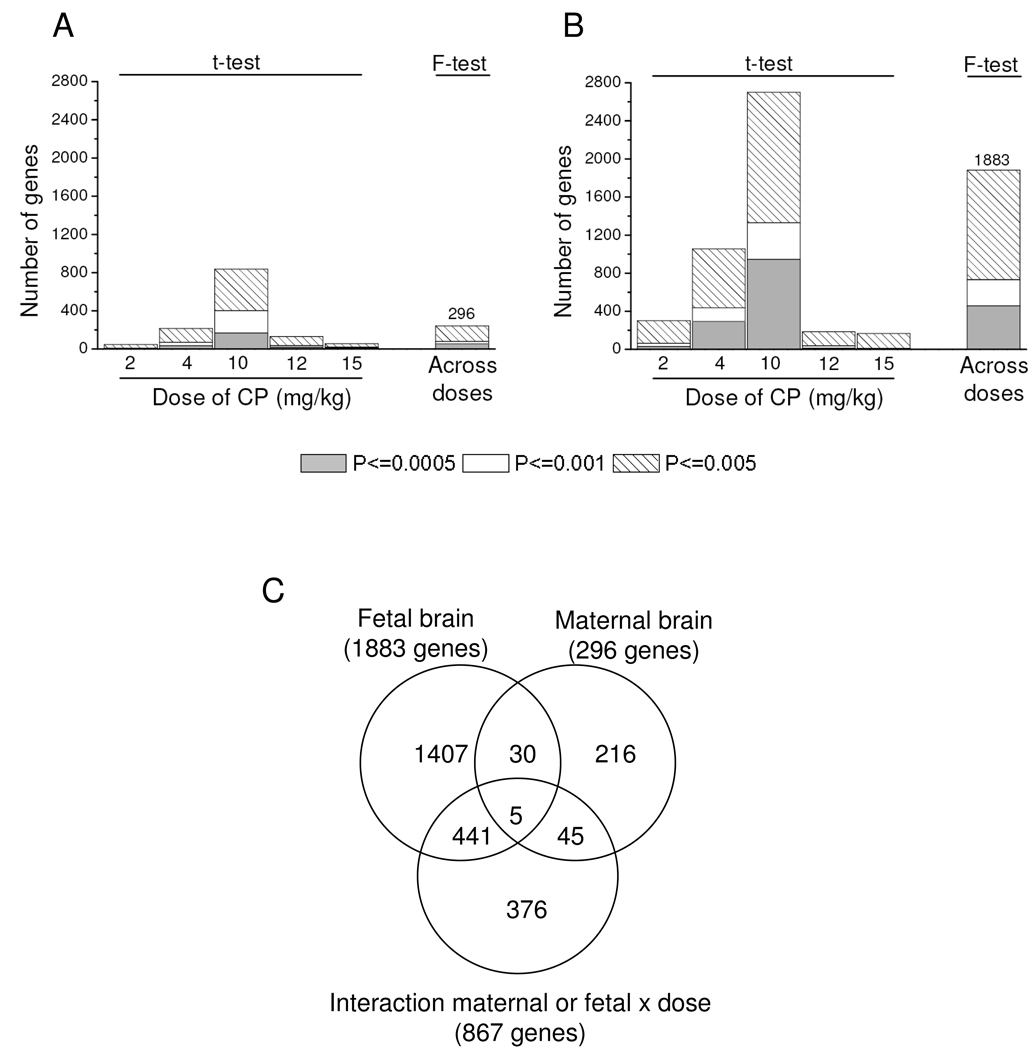
As shown in Fig. 4, we examined the impact of CP doses on gene expression in maternal (Fig 4A) and fetal brain (Fig 4B). Comparing each dose group to control, we observed a non-monotonic dose response in the amount of genes altered with each dose, with peak effects occurring in the 10mg/kg dose group for both maternal (Fig 4A) and fetal (Fig 4B) brains. This relationship was observed independently of the P-value chosen for CP assessments for this assessment. To investigate possible biological processes and pathways altered by CP treatment, we identified DEGs impacted across all doses of CP for maternal and fetal brain (F-test). In addition, to evaluate genes differentially impacted by CP between dam and fetal brain, we examined the interaction of maternal-fetal with CP doses. Using a cutoff of P ≤ 0.005, we identified 296, 1883 and 867 DEG in the maternal brain, fetal brain and interaction term (interaction of maternal-fetal with CP doses), respectively (Fig. 4C). As displayed in Fig. 4C, the gene lists obtained for CP effects in maternal and fetal brains were unique with approximately <10% of genes (35 genes) altered in the maternal brain observed to overlap with genes altered by CP in the fetal brain.
Fig. 4.
Number of genes whose expressions were altered at different P values in the maternal (A) and fetal (B) brain after exposure to increasing doses of CP from GD 6 to GD 17. For details on the statistical model employed, please refer to the text. In C, a Venn diagram illustrates the number of differentially expressed genes (P ≤ 0.005) across CP doses in the in the maternal brain, across CP doses in the fetal brain and considering the interaction of maternal-fetal with CP doses.
GO analysis of CP-induced DEG
GO analysis was conducted using MAPPFinder to identify enriched biological processes within DEG for the three different gene lists (maternal across doses, fetal across doses, interaction of maternal-fetal with CP doses).
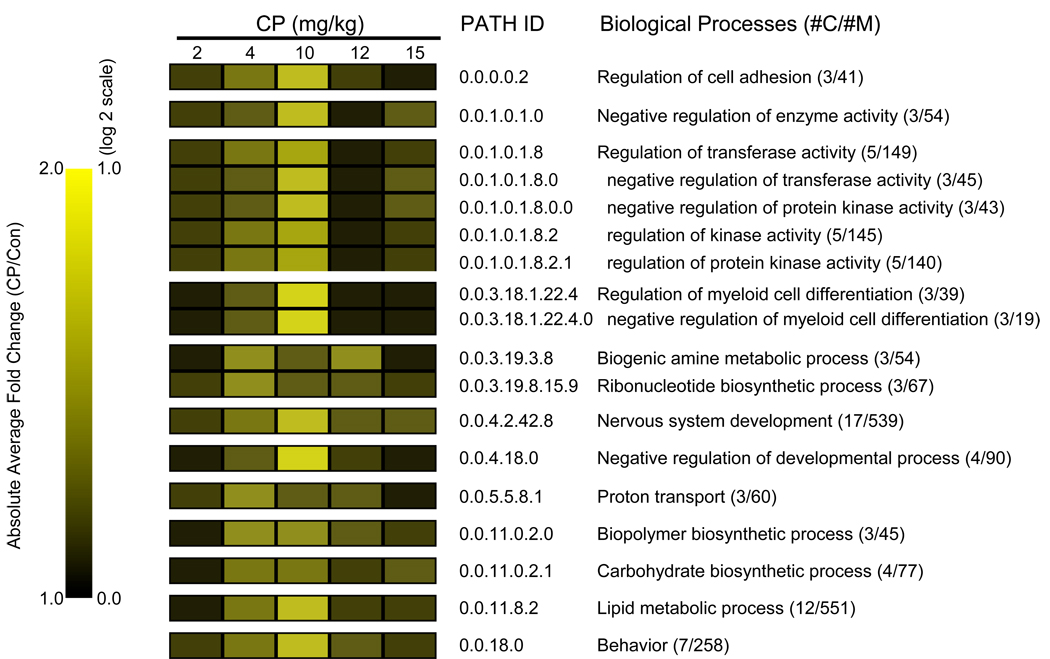
In Fig. 5–Fig. 6, we present quantitative changes in selected enriched GO biological process found to be altered in maternal (Fig. 5) and fetal brain (Fig. 6). In maternal brain (Fig. 5), CP altered the expression of genes involved in biological processes, such as cell adhesion, enzymatic activity (transferase and kinase), regulation of myeloid cell differentiation, metabolism (biogenic amine, ribonucleotide, carbohydrate, lipid), nervous system development, proton transport, behavior. GO-Quant analysis showed a monotonic dose-response relationship for 2, 4 and 10 mg/kg, with the dose of 10 mg/kg having the great quantitative impact across genes within enriched processes. At 12 and 15 mg/kg dose groups, the responses were lower than the peak 10mg/kg. For example, in 17 genes associated with “nervous system development”, we observed absolute average fold changes of 1.1, 1.4, 1.7, 1.2 and 1.2 for the CP doses of 2, 4, 10, 12 and 15mg/kg, respectively.
Fig. 5.
Quantitative analysis of gene expression-linked GO biological processes impacted by gestational CP treatment in the maternal brain. MAPPFinder was used to link the gene expression data to the GO hierarchy and establish associations between treatment and the affected GO terms. The cut-off criteria for the GO analysis results were Z score ≥ 2.0, P ≤ 0.05 and at least 3 genes changed in each category. The heat-map shows the average absolute magnitude in change within enriched subsets of biological process categories, calculated by GO-Quant. #C: number of genes changed; #M: number of genes linked to each category that was detected in the array.
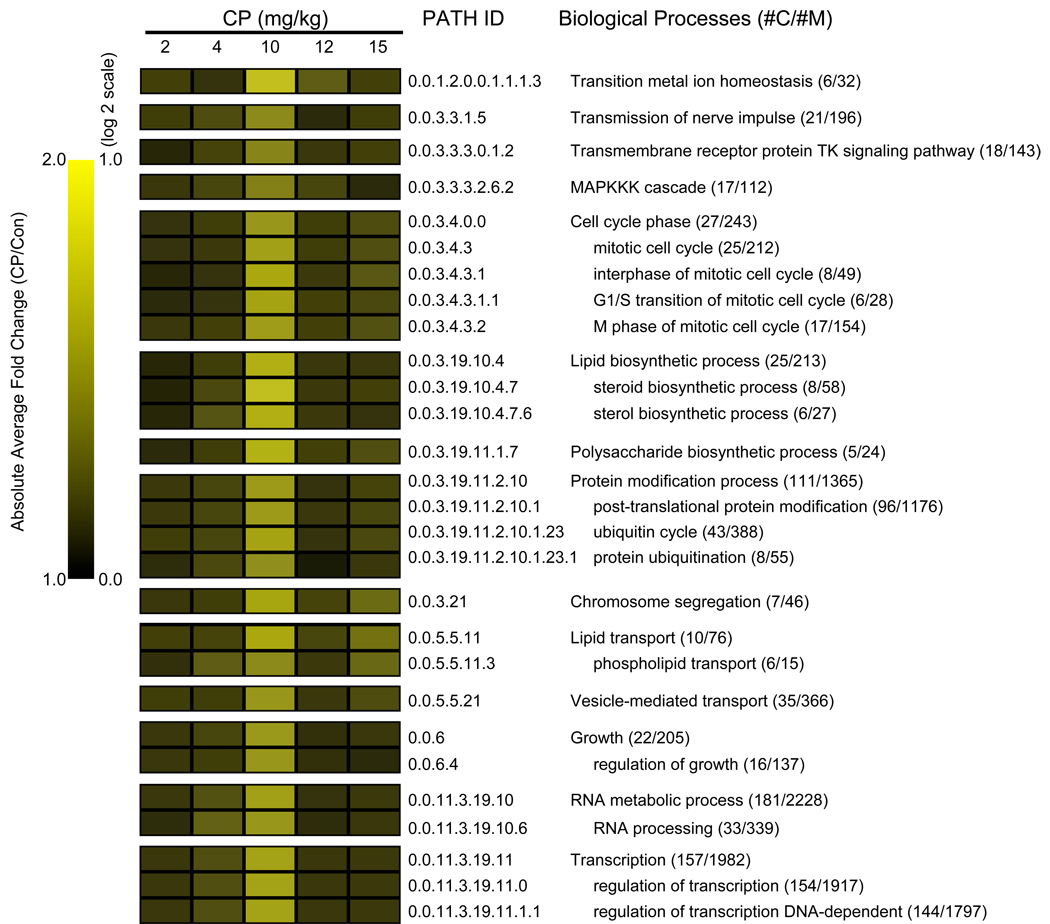
Fig. 6.
Quantitative analysis of gene expression-linked GO biological processes impacted by gestational CP exposure in the fetal brain. MAPPFinder was used to link the gene expression data to the GO hierarchy and establish associations between treatment and the affected GO terms. The cut-off criteria for the GO analysis results were Z score ≥ 2.0, P ≤ 0.05 and at least 5 genes changed in each category. The heat-map shows the average absolute magnitude in change within enriched subsets of biological process categories, calculated by GO-Quant. #C: number of genes changed; #M: number of genes linked to each category that was detected in the array.
In the fetal brain (Fig. 6), we identified enrichment of GOIDs related to ionic homeostasis, signaling and signal transduction (transmission of nerve impulse, tyrosine kinase signaling pathway, MAPKKK cascade), cell cycle, metabolism (lipid, polysaccharide, protein modification, RNA), growth, transport (lipid, vesicle-mediated), transcription. Again, similar to maternal brain, the greatest changes were observed in the 10 mg/kg dose group. In 27 genes associated with “cell cycle phase”, the absolute average fold changes were 1.1, 1.1, 1.5, 1.1, 1.2 for the CP doses of 2, 4, 10, 12 and 15mg/kg CP, respectively.
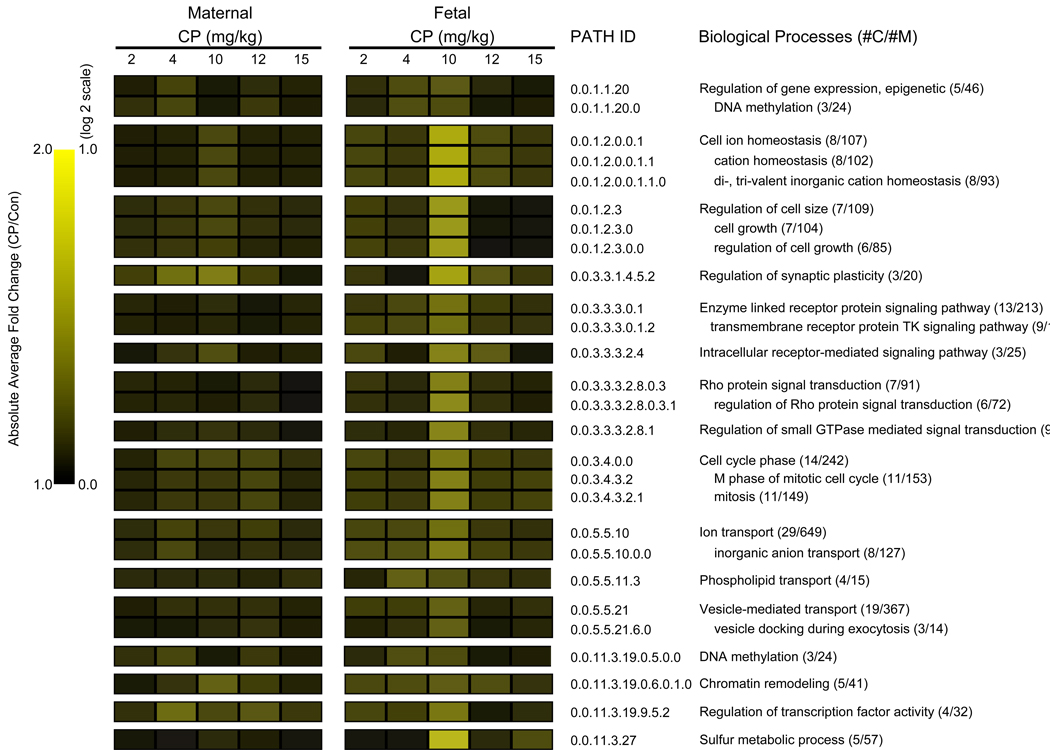
In addition, we conducted GO analysis for genes identified to be altered differentially by CP between maternal and fetal brains (interaction term) (Fig. 7). Enriched GOIDs included regulation of gene expression - epigenetic, ionic homeostasis, developmental processes (regulation of cell size, growth, regulation of synaptic plasticity), signaling and signal transduction (tyrosine kinase signaling, Rho protein signal transduction, small GTPase mediated signal transduction), cell cycle, transport (ion, vesicle-mediated transport, phospholipids), metabolism (sulfur metabolic process), transcription. In general, quantitative changes within these enriched GOIDs suggested the 10 mg/kg to be the peak effect dose group for all GOIDs, with exceptions in genes related to GOIDs “epigenetic mechanisms of regulation of gene expression” and “enzyme linked receptor protein signaling pathway”, which showed similar quantitiative changes in the 4 and 10mg/kg group. In general, the degree of CP-effects were greater in fetal versus maternal brain at the 10mg/kg dose group within these enriched GOIDs.
Fig. 7.
Quantitative analysis of GO biological processes found to be enriched within genes that showed differential expression pattern in the maternal and fetal brain across CP doses. MAPPFinder was used to link the gene expression data to the GO hierarchy and establish associations between treatment and the affected GO terms. The cut-off criteria for the GO analysis results were Z score ≥ 2.0, P ≤ 0.05 and at least 3 genes changed in each category. The heat-map shows the average absolute magnitude in change within enriched subsets of biological process categories, calculated by GO-Quant. #C: number of genes changed; #M: number of genes linked to each category that was detected in the array.
KEGG pathways enriched within CP-induced DEG
We used DAVID to identify enriched KEGG pathways within DEG impacted by CP. Pathways found to be enriched in the maternal brain included adherens junction, axon guidance, ErbB signaling pathway, GnRH signaling pathway, Jak-STAT signaling pathway (Table 2). In the fetal brain, pathways related to carbon fixation, cell cycle (cell cycle, p53 signaling pathway), long-term potentiation, melanogenesis, neurodegenerative diseases (neurodegenerative diseases, amyotrophic lateral sclerosis), Wnt signaling and diverse types of tumor-related pathways (glioma, colorectal cancer, acute myeloid leukemia, prostate cancer) were identified to be enriched (Table 3).
Table 2.
KEGG pathways found to be enriched in the maternal brain after gestational exposure to CP.
| Pathway | Genes | P | |
|---|---|---|---|
| All clusters | |||
| mmu04520 | Adherens junction | Csnk2a1, Ctnna1, Egfr, Fert2, Fyn, Iggap1, Mapk1, Mapk7, Pvrl1, Ssx21p, Tcf7l2, Wasf1, Wasf3 |
0.03 |
| mmu04360 | Axon guidance | Ablim1, Cfl2, Efna1, Efnb2, Fyn, Limk1, Mapk1, Nck1, Ntng1, Plxna3, Rnd1, Sema3b, Sema3d, Sema3f, Sema4b, Sema5b, Sema6d, Unc5a |
0.00 |
| mmu04012 | ErbB signaling pathway | Braf, Camk2a, Camk2d, Cbl, Crk, Egfr, Eif4ebp1, Hbegf, Mapk1, Mapk10, Nck1, Pak7, Pik3cd, Pik3r1, Sos1 |
0.02 |
| mmu04912 | GnRH signaling pathway | Cacna1c, Calm3, Camk2a, Camk2d, Egfr, Gnas, Hbegf, Itpr, Mapk1, Mapk10, Mapk3, Pla2g4e, Plcb4, Ptk2b, Sos1 |
0.02 |
| mmu04630 | Jak-STAT signaling pathway | Clcf1, Cntfr, Gh, Il11ra1, Il11ra2, Il22, Il6st, Iltifb, Osmr, Pik3r1, Prl, Sos1, Spred1, Stam2, Stat2 |
0.00 |
DAVID Bioinformatics Resources (NIAID/NIH) was used for this analysis and it was conducted using all probes that reached statistical significance across doses as well as probes belonging to each one of the 5 different clusters. A pathway was considered to be significantly enriched if it presented at least 3 differentially expressed genes and a P ≤ 0.05 for the association between the significantly changed genes and the pathway. No Kegg pathway was enriched in the clusters.
Table 3.
KEGG pathways found to be enriched in the fetal brain after gestational exposure to CP.
| Pathway | Genes | P | |
|---|---|---|---|
| All clusters | |||
| mmu05221 | Acute myeloid leukemia | Akt2, Akt3, Ccnd1, Grb2, Kit, Lef1, Rps6kb1, Rps6kb2, Runx1t1, Tcf7l2 |
0.03 |
| mmu05030 | Amyotrophic lateral sclerosis | Als2, Bcl2, Ppp3ca, Sod1, Trp53 | 0.02 |
| mmu00710 | Carbon fixation | Aldoart1, Aldoc, Gpt2, Mdh1, Me3, Tkt | 0.01 |
| mmu04110 | Cell cycle | Ccnb3, Ccnd1, Ccnd2, Cdc23, Cdc27, Cdc6, Cdkn1a, Chek1, Crebbp, Gadd45g, Rbl1, Skp2, Tgfb2, Trp53, Ywhaz |
0.02 |
| mmu0521 | Colorectal cancer | Akt2, Akt3, Bcl2, Ccnd1, Dvl1, Fzd1, Fzd3, Grb2, Lef1, Mapk9, Tcf7l2, Tgfbr1, Tgfb2, Trp53 |
0.01 |
| mmu05214 | Glioma | Akt2, Camk2g, Ccnd1, Cdkn1a, Grb2, Igf1, Tgfa, Trp53 | 0.04 |
| mmu04720 | Long-term potentiation | Atf4, Camk2b, Camk2g, Gnaq, Gria1, Gria2, Grm5, Ppp3ca, Ppp3cb |
0.03 |
| mmu04916 | Melanogenesis | Adcy2, Camk2b, Camk2g, Creb1, Crebbp, Dvl1, Fzd1, Fzd3, Gnaq, 0.02 Kit, Kitl, Lef1, Pomc, Tcf7l2 |
|
| mmu01510 | Neurodegenerative diseases | Als2, Bcl2, Crebbp, Grb2, Hspa5, Htt, Prnp, Sod1, Vapb | 0.00 |
| mmu04115 | p53 signaling pathway | Apaf1, Ccnb2, Ccnd1, Ccnd2, Cdkn1a, Chek1, Ddb2, Gadd45g, Igf1, Steap3, Trp53 |
0.02 |
| mmu05215 | Prostate cancer | Akt2, Akt3, Atf4, Bcl2, Ccnd1, Cdkn1a, Creb1, Crebbp, Grb2, Hsp90b1, Igf1, Lef1, Tcf7l2, Tgfa, Trp53 |
0.01 |
| mmu04310 | Wnt signaling pathway | Camk2b, Ccnd1, Ccnd2, Ctbp2, Daam1, Dlv1, Fbxw11, Fzd1, Fzd3, Lef1, Porcn, Ppp3ca, Ppp3cb, Prickle2, Senp2, Tbl1x, Trp53 |
0.00 |
| Cluster A | |||
| mmu05030 | Amyotrophic lateral sclerosis | Als2, Ppp3ca, Sod1, Trp53 | 0.03 |
| mmu00710 | Carbon fixation | Aldoc, Aldoart1, Me3, Tkt | 0.05 |
| mmu00030 | Pentose phosphate pathway | Aldoc, Aldoart1, Dera, Tkt | 0.05 |
| Cluster B | |||
| mmu00100 | Biosynthesis of steroids | Lss, Sc5d, Sc4mol, Sqle | 0.02 |
| mmu04110 | Cell cycle | Ccnb2, Ccnd1, Ccnd2, Cdc6, Cdc23, Chek1, Crebbp, Rbl1, Skp2 | 0.00 |
| mmu05210 | Colorectal cancer | Akt3, Ccnd1, Fzd1, Fzd3, Grb2, Mapk9, Tgfbr1 | 0.01 |
| mmu04910 | Insulin signaling pathway | Acaca, Akt3, Grb2, Mapk9, Pp1r3b, Ppp1r3c, Prkar2a, Prkar2b | 0.04 |
| mmu01510 | Neurodegenerative diseases | Crebbp, Grb2, Htt, Hspa5 | 0.06 |
| Cluster C | |||
| -- | |||
| Cluster D | |||
| mmu04730 | Long-term depression | Igf1, Gria2, Grm5, Npr2, Prkg2 | 0.04 |
| mmu04720 | Long-term potentiation | Atf4, Camk2g, Gria2, Grm5, Ppp3cb | 0.03 |
| mmu00350 | Tyrosine metabolism | Esco1, Esco2, Hemk1, Prmt6 | 0.05 |
DAVID Bioinformatics Resources (NIAID/NIH) was used for this analysis and it was conducted using all probes that reached statistical significance across doses as well as probes belonging to each one of the 4 different clusters. A pathway was considered to be significantly enriched if it presented at least 3 differentially expressed genes and a P ≤ 0.05 for the association between the significantly changed genes and the pathway
Cluster-based GO and pathway analysis of CP-induced DEG
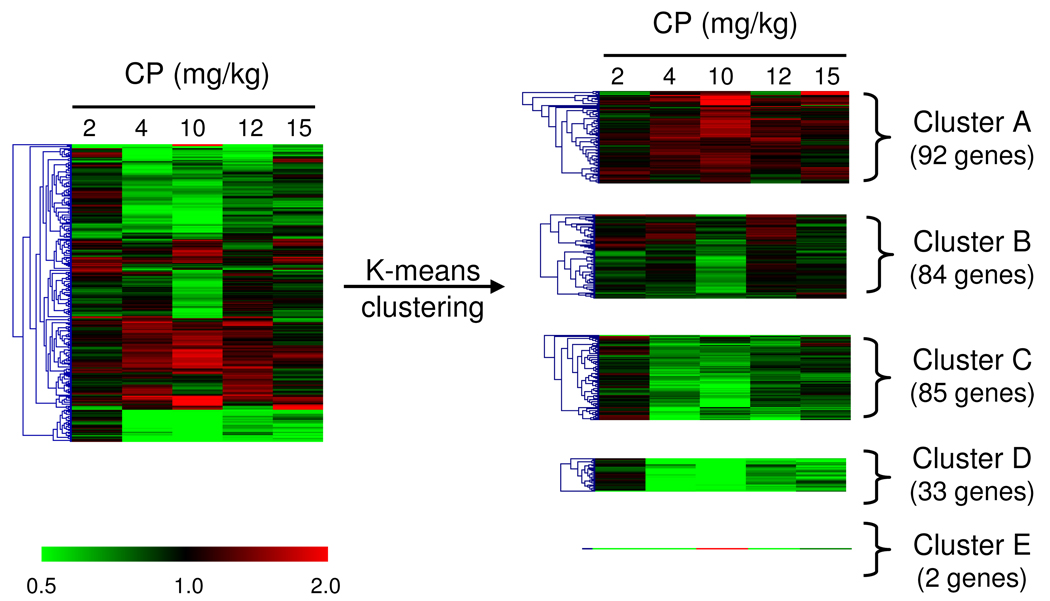
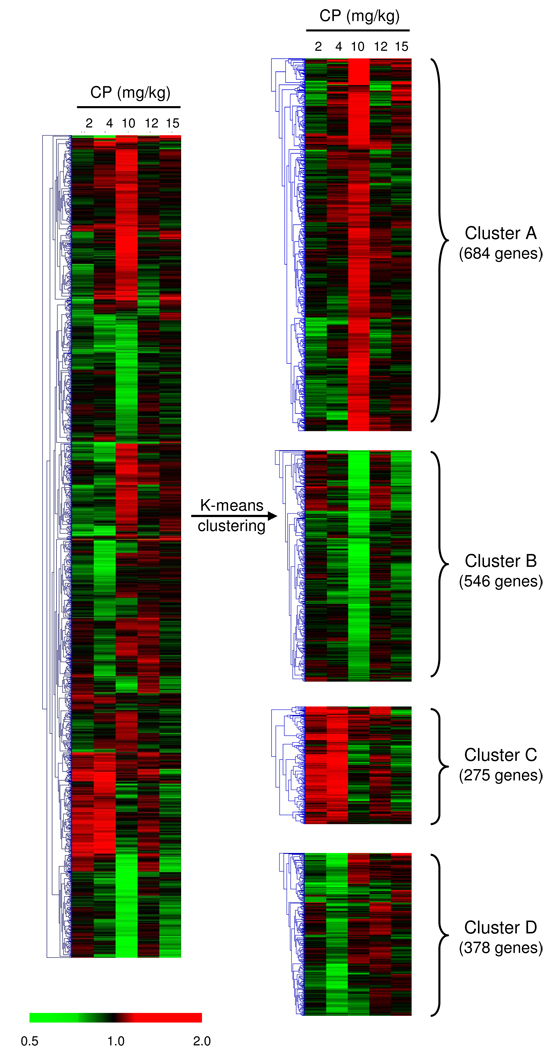
Considering the non-monotonic nature of the CP effect on gene expression we decided to use k-means clustering to split our DEG in different groups of genes based on similarity of expression patterns across doses. After clustering the DEG list into 3–10 nodes, we found that the best approach was to group the genes into 5 and 4 clusters for the maternal and fetal brain, respectively (Fig. 8 and Fig. 9) and then to conduct GO and pathway analysis.
Fig. 8.
Hierarchical cluster analysis (left panel) of the 296 genes that were differentially expressed in the maternal brain (P ≤ 0.005) after gestational exposure to CP (GD6-17). Data are mean fold changes for each treatment group. The right panel shows the division of these genes, by K-means clustering analysis, in five clusters describing the pattern of expression across doses. Cluster A shows genes primarily up-regulated in the 10 mg/kg group; Cluster B, genes down-regulated in the 10 mg/kg group; Cluster C, genes down-regulated primarily in the 4 and 10 mg/kg groups; Cluster D, genes down-regulated across doses of 4 to 15 mg/kg and Cluster E shows 2 genes (Prl and Pomc) up-regulated in the 10 mg/kg group and down-regulated in the other groups.
Fig. 9.
Hierarchical cluster analysis (left panel) of the 1883 genes that were differentially expressed in the fetal brain (P ≤ 0.005) after gestational exposure to CP. Data are mean fold changes for each treatment group. The right panel shows the division of these genes, by K-means clustering analysis, in four clusters describing the pattern of expression across doses. Cluster A shows genes primarily up-regulated in the 10 mg/kg group; Cluster B, genes down-regulated primarily in the 10 mg/kg group; Cluster C, genes up-regulated primarily in the 2 and 4 mg/kg groups; Cluster D, genes down-regulated primarily in the 4 mg/kg group.
As presented in Fig. 8, the 296 DEG from the maternal brain were divided into clusters A (92 genes primarily up-regulated at the 10 mg/kg), B (84 genes down-regulated primarily in the 10 mg/kg group), C (85 genes down-regulated primarily in the 4 and 10 mg/kg groups), D (33 genes down-regulated in the 4, 10, 12 and 15 mg/kg groups ) and E (2 genes – Pomc and Prl-down-regulated in all doses except for 10 mg/kg). GO and pathway analysis (MAPPFFinder, DAVID) were conducted within each cluster (Table 4). Table 4 summarizes the biological processes enriched within each cluster. In cluster A, GO categories such as cellular developmental process, cell differentiation, embryonic morphogenesis and DNA processes were identified to be overrepresented. Genes within cluster B showed enrichment of metabolism (lipid and phosphorus) and behavior categories. Genes within cluster C showed enrichment of GO categories involved in regulation of developmental process, cell adhesion, nervous system development, neurogenesis, lipid metabolism, membrane lipid metabolic process, vesicle-mediated transport, secretory pathway, microtubule-based process and synaptic transmission. No GO biological process was enriched in cluster D and no KEGG pathway was enriched in any of the clusters based on our cut-off criteria (3 DEG, P ≤ 0.05).
Table 4.
Summary of the biological processes enriched in each cluster of genes derived from the differentially expressed genes in the maternal brain after gestational CP treatment. MAPPFinder was used to identify the enrichement. There was no enriched biological process for cluster D.
| Cluster A | Cluster B | Cluster C | |
|---|---|---|---|
| Development | |||
| Developmental process | X | X | |
| Cell adhesion | X | ||
| Cell differentiation | X | ||
| Embryonic morphogenesis | X | ||
| Nervous system development | X | ||
| Neurogenesis | X | ||
| Metabolism | |||
| Lipid metabolism | X | X | |
| Membrane lipid metabolic process | X | ||
| Phosphorus metabolism | X | ||
| DNA/RNA processes | |||
| DNA packaging | X | ||
| Establishment and/or maintenance of chromatin architecture | X | ||
| Transport | |||
| Vesicle-mediated transport | X | ||
| Secretory pathway | X | ||
| Others | |||
| Behavior | X | ||
| Synaptic transmission | X | ||
| Microtubule-based process | X |
The 1883 DEG identified to be impacted by CP in the fetal brain clustered into 4 groups: A (684 genes primarily upregulated in the 10 mg/kg group), B (546 genes mainly downregulated at 10 mg/kg); C (275 genes primarily upregulated in the 2 and 4 mg/kg groups) and D (378 genes mainly downregulated in the 4 mg/kg groups) (Fig. 9). As described for the maternal brain, GO and pathway analysis (MAPPFinder, DAVID) were conducted within each cluster (Table 5). In cluster A, enriched GOIDs included, ion homeostasis, signaling and signal transduction (regulation of signal transduction, regulation of action potential, MAPKKK cascade, JNK cascade), cell cycle (G1/S transition of mitotic cell cycle), myelination and metabolism (lipid, carbohydrate, sulfur) (Table 5) whereas enriched KEGG pathways included amyotrophic lateral sclerosis, carbon fixation and pentose phosphate pathway (Table 3). In cluster B, enriched GOIDs were signaling and signal transduction (ER-nuclear signaling pathway, smoothened signaling pathway, tyrosine kinase signaling), multiple categories of cell cycle, cell division, developmental process (cell adhesion, cell migration, embryonic development), metabolism (carbohydrate, lipid, ubiquitin cycle), DNA and RNA processes (chromatin modification, transcription) as well as some categories of transport and behavior (locomotion) (Table 5); enriched KEGG pathways included biosyntesis of steroids, cell cycle, colorectal cancer, insulin signaling pathway and neurodegenerative diseases (Table 3). In cluster C, cell division, translation, general biological processes (cellular component organization and biogenesis, macromolecular complex assembly, membrane organization and biogenesis) and transport (vesicle-mediated, ion) were the enriched GOIDs (Table 5) and no KEGG pathway was shown to be enriched. Finally, cluster D contained GOIDs related to transmission of nerve impulse, chromatin modification, RNA processing and lymphocyte differentiation and activation (Table 5); enriched KEGG pathways included long-term depression, long-term potentiation and tyrosine metabolism (Table 3).
Table 5.
Summary of the biological processes enriched in each cluster of genes derived from the differentially expressed genes in the fetal brain after gestational CP exposure. MAPPFinder was used to identify the enrichement.
| Cluster A | Cluster B | Cluster C | Cluster D | |
|---|---|---|---|---|
| Signaling | ||||
| Regulation of signal transduction | X | |||
| ER-nuclear signaling pathway | X | |||
| Transmembrane receptor tyrosine kinase signaling | X | |||
| MAPKKK cascade | X | |||
| JNK cascade | X | |||
| Smoothened signaling pathway | X | |||
| Transmission of nerve impulse and/or synaptic transmission | X | X | ||
| Cell cycle | X | X | ||
| Cell division | X | X | ||
| Development | ||||
| Cell adhesion | X | |||
| Cell migration | X | |||
| Cell motility | X | |||
| Embryonic development | X | |||
| Membrane organization and biogenesis | X | |||
| Myelination | X | |||
| Metabolism | ||||
| Carbohydrate metabolism | X | X | ||
| Lipid metabolism | X | X | ||
| Protein modification, ubiquitin cycle | X | |||
| Sulfur metabolism | X | |||
| DNA/RNA processes | ||||
| Chromatin modification or remodeling | X | X | ||
| Chromosome segregation | X | |||
| Nucleoside metabolism | X | |||
| RNA processing | X | |||
| Transcription | X | |||
| Translation | X | |||
| Transport | ||||
| Electron transport | ||||
| Endocytosis | X | |||
| Exocytosis | ||||
| Ion transport | X | X | ||
| Lipid transport | X | |||
| Secretory pathway | X | |||
| Vesicle-mediated transport | X | |||
| Others | ||||
| Ionic homeostasis | X | |||
| Locomotion | X | |||
| T cell differentiation / activation | X |
It is noteworthy that, in general, 50–60% of our DEG were linked to a GO term in the MAPPFinder database except for fetal brain cluster C. From the 275 genes present in this cluster, only 20 have been linked to a GO term in the MAPPFinder database. A closer look at this genelist showed that 125 probes have not even been annotated. As a result of this poor annotation and GO linking in the MAPPFinder database, this cluster could not be adequately characterized. Since this cluster is of particular interest because it contains genes shown to be differentially expressed in low CP doses we searched each individual gene in the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/gene). We observed that this cluster contains genes involved in axon guidance (Ank3, Myh10, Nfasc, Nrp1), cell cycle (6720463M24Rik, Nasp, Steap3, Zfp830), cytoskeleton organization (Cttnbp2, Diap1, Myh10, Ttl), nervous system development (Med1, Mtss1, Myh10), proteolysis (4931433A01Rik, Adamts9, Adamts18), regulation of growth (Brd8, Socs2), regulation of apoptosis (Camk1d, Steap3), signal transduction (Fgd3, Itpkb, Mtss1, Pde10a, Rgs20, Socs2, Tnc), synapse organization (Ank3, Nfasc), transcription (A730035I17Rik, Akna, Brd8, Cux1, Etv3, Med1, Psmd9, Tle4, Zmiz1), translation (Eif2ak1, Eif3c, Eif4g3).
Dose-effect of CP on genes belonging to “cell cycle” and “nervous system development” GO categories
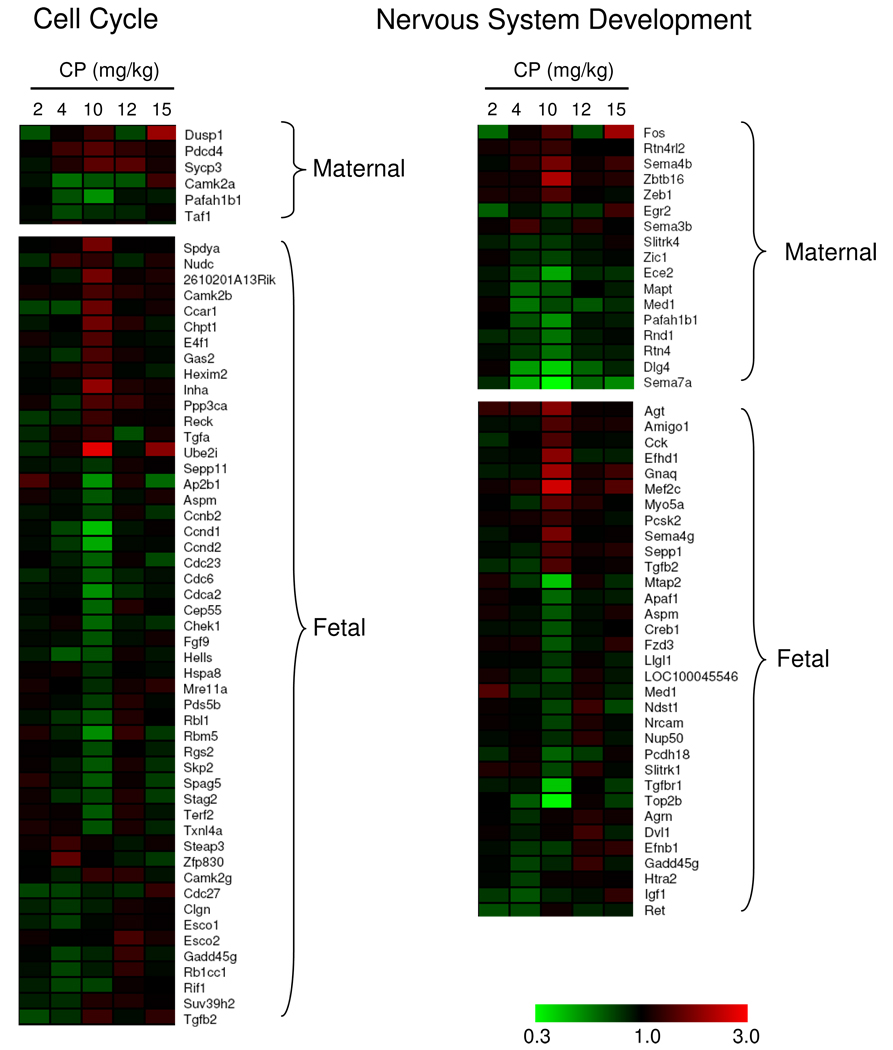
To illustrate the dose-effect of CP on gene expression at the gene level, two GOIDs shown to be enriched in our dataset were selected: “cell cycle” and “nervous system development” (Fig. 10).
Fig. 10.
Differentially expressed genes (P ≤ 0.005) associated with the GOIDs “cell cycle” and “nervous system development”. MAPPFinder was used to filter the genes belonging to each one of these categories. Upper panels show genes found to be significantly altered in the maternal brain and bottom panels show genes found to be significantly altered in the fetal brain.
The “cell cycle” GOID has 602 genes linked to it in the MAPPFinder database. From those, 6 and 50 were significantly altered in the maternal and fetal brain, respectively. In the maternal brain (Fig. 10, top lef panel), 3 genes were primarily up-regulated (Dusp1, Pdcd4, Sycp3) and 3 down-regulated (Camk2a, Pafah1b1, Taf1). In the fetal brain (Fig. 10, bottom left panel), from the 50 DEG, 14 of them were primarily up-regulated at the 10 mg/kg dose (Spdya, Nudc, 2610201A13Rik, Camk2b, Ccar1, Chpt1, E4f1, Gas2, Hexim2, Inha, Ppp3ca, Reck, Tgfa, Ube2i), 24 of them were primarily down-regulated at 10 mg/kg (Sepp11, Ap2b1, Aspm, Ccnb2, Ccnd1, Ccnd2, Cdc23, Cdc6, Cdca2, Cep55, Chek1, Fgf9, Hells, Hspa8, Mre11a, Pds5b, Rbl1, Rbm5, Rgs2, Skp2, Spag5, Stag2, Terf2, Txnl4a). For the others, no clear peak effect can be observed and their expression altered in both directions depending on the dose group.
In the “nervous system development” GOID which has 539 genes linked in the MAPPFinder database, 17 genes were shown to be significantly altered in the maternal brain and 33 in the fetal brain. The only gene commonly altered both in the maternal and fetal brain was Med1 that has been described to play a role in transcription and brain development. In the maternal brain (Fig. 10, top right panel), genes that were significantly altered by CP treatment are involved in behaviors such as locomotory behavior, learning and memory (Dlg4, Pafah1b1, Zic1) as well as rhythmic behavior (Egr2); response to stimulus (Fos); cytoskeleton organization (Pafah1b1); signaling (Rnd1); cell differentiation (Pafah1b1, Sema3b, Sema4b, Sema7a, Zbtb16, Zic1); axonogenesis (Rtn4, Rtn4rl2) and apoptosis (Rtn4, Zbtb16). The peak effect for 5 of them was an up-regulation at the 10mg/kg dose (Fos, Rtn4rl2, Sema4b, Zbtb16, Zeb1) whereas for the others the peak effect was a down-regulation primarily at the 4 and 10 mg/kg doses. In the fetal brain (Fig. 10, bottom right panel), CP exposure altered the expression of genes involved in multiple processes underlying nervous system development, such as cell proliferation (Agt, Tgfb2), migration (Cck, Efhd1, Ret, Top2b), differentiation (Amigo1, Aspm, Efnb1, Htra2, Igf1, Mef2c, Sema4g, Tgfb2, Tgfbr1), axonogenesis (Amigo1, Cck, Creb1, Efnb1, Nrcam, Slitrk1, Tgfb2, Top2b), synaptogenesis (Dvl1, Myo5a), myelination (Amigo1, Egr2, Myo5a) and apoptosis (Apaf1, Htra2, Mef2c, Tfgb2, Tgfbr1). From the 33 genes, 11 of them had a peak effect of up-regulation in the 10 mg/kg dose (Agt, Amigo1, Cck, Efhd1, Gnaq, Mef2c, Myo5a, Pcsk2, Sema4g, Sepp1, Tgfb2) whereas 22 of them had a peak effect of down-regulation in the 4 and/or 10 mg/kg doses (Agrn, Apaf1, Aspm, Creb1, Dvl1, Efnb1, Fzd3, Gadd45g, Htra2, Igf1, Llgl1, Loc100045546, Med1, Mtap2, Ndst1, Nrcam, Nup50, Pcdh18, Ret, Slitrk1, Tgfbr1, Top2b).
DISCUSSION
Dose-effect relationship between CP and brain AChE activity
Using a systems-based analysis, this study was designed to evaluate the effect of gestational exposure to CP on gene expression. We investigated CP doses with differential inhibition on maternal and fetal brain AChE. We observed dose-dependent effects on AChE inhibition with a LOAEL for maternal brain at 10 mg/kg CP and with up to a 60% inhibition in the 15mg/kg group (highest dose tested, 24 h after last treatment). In the fetal brain, significant inhibition of AChE was observed at 24 h after the last CP exposure, with the LOAEL and peak AChE inhibition occurring in the 10 mg/kg dose group (∼35% inhibition). Higher inhibition of AChE in the maternal brain compared to the fetal brain is in accordance with other studies that evaluated the degree of cholinesterase inhibition in the fetal and maternal brain (Chanda et al., 1995; Chanda and Pope, 1996; Lassiter et al., 1998; Mattsson et al., 2000). It has been proposed that AChE in the fetus is able to recover more quickly between dosages than AChE in the mature adult brain (Lassiter et al., 1998). Surprisingly, in our study, AChE inhibition in the fetal brain was observed only at 24h after the last CP treatment and not at the earlier timepoint (6h). In rats, peak brain cholinesterase inhibition has been suggested to occur between 5 to 10 h following the last dosage (7 mg/kg of CP, GD14–18, daily oral gavage), with minimal changes in AChE activity occurring at 24h, suggesting full recovery in the fetal brain (Lassiter et al., 1998). Another study suggests inhibition in rats to occur at least three days after the last dosage (3 or 7 mg/kg of CP, GD6–20, daily oral gavage) (Richardson and Chambers, 2004). Another surprising result was the value of the maternal and fetal LOAEL based on AChE inhibition: 10 mg/kg. In studies carried out with rats, significant brain cholinesterase inhibition has been described to occur in lower doses (3 or 7 mg/kg) (Lassiter et al., 1998; Richardson and Chambers, 2004). However, in mice, lack of fetal brain AChE inhibition 24 h after the last CP exposure has been reported in levels lower than 10mg/kg (3 and 6 mg/kg, GD15–18, daily oral gavage), supporting our observations (Ricceri et al., 2006). These discrepancies illustrate that different experimental designs (animal species, route of administration, gestational days of exposure, different laboratories, etc) may impact observations of cholinesterase inhibition and highlight the importance of having cholinesterase values parallel with our toxicogenomic assessments (obtained from the same litters) for a comparable context across previous studies.
Dose-effect relationship between CP and toxicogenomic alterations
In this microarray study, 296 and 1883 DEGs were identified to be impacted by CP in the maternal and fetal brain, repectively (P ≤ 0.005) (Fig. 4). Only 35 genes were commonly impacted (Fig. 4C) in both maternal and fetal brain. These 35 genes were distributed sparsely among several diverse biological functions (results not shown). Such lists of DEG resulted in unique lists of enriched GO categories and pathways. Clear differences were observed not only in specific gene changes, but also in the corresponding GO categories/pathways represented between maternal and fetal brain. Despite these differences, it was significant to note the similarity in the shape of the dose-effect curves. This is revealed by our intial analysis using t-tests between each dose and its comparative control (Fig. 4A), as well as later analyses using GO-Quant at the GO/pathway level, the greater efficacy of the 10 mg/kg dose in altering gene expression in both the maternal and fetal brains was translated. Our results suggest the existence of an inverted U dose-effect relationship for the CP influence on gene expression both in the developing and mature brain which does not follow the AChE inhibition dose-effect relationship. In general, gene expression increased in a dose-dependent manner up to the 10 mg/kg and then decreased. The 4 mg/kg dose had a greater impact on gene expression than 12 and 15 mg/kg, which, in turn, had a greater impact on AChE activity. In recent published studies assessing gene expression alterations following CP exposure/treatment (Betancourt et al., 2006; Mense et al., 2006; Slotkin and Seidler, 2007; Slotkin et al., 2007; Slotkin and Seidler, 2008; Slotkin et al., 2008; Slotkin and Seidler, 2009; Stapleton and Chan, 2009), only two of them were conducted with more than one dose (Betancourt et al., 2006; Stapleton and Chan, 2009) and only one employed a genome-wide analysis, in vivo (Stapleton and Chan, 2009). The later study investigated gene expression in the forebrain of Male Fischer 344 adult rats 4 days following acute exposure to 0.5, 1.0, 5, 10, 30, and 50 mg/kg CP and, corroborating our results, the authors reported that, for up-regulated genes, the number of probes meeting their criteria for analysis gradually increased as the dose of CP escalated and reached a peak of 139 at 5 and 10 mg/kg then decreased to less than 20 at 30 and 50 mg/kg (Stapleton and Chan, 2009). No information on AChE activity inhibition was provided. Interestingly, it is worth noting that non-monotonic dose-effect relationship has also been reported in a behavioral study that investigated the cognitive performance of rats after gestational exposure (GD 17–20) to CP (1 and 5 mg/kg, sc) (Levin et al., 2002). Since only the lower dose negatively impacted the cognitive performance of female offspring, the authors suggested that the cognitive impairment is offset by higher exposures that cause greater inhibition of fetal brain AChE (and, consequently, increase acetylcholine availability), which would indicate that noncholinergic mechanisms would be more important at low exposure levels.
GO categories and KEGG pathways enriched within CP-induced DEG with similar expression profile
Due to the complexity within the dose-effect relationship between CP and gene expression, the use of K-means clustering to group genes with similar expression profile followed by the GO and pathway analysis in separated clusters seemed a reasonable approach to evaluate the impact of CP doses with differential inhibition of brain AChE on gene expression.
In the maternal brain, genes that had a peak effect (upregulated) in the 10 mg/kg group (cluster A) were found to be involved in developmental and DNA processes; genes that had a peak effect of down-regulation in the 10 mg/kg group (cluster B) were found to be involved in metabolism (lipid, phosphorus) and behavior; genes that were primarily downregulated in 4 and 10 mg/kg (Cluster C), were involved in GO categories such as regulation of developmental process, cell adhesion, nervous system development, neurogenesis, lipid metabolism, secretory pathway, microtubule-based process and synaptic transmission. Interestingly, despite the differences regarding animal species and gender, CP treatment duration, region sampled and statistical approach the study from Stapleton and Chan (Stapleton and Chan, 2009) described in the previous subsection also resulted in the enrichment of categories/pathways such as nervous system development, cell adhesion, synaptic transmission, long-term potentiation.
In terms of the amount of DEGs identified with CP exposure, fetal brain was impacted to a greater degree than the maternal brain (Fig. 4). Moreover, in the fetal brain we observed the presence of genes that were specifically impacted by doses of CP that did not impact AChE activity (2 and 4 mg/kg) (Fig. 9). They were shown to be involved in cell division, translation, and general biological processes (cluster C); and lymphocyte differentiation and activation, transmission of nerve impulse, chromatin modification, RNA processing, long-term potentiation, long-term depression and tyrosine metabolism (cluster D). Unfortunately, a better characterization of cluster C was compromised by the poor annotation of genes belonging to this cluster as well as poor linking of the annotated genes to GO categories in the MAPPFinder database but a gene-based search revealed that the annotated genes belonging to this category have been linked in developmental processes (axon guidance, cytoskeleton organization, regulation of growth, regulation of apoptosis, synapse organization), cell cycle, signal transduction, proteolysis, transcription and translation.
The approach used to analyze our data was sensitive to reveal the enrichment of GO categories and/or pathways consistent with morphological, biochemical and molecular endpoints that have been described in the neurotoxicity and developmental neurotoxicity of CP, such as cell division and proliferation, axonogenesis, synaptogenesis, DNA and RNA synthesis, synaptic transmission, cell signaling cascades, nuclear transcription factors (Song et al., 1997; Song et al., 1998; Crumpton et al., 2000b; Crumpton et al., 2000a; Dam et al., 2000; Garcia et al., 2001; Levin et al., 2001; Schuh et al., 2002; Dam et al., 2003; Qiao et al., 2003; Eaton et al., 2008). It is notheworthy that most of the mentioned categories were found to have genes differentially expressed not only in the groups treated with doses above the threshold for AChE inhibition but also in the low dose groups (2 and/or 4 mg/kg). Even though alterations in gene expression are not necessarily reflecting alterations in protein content, our study suggest that at least at the gene expression level, doses that do not impact AChE activity may influence the expression of genes that may, directly or indirectly, play a role in the nervous system development and functioning.
A surprising finding in our study was the the lack of enriched categories related to oxidative stress that has been suggested as one of the CP mechanisms of toxicity in rats postanatally exposed to CP (1 mg/kg, s.c., PND 1–4) and in vitro models (Crumpton et al., 2000b; Slotkin and Seidler, 2007; Slotkin and Seidler, 2009). Even though our study was conducted with mice gestationally exposed to CP (i.e., different species and different regimen of exposure) it should be considered that lack of enrichment of a category does not mean that genes related to a process have not been altered. As a matter of fact, from the 43 genes related to the GO category “response to oxidative stress”, 5 of them (Als2, Epas1, Prnp, Sod1 and Txnrd2) were differentially expressed in the fetal brain after CP exposure at the conservative P-value of P ≤ 0.005. Moreover, the definition of oxidative stress is variable and of course this may influence genes that are linked to this process. For instance, in the GO project, the genes encoding glutathione reductase (Gsr), glutathione synthetase (Gss), and glutathione transferases (Gst) are not linked to the category “response to oxidative stress”. In our dataset, Gstz1 was differentially expressed. From those 6 genes that were differentially expressed, except for Prnp all the other 5 of them were in cluster A, i.e., they were primarily up-regulated in the 10 mg/kg dose group.
CP exposure and cell cycle
Multiple GO subcategories and pathways related to cell cycle were found to be enriched, especially in the fetal brain. Fifty genes linked to the “cell cycle” category in the MAPPFinder database and 15 linked in the KEGG pathway “cell cycle” were identified to be significantly altered in the fetal brain. Alterations in proliferation pahtways are implicated in several neurodevelopmental disorders including neural tube defects and cleft palate as well as more subtle structural and functional alterations. Multiple studies suggest disturbances in cell division to be a relevant mechanism of CP toxicity in rodents (Roy et al., 1998; Guizzetti et al., 2005). Here, we support previous observations of CP-induced alterations in cell proliferation, providing evidence of disrupted expression of genes involved in cell cycle regulation in the fetal brain (and to a lesser extent in the maternal brain) (Figure 10). In the fetal brain (Figure 10 bottom left), we observed clusters of up and down-regulated genes (13 and 24 genes) showing the greatest degree of change within the 10mg/kg dose group. In particular, down-regulated genes included key cyclins, critical in promoting transition through G1-S (Ccnd1, Ccnd2) and G2-M (Ccnb2) checkpoints, members involved in mitosis (Cdc23, Aspm), and genes associated with maintaining proper DNA replication (Cdc6, Chk1, Hells, Pds5b). Previous studies indicate alteration in these genes from this cluster to be linked with defects in developmental growth and central nervous system alterations. For example, Ccnd2 knockout mice show reduced body size, reduced cell number in the retina and neurological impairment (Sicinski et al., 1995). While Aspm expressed specifically during prenatal cerebral cortical neurogenesis is implicated in the size and evolution of the cerebral cortex (Fish et al., 2006) and furthermore, mutations are associated with human primary microcephaly. In the fetal brain, 10 genes in particular were shown to be primarily downregulated (fold change >1.2X) with 2 and/or 4mg/kg, doses reflective of sub-AChE inhibition. Those genes included acetyltransferases, Esco1 and Esco2 involved in S-phase sister chromatid cohesion and Gadd45g, a mediator of cell cycle arrest and activator of the p38/JNK pathway. Here, in this study, we propose multiple targets related to cell cycle regulation in the fetuses that may underlie previous observations of disrupted cellular proliferation which occurred in parallel with CP-induced developmental neurotoxicity.
CP exposure and neurodevelopment
In the present study, CP exposure impacted the expression of genes that are involved in the development of the nervous system. Such an impact resulted in the enrichment of categories and pathways in the fetal brain such as cell adhesion, cell migration, myelination, Wnt signaling pathway, smoothened signaling pathway, transmission of nerve impulse, neurodegenerative diseases (a pathway that contains genes known to be involved in neurodevelopment) and long-term potentiation (Table 3).
As observed with cell cycle-related genes, genes listed under the GOID “Nervous System Development” of MAPPFinder database and that were differentially expressed in our study have also been linked with neurodevelopmental and neurological disorders. From the 22 genes shown to have a peak effect of down-regulation in the 4 and/or 10 mg/kg groups (Fig 10) at least 12 of them (Apaf1, Aspm, Creb1, Fzd3, Mtap2, Ndst1, Nrcam, Slitrk1, Ret, Igf1, Htra2, Dvl1) lead to morphological and/or functional encephalic alterations when their expressions are decreased in mice models. Specifically, alterations in six of these genes in knockout mouse models reflect CP-induced neurotoxicity-phenotypes including decreased dendritic length described in Mtap2-deficient mice (Harada et al., 2002); altered axonal outgrowth described in FZD3 (Wang et al., 2006) and Igf1-knockout mice (Beck et al., 1995); altered neuronal migration described in Creb (Aguado et al., 2009) and Ndst1 (Grobe et al., 2005); behavioral alterations such as anxiety and depression described in Slitrk1-deficient-mice (Katayama et al., 2008) and Nrcam-deficient mice (Aonurm-Helm et al., 2008); decreased dopaminergic neurotransmission described in Ret-deficient mice (Kramer et al., 2007). Our results suggest CP may disrupt several aspects of cell cycle regulation and nervous system development in the fetal brain and suggest alterations in the listed genes may underlie previous phenotypes described in its neurodevelopmental toxicity.
CP exposure and signaling disturbances
Signaling-related categories were also identified to be overrepresented in our dataset. In the fetal brain, MAPK, Wnt, smoothened (hedgehog), transmembrane receptor tyrosine kinase and insulin signaling pathways were identified to be enriched as well as synaptic transmission. From those, “synaptic transmission” was found to be enriched among genes down-regulated in low dose groups (fetal brain, cluster D, Fig. 9). Signaling pathways, MAPKKK, adenylate cyclase and insulin have been described to be disrupted in various tissues after CP exposure (Auman et al., 2000; Meyer et al., 2003; Caughlan et al., 2004; Mense et al., 2006; Saulsbury et al., 2008; Stapleton and Chan, 2009). Two recent CP genomic studies in rats have reported alterations in the Wnt signaling pathway in the brain of both neonates (Slotkin et al., 2008) and adults (Stapleton and Chan, 2009). The function of the Wnt family of signaling proteins during developmental processes in many different species and organs has been well established. There are at least 3 different Wnt pathways: the canonical pathway (implicated in cell fate decisions and may play a role in synaptogenesis); the planar cell polarity (PCP) pathway (leads to remodeling of the cytoskeleton and is implicated in cell polarity and dendritic arborization); and the Wnt-Ca2+ signalling pathway (implicated in cell fate and cell movement) (Ciani and Salinas, 2005). In the present study, CP altered the expression of 2 genes encoding frizzled Wnt receptors (Fzd1, Fzd3), a modulator of Wnt proteins processing (Procn) as well as 14 downstream genes in all Wnt pathways (Table 3). From those, 9 participate in the canonical pathway (Ccnd1, Ccnd2, Ctbp2, Dvl1, Fbxw11, Lef1, Senp2, Tbl1x, Trp53), 3 in the PCP pathway (Prickle2, Daam1, Dvl1) and 3 in the Wnt-Ca2+ pathway (Camk2b, Ppp3ca, Ppp3cb). In the developing brain, Wnt signaling plays a role in the formation and modulation of neural circuits regulating cellular functions such as neuronal migration, neuronal polarization, axon guidance, dendrite development and synapse formation (Salinas and Zou, 2008), which are known to be disrupted after developmental CP exposure.
Disturbances in growth factors signaling have also been investigated in the CP-induced neurotoxicity (Betancourt et al., 2006; Betancourt et al., 2007; Slotkin et al., 2007; Slotkin et al., 2008). Fibroblast growth factor (FGF) signaling was shown to be altered in neonatal rats exposed to CP 1 mg/kg from PND 1–4 (Slotkin et al., 2007). In addition, nerve growth factor (NGF) and brain-derived neurotrophic factor (BNDF) signaling were altered in brain regions of rats exposed to 4 or 6 mg/kg of CP from PND 10–20 (Betancourt et al., 2007). In the present study, the GO category “transmembrane receptor tyrosine kinase signaling” was shown to be enriched in the fetal brain, primarily in the 10 mg/kg dose group (Fig. 7, Table 5). This GOID reflects the series of molecular signals generated as a consequence of a transmembrane receptor tyrosine kinase binding to its physiological ligands. Despite the diversity of the physiological ligands, growth factors are part of them (BDNF, EGF, IGF, FGF, NGF) and even though in the present study no specific growth factor category/pathway has been shown to be enriched, disturbances in growth factors signaling transduction may have occurred. Eighteen genes were identified to be linked in the MAPPFinder database to the GOID “transmembrane receptor tyrosine kinase signaling” and significantly altered by CP in the fetal brain (Fig. 6, specific genes not shown), including growth factors Tgfα, Fgf9 and Igf1, receptors Agrn, Kdr, Kit and multiple genes involved in the signal transduction of tyrosine kinase receptors (Alk, Cryab, Dvl1, Grb10, Ncoa4, Ndst1, Phip, Rgs2, Slc2a8, Sparc, Timp3, Zfp106). Except for the genes Alk, Cryab, Slc2a8, Tgfa and Timp3, all the other genes, including the Fgf9, Igf1 and the receptors presented peak effects of down-regulation in the 4 and/or 10 mg/kg dose groups.
CP exposure and protein metabolism
The ubiquitin-proteasome system (UPS) is a highly conserved cellular pathway responsible for the metabolism of the majority of intracellular proteins. It plays essential roles in maintaining almost every aspect of cellular activities including gene transcription, protein translation, cell development, signal transduction, and protein quality control. Disruption of this system, therefore, can have significant downstream effects on critical cellular functions, impacting susceptibility and development of diseases. In our dataset, “protein modification” as well as “ubiquitin cycle” GOIDs were enriched in the fetal brain and GO-Quant analysis showed that exposure to 10 mg/kg of CP had the highest impact on genes belonging to these categories (Fig. 6). Moreover, the GO analysis conducted in separated clusters of genes (Table 5) also pointed that “ubiquitin cycle” GOID was enriched in cluster B, which was composed by genes primarily down-regulated in the 10 mg/kg group. Most of the 43 genes shown to be differentially expressed in this pathway across CP doses (Fig 6) represented ubiquitin-protein ligases (E3) but ubiquitin-conjugating enzymes (E2) and protein deubiquination enzymes were also altered (data not shown). Even though a higher number of down-regulated genes were observed in the three classes we also observed up-regulated genes. Genes with a peak effect on the 10 mg/kg group and that were down-regulated included Cbx8, Cdc23, Cnot4, Dzip3, Fbxo30, Hecw1, Rnf128, Senp7, Shprh, Skp2, Trip12 (E3), Aktip, Arih1, Det1 (E2) and Usp33, Usp9x (deubiquination). The up-regulated genes included Fbxo3, Fbxo45, Huwe1, Senp2, Rnf144b, Ubox5, Znrf1 (E3), Arih2, Ube21 (E2) and Otud7b, Usp2, Usp7 (deubiquitination). This result suggests that a number of protein targets may have been impacted by the exposure to CP due to its effect on the UPS.
Others GOIDs related to protein metabolism enriched in the fetal brain cluster B (genes down-regulated in the 10 mg/kg group) were “secretory pathway” and “ER-nuclear signaling pathway”, which represents molecular signals that conveys information from the endoplasmatic reticulum (ER) to the nucleus in response to unfolded proteins, ER overload or SREBP (Sterol Regulatory Element Binding Proteins).
Final considerations
There are some limitations to the current work. First, performing transcriptomic analysis at a single time point post-treatment does not allow for the identification to examine temporal effects of CP treatment on gene expression. The gene expression profiles identified in this study may represent a mixed pattern of toxic responses, repair processes and adaptative responses. Secondly, the use of the whole brain to extract the RNA could have masked some results because of the particularities of each brain region on maturation stage, cell composition and neurochemistry. Despite these limitations, this study serves as a genome-wide look at how CP impacts maternal and fetal brain and confirms the utility of toxicogenomic studies to screen for neurotoxicants, illustrating a pathway-based analysis to inform the dose-effect relationship of neurotoxicants on gene expression.
In summary, in the present study, using a systems-based approach, we showed that the dose-effect relationship of CP on gene expression, both at the gene and pathway levels, was non-monotonic and not necessarily related to brain AChE activity inhibition. The largest number of DEGs in the maternal and fetal brain was observed in the 10 mg/kg dose group and this dose also had the higher impact on biological processes found to be enriched across CP doses. However, sub-anticholinesterasic doses of CP were also shown to impact maternal and fetal brain. In the maternal brain, the dose of 4 mg/kg influenced GO categories and pathways such as cell adhesion, behavior, lipid metabolism, long-term potentiation, nervous system development, neurogenesis, secretory pathway, synaptic transmission. In the fetal brain, which was greatly impacted by CP exposure, sub-anticholinesterasic doses of CP (2 and/or 4 mg/kg) altered cell division, translation, transmission of nerve impulse, chromatin modification, long-term potentiation. In addition, some genes involved in nervous system development and signaling were shown to be specifically influenced by these lower CP doses. Our approach was sensitive to reflect the diversity of effects induced by CP and retrieved the enrichment of GO categories and pathways terms known to be disrupted by CP. Moreover, it highlighted additional consequences of CP neurotoxicity, such as disturbance of the ubiquitin proteasome system. In the future, additional kinetic and temporal analysis of AChE effects will be phenotypically linked with our omic analysis.
ACKNOWLEDGMENTS
This work was supported in part by the National Institute of Environmental Health Sciences (NIEHS) (Toxicogenomics, U10 ES 11387 and R01-ES10613), the US Environmental Protection Agency-NIEHS UW Center for Child Environmental Health Risks Research (EPA R826886 and NIEHS 1PO1ES09601), National Institute of Child Health and Human Development (NICHD) and the UW NIEHS Center for Ecogenetics and Environmental Health (5 P30 ES07033). EG Moreira was a post-doctoral fellow from CNPq-Brazil (200213/2008-3).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICTS OF INTEREST
The authors state that they have no conflicts of interest.
REFERENCES
- Aguado F, Diaz-Ruiz C, Parlato R, Martinez A, Carmona MA, Bleckmann S, Urena JM, Burgaya F, del Rio JA, Schutz G, Soriano E. The CREB/CREM transcription factors negatively regulate early synaptogenesis and spontaneous network activity. J Neurosci. 2009;29:328–333. doi: 10.1523/JNEUROSCI.5252-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aonurm-Helm A, Jurgenson M, Zharkovsky T, Sonn K, Berezin V, Bock E, Zharkovsky A. Depression-like behaviour in neural cell adhesion molecule (NCAM)-deficient mice and its reversal by an NCAM-derived peptide, FGL. Eur J Neurosci. 2008;28:1618–1628. doi: 10.1111/j.1460-9568.2008.06471.x. [DOI] [PubMed] [Google Scholar]
- Auman JT, Seidler FJ, Slotkin TA. Neonatal chlorpyrifos exposure targets multiple proteins governing the hepatic adenylyl cyclase signaling cascade: implications for neurotoxicity. Brain Res Dev Brain Res. 2000;121:19–27. doi: 10.1016/s0165-3806(00)00021-3. [DOI] [PubMed] [Google Scholar]
- Beck KD, Powell-Braxton L, Widmer HR, Valverde J, Hefti F. Igf1 gene disruption results in reduced brain size, CNS hypomyelination, and loss of hippocampal granule and striatal parvalbumin-containing neurons. Neuron. 1995;14:717–730. doi: 10.1016/0896-6273(95)90216-3. [DOI] [PubMed] [Google Scholar]
- Betancourt AM, Burgess SC, Carr RL. Effect of developmental exposure to chlorpyrifos on the expression of neurotrophin growth factors and cell-specific markers in neonatal rat brain. Toxicol Sci. 2006;92:500–506. doi: 10.1093/toxsci/kfl004. [DOI] [PubMed] [Google Scholar]
- Betancourt AM, Filipov NM, Carr RL. Alteration of neurotrophins in the hippocampus and cerebral cortex of young rats exposed to chlorpyrifos and methyl parathion. Toxicol Sci. 2007;100:445–455. doi: 10.1093/toxsci/kfm248. [DOI] [PubMed] [Google Scholar]
- Bigbee JW, Sharma KV, Chan EL, Bogler O. Evidence for the direct role of acetylcholinesterase in neurite outgrowth in primary dorsal root ganglion neurons. Brain Res. 2000;861:354–362. doi: 10.1016/s0006-8993(00)02046-1. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brimijoin S, Koenigsberger C. Cholinesterases in neural development: new findings and toxicologic implications. Environ Health Perspect. 1999;107 Suppl 1:59–64. doi: 10.1289/ehp.99107s159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughlan A, Newhouse K, Namgung U, Xia Z. Chlorpyrifos induces apoptosis in rat cortical neurons that is regulated by a balance between p38 and ERK/JNK MAP kinases. Toxicol Sci. 2004;78:125–134. doi: 10.1093/toxsci/kfh038. [DOI] [PubMed] [Google Scholar]
- Chanda SM, Harp P, Liu J, Pope CN. Comparative developmental and maternal neurotoxicity following acute gestational exposure to chlorpyrifos in rats. J Toxicol Environ Health. 1995;44:189–202. doi: 10.1080/15287399509531954. [DOI] [PubMed] [Google Scholar]
- Chanda SM, Pope CN. Neurochemical and neurobehavioral effects of repeated gestational exposure to chlorpyrifos in maternal and developing rats. Pharmacol Biochem Behav. 1996;53:771–776. doi: 10.1016/0091-3057(95)02105-1. [DOI] [PubMed] [Google Scholar]
- Ciani L, Salinas PC. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat Rev Neurosci. 2005;6:351–362. doi: 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- Crumpton TL, Seidler FJ, Slotkin TA. Developmental neurotoxicity of chlorpyrifos in vivo and in vitro: effects on nuclear transcription factors involved in cell replication and differentiation. Brain Res. 2000a;857:87–98. doi: 10.1016/s0006-8993(99)02357-4. [DOI] [PubMed] [Google Scholar]
- Crumpton TL, Seidler FJ, Slotkin TA. Is oxidative stress involved in the developmental neurotoxicity of chlorpyrifos? Brain Res Dev Brain Res. 2000b;121:189–195. doi: 10.1016/s0165-3806(00)00045-6. [DOI] [PubMed] [Google Scholar]
- Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- Dam K, Seidler FJ, Slotkin TA. Chlorpyrifos exposure during a critical neonatal period elicits gender-selective deficits in the development of coordination skills and locomotor activity. Brain Res Dev Brain Res. 2000;121:179–187. doi: 10.1016/s0165-3806(00)00044-4. [DOI] [PubMed] [Google Scholar]
- Dam K, Seidler FJ, Slotkin TA. Transcriptional biomarkers distinguish between vulnerable periods for developmental neurotoxicity of chlorpyrifos: Implications for toxicogenomics. Brain Res Bull. 2003;59:261–265. doi: 10.1016/s0361-9230(02)00874-2. [DOI] [PubMed] [Google Scholar]
- Eaton DL, Daroff RB, Autrup H, Bridges J, Buffler P, Costa LG, Coyle J, McKhann G, Mobley WC, Nadel L, Neubert D, Schulte-Hermann R, Spencer PS. Review of the toxicology of chlorpyrifos with an emphasis on human exposure and neurodevelopment. Crit Rev Toxicol 38 Suppl. 2008;2:1–125. doi: 10.1080/10408440802272158. [DOI] [PubMed] [Google Scholar]
- Ellman GL, Courtney KD, Andres V, Jr, Feather-Stone RM. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci U S A. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia SJ, Seidler FJ, Crumpton TL, Slotkin TA. Does the developmental neurotoxicity of chlorpyrifos involve glial targets? Macromolecule synthesis, adenylyl cyclase signaling, nuclear transcription factors, and formation of reactive oxygen in C6 glioma cells. Brain Res. 2001;891:54–68. doi: 10.1016/s0006-8993(00)03189-9. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobe K, Inatani M, Pallerla SR, Castagnola J, Yamaguchi Y, Esko JD. Cerebral hypoplasia and craniofacial defects in mice lacking heparan sulfate Ndst1 gene function. Development. 2005;132:3777–3786. doi: 10.1242/dev.01935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizzetti M, Pathak S, Giordano G, Costa LG. Effect of organophosphorus insecticides and their metabolites on astroglial cell proliferation. Toxicology. 2005;215:182–190. doi: 10.1016/j.tox.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Harada A, Teng J, Takei Y, Oguchi K, Hirokawa N. MAP2 is required for dendrite elongation, PKA anchoring in dendrites, and proper PKA signal transduction. J Cell Biol. 2002;158:541–549. doi: 10.1083/jcb.200110134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Katayama K, Yamada K, Ornthanalai VG, Inoue T, Ota M, Murphy NP, Aruga J. Slitrk1-deficient mice display elevated anxiety-like behavior and noradrenergic abnormalities. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.97. [DOI] [PubMed] [Google Scholar]
- Kramer ER, Aron L, Ramakers GM, Seitz S, Zhuang X, Beyer K, Smidt MP, Klein R. Absence of Ret signaling in mice causes progressive and late degeneration of the nigrostriatal system. PLoS Biol. 2007;5:e39. doi: 10.1371/journal.pbio.0050039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassiter TL, Padilla S, Mortensen SR, Chanda SM, Moser VC, Barone S., Jr Gestational exposure to chlorpyrifos: apparent protection of the fetus? Toxicol Appl Pharmacol. 1998;152:56–65. doi: 10.1006/taap.1998.8514. [DOI] [PubMed] [Google Scholar]
- Levin ED, Addy N, Baruah A, Elias A, Christopher NC, Seidler FJ, Slotkin TA. Prenatal chlorpyrifos exposure in rats causes persistent behavioral alterations. Neurotoxicol Teratol. 2002;24:733–741. doi: 10.1016/s0892-0362(02)00272-6. [DOI] [PubMed] [Google Scholar]
- Levin ED, Addy N, Nakajima A, Christopher NC, Seidler FJ, Slotkin TA. Persistent behavioral consequences of neonatal chlorpyrifos exposure in rats. Brain Res Dev Brain Res. 2001;130:83–89. doi: 10.1016/s0165-3806(01)00215-2. [DOI] [PubMed] [Google Scholar]
- Mattsson JL, Maurissen JP, Nolan RJ, Brzak KA. Lack of differential sensitivity to cholinesterase inhibition in fetuses and neonates compared to dams treated perinatally with chlorpyrifos. Toxicol Sci. 2000;53:438–446. doi: 10.1093/toxsci/53.2.438. [DOI] [PubMed] [Google Scholar]
- Mense SM, Sengupta A, Lan C, Zhou M, Bentsman G, Volsky DJ, Whyatt RM, Perera FP, Zhang L. The common insecticides cyfluthrin and chlorpyrifos alter the expression of a subset of genes with diverse functions in primary human astrocytes. Toxicol Sci. 2006;93:125–135. doi: 10.1093/toxsci/kfl046. [DOI] [PubMed] [Google Scholar]
- Meyer A, Seidler FJ, Cousins MM, Slotkin TA. Developmental neurotoxicity elicited by gestational exposure to chlorpyrifos: when is adenylyl cyclase a target? Environ Health Perspect. 2003;111:1871–1876. doi: 10.1289/ehp.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope CN. Organophosphorus pesticides: do they all have the same mechanism of toxicity? J Toxicol Environ Health B Crit Rev. 1999;2:161–181. doi: 10.1080/109374099281205. [DOI] [PubMed] [Google Scholar]
- Qiao D, Seidler FJ, Tate CA, Cousins MM, Slotkin TA. Fetal chlorpyrifos exposure: adverse effects on brain cell development and cholinergic biomarkers emerge postnatally and continue into adolescence and adulthood. Environ Health Perspect. 2003;111:536–544. doi: 10.1289/ehp.5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricceri L, Venerosi A, Capone F, Cometa MF, Lorenzini P, Fortuna S, Calamandrei G. Developmental neurotoxicity of organophosphorous pesticides: fetal and neonatal exposure to chlorpyrifos alters sex-specific behaviors at adulthood in mice. Toxicol Sci. 2006;93:105–113. doi: 10.1093/toxsci/kfl032. [DOI] [PubMed] [Google Scholar]
- Richardson JR, Chambers JE. Neurochemical effects of repeated gestational exposure to chlorpyrifos in developing rats. Toxicol Sci. 2004;77:83–90. doi: 10.1093/toxsci/kfh014. [DOI] [PubMed] [Google Scholar]
- Roy TS, Andrews JE, Seidler FJ, Slotkin TA. Chlorpyrifos elicits mitotic abnormalities and apoptosis in neuroepithelium of cultured rat embryos. Teratology. 1998;58:62–68. doi: 10.1002/(SICI)1096-9926(199808)58:2<62::AID-TERA7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Salinas PC, Zou Y. Wnt signaling in neural circuit assembly. Annu Rev Neurosci. 2008;31:339–358. doi: 10.1146/annurev.neuro.31.060407.125649. [DOI] [PubMed] [Google Scholar]
- Saulsbury MD, Heyliger SO, Wang K, Round D. Characterization of chlorpyrifos-induced apoptosis in placental cells. Toxicology. 2008;244:98–110. doi: 10.1016/j.tox.2007.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh RA, Lein PJ, Beckles RA, Jett DA. Noncholinesterase mechanisms of chlorpyrifos neurotoxicity: altered phosphorylation of Ca2+/cAMP response element binding protein in cultured neurons. Toxicol Appl Pharmacol. 2002;182:176–185. doi: 10.1006/taap.2002.9445. [DOI] [PubMed] [Google Scholar]
- Sicinski P, Donaher JL, Parker SB, Li T, Fazeli A, Gardner H, Haslam SZ, Bronson RT, Elledge SJ, Weinberg RA. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- Slotkin TA, Seidler FJ. Comparative developmental neurotoxicity of organophosphates in vivo: transcriptional responses of pathways for brain cell development, cell signaling, cytotoxicity and neurotransmitter systems. Brain Res Bull. 2007;72:232–274. doi: 10.1016/j.brainresbull.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Seidler FJ. Developmental neurotoxicants target neurodifferentiation into the serotonin phenotype: Chlorpyrifos, diazinon, dieldrin and divalent nickel. Toxicol Appl Pharmacol. 2008;233:211–219. doi: 10.1016/j.taap.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Seidler FJ. Oxidative and excitatory mechanisms of developmental neurotoxicity: transcriptional profiles for chlorpyrifos, diazinon, dieldrin, and divalent nickel in PC12 cells. Environ Health Perspect. 2009;117:587–596. doi: 10.1289/ehp.0800251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Seidler FJ, Fumagalli F. Exposure to organophosphates reduces the expression of neurotrophic factors in neonatal rat brain regions: similarities and differences in the effects of chlorpyrifos and diazinon on the fibroblast growth factor superfamily. Environ Health Perspect. 2007;115:909–916. doi: 10.1289/ehp.9901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Seidler FJ, Fumagalli F. Targeting of neurotrophic factors, their receptors, and signaling pathways in the developmental neurotoxicity of organophosphates in vivo and in vitro. Brain Res Bull. 2008;76:424–438. doi: 10.1016/j.brainresbull.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Song X, Seidler FJ, Saleh JL, Zhang J, Padilla S, Slotkin TA. Cellular mechanisms for developmental toxicity of chlorpyrifos: targeting the adenylyl cyclase signaling cascade. Toxicol Appl Pharmacol. 1997;145:158–174. doi: 10.1006/taap.1997.8171. [DOI] [PubMed] [Google Scholar]
- Song X, Violin JD, Seidler FJ, Slotkin TA. Modeling the developmental neurotoxicity of chlorpyrifos in vitro: macromolecule synthesis in PC12 cells. Toxicol Appl Pharmacol. 1998;151:182–191. doi: 10.1006/taap.1998.8424. [DOI] [PubMed] [Google Scholar]
- Stapleton AR, Chan VT. Subtoxic chlorpyrifos treatment resulted in differential expression of genes implicated in neurological functions and development. Arch Toxicol. 2009;83:319–333. doi: 10.1007/s00204-008-0346-2. [DOI] [PubMed] [Google Scholar]
- USEPA. Interim reregistration interim elegibility decision for chlorpyrifos. United States Environmental Protection Agency. 2002 [Google Scholar]
- Wang Y, Zhang J, Mori S, Nathans J. Axonal growth and guidance defects in Frizzled3 knock-out mice: a comparison of diffusion tensor magnetic resonance imaging, neurofilament staining, and genetically directed cell labeling. J Neurosci. 2006;26:355–364. doi: 10.1523/JNEUROSCI.3221-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Howard A, Bruun D, Ajua-Alemanj M, Pickart C, Lein PJ. Chlorpyrifos and chlorpyrifos-oxon inhibit axonal growth by interfering with the morphogenic activity of acetylcholinesterase. Toxicol Appl Pharmacol. 2008;228:32–41. doi: 10.1016/j.taap.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Griffith WC, Hanspers K, Dillman JF, 3rd, Ong H, Vredevoogd MA, Faustman EM. A system-based approach to interpret dose- and time-dependent microarray data: quantitative integration of gene ontology analysis for risk assessment. Toxicol Sci. 2006;92:560–577. doi: 10.1093/toxsci/kfj184. [DOI] [PubMed] [Google Scholar]
- Yu X, Hong S, Moreira EG, Faustman EM. Improving in vitro Sertoli cell/gonocyte co-culture model for assessing male reproductive toxicity: Lessons learned from comparisons of cytotoxicity versus genomic responses to phthalates. Toxicol Appl Pharmacol. 2009;239:325–336. doi: 10.1016/j.taap.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]