Summary
Melanomas are highly heterogeneous tumors, but the biological significance of their different subpopulations is not clear. Using the H3K4 demethylase JARID1B (KDM5B/PLU-1/RBP2-H1) as a biomarker, we have characterized a small subpopulation of slow-cycling melanoma cells that cycle with doubling times of >4 weeks within the rapidly proliferating main population. Isolated JARID1B-positive melanoma cells give rise to a highly proliferative progeny. Knock-down of JARID1B leads to an initial acceleration of tumor growth followed by exhaustion which suggests that the JARID1B-positive subpopulation is essential for continuous tumor growth. Expression of JARID1B is dynamically regulated and does not follow a hierarchical cancer stem cell model because JARID1B-negative cells can become positive and even single melanoma cells irrespective of selection are tumorigenic. These results suggest a new understanding of melanoma heterogeneity with tumor maintenance as a dynamic process mediated by a temporarily distinct subpopulation.
Introduction
Malignant melanoma is an aggressive tumor of neuroectodermal origin that can be cured if excised in an early stage, however, once disseminated to distant organs, the median survival of melanoma patients drops below 9 months (Gogas et al., 2007). The vast intratumoral heterogeneity of melanoma paired with the potential for continuous tumor self-renewal previously led to the question of whether melanomas follow the cancer stem cell (CSC) model with a melanoma stem cell on top of a tumor differentiation pyramid (Reya et al., 2001; Zabierowski and Herlyn, 2008). Since the initial validation of the CSC model for acute myeloid leukemia (Bonnet and Dick, 1997), CSCs have been identified in various solid tumors, e.g. breast (Wright et al., 2008) or brain cancer (Singh et al., 2003). We previously reported that the B cell marker CD20 is indicative for self-renewal of melanoma spheres after propagation in stem cell medium (Fang et al., 2005). Subsequently, other markers, e.g. CD133 (Monzani et al., 2007) or ABCB5 (Schatton et al., 2008), have been used to characterize stem-like subpopulations in melanomas with frequencies broadly ranging between ~0.0001% and 0.1% of the total population depending on the (surface) marker and experimental methods used. However, a recent seminal publication pointed out that modifications to xenotransplantation assays, which currently represent the standard assay to assess tumor self-renewal (Clarke et al., 2006), can dramatically increase the frequency of tumor-initiating/melanoma stem cells up to 25% of unsorted cells, i.e. independent from any putative marker (Quintana et al., 2008). Besides the conclusion that basically every melanoma cell could initiate a tumor if the host system is susceptible enough, this new finding refreshed the ongoing discussion about suitable experimental models, the definition, and, finally, the existence of melanoma stem cells (Adams and Strasser, 2008). Melanomas may not be hierarchically organized into subpopulations of tumorigenic and non-tumorigenic cells and the CSC model might not account for melanoma heterogeneity. However, it remains unanswered if, within an established tumor microenvironment, continuous tumor maintenance is similarly assured by each individual melanoma cell or if distinct subpopulations are more suited as a resource for replenishment. In the latter scenario, the potential to continuously maintain tumors might be independent of the capacity to initiate new tumors in host organisms and might not follow a unidirectional CSC model, particularly when the considerable plasticity and heterogeneity of melanomas are taken into consideration.
JARID1B (KDM5B/PLU-1/RBP2-H1, Lu et al., 1999; Roesch et al., 2005; Vogt et al., 1999) is a member of the highly conserved family of jumonji/ARID1 (JARID1) histone 3 K4 (H3K4) demethylases which are involved in tissue development, cancer, and normal stem cell biology (Christensen et al., 2007; Iwase et al., 2007; Klose et al., 2007; Yamane et al., 2007). In normal adult tissues, JARID1B is marginally expressed with dramatic peak expression levels in regenerative tissues like testis and bone marrow (Roesch et al., 2005; Vogt et al., 1999). In cancer, JARID1B functions as a transcriptional regulator of oncogenes, e.g. BRCA1 in breast cancer, via direct interaction with promoter sites (Scibetta et al., 2007; Tan et al., 2003). Depending on the cancer context, JARID1B is associated with either positive (melanoma) or negative (breast cancer) cell cycle control (Roesch et al., 2006; Roesch et al., 2008; Scibetta et al., 2007; Yamane et al., 2007). In melanocytic tumors, JARID1B is highly expressed in benign nevi, which typically are characterized by oncogene-induced senescence. However, in aggressive primary melanomas and melanoma metastases, there are only single cells with high JARID1B expression [~5%–10% of the total population, (Roesch et al., 2005)].
Given JARID1B’s potential role in stem cell biology and the low percentage of melanoma cells expressing JARID1B within the bulk population, we asked, (1) if the JARID1B-positive subpopulation has an enhanced potential for maintenance of the entire tumor, and (2) if/how this subpopulation differs from the current definition of a CSC. Our findings argue against a unidirectional CSC model in melanomas leading to an alternative concept of tumor heterogeneity with dynamic subpopulations of tumor-maintaining cells.
Results
Identification of JARID1B as a marker for slow-cycling melanoma cells
JARID1B is highly expressed in a small percentage of melanoma cells in advanced patients’ tumors (Roesch et al., 2005); a representative example is shown in Figure 1A. Similar expression can be observed in melanoma cell lines. Independent of the cell line analyzed, immunostaining showed broad “intraculture” heterogeneity of JARID1B. Under conventional culture conditions, some cells expressed high levels of JARID1B while most expressed medium to low levels (Figure 1 A). In contrast, human embryonic stem cell medium (hESCM4)-derived melanoma spheres displayed a more distinct JARID1B heterogeneity, i.e. small-sized highly JARID1B-positive cells being surrounded by a JARID1B-negative bulk population. Using digital quantitation of pseudocolored immunosections, an average of 4.8% JARID1B-positive cells was present across randomly selected sphere sections (10 representative images out of 5 different melanoma cell lines, Figure 1A and Figure S1A). This pattern was similar to that observed in patients’ tumors suggesting that growth as spheres in hESCM4 better recapitulates the phenotype observed in vivo. Notably, JARID1B-positive cells mostly lacked expression of the proliferation marker Ki-67 in both cultured cells and patient tumors (Figure 1B, Figure S1B), suggesting a correlation between slow proliferation and JARID1B expression.
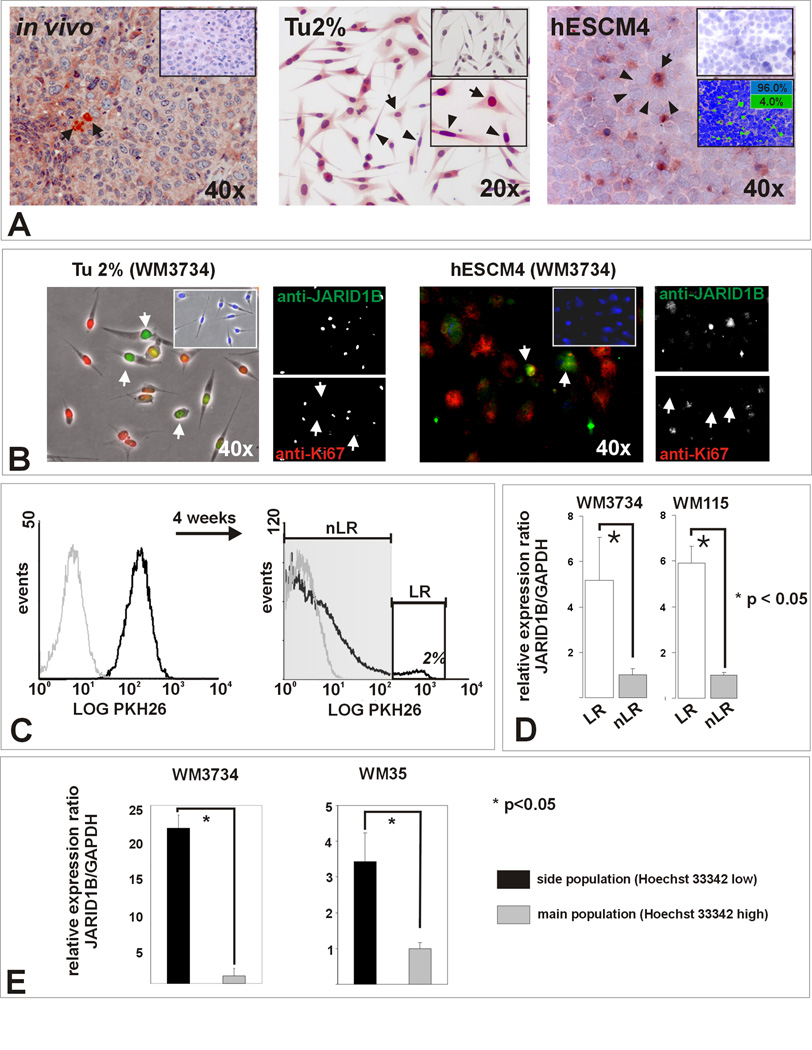
Figure 1. Melanomas contain a subpopulation of slow-cycling cells characterized by increased JARID1B expression.
(A) Left panel: Representative example of a vertical growth phase melanoma after immunostaining for JARID1B. Single positive cells (arrows) with predominantly nuclear and minor cytoplasmic staining are surrounded by negative cells. Middle: Single adherent WM3734 melanoma cells show high JARID1B (arrows) while the bulk of cells shows median or low expression (arrowheads) when grown in conventional medium (Tu2%). Isotype control (top) and higher magnification (bottom) are shown as inserts. Right: More distinct JARID1B staining was seen in cryosections of WM3734 melanoma spheres (hESCM4). Digital quantitation after pseudocoloring revealed a relative frequency of 4% JARID1B-immunopositive (green) and 96% immunonegative cells (blue) for this section (see also Figure S1). (B) Anti- JARID1B-positive cells (Alexa Fluor® 488, green) are predominately negative for the proliferation marker Ki-67 (Alexa Fluor® 568, red). The result was reproduced in WM115 cells (not shown). (C) Flow cytometry analysis of WM3734 melanoma sphere cells after incubation with PKH26 (black line) compared to unlabeled control cells (grey line). After 4 weeks in hESCM4, 2% of the cells retained the maximum label (label-retaining ‘LR’ cells, white square). (D) LR cells displayed significantly enhanced JARID1B expression compared to non label-retaining (nLR) cells in QPCR. (E) QPCR screening of side population cells from WM3734 and WM35 melanoma sphere cells showing an upregulation of JARID1B compared to the main populations.
To further explore a possible connection between JARID1B and slow-cycling cells we used retention of the membrane dye PKH26 as a marker for melanoma cells with a low doubling rate. The sphere model was chosen because hESCM4 better separates the JARID1B-positve subpopulation from the bulk. The average doubling time of unsorted WM3734 cells is ~48 hrs. Only 2% of WM3734 cells retained the maximum amount of label after 4 weeks suggesting that those cells had not divided (Figure 1C). When assessed for JARID1B, PKH26 label-retaining cells expressed higher levels than non-label-retaining bulk cells (p<0.05, t-test, Figure 1D). Following the observation that slow-cycling tumor cells show increased Hoechst 33342 efflux (Goodell et al., 1996), we applied side population analysis to WM3734 and WM35 spheres. Subsequent QPCR revealed a significant upregulation of JARID1B in side population cells (Figure 1E, p<0.05, t-test). Genome-wide expression profiling of label-retaining and side population cells from WM3734, WM1976, WM115, and WM 3248 melanoma cell lines (not shown) suggested that other jumonji family members, e.g. JMJD1A, could be also differentially regulated; however, the regulation of JARID1B appeared most consistent. Our data indicate that even within highly proliferative melanomas, a JARID1B-positive subpopulation resides in a slow-cycling state and melanoma heterogeneity might also apply to the level of cell proliferation.
Slow-cycling melanoma cells form a distinct JARID1B-positive subpopulation in vitro and in vivo
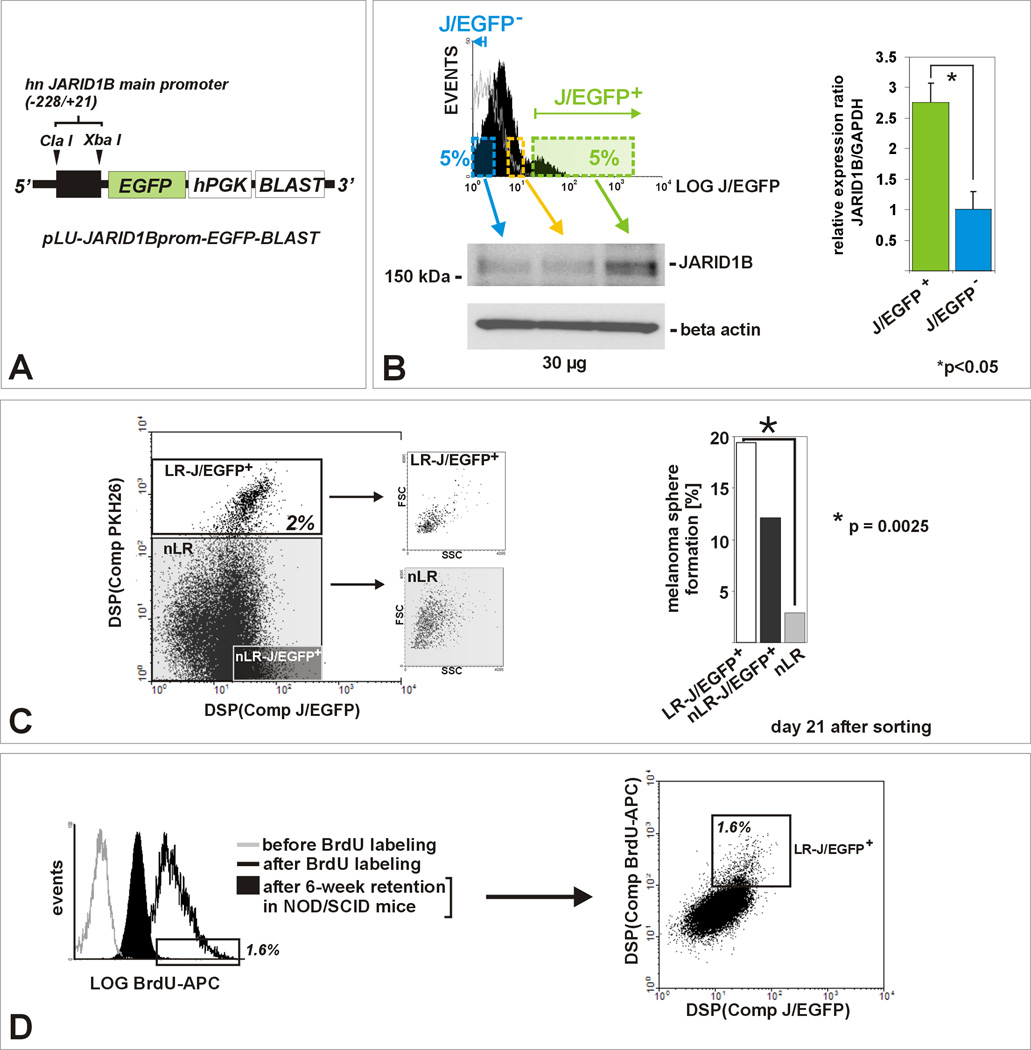
For isolation of live JARID1B-positive cells, we cloned a lentiviral construct, pLU-JARID1Bprom-EGFPBlast, which drives cytoplasmic EGFP expression controlled by the co-cloned human JARID1B promoter (abbreviated J/EGFP; Figure 2A, B; Figure S2). When we analyzed stably infected WM3734JARID1Bprom-EGFP melanoma cells that had been pre-labeled with PKH26 four weeks prior, the PKH26 label-retaining cell population displayed as a distinct, almost completely J/EGFP-positive subpopulation (Figure 2C). This population was enriched for small-sized cells as reported to be typical for melanoma cells with increased stemness (Grichnik et al., 2006). Limited (single cell) dilution assays in hESCM4, which requires self-renewal for continuous expansion as spheres (Fang et al., 2005), revealed significantly increased sphere formation capacity of the label-retaining-J/EGFP-double positive subpopulation after 21 days (Figure 2C, p=0.0025, Fisher’s exact test). Of note, non label-retaining but J/EGFP-positive cells also self-renewed into spheres, a finding which could be explained by the dynamics of the JARID1B phenotype, as shown later (see Figure 6). Most of the non label-retaining-J/EGFP-negative cells died after 3 weeks of culture in hESCM4. To exclude delayed sphere formation, the plates were periodically re-assessed (additional 3 weeks, not shown). We next xenotransplanted WM3734JARID1Bprom-EGFP melanoma cells into NOD/LtSscidIL2Rγnull mice and labeled the developing tumors with intraperitoneally and orally administered BrdU for 12 days (n=5). After an additional 6 weeks of tumor growth and BrdU dilution into subsequent daughter cells, BrdU label-retaining-J/EGFP-positive cells were identified as a distinct subpopulation and at a similar percentage (1–2%) as seen before in vitro (Figure 2D, 1C).
Figure 2. JARID1B is a biomarker for the label-retaining subpopulation in melanoma.
(A) Lentiviral pLU-JARID1Bprom-EGFP-Blast. (B) Flow cytometric determination of the J/EGFP-positive subpopulation, exemplarily shown for WM3734JARID1Bprom-EGFP melanoma spheres grown in hESCM4 (see also Figure S2). QPCR and immunoblots of sorted populations showed a significant correlation between exogenous EGFP and endogenous JARID1B expression (p<0.05, t-test). The 5%-threshold was based on our in vitro and in vivo observations on endogenous JARID1B expression frequency (Figure 1). (C) PKH26 label-retention of WM3734JARID1Bprom-EGFP sphere cells in vitro. Left: LR cells grouped as a distinct J/EGFP-positive subpopulation (LR/JEGFP-positive, white square) and were enriched for small-sized cells. Right: LR-J/EGFP-positive cells (white bar) showed the highest capacity to re-form spheres. Depicted is one representative out of three independent experiments. (D) Left: Anti-BrdU-APC flow cytometry of dissociated tumor cells revealed that 6 weeks after BrdU incubation the majority of cells diluted out the BrdU (filled black). Unlabeled (grey line) and freshly BrdU-labeled cells (black line) served as controls. The threshold was set to the peak APC intensity of freshly BrdU-labeled cells. Right: In vivo BrdU-labeled cells grouped as a distinct J/EGFP-positive subpopulation (LR/JEGFP-positive, white square, 1.6% of total population).
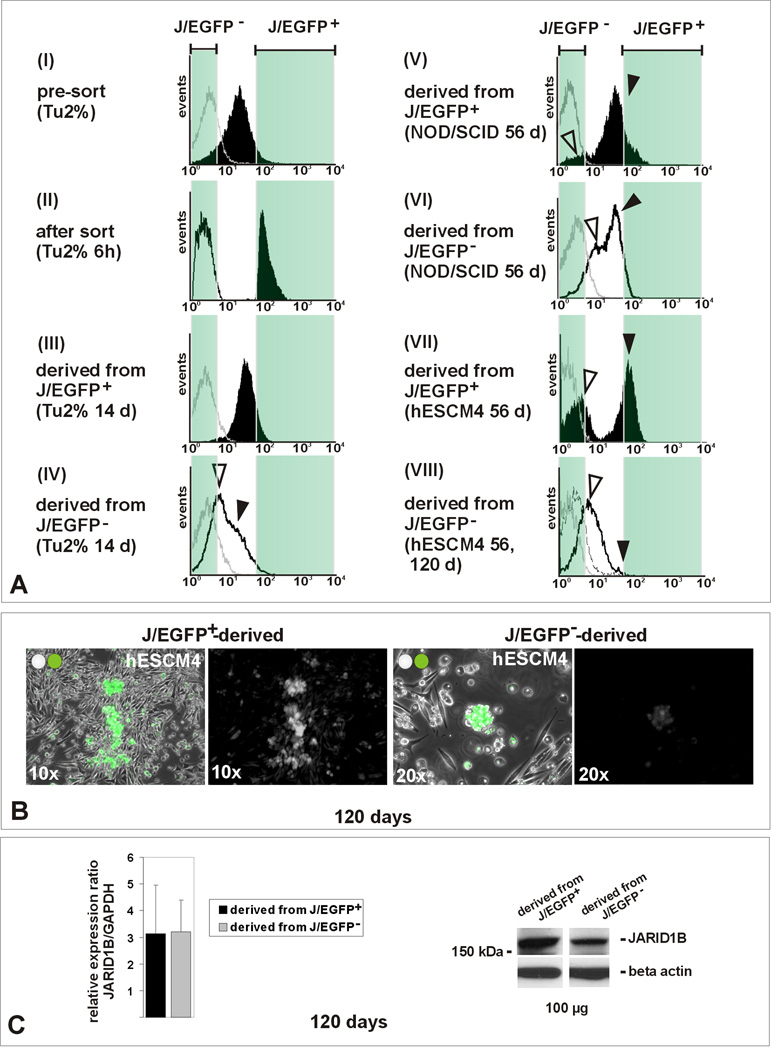
Figure 6. The JARID1B phenotype is dynamic (see also Figure S6).
(A) WM3734JARID1Bprom-EGFP cells were sorted according to J/EGFP levels as described before using maximum and minimum thresholds set at 5%. Untreated parental cells were used as a background control (grey line). Panel I: typical histogram pre-sort in conventional medium (Tu2%). Panel II: isolated J/EGFP-positive (filled black) and J/EGFP-negative cells (black line) 6 hours after FACS. Panel III: during subsequent culturing in Tu2% medium, the J/EGFP-positive subpopulation re-established a heterogeneous progeny of J/EGFP-positive and – negative cells (filled black). Panel V: same result in vivo after injection into NOD/LtSscidIL2Rγnull mice (filled black). Panels IV and VI: also J/EGFP-negative cells gave rise to a heterogeneous progeny including J/EGFP-positive cells (black line). Panel VII: in hESCM4, the J/EGFP-positive-derived progeny maintained a high number of J/EGFP-positive cells (filled black). Panel VIII: also the reversion of J/EGFP-negative to -positive cells was considerably decelerated (50-day line dashed, 120-day line black). Black and white arrowheads indicate developing positive and negative subpopulations. Depicted tumor histograms resulted from injection of 100 J/EGFP-positive or –negative cells, respectively. Shown are representative histograms from at least 3 analyses. (B) Immunofluorescence of cultures originating from J/EGFP-positive (left) and J/EGFP-negative cells (right) after 120 days in hESCM4. (C) Similar overall JARID1B expression in J/EGFP-positive and -negative-derived progenies after long-term culturing as determined by QPCR and immunoblotting.
The slow-cycling JARID1B-positive subpopulation shows increased in vitro self-renewal
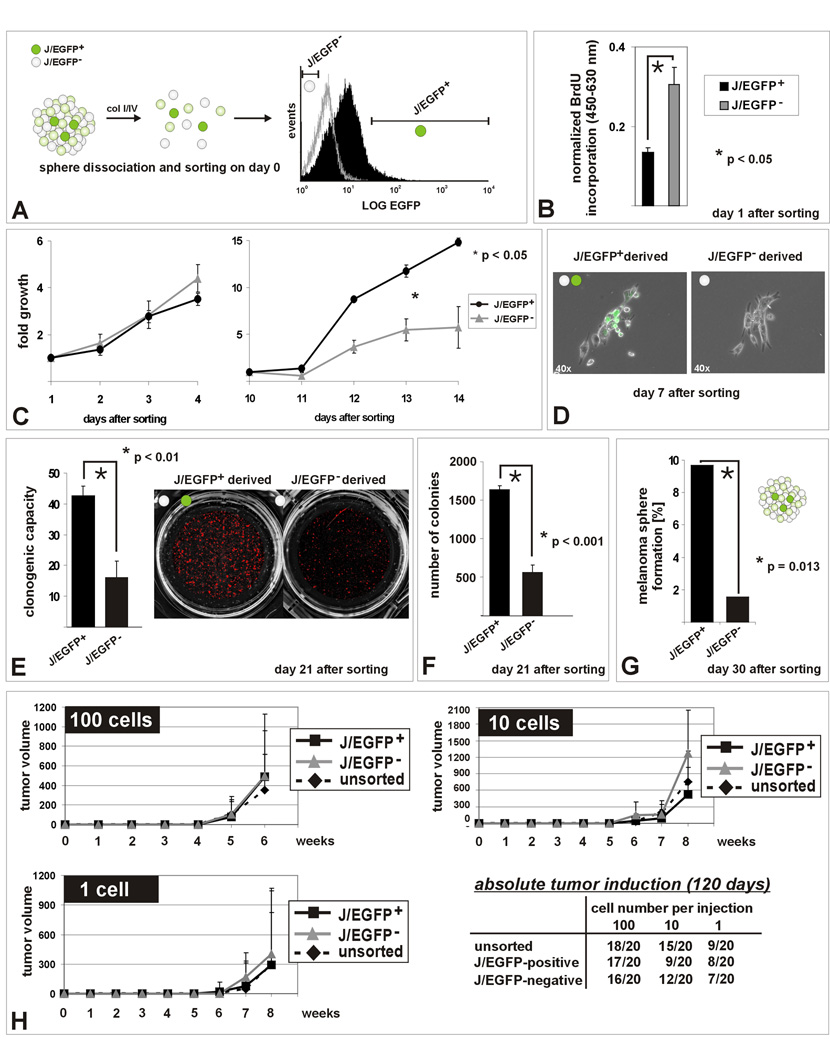
As illustrated in Figure 3A, WM3734JARID1Bprom-EGFP melanoma spheres were dissociated and sorted according to J/EGFP-expression levels. Twenty-four hours after sorting, microscopic evaluation showed similar numbers of healthy appearing single cells in both populations. Low BrdU incorporation confirmed that isolated J/EGFP-positive cells remained in a slow-cycling state (Figure 3B, p<0.05, t-test). Slow-cycling cells incorporate less BrdU compared to bulk cells but once incorporated they retain it longer as shown in Figure 2. No significant difference in proliferation was observed between J/EGFP-positive and - negative cells within the first days after sorting and re-seeding in hESCM4 (Figure 3C and Figure S3A). However, after day 10, there was a significant boost in the J/EGFP-positive-derived progeny (p<0.05, ANOVA). Interestingly, under conventional culture conditions, the difference in proliferation was less distinct (Figure S3B). Seven days after sorting, J/EGFP-positive cells started to resemble the original culture heterogeneity of J/EGFP-positive and -negative cells (Figure 3D). The enhanced capacity of J/EGFP-positive cells to expand and to form a heterogeneous progeny was confirmed by clonogenic assays in which sorted cells were seeded at clonal density and grown for 21 days in hESCM4 (Figure 3E, p<0.01, t-test). J/EGFP-positive sphere cells again formed more and larger colonies in soft agar than J/EGFP-negative cells (Figure 3F, p<0.001, t-test), indicating increased anchorage-independent growth. Interestingly, J/EGFP-negative cells formed the first visible colonies, but after 1–2 weeks growth exhausted, whereas colonies derived from J/EGFP-positive cells accelerated growth. Finally, J/EGFP-positive cells re-formed more spheres in single cell dilution assays additionally suggesting increased self-renewal capacity (Figure 3G, p=0.013, Fisher’s exact test). Single-seeded J/EGFP-negative cells died or did not form spheres.
Figure 3. In vitro self renewal capacity of the JARID1B-positive subpopulation.
(A) Methodical scheme. (B) Decreased BrdU incorporation into J/EGFP-positive cells 24 hours after separation from spheres (hESCM4) as determined by ELISA after pulsed BrdU exposition. (C) MTS assays of sorted J/EGFP-positive and –negative cells in hESCM4 (see also Figure S3). After day 10, the progeny of J/EGFP-positive cells proliferated significantly faster and (D) microscopically consisted of J/EGFP-positive and an increasing number of J/EGFP-negative cells. (E) Enhanced clonogenicity of single J/EGFP-positive cells. (F) J/EGFP-positive cells had both a higher potential to form 3D-colonies in 0.35% hESCM4-soft agar and (G) to self-renew again into heterogeneous melanoma spheres in limited dilution assays (hESCM4). Shown are representative results from at least two independent experiments. (H) Xenotransplantation growth curves after subcutaneous injection of 100, 10 or 1 WM3734JARID1Bprom-EGFP melanoma cell into NOD/LtSscidIL2Rγnull mice. Growth curves were stopped when the first mouse of the respective series exceeded a tumor size >1000 mm3. Also during the subsequent observation of remaining mice, no differences were seen.
Together with the expression studies in melanoma patient specimens and cell lines (Figure 1 and Figure S1), our data suggest a JARID1B-expressing subpopulation that remains in a slow-cycling state, but when released from its microenvironment, it can give rise to a rapidly proliferating progeny that reconstitutes the parental heterogeneity of JARID1B-positive and –negative cells. Particularly when the culture conditions supported the survival of cells with inherent self-renewal potential (hESCM4), single-seeded J/EGFP-positive cells were superior to single J/EGFP-negative cells regarding their capacity to repopulate.
The JARID1B-positive phenotype is not a prerequisite for tumor initiation in vivo
Titrated xenotransplantation assays were done in NOD/LtSscidIL2Rγnull mice according to the improved protocol published recently (Quintana et al., 2008). We subcutaneously injected 100, 10 or 1 cell from the FACS-isolated J/EGFP-positive vs. -negative subpopulation cultured in conventional medium (each n=20, Figure 3H). Unsorted cells were injected as controls. The absolute tumor initiation rate of J/EGFP-positive and -negative cells was nearly identical in line with the observations made by Quintana et al. We found in all titration steps that J/EGFP-negative cells started to grow earlier. Although not a statistically significant difference, this is consistent with our in vitro observations of colony formation assays.
Knockdown of JARID1B leads to in vitro exhaustion of melanoma cells
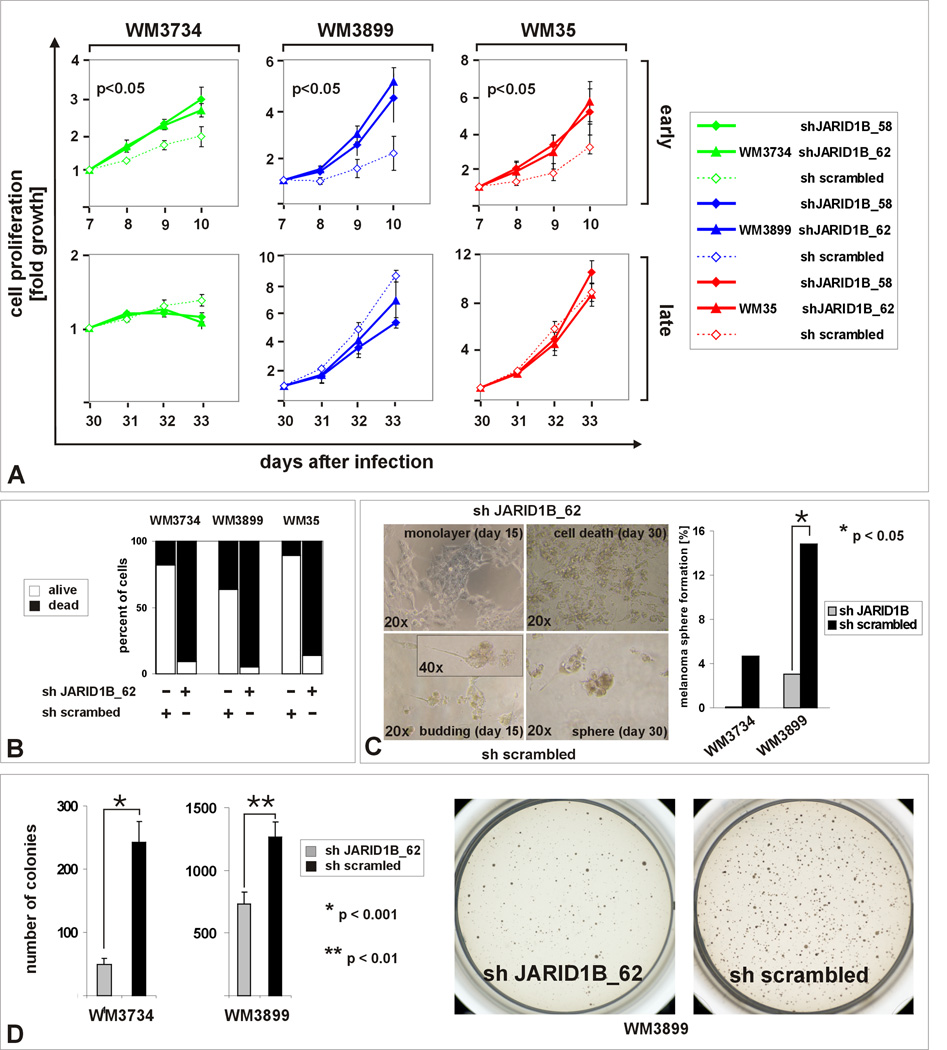
Given the paradox of an increased in vitro self-renewal capacity without any effect on in vivo tumor initiation, we asked whether JARID1B could still be important for the continuous growth of melanomas. JARID1B may not be required for tumor initiation but in the maintenance of established tumors thus reflecting two different biological processes. To address this, JARID1B was stably knocked down in WM3734, WM35, and WM3899 melanoma cells (Figure 4) and in primary foreskin melanocytes (FOM) as control (Figure S4H–J). JARID1B knockdown efficiency was validated as summarized in Figure S4A–F. Two shRNA clones targeting different mRNA regions of JARID1B were selected for analysis. Off target effects were excluded by computerized analysis (http://rnai.cs.unm.edu/offTarget). Unspecific effects due to knockdown or secondary regulation of other jumonji/ARID family members were excluded by cDNA microarrays (not shown). As expected from our previous data on JARID1B-mediated cell cycle arrest (Roesch et al., 2006), JARID1B knockdown was followed by an increase in proliferation starting on day 7 after infection. However, after day 10, cell proliferation flattened although the cells were still subconfluent (Figure 4A and Figure S4D, E, p<0.05, ANOVA). This effect seemed to occur independently of the biological and genetic backgrounds of the cell lines (Suppl. Experimental Procedures). Knockdown of different JARID1B mRNA regions showed similar results. Proliferation and pigmentation of normal melanocytes, on the other hand, remained unaffected (Figure S4).
Figure 4. In vitro exhaustion after knockdown of JARID1B.
(A) MTS proliferation assays between days 7–10 and days 30–33 after JARID1B knock down (clones 58 and 62, see also Figure S4). Experiments were done in three independent approaches in conventional medium (Tu2%). (B) Exposure of JARID1B knocked down cells (5×104 cells) to hESCM4 resulted in a strong increase of the number of trypan blue-positive cells after 4–6 weeks. (C) Limited (single cell) dilution assays determined the reduction of melanoma sphere formation in JARID1B knockdown cells (right panel). The left panel shows the corresponding cell morphologies. (D) Reduced 3D colony formation after JARID1B knockdown in Tu2% softagar. Depicted are representative results from at least 3 independent experiments.
After changing the culture conditions towards hESCM4, control cells still formed viable spheres within 21 days, whereas knockdown of JARID1B led to exhaustion after 28 (WM3899), 37 (WM3734), and 39 (WM35) days, respectively, with 86–95% cell death suggesting that in JARID1B knockdown cultures the capacity for continuous growth was lost (Figure 4B). Cell exhaust was confirmed for JARID1B knocked-down WM3928MP primary melanoma cells (Figure S4K, L). Furthermore, 30 days after seeding single cells in 96-well plates in hESCM4, the decrease in sphere formation of WM3734 and WM3899 JARID1B knockdown cells could be quantified (Figure 4C, p<0.05, Fisher’s exact test). Together with the observation that spheres were predominantly generated from J/EGFP-positive cells (Figure S2A, III) this indicates that JARID1B defines a subpopulation which is involved in self-renewal of spheres. As seen with J/EGFP-negative cells (Figure 3F), JARID1B knockdown lines also formed fewer soft agar colonies after 2–3 weeks (Figure 4D, p<0.001 and p<0.01, t-test).
JARID1B is required for continuous growth of xenografted melanoma and for metastatic progression
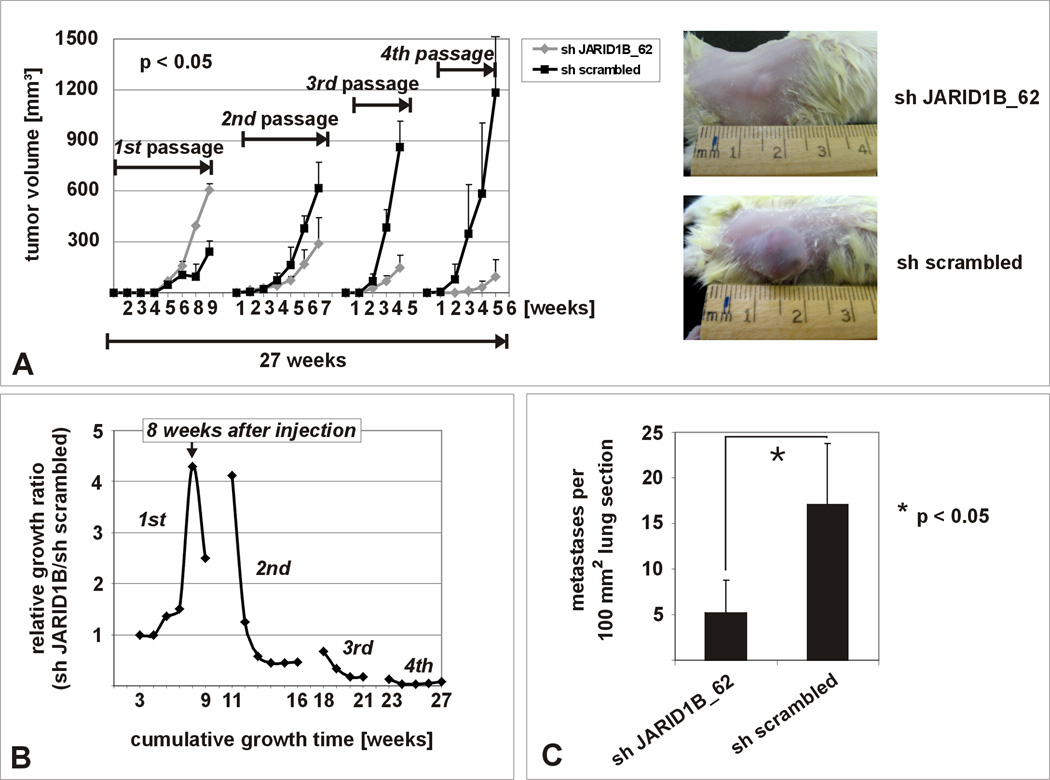
Serial xenotransplantation assays allow assessment of long-term growth of implanted tumor cells without temporary restrictions due to maximum tumor size (Clarke et al., 2006) and, therefore, were applied to investigate the influence of JARID1B knockdown on in vivo tumor growth. In the first passage, there was higher proliferation rate of JARID1B knockdown cells compared to the control (Figure 5A, p<0.05, ANOVA) which mimicked the proliferation pattern seen in vitro (Figure 4A) and the results from single injected J/EGFP-negative cells (Figure 3H). Strikingly, in the subsequent three in vivo passages (Figure 5A and Figure S5), JARID1B knockdown cells gradually lost their potential to expand (p<0.05, ANOVA). When tumor growth was displayed as a relative ratio normalized to the sh scrambled control (Figure 5B), it became apparent that over the total incubation time of 27 weeks in vivo, the proliferation of JARID1B knockdown cells peaked and then steadily exhausted as predicted by our in vitro experiments. Using WM3899 cells, which are known to spontaneously metastasize to the lungs after subcutaneous xenotransplantation, we found a reduction of metastases by JARID1B knockdown cells, as determined by computerized quantitation of histological sections (Figure 5C, p<0.05, t-test). The process of continuous tumor maintenance can only be seen in long-term experiments and seems to be independent of the initial tumor formation.
Figure 5. In vivo exhaustion after knockdown of JARID1B.
(A) In vivo tumor growth is exhausted after 4 passages of serial xenotransplantation of JARID1B knockdown WM3734 cells in NOD/LtSscidIL2Rγnull mice (n=5 per sample) compared to the control (see also Figure S5). (B) Normalized growth ratio (tumor volumes of sh JARID1B tumors divided by volumes of sh scrambled tumors) over the cumulative growth phase of 27 weeks. (C) Significant decrease of pulmonary metastasis after subcutaneous injection of 5×105 JARID1B knocked down WM3899 cells into NOD/LtSscidIL2Rγnull mice (n=5 per sample). Shown is one representative from two independent experiments.
The JARID1B phenotype is dynamic
The CSC concept postulates a unidirectional hierarchy of tumor cells (Reya et al., 2001). Long-term culture after FACS indeed confirmed that the J/EGFP-positive subpopulation induces a heterogeneous daughter population consisting of J/EGFP-positive and –negative cells. Fourteen days after FACS and re-seeding in conventional medium, the J/EGFP-positive-derived progeny consisted of J/EGFP-positive cells and an increasing number of J/EGFP-negative cells (Figure 6A, III). Development of a heterogeneous tumor population was also seen in vivo 56 days after J/EGFP-positive cells (100, 10 or 1) had been xenotransplanted (Figure 6A, V). However, after 14 days in vitro or 56 days in vivo, also J/EGFP-negative cells gave rise to a heterogeneous progeny including J/EGFP-positive cells (Figure 6A, IV and VI), even when derived from a single J/EGFP-negative cell. The re-establishing culture heterogeneity in both progenies resulted in a balanced overall JARID1B expression (Figure 6C). When sorted cells were cultured in hESCM4 rather than conventional medium, the interconversion of phenotypes was considerably decelerated (Figure 6A, VII and VIII). Even after 120 days, only a few J/EGFP-positive cells were found in cultures derived from J/EGFP-negative cells (Figure 6A, VII and B). Accordingly, J/EGFP-positive cells seeded in hESCM4 maintained a higher number of J/EGFP-positive cells. Daughter cultures from both J/EGFP-positive or –negative cells could be cultured for several months without exhaustion which suggests that the self renewal function of second generation J/EGFP-positive cells is also reversible. Since conventional culture conditions seemed to allow a higher dynamics of the JARID1B-positive phenotype, we repeated clonogenic and colony formation assays of sorted cells in conventional medium. Although J/EGFP-positive cells still showed increased colony formation, now the difference from J/EGFP-negative cells was clearly decreased compared to hESCM4 (Figure S3C, D vs. Figure 3). Next to soluble factors from the culture medium, we identified oxygen as significant environmental factor for the dynamic regulation of JARID1B (Figure S6). In agreement with a recent report in HepG2 hepatocellular carcinoma and U87 glioblastoma-astrocytoma cells (Xia et al., 2009), JARID1B expression in melanoma cells rapidly enhanced at low oxygen (3 days, 1% pO2) and steadily reverted to normal expression intensity and frequency after 10 to 14 days of conventional culture at atmospheric oxygen. At last, we observed that J/EGFP-negative cells that were grown as dense patches usually developed a more distinct J/EGFP-positive subpopulation than J/EGFP-negative cells that were kept at lower density (not shown). Finally, the dynamics of the JARID1B phenotype also explains the relatively high sphere forming capacity of non-label-retaining but J/EGFP-positive cells (shown in Figure 2C). These cells most likely acquired the J/EGFP-associated self-renewal potential after PKH-labeling.
JARID1B affects Jagged 1/Notch 1-signaling in melanoma cells
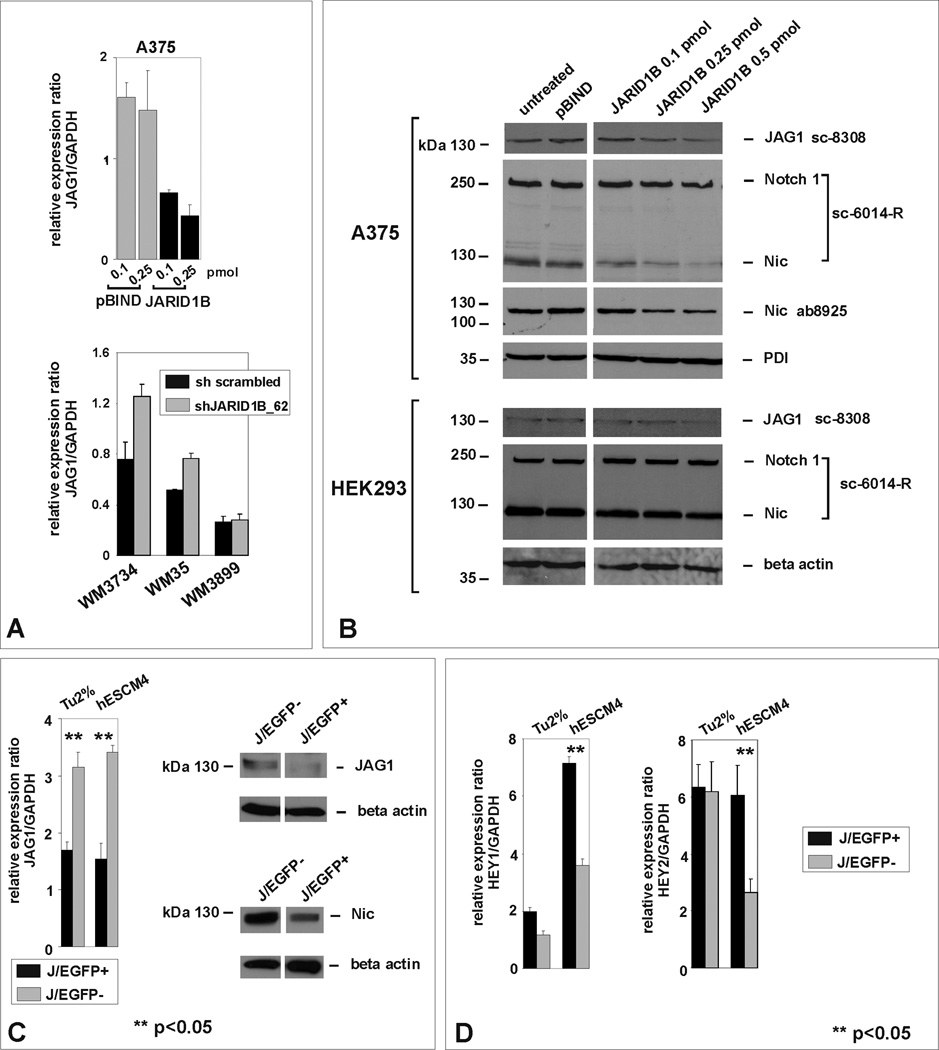
The bidirectional Notch signaling pathway is known to maintain neural progenitors and melanocyte stem cells (Moriyama et al., 2006) and, as we reported earlier, also propagates melanoma progression (Balint et al., 2005; Liu et al., 2006; Pinnix et al., 2009). Besides, we showed that JARID1B transcriptionally represses the Notch ligand Jagged 1 (JAG1) through direct interaction with its promoter (Roesch et al., 2008). In low JARID1B expressing A375-SM melanoma cells (Roesch et al., 2005), transient transfection with JARID1B leads to a concentration-dependent downregulation of JAG1, whereas stable knock down in highly JARID1B-expressing WM3734 and WM35 melanoma cells is followed by JAG1 upregulation (Figure 7A, B). The JARID1B-mediated repression of JAG1 in A375-SM cells was followed by reduced cleavage of Notch 1 into its active form, Nic, while leaving the overall expression of Notch 1 unchanged (Figure 7B). Notch cleavage in non-melanocytic control cells, HEK293, was not affected by JARID1B (Figure 7B). Since in experimentally “homogenized” cultures (by transfection or lentiviral infection), the bidirectional nature of Notch signaling could be masked, we next switched to endogenously JARID1B-positive vs. –negative cells (J/EGFP FACS). Again, high JARID1B expression was associated with low JAG1, in both adherent and sphere cultures (Figure 7C). Particularly in spheres, high JARID1B/low JAG1 was associated with high HEY1 and HEY2 expression, which are both common Notch downstream targets (Figure 7D, p<0.05, t-test). The inverse expression of JARID1B, JAG1, and HEY1/2 amongst neighboring cells is reminiscent of the concept of lateral inhibition where reciprocal stimulation/inhibition of Notch signaling dynamically leads to the maintenance of neuroectodermal stem cells (Kageyama et al., 2008). During differentiation of neuroprogenitors, Nic levels were reported to oscillate resulting in a steadily increasing and decreasing activation of downstream targets (Kageyama et al., 2009). Thus, we suggest that together with JAG1/Notch 1, JARID1B is part of a complex dynamic program of stemness regulation in melanoma.
Figure 7. JARID1B affects Jagged 1/Notch 1 signaling.
(A) QPCR of A375-SM melanoma cells transiently transfected with pBIND-JARID1B vs. empty vector control (upper panel) and of JARID1B-knocked down WM3734, WM35, and WM3899 cells vs. sh scrambled control (lower panel). (B) Immunoblotting of JARID1B-transfected A375-SM melanoma and HEK293 control cells with antibodies against JAG1, uncleaved and cleaved Notch 1. Notch cleavage was confirmed by a second antibody. (C) Differential JAG1 expression and Notch cleavage in FACS-isolated J/EGFP-positive and -negative cells. (D) Differential expression of HEY1 and 2 in J/EGFP-positive and -negative cells.
Discussion
JARID1B has recently been identified as a H3K4 demethylase (Christensen et al., 2007; Yamane et al., 2007). H3K4 methylation has been proposed as a critical component in regulating gene expression and cellular identities and as an epigenetic mark in embryonic stem cells (Guenther et al., 2007). Demethylation of H3K4 by JARID1B was shown to play a role in cell fate decisions of embryonic stem cells by blockage of terminal differentiation (Dey et al., 2008). In cancer, it is known that demethylation of H3K4 supports transformation from hematopoietic precursors to leukemia stem cells via regulation of the developmental Hox gene family (Krivtsov and Armstrong, 2007). As previously suggested for other chromatin remodeling factors such as HDACs or the histone methylase SUV39H1 (Macaluso et al., 2006), also JARID1B maybe part of a multi-molecular complex consisting of DNA, histones, transcription factors such as PAX9, FOXG1B, or LMO2, and pRB (Roesch et al., 2008; Tan et al., 2003). Former studies by us indicated that JARID1B can be actively involved in the maintenance of the slow-cycling state in melanoma via stabilization of pRB (Ser795)-mediated cell cycle control (Roesch et al., 2006). Interestingly, stabilization of hypophosphorylated pRB is usually understood as a tumor-suppressive mechanism because of its anti-proliferative effect, but in the long run, slow proliferation can be also associated with tumor maintenance as we suggest here. Thus, JARID1B may have a dual role over time, immediately anti-proliferative but long-term tumor maintaining.
JARID1B is overexpressed in breast cancer (Lu et al., 1999) raising the question of whether it determines also in epithelial malignancies a population with increased stemness capacity. Our unpublished observations from head and neck squamous cell carcinoma (HNSCC) revealed a broad JARID1B immunoreactivity throughout the entire epithelial portion and, thus, in a pattern very different from that of melanoma. A possible explanation for this discrepancy is provided by publications on another family member, JARID2. JARID2 can be expressed in an undulating fashion. It is strongly expressed in embryonic stem cells, decreases when cells proliferate and tissues expand, but increases again after terminal differentiation (Takeuchi et al., 2006). Thus, the observed immunostaining pattern of HNSCCs could reflect a high expression of JARID1B also in differentiated carcinoma cells. Since epithelial carcinomas generally harbor more differentiated cells than melanomas, it was also not surprising that sorted label-retaining esophageal carcinoma cells showed decreased stemness (differentiated cells were less clonogenic, not shown). Consequently, to unravel the role of JARID1B as a marker for epithelial CSCs, further biological markers are needed to exclude the differentiated progeny.
According to the traditional CSC concept, tumor initiation is regarded as an exclusive characteristic of CSCs (Clarke et al., 2006). Thus, our finding that tumor initiation in immunodeficient animals is independent of JARID1B expression challenges the role of JARID1B as a classic CSC marker. In highly susceptible xenotransplantation assays like the one we used, the ratio of tumor initiating cells can increase up to 25% of unsorted cells (Quintana et al., 2008). This suggests that either virtually every melanoma cell is a CSC because it can induce de novo tumors in xenograft assays irrespective of any known stem cell marker, or that melanoma is not hierarchically organized into subpopulations of tumorigenic and non-tumorigenic cells and the CSC model does not apply. However, tumor initiation in xenograft models could be more dependent on secondary factors, such as the adaptation to host-derived growth factors (Adams and Strasser, 2008). For example, congenic transplantation of murine lymphoma cells revealed that tumor initiation is not necessarily restricted to a minority population (Kelly et al., 2007). Thus, it is generally questionable whether the ability of melanoma cells to initiate de novo tumors really reflects the same potential as to maintain growth within an already established tumor microenvironment. Particularly the normalized growth data in Figure 5B support that this assay measures more the gradual loss of tumor growth than the loss of tumor initiation.
The Weinberg group recently observed a dependency of self-renewal on exogenous factors. Treatment of immortal human mammary epithelial cells with recombinant TGFβ could induce the stem like CD44high/CD24low phenotype known from breast CSCs (Mani et al., 2008). Ours and Weinberg’s data suggest that at least some stem-like cells from solid tumors may actually not be static entities, but rather tumor cells that transiently acquire stemness properties depending on the tumor context. This supports a model of dynamic stemness in which at any given point of time a slow-cycling, self-renewing but dynamic subpopulation of tumor cells exists among the bulk of tumor cells. This subpopulation might continually arise or disappear (‘moving target’). Certainly, improved assays, e.g. humanized mouse models, are critically needed to address these questions in the future.
Since the study by Mani et al. suggested epithelial-mesenchymal transition (EMT) as an alternative stemness-associated mechanism, we asked if also JARID1B-positive melanoma cells show EMT or, at least, an EMT-like phenotype (because of their neuroectodermal origin, melanoma cells may not undergo classic EMT). However, none of our genome-wide profiling experiments (1. JARID1B knockdown vs. scrambled, 2. transient JARID1B overexpression vs. mock, and 3. J/EGFP-positive vs. J/EGFP-negative) detected a consistent classic EMT signature in correlation with high JARID1B expression. We also did not find a consistent overlap with previously suggested stemness markers, like CD133 (Monzani et al., 2007) and p75/NGFR (Wong et al., 2006). A trend towards higher expression of CD20 (Fang et al., 2005) in J/EGFP-positive cells was assumed but due to the low overall expression frequency (0.7’2%), there was a lack of experimental consistency in replicate analyses (not shown).
The existence of a slow-cycling subpopulation is of high clinical importance because almost all current therapeutic regimens predominantly target the rapidly proliferating tumor bulk (Blagosklonny, 2005). Indeed, we observed that anti-cancer therapies in vitro (cytotoxic or BRAFV600E-targeted) uniformly result in an enrichment for JARID1B-positive cells (data not shown). Thus, we postulate that targeting of the slow-cycling subpopulation, e.g. by inhibition of its H3K4 demethylase activity, in combination with a conventional debulking strategy could help eradicate all melanoma cells and increase the low therapy response rate in malignant melanoma.
Experimental Procedures
Melanoma samples, cell lines, and lentiviral constructs
Human melanoma tissues and cell lines were obtained in accordance with consent procedures approved by the Internal Review Boards of the University of Pennsylvania School of Medicine, The Wistar Institute, and the Regensburg University Medical Center (see also Suppl. Experimental Procedures). Cells were isolated and maintained in 2% FBS-substituted melanoma medium (Tu2%) as previously described (Satyamoorthy et al., 1997). Melanoma spheres were propagated in mouse embryonic fibroblast (MEF)–conditioned human embryonic stem cell medium as reported (hESCM4, Fang et al., 2005). Spheres were dissociated by collagenase I/IV (Sigma) and mechanical treatment, i.e. 200–250 units per ml DMEM (Cellgro) at 37 °C for 10 min plus subsequent pipetting. Cell viability was assured microscopically and by 7-AAD dead cell exclusion as explained in Suppl. Experimental Procedures. The consistency of cellular genotypes was confirmed by DNA fingerprinting using Coriell's microsatellite kit. The lentiviral vector constructs for stable knockdown of JARID1B and the scrambled control were purchased from Sigma. Lentiviral pLU-CMV-Blast and pLU-CMV-EGFP (Wistar Libraries Core Facility) were used to clone pLU-JARID1Bprom-EGFP-Blast and pLU-CMV-EGFP-Blast. The JARID1B main promoter was PCR-cloned from human genomic DNA (Promega) and verified by DNA sequencing as described in Suppl. Experimental Procedures. Selection of positive clones was done by treatment with puromycin or blasticidin.
Detection of JARID1B
JARID1B-immunohistochemistry and -immunofluorescence microscopy were done with a polyclonal rabbit anti-JARID1B antibody (Roesch at al., 2005). For immunoblotting, we used anti-JARID1B aa 784–883 No. 2226.00.02 from Strategic Diagnostics. Quantitative real time RT-PCR was done in an ABI PRISM 7000 using own primers or primers from the Harvard primer bank http://pga.mgh.harvard.edu/primerbank (see also Suppl. Experimental Procedures).
Flow cytometry and FACS of J/EGFP signals
For detection of JARID1B promoter-driven EGFP signals, adherent WM3734JARID1Bprom-EGFP cells were harvested with 0.05% trypsin and spheres were dissociated as described before. Dead cells were excluded by staining for 7-AAD. All experiments were performed by the Wistar Flow Cytometry Facility using an EPICS XL instrument (Beckman-Coulter) and a Cytomation MoFlo cytometer (DakoCytomation). After sorting, aliquots were microscopically checked and were cultured for a short time to exclude disproportional enrichment of debris or apoptotic cells.
Cell proliferation and colony formation assays
Cell proliferation was quantified by MTS (Promega) and BrdU incorporation assays (Roche) according to the manufacturers’ protocols and confirmed by manual cell counts using a hematocytometer. Anchorage-independent colony formation was measured in 0.35% soft agar assays (see also Suppl. Experimental Procedures).
In vitro self-renewal assay
Quantitation of melanoma sphere self-renewal was done by limited (single cell) dilution assays. Briefly, cells were seeded at a ratio of 0.5 cell per well in 96-well plates to avoid doublets. Using an Olympus CKX41SF phase contrast microscope, wells containing one live cell were marked after 2 hours. Development of spheres was assessed after 20–30 days.
In vivo self-renewal and metastasis assays
Titrated xenotransplantation of J/EGFP-sorted cells was done for 100, 10 and 1 cell per injection (5 mice per sample, 4 injections per mouse). 100 and 10 cell dilutions were based on FACS counts and were verified microscopically. Preparation of single cell injections was performed as published elsewhere (Quintana et al., 2008). For serial xenotransplantation, 104 cells were subcutaneously injected at a Matrigel®/Tu2% ratio of 1:1 (5 mice per sample). One passage comprised: injection, tumor growth, tumor dissection, cell isolation (mechanical and collagenase I/IV treatment) and melanoma cell purification (3-day-puromycin treatment confirmed by melanoma-specific MCAM flow cytometry). Verification of knockdown was done by QPCR before re-injection and immunohistochemistry of tumor sections. Tumor growth was measured weekly using a caliper and was terminated when the first tumor of the series reached 1000 mm3. For induction of spontaneous pulmonary metastasis, 5 × 105 WM3899 JARID1B knockdown melanoma cells were injected subcutaneously into NOD/LtSscidIL2Rγnull mice (5 mice per sample) and incubated for five weeks. Metastases were microscopically determined as described in Suppl. Experimental Procedures. All animal experiments were performed in accordance with Wistar IACUC protocol 111983 in NOD/LtSscidIL2Rγnull mice.
In vitro and in vivo label-retaining (LR) assays
In vitro LR of cells was done using the PKH26 Red Fluorescent Cell Linker Kit for general membrane labeling (Sigma). 100% labeling efficiency was reached with 106 dissociated sphere cells incubated in 100 µl of 1 µM PKH26. LR was determined by flow cytometry and fluorescence microscopy. For in vivo BrdU-LR, 104 cells were xenotransplanted into NOD/LtSscidIL2Rγnull mice (n=10). Five of the mice were given a single i.p. injection of 1.5 mg BrdU in DPBS and were subsequently maintained on 1 mg/ml BrdU in the drinking water for 12 days. 5 mice served as controls. Tumors were grown for an additional 6 weeks. BD Pharmingen’s APC BrdU Flow Kit was used for detection.
Statistics
To determine the statistical significance of growth curves, ANOVA for repeated measures was applied and confirmed by Student’s t-test. Differences in limited dilution assays were statistically determined using Fisher’s Exact Test. For all other experiments, the Student’s t-test was used. A p-value of less than 0.05 was considered significant. As software tools, SAS version 9.2 using Proc Freq and Microsoft Excel were used.
Supplementary Material
Acknowledgments
We thank J Hayden and F Keeney (Wistar Microscopy Facility), DC Schultz (Wistar Protein Expression Facility), JS Faust, D Ambrose, and D Hussey (Wistar Flow Cytometry Facility) for technical support; L Li, L Kuenzel, T Schifferstein, E Herschberger, and A Mueller for technical assistance, JT Lee for valuable discussion, and DE Elder (Hospital of the University of Pennsylvania) for providing patient material. The animals were housed at Wistar’s Animal Facility. The research was funded by NIH grants P50 CA 093372, P01 CA 025874, R01 CA 047159, R01 CA 076674, P30 CA10815 and by the DFG grant R3577/2-1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams JM, Strasser A. Is tumor growth sustained by rare cancer stem cells or dominant clones? Cancer Res. 2008;68:4018–4021. doi: 10.1158/0008-5472.CAN-07-6334. [DOI] [PubMed] [Google Scholar]
- Balint K, Xiao M, Pinnix CC, Soma A, Veres I, Juhasz I, Brown EJ, Capobianco AJ, Herlyn M, Liu ZJ. Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. J Clin Invest. 2005;115:3166–3176. doi: 10.1172/JCI25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV. Why therapeutic response may not prolong the life of a cancer patient: selection for oncogenic resistance. Cell Cycle. 2005;4:1693–1698. doi: 10.4161/cc.4.12.2259. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- Dey BK, Stalker L, Schnerch A, Bhatia M, Taylor-Papidimtriou J, Wynder C. The histone demethylase KDM5b/JARID1b plays a role in cell fate decisions by blocking terminal differentiation. Mol Cell Biol. 2008;17:5312–5327. doi: 10.1128/MCB.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, Van Belle PA, Xu X, Elder DE, Herlyn M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65:9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- Gogas HJ, Kirkwood JM, Sondak VK. Chemotherapy for metastatic melanoma: time for a change? Cancer. 2007;109:455–464. doi: 10.1002/cncr.22427. [DOI] [PubMed] [Google Scholar]
- Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grichnik JM, Burch JA, Schulteis RD, Shan S, Liu J, Darrow TL, Vervaert CE, Seigler HF. Melanoma, a tumor based on a mutant stem cell? J Invest Dermatol. 2006;126:142–153. doi: 10.1038/sj.jid.5700017. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Niwa Y, Shimojo H. Rhythmic gene expression in somite formation and neural development. Mol Cells. 2009;27:497–502. doi: 10.1007/s10059-009-0068-1. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Shimojo H, Imayoshi I. Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat Neurosci. 2008;11:1247–1251. doi: 10.1038/nn.2208. [DOI] [PubMed] [Google Scholar]
- Kelly PN, Dakic A, Adams JM, Nutt SL, Strasser A. Tumor growth need not be driven by rare cancer stem cells. Science. 2007;317:337. doi: 10.1126/science.1142596. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG., Jr The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Liu ZJ, Xiao M, Balint K, Smalley KS, Brafford P, Qiu R, Pinnix CC, Li X, Herlyn M. Notch1 signaling promotes primary melanoma progression by activating mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt pathways and up-regulating N-cadherin expression. Cancer Res. 2006;66:4182–4190. doi: 10.1158/0008-5472.CAN-05-3589. [DOI] [PubMed] [Google Scholar]
- Lu PJ, Sundquist K, Baeckstrom D, Poulsom R, Hanby A, Meier-Ewert S, Jones T, Mitchell M, Pitha-Rowe P, Freemont P, et al. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J Biol Chem. 1999;274:15633–15645. doi: 10.1074/jbc.274.22.15633. [DOI] [PubMed] [Google Scholar]
- Macaluso M, Montanari M, Giordano A. Rb family proteins as modulators of gene expression and new aspects regarding the interaction with chromatin remodeling enzymes. Oncogene. 2006;25:5263–5267. doi: 10.1038/sj.onc.1209680. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzani E, Facchetti F, Galmozzi E, Corsini E, Benetti A, Cavazzin C, Gritti A, Piccinini A, Porro D, Santinami M, et al. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur J Cancer. 2007;43:935–946. doi: 10.1016/j.ejca.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Moriyama M, Osawa M, Mak SS, Ohtsuka T, Yamamoto N, Han H, Delmas V, Kageyama R, Beermann F, Larue L, et al. Notch signaling via Hes1 transcription factor maintains survival of melanoblasts and melanocyte stem cells. J Cell Biol. 2006;173:333–339. doi: 10.1083/jcb.200509084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinnix CC, Lee JT, Liu ZJ, McDaid R, Balint K, Beverly LJ, Brafford PA, Xiao M, Himes B, Zabierowski SE, et al. Active Notch1 confers a transformed phenotype to primary human melanocytes. Cancer Res. 2009;69:5312–5320. doi: 10.1158/0008-5472.CAN-08-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Roesch A, Becker B, Meyer S, Wild P, Hafner C, Landthaler M, Vogt T. Retinoblastoma-binding protein 2-homolog 1: a retinoblastoma-binding protein downregulated in malignant melanomas. Mod Pathol. 2005;18:1249–1257. doi: 10.1038/modpathol.3800413. [DOI] [PubMed] [Google Scholar]
- Roesch A, Becker B, Schneider-Brachert W, Hagen I, Landthaler M, Vogt T. Re-expression of the retinoblastoma-binding protein 2-homolog 1 reveals tumor-suppressive functions in highly metastatic melanoma cells. J Invest Dermatol. 2006;126:1850–1859. doi: 10.1038/sj.jid.5700324. [DOI] [PubMed] [Google Scholar]
- Roesch A, Mueller AM, Stempfl T, Moehle C, Landthaler M, Vogt T. RBP2-H1/JARID1B is a transcriptional regulator with a tumor suppressive potential in melanoma cells. Int J Cancer. 2008;122:1047–1057. doi: 10.1002/ijc.23211. [DOI] [PubMed] [Google Scholar]
- Satyamoorthy K, DeJesus E, Linnenbach AJ, Kraj B, Kornreich DL, Rendle S, Elder DE, Herlyn M. Melanoma cell lines from different stages of progression and their biological and molecular analyses. Melanoma Res. 1997;7 Suppl 2:S35–S42. [PubMed] [Google Scholar]
- Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM, Weishaupt C, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scibetta AG, Santangelo S, Coleman J, Hall D, Chaplin T, Copier J, Catchpole S, Burchell J, Taylor-Papadimitriou J. Functional analysis of the transcription repressor PLU-1/JARID1B. Mol Cell Biol. 2007;27:7220–7235. doi: 10.1128/MCB.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Takeuchi T, Watanabe Y, Takano-Shimizu T, Kondo S. Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev Dyn. 2006;235:2449–2459. doi: 10.1002/dvdy.20851. [DOI] [PubMed] [Google Scholar]
- Tan K, Shaw AL, Madsen B, Jensen K, Taylor-Papadimitriou J, Freemont PS. Human PLU-1 Has transcriptional repression properties and interacts with the developmental transcription factors BF-1 and PAX9. J Biol Chem. 2003;278:20507–20513. doi: 10.1074/jbc.M301994200. [DOI] [PubMed] [Google Scholar]
- Vogt T, Kroiss M, McClelland M, Gruss C, Becker B, Bosserhoff AK, Rumpler G, Bogenrieder T, Landthaler M, Stolz W. Deficiency of a novel retinoblastoma binding protein 2-homolog is a consistent feature of sporadic human melanoma skin cancer. Lab Invest. 1999;79:1615–1627. [PubMed] [Google Scholar]
- Wong CE, Paratore C, Dours-Zimmermann MT, Rochat A, Pietri T, Suter U, Zimmermann DR, Dufour S, Thiery JP, Meijer D, et al. Neural crest-derived cells with stem cell features can be traced back to multiple lineages in the adult skin. J Cell Biol. 2006;175:1005–1015. doi: 10.1083/jcb.200606062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MH, Calcagno AM, Salcido CD, Carlson MD, Ambudkar SV, Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24− and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10:R10. doi: 10.1186/bcr1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Lemieux ME, Li W, Carroll JS, Brown M, Liu XS, Kung AL. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci U S A. 2009;106:4260–4265. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Zabierowski SE, Herlyn M. Melanoma stem cells: the dark seed of melanoma. J Clin Oncol. 2008;26:2890–2894. doi: 10.1200/JCO.2007.15.5465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.