Abstract
Sphingolipids comprise a highly diverse and complex class of molecules that serve not only as structural components of membranes but also as signaling molecules. To understand the differential role of sphingolipids in a regulatory network, it is important to use specific and quantitative methods. We developed a novel LC-MS/MS method for the rapid, simultaneous quantification of sphingolipid metabolites, including sphingosine, sphinganine, phyto-sphingosine, di- and trimethyl-sphingosine, sphingosylphosphorylcholine, hexosylceramide, lactosylceramide, ceramide-1-phosphate, and dihydroceramide-1-phosphate. Appropriate internal standards (ISs) were added prior to lipid extraction. In contrast to most published methods based on reversed phase chromatography, we used hydrophilic interaction liquid chromatography and achieved good peak shapes, a short analysis time of 4.5 min, and, most importantly, coelution of analytes and their respective ISs. To avoid an overestimation of species concentrations, peak areas were corrected regarding isotopic overlap where necessary. Quantification was achieved by standard addition of naturally occurring sphingolipid species to the sample matrix. The method showed excellent precision, accuracy, detection limits, and robustness. As an example, sphingolipid species were quantified in fibroblasts treated with myriocin or sphingosine-kinase inhibitor. In summary, this method represents a valuable tool to evaluate the role of sphingolipids in the regulation of cell functions.
Keywords: HILIC, high throughput, ESI, free sphingoid base, methylated sphingoid base, hexosylceramide, lactosylceramide, sphingosylphosphorylcholine, ceramide-1-phosphate
Sphingolipids comprise a highly diverse and complex class of molecules that serve not only as structural components of cellular membranes but also as bioactive compounds with crucial biological functions (1). Some metabolites, including ceramide, sphingosine (SPH), and SPH-1-phosphate (S1P), have been shown to be involved in different cell functions such as proliferation, differentiation, growth arrest, and apoptosis (2). Especially the counter-regulatory functions of ceramide and S1P, resembling the sphingolipid rheostat, indicate that not only a single metabolite concentration but rather the relative levels of these lipids are important to determine the cell fate (2–5). Sphingolipids are associated with several diseases such as cancer, obesity, and atherosclerosis (1, 2, 6–9). Structural diversity and inter-conversion of these sphingolipid metabolites represent technical challenges. Nevertheless, to understand the differential role of sphingolipids in a regulatory network, it is imperative to use specific and quantitative methods.
During the last decade, LC-MS/MS has become a powerful tool for sphingolipid analysis (10–21). However, either these methods do not cover a broad spectrum of sphingolipid metabolites or they show disadvantages like laborious sample preparation, time-consuming LC-separation, or separation of analytes and internal standards (ISs).
Therefore, we applied, as previously described for lysophosphatidic acid and sphingoid base phosphates, hydrophilic interaction chromatography (HILIC) coupled to MS (18) to achieve coelution of sphingolipid species and their ISs. We present a fast and simple LC-MS/MS method for the quantification of hexosylceramide (HexCer), lactosylceramide (LacCer), sphingosine (SPH), sphinganine (SPA), phyto-SPH (PhytoSPH), di- and trimethyl-SPH (Di-; TrimetSPH), sphingosylphosphorylcholine (SPC), ceramide-1-phosphate (Cer1P), and dihydroceramide-1-phosphate (dhCer1P). This method was validated and applied to fibroblasts treated with myriocin and a SPH-kinase inhibitor (SKI), respectively.
MATERIALS AND METHODS
Chemicals and solutions
Butanol, methanol (HPLC grade), and formic acid (98–100%, for analysis) were purchased from Merck (Darmstadt, Germany). Water was obtained from B. Braun (Melsungen, Germany). Ammonium formate (Fluka, Buchs, Switzerland), citric acid monohydrate, and disodium hydrogenphosphate (Merck) were of the highest analytical grade available. S1P (d18:1) C17-SPH (d17:1); SPH (d18:1) SPA (d18:0); C17-SPC (d17:1); N,N-dimethyl-SPH (d18:1); N,N,N-trimethyl-SPH (d18:1); PhytoSPH (t18:0); SPC (d18:1), C12:0-glucosylceramide; C16:0-glucosylceramide; C24:1-galactosylceramide; C12:0-LacCer; C16:0-LacCer; C24:0-LacCer; C12:0- Cer1P; C16:0-Cer-1-phosphate and C24:0- Cer1P were purchased from Avanti Polar Lipids (Alabaster, AL) with purities higher than 99%. 13C2D2-SPH-1-phosphate (d18:1) was purchased from Toronto Research Chemicals (Toronto, Canada). Stock solutions of individual sphingolipid compounds at a concentration of 1 mg/ml were prepared in methanol and stored at −20°C. Working solutions of the desired concentrations were prepared by dilution in methanol. Myriocin and SPH-kinase-inhibitor [2-(p-hydroxyanilino)-4-(p-chlorphenyl) thiazole] were purchased from Calbiochem (San Diego, CA).
Cell culture
Primary human skin fibroblasts were cultured as described previously (22) in Dulbecco's modified Eagle's medium supplemented with L-glutamine and 10% fetal calf serum in a humidified 5% CO2 atmosphere at 37°C. For lipid analysis, cells were seeded into 6-well plates and grown to confluence. Cells were rinsed two times with ice-cold PBS and either lysed in 0.2% SDS in water or scraped in PBS. Subsequently, samples were subjected to centrifugation at 240 g for 7 min, and the resulting pellet was homogenized in distilled water by sonication. Fibroblasts treated with myriocin or SKI (Calbiochem) were lysed in 0.2% SDS (Fig. 4). Aliquots of the cell homogenates were taken for protein determination. Protein concentrations were measured using bicinchoninic acid as described previously (23).
Fig. 4.
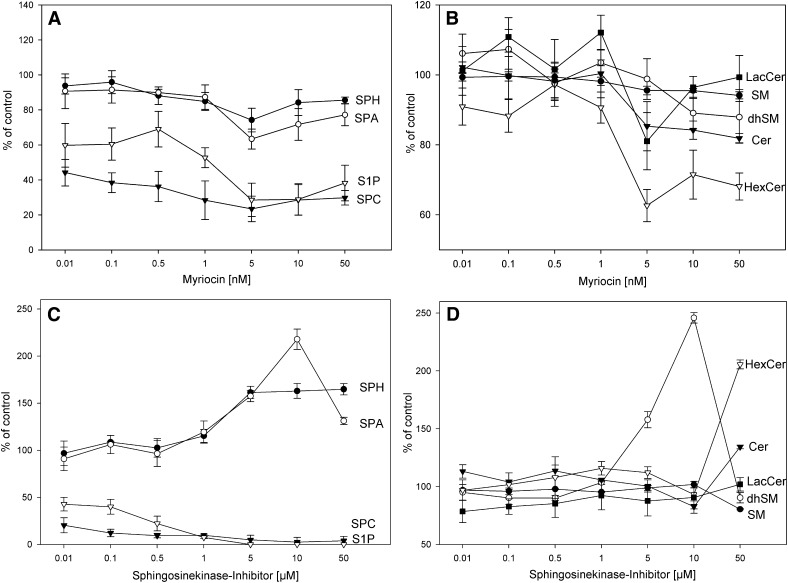
The effect of myriocin and SKI on intracellular sphingolipids in primary human skin fibroblasts. Cells were treated with increasing concentrations of myriocin (A + B) and SKI (C + D) for 24 h, respectively. SPH (closed circle), SPA (open circle), SPC (closed triangle), S1P (open triangle), HexCer (open triangle), and LacCer (closed square) were quantified by LC-MS/MS; Cer (closed triangle), SM (closed circle), and dhSM (open circle) were quantified by flow injection analysis (ESI-MS/MS). Values represent the mean ± SD of three independent samples.
Sample preparation
Unless otherwise indicated, aliquots of 100 µg protein from the fibroblast homogenates were used for sphingolipid analysis. A total of 20 µL of an IS mixture containing 20 ng SPH d17:1, 2 ng SPC d17:1, 20 ng GluCer 12:0, 20 ng LacCer 12:0, and 20 ng Cer1P 12:0 was added prior to lipid extraction. We applied a butanolic extraction procedure described by Baker et al. (24). In brief, 500 µl cell homogenate corresponding to 100 µg of cellular protein were mixed with 60 µL of a buffer containing 200 mM citric acid and 270 mM disodium hydrogenphosphate (pH 4). Extraction was performed with 1 ml of 1-butanol and 500 µL of water-saturated 1-butanol. The recovered butanol phase was evaporated to dryness under reduced pressure. The residue was redissolved in 200 µL ethanol.
Sphingolipid analysis by LC-MS/MS
Sphingolipid analysis was performed by LC-MS/MS. The HPLC equipment consisted of a 1200 series binary pump (G1312B), a 1200 series isocratic pump (G1310A), and a degasser (G1379B) (Agilent, Waldbronn, Germany) connected to an HTC Pal autosampler (CTC Analytics, Zwingen, Switzerland). A hybrid triple quadrupole linear ion trap mass spectrometer API 4000 Q-Trap equipped with a Turbo V source ion spray operating in positive ESI mode was used for detection (Applied Biosystems, Darmstadt, Germany). High purity nitrogen was produced by a nitrogen generator NGM 22-LC/MS (cmc Instruments, Eschborn, Germany).
Gradient chromatographic separation was performed on an Interchim (Montlucan, France) HILIC silica column (50 × 2.1 mm) with a 1.8 µm particle size equipped with a 0.5 µm prefilter (Upchurch Scientific, Oak Harbor, WA). The injection volume was 2 µL and the column was maintained at 50°C. The mobile phase consisted of water containing 0.2% formic acid and 200 mM ammonium formate (eluent A) and acetonitrile containing 0.2% formic acid (eluent B). Gradient elution was performed with 100% B for 0.1 min, a step to 90% B until 0.11 min, a linear increase to 50% B until 2.5 min, 50% B until 3.5 min, and reequilibration from 3.51 to 4.5 min with 100% B. The flow rate was set to 800 µl/min. To minimize contamination of the mass spectrometer, the column flow was directed only from 1.0 to 3.0 min into the mass spectrometer using a diverter valve. Otherwise, methanol with a flow rate of 250 µl/min was delivered into the mass spectrometer.
The turbo ion spray source was operated in the positive ionization mode using the following settings: ion spray voltage = 5,500 V, ion source heater temperature = 400°C, source gas 1 = 40 psi, source gas 2 = 35 psi, and curtain gas setting = 20 psi. Analytes were monitored in the multiple reaction monitoring (MRM) mode, mass transitions and MS parameters are shown in Table 1. Quadrupoles Q1 and Q3 were working at unit resolution.
TABLE 1.
MS parameter and LOD of sphingolipids studied
| Sphingolipid | [M+H] m/z | MRM | IS (MRM) | CE (V) | RT (min) | LOD (fmol) on column |
|---|---|---|---|---|---|---|
| SPH | 300.3 | 300.3→282.2 | C17 SPH (286.3→268.2) | 17 | 1.04 | 7.3 |
| 300.3→252.2 | C17 SPH (286.3→238.2) | 25 | 1.04 | 7.7 | ||
| SPA | 302.3 | 302.3→284.2 | C17 SPH (286.3→268.2) | 21 | 1.04 | 6.1 |
| 302.3→254.2 | C17 SPH (286.3→238.2) | 29 | 1.04 | 8.8 | ||
| PhytoSPH | 318.4 | 318.4→282.2 | C17 SPH (286.3→268.2) | 23 | 1.06 | 24.2 |
| DimetSPH | 328.4 | 328.4→280.3 | C17 SPH (286.3→268.2) | 29 | 1.02 | 0.2 |
| TrimetSPH | 342.4 | 342.4→60.1 | C17 SPH (286.3→268.2) | 49 | 1.05 | 0.1 |
| SPC | 465.3 | 465.3→184 | C17 SPC (451.3→184) | 31 | 1.75 | 4.9 |
| HexCer | var. | M+H→264.3 | C12 GluCer (644.5→264.3) | 55 | 0.80 | 0.6 |
| LacCer | var. | M+H→264.3 | C12 LacCer (806.6→264.3) | 65 | 0.94 | 2.7 |
| Cer1P | var. | M+H→264.3 | C12 Cer1P (562.4→264.3) | 39 | 1.29 | 6.3 |
| dhCer1P | var. | Neutral loss 98 | C12 Cer1P (562.4→262.3) | 29 | 1.29 | 53.6 |
CE, collision energy; RT, retention time.
Calibration and quantification
Calibration was achieved by standard addition of naturally occurring sphingolipid species (S1P, GluCer 16:0, GalCer 24:1, LacCer 16:0 and 24:0, Cer1P 16:0 and 24:0, SPH, SPA, SPC, DimetSPH, TrimetSPH, PhytoSPH). A 6 point calibration was performed by adding the indicated amounts (0–300 pmol) of a combined sphingolipid standard mixture to matrix samples. Calibration curves were calculated by linear regression without weighting.
Data analysis was performed with Analyst software 1.4.2. (Applied Biosystems). The data were exported to Excel spreadsheets and further processed by self-programmed Excel macros that sorted the results, calculated the analyte/IS peak area ratios, generated calibration lines, and calculated sample concentrations. Where necessary, isotopic overlap of the species was corrected based on theoretical isotope distribution according to principles described previously (25). Analytes and their corresponding ISs are shown in Table 1.
Analysis of S1P, ceramide, and sphingomyelin
S1P was analyzed by LC-MS/MS as described previously (18). Ceramide and sphingomyelin species were analyzed by flow injection analysis ESI-MS/MS (25, 26).
RESULTS
Sphingolipid fragmentation
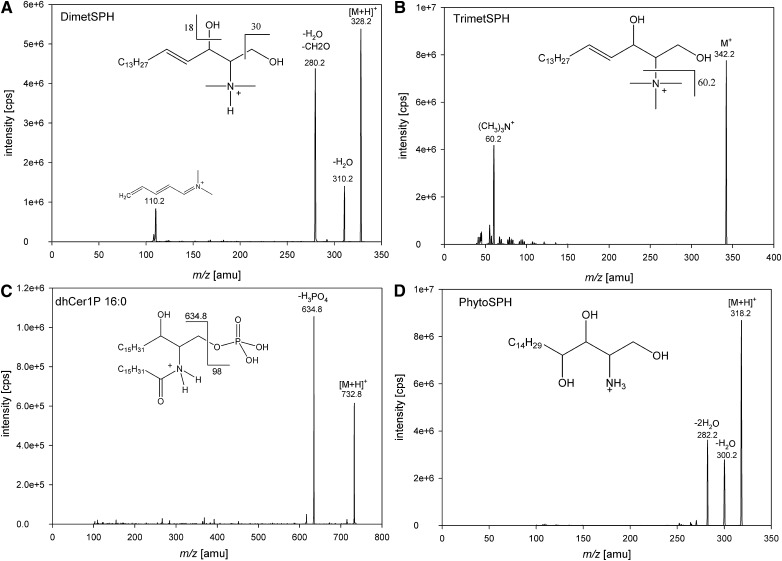
To analyze various sphingolipid classes, we applied ESI in the positive ion mode and acquired product ion spectra. The fragmentation patterns obtained were in accordance to previous studies for SPH, SPA, Cer1P, and glycosylated ceramide species (Table 1) (12, 16, 19, 21, 27, 28). Glycosylated ceramides displayed [M+H]+ ions as well as [M+H-H2O]+ ions generated by in-source fragmentation (data not shown). Because [M+H]+ ions exhibited much higher intensities, we did not use [M+H-H2O]+ for further analysis of glycosylated ceramides. As expected, SPC showed only one intense fragment ion at m/z 184 due to the loss of the phosphocholine head group (29). DimetSPH showed beside fragments resulting from a loss of one water molecule (m/z 310) or one water molecule and a formaldehyde molecule (m/z 280), and an ion at m/z 110, possibly a conjugated iminium ion (Fig. 1A). TrimetSPH showed only one intense fragment representing a trimethylammonium-ion at m/z 60 (Fig. 1B). In contrast to Cer1P species showing a sphingoid base fragment, dihydro-Cer-1P displayed a neutral loss of phosphoric acid in positive ion mode (Fig. 1C). Collision-induced dissociation of PhytoSPH showed two prominent fragment ions resulting from the loss of one and two water molecules (Fig. 1D).
Fig. 1.
Product ion spectrum and proposed fragmentation of DimetSPH (A), TrimetSPH (B), dhCer1P (C), and PhytoSPH (D) in positive ion mode.
HILIC of sphingolipids
Due to the relatively low level of the selected sphingolipids in crude lipid extracts, a direct analysis using “shotgun approaches” may be hampered by signal suppression caused by other matrix components (12, 19, 27). Therefore, we decided to establish an HPLC separation of sphingolipids with a short analysis time and coelution of analyte and IS. The latter is of major importance to compensate for matrix effects and varying ionization efficiencies, especially during gradient elution. Because reversed phase chromatography shows chain length-dependent separation, coelution of analytes and ISs may not be accomplished (13, 16, 19, 27). Classical normal phase chromatography offers polar head group-specific separation but may be impaired by limited reproducibility and insufficient peak shapes. Moreover, the use of apolar solvents may not provide optimal ionization conditions for ESI. Hence, we established an LC separation based on HILIC, which shows lipid head group selectivity along with the use of polar solvents. Using a sub-2 μm particle size, we achieved baseline separation for all sphingolipid classes within 2 min and 4.5 min total run time including reequilibration (Figs. 2 and 3). Gradient elution was performed with a mixture of acetonitrile and water including 0.2% formic acid and 200 mM ammonium formate. Addition of formic acid improved the ionization efficiency; an optimum was found at 0.2%. For optimum performance and reproducibility, it is recommended to use at least a concentration of 10 mmol/L ammonium formate in the mobile phase. Therefore, 200 mmol/L buffer and 0.2% formic acid were added to mobile phase A and 0.2% formic acid to mobile phase B.
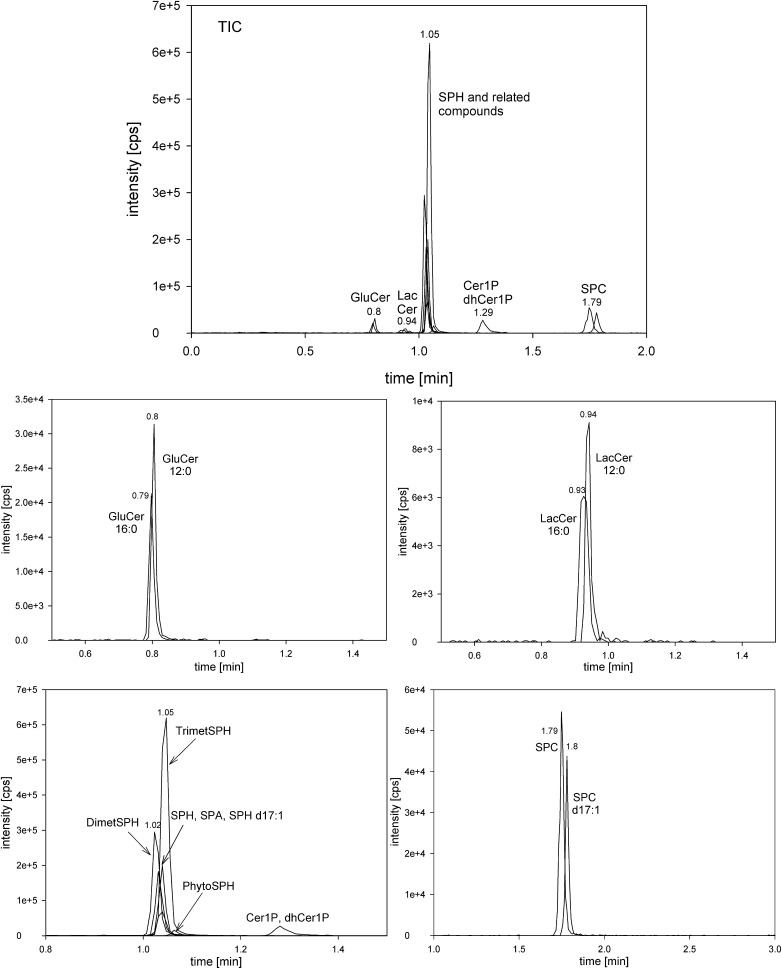
Fig. 2.
Chromatogram of a sphingolipid standard mixture. Displayed are MS/MS transitions representative of distinct sphingolipid classes and their respective ISs.
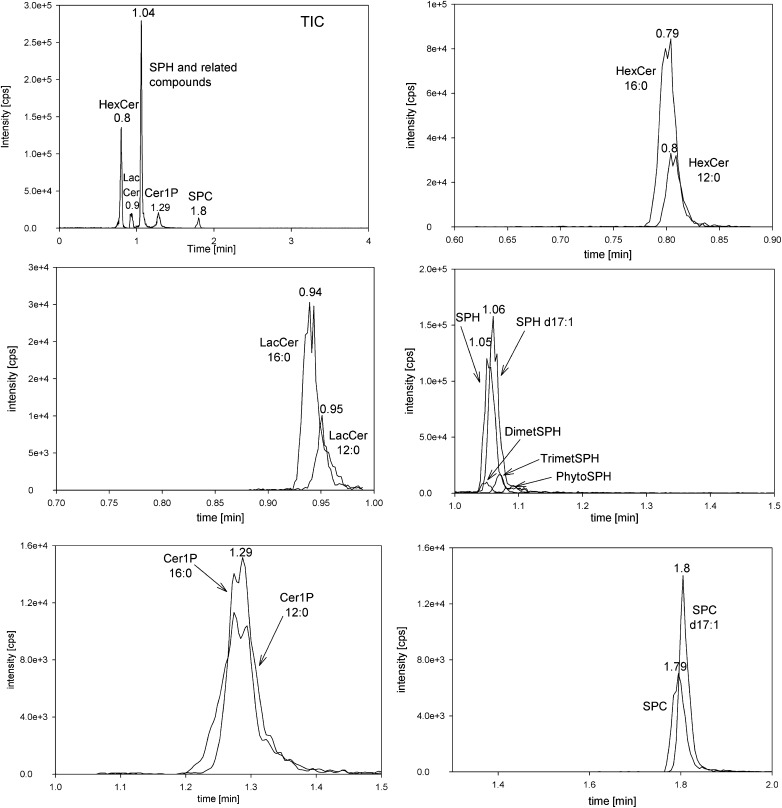
Fig. 3.
Chromatogram of a fibroblast sample. Displayed is a representative mass chromatogram obtained from a human skin fibroblast lipid extract.
Because numerous MS transitions are required to cover the naturally occurring sphingolipid species and their ISs, we split the MS program into four periods: 0–0.75 min (HexCer), 0.75–0.89 (LacCer), 0.89–1.5 (SPH and related compounds), and 1.5–4.5 (SPC) (Fig. 2).
Extraction efficiency and matrix effects
To analyze polar sphingolipids from one lipid extract, we tested a butanolic extraction previously described for S1P analysis (18). The extraction efficiency was determined in fibroblast homogenate by adding a sphingolipid standard mixture before and after extraction (Table 2). Mean recoveries were between 60 and 70% and did not vary with concentration of standard added.
TABLE 2.
Recovery and matrix effects
| Analyte | Spiked amount (pmol) | Recovery (SD) (n = 4) | Matrix effect (n = 4) |
|---|---|---|---|
| GluCer 16:0 | 50 | 66 ± 3 | 107 ± 4 |
| 250 | 62 ± 3 | 100 ± 5 | |
| GalCer 24:1 | 50 | 62 ± 3 | 108 ± 5 |
| 250 | 60 ± 2 | 101 ± 4 | |
| LacCer 16:0 | 50 | 59 ± 2 | 116 ± 3 |
| 250 | 62 ± 3 | 117 ± 4 | |
| LacCer 24:0 | 50 | 60 ± 2 | 113 ± 3 |
| 250 | 61 ± 3 | 103 ± 5 | |
| Cer1P 16:0 | 30 | 68 ± 4 | 100 ± 4 |
| 150 | 64 ± 3 | 106 ± 5 | |
| Cer1P 24:0 | 30 | 64 ± 3 | 105 ± 4 |
| 150 | 62 ± 6 | 112 ± 5 | |
| dhCer1P 16:0 | 10 | 64 ± 7 | 106 ± 10 |
| 50 | 61 ± 2 | 106 ± 8 | |
| dhCer1P 24:0 | 10 | 67 ± 5 | 109 ± 6 |
| 50 | 62 ± 7 | 105 ± 3 | |
| SPH | 60 | 72 ± 4 | 100 ± 5 |
| 300 | 72 ± 3 | 111 ± 3 | |
| SPA | 30 | 68 ± 3 | 100 ± 5 |
| 150 | 66 ± 3 | 104 ± 4 | |
| PhytoSPH | 10 | 69 ± 7 | 103 ± 6 |
| 50 | 70 ± 2 | 105 ± 4 | |
| DimetSPH | 6 | 69 ± 3 | 120 ± 6 |
| 30 | 63 ± 5 | 108 ± 7 | |
| TrimetSPH | 6 | 71 ± 2 | 100 ± 3 |
| 30 | 64 ± 5 | 113 ± 7 | |
| SPC | 20 | 67 ± 8 | 106 ± 2 |
| 100 | 70 ± 7 | 101 ± 4 |
Values represent the percent recovery of standards spiked before and after extraction to examine extraction efficiencies. Matrix effects are calculated from fibroblast lipid extracts corresponding to 50 µg of cellular protein spiked after extraction (corrected by endogenous sphingolipid concentrations) in percent of the same sphingolipid standard mixture used as spike. Each value represents the average of four determinations ± SD.
We assessed matrix effects by analyzing a standard mixture in methanol and also spiked into fibroblast lipid extracts (Table 2). Addition of fibroblast cell extract either did not influence or slightly increased the signals up to 20%.
Quantification of sphingolipid species
To compensate for variations in sample preparation and ionization efficiency, a set of non-naturally occurring sphingolipids, GluCer 12:0, LacCer 12:0, SPH d17:1, Cer1P 12:0, and SPC d17:1, was added as ISs prior to extraction. The ratio between analyte and IS was used for quantification as indicated in Table 1. We generated calibration lines by addition of different concentrations of naturally occurring sphingolipids to human skin fibroblasts (Table 3). For glycosylated ceramide species, a possible chain length dependency was addressed by generating two independent calibration lines with a short-chain (16:0) and a long-chain fatty acid (24:0). The obtained standard curves were linear in the tested calibration range. Additional evidence for the specificity of the method is derived from the fact that both mass transitions used for SPH and SPA analysis (Table 1) revealed similar results (data not shown).
TABLE 3.
Calibration data of different sphingolipids
| Sphingolipid | Calibration range (pmol) | IS added (pmol) | Slope (mean ± SD) | Correlation coefficient (mean ± SD) |
|---|---|---|---|---|
| GluCer 16:0 | 25–250 | 31.1 | 59.3 ± 5.6 | 0.997 ± 0.003 |
| GalCer 24:1 | 25–250 | 31.1 | 56.1 ± 3.5 | 0.998 ± 0.001 |
| LacCer 16:0 | 25–250 | 24.8 | 123.6 ± 7.8 | 0.996 ± 0.002 |
| LacCer 24:0 | 25–250 | 24.8 | 107.0 ± 6.0 | 0.994 ± 0.002 |
| Cer1P 16:0 | 15–150 | 35.6 | 153.5 ± 8.2 | 0.997 ± 0.002 |
| Cer1P 24:0 | 15–150 | 35.6 | 134.1 ± 6.2 | 0.994 ± 0.001 |
| SPH | 30–300 | 70.1 | 25.0 ± 1.3 | 0.996 ± 0.002 |
| SPA | 15–150 | 70.1 | 18.6 ± 2.4 | 0.998 ± 0.001 |
| PhytoSPH | 10–100 | 70.1 | 9.1 ± 0.5 | 0.996 ± 0.002 |
| DimetSPH | 0.3–3 | 70.1 | 70.6 ± 4.5 | 0.999 ± 0.001 |
| TrimetSPH | 0.3–3 | 70.1 | 68.7 ± 7.1 | 0.996 ± 0.004 |
| SPC | 10–100 | 4.4 | 41.0 ± 3.2 | 0.996 ± 0.003 |
Calibration lines were generated by plotting the ratios of the areas analyte to IS against the spiked concentrations (pmol). Each value represents the average of four determinations ± SD.
Due to coelution, monounsaturated species exhibit an overlap of the M+2 isotope peak with the corresponding saturated species. To correct this overlap, we applied a previously described algorithm based on calculated isotope distributions (25). To test this procedure, we added increasing amounts of GluCer 24:1 (m/z 810.7) to fibroblast homogenate and calculated analyte to IS ratios of GluCer 24:0 (m/z 812.7) with and without isotope correction. Whereas the GluCer 24:0 to IS ratio increased almost 2-fold upon the addition of 200 pmol GluCer 24:1 without correction, no significant increase was detected after correction of isotope overlap (Table 4).
TABLE 4.
Correction of isotope overlap
| GluCer 24:1 spiked (pmol) | Ratio GluCer 24:1/IS | Uncorrected ratio GluCer 24:0/IS | Corrected ratio GluCer 24:0/IS |
|---|---|---|---|
| 0 | 2.86 ± 0.16 | 1.32 ± 0.12 | 1.18 ± 0.08 |
| 25 | 4.41 ± 0.24 | 1.53 ± 0.11 | 1.28 ± 0.07 |
| 50 | 5.99 ± 0.39 | 1.71 ± 0.16 | 1.35 ± 0.07 |
| 100 | 8.43 ± 0.44 | 1.84 ± 0.16 | 1.30 ± 0.08 |
| 150 | 10.58 ± 0.52 | 2.06 ± 0.23 | 1.36 ± 0.12 |
| 200 | 16.66 ± 0.99 | 2.47 ± 0.26 | 1.33 ± 0.15 |
Fibroblast homogenates (100 µg cellular protein) were spiked with increasing amounts of GluCer 24:1. Values represent peak area ratios of GluCer 24:1 and 24:0 to GluCer 12:0. The GluCer 24:0 peak area ratios are shown before and after isotope correction. The displayed values are mean of three independent samples.
Assay characteristics
Assay accuracy was calculated using three spiked fibroblast lipid extracts at different concentrations, covering the entire calibration range. Accuracy was found between 90 and 110% (Table 5).
TABLE 5.
Intraday and interday precisions and accuracy
| Sphingolipid species | Protein (µg) | Intraday (pmol ± SD) | CV (%) | Interday (pmol ± SD) | CV (%) | Spiked (pmol) | Accuracy (%) |
|---|---|---|---|---|---|---|---|
| HexCer 16:0 | 25 | 2.6 ± 0.1 | 4.13 | 2.2 ± 0.2 | 7.4 | 25 | 110 ± 8 |
| 50 | 5.1 ± 0.3 | 5.14 | 5.0 ± 0.2 | 3.1 | 150 | 109 ± 5 | |
| 100 | 10.1 ± 0.8 | 7.89 | 9.3 ± 0.6 | 7 | 250 | 104 ± 5 | |
| HexCer 22:0 | 25 | 2.5 ± 0.1 | 5.7 | 3.0 ± 0.2 | 5.7 | ||
| 50 | 5.1 ± 0.4 | 8.7 | 6.1 ± 0.4 | 5.5 | |||
| 100 | 10.3 ± 0.4 | 4.1 | 10.7 ± 0.6 | 5.4 | |||
| HexCer 24:0 | 25 | 7.8 ± 0.4 | 5.3 | 8.6 ± 0.4 | 4.2 | ||
| 50 | 15.3 ± 0.7 | 4.6 | 17.6 ± 1.0 | 5.1 | |||
| 100 | 30.3 ± 1.1 | 3.7 | 33.4 ± 0.8 | 2.4 | |||
| HexCer 24:1 | 25 | 5.1 ± 0.4 | 6.9 | 5.5 ± 0.4 | 4.3 | 25 | 109 ± 7 |
| 50 | 10.0 ± 0.6 | 6.1 | 12.1 ± 0.7 | 6.2 | 150 | 108 ± 5 | |
| 100 | 19.9 ± 0.6 | 2.9 | 20.3 ± 0.8 | 3.9 | 250 | 104 ± 2 | |
| LacCer 16:0 | 25 | 1.1 ± 0.1 | 9 | 1.4 ± 0.1 | 7.3 | 25 | 97 ± 4 |
| 50 | 2.6 ± 0.1 | 5 | 2.6 ± 0.2 | 6.8 | 150 | 100 ± 6 | |
| 100 | 4.5 ± 0.6 | 13 | 4.2 ± 0.4 | 8.6 | 250 | 99 ± 6 | |
| LacCer 22:0 | 25 | 0.66 ± 0.08 | 11.9 | 0.70 ± 0.08 | 11.3 | ||
| 50 | 1.4 ± 0.1 | 8.1 | 1.5 ± 0.13 | 9.1 | |||
| 100 | 2.6 ± 0.2 | 7.3 | 2.6 ± 0.24 | 9.4 | |||
| LacCer 24:0 | 25 | 1.7 ± 0.1 | 7.8 | 2.3 ± 0.1 | 6.1 | 25 | 95 ± 5 |
| 50 | 3.8 ± 0.2 | 5.9 | 4.8 ± 0.3 | 7.2 | 150 | 97 ± 5 | |
| 100 | 7.3 ± 0.9 | 11.8 | 7.8 ± 0.8 | 9.9 | 250 | 95 ± 5 | |
| LacCer 24:1 | 25 | 1.2 ± 0.05 | 4.4 | 1.5 ± 0.1 | 6.5 | ||
| 50 | 2.9 ± 0.2 | 8.5 | 3.6 ± 0.2 | 6.7 | |||
| 100 | 5.4 ± 0.7 | 12 | 5.7 ± 0.5 | 9.2 | |||
| SPH | 25 | 2.5 ± 0.1 | 4.3 | 2.1 ± 0.2 | 9.7 | 30 | 95 ± 5 |
| 50 | 5.1 ± 0.5 | 9.4 | 4.6 ± 0.5 | 10.9 | 180 | 97 ± 6 | |
| 100 | 8.7 ± 0.7 | 8.3 | 8.5 ± 0.7 | 7.9 | 300 | 95 ± 5 | |
| SPA | 25 | n.d. | 13.3 | 0.81 ± 0.06 | 7.4 | 15 | 104 ± 8 |
| 50 | n.d. | 90 | 107 ± 5 | ||||
| 100 | 0.62 ± 0.08 | 150 | 99 ± 2 | ||||
| PhytoSPH | 25 | n.d. | 7.5 | 2.2 ± 0.2 | 8.6 | 10 | 97 ± 9 |
| 50 | n.d. | 60 | 96 ± 9 | ||||
| 100 | 2.4 ± 0.2 | 100 | 97 ± 5 | ||||
| DimetSPH | 25 | 0.12 ± 0.01 | 9.3 | 0.13 ± 0.01 | 11.2 | 0.3 | 108 ± 9 |
| 50 | 0.27 ± 0.03 | 9.7 | 0.24 ± 0.03 | 9.4 | 1.8 | 96 ± 9 | |
| 100 | 0.44 ± 0.03 | 4.8 | 0.44 ± 0.03 | 6.6 | 3 | 90 ± 5 | |
| TrimetSPH | 25 | 0.12 ± 0.01 | 9.3 | 0.11 ± 0.01 | 11 | 0.3 | 93 ± 8 |
| 50 | 0.22 ± 0.03 | 11.9 | 0.23 ± 0.03 | 11.6 | 1.8 | 109 ± 7 | |
| 100 | 0.37 ± 0.03 | 8.4 | 0.45 ± 0.04 | 9.7 | 3 | 95 ± 5 | |
| SPC | 25 | n.d. | 11.0 | 0.64 ± 0.06 | 9.7 | 10 | 111 ± 8 |
| 50 | n.d. | 60 | 107 ± 5 | ||||
| 100 | 0.58 ± 0.06 | 100 | 109 ± 4 | ||||
| Cer1P 16:0 | 25 | n.d. | 6.4 | 1.5 ± 0.1 | 6.8 | 15 | 109 ± 8 |
| 50 | 1.3 ± 0.09 | 8.0 | 3.5 ± 0.3 | 7.5 | 90 | 106 ± 7 | |
| 100 | 3.2 ± 0.02 | 150 | 101 ± 4 | ||||
| Cer1P 24:0 | 25 | n.d. | 8.4 | 5.5 ± 0.5 | 9.3 | 15 | 107 ± 8 |
| 50 | n.d. | 90 | 110 ± 9 | ||||
| 100 | 4.9 ± 0.4 | 150 | 103 ± 6 | ||||
| dhCer1P 24:0 | 25 | n.d. | 8.1 | 5.9 ± 0.5 | 8.1 | 5 | 92 ± 5 |
| 50 | n.d. | 30 | 102 ± 7 | ||||
| 100 | 6.4 ± 0.5 | 50 | 100 ± 5 |
The displayed values are mean concentrations in pmol and the CV of human skin fibroblast lipid extracts corresponding to 25, 50, and 100 µg of cellular protein. A pool of fibroblast homogenates was aliquoted, and lipid extracts were analyzed in series for intraday precision (n = 6) and on 6 different days for interday precision (n = 6). Accuracy is displayed as the mean of the assayed concentration (corrected by endogenous sphingolipid concentrations in human skin fibroblasts) in percent of the spiked concentration. Each value represents the average of three determinations ± SD.
Precision was determined in three fibroblast samples containing 25, 50, and 100 µg of cellular protein (Table 5). Coefficients of variation (CVs) were below 10% for most species for both intraday and interday precision (Table 5).
Because no analyte free matrix was available, we calculated the limit of detection (LOD), defined as a signal to noise ratio of 3. Whereas for most of the analyzed sphingolipid classes, <10 fmol is sufficient for quantification, PhytoSPH and dhCer1P displayed a LOD up to 50 fmol on column (Table 1).
Preparation of cell culture samples and sample stability
Because a main application of this method is the analysis of cultured cells, we tested different methods to harvest the cells. First, a precursor ion scan of m/z 264 was applied to check which HexCer, LacCer, and Cer1P species are found in primary human skin fibroblasts. For both HexCer and LacCer, we found 16:0, 22:0, 23:0, 24:0, and 24:1 species; for Cer1P, only 16:0 was detected. To compare sample preparations, fibroblasts were either scraped in PBS and homogenized in water by sonication or lysed in 0.2% SDS. Both sample preparations did not differ in their ionization response, because IS signals were similar (data not shown). Cells lysed in water showed about 10% higher HexCer and PhytoSPH levels as well as slightly decreased LacCer 16:0, 22:0, 24:0, and 15% decreased SPH level (Table 6). For reproducibility, SDS showed advantages compared with water, which gave higher SDs.
TABLE 6.
Lipid extracts prepared from fibroblasts either homogenized in water by sonication or lysed in 0.2% SDS
| HexCer | LacCer | SPH | SPA | PhytoSPH | Cer1P | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fatty acid | 16:0 | 22:0 | 23:0 | 24:1 | 24:0 | 16:0 | 22:0 | 23:0 | 24:1 | 24:0 | 16:0 | |||
| Water | 253 ± 13 | 129.1 ± 7.0 | 41.2 ± 3.7 | 290 ± 16 | 315 ± 19 | 42.2 ± 3.3 | 50.6 ± 7.1 | 16.1 ± 2.9 | 1356 ± 13 | 94.5 ± 2.4 | 72.8 ± 7.1 | 15.8 ± 2.1 | 20.7 ± 0.1 | 3.7 ± 0.5 |
| 0.2%SDS | 233 ± 5 | 119.7 ± 0.4 | 36.0 ± 1.5 | 265 ± 4 | 278 ± 10 | 46.6 ± 1.1 | 55.0 ± 6.2 | 17.8 ± 1.9 | 136 ± 9 | 110.5 ± 9.5 | 84.6 ± 4.4 | 16.1 ± 1.6 | 17.4 ± 0.7 | 4 ± 0.5 |
The displayed values are mean (pmol/mg cellular protein) ± SD of three independent samples.
Next, we tested the stability of the homogenates. Fibroblast homogenates prepared either in water or SDS were frozen immediately or after 6 h at room temperature. Storage at room temperature showed no effect on most sphingolipid levels, except a slight increase of SPH and Cer1P in SDS and PhytoSPH in water (Table 7).
TABLE 7.
Sample stability
| Percent change after 6 h RT | HexCer | LacCer | SPH | SPA | PhytoSPH | Cer1P |
|---|---|---|---|---|---|---|
| Water | 98 ± 1 | 105 ± 6 | 100 ± 3 | 96 ± 5 | 113 ± 9 | 98 ± 7 |
| SDS | 101 ± 7 | 96 ± 9 | 111 ± 2 | 101 ± 8 | 97 ± 5 | 113 ± 3 |
Fibroblasts, either homogenized in water by sonication or lysed in 0.2% SDS, were stored immediately at −80°C or for 6 h at room temperature. The displayed values are percent of the immediately stored fibroblast cell homogenates. The displayed values are the mean ± SD of three independent samples.
Analysis of fibroblasts treated with myriocin/SKI
To test feasibility of this novel method, we treated primary human skin fibroblasts either with myriocin, an inhibitior of serine-palmitoyl transferase (30) or SKI (30–33). To obtain the full range of sphingolipid concentrations, we additionally analyzed S1P, sphingomyelin, and ceramide species using previously described methods (18, 25, 26, 28).
Myriocin decreased cellular S1P and SPC levels at subnanomolar concentrations to 60% and 40% of the untreated control (Fig. 4A). The other analyzed sphingolipid classes showed only minor changes upon treatment with myriocin up to 1 nM (Fig. 4A, B). The most pronounced effects were observed at 5 nM myriocin, with decreased Cer, HexCer, LacCer, and free sphingoid bases concentrations and a further decline of S1P and SPC level.
SKI treatment of fibroblasts at nanomolar concentrations decreased S1P and SPC by more than 50% (Fig. 4C). Micromolar concentrations of SKI resulted in S1P below and the SPC concentration close to the LOD and led to a pronounced increase in the level of the free sphingoid base. Interestingly, increased levels of SPA were paralleled by dihydro-SM (Fig. 4C, D). SKI treatment in the pharmacological range (0.5–5 µM) (34) did not change Cer and SM levels significantly (Fig. 4). Surprisingly, SKI treatment decreased LacCer at low concentrations.
Taken together, these data show that drug treatments that affect enzymes involved in sphingolipid metabolism may not affect only the targeted metabolites but also the whole pathway.
DISCUSSION
Sphingolipid metabolism consists of a dynamic network of molecules, including important bioactive signaling molecules (1–9). Therefore, to understand the function of sphingolipids, it is necessary to assess a sphingolipid profile instead of one single metabolite.
Although analysis of the “sphingolipidome” by shotgun approaches has been recently demonstrated for yeast (35), an analysis of a more complex sphingolipid pattern in mammalian systems may be hampered, especially for minor metabolites, by signal-suppressing matrix effects or lack of sensitivity (12, 19, 27).
In this study, we present a novel LC-MS/MS method to quantify various sphingolipid species from cultured cells. In contrast to most previous methods using reversed-phase chromatography (10, 11, 15, 16, 19, 21, 27), we applied HILIC, which allows coelution of analytes and non-natural occurring ISs. This is a key feature of LC-based MS methods, because matrix effects and ionization response may vary during LC separation, especially when gradient methods are used. Consequently, only coelution of analytes with adequate ISs may compensate for these effects and prevent misquantification. Due to the coelution of multiple species, an isotopic overlap of species is possible. Therefore, we corrected peak areas according to principles described previously (25) to avoid an overestimation of species.
Further advantages of our method are a short analysis time of 4.5 min per sample and a simple liquid-liquid extraction as sample preparation. Because the presented method uses the same butanolic extraction and LC components as a previously described method for S1P and lysophosphatidic acid analysis (18), it is possible to analyze both sets of analytes from one extract. Consequently, one can achieve with two straightforward liquid-liquid extractions (Bligh and Dyer and butanol) a full coverage of the main sphingolipid metabolites (25, 26) as well as glycerophospholipids (22, 25, 36) and cholesterol/cholesteryl ester (37). Calibration was performed in the sample matrix by addition of naturally occurring species prior to lipid extraction. This allows compensation for potential matrix effects on ionization and extraction efficiency as well as for small retention time differences observed between short-chain and very long-chain species. Moreover, a full validation was performed according to U.S. Food and Drug Administration guidelines (28). This extensive validation showed excellent precision, accuracy, and sensitivity for all analyzed sphingolipid classes.
First, applications of this method showed that sample preparation methods may influence sphingolipid levels, particularly HexCer and free sphingoid bases. Due to reproducibility and handling reasons, we prefer a direct lysis of cultured cells with 0.2% SDS instead of scraping cells. However, immediate freezing of the samples until analysis is advisable. Finally, treatment of fibroblasts with myriocin and SKI demonstrated the importance of methods covering multiple instead of single sphingolipid metabolites because treatment affected not only direct metabolites but almost the whole pathway including unexpected concentration changes of some species.
In summary, we could show that LC-MS/MS-based sphingolipid profiling using HILIC may provide a powerful tool to understand regulatory and metabolic mechanisms involved in cellular sphingolipid homeostasis. Similar as previously shown for glycerophospholipid metabolism (22), this method can be also used for metabolic profiling using stable isotope labeled precursors.
Acknowledgments
We thank Jolante Aiwanger, Doreen Müller, and Simone Peschel for excellent technical assistance.
Footnotes
Abbreviations:
- Cer
- ceramide
- Cer1P
- ceramide-1-phosphate
- CV
- coefficient of variation
- dhCer1P
- dihydroceramide-1-phosphate
- DimetSPH
- dimethyl-sphingosine
- GluCer
- glucosyl-ceramide
- HexCer
- hexosyl-ceramide
- HILIC
- hydrophilic interaction chromatography
- IS
- internal standard
- LacCer
- lactosyl-ceramide
- LOD
- limit of detection
- MRM
- multiple reaction monitoring
- PhytoSPH
- phyto-sphingosine
- SKI
- sphingosine-kinase inhibitor
- SPA
- sphinganine
- SPC
- sphingosylphosphorylcholine
- SPH
- sphingosine
- S1P
- sphingosine-1-phosphate
- TrimetSPH
- trimethyl-sphingosine
This work was supported by the seventh framework program of the EU-funded “LipidomicNet” (proposal number 202272) and “eurIPFnet” (proposal number 20224) as well as the BMBF network project “Systems Biology Consortium on Metabotypes (SysMBo).”
REFERENCES
- 1.Lahiri S., Futerman A. H. 2007. The metabolism and function of sphingolipids and glycosphingolipids. Cell. Mol. Life Sci. 64: 2270–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartke N., Hannun Y. A. 2009. Bioactive sphingolipids: metabolism and function. J. Lipid Res. 50 (Suppl.): S91–S96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spiegel S., Kolesnick R. 2002. Sphingosine 1-phosphate as a therapeutic agent. Leukemia. 16: 1596–1602. [DOI] [PubMed] [Google Scholar]
- 4.Spiegel S., Milstien S. 2003. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol. 4: 397–407. [DOI] [PubMed] [Google Scholar]
- 5.Taha T. A., Mullen T. D., Obeid L. M. 2006. A house divided: ceramide, sphingosine, and sphingosine-1-phosphate in programmed cell death. Biochim. Biophys. Acta. 1758: 2027–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liliom K., Sun G., Bunemann M., Virag T., Nusser N., Baker D. L., Wang D. A., Fabian M. J., Brandts B., Bender K., et al. 2001. Sphingosylphosphocholine is a naturally occurring lipid mediator in blood plasma: a possible role in regulating cardiac function via sphingolipid receptors. Biochem. J. 355: 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wymann M. P., Schneiter R. 2008. Lipid signalling in disease. Nat. Rev. Mol. Cell Biol. 9: 162–176. [DOI] [PubMed] [Google Scholar]
- 8.Merrill A. H., Jr., Stokes T. H., Momin A., Park H., Portz B. J., Kelly S., Wang E., Sullards M. C., Wang M. D. 2009. Sphingolipidomics: a valuable tool for understanding the roles of sphingolipids in biology and disease. J. Lipid Res. 50 (Suppl.): S97–S102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowart L. A. 2009. Sphingolipids: players in the pathology of metabolic disease. Trends Endocrinol. Metab. 20: 34–42. [DOI] [PubMed] [Google Scholar]
- 10.Berdyshev E. V., Gorshkova I. A., Garcia J. G., Natarajan V., Hubbard W. C. 2005. Quantitative analysis of sphingoid base-1-phosphates as bisacetylated derivatives by liquid chromatography-tandem mass spectrometry. Anal. Biochem. 339: 129–136. [DOI] [PubMed] [Google Scholar]
- 11.Butter J. J., Koopmans R. P., Michel M. C. 2005. A rapid and validated HPLC method to quantify sphingosine 1-phosphate in human plasma using solid-phase extraction followed by derivatization with fluorescence detection. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 824: 65–70. [DOI] [PubMed] [Google Scholar]
- 12.Lieser B., Liebisch G., Drobnik W., Schmitz G. 2003. Quantification of sphingosine and sphinganine from crude lipid extracts by HPLC electrospray ionization tandem mass spectrometry. J. Lipid Res. 44: 2209–2216. [DOI] [PubMed] [Google Scholar]
- 13.Mano N., Oda Y., Yamada K., Asakawa N., Katayama K. 1997. Simultaneous quantitative determination method for sphingolipid metabolites by liquid chromatography/ionspray ionization tandem mass spectrometry. Anal. Biochem. 244: 291–300. [DOI] [PubMed] [Google Scholar]
- 14.Markham J. E., Jaworski J. G. 2007. Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 21: 1304–1314. [DOI] [PubMed] [Google Scholar]
- 15.Murph M., Tanaka T., Pang J., Felix E., Liu S., Trost R., Godwin A. K., Newman R., Mills G. 2007. Liquid chromatography mass spectrometry for quantifying plasma lysophospholipids: potential biomarkers for cancer diagnosis. Methods Enzymol. 433: 1–25. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt H., Schmidt R., Geisslinger G. 2006. LC-MS/MS-analysis of sphingosine-1-phosphate and related compounds in plasma samples. Prostaglandins Other Lipid Mediat. 81: 162–170. [DOI] [PubMed] [Google Scholar]
- 17.Yoo H. H., Son J., Kim D. H. 2006. Liquid chromatography-tandem mass spectrometric determination of ceramides and related lipid species in cellular extracts. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 843: 327–333. [DOI] [PubMed] [Google Scholar]
- 18.Scherer M., Schmitz G., Liebisch G. 2009. High-throughput analysis of sphingosine 1-phosphate, sphinganine 1-phosphate, and lysophosphatidic acid in plasma samples by liquid chromatography-tandem mass spectrometry. Clin. Chem. 55: 1218–1222. [DOI] [PubMed] [Google Scholar]
- 19.Haynes C. A., Allegood J. C., Park H., Sullards M. C. 2009. Sphingolipidomics: methods for the comprehensive analysis of sphingolipids. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2696–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farwanah H., Wirtz J., Kolter T., Raith K., Neubert R. H., Sandhoff K. 2009. Normal phase liquid chromatography coupled to quadrupole time of flight atmospheric pressure chemical ionization mass spectrometry for separation, detection and mass spectrometric profiling of neutral sphingolipids and cholesterol. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2976–2982. [DOI] [PubMed] [Google Scholar]
- 21.Shaner R. L., Allegood J. C., Park H., Wang E., Kelly S., Haynes C. A., Sullards M. C., Merrill A. H., Jr 2009. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J. Lipid Res. 50: 1692–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Binder M., Liebisch G., Langmann T., Schmitz G. 2006. Metabolic profiling of glycerophospholipid synthesis in fibroblasts loaded with free cholesterol and modified low density lipoproteins. J. Biol. Chem. 281: 21869–21877. [DOI] [PubMed] [Google Scholar]
- 23.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150: 76–85. [DOI] [PubMed] [Google Scholar]
- 24.Baker D. L., Desiderio D. M., Miller D. D., Tolley B., Tigyi G. J. 2001. Direct quantitative analysis of lysophosphatidic acid molecular species by stable isotope dilution electrospray ionization liquid chromatography-mass spectrometry. Anal. Biochem. 292: 287–295. [DOI] [PubMed] [Google Scholar]
- 25.Liebisch G., Lieser B., Rathenberg J., Drobnik W., Schmitz G. 2004. High-throughput quantification of phosphatidylcholine and sphingomyelin by electrospray ionization tandem mass spectrometry coupled with isotope correction algorithm. Biochim. Biophys. Acta. 1686: 108–117. [DOI] [PubMed] [Google Scholar]
- 26.Liebisch G., Drobnik W., Reil M., Trumbach B., Arnecke R., Olgemoller B., Roscher A., Schmitz G. 1999. Quantitative measurement of different ceramide species from crude cellular extracts by electrospray ionization tandem mass spectrometry (ESI-MS/MS). J. Lipid Res. 40: 1539–1546. [PubMed] [Google Scholar]
- 27.Bielawski J., Szulc Z. M., Hannun Y. A., Bielawska A. 2006. Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods. 39: 82–91. [DOI] [PubMed] [Google Scholar]
- 28.U.S. Department of Health and Human Services, Food and Drug Administration. (2001). Guidance for Industry Bioanalytical Method Validation. Fed. Regist. 66 (100), 28526 (Docket No. 98D-1195). [Google Scholar]
- 29.Brugger B., Erben G., Sandhoff R., Wieland F. T., Lehmann W. D. 1997. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc. Natl. Acad. Sci. USA. 94: 2339–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyake Y., Kozutsumi Y., Nakamura S., Fujita T., Kawasaki T. 1995. Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem. Biophys. Res. Commun. 211: 396–403. [DOI] [PubMed] [Google Scholar]
- 31.Glaros E. N., Kim W. S., Wu B. J., Suarna C., Quinn C. M., Rye K. A., Stocker R., Jessup W., Garner B. 2007. Inhibition of atherosclerosis by the serine palmitoyl transferase inhibitor myriocin is associated with reduced plasma glycosphingolipid concentration. Biochem. Pharmacol. 73: 1340–1346. [DOI] [PubMed] [Google Scholar]
- 32.Hojjati M. R., Li Z., Zhou H., Tang S., Huan C., Ooi E., Lu S., Jiang X. C. 2005. Effect of myriocin on plasma sphingolipid metabolism and atherosclerosis in apoE-deficient mice. J. Biol. Chem. 280: 10284–10289. [DOI] [PubMed] [Google Scholar]
- 33.Cheon S., Song S. B., Jung M., Park Y., Bang J. W., Kim T. S., Park H., Kim C. H., Yang Y. H., Bang S. I., et al. 2008. Sphingosine kinase inhibitor suppresses IL-18-induced interferon-gamma production through inhibition of p38 MAPK activation in human NK cells. Biochem. Biophys. Res. Commun. 374: 74–78. [DOI] [PubMed] [Google Scholar]
- 34.French K. J., Upson J. J., Keller S. N., Zhuang Y., Yun J. K., Smith C. D. 2006. Antitumor activity of sphingosine kinase inhibitors. J. Pharmacol. Exp. Ther. 318: 596–603. [DOI] [PubMed] [Google Scholar]
- 35.Ejsing C. S., Sampaio J. L., Surendranath V., Duchoslav E., Ekroos K., Klemm R. W., Simons K., Shevchenko A. 2009. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA. 106: 2136–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liebisch G., Drobnik W., Lieser B., Schmitz G. 2002. High-throughput quantification of lysophosphatidylcholine by electrospray ionization tandem mass spectrometry. Clin. Chem. 48: 2217–2224. [PubMed] [Google Scholar]
- 37.Liebisch G., Binder M., Schifferer R., Langmann T., Schulz B., Schmitz G. 2006. High throughput quantification of cholesterol and cholesteryl ester by electrospray ionization tandem mass spectrometry (ESI-MS/MS). Biochim. Biophys. Acta. 1761: 121–128. [DOI] [PubMed] [Google Scholar]