Abstract
Influenza is a contagious, acute respiratory disease that is a major cause of morbidity and mortality throughout the world. CD4 T cells play an important role in the immune response to this pathogen through the secretion of antiviral cytokines, and by providing help to CD8 T cells and B cells to promote the development of immunological memory and neutralizing antibody responses. Despite these well-defined roles in the anti-influenza response, our understanding of CD4 T-cell diversity and specificity remains limited. In the study reported here, overlapping peptides representing 5 different influenza viral proteins were used in EliSpot assays to enumerate and identify the specificity of anti-influenza CD4 T cells directly ex vivo following infection of mice with influenza virus, using two strains that express unrelated MHC class II molecules. These experiments evaluated whether the reactivity of CD4 T cells generally tracked with particular influenza proteins, or whether MHC preferences were the predominant factor dictating anti-CD4 T-cell specificity in the primary immune response. We made the unexpected discovery that the distribution of CD4 T-cell specificities for different influenza proteins varied significantly depending on the single class II molecule expressed in vivo. In SJL mice, the majority of epitopes were specific for the HA protein, while the NP protein dominated the response in C57BL/10 mice. Given the diversity of human MHC class II molecules, these findings have important implications for the ability to rationally design a vaccine that will generate a specific CD4 T-cell immune response that is effective across diverse human populations.
Introduction
Influenza virus is an enveloped, negative, single-stranded RNA virus that infects a wide range of avian and mammalian species (19,35,58). In humans it causes annual epidemics, leading to significant morbidity and mortality (60,61). Although there are currently two types of licensed influenza vaccines, the burden of influenza disease remains significant (2). In recent years, influenza virus has been associated with around 40,000 underlying respiratory and circulatory deaths, and around 290,000 pneumonia and circulatory hospitalizations in the United States annually, with a disproportionate burden of disease in children and the elderly (60,61). This continued high burden of disease despite the availability of an effective vaccine may be in part due to the need for yearly redesign and manufacture of the vaccine, with resulting lower-than-optimal vaccination rates (3), and a yearly risk of mismatch between the vaccination and circulating strains (19,40). The recent human pandemic with a novel strain of H1N1 influenza, for which there is apparently little pre-existing antibody-mediated immunity in the majority of humans, has accelerated the interest in production of vaccines that elicit a broadly cross-protective immune response (4,17,26,45).
Major strides in understanding the protective immune response to influenza have been made in recent years, and it is now thought that immunity is the result of a combination of both the innate and adaptive immune responses. Early immunity is mediated primarily by the innate response, with important roles for antiviral cytokines such as interferons in limiting early viral replication (25,31,39,58). Once intracellular infection is established, immune control and clearance largely depend on the detection and elimination of infected host cells by the adaptive immune response (19,35,58). Cytotoxic CD8 T cells home to respiratory sites where virus replication is localized and eliminate infected cells (18,19,31,35,40,58,59). Antigen-specific B cells secrete neutralizing antibodies to HA and NA, which provide the most significant source of protection from future infections (18,19,31,35,40,58,59). The role of CD4 T cells in this adaptive response is multifaceted, and includes providing help to promote B-cell isotype switching and affinity maturation, and expansion of CD8 effector and memory cells (8,10,18,35,56,57,59). CD4 T cells also participate directly in viral clearance through the secretion of antiviral cytokines, and possibly through direct, cell-mediated cytotoxicity (10,11,18,35,57).
In order to further understand the immune response to influenza and develop improved influenza vaccination strategies, it is necessary to first gain insight into what controls the specificity and diversity of T-cell epitopes elicited upon infection and vaccination with this virus. Recently, there have been great strides in understanding the epitopes important in the CD8 T-cell response to influenza (6,7,16,59,65,66). However, an understanding of the specificity of the CD4 immune response to the influenza virus has lagged behind. Although it is thought that the CD4 T-cell response is more diverse than the CD8 T-cell response, questions about the breadth of this diversity remain (15,46,47). In addition, the distribution of epitopes among the various influenza proteins needs to be better understood, as there is now evidence that the specificity of CD4 T cells and antibody-producing B cells may be linked (54). Thus it is possible that HA- and NA-specific CD4 T cells may provide the most potent help for production of neutralizing influenza-specific antibodies.
CD4 T cells recognize peptides that are loaded onto the MHC class II molecule in the acidic endosomal compartments of antigen-presenting cells. Based on previous studies using model antigens and from biochemical identification of peptides that are presented by class II molecules (13,20,24,38,48,64), one might predict that membrane-bound proteins such as HA and NA would have preferential access to the MHC class II antigen-processing compartment. However, the abundant viral protein synthesis that occurs within the infected dendritic cells that prime CD4 T cells, as well as the intracellular process of autophagy (14,34,36,41), may increase the access of cytosolic and nuclear antigens such as NS1 and NP to class II molecules. With regard to the breadth of the CD4 T-cell response, our previous studies with exogenous protein antigens suggest that DM editing of peptide loading onto class II dramatically limits the peptide epitopes presented, thus limiting the diversity of the responding CD4 T cells (32,33,51,52). Here, too, the paradigms established through the study of model protein antigens might not be sufficient to predict that observed with viral infection, because the abundant viral protein synthesis in infected dendritic cells may override DM editing.
In order to further advance the understanding of the specificity and diversity of MHC class II-restricted epitopes in influenza, we sought to comprehensively and empirically evaluate the peptide diversity and immunodominance hierarchy of CD4 T cells elicited in the primary response to influenza infection. We used two strains of mice (C57BL/10 and SJL) that express single class II molecules (I-Ab or I-As) respectively, and asked first, whether these strains displayed the same overall reactivity to different influenza viral proteins, and second, to what extent the overall pattern of CD4 T-cell reactivity to different viral proteins was influenced by the particular class II molecule expressed. For epitope identification, we obtained panels of overlapping peptides representing the entire sequences of the HA, NA, NP, NS1, and M1 proteins, and tested these peptides using enzyme-linked cytokine immunospot (EliSpot) assays to directly enumerate peptide-specific CD4 T cells isolated from the spleens of mice following infection with live influenza virus. The results of these experiments demonstrated a relatively low degree of diversity in the influenza-specific CD4 T-cell immune response restricted to the I-Ab and I-As MHC class II molecules, with a strikingly different response distribution among influenza proteins, depending on the MHC class II molecule presenting the peptide to the CD4 T cell.
Materials and Methods
Virus production
Embryonated eggs, purchased from SPAFAS Inc. (North Franklin, CT), were incubated at 70°F and 100% humidity for 9 d. The allantoic cavity was then infected with 100 μL of the A/New Caledonia/20/99 human influenza virus (H1N1), generously provided by Dr. John Treanor at the University of Rochester, at 103 50% egg-infective doses (EID50) per milliliter. The eggs were incubated at 37°C for 48 h, and then for 24 h at 4°C. The allantoic fluid was harvested under sterile conditions and centrifuged at 2000g for 20 min at 4°C in order to remove particulate matter. Aliquots were frozen at −70°C. Virus titer was determined by infecting embryonated eggs with serial dilutions of the harvested virus, and harvesting and immediately titrating the resulting fluid by a hemagglutination assay of chicken red blood cells according to the procedure recommended by the 2005–2006 World Health Organization bulletin for the identification of influenza virus isolates from subjects with influenza.
Influenza infection
Female C57BL/10 (“B10”), SJL, and B10.S mice were purchased from Jackson Laboratories (Bar Harbor, ME), and maintained in the pathogen-free facility at the University of Rochester Medical Center according to institutional guidelines. Mice that were on average 3 mo of age were anesthetized by intraperitoneal (IP) injection of tribromoethanol (provided by Dr. David Topham at the University of Rochester), at a dose of 200–300 μL per mouse. They were then infected intranasally with A/New Caledonia/20/99 influenza (H1N1) at a dose of 20,000–50,000 EID50 per mouse in 30 μL phosphate-buffered saline (PBS), as was previously described (46,47). At 8–10 d post-infection the mice were sacrificed and spleens were excised and used as a source of CD4 T cells for EliSpot analysis. Syngeneic splenocytes from uninfected mice were used as a source of antigen-presenting cells for these assays.
Cell purification
Spleens were excised, gently disrupted to yield a single-cell suspension, and the resulting splenocyte populations were depleted of red blood cells using ACK lysis buffer (0.15 M NH4Cl, 1 mM KHCO3, and 0.1 mM NA2-EDTA in H2O, pH 7.2–7.4). The remaining cells were washed and incubated with a pool of monoclonal antibody supernatants specific for CD8 T cells and APC at a concentration of 2 × 107 cells/mL. These cell lines were obtained from the American Type Culture Collection, and included 3.155 (anti-CD8), RA3/3A1/6.1 (anti-B220), and M5/114 (anti-I-Ab-expressing cells) for C57BL/10 mice, and 3.155 (anti-CD8), RA3-3A1/6.1 (anti-B220), and 10.2.16 (anti-I-As-expressing cells) for SJL/B10.S mice. Syngeneic splenocytes from uninfected mice were depleted of T cells using supernatant from the J1j.10 cell line (anti-Thy-1.2). Following incubation with the above antibodies, the cells were resuspended in complement (Low Tox M; Cedarlane Laboratories, Burlington, NC) at a concentration of 2 × 107 cells/mL and incubated at 37°C for 30 min. Dead cells were removed by density gradient centrifugation with Lympholyte-M (Cedarlane Laboratories). The remaining cells were washed and used in EliSpot assays. The composition of the resulting cell populations was assessed by staining for expression of the CD4, CD8, and MHC class II cell surface markers to quantify the percentage of each cell type present following the enrichment process.
EliSpot assays
For the assays, 96-well EliSpot plates (Millipore, Billerica, MA) were coated with 50 μL of purified rat anti-mouse IL-2 (BD Biosciences, San Jose, CA) prepared at a concentration of 2 μg/μL in PBS, and were incubated at room temperature for at least 2 h. The antibody was removed and the plates were washed three times with cell culture media. Media from the last wash was left on for at least 1 h at room temperature to block nonspecific interactions. APC isolated from syngeneic mice were plated at a concentration of 500,000 cells/well, while CD4-enriched T cells were plated at several concentrations, ranging from 50,000–300,000 cells/well, and peptide was added at a final concentration of either 2 or 10 μM. The plates were incubated at 37°C in 5% CO2 for 16–18 h. The cells were then removed and the plates were washed with PBS containing 0.1% Tween-20 (EliSpot wash buffer). Then 50 μL of a 2 μg/μL biotinylated rat anti-mouse IL-2 solution (BD Biosciences) was added and the plates were incubated at room temperature for 30 min. Following repeat washing with EliSpot wash buffer, alkaline phosphatase-conjugated streptavidin (Jackson ImmunoResearch, West Grove, PA) was added at a dilution of 1:1000. The plates were again incubated at room temperature for 30 min and washed with EliSpot wash buffer. Substrate was prepared with the Vector Blue Substrate Kit III (Vector Laboratories, Burlingame, CA) in 100 mM Tris (pH 8.2), and the plates were developed for approximately 5 min before the reaction was stopped with cold water. Following drying, the plates were analyzed on an Immunospot Reader Series 2A using Immunospot software (Cellular Technology Ltd., Cleveland, OH), version 2. Data were calculated and presented as cytokine EliSpots per million CD4 T cells, with background values subtracted.
Flow cytometry
Aliquots of cells obtained before and after CD4 T-cell enrichment were placed into wells of a 96-well plate, centrifuged, and resuspended in FACS buffer (PBS, 2.5% FBS, and 0.1% sodium azide). Fc block (CD16/CD32; BD Biosciences) was added at a 1:100 dilution and the cells were incubated on ice for 10 min. The cells were centrifuged and resuspended in FACS buffer (negative control), a 1:100 dilution of CD4-fluorescein isothiocyanate (CD4-FITC clone RM4-4; BD Biosciences), or a 1:100 dilution of CD8a-FITC (Ly-2 clone 53-6.7; eBioscience, San Diego, CA). Following 15 min of incubation on ice, the cells were washed with FACS buffer and analyzed on a BD FACS Scan flow cytometer. Data were analyzed using Cell Quest software.
Peptides
We used 17-mer peptides overlapping by 11 amino acids to encompass the entire sequences of the HA and NA proteins from the A/New Caledonia/20/99 influenza virus (H1N1), the NS1 sequence from the A/New York/444/2001 influenza virus (H1N1), and the NP and M1 sequences from A/New York/348/2003 influenza virus (H1N1). The following reagents were obtained through the National Institutes of Health (NIH) Biodefense and Emerging Infections Research Resources Repository, National Institute of Allergy and Infectious Diseases (NIAID), NIH: Peptide Arrays, Influenza Virus A/New Caledonia/20/1999 (H1N1) Hemagglutinin Protein, NR-2602; Neuraminidase Protein, NR-2606; Peptide Array, Influenza Virus A/New York/444/2001 (H1N1) Nonstructural Protein 1, NR-2612; Peptide Array, Influenza Virus A/New York/348/2003 (H1N1) Nucleocapsid Protein, NR-2611; and Matrix Protein 1, NR-2613. The A/New York/348/2003 amino acid sequences for NP and M1 are completely conserved, and the A/New York/444/2001 NS1 amino acid sequenc is >99% conserved when compared to their respective amino acid sequences from the A/New Caledonia/20/99 influenza virus. The peptides were reconstituted to 10 mM in PBS, with or without added dimethyl sulfoxide for hydrophobic peptides, and 1 mM dithiothreitol for cysteine-containing peptides. Stocks were stored at −20°C, and working stocks at concentrations of 1 mM or 100 μM were prepared in Dulbecco's modified Eagle's medium (Invitrogen Corp., Carlsbad, CA), filter sterilized, and also stored at −20°C. The final concentrations of the individual peptides used in the EliSpot assays were either 2 μM (pools containing all peptides for a given protein) or 10 μM (single-peptide assays or peptide-pooling matrices).
Results
The overall goal of these studies was to comprehensively and empirically evaluate CD4 T-cell specificity in peripheral lymphoid tissues following primary infection with influenza virus. We had a particular interest in determining whether CD4 T cells reactive to a subset of influenza proteins dominate the response, or if the response was equally distributed among the viral proteins expressed. Mouse strains expressing only single I-A MHC class II molecules were used to determine the impact of MHC class II allele variability on the distribution of specificities, and to simplify the determination of CD4 T-cell MHC restriction assignment. To accomplish this goal, C57BL/10 (I-Ab) or SJL (I-As) mice were infected intranasally with A/New Caledonia/20/99 influenza virus in PBS. This strain of influenza was isolated from humans and has been included in clinical vaccines in recent years (1,22). Although it has not been adapted to mice, it replicates in the respiratory tract after intranasal infection of mice and is cleared within 10 d of infection (data not shown). At 8–10 d after inoculation with live virus, the mice were sacrificed and single-cell suspensions were prepared from the spleens. CD4 T cells were purified and then used in an IL-2 EliSpot assay, with syngeneic splenocytes depleted of T cells used as APC, and synthetic peptide used as antigen. Representative flow cytometry data demonstrating cell purity are shown in Supplementary Fig. 1 (see online supplementary material at http://www.liebertonline.com). The CD4 T cells used were completely depleted of CD8 T cells (<1%), but had variable (15–40%) class II-positive cells remaining, mostly accounted for by B cells that are typically more resistant to complement-mediated lysis.
To evaluate the specificity of CD4 T cells elicited in response to primary influenza virus infection, we first obtained panels of 17-mer peptides overlapping by 11 amino acids that comprised the entire translated sequences of the HA, NA, NP, NS1, and M1 proteins. The protein length and number of peptides in each of these sets are shown in Supplementary Table 1 (see online supplementary material at http://www.liebertonline.com). These proteins were chosen because together they encompass approximately half of the influenza genome. Importantly, they represent the major subcellular localizations of influenza virus proteins in infected cells, allowing us to determine if protein localization has any impact on the overall distribution of the epitopes identified. HA and NA are transmembrane proteins expressed both in infected cells and in the influenza virion. NP and NS1 are both expressed within the cytosol and nucleus of infected cells. While NP is expressed both in the virion and within the host cells, NS1 is excluded from the virion particle. The M1 matrix protein is the most abundant virion protein, and is expressed in cytosol and nuclei of infected cells, playing a major role in viral budding (9,23,42,43). To evaluate if each of these proteins gained access to class II molecules and elicited CD4 T cells, these overlapping 17-mer synthetic peptides were tested for their ability to recall primed CD4 T cells isolated from previously infected mice using an EliSpot assay. The EliSpot assay has several advantages for epitope discovery, most notably that it allows rapid screening of multiple (>100) potential epitopes, and allows direct enumeration of antigen-specific T cells immediately ex-vivo, without the need for further T-cell proliferation or persistence in culture (5,21,29). We have found that the patterns of epitope distribution identified by IL-2 are similar to those identified by production of IFN-γ, but chose IL-2 for all studies reported here because some strains of mice produce fewer gamma-producing cells than IL-2-producing cells [(46) and unpublished data], allowing IL-2 to be considered a more CD4-inclusive cytokine. Because the strains chosen express a single MHC class II molecule, the MHC restriction of any peptides that elicit a positive response will be known, and thus any discovered epitopes could potentially be used in subsequent studies to derive tetramers to label antigen-specific CD4 T cells, and more fully characterize the functional properties of these responding cells.
In order to determine whether the reactivity of CD4 T cells predictably tracked with particular influenza proteins, initial experiments tested CD4 T-cell reactivity to pools of peptides encompassing the entire sequences of the NP, NS1, M1, NA, and HA viral proteins. Overlapping peptides representing each protein were pooled together in a single sample and added to the cytokine EliSpot assay. We expected the two different MHC class II molecules to select for different influenza-derived peptides, as MHC molecules of different alleles have quite distinct peptide-binding motifs. However, if immunogenicity was primarily determined by a property of the viral protein, such as the protein's abundance or localization within an infected cell, we reasoned that the overall distribution of epitopes among the proteins should be similar between the two mouse strains. On the other hand, if selectivity of the individual MHC class II molecules determined specificity, a different distribution of epitopes among these proteins would be expected.
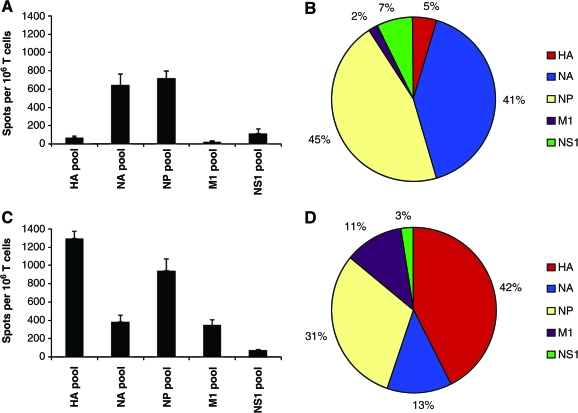
When CD4 T-cell specificity was examined using this method, dramatic differences were noted in the distribution of responses among the different influenza proteins in the two mouse strains examined (Fig. 1). In the C57BL/10 strain (Fig. 1A and 1B), the predominant CD4 T-cell specificity was for NP and NA. Strikingly, there were very few CD4 T cells specific for HA and M1 in this strain, and only a modest number of CD4 T cells specific for the small NS1 protein. In contrast, in the SJL strain (Fig. 1C and D) the epitopes were distributed primarily among the HA and NP proteins, with readily detectable CD4 T-cell responses specific for NA and M1, but with very few cells recruited by peptides representing NS1. Together, these findings suggested that the reactivity of CD4 T cells was not determined to a significant extent by the localization of the protein within the infected cell, as both the NP protein (cytosolic and nuclear) and the HA and NA (membrane-associated) proteins recruited significant numbers of CD4 T cells, depending on the strain being examined. In addition, these findings suggested that the epitope distribution among different viral proteins could be strongly influenced by MHC class II polymorphism.
FIG. 1.
Analyzing anti-influenza CD4 T-cell reactivity toward different viral proteins using pools of synthetic peptides representing the entire protein sequences of HA, NA, NP, M1, and NS1. Individual 17-mer peptides representing the entire sequences of the above proteins were pooled such that each peptide in the pools was at a final concentration of 2 μM. CD4 T-cell-enriched populations from previously infected mice were plated with the indicated peptide pool and the number of reactive cells was quantified by IL-2 EliSpots, as described in the materials and methods section. Data are shown as the average number of spots per 106 CD4-enriched T cells, with error bars representing the standard error over all experiments. Reactivity of CD4 T cells from C57BL/10 (“B10”) mice are shown in panels A and B, while the reactivity of CD4 T cells from SJL mice are shown in panels C and D. (Color image is available online at www.liebertonline.com/vim.)
To further explore what accounted for the strain-dependent differences seen in the distribution of the CD4 T-cell specificities among the influenza proteins, the epitopes within the HA, NS1, NP, NA, and M1 proteins were directly identified and quantified in both strains. When screening for epitopes in smaller proteins (NS1 and M1), peptides were tested individually, with any peptide eliciting on average >30 CD4 T cells per 1 × 106 cells above background considered positive. In order to quickly screen larger proteins (HA, NA, and NP), pools of non-overlapping peptides were created and organized into a matrix design as described by Tobery and Caulfield (62,63). Here, non-overlapping peptides were pooled such that an epitope would trigger intersecting rows and columns as positive, allowing us to more rapidly localize epitopes within these larger proteins. Shown in Supplementary Fig. 2B and D are examples of the peptide-pooling matrix analyses performed with peptides representing the NP protein, with layouts of the peptide-pooling matrix shown (the numbers indicate an individual peptide's position in the matrix) (see online supplementary material at http://www.liebertonline.com). Shown in Supplementary Fig. 2A and C are examples of NP-specific EliSpots obtained using these matrices, with the results presented as the number of antigen-dependent IL-2 spots above background detected per 1 × 106 CD4 T cells (see online supplementary material at http://www.liebertonline.com). Any pool that elicited on average >40 CD4 T cells per 1 × 106 cells over background was considered for further analysis, while negative pools were presumed to have only very weak or no CD4 T-cell epitopes. Peptides contained in pools within negative rows or columns were eliminated from further consideration, and the other peptides were tested individually in subsequent studies.
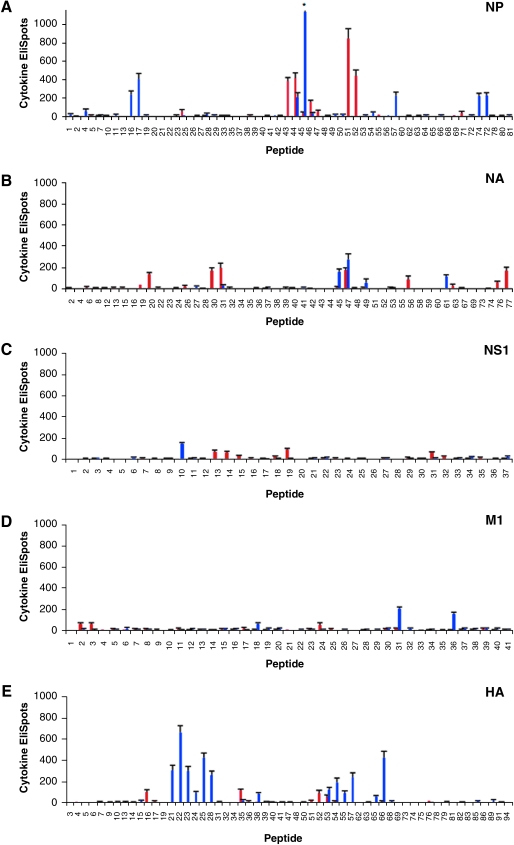
Shown in Fig. 2A through E are the results of CD4 cytokine EliSpots using individual peptides from each of the influenza virus proteins tested. Epitopes that elicited on average between 30 and 150 IL-2 spots per 1 × 106 CD4 T cells were considered subdominant, while peptides that elicited greater than 150 but less than 300 IL-2 spots per 1 × 106 T cells were considered dominant, and peptides that elicited greater than 300 IL-2 spots per 1 × 106 T cells were considered to be “super-dominant.” All peptides were tested in at least 2 individual experiments, and positive peptides were tested a greater number of times (Table 1).
FIG. 2.
Screening of candidate individual epitopes in the NP, NA, NS1, M1, and HA proteins using EliSpot assays. Panels A–E show the peptides in the C57BL/10 (red) and SJL (blue) mouse strains screened as individual peptides in the NP, NA, NS1, M1, and HA proteins, as indicated in each panel. Peptides were used at a final concentration of 10 μM, and the number of reactive CD4 T cells was quantified by the number of IL-2 EliSpots per 106 CD4 T cells, as described in the Materials and Methods section. All peptides were tested in at least two individual experiments, with the data presented as the average number of spots per 106 CD4-enriched T cells, and error bars representing the standard error over all experiments. Note that the standard error for peptide NP 46 in SJL mice in panel A is not fully depicted on the graph shown, as indicated by an asterisk, but is equal to 130 spots per 106 CD4 T cells. Peptides were determined to be negative using either peptide-pooling matrices or by screening individual peptides.
Table 1.
I-Ab and I-As Restricted Epitopes
| I-Ab restricted epitopes | Spots per 106 T cells | ± SE (n) | I-As restricted epitopes | Spots per 106 T cells | ± SE (n) |
|---|---|---|---|---|---|
| HA 16: 90 KESWSYIVETPNPENGT 106 | 100 | 23 (13) | HA 21: 120 EQLSSVSSFERFEIFPK 136 | 306 | 53 (10) |
| HA 35: 203 NQRALYHTENAYVSVVS 219 | 119 | 14 (25) | HA 22: 126 SSFERFEIFPKESSWPN 142 | 663 | 66 (16) |
| HA 52: 304 SSLPFQNVHPVTIGECP 320 | 93 | 29 (5) | HA 23: 132 EIFPKESSWPNHTVTGV 148 | 300 | 48 (10) |
| HA 53: 310 NVHPVTIGECPKYVRSA 326 | 56 | 21 (3) | HA 24: 138 SSWPNHTVTGVSASCSH 154 | 91 | 19 (10) |
| NA 19: 108 IGSKGDVFVIREPFIS 123 | 34 | 5 (5) | HA 25: 144 TVTGVSASCSHNGKSSF 160 | 426 | 48 (14) |
| NA 20: 113 DVFVIREPFISCSHLE 128 | 134 | 19 (17) | HA 28: 162 RNLLWLTGKNGLYPNLS 178 | 263 | 40 (10) |
| NA 30: 171 NSKFESVAWSASACHDG 187 | 168 | 31 (10) | HA 38: 221 HYSRRFTPEIAKRPKVR 237 | 84 | 13 (10) |
| NA 31: 177 VAWSASACHDGMGWLTI 193 | 195 | 46 (10) | HA 53: 310 NVHPVTIGECPKYVRSA 326 | 123 | 25 (10) |
| NA 47: 273 NFHYEECSCYPDTGTVM 289 | 174 | 25 (19) | HA 54: 316 IGECPKYVRSAKLRMVT 332 | 191 | 46 (10) |
| NA 56: 327 RPKDGEGSCNPVTVDGA 343 | 86 | 33 (4) | HA 55: 322 YVRSAKLRMVTGLRNIP 338 | 93 | 20 (10) |
| NA 76: 446 FCGVNSDTANWSWPDGA 462 | 61 | 12 (3) | HA 57: 334 LRNIPSIQSRGLFGAIA 350 | 243 | 40 (10) |
| NA 77: 452 DTANWSWPDGAELPFTI 468 | 170 | 30 (13) | HA 65: 380 DQKSTQNAINGITNKVN 396 | 61 | 8 (10) |
| NP 25: 145 DTTYQRTRALVRTGMDP 161 | 56 | 18 (2) | HA 66: 386 NAINGITNKVNSVIEKM 402 | 428 | 60 (14) |
| NP 44: 258 FLARSALILRGSVAHKS 274 | 381 | 41 (10) | NA 45: 261 GKVTKSIELNAPNFHYE 277 | 161 | 23 (15) |
| NP 45: 264 LILRGSVAHKSCLPACV 280 | 418 | 54 (10) | NA 47: 273 NFHYEECSCYPDTGTVM 289 | 272 | 54 (4) |
| NP 46: 270 VAHKSCLPACVYGPAVA 286 | 33 | 13 (5) | NA 49: 285 TGTVMCVCRDNWHGSNR 301 | 55 | 35 (4) |
| NP 47: 276 LPACVYGPAVASGYDFE 292 | 157 | 15 (20) | NA 61: 357 VWIGRTKSNRLRKGFEM 373 | 112 | 19 (9) |
| NP 48: 282 GPAVASGYDFEKEGYSL 298 | 53 | 17 (7) | NP 4: 19 RQNATEIRASVGRMIGG 35 | 57 | 25 (6) |
| NP 52: 306 LLQTSQVYSLIRPNENP 322 | 848 | 106 (16) | NP 16: 91 KTGGPIYKRVDGKWVRE 107 | 230 | 45 (10) |
| NP 53: 312 VYSLIRPNENPAHKSQL 328 | 438 | 66 (10) | NP 17: 97 YKRVDGKWVRELVLYDK 113 | 403 | 61 (14) |
| NP 71: 420 FDKTTIMAAFTGNTEGR 436 | 39 | 16 (4) | NP 45: 264 LILRGSVAHKSCLPACV 280 | 208 | 49 (4) |
| M1 2: 7 VETYVLSIVPSGPLKAE 23 | 66 | 11 (7) | NP 46: 270 VAHKSCLPACVYGPAVA 286 | 1139 | 130 (17) |
| M1 3: 13 SIVPSGPLKAEIAQRLE 29 | 65 | 12 (12) | NP 47: 276 LPACVYGPAVASGYDFE 292 | 33 | 9 (8) |
| M1 24: 139 TTESAFGLICATCEQIA 155 | 55 | 20 (6) | NP 55: 324 HKSQLVWMACNSAAFED 340 | 38 | 8 (8) |
| NS1 13: 72 ESDEAFKMTMASALASR 88 | 71 | 17 (12) | NP 58: 342 RVSSFIRGTRVLPRGKL 358 | 220 | 41 (10) |
| NS1 14: 78 KMTMASALASRYLTDMT 94 | 63 | 12 (13) | NP 74: 438 SDMRAEIIKMMESARPE 454 | 217 | 37 (10) |
| NS1 19: 108 KQKVAGPLCVRMDQAIM 124 | 91 | 14 (21) | NP 75: 444 IIKMMESARPEEVSFQG 460 | 224 | 31 (14) |
| NS1 31: 180 VLIGGLEWNDNTVRVSE 196 | 62 | 9 (14) | M1 18: 103 LKREITFHGAKEIALSY 119 | 63 | 11 (10) |
| M1 31: 181 LASTTAKAMEQMAGSSE 197 | 208 | 19 (10) | |||
| M1 36: 210 RQMVQAMRAIGTHPSSS 226 | 157 | 18 (7) | |||
| NS1 10: 54 IETATCVGKQIVERILK 70 | 144 | 16 (2) |
Summary of CD4 peptide epitopes identified in C57BL/10 mice (left) and in SJL mice (right) in response to primary influenza infection. Indicated for each peptide epitope are the average spot count per million CD4 T cells, the standard error (SE), and the number of experiments in which the peptide was tested (n). Major epitopes are indicated in bold. Underlined residues represent the core epitope as determined by use of truncated peptides (not shown).
The results of epitope mapping using CD4 T cells obtained from C57BL/10 mice revealed that the diversity of the repertoire was very limited. Most of the proteins that were studied contained only a limited number of epitopes; however, while some proteins contained only subdominant epitopes, others had several highly dominant epitopes that accounted for the majority of the anti-influenza reactivity in the CD4 T-cell repertoire. When candidate positive NP peptides were screened individually (Fig. 2A), nine epitopes were identified, of which four were “superdominant.” Two of these superdominant epitopes were in overlapping peptides, and thus likely belong to a single unique CD4 T-cell epitope. Individual peptide screening of the NA protein revealed that the response observed in the large peptide pools could be accounted for by eight epitopes out of a total of 78 peptides tested, with the responses ranging from around 30 to 200 recruited CD4 T cells (Fig. 2B). In NS1 (Fig. 2C), four subdominant epitopes were identified, each of which recruited approximately 80 CD4 T cells per million cells, while in M1 (Fig. 2D), three minor epitopes were discovered that recruited around 50 CD4 T cells per million cells. A similar pattern of only four epitopes was observed for HA, all of which elicited on average less than 150 CD4 T cells per million cells (Fig. 2E). Based on these data, we concluded that there was a narrow distribution of epitopes restricted to the I-Ab MHC class II molecule in C57BL/10 mice that accounted for the overall reactivity detected in the pools, with less than 15 epitopes eliciting CD4 T cells at a frequency of greater than 100 per million cells tested, or 0.01%. The CD4 T-cell response specific for NP and NA dominated the immune response to influenza, in large part due to the strongly immunodominant epitopes present in these proteins.
When we examined the CD4 T-cell specificity from I-As-restricted cells obtained from SJL mice, we again found that the diversity of the repertoire was quite limited. Similarly to C57BL/10 mice, candidate NP epitopes were individually screened and 10 peptide epitopes were identified, many of which were dominant or superdominant (Fig. 2A). Several of these (16/17, 45/46, and 74/75) were present in overlapping peptides and thus likely represent a single CD4 T-cell epitope (Table 1). However, in contrast to what was found in C57BL/10 mice, 13 peptide epitopes were identified in HA, four of which elicited greater than 300 CD4 T cells per million cells (Fig. 2E). In screening for reactivity towards NA in SJL mice, only four epitopes were discovered, and these elicited a cytokine response from only a modest number of CD4 cells (Fig. 2B). CD4 T cells specific for NS1 and M1 were identified by screening individual peptides, with only one subdominant epitope identified in NS1 that elicited on average around 150 spots per million CD4 T cells (Fig. 2C). In M1, there was one subdominant and two dominant epitopes present (Fig. 2D), all of which elicited less than 300 CD4 T cells per million cells. Based on these data, we were able to conclude that similarly to the case in C57BL/10 mice, there was also a relatively narrow distribution of epitopes restricted to the I-As molecule in SJL mice. However, in this strain the CD4 T-cell response specific for HA dominated the immune response to influenza, again in large part due to the strongly immunodominant epitopes present.
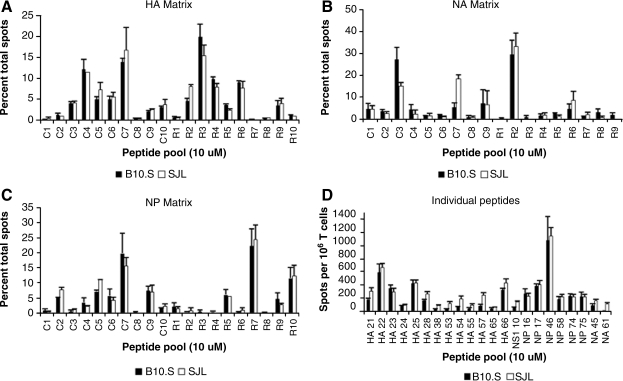
In order to ensure that the differences in CD4 T-cell protein specificity noted between the two mouse strains examined were not the result of differences in the genetic background of the mice leading to, for example, different rates of influenza clearance in vivo, we obtained and infected B10.S mice with A/New Caledonia influenza. B10.S mice are MHC congenic with C57BL/10 mice, and like SJL mice, they express the H-2s haplotype genes and thus the I-As protein. Screening EliSpot assays were done in these mice using the same matrices previously used to discover epitopes in the SJL mouse strain. No striking differences were found in the patterns of positive and negative pools restricted to the I-As molecule in the HA, NA, or NP matrices (Fig. 3A, B, and C, respectively). Also, select I-As-restricted epitopes were screened in parallel in SJL and B10.S mice, with no major dissimilarities in the immunodominance pattern of positive epitopes noted (Fig. 3D). Overall, these results indicate that the differences in CD4 T-cell protein specificity found between the C57BL/10 and SJL mouse strains are the result of the MHC molecule expressed, rather than the result of differences in non-MHC-related genetic background genes that might affect factors such as viral gene expression or influenza growth in vivo.
FIG. 3.
Comparison of the HA, NA, and NP peptide-pooling matrices and individual peptides in the SJL and B10.S mouse strains. HA (A), NA (B), and NP (C) peptide-pooling matrices were screened in EliSpot assays using CD4-enriched T cells obtained from previously infected SJL (white bars) and B10.S (black bars) mice, with each peptide in the pooling matrix at a final concentration of 10 μM. The pattern of positive and negative peptide pools was compared between the two mouse strains. Data are presented as the percentage of total spots, with error bars representing the standard error over at least two individual assays. Panel D shows a comparison of select I-As-restricted epitopes in an EliSpot assay using CD4-enriched T cells isolated from previously infected SJL (white bars) and B10.S (black bars) mouse strains in parallel. Data are presented as the average number of spots per 106 CD4-enriched T cells, with error bars representing the standard error over all assays performed.
Based on these results, we concluded that there was a relatively low degree of diversity in the primary influenza-specific CD4 T-cell response restricted to both the I-Ab and I-As MHC class II molecules. Table 1 summarizes the epitopes identified in both strains, and when known, the core epitopes as determined through the use of truncated peptides (data not shown). All of the proteins that were screened had only a limited number of epitopes, of which several were strongly immunodominant. Given the limited diversity of the overall response, it was these strongly immunodominant epitopes that determined the overall protein specificity of the responding CD4 T cells. Interestingly, this distribution was quite distinct in the two mouse strains examined in this study, depending on the MHC class II molecule presenting the peptides in the host.
Discussion
In the studies reported here, we have comprehensively identified the major CD4 T-cell peptide epitopes contained in NP, NS1, M1, HA, and NA proteins elicited upon primary infection with influenza in C57BL/10 and SJL mice. We initiated these studies in order to better understand the diversity and specificity of the primary CD4 immune response to influenza virus. In theory, there are multiple factors that could drive the distribution of CD4 T cells specific for different proteins in this virus. These factors include the representation of specificities in the naive T-cell repertoire, the overall size of the protein, the protein's abundance in the infected cell, access of the protein to the MHC class II peptide loading compartment, or the ability of the derived peptides to bind to a given MHC class II molecule. Prior to initiating these studies, we hypothesized that the primary immune response to influenza would be driven by a combination of the abundance of the protein within the infected cell and access to the MHC class II peptide-loading compartment. Hence we speculated that the immune response would be skewed in favor of the both abundant and membrane-associated HA and NA proteins. However, the results obtained led us to conclude that this is not the case. Instead, the primary CD4 T-cell immune repertoire restricted to the I-Ab and I-As MHC class II molecules had specificities for most of the viral proteins tested. Epitopes from the NS1 protein did not contribute significantly to the specificity of CD4 T cells in either SJL or in B10 mice. One might speculate that this failure of NS1 to elicit T cells might be due to the lack of its presence in the virion particles that are taken up by APC. However, in previously published studies in HLA-DR1 transgenic mice, we found the NS1 protein to be highly immunogenic (47), suggesting that the failure to detect epitopes in the strains studied here reflects a lack of suitable I-Ab- or I-As-binding motifs in peptides derived from this protein.
CD4 T cells responding to the influenza virus displayed a low degree of epitope diversity, with specificities that varied significantly between strains that expressed these alternate MHC class II molecules. The total number of CD4 T-cell epitopes present at >100 cytokine-producing cells per million CD4 T cells (or representing >0.01% of the total CD4 T cells tested) within the five proteins examined in this report was approximately 12 in C57BL/10 mice and 22 in SJL mice. If we assume that the remaining three large nuclear proteins (PB1, PB2, and PA) contain a similar number of epitopes, we can estimate that the total influenza CD4 T-cell repertoire encompasses approximately 20–35 epitopes. This modest value is in agreement with findings of an earlier study that analyzed the epitope specificity of lung-infiltrating CD4 T cells in response to the X-31 (H3N2) strain of influenza in C57BL/6 mice (15), and is within the range of diversity estimated for CD8 T-cell responses (16,65,66).
The finding that the pattern of epitope distribution among viral proteins varied significantly between the two strains studied here suggests to us that for CD4 T-cell responses in these mice, it may be MHC class II binding preferences rather than a characteristic of the viral protein that is driving the apparent distribution of epitopes across the different influenza proteins. There is some evidence to support the idea that the I-Ab MHC class II molecule may be somewhat atypical in its peptide-binding preferences. The crystal structure of this molecule has been determined in association with the human CLIP fragment (Ii 81–104), and an Eα peptide variant (Eα 54–66), and was noted to have smaller P4 and P6 pockets, and to favor the binding of more hydrophobic residues in the peptide binding groove than other I-A molecules (37,67). In addition, this class II molecule has been shown to bind to the CLIP peptide with very high affinity (53), which may increase the dependence of the peptide-binding process on the H2-DM molecule (12,32,33,51). In peptide elution studies utilizing automated electrospray ionization tandem mass spectrometry with computer assisted interpretation of tandem mass spectra data, 128 distinct peptide sequences were found associated with the I-Ab molecule, with about 100 additional peptide ions that were isolated, but were unable to be identified (20). This is in contrast to a similar study completed by Hunt et al., who found that between 650 and 2000 different peptides were found to be associated with the I-Ad molecule using reversed-phase HPLC and the combination of microcapillary HPLC-electrospray ionization–tandem mass spectrometry (24). Overall, these data support the possibility that I-Ab may be particularly selective in its peptide-binding characteristics. There is less information known about the I-As MHC class II molecule and its interaction with antigenic peptides. This class II molecule, unlike I-Ab, does not have particularly high affinity for CLIP (dissociation time <10 h; F. Chaves and A. Sant, unpublished data), making it less likely that this property alone is responsible for highly selective peptide capture. However, this allele is known to be associated with susceptibility to autoimmune diseases (27,28), and does have two very rare polymorphisms in amino acids that result in abrogated peptide backbone hydrogen bond formation at the peptide's carboxy-terminus (28). This alteration in hydrogen bonds may lead to more reliance on strong anchor–pocket interactions for stable peptide binding (50). Although our previous studies indicate that immunodominant CD4 T-cell epitopes possess very stable interactions with their presenting MHC class II molecule (32,51), we have not yet determined if the same is true for epitopes derived from live influenza infection. We have not evaluated the dissociation rates of the immunodominant I-Ab- and I-As-restricted peptides identified here. Using a recently published neural network algorithm designed for MHC class II molecules (44), the major peptide epitopes identified here are predicted to have only modest affinities for their respective MHC class II ligands, with the epitope contained within NP (306–328), with the core YSIRPNEN predicted to be the highest-affinity class II:peptide complex.
If the MHC class II molecules tested in this study are indeed very selective in their peptide binding, it may explain the overall low diversity of epitopes found, as well as why the distribution of CD4 T cells specific for the influenza proteins tested seems to vary as a function of the MHC molecule rather than as a function of protein localization or abundance. It is important to consider that there may be a wide range of total diversity in CD4 T-cell responses, depending on the class II molecule expressed. Our previously published studies analyzing primary CD4 responses to live influenza virus infection in HLA-DR1 transgenic mice (46,47) indicate that in responses restricted to this class II allele, the CD4 T-cell response is highly diverse and includes more than 150 different peptide specificities. Preliminary experiments (not shown) analyzing the CD4 T-cell response in H-2d-mice expressing both the I-Ad and I-Ed class II molecules suggest that, similarly to HLA-DR1 transgenic mice, the response is characterized by a high degree of diversity, which we estimate will include more than 120 different CD4 T-cell specificities. In both of these strains, the responses were widely distributed among all of the viral proteins tested, with many minor epitopes and many fewer “superdominant” epitopes than were observed in the SJL and C57BL/10 strains of mice. Therefore we speculate that the overall specificity of CD4 T cells for different proteins in influenza may in some cases be dictated by the selectivity of MHC-peptide binding, rather than by characteristics of viral protein expression or access to the MHC peptide-loading compartment. If this is the case, the practice of drawing conclusions about an overall pattern of immunodominance based on trends noted in a single mouse strain could be flawed. Instead, it may be necessary to perform more comprehensive animal studies prior to coming to any generalized conclusions regarding the relative immunogenicity of particular proteins in pathogenic organisms.
It is interesting to consider the implications of the results of our studies for the human immune response to influenza. The human MHC class II consists of multiple isotypes (HLA-DR, HLA-DQ, and HLA-DP), and for some haplotypes there may be up to two different DR beta chains expressed (49). The number of class II protein products expressed by any given individual is extended even more by heterozygosity at most loci, leading to the possible expression of up to 12 different class II molecules by some individuals (30,55). It is not known whether any human class II molecules will be as selective as the murine counterparts studied here. Regardless, our results suggest the possibility that in the highly complex MHC class II genotypes in humans, there could be quite unequal utilization of some of the MHC molecules, with those MHC molecules able to bind to a more diverse set of peptides perhaps being utilized to a greater extent than the others. This suggests the possibility that the characteristics of the CD4 T-cell repertoire to any given pathogen, including influenza, may be highly dependent on the array of MHC class II molecules in a given individual; simply expressing a particular allele may not indicate significant representation of those specificities in the host. Better understanding of the issues detailed here is a prerequisite for the rational design of epitope-based vaccines that will promote a specific, CD4 T-cell-mediated immune response.
Supplementary Material
Acknowledgments
This work was supported by grant HHSN266200700008C to A.J. Sant from the National Institutes of Health. It was also supported by T32 AI007464 from the National Institutes of Health and the Bradford Fellowship, provided by the University of Rochester Department of Pediatrics to J.L. Nayak.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.World Health Organization. New WHO influenza vaccine recommendations for 2005–2006 season, northern hemisphere. Euro Surveill. 2005;10:E050217–E050211. [PubMed] [Google Scholar]
- 2.American Academy of Pediatrics. Prevention of influenza: recommendations for influenza immunization of children, 2007–2008. Pediatrics. 2008;121:e1016–e1031. doi: 10.1542/peds.2008-0160. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Influenza vaccination coverage among children and adults—United States, 2008–2009 influenza season. MMWR Morb Mortal Wkly Rep. 2009;58:1091–1095. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb Mortal Wkly Rep. 2009;58:521–524. [PubMed] [Google Scholar]
- 5.Anthony DD. Lehmann PV. T-cell epitope mapping using the ELISPOT approach. Methods. 2003;29:260–269. doi: 10.1016/s1046-2023(02)00348-1. [DOI] [PubMed] [Google Scholar]
- 6.Belz GT. Stevenson PG. Doherty PC. Contemporary analysis of MHC-related immunodominance hierarchies in the CD8 T cell response to influenza A viruses. J Immunol. 2000;165:2404–2409. doi: 10.4049/jimmunol.165.5.2404. [DOI] [PubMed] [Google Scholar]
- 7.Belz GT. Xie W. Altman JD. Doherty PC. A previously unrecognized H-2D(b)-restricted peptide prominent in the primary influenza A virus-specific CD8(+) T-cell response is much less apparent following secondary challenge. J Virol. 2000;74:3486–3493. doi: 10.1128/jvi.74.8.3486-3493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belz GT. Wodarz D. Diaz G, et al. Compromised influenza virus-specific CD8(+)-T-cell memory in CD4(+)-T-cell-deficient mice. J. Virol. 76:12388–12393. doi: 10.1128/JVI.76.23.12388-12393.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulo S. Akarsu H. Ruigrok RWH. Baudin F. Nuclear traffic of influenza virus proteins and ribonucleoprotein complexes. Virus Res. 2007;124:12–21. doi: 10.1016/j.virusres.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Brown DM. Román E. Swain SL. CD4 T cell responses to influenza infection. Semin Immunol. 2004;16:171–177. doi: 10.1016/j.smim.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Brown DM. Dilzer AM. Meents DL. Swain SL. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J Immunol. 2006;177:2888–2898. doi: 10.4049/jimmunol.177.5.2888. [DOI] [PubMed] [Google Scholar]
- 12.Busch R. Rinderknecht CH. Roh S, et al. Achieving stability through editing and chaperoning: regulation of MHC class II peptide binding and expression. Immunol Rev. 2005;207:242–260. doi: 10.1111/j.0105-2896.2005.00306.x. [DOI] [PubMed] [Google Scholar]
- 13.Chicz RM. Urban RG. Gorga JC, et al. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J Exp Med. 1993;178:27–47. doi: 10.1084/jem.178.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crotzer VL. Blum JS. Cytosol to lysosome transport of intracellular antigens during immune surveillance. Traffic. 2008;9:10–16. doi: 10.1111/j.1600-0854.2007.00664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crowe SR. Miller SC. Brown DM, et al. Uneven distribution of MHC class II epitopes within the influenza virus. Vaccine. 2006;24:457–467. doi: 10.1016/j.vaccine.2005.07.096. [DOI] [PubMed] [Google Scholar]
- 16.Crowe SR. Miller SC. Woodland DL. Identification of protective and non-protective T cell epitopes in influenza. Vaccine. 2006;24:452–456. doi: 10.1016/j.vaccine.2005.07.090. [DOI] [PubMed] [Google Scholar]
- 17.Dawood FS. Jain S. Finelli L, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 18.Doherty PC. Topham DJ. Tripp RA, et al. Effector CD4 and CD8 T-cell mechanisms in the control of respiratory virus infections. Immunol Rev. 1997;159:105–117. doi: 10.1111/j.1600-065x.1997.tb01010.x. [DOI] [PubMed] [Google Scholar]
- 19.Doherty PC. Turner SJ. Webby RG. Thomas PG. Influenza and the challenge for immunology. Nat Immunol. 2006;7:449–455. doi: 10.1038/ni1343. [DOI] [PubMed] [Google Scholar]
- 20.Dongre AR. Kovats S. deRoos P, et al. In vivo MHC class II presentation of cytosolic proteins revealed by rapid automated tandem mass spectrometry and functional analyses. Eur J Immunol. 2001;31:1485–1494. doi: 10.1002/1521-4141(200105)31:5<1485::AID-IMMU1485>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 21.Geginat G. Schenk S. Skoberne M, et al. A novel approach of direct ex vivo epitope mapping identifies dominant and subdominant CD4 and CD8 T cell epitopes from Listeria monocytogenes. J Immunol. 2001;166:1877–1884. doi: 10.4049/jimmunol.166.3.1877. [DOI] [PubMed] [Google Scholar]
- 22.Gerdil C. The annual production cycle for influenza vaccine. Vaccine. 2003;21:1776–1779. doi: 10.1016/s0264-410x(03)00071-9. [DOI] [PubMed] [Google Scholar]
- 23.Hale BG. Randall RE. Ortín J. Jackson D. The multifunctional NS1 protein of influenza A viruses. J Gen Virol. 2008;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 24.Hunt DF. Michel H. Dickinson TA, et al. Peptides presented to the immune system by the murine class II major histocompatibility complex molecule I-Ad. Science. 1992;256:1817–1820. doi: 10.1126/science.1319610. [DOI] [PubMed] [Google Scholar]
- 25.Hussell T. Cavanagh MM. The innate immune rheostat: influence on lung inflammatory disease and secondary bacterial pneumonia. Biochem Soc Trans. 2009;37:811–813. doi: 10.1042/BST0370811. [DOI] [PubMed] [Google Scholar]
- 26.Itoh Y. Shinya K. Kiso M, et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460:1021–1025. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jane-wit D. Yu M. Edling AE, et al. A novel class II-binding motif selects peptides that mediate organ-specific autoimmune disease in SWXJ, SJL/J, and SWR/J mice. J Immunol. 2002;169:6507–6514. doi: 10.4049/jimmunol.169.11.6507. [DOI] [PubMed] [Google Scholar]
- 28.Kalbus M. Fleckenstein BT. Offenhäusser M, et al. Ligand motif of the autoimmune disease-associated mouse MHC class II molecule H2-A(s) Eur J Immunol. 2001;31:551–562. doi: 10.1002/1521-4141(200102)31:2<551::aid-immu551>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 29.Karlsson AC. Martin JN. Younger SR, et al. Comparison of the ELISPOT and cytokine flow cytometry assays for the enumeration of antigen-specific T cells. J Immunol Methods. 2003;283:141–153. doi: 10.1016/j.jim.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Karp DR. Teletski CL. Jaraquemada D, et al. Structural requirements for pairing of alpha and beta chains in HLA-DR and HLA-DP molecules. J Exp Med. 1990;171:615–628. doi: 10.1084/jem.171.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohlmeier JE. Woodland DL. Immunity to respiratory viruses. Annu Rev Immunol. 2009;27:61–82. doi: 10.1146/annurev.immunol.021908.132625. [DOI] [PubMed] [Google Scholar]
- 32.Lazarski CA. Chaves FA. Jenks SA, et al. The kinetic stability of MHC class II:peptide complexes is a key parameter that dictates immunodominance. Immunity. 2005;23:29–40. doi: 10.1016/j.immuni.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Lazarski CA. Chaves FA. Sant AJ. The impact of DM on MHC class II-restricted antigen presentation can be altered by manipulation of MHC-peptide kinetic stability. J Exp Med. 2006;203:1319–1328. doi: 10.1084/jem.20060058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee HK. Iwasaki A. Autophagy and antiviral immunity. Curr Opin Immunol. 2008;20:23–29. doi: 10.1016/j.coi.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis DB. Avian flu to human influenza. Annu Rev Med. 2006;57:139–154. doi: 10.1146/annurev.med.57.121304.131333. [DOI] [PubMed] [Google Scholar]
- 36.Li P. Gregg JL. Wang N, et al. Compartmentalization of class II antigen presentation: contribution of cytoplasmic and endosomal processing. Immunol Rev. 2005;207:206–217. doi: 10.1111/j.0105-2896.2005.00297.x. [DOI] [PubMed] [Google Scholar]
- 37.Liu X. Dai S. Crawford F, et al. Alternate interactions define the binding of peptides to the MHC molecule IA(b) Proc Natl Acad Sci USA. 2002;99:8820–8825. doi: 10.1073/pnas.132272099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loss GE. Elias CG. Fields PE, et al. Major histocompatibility complex class II-restricted presentation of an internally synthesized antigen displays cell-type variability and segregates from the exogenous class II and endogenous class I presentation pathways. J Exp Med. 1993;178:73–85. doi: 10.1084/jem.178.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGill J. Heusel JW. Legge KL. Innate immune control and regulation of influenza virus infections. J Leukoc Biol. 2009;86:803–812. doi: 10.1189/jlb.0509368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McMurry JA. Johansson BE. De Groot AS. A call to cellular humoral arms: enlisting cognate T cell help to develop broad-spectrum vaccines against influenza A. Human Vaccines. 2008;4:148–157. doi: 10.4161/hv.4.2.5169. [DOI] [PubMed] [Google Scholar]
- 41.Münz C. Enhancing immunity through autophagy. Annu Rev Immunol. 2009;27:423–449. doi: 10.1146/annurev.immunol.021908.132537. [DOI] [PubMed] [Google Scholar]
- 42.Nayak DP. Hui EK. Barman S. Assembly and budding of influenza virus. Virus Res. 2004;106:147–165. doi: 10.1016/j.virusres.2004.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nayak DP. Balogun RA. Yamada H, et al. Influenza virus morphogenesis and budding. Virus Res. 2009;143:147–161. doi: 10.1016/j.virusres.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nielsen M. Lund O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinformatics. 2009;10:296–306. doi: 10.1186/1471-2105-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peiris JS. Poon LL. Guan Y. Emergence of a novel swine-origin influenza A virus (S-OIV) H1N1 virus in humans. J Clin Virol. 2009;45:169–173. doi: 10.1016/j.jcv.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richards KA. Chaves FA. Krafcik FR, et al. Direct ex vivo analyses of HLA-DR1 transgenic mice reveal an exceptionally broad pattern of immunodominance in the primary HLA-DR1-restricted CD4 T-cell response to influenza virus hemagglutinin. J Virol. 2007;81:7608–7619. doi: 10.1128/JVI.02834-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richards KA. Chaves FA. Sant AJ. Infection of HLA-DR1 transgenic mice with a human isolate of influenza a virus (H1N1) primes a diverse CD4 T-cell repertoire that includes CD4 T cells with heterosubtypic cross-reactivity to avian (H5N1) influenza virus. J Virol. 2009;83:6566–6577. doi: 10.1128/JVI.00302-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson JH. Delvig AA. Diversity in MHC class II antigen presentation. Immunology. 2002;105:252–262. doi: 10.1046/j.0019-2805.2001.01358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rollini P. Mach B. Gorski J. Linkage map of three HLA-DR beta-chain genes: evidence for a recent duplication event. Proc Natl Acad Sci USA. 1985;82:7197–7201. doi: 10.1073/pnas.82.21.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sant AJ. Beeson C. McFarland B, et al. Individual hydrogen bonds play a critical role in MHC class II:peptide interactions: implications for the dynamic aspects of class II trafficking and DM-mediated peptide exchange. Immunol Rev. 1999;172:239–253. doi: 10.1111/j.1600-065x.1999.tb01369.x. [DOI] [PubMed] [Google Scholar]
- 51.Sant AJ. Chaves FA. Jenks SA, et al. The relationship between immunodominance, DM editing, and the kinetic stability of MHC class II:peptide complexes. Immunol Rev. 2005;207:261–278. doi: 10.1111/j.0105-2896.2005.00307.x. [DOI] [PubMed] [Google Scholar]
- 52.Sant AJ. Chaves FA. Krafcik FR, et al. Immunodominance in CD4 T-cell responses: implications for immune responses to influenza virus and for vaccine design. Expert Rev Vaccines. 2007;6:357–368. doi: 10.1586/14760584.6.3.357. [DOI] [PubMed] [Google Scholar]
- 53.Sette A. Southwood S. Miller J. Appella E. Binding of major histocompatibility complex class II to the invariant chain-derived peptide, CLIP, is regulated by allelic polymorphism in class II. J Exp Med. 1995;181:677–683. doi: 10.1084/jem.181.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sette A. Moutaftsi M. Moyron-Quiroz J, et al. Selective CD4+ T cell help for antibody responses to a large viral pathogen: deterministic linkage of specificities. Immunity. 2008;28:847–858. doi: 10.1016/j.immuni.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stern LJ. Calvo-Calle JM. HLA-DR: molecular insights and vaccine design. Curr Pharm Des. 2009;15:3249–3261. doi: 10.2174/138161209789105171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun JC. Williams MA. Bevan MJ. CD4 T cells are required for the maintenance, not programming, of memory CD8 T cells after acute infection. Nat. Immunol. 2004;5:927–933. doi: 10.1038/ni1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swain SL. Agrewala JN. Brown DM, et al. CD4 T-cell memory: generation and multi-faceted roles for CD4 T cells in protective immunity to influenza. Immunol Rev. 2006;211:8–22. doi: 10.1111/j.0105-2896.2006.00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tamura S-I. Kurata T. Defense mechanisms against influenza virus infection in the respiratory tract mucosa. Jpn J Infect Dis. 2004;57:236–247. [PubMed] [Google Scholar]
- 59.Thomas PG. Keating R. Hulse-Post DJ. Doherty PC. Cell-mediated protection in influenza infection. Emerg Infect Dis. 2006;12:48–54. doi: 10.3201/eid1201.051237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson WW. Shay DK. Weintraub E, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 61.Thompson WW. Shay DK. Weintraub E, et al. Influenza-associated hospitalizations in the United States. JAMA. 2004;292:1333–1340. doi: 10.1001/jama.292.11.1333. [DOI] [PubMed] [Google Scholar]
- 62.Tobery TW. A simple and efficient method for the monitoring of antigen-specific T cell responses using peptide pool arrays in a modified ELISpot assay. J Immunol Methods. 2001;254:59–66. doi: 10.1016/s0022-1759(01)00397-0. [DOI] [PubMed] [Google Scholar]
- 63.Tobery TW. Caulfield MJ. Identification of T-cell epitopes using ELISpot and peptide pool arrays. Methods Mol Med. 2004;94:121–132. doi: 10.1385/1-59259-679-7:121. [DOI] [PubMed] [Google Scholar]
- 64.Weiss S. Bogen B. MHC class II-restricted presentation of intracellular antigen. Cell. 1991;64:767–776. doi: 10.1016/0092-8674(91)90506-t. [DOI] [PubMed] [Google Scholar]
- 65.Yewdell JW. Bennink JR. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
- 66.Zhong W. Reche PA. Lai C-C, et al. Genome-wide characterization of a viral cytotoxic T lymphocyte epitope repertoire. J Biological Chem. 2003;278:45135–45144. doi: 10.1074/jbc.M307417200. [DOI] [PubMed] [Google Scholar]
- 67.Zhu Y. Rudensky AY. Corper AL, et al. Crystal structure of MHC class II I-Ab in complex with a human CLIP peptide: prediction of an I-Ab peptide-binding motif. J Mol Biol. 2003;326:1157–1174. doi: 10.1016/s0022-2836(02)01437-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.