Abstract
Background
Short-acting inhaled β2-agonists such as albuterol are used for bronchodilation and are the mainstay of asthma treatment worldwide. There is significant variation in bronchodilator responsiveness to albuterol not only between individuals but also across racial/ethnic groups. The β2-adrenergic receptor (β2AR) is the target for β2-agonist drugs. The enzyme S-nitrosoglutathione reductase (GSNOR), which regulates levels of the endogenous bronchodilator S-nitrosoglutathione, has been shown to modulate the response to β2-agonists.
Objective
We hypothesized that there are pharmacogenetic interactions between GSNOR and β2AR gene variants which are associated with variable response to albuterol.
Methods
We performed family-based analyses to test for association between GSNOR gene variants and asthma and related phenotypes in 609 Puerto Rican and Mexican families with asthma. In addition, we tested these subjects for pharmacogenetic interaction between GSNOR and β2AR gene variants and responsiveness to albuterol using linear regression. Cell transfection experiments were performed to test the potential effect of the GSNOR gene variants.
Results
Among Puerto Ricans, several GSNOR SNPs and a haplotype in the 3′UTR were significantly associated with increased risk for asthma and lower bronchodilator responsiveness (p = 0.04 to 0.007). The GSNOR risk haplotype affects expression of GSNOR mRNA and protein, suggesting a gain of function. Furthermore, gene-gene interaction analysis provided evidence of pharmacogenetic interaction between GSNOR and β2AR gene variants and the response to albuterol in Puerto Rican (p = 0.03), Mexican (p = 0.15) and combined Puerto Rican and Mexican asthmatics (p = 0.003). Specifically, GSNOR+17059*β2AR+46 genotype combinations (TG+GG*AG and TG+GG*GG) were associated with lower bronchodilator response.
Conclusion
Genotyping of GSNOR and β2AR genes may be a useful in identifying Latino subjects, who might benefit from adjuvant therapy for refractory asthma.
Keywords: Asthma, Bronchodilator responsiveness, GSNO Reductase, β2-Adrenergic Receptor, Latinos, Gene-gene interaction, Polymorphisms, Pharmacogenetics
Introduction
Asthma is a common but complex respiratory ailment caused by the interaction of genetic and environmental factors. Short-acting inhaled β2-agonists (albuterol) are the mainstay of asthma treatment worldwide.(1, 2) Furthermore, among low income and minority populations in the U.S., albuterol is more likely to be the only medication used for asthma regardless of asthma severity.(3, 4)
There is substantial variation in bronchodilator responsiveness between individuals and racial/ethnic groups.(2, 5-7) Although a significant proportion of the individual and population variance is assumed to be due to environmental factors and measurement errors, previous investigators have demonstrated that there are strong genetic effects influential to bronchodilator responsivenss and these studies indicate that a clinically significant fraction of the total population variance is genetic in origin.(6, 8) A search for genetic determinants of responsiveness to albuterol have yielded a large number of studies, most of which have focused on genetic polymorphisms within the β2- adrenergic receptor (β2AR) gene.(9, 10) However, polymorphisms in the β2AR gene have been inconsistently associated with bronchodilatory response to β-agonists and with tachyphylaxis.(9, 10) The most commonly studied functional polymorphism in the β2AR gene is SNP +A46G, which results in a change in amino acid sequence from arginine to glycine at codon 16. Although this SNP was initially associated with increased response to albuterol(11, 12), it has subsequently also been associated with tachyphylaxis from long-term use of β2-agonists.(13) Furthermore, we and others have previously reported that pharmacogenetic associations with β2-agonists vary by population.(7, 8, 11, 14, 15) The fact that results vary within and between populations is suggestive of complex gene-environment and/or gene-gene interactions which may help to explain discordant results.
Nitric Oxide (NO) bioactivity in vivo is mediated primarily through the covalent modification of cysteine sulfur by NO to form S-nitrosothiols (SNOs). S-nitrosogluathione (GSNO), is the predominant SNO present in the airway lining fluid of the healthy lung and is a potent endogenous bronchodilator.(16) Children with severe asthma have decreased SNO content in the airway lining fluid(17) and adults with mild asthma undergoing segmental airway antigen challenge have evidence of accelerated SNO breakdown.(18) GSNO reductase (GSNOR) has been identified as a key regulator of SNO levels in the lung and airway reactivity in mice(19) and humans.(20) In a murine model of allergic asthma, deletion of the GSNOR gene resulted in increased lung SNO concentrations and protection from allergen challenge. The airways of GSNOR-deficient (GSNOR-/-) mice were also protected from β2-adrenergic receptor desensitization under basal conditions, whereas supplementation of GSNO attenuated β2 receptor desensitization in wild type mice. The molecular basis by which GSNO inhibits β2AR desensitization may relate to inhibition of G-protein coupled receptor kinases (GRKs), thereby preventing β2AR phosphorylagtion and downregulation.(21) More recently, we observed increased GSNOR activity and low lung SNO content in subjects with mild asthma. The increased GSNOR activity correlated inversely with methacholine PC20 suggesting that GSNOR is a key regulator of airway hyperresponsiveness in asthma.(20)
Work done previously by Wu et al. and in our lab has shown associations between genetic variants in the GSNOR gene and asthma(22, 23). Based upon the compelling results in mice(19) and human(20) and prior analyses of pharmacogenetic associations for β2AR in Latinos(11, 14), we hypothesized that gene-gene interactions between genetic variants in the GSNOR and β2AR genes may modify bronchodilator responsiveness to albuterol. We investigated this hypothesis in Mexican and Puerto Rican asthma trios participating in the Genetics of Asthma in Latino Americans (GALA) study.(5, 11) We chose to study these populations because U.S. vital statistics indicate that asthma prevalence, morbidity, and mortality are highest in Puerto Ricans and lowest in Mexicans.(24, 25)
Methods
Subjects
A total of 609 Latino asthmatic trios (probands and their biological parents; total n = 1827) with complete spirometry data were analyzed in this study. Out of 609 trios, 273 were Mexican and 336 were Puerto Rican trios. Detailed recruitment criteria and subject characteristics are described elsewhere.(5, 11) Briefly, asthmatic trios of Puerto Rican or Mexican ethnicity were recruited from the New York City, Puerto Rico, San Francisco Bay Area and Mexico City as part of the Genetics of Asthma in Latino Americans (GALA) study. Ethnicity was self-reported. In GALA, asthmatic subjects were enrolled only if both biological parents and all four biological grandparents were of the same ethnic backgrounds - Puerto Rican in New York and Puerto Rico and Mexican in San Francisco and Mexico City. Additional inclusion criteria for asthmatic patient were age between 8-40 years, a current physician diagnosis of asthma, and presence of asthma symptoms (wheezing, cough, or shortness of breath) over the last two years. The patient had to be clinically stable and without symptoms or upper respiratory tract infection for at least 6 weeks prior to participating in the study. Recruitment criteria were identical at each site. Total plasma IgE was measured in duplicate using Uni-Cap technology (Pharmacia, Kalamazoo, MI). Local Institutional Review Boards (IRBs) approved all the studies, and all subjects provided written, age-appropriate informed consent or assent.
Pulmonary Function Tests
Pulmonary function test results were expressed as a percentage of the predicted normal value using age-adjusted prediction equations from Hankinson.(26) A quantitative measure of bronchodilator responsiveness was calculated as ΔFEV1 (change in forced expiratory volume in one second) which is the relative percent change in baseline FEV1 (Pre-FEV1) after administration of 180μg (2 puffs) for subjects < 16 years old, and 360μg (4 puffs) for subjects ≥ 16 years old for the GALA and SAGE studies. Details of the pulmonary function tests are described elsewhere.(5, 11)
SNP Discovery
Sequencing of the GSNOR gene was performed on 48 unrelated asthmatic subjects, 24 of Mexican and 24 of Puerto Rican ethnicity (96 chromosomes). This number of subjects provides greater than 90% power to detect variants with an allele frequency of 5% or more. All exons, exon-intron boundaries, and two potential promoter regions (-711 bp to -113 bp and +2933 bp to +3531 bp predicted by the program GENSCAN; http://genes.mit.edu/GENSCAN.html) of the GSNOR gene were sequenced (GenBank Accession No. AY987960). Sequencing was performed using the Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, California) and ABI Prism 3700 sequencher (Applied Biosystems, Foster City, CA). The degree of linkage disequilibrium (LD) was estimated by using the r2 statistic.(27)
Genotyping
The five GSNOR SNPs (+3207 in the potential promoter region and +16701, +16777, +17059 and +17250 in the 3′UTR region) were further selected for genotyping in the entire GALA cohort based on their allele frequency (minor allele frequency > 5%), location (exonic or in potential promoter region) and the haplotype structure of the GSNOR gene. All five SNPs were genotyped using the AcycloPrime-FP™ (PerkinElmer) method.(28) Detailed information for SNP genotyping are provided in the online supplement. Genetic variant +A46G in the β2AR gene, which corresponds to amino acid change from Arg to Gly at codon 16, was previously genotyped and AA genotype has been shown to be associated with bronchodilator responsiveness in Puerto Rican, but not in Mexican asthmatics.(11, 14)
Statistical Analysis
Mendelian inconsistencies were identified using PEDCHECK.(29) A total of 3 families were excluded due to Mendelian inconsistencies (0.5%, 3/609). Families with Mendelian inconsistencies were excluded from further analysis including Hardy-Weinberg Equilibrium (HWE) and family-based association tests. The HWE was calculated by means of χ2 goodness-of-fit tests, separately for Puerto Ricans and Mexicans. The HWE was calculated separately for parents and probands within Puerto Ricans and Mexicans. FBAT(30) and HaploFBAT(31) were used to perform transmission disequilibrium tests to assess the association between individual SNPs and haplotypes, respectively, with asthma and quantitative measures of asthma-related phenotypes in the GALA families. The minor homozygotes and heterozygotes were combined for the analysis as the minor allele frequency of most of the genotyped GSNOR SNP is low. The quantitative phenotypes analyzed include baseline lung function (measured as Pre-FEV1, % of predicted) and bronchodilator responsiveness (measured as ΔFEV1, relative % of predicted). Odds ratios (OR) and their 95% confidence intervals for associations between individual SNPs and asthma were estimated using the program UNPHASED.(32)
Gene-gene interactions for bronchodilator responsiveness were tested using non-parametric linear regression models. An interaction term (cross product of SNPs in the GSNOR and β2AR genes) was included in the regression model along with the main effects and confounding variables such as age, sex, Pre-FEV1, asthma duration, log transformed body mass index (log10 BMI), steroid use and regular or as needed bronchodilator use to test for association with ΔFEV1. Genetic association analyses may be confounded by population stratification. Therefore, the interaction analyses were also adjusted for individual ancestral proportions estimated using 107 unlinked ancestry informative markers (AIMs) using the method described elsewhere.(33, 34) In addition, the interaction analysis performed on combined Mexican and Puerto Rican asthmatics was also adjusted for Latino ethnicity. All analyses were performed using the statistical software package Stata/SE 9.0 (Stata Corp., College Station, TX).
Correction for Multiple Testing
The correction for multiple testing was performed using SNPSpD (an effective Bonferroni-type correction).(35) Each phenotype (asthma, Pre-FEV1 and ΔFEV1) in the present study was tested for 5 SNPs. There is significant LD between these SNPs and therefore the program SNPSpD calculated these 5 SNPs to be equivalent to 2 independent SNPs. Thus, the α for GSNOR association tests was set at 0.008 [0.05/(2*3)], where 2 is the number of independent SNPs and 3 is the number of phenotypes tested. Since Mexicans and Puerto Ricans are two separate populations and could have different risk alleles; we did not correct the analysis within populations for two tests. The association analyses were performed separately in all asthmatics and on those asthmatics that were using albuterol as their only asthma medication. However, the latter group is not an independent cohort, rather a subset of all asthmatics. Therefore analyses within all asthmatics and the subgroup of asthmatics using albuterol as their only asthma medication were not corrected for two tests.
Constructs and Cell Transfections
The full length GSNOR 3′UTR was amplified from human cDNA. The resulting PCR products were cloned into both forward and reverse orientations in the pGL3 plasmid containing an SV40 promoter upstream of the firefly luciferase gene (Promega Corporation, Madison, WI). Site directed mutagenesis of the 3′UTR was performed using QuikChange® II XL Site Directed Mutagenesis Kit (Stratagene, CA) according to the manufacturer's instructions. Individual mutants mimicking the SNPs associated with asthma in our patient population were generated by substituting C+16701→T, A+17059→C and T+17250→C (data not shown). Another mutant was created containing the three individual mutants to mimic the GSNOR haplotype associated with asthma. The mutated fragments were amplified and ligated into the pGL3 promoter vector. The luciferase reporter gene constructs were transiently transfected into HEK293 cells with Lipofectamine 2000 (Invitrogen; Carlsbad CA) according to the manufacturer's protocol. Specific substrates were used to measure the activities of the firefly and Renilla luciferase with a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA). Firefly luminescence was normalized to Renilla luminescence and reported as relative luciferase units (RLU). Differences among cell transfection groups were evaluated using a student's t-test. A p value of < 0.05 was considered statistically significant. Details of the constructs and the cell transfection assays are provided in the online supplement.
Results
Characteristics of the Subjects
Demographic and clinical characteristics of all probands with asthma that were analyzed for this study are shown in Table 1. Although the GALA cohort has a total of 686 trios with asthma (probands and their biological parents; total n = 1827), this study only included 609 trios that had probands with complete spirometry data (Mexican: n = 273 trios, Puerto Rican: n = 336 trios). The median age of the Mexican and Puerto Rican asthmatic probands was 13 and 12 years, respectively. The median Pre-FEV1 and ΔFEV1 were 89% and 7.4%; 83% and 4.9% in Mexican and Puerto Rican asthma subjects, respectively.
Table 1.
Demographic and clinical characteristics of Puerto Rican and Mexican subjects with asthma from the GALA families.
| Puerto Ricans (n = 336) | Mexicans (n = 273) | |
|---|---|---|
| Characteristic | ||
| Age (yrs) * | 12 (10:15) | 13 (11:20) |
| Sex (% male) | 56.7% | 53.0% |
| BMI (kg/m2) * | 21.3 (17:26) | 23.7 (20:28) |
| Mild Asthma (%) | 32.6% | 32.9% |
| Serum IgE (IU/mL) *# | 254 (95:632) | 246 (100:591) |
| Baseline spirometry | ||
| Pre-FEV1* | 83 (74:93) | 89 (77:100) |
| Pre-FEV1 < 80% | 38% | 30% |
| Bronchodilator responsiveness | ||
| ΔFEV1 (relative % predicted) * | 4.9 (0.6:10) | 7.4 (4:13) |
Values are expressed as median (25th:75th percentile).
Serum IgE levels were missing for 33 Puerto Rican and 20 Mexican subjects.
Resequencing of the GSNOR Gene
A total of 31 SNPs were identified in the GSNOR gene by resequencing the exons, exon-intron boundaries and two potential promoter regions in 24 Mexican and 24 Puerto Rican asthmatics (see materials and methods and Table 1S in the online supplement). Only 13 of these 31 SNPs had an allele frequency greater than 5% in Mexicans and Puerto Ricans. For both Mexicans and Puerto Ricans, we observed tight LD between the SNPs in the GSNOR gene (see Figure 1S in the online supplement). Five GSNOR SNPs were selected for further genotyping in the GALA cohort based on a minor allele frequency > 5% in Mexicans or Puerto Ricans, location (exonic or in potential promoter region) and linkage disequilibrium pattern. These SNPs included: +3207 in the potential promoter region, +16701, +16777, +17059 and +17250 in the 3′UTR region.
Association between GSNOR Genetic Variants and Asthma
The genotyping call rates for all five SNPs were > 95% and all SNPs were in HWE in Mexican and Puerto Rican parents (p > 0.05). Among Puerto Ricans, single locus family-based analyses demonstrated that several GSNOR SNPs were significantly associated with increased risk for asthma (Table 2). Associated genetic variants included: CT +TT genotypes of SNP +3207 in the promoter region (p = 0.05) and genotypes GA+AA, TG+GG and AG+GG of SNPs +16701, +17059 and 17250, respectively in the 3′UTR (p = 0.04 to 0.007), and the 5 SNP haplotype T.A.G.G.G. with a frequency of 10% (p = 0.02) (Table 2 and 3). Only the association between SNP +17059 and asthma remained significant after correction for multiple testing (p = 0.007, adjusted α = 0.008). No association was found between GSNOR SNPs or haplotypes with Pre-FEV1 and ΔFEV1. However, since steroids can blunt the effect of GSNOR,(36) the subjects were stratified by steroid use and the analysis was repeated in those subjects who were using albuterol as their only asthma medication. Although we had a significant reduction in sample size (n = 168) and therefore loss of statistical power, we observed that several GSNOR SNPs were significantly associated with ΔFEV1 (p = 0.04 to 0.01) in Puerto Rican subjects who were using albuterol as their only asthma medication before correction for multiple testing (Table 2). A marginal association was also observed between the five SNP haplotype and ΔFEV1 (p = 0.06) in this subgroup analysis (Table 3). Among Mexicans we did not find any association between GSNOR SNPs and asthma or related phenotypes including ΔFEV1 either in the analysis including all asthmatics or analysis stratified by steroid use.
Table 2.
Association of the GSNOR SNPs and 5 SNP haplotypes with asthma and related phenotypes in Puerto Rican asthma families before correction for multiple testing. A stratified analysis was also performed for those subjects who were using albuterol as their only asthma medication.
| SNP Genotype (Allele Frequency, Allele) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| +3207 CT+TT/CC (0.26, T) | +16701 GA+AA/GG (0.10, A) | +16777 TG+GG/TT (0.37, G) | +17059 TG+GG/TT (0.11, G) | +17250 AG+GG/AA (0.16, G) | ||||||
| p-value | OR (95% CI) or Mean (SD) | p-value | OR (95% CI) or Mean (SD) | p-value | OR (95% CI) or Mean (SD) | p-value | OR (95% CI) or Mean (SD) | p-value | OR (95% CI) or Mean (SD)) | |
| All Puerto Rican Subjects (n = 336) | ||||||||||
| Asthma | 0.05 (+) | 1.17 (1.03 - 1.57) | 0.04 (+) | 1.35 (1.05 - 2.02) | 0.42 (+) | 1.08 (0.97 - 1.45) | 0.007 (+) | 1.41 (1.10 - 2.10) | 0.01 (+) | 1.40 (1.15 - 1.97) |
| Pre-FEV1 | 0.23 (-) | 83.6 (17.4)/84.9 (16.4) | 0.88 (-) | 84.2 (19.6)/84.5 (16.3) | 0.80 (+) | 83.9 (17.9)/84.7 (16.2) | 0.82 (+) | 85.7 (21.5)/83.6 (15.6) | 0.27 (+) | 85.2 (20.1)/84.1 (15.6) |
| ΔFEV1 | 0.13 (-) | 5.2 (12.9)/7.4 (10.2) | 0.85 (+) | 7.2 (14.2)/6.4 (13.2) | 0.39 (-) | 6.5 (15.0)/7.1 (10.6) | 0.93 (+) | 7.5 (14.4)/6.5 (13.0) | 0.69 (-) | 7.2 (13.3)/6.4 (13.4) |
| Puerto Rican Subjects using albuterol only (n = 168) | ||||||||||
| Asthma | 0.19 (+) | 1.18 (0.93 - 1.78) | 0.20 (+) | 1.38 (0.89 - 2.52) | 0.50 (+) | 1.13 (0.90 - 1.70) | 0.09 (+) | 1.44 (0.94 - 2.54) | 0.02 (+) | 1.68 (1.15 - 2.78) |
| Pre-FEV1 | 0.96 (+) | 88.1 (15.4)/85.9 (14.0) | 0.24 (+) | 92.5 (17.5)/85.2 (14.1) | 0.90 (+) | 86.8 (17.0)/86.2 (13.0) | 0.11 (+) | 93.3 (17.7)/84.4 (13.9) | 0.17 (+) | 91.8 (17.1)/84.5 (14.0) |
| ΔFEV1 | 0.01 (-) | 3.4 (7.2)/7.3 (10.3) | 0.03 (-) | 2.9 (6.4)/7.1 (15.4) | 0.49 (-) | 6.4 (16.4)/6.7 (10.7) | 0.01 (-) | 2.9 (6.3)/7.4 (15.5) | 0.01 (-) | 3.9 (6.9)/7.3 (15.9) |
“+” and “-” indicate direction of association. “+” indicates increased risk for asthma or higher value of the quantitative phenotypes Pre-FEV1 and ΔFEV1 whereas “-” indicates decrease risk for asthma and lower value of the quantitative phenotypes. p < 0.1 are shown in bold.
Table 3.
Association of the GSNOR 5 SNP haplotypes with asthma and related phenotypes in Puerto Rican asthma families before correction for multiple testing. A stratified analysis was also performed for those subjects who were using albuterol as their only asthma medication.
| 5 SNP Haplotype (Frequency) | ||||||
|---|---|---|---|---|---|---|
| C.G.T.T.A (0.63) | T.G.G.T.A (0.14) | T.A.G.G.G (0.10) | C.G.G.T.A (0.06) | C.G.G.T.G (0.05) | ||
| p-value | ||||||
| All Puerto Rican Subjects (n = 336) | ||||||
| Asthma | 0.67 (-) | 0.71 (+) | 0.02 (+) | 0.38 (-) | 0.89 (-) | |
| Pre-FEV1 | 0.50 (+) | 0.26 (-) | 0.74 (-) | 0.64 (-) | 0.55 (+) | |
| ΔFEV1 | 0.53 (+) | 0.11 (-) | 0.59 (+) | 0.90 (+) | 0.35 (-) | |
| Puerto Rican Subjects using albuterol only (n = 168) | ||||||
| Asthma | 0.73 (-) | 0.65 (+) | 0.17 (+) | 0.21 (-) | 0.32 (+) | |
| Pre-FEV1 | 0.76 (-) | 0.19 (-) | 0.10 (+) | 0.56 (+) | 0.44 (-) | |
| ΔFEV1 | 0.06 (+) | 0.42 (-) | 0.19 (-) | 0.40 (-) | 0.34 (-) | |
“+” and “-” indicate direction of association. “+” indicates increased risk for asthma or higher value of the quantitative phenotypes Pre-FEV1 and ΔFEV1 whereas “-” indicates decrease risk for asthma and lower value of the quantitative phenotypes. p < 0.1 are shown in bold.
GSNOR and β2AR Gene-Gene Interaction
We performed gene-gene interaction analysis between the genetic variants of the GSNOR and β2AR genes for association with bronchodilator responsiveness using linear regression models. To avoid multiple testing issues, we only analyzed the gene-gene interaction between GSNOR +17059 and β2AR +46 SNPs. We selected GSNOR +17059 SNP as it showed the most significant association with bronchodilator responsiveness in our single gene analysis of GSNOR and SNP +46 (Arg16Gly) is one of the two most extensively studied SNPs for the β2AR gene and is in strong LD with the other most studied β2AR SNP +79 (Gln27Glu).
An interaction term of cross product between GSNOR +17059 and β2AR +46 genotypes was generated to test gene-gene interaction between β2AR and GSNOR genes (Table 4). Specifically, we paired the subject's genotype of GSNOR +17059 with genotype of β2AR +46 to determine if the presence of risk genotype for both the genes in a subject modified the risk for asthma differently than each gene alone. The association between the interaction term and ΔFEV1 was tested using linear regression models. The linear regression models also included the two main effects SNPs GSNOR +17059 and β2AR +46 and other confounding variables as listed in the methods section. A significant association was observed between the interaction term and ΔFEV1 for Puerto Ricans (p = 0.03) and a borderline association for Mexicans (p = 0.15). The association between the interaction term and ΔFEV1 became more significant in the combined analysis of Puerto Rican and Mexican asthmatic subjects (p = 0.003) suggesting that the direction of association is in the same direction in Mexicans and Puerto Ricans. Table 5 shows the mean ΔFEV1 stratified by interaction term after adjusting for main effects and confounders in Puerto Rican, Mexican and Puerto Rican and Mexican combined asthmatic subjects. Puerto Rican and Mexican asthmatics subjects, who had the GSNOR +17059 TG+GG genotype and either the β2AR +46 AG or GG genotypes (interaction term 1 and 2 respectively), had significantly lower bronchodilator responsiveness (mean ΔFEV1 relative % predicted = 7.53 and 3.9 respectively) compared to those subjects that had the GSNOR +17059 TT genotype and any of the β2AR +46 genotype or GSNOR +17059 TG or GG risk genotype and β2AR +46 AA genotype (interaction term 0, mean ΔFEV1 relative % predicted = 11.15) (Table 5). These results suggest a gene-gene interaction between GSNOR and β2AR genes for bronchodilator drug responsiveness and that specific combinations of GSNOR and β2AR genotypes are associated with lower ΔFEV1 in both Puerto Rican and Mexican asthmatics.
Table 4.
The interaction term, the cross-product between GSNOR +17059 and β2AR +46 genotypes and its coding for gene-gene interaction analysis.
| GSNOR +17059 (T/G) genotypes | GSNOR +17059 (T/G) genotype coding | β2AR +46 (A/G) genotypes | β2AR +46 (A/G) genotype coding | Interaction term GSNOR+17059*β2AR+46 | Interaction term GSNOR+17059*β2AR+46 coding | No. of Subjects (Puerto Ricans and Mexicans combined) |
|---|---|---|---|---|---|---|
| TT | 0 | AA | 0 | TT*AA | 0*0 = 0 | 90 |
| TG+GG | 1 | AA | 0 | TG+GG*AA | 1*0 = 0 | 18 |
| TT | 0 | AG | 1 | TT*AG | 0*1 = 0 | 193 |
| TG+GG | 1 | AG | 1 | TG+GG*AG | 1*1 = 1 | 43 |
| TT | 0 | GG | 2 | TT*GG | 0*2 = 0 | 136 |
| TG+GG | 1 | GG | 2 | TG+GG*GG | 1*2 = 2 | 32 |
Table 5.
Mean ΔFEV1 (relative % predicted) for each interaction term adjusted for two main effects and potential confounders including age, sex, Pre-FEV1, asthma duration, log transformed body mass index (log10 BMI), steroid use, regular or as needed bronchodilator use and individual ancestral proportions in Puerto Rican, Mexican and Puerto Rican and Mexican combined asthmatic subjects. The table also shows number of asthmatic subjects for each interaction term category (n), standard error (Std. error) for the mean, 95% confidence interval lower and upper bounds (lb, ub) and p-value for the association of interaction terms with ΔFEV1 (interaction term “0” is the reference).
| Interac-tion term* | Puerto Ricans | Mexicans | Puerto Ricans and Mexicans combined | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Mean ΔFEV1 | Std. error | lb | ub | p-value | n | M ean ΔFEV1 | Std. error | lb | ub | p-value | n | Mean ΔFEV1 | Std. error | lb | ub | p-value | |
| 0 | 225 | 8.92 | 0.82 | 7.31 | 10.54 | ref | 212 | 11.15 | 0.66 | 9.85 | 11.88 | ref | 437 | 10.04 | 0.55 | 8.96 | 11.11 | ref |
| 1 | 33 | 4.60 | 1.59 | 1.48 | 7.73 | 0.009 | 10 | 7.53 | 2.27 | 3.06 | 11.30 | 0.15 | 43 | 5.29 | 1.33 | 2.67 | 7.90 | 0.002 |
| 2 | 22 | 0.28 | 3.53 | -6.66 | 7.22 | 0.02 | 10 | 3.90 | 4.74 | -5.44 | 12.13 | 0.12 | 32 | 0.54 | 2.87 | -5.10 | 6.17 | 0.001 |
Interaction term “0” corresponds to the genotype combinations TT*AA, TG+GG*AA, TT*AG or TT*GG; interaction term “1” corresponds to genotype combination TG+GG*AG; and interaction term “2” corresponds to genotype combination TG+GG*GG as calculated in Table 3.
Functional Analysis of GSNOR Risk Haplotype
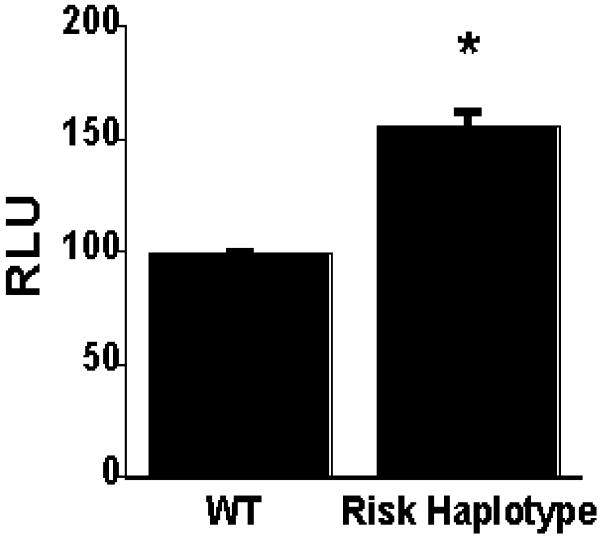
The 3′UTR of a gene is important for mRNA maturation and stability. Therefore, we reasoned that the GSNOR 3′UTR SNPs may affect expression levels of GSNOR mRNA and protein. To show that the GSNOR 3′UTR construct was functional, we cloned the 3′UTR into the PGL plasmid in both forward and reverse orientation. Only the 3′UTR inserted in the forward orientation demonstrated luciferase activity in HEK cells (data not shown). Moreover, the luciferase reporter activity from the construct containing the risk haplotype of GSNOR 3′UTR, was significantly higher than that from the wild type haplotype (Figure 1). These data suggest that the risk haplotype of GSNOR 3′UTR is a gain of function polymorphism.
Figure 1.
Luciferase activities in HEK293 transfected cells. HEK293 cells were transiently transfected with the GSNOR wild type (WT) and risk haplotype vectors. Cells were harvested 48 hours after transfection. Protein extracts from cells were analyzed in a luciferase assay. The increased luciferase activity of the GSNOR 3′UTR risk haplotype construct was statistically significant (p < 0.009 vs. wild type)*. Values are means ± standard error; n = 6 per group. (Relative luciferase units, RLU) reported were calculated by normalizing firefly luminescence to Renilla luminescence.
Discussion
In this study, we identified SNPs in the promoter and the 3′UTR region of the GSNOR gene that are associated with increased risk of asthma in Puerto Rican subjects, as well as gene-gene interactions between GSNOR and β2AR that predict responsiveness to albuterol in both Mexicans and Puerto Ricans. Our functional studies further suggest that the GSNOR 3′ UTR risk haplotype may represent a gain of function mutation, consistent with our discovery of increased GSNOR activity in mild asthmatics.(20)
The results of association analysis between SNPs in the GSNOR gene and asthma and related phenotypes in our Puerto Rican subjects support the importance of GSNOR in asthma and responsiveness to albuterol. Also it is interesting to note that the impact of GSNOR on response to albuterol appears to be ameliorated by use of inhaled corticosteroids. These results are consistent with previous work published by our group and by that of Gaston and colleagues showing SNOs protect from β2 agonist desensitization(21) and steroids decrease GSNOR protein expression(36) If decreased GSNOR expression results in decreased GSNOR activity, then steroid therapy may increase SNO bioavailability and decrease desensitization to β2 agonists in asthma. We did not find an association between GSNOR genetic variants and IgE levels or emergency department visits (data not shown) in either Mexican or Puerto Rican asthmatics. This is in agreement with the murine model of allergic asthma in GSNOR deficient mice that shows uncoupling of airway hyperresponsiveness and airway inflammation. No differences in levels of interleukin 13, IgE, mucus cell hyperplasia or other markers of inflammation between wild type and GSNOR deficient mice were observed.(19)
Although only one of the associations (SNP +17059 and asthma) between SNPs in the GSNOR gene and asthma and response to albuterol remained significant after correction for multiple testing, we performed multiple tests based on strong a priori hypothesis that GSNOR may play a role in asthma and bronchodilator responsiveness based on published literature.(19, 23) Therefore, it may not be appropriate to disregard prior information on the role of GSNOR in asthma by using overly conservative correction methods for multiple testing such as Bonferroni correction. Wu et al previously described an association between the GSNOR gene and asthma in Mexicans.(23) There may be several reasons why we did not find an association between GSNOR genetic variants and asthma in our Mexican subjects. The study by Wu et al included 532 Mexican case-parent asthma trios and therefore had higher statistical power compared to our study which only analyzed 273 Mexican trios. In addition, 4 out of 5 SNPs analyzed in the two studies are different due to differences in selection criteria for SNPs for genotyping. Four of the 5 SNPs genotyped by Wu et al are intronic whereas SNPs analyzed in our study are in the promoter and 3′ UTR region of the GSNOR gene. The SNP +3207 in the promoter region that was genotyped by Wu et al and also in our study was not associated with asthma in Mexican subjects in either study.(23)
We have previously provided evidence for an ethnic-specific association of β2AR Gly allele at codon 16 with lower bronchodilator responsiveness in our Puerto Rican asthmatics, but not in Mexican asthmatics.(11, 14) Herein, we found evidence of a pharmacogenetic gene-gene interaction between genetic variants of the GSNOR and β2AR genes, which results in lower responsiveness to albuterol in both Mexican and Puerto Rican asthmatics. The interaction between the genetic variants of GSNOR and β2AR genes is consistent with the emerging molecular and mechanistic understanding that these genes share a common signal-transduction pathway which mediates bronchodilator responsiveness (i.e. β2AR signaling).(21) The evidence of gene-gene interaction between GSNOR and β2AR genes can help explain inconsistency in association of β2AR genetic variants with responsiveness to albuterol in previous studies. These results suggest that two-locus genotype analysis of GSNOR and β2AR may be a better predictor of responsiveness to albuterol than single locus analysis.
We studied gene-gene interaction between GSNOR and β2AR for association with acute bronchodilator responsiveness to albuterol. In our Mexican and Puerto Rican GALA cohort we did not study the effect of chronic use of short acting β2-agonists and therefore we could not assess the effect of tachyphylaxis. It has been shown in some studies that the β2AR +46 AA genotype (Arg/Arg at codon 16), which is associated with better acute response to bronchodilator, may result in poor long term response due to desensitization of β2AR (tachyphylaxis). In mice, tracheal rings from GSNOR knockout mice maintain relaxation upon repeated exposure to the bronchodilator isoproterenol, whereas wild type mice demonstrate a decrease in responsiveness to treatment.(19) Protection from desensitization appears to occur through S-nitrosylation of G-protein kinase 2 (GRK2) by SNO(21), which results in decreased GRK2-mediated downregulation of β2AR by its phosphorylation. Therefore it would be interesting to extend these gene-gene interaction analyses between GSNOR and β2AR to randomized clinical studies of long-term bronchodilator use.
Given the strength of the association in the Latino population, a specific two-locus genotype may indicate the need for individualized pharmacologic therapy with GSNOR inhibitors and β2AR agonists, or additional adjuvant therapy. Likewise, Salpeter et. al. suggested that African American asthmatics receive additional therapy above and beyond therapies recommended by National Asthma Guidelines.(37) Patients with the combined risk alleles at the GSNOR and β2AR loci may represent a subgroup of asthmatics that are at risk for worse asthma outcomes. Further studies need to be performed to determine if genotyping at these loci can be used to tailor pharmacologic therapy to this patient subset in the future.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the families and the patients for their participation. The authors would also like to thank the numerous health care providers and community clinics for their support and participation in the GALA Study. The authors would like to especially thank Jeffrey M. Drazen, M.D., Scott Weiss, M.D., Ed Silverman, M.D., Ph.D., Homer A. Boushey, M.D. and Jean G. Ford, M.D. for all of their effort towards the creation of the GALA Study.
Supported By: This work was supported by National Institutes of Health (HL078885, HL088133, U19 AI077439, ES015794), Flight Attendant Medical Research Institute (FAMRI), and the RWJ Amos Medical Faculty Development Award to EGB), HL086887 to LGQ, National Institute of Environmental Health Sciences 5U19-ES012496 and NHLBI R01HL96673-01 to JSS, American Thoracic Society “Breakthrough Opportunities in Lung Disease” (BOLD) Award and Tobacco-Related Disease Research Program New Investigator Award (15KT-0008) to SC, the Ernest S. Bazley Grant to PCA, and the Sandler Center for Basic Research in Asthma and the Sandler Family Supporting Foundation.
References
- 1.Nelson HS. Beta-adrenergic bronchodilators. N Engl J Med. 1995;333:499–506. doi: 10.1056/NEJM199508243330807. [DOI] [PubMed] [Google Scholar]
- 2.Palmer LJ, Silverman ES, Weiss ST, Drazen JM. Pharmacogenetics of asthma. Am J Respir Crit Care Med. 2002;165:861–866. doi: 10.1164/ajrccm.165.7.2109096. [DOI] [PubMed] [Google Scholar]
- 3.Eggleston PA, Malveaux FJ, Butz AM, Huss K, Thompson L, Kolodner K, Rand CS. Medications used by children with asthma living in the inner city. Pediatrics. 1998;101:349–354. doi: 10.1542/peds.101.3.349. [DOI] [PubMed] [Google Scholar]
- 4.Finkelstein JA, Lozano P, Farber HJ, Miroshnik I, Lieu TA. Underuse of controller medications among medicaid-insured children with asthma. Arch Pediatr Adolesc Med. 2002;156:562–567. doi: 10.1001/archpedi.156.6.562. [DOI] [PubMed] [Google Scholar]
- 5.Burchard EG, Avila PC, Nazario S, Casal J, Torres A, Rodriguez-Santana JR, Toscano M, Sylvia JS, Alioto M, Salazar M, et al. Lower bronchodilator responsiveness in puerto rican than in mexican subjects with asthma. Am J Respir Crit Care Med. 2004;169:386–392. doi: 10.1164/rccm.200309-1293OC. [DOI] [PubMed] [Google Scholar]
- 6.Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br Med Bull. 2000;56:1054–1070. doi: 10.1258/0007142001903535. [DOI] [PubMed] [Google Scholar]
- 7.Naqvi M, Thyne S, Choudhry S, Tsai HJ, Navarro D, Castro RA, Nazario S, Rodriguez-Santana JR, Casal J, Torres A, et al. Ethnic-specific differences in bronchodilator responsiveness among african americans, puerto ricans, and mexicans with asthma. J Asthma. 2007;44:639–648. doi: 10.1080/02770900701554441. [DOI] [PubMed] [Google Scholar]
- 8.Palmer LJ, Celedon JC, Chapman HA, Speizer FE, Weiss ST, Silverman EK. Genome-wide linkage analysis of bronchodilator responsiveness and post-bronchodilator spirometric phenotypes in chronic obstructive pulmonary disease. Hum Mol Genet. 2003;12:1199–1210. doi: 10.1093/hmg/ddg125. [DOI] [PubMed] [Google Scholar]
- 9.Contopoulos-Ioannidis DG, Manoli EN, Ioannidis JP. Meta-analysis of the association of beta2-adrenergic receptor polymorphisms with asthma phenotypes. J Allergy Clin Immunol. 2005;115:963–972. doi: 10.1016/j.jaci.2004.12.1119. [DOI] [PubMed] [Google Scholar]
- 10.Yu IW, Bukaveckas BL. Pharmacogenetic tests in asthma therapy. Clin Lab Med. 2008;28:645–665. doi: 10.1016/j.cll.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Choudhry S, Ung N, Avila PC, Ziv E, Nazario S, Casal J, Torres A, Gorman JD, Salari K, Rodriguez-Santana JR, et al. Pharmacogenetic differences in response to albuterol between puerto ricans and mexicans with asthma. Am J Respir Crit Care Med. 2005;171:563–570. doi: 10.1164/rccm.200409-1286OC. [DOI] [PubMed] [Google Scholar]
- 12.Martinez FD, Graves PE, Baldini M, Solomon S, Erickson R. Association between genetic polymorphisms of the beta2-adrenoceptor and response to albuterol in children with and without a history of wheezing. J Clin Invest. 1997;100:3184–3188. doi: 10.1172/JCI119874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Israel E, Drazen JM, Liggett SB, Boushey HA, Cherniack RM, Chinchilli VM, Cooper DM, Fahy JV, Fish JE, Ford JG, et al. The effect of polymorphisms of the beta(2)-adrenergic receptor on the response to regular use of albuterol in asthma. Am J Respir Crit Care Med. 2000;162:75–80. doi: 10.1164/ajrccm.162.1.9907092. [DOI] [PubMed] [Google Scholar]
- 14.Tsai HJ, Shaikh N, Kho JY, Battle N, Naqvi M, Navarro D, Matallana H, Lilly CM, Eng CS, Kumar G, et al. Beta 2-adrenergic receptor polymorphisms: Pharmacogenetic response to bronchodilator among african american asthmatics. Hum Genet. 2006;119:547–557. doi: 10.1007/s00439-006-0169-2. [DOI] [PubMed] [Google Scholar]
- 15.Burchard EG, Ziv E, Coyle N, Gomez SL, Tang H, Karter AJ, Mountain JL, Perez-Stable EJ, Sheppard D, Risch N. The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med. 2003;348:1170–1175. doi: 10.1056/NEJMsb025007. [DOI] [PubMed] [Google Scholar]
- 16.Stamler JS. S-nitrosothiols in the blood: Roles, amounts, and methods of analysis. Circ Res. 2004;94:414–417. doi: 10.1161/01.RES.0000122071.55721.BC. [DOI] [PubMed] [Google Scholar]
- 17.Gaston B, Sears S, Woods J, Hunt J, Ponaman M, McMahon T, Stamler JS. Bronchodilator s-nitrosothiol deficiency in asthmatic respiratory failure. Lancet. 1998;351:1317–1319. doi: 10.1016/S0140-6736(97)07485-0. [DOI] [PubMed] [Google Scholar]
- 18.Dweik RA, Comhair SA, Gaston B, Thunnissen FB, Farver C, Thomassen MJ, Kavuru M, Hammel J, Abu-Soud HM, Erzurum SC. No chemical events in the human airway during the immediate and late antigen-induced asthmatic response. Proc Natl Acad Sci U S A. 2001;98:2622–2627. doi: 10.1073/pnas.051629498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Que LG, Liu L, Yan Y, Whitehead GS, Gavett SH, Schwartz DA, Stamler JS. Protection from experimental asthma by an endogenous bronchodilator. Science. 2005;308:1618–1621. doi: 10.1126/science.1108228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Que LG, Yang Z, Stamler JS, Lugogo NL, Kraft M. S-nitrosoglutathione reductase: An important regulator in human asthma. Am J Respir Crit Care Med. 2009;180:226–231. doi: 10.1164/rccm.200901-0158OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whalen EJ, Foster MW, Matsumoto A, Ozawa K, Violin JD, Que LG, Nelson CD, Benhar M, Keys JR, Rockman HA, et al. Regulation of beta-adrenergic receptor signaling by s-nitrosylation of g-protein-coupled receptor kinase 2. Cell. 2007;129:511–522. doi: 10.1016/j.cell.2007.02.046. [DOI] [PubMed] [Google Scholar]
- 22.Choudhry S, Liu L, Gonzalez Burchard E. Genetic association of s-nitroso-glutathione reductase with asthma. Nitric Oxide-Biology and Chemistry. 2006;14:A64–65. [Google Scholar]
- 23.Wu H, Romieu I, Sienra-Monge JJ, Estela Del Rio-Navarro B, Anderson DM, Jenchura CA, Li H, Ramirez-Aguilar M, Del Carmen Lara-Sanchez I, London SJ. Genetic variation in s-nitrosoglutathione reductase (gsnor) and childhood asthma. J Allergy Clin Immunol. 2007;120:322–328. doi: 10.1016/j.jaci.2007.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beckett WS, Belanger K, Gent JF, Holford TR, Leaderer BP. Asthma among puerto rican hispanics: A multi-ethnic comparison study of risk factors. Am J Respir Crit Care Med. 1996;154:894–899. doi: 10.1164/ajrccm.154.4.8887582. [DOI] [PubMed] [Google Scholar]
- 25.Lara M, Akinbami L, Flores G, Morgenstern H. Heterogeneity of childhood asthma among hispanic children: Puerto rican children bear a disproportionate burden. Pediatrics. 2006;117:43–53. doi: 10.1542/peds.2004-1714. [DOI] [PubMed] [Google Scholar]
- 26.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general u.S. Population. Am J Respir Crit Care Med. 1999;159:179–187. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- 27.Ott J. Analysis of human genetic linkage. Baltimore; 1999. [Google Scholar]
- 28.Chen X, Levine L, Kwok PY. Fluorescence polarization in homogeneous nucleic acid analysis. Genome Research. 1999;9:492–498. [PMC free article] [PubMed] [Google Scholar]
- 29.O'Connell JR, Weeks DE. Pedcheck: A program for identification of genotype incompatibilities in linkage analysis. American Journal of Human Genetics. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19(Suppl 1):S36–42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 31.Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: Application to asthma genetics. Genet Epidemiol. 2004;26:61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- 32.Dudbridge F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered. 2008;66:87–98. doi: 10.1159/000119108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai HJ, Kho JY, Shaikh N, Choudhry S, Naqvi M, Navarro D, Matallana H, Castro R, Lilly CM, Watson HG, et al. Admixture-matched case-control study: A practical approach for genetic association studies in admixed populations. Hum Genet. 2006;118:626–639. doi: 10.1007/s00439-005-0080-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choudhry S, Coyle NE, Tang H, Salari K, Lind D, Clark SL, Tsai HJ, Naqvi M, Phong A, Ung N, et al. Population stratification confounds genetic association studies among latinos. Hum Genet. 2006;118:652–664. doi: 10.1007/s00439-005-0071-3. [DOI] [PubMed] [Google Scholar]
- 35.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carraro S, Palmer L, DeRonde K, Brown-Steinke K, Zaman K, Que LG, Stamler JS, Gaston B. Upregulation of the asthma enzyme, s-nitrosoglutathione reductase by interleukin 13. Proceedings of the American Thoracic Society. 2006;3 [Google Scholar]
- 37.Salpeter SR, Buckley NS, Ormiston TM, Salpeter EE. Meta-analysis: Effect of long-acting beta-agonists on severe asthma exacerbations and asthma-related deaths. Ann Intern Med. 2006;144:904–912. doi: 10.7326/0003-4819-144-12-200606200-00126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.