Abstract
Capsule expression in Neisseria meningitidis is encoded by the cps locus comprised of genes required for biosynthesis and surface translocation. Located adjacent to the gene encoding the polysialyltransferase in serogroups expressing sialic acid-containing capsule, NMB0065 is likely a member of the cps locus, but it is not found in serogroups A or X that express non-sialic acid capsules. To further understand its role in CPS expression, NMB0065 mutants were created in the serogroups B, C and Y strains. The mutants were as sensitive as unencapsulated strains to killing by normal human serum, despite producing near wild-type levels of CPS. Absence of surface expression of capsule was suggested by increased surface hydrophobicity and confirmed by immunogold electron microscopy, which revealed the presence of large vacuoles containing CPS within the cell. GC-MS and NMR analyses of purified capsule from the mutant revealed no apparent changes in polymer structures and lipid anchors. Mutants of NMB0065 homologues in other sialic acid CPS expressing meningococcal serogroups had similar phenotypes. Thus, NMB0065 (CtrG) is not involved in biosynthesis or lipidation of sialic acid-containing capsule but encodes a protein required for proper coupling of the assembly complex to the membrane transport complex allowing surface expression of CPS.
Keywords: CAPSULE, SIALIC ACID, CAPSULAR POLYSACCHARIDE, NEISSERIA MENINGITIDIS
1. Introduction
Neisseria meningitidis is a Gram-negative encapsulated bacterium and is an important cause of septicemia and meningitis in humans [1]. Capsular polysaccharide (CPS) plays a crucial role in virulence by enabling the bacterium to evade complement-mediated and phagocytic killing and is the basis for immunological serogrouping. Thirteen serogroups have been described, six of which cause the majority of invasive disease (A, B, C, Y, X, and W-135) [2]. Four serogroups (B, C, Y and W-135) express capsules containing sialic acid. Serogroups B and C are homopolymers of N-acetylneuraminic acid in an α2→8 or an α2→9 linkage, respectively [3]. Serogroups Y and W135 are composed of alternating disaccharide repeat units of sialic acid and D-glucose or D-galactose, respectively [4]. Serogroup A is composed of (α1→6)-linked N-acetylmannosamine-1-phosphate [5], while serogroup X expresses (α1→4) linked N-acetyl-D-glucosamine 1-phosphate [6]. Like other pathogenic group II capsule-expressing bacteria, such as Escherichia coli K1, the genetic organization of the meningococcal capsule polysaccharide (cps) locus is comprised of three regions (Fig. 1). The meningococcal cps locus is a 24-kb virulence island of low G+C content [7]. Region A (synA-D) encodes proteins involved in sialic acid synthesis and elongation of the sialic acid polymer. The first three genes (synA-C) are highly homologous among sialic acid capsule-expressing serogroups, while the gene encoding the capsule polymerase is serogroup-specific [8]. Regions C (ctrA-D) encode proteins forming the capsule transport apparatus and are required for export of capsule polymers through the inner and outer membranes. CtrB/C/D belong to the superfamily of ATP-dependent (ABC) transport cassette, while CtrA is the designated outer membrane porin. Region B contains two genes, ctrE (lipA) and ctrF (lipB), which are also required for export of lipidated polymer to the meningococcal outer membrane [9].
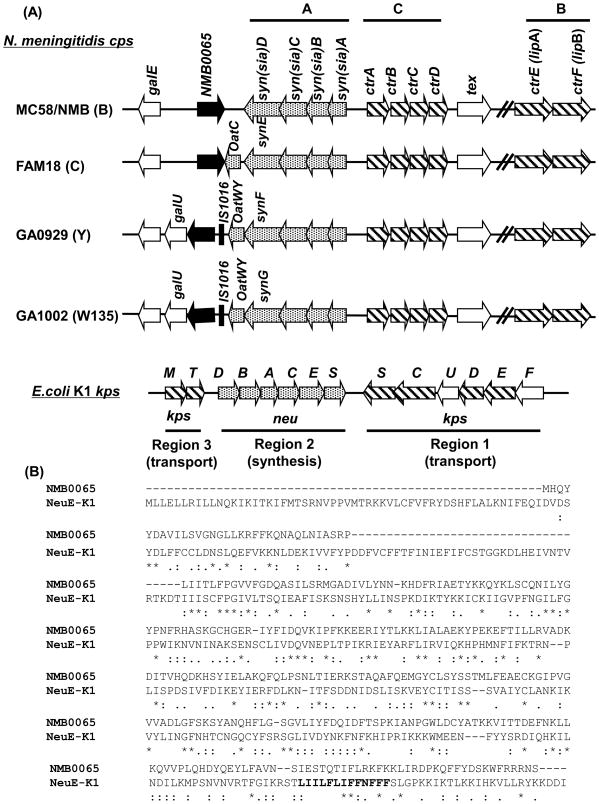
Fig. 1.
(A) Genetic organization of the N. meningitidis capsule gene complex (cps) and the E. coli K1 capsule (kps) locus. The genes responsible for capsular biosynthesis (dotted arrows, syn (sia)ABCD), capsule transport (hatched arrows, ctrABCDEF; ctrEF were formerly known as lipA and lipB), and the E. coli K1 homologues of each are shown. The location of NMB0065 (black arrow) within the cps complex in the serogroup B strain NMB was similar to that of the published serogroup B MC58 genome sequence[45]. An IS1016-like transposase (indicated as a black bar) is present upstream of the NMB0065 homologue in serogroups Y and W135. OatC and OatWY are O-acetyltransferases specific for capsules of serogroup C and groups Y and W135, respectively. (B) The Clustal W protein sequence alignment of NMB0065 and NeuE [46]. The predicted polyprenyl-binding motif (PIRS) motif sequence in NeuE is highlighted in bold.
Located immediately downstream and transcribed in the opposite direction of synD (Fig. 1), the gene NMB0065 has been implicated as a member of the cps locus. A random Tn10 transposon mutant that was unable to induce septicemia in an infant rat model was mapped to the NMB0065 gene [10]. In addition, Himar1 mariner random mutagenesis of a serogroup C strain, 8013, identified a mutation in the serogroup C homologue of NMB0065 that caused increased susceptibility to complement-mediated lysis and reduced capsule expression [11]. These data suggest that NMB0065 may be an uncharacterized determinant of CPS biosynthesis or CPS transport. This study was conducted to understand the role of NMB0065 in meningococcal CPS expression.
2. Materials and methods
2.1. Bacterial strains, plasmids and media
The strains and plasmids used in this study are listed in Table 1. All meningococcal strains were grown on GC base agar (Difco) supplemented with 0.4 % glucose and 0.68 mM Fe(NO3)3 at 37°C with 5% CO2. Meningococcal mutants with kanamycin selection were grown on brain heart infusion base agar (Becton Dickinson) containing 1.25% fetal bovine serum (Gibco-BRL). Liquid cultures were grown in GC broth with the same supplements and 0.43% NaHCO3. E. coli strains were grown in Luria-Bertani (LB) broth. Antibiotics (μg/mL) used for meningococci were: kanamycin, 80; erythromycin, 3, and for E. coli were: kanamycin, 50; ampicillin, 50; erythromycin, 350. Monoclonal antibodies against the meningococcal serogroup B capsule (2-2-B), the serogroup C capsule (4-2-C) and the serogroup Y capsule (5-2-Y) were generous gifts of Wendell Zollinger (Walter Reed Army Institute of Research).
TABLE 1.
Strains and plasmids used in this study.
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| N. meningitides | ||
| NMB | B:2b:P1.2,5:L2 (CDC 8201085) (serogroup B) | [47] |
| M7 | synA::Tn916, unencapsulated derivative of NMB | [47] |
| RH2.2 | NMB with chromosomal NMB0065::aphA3 mutant | This study |
| RH4.1 | NMB0065::aphA3 mutant carrying pRH18 | This study |
| RH3.1 | FAM18 (serogroup C) with chromosomal NMB0065::aphA3 mutant | This study |
| FAM18::M7 | Unencapsulated derivative of FAM18 | This study |
| RH5 | GA0929 (serogroup Y) with chromosomal NMB0065::aphA3 mutant | This study |
| GA0929::M7 | Unencapsulated derivative of GA0929 | This study |
| E. coli | ||
| Top10 | Cloning strain | Invitrogen |
| Plasmids | ||
| pCR2.1 | TA cloning vector | Invitrogen |
| pUC19 | Cloning vector | Invitrogen |
| pUC18k | Source of aphA3 cassette | [12] |
| pYT250 | Meningococcal shuttle vector | [48] |
| pYT328 | lacZ fusion vector | [13] |
| pBADphoA | Vector for construction of phoA fusion | [49] |
| pRH5 | pUC19 with XbaI/KpnI fragment of internal fragment of NMB0065 amplified with primers RH014 and RH019 in pCR2.1 | This study |
| pRH6 | SmaI digested aphA3 cassette inserted into the BstBI site of pRH5 | This study |
| pRH18 | The KpnI/XbaI fragment of 5′ region and full length of NMB0065 amplified with primers RH035 and RH040 in pCR2.1 cloned into pYT250 shuttle vector | This study |
| pRH11 | pUC19 with the XbaI/KpnI fragment of NMB0065 fragment amplified with primers RH014 and RH019 from FAM18 | This study |
| pRH12 | SmaI digested aphA3 cassette inserted into the BstBI site of pRH11 | This study |
| pRH13 | pUC19 with XbaI/KpnI fragment of NMB0065 amplified with primers RH030 and RH031 from GA0929 | This study |
| pRH15 | pRH13 with engineered NaeI site within the NMB0065 fragment | This study |
| pRH23 | SmaI digested aphA3 cassette inserted into the NaeI site of pRH15 | This study |
| pYT415 | Full length NMB0065 in KpnI site of pBADphoA | This study |
| pYT416 | Full length NMB0065 in HindIII site of pBADphoA | This study |
| pYT417 | CtrC1–145 in KpnI site of pBADphoA | This study |
| pYT427 | A 397-bp ctrG::lacZ fusion in pYT328 | This study |
2.2. Construction of NMB0065 mutants and ctrG::lacZ reporter strain
An internal coding region was amplified by PCR with primers RH014 (5′-CAATATTATGACGCAGTAATTTTATCGG-3′) and RH019 (5′-CCGGATTTGCTATTTTTGGG-3′) with NMB chromosomal DNA as the template. The resulting PCR fragment was cloned into pCR2.1 (Invitrogen) and sub-cloned into pUC19 using KpnI/XbaI digestion to yield pRH5. A SmaI fragment of pUC18K containing the aphA3 cassette [12] was inserted into the unique BstBI site of pRH5 blunted with Klenow DNA polymerase (New England Biolabs) to yield pRH6 with the aphA3 cassette inserted at 285 bp of the 927-bp NMB0065 coding sequence. pRH6 was used to transform wild-type NMB cells and kanamycin-resistant colonies were selected. The mutation was confirmed by PCR using an aphA3-specific primer (KanA: 5′-CTTAGCAGGAGACATTCCTTCCG-3′) and a NMB0065-specific primer (RH035: 5′-CTTTGATAGATTGATAATAATGGTTG-3′).
Similar mutations were also constructed in the serogroups C and Y strains. Primers RH014 and RH019 were used to amplify the serogroup C homologue from FAM18 chromosomal DNA and primers RH030 (5′-GATATTGTTATTCTATCATTAGG-3′) and RH031 (5′-CAACAAAGAATTGCTTAGC-3′) were used for serogroup Y (GA0929). Construction of the FAM18 mutation was performed as described above. A unique NaeI site was engineered into the serogroup Y (GA0929) NMB0065 homologue (pRH13) using Quickchange site-directed mutagenesis (Stratagene) with primers RH038 (5′-GCACTGGCTAATCGCCGGCCACATAAAGATTTAGTAC-3′) and RH039 (5′-GTACTAAATCTTTATGTCGGCCGGCGATTAGCCAGTGC-3′) to create pRH15. The SmaI released aphA3 cassette from pUC18k was cloned into the blunt NaeI site to create pRH23. Transformation of the parent strains (FAM18 and GA0929) was performed as described above.
A 397-bp fragment of the NMB0065 promoter region was obtained by PCR amplification using primers RH035-ER (5′-cggaattcCTTTGATAGATTGATAATAATGGTTG-3′) and RH014-R-ER (5′-cggaattcCCGATAAAATTACTGCGTCATAATATTG-3′, EcoRI site underlined), and chromosomal DNA of serogroup C strain FAM18 as template. The PCR product was digested with EcoRI and the released insert was purified and cloned into the EcoRI site of pYT328 [13] to generate transcriptional fusion to the lacZ gene that is flanked by meningococcal sequences of NMB0428 and NMB0430. Correct orientation of the promoter relative to the lacZ gene was confirmed by colony PCR using a lacZ 5′ outward primer, YT168, and a forward primer within the cloned promoter fragment. The resulting plasmid, pYT427, was confirmed by sequencing analysis and then digested with NcoI for linearization and transformed into N. meningitidis NMB. Transformants (NMB427) were selected on GC/erm plates and the integration of the lacZ fusion via homologous recombination into an irrelevant intergenic region verified by PCR.
2.3. Complementation of the serogroup B NMB0065 mutant
Primers RH035 (5′-CTTTGATAGATTGATAATAATGGTTG-3′) and RH040 (5′-CGCTTTATATTAAATCACCTTTCTCAACC-3′) were used to amplify a 1,324-bp fragment from strain NMB that contains 362-bp upstream sequence of the NMB0065 start codon and the entire coding sequence of NMB0065. The PCR product was cloned into pCR2.1 and the insert was released by KpnI/XbaI digestion and subsequently cloned into the KpnI/XbaI site of the meningococcal shuttle vector pYT250 [14] to yield pRH18. The plasmid was methylated with HaeIII methylase (New England Biolabs) according to manufacturer’s instructions and used to transform the NMB0065::aphA3 mutant (RH2.2). Kanamycin-resistant/erythromycin-resistant colonies were selected and analyzed by PCR. The presence of the original mutation and an intact copy of the complemented gene on the plasmid were confirmed with chromosome-specific and vector-specific primers, respectively. The complemented mutant strain is referred to as RH4.1.
2.4. NMB0065-phoA fusions and PhoA activity assay
The coding sequence of NMB0065 was PCR amplified with primers B65-F-KpnI (5′-GGGGTACCGCATCAATATTATGACGCAG-3′) and B65-R-KpnI (5′-AAGGTACCCTATTTCTTCTTCTAAACCATTTAG-3′) (KpnI underlined). The PCR product was digested with KpnI, and inserted at the 5′ end of phoA in KpnI-cleaved pBAD-phoA [15]. Colonies were screened by PCR using primers B65-F-KpnI and phoA-5-Rev (5′-GCAGAGCGGCAGTCTGATCA-3′) to identify the correctly orientated NMB0065 fragment and the in-frame fusion with phoA of the resulting plasmid, pYT415, was confirmed by sequencing. Similarly, a second phoA fusion in which the NMB0065 fragment was cloned to the 3′ end of phoA was constructed by cloning the NMB0065 coding sequence into the HindIII site of pBAD-phoA to yield pYT416. PCR amplification was performed with primers B65-F-HdIII (5′-TTAAAAGCTTGCATCAATATTATGACGCA-3′) and B65-R-HdIII (5′-TTTTAAGCTTCAACTATTTCTTCTTCTAAACC-3′) (HindIII site underlined). A positive fusion control, pYT417, was made by cloning the N-terminal 145 amino acid residues of CtrC and fused with the 5′ end of phoA in pBADphoA. The ctrC fragment containing three transmembrane domains with the C-terminus ended in periplasm was obtained by PCR using primers C145-F-KpnI (5′-CGGGGTACCGAAAGCCTTGCATAAAACATC-3′) and C145-R-KpnI (5′-CGGGGTACCGGCATTTCAATCCAG-3′).
DH5α strains carrying the phoA fusion plasmids or the empty vector were grown in LB broth supplemented with 0.2% glucose and induced with 0.2% arabinose. Aliquots (100 μl) of overnight culture were mixed with 900 μl of 0.2 M Tris, pH 8, chloroform (100 μl) and 0.1% SDS (50 μl), and the cells were permeabilized with pipetting. The PhoA activity was measured by incubation with p-nitrophenyl phosphate (50 μl, pNPP, Sigma liquid substrate system) at 37°C for 50 minutes. Absorbances at 420 nm and 550 nm were measured after adding 50 μl of 0.1 M EDTA. The activity was calculated as unit = 1000*(OD420−1.75*OD550)/(OD600*time*volume of cells) [16].
2.5. Western blots and subcellular fractionation
Cells from cultures used in the PhoA activity assays were collected by centrifugation. Equal numbers of cells corresponding to absorbance of 0.1 at OD600 were boiled in SDS-PAGE sample buffer and proteins separated on a 10% SDS-PAGE. Resolved proteins were transferred to nitrocellulose membranes using a semi-dry transfer apparatus (BioRad). The membranes were blocked in blocking buffer (5% non-fat dry milk in 1× TBS) at 4°C overnight, and then incubated with the monoclonal anti-PhoA antibody, (Chemicon, 1:10,000 dilution in 1× TBS with 0.1% Tween 20) at RT for 1 hr. After two 10-min washes, membranes were incubated with anti-mouse IgG-Horseradish peroxidase conjugate (Pierce) (diluted 1:25,000) for 1 hr at RT. After three washes, the membranes were developed with the ECL immunoblot system (Pierce). The soluble protein and total membrane protein fractions were separated by the method of Clark et al. [17] as previously described [18] from 50 ml culture of the induced pYT415-containing strain. Protein concentration was determined by BCA protein assay (Pierce) with BSA as standard.
2.6. Capsule ELISA, hydrophobic interaction chromatography and Serum bactericidal assay
The whole cell ELISA were performed as previously described [19] using monoclonal antibodies 2-2-B, 4-2-C and 5-2-Y for serogroups B, C and Y, respectively. The bacteria in 96-well plates were dried overnight, resulting in bacterial lysis and access to intracellular compartments of the cell, thus the assay detected total capsule. The lysate obtained by repeated freeze-thaw of bacteria gave similar results. The procedure of hydrophobicity test has been described previously [9].
A microdilution serum bactericidal assay was performed as described previously [20]. Briefly, cultures at mid-log phase were diluted in HEPES/MEM, pH 7.3, to 1 × 105 cfu/mL. Normal human serum was added to final concentrations of 10 or 25%. After incubation at 37°C for 30 min, 10 μL (in duplicate) of each well were plated and colonies were counted after overnight growth. Percentage survival was calculated by comparing to a no-serum control sample for each strain.
2.7. Electron microscopy
Liquid grown cultures were fixed in 2.5% glutaraldehyde for 2 hours. Thin sections of Epon-embedded samples on nickel 200-mesh grids coated with Formvar and carbon were etched with 0.1% H2O2, followed by a treatment with 1% metaperiodate. The grids were washed with PBS three times and incubated with 50 mM glycine in PBS, followed by incubation in the blocking solution (5% BSA, 0.1% gelatin, 5% normal mouse serum in PBS). After washing in 0.1% BSA-c (acylated BSA, Aurion) in PBS, the grids were incubated in primary antibody (2-2-B, 1:25 in 0.1% BSA-c) overnight at 4°C. Again the grids were washed and incubated for 1.5 hours at room temperature with the secondary antibody (10 nm gold-conjugated goat anti-mouse IgM, 1:20). The grids were washed, incubated with 2.5% glutaraldehyde in 0.1 M phosphate buffer and counterstained with lead citrate. The grids were viewed on a Hitachi H-7500 transmission electron microscope.
2.8. CPS purification, phospholipases treatment, electrophoresis and staining
Capsular polysaccharide was purified from strains NMB and RH2.2 according to the protocol of Gotschlich [21], modified as follows. Cells from overnight cultures were lysed by addition of 10% Cetavlon (hexadecyltrimethyl ammonium bromide) to a final concentration of 1% (w/v). The precipitate and bacterial debris were collected by centrifugation (11,000 × g for 15 min) and then resuspended in distilled water. To dissociate the polysaccharide-Cetavlon complex, 1 volume of 2 M CaCl2 was added, and the mixture was allowed to stir for 16 hours. Absolute ethanol was added to a final concentration of 25% to precipitate nucleic acids. After 2 hours, the precipitated nucleic acids were removed by centrifugation (25,000 × g for 20 min). The ethanol concentration of the supernatant was raised to 80% (v/v) to precipitate polysaccharide. This polysaccharide was collected by centrifugation (2,000 × g for 10 min), washed with absolute ethanol and resuspended in distilled water. This solution was incubated with 50 mg/mL DNase I and 100 mg/mL RNase A at 37°C for 2 hr, followed by incubation with 50 μg/mL proteinase K in 10 mM MgSO4 buffer at room temperature for 16 hr. The solution was dialyzed against water in a 3.5-kDa cut-off dialysis bag (Spectrum). After 24 hr dialysis, the buffer was changed to CTAB removal buffer (10 g NaCl, 1.8 g Tris base, 400 mL ethanol per liter) and the sample was dialyzed for additional 48 hr with one change. Following a further 48 hr dialysis against distilled water, the dialysate was lyophilized. Treatment procedures of purified CPS with phospholipase C to release the phospholipid have been described previously [9].
The deoxycholic acid (DOC)-containing polyacrylamide gel electrophoresis with Laemmli buffer system (DOC-PAGE) [22] and capsule visualization with Alcian Blue was used to analyze phospholipase treated CPS samples as previously described [9].
2.9. NMR and GC-MS analysis
The purified CPS samples were initially washed with 9:1 ethanol-water-mix to remove phosholipids. The glycosyl composition analysis of the purified CPS (CPS) samples was done by the preparation and gas chromatograph-mass spectrometry (GC-MS) of trimethylsilyl (TMS) methyl glycosides. In brief, the samples were methanolyzed (methanolic 1 M HCl at 80°C for 18 hr) to methyl glycosides followed by re-N-acetylation (pyridine and acetic anhydride in presence of methanol at 100°C for 1 hr). Finally, the re-N-acetylated sugars were derivatized using Tri-Sil reagent (Pierce) (80°C for 30 min) to give TMS methyl glycosides. The TMS methyl glycosides were analyzed on a Hewlett-Packard mass spectrometer (HP 5890) interfaced with a mass selective detector from HP (5970 MSD) using a DB-1 (30m × 0.25mm × 0.25 micrometer).
Nuclear magnetic resonance (NMR) analysis of the CPS samples was done using a 600 MHz Varian spectrometer. Each sample was initially exchanged twice with 99.8% Deuterium oxide (D2O) (Aldrich) and then was finally dissolved in 100% D2O (Cambridge Isotope Lab). The 1D proton and 2D proton-carbon correlation spectra were collected using standard pulse sequence provided by Varian.
3. Results
3.1 Genomic location and characterization of NMB0065
The open reading frame of NMB0065 is adjacent to but transcribed convergently from the biosynthesis cassette synA-D (Fig. 1A) in the serogroup B strain NMB. The serogroup C (FAM18) homologue of NMB0065 was 96% identical to the NMB sequence at the nucleotide level, while the serogroup Y (GA0929) and W-135 (GA1002) homologues were both 61% identical to the NMB sequence. However, the serogroup Y and W-135 NMB0065 homologues were 99% identical to each other and their transcriptional orientations were opposite to those of serogroups B and C (Fig. 1A). Interestingly, no homologue of NMB0065 is found in the serogroup A (Z2491) genome sequence [23] or serogroup X cps region [24]. Although homologous to many hypothetical proteins with unknown functions, the best characterized homolog is the NeuE protein encoded within the polysialic acid capsule locus of K1 E. coli [25–27]. Sequence alignment between the two proteins revealed the absence of the N-terminal ~90 residues of NeuE in NMB0065 (Fig. 1B) and no highly conserved motifs can be recognized despite similarity throughout the remaining sequence,
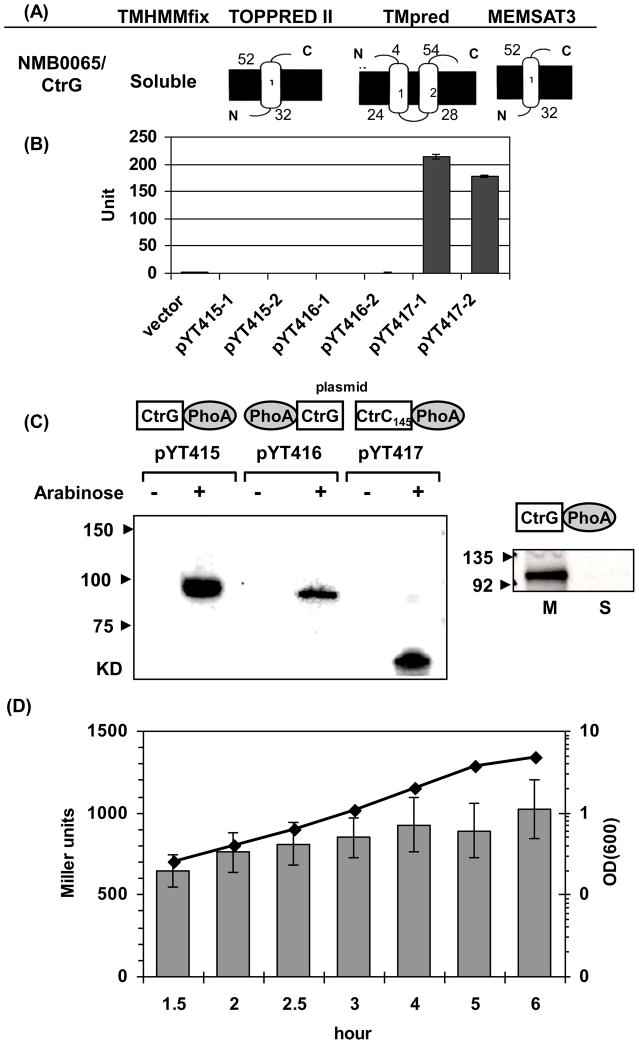
The NMB0065 protein is predicted to be a 318-aa protein of 37 KDa and contains no secretion leader sequence. Analyses of the protein sequence using several topology prediction algorithms [28–30] suggested that it is ether a soluble protein, contains a single transmembrane (TM) domain with its C-terminus projected into the periplasm, or contains two TM domains with both termini located in periplasm (Fig. 2A). To examine the topology of NMB0065, both N-terminal and C-terminal PhoA fusions of the full-length NMB0065 were constructed and examined in E. coli. No PhoA activity was detected for either fusion; while a positive control of CtrC-PhoA fusion yielded activity (Fig. 2B). Expression of the fusion proteins with an expected molecular weight was confirmed by Western blots using anti-PhoA monoclonal antibody (Fig. 2C). Thus, the data suggested that NMB0065 is a soluble protein located in the cytoplasm. To assess whether NMB0065 peripherally associated with the inner membrane, the soluble protein fraction and total membrane protein fraction of the E. coli strain expressing the C-terminal PhoA fusion (pYT415) was prepared. Western blot against PhoA antibody showed that NMB0065-PhoA protein exclusively associated with the membrane, indicating that NMB0065 likely associated peripherally with the cytoplasmic side of the inner membrane.
Fig. 2.
(A) Schematic topology predictions of NMB0065 using four algorithms. The inner membranes are shown as thick black lines and the residue numbers of the predicted transmembrane segments are indicated. (B) PhoA activity assays. Two independent plasmids for each construct were examined together with the empty vector control. The error bars represent the standard deviation of triplicate measurements. This is a representative of two independent experiments. (C) Western blots with anti-PhoA monoclonal antibody. Expression of PhoA fusion proteins in each plasmid construct (configurations shown above the plasmid name) was assessed with and without induction by 0.2% arabinose. Equal numbers of cells were examined in each sample (Left panel). Whole cell lysate of the E. coli strain expressing the NMB0065-PhoA fusion was separated into soluble protein fraction (S) and total membrane fraction (M) and probed with anti-PhoA antibody. Equal amount of proteins (20 μg) were loaded in each lane (Right panel). (D) The expression of NMB0065 is not growth phase dependent. The NMB0065::lacZ reporter integrated at a permissive locus in strain NMB427 was grown in GC broth and samples collected at various growth phases. Data presented are the mean values and standard deviations of four independent cultures. The line graph shows the OD600 values, while the corresponding β-galactosidase activities in Miller units were shown as gray bars. No significant difference in transcription was noted from the exponential to the early stationary phase.
The genetic location and orientation of the coding sequence suggested that NMB0065 is most likely independently transcribed. To investigate its transcription profile, we generated a single copy lacZ fusion integrated at a permissive chromosomal locus, and monitored its transcriptional activity throughout the growth phase. As shown in Fig. 2D, NMB0065 was constitutively expressed with no apparent growth phase dependent changes.
3.2. The meningococcal NMB0065 mutant was sensitive to complement-mediated killing of normal human serum (NHS)
Whole-cell ELISAs that detect total CPS were performed with an anti-serogroup B capsular monoclonal antibody and the NMB0065 mutant produced a level of CPS similar to the wild-type parent (90.6±16.3%). Multiple independent NMB0065 transformants were assayed, and similar results were obtained (data not shown). Likewise, the NMB0065 mutants of a serogroup C strain (FAM18) and a serogroup Y strain (GA0929) yielded near wild type levels of CPS compared to the corresponding parental strains (93.5±20.6% and 125.5±12.0%, respectively).
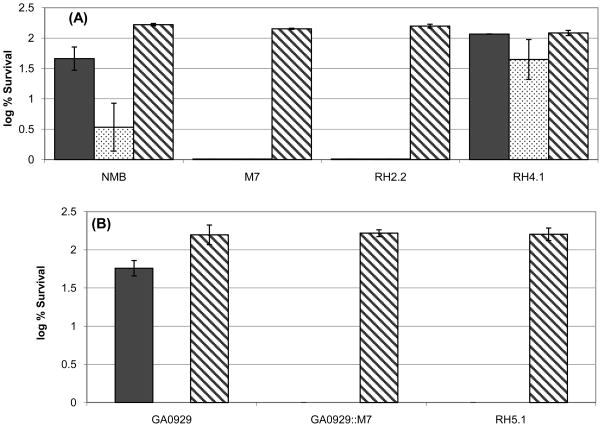
Encapsulated meningococcal strains can resist complement-mediated killing of NHS [20] as the unencapsulated mutant (M7) was completely killed at 10% NHS, while the wild type parent strain survived. The NMB0065 mutant was completely killed by 10% NHS, despite producing near wild-type levels of total CPS (Fig. 3A). The bactericidal activity of human serum against M7 and the NMB0065 mutant was due to complement-mediated lysis, as all strains survived in heat-inactivated serum. Thus, the CPS produced by the NMB0065 mutant failed to protect meningococci against the complement-mediated bactericidal activity of NHS. Data obtained for the serogroup Y and C mutants were similar to the serogroup B observations (Fig. 3B and data not shown).
Fig. 3.
Bactericidal activity of normal human serum (NHS) against serogroup B and Y meningococci. Serum bactericidal assays were performed with serogroup B, and Y parent strains (NMB and GA0929), the corresponding NMB0065 mutant strains (RH2.2 and RH5.1) and the complemented serogroup B RH4.1 strain using 10% (black bars) and 25% (dotted bars) of NHS. The unencapsulated biosynthesis-deficient mutants were used as negative controls. Heat-inactivated serum (56°C for 30 min, 25%, hatched bars) was tested to confirm that serum bactericidal activity was due to complement-mediated lysis. Panel A, serogroup B (NMB), panel B, serogroup Y (GA0929). Data presented are the averages and standard deviations of two independent experiments.
Complementation was performed to ensure that the observed NHS sensitivity of the NMB0065 mutant was caused by the mutation. A meningococcal shuttle vector carrying a fragment that contains the 363-bp upstream sequence and the entire coding region of NMB0065 from strain NMB was transferred into the NMB0065 mutant (RH2.2) by transformation. As shown in Fig. 3A, resistance to killing by NHS was restored by complementation of the NMB0065 mutation (strain RH4.1) to higher levels than those of the wild type, possibly due to an elevated expression of NMB0065 from the complementation plasmid. Thus, these data confirmed that the loss of serum resistance was caused by the NMB0065 mutation.
3.3. CPS produced by the NMB0065 mutant was not expressed on the cell surface
The serum sensitivity of the NMB0065 mutant suggested that the CPS was likely not transported to the cell surface. Since the meningococci expressing highly anionic polysialic acid capsule display more hydrophilic surface compared to unencapsulated meningococci [9]. Surface hydrophobicity assay with Octyl-sepharose column was used to assess whether capsule was present on the cell exterior [31]. For the encapsulated strain, ~40% of cells (39.0±7.6%) retained on the Octyl-sepharose column, while absorption onto the column was near 100% for the unencapsulated strain M7 (97.9±1.0%), demonstrating increased hydrophobicity of unencapsulated meningococci. The NMB0065 mutant behaved similarly (90.5±6.3%) to the unencapsulated M7 strain and this enhanced hydrophobicity can be reversed by complementation (22.0±4.0%). These results suggested that there was limited or no CPS present on the surface of the NMB0065 mutant. Likewise, the serogroup C and Y NMB0065 mutants displayed enhanced surface hydrophobicity when compared to the wild type parent strains (92.7±9.7% vs. 42.9±7.1% and 98.0±2.7% vs. 13.5±6.5%, respectively). Thus, consistent with the results of the serum bactericidal assays, little or no CPS was expressed on the cell surface in these NMB0065 mutants.
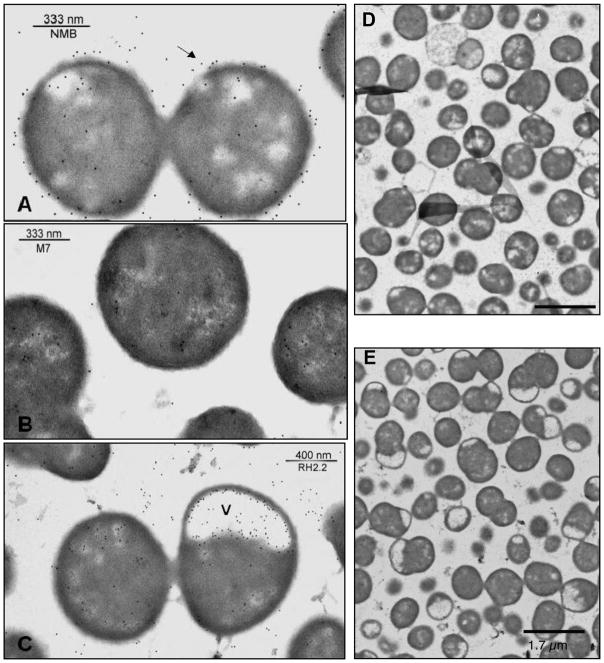
Immunogold labeling electron microscopy with the serogroup B-specific 2-2-B monoclonal antibody was conducted to further assess the location of the capsule of the NMB0065 mutant. As shown in Fig. 4, surface labeling of the antibody was demonstrated in the wild type serogroup B parent strain NMB, but not the capsule-deficient mutant M7, or the NMB0065 mutant (panel A versus panels B and C). Immunogold staining of cell cross-sections revealed large electron-translucent zones, or lacunae, at the poles of the NMB0065 mutant that were labeled with the gold particles (Fig. 4, panels C and E). Electron-translucent zones similar to those seen in the NMB0065 mutant have been observed in many capsule transport-deficient mutants of E. coli K1 and K5 strains [32–34], as well as N. meningitidis [9]. Immunogold labeling of CPS was abundant within the electron-translucent areas of the NMB0065 mutant. The complemented mutant showed both capsule labeling on the surface of the cell and the presence of small lacunae in some cells (data not shown). However, sufficient capsular material was present on the cell surface to restore serum resistance (Fig. 3) and altered the hydrophobicity profile of the bacterial surface.
Fig. 4.
Electron photomicrographs of immunogold-labeled meningococcal cell sections. Panel (A) shows the wild-type parental strain NMB, (B) shows the unencapsulated mutant (synA) M7, and (C) shows the NMB0065 mutant. Labeling was performed with the serogroup B capsule-specific monoclonal antibody, 2-2-B, as the primary antibody, and the 10-nm gold-conjugated anti-immunoglobulin G/M antibody was used as the secondary antibody. The arrows indicate the capsule surrounding the wild-type strain but not the M7 or the NMB0065 mutant. The (v) indicates electron-translucent vacuoles containing internalized capsule of the NMB0065 mutant. Overviews of the wild type strain and the NMB0065 mutant are shown in Panels D and E, respectively.
3.4 The CPS structure of the NMMB0065 mutant was identical to that of the wild-type parent
CPS was purified to determine whether the CPS structure of the NMB0065 mutant was different from that produced by the wild-type parent. Purified capsular polysaccharides were treated with phospholipase C, which removes the glycerophospholipid anchor of capsule polymers [9], and then separated on DOC-PAGE along with the untreated CPS control. The presence of diacylglycerophosphatidic acid on CPS causes the formation of larger micelles that migrate more slowly during electrophoresis compared with polymers that are not lipidated. Phospholipase C digestion resulted in the release of the lipid moiety and the observed pattern for the NMB0065 mutant was identical to that of the wild type polysaccharide (data not shown). Furthermore, comparison of the nonlipidated capsule polymers resolved by DOC-PAGE revealed no discernible difference in polymer length between the mutant and the wild type parent. Together, these data suggest that the polysaccharide of the NMB0065 mutant was lipidated and that the NMB0065 protein did not control the length of the sialic acid polymer.
A combination of GC-MS and NMR analyses were performed to gain further detailed comparison of the polysaccharide structures. After extraction of the CPSs with 9:1 ethanol-water to remove phospholipids, compositional analysis of the methanolyzed and trimethylsilyated CPSs using GC-MS was performed and revealed identical chromatographs (data not shown). The major sugar was found, as expected, to be N-acetylneuraminic acid (NANA). Proton NMR analysis was performed on the same purified CPS samples after exchanging with Deuterium oxide (D2O). NMR spectra showed no major differences between the wild type and mutant capsules, and, once again, NANA was the major constituent in both spectra (data not shown). In combination, these data indicate that there was no difference in the structures of the capsules of the serogroup B wild type strain and the NMB0065 mutant.
4. Discussion
Encapsulated N. meningitidis expressing sialic acid capsules of serogroups B, C, Y or W-135 remain a major cause of epidemic meningitis and septicemia globally [1]. Capsular polysaccharides are the outermost structure of the bacterial surface and as such play a key role in interactions between the pathogen and the host [35, 36]. Sialic acid capsules expressed by meningococci permit evasion of host responses such as complement-mediated bacteriolysis and phagocytosis [20, 35] and are important in bacterial transmission and colonization [37–40].
The meningococcal NMB0065 was first identified in mutational analyses as required for septicemia in an infant rat model [10]. Another mutagenesis study searching for genes conferring resistance to complement-mediated killing identified a mutant of the serogroup C NMB0065 homologue, and its apparently reduced capsule expression may account for the serum sensitivity [11]. These data are consistent with NMB0065 being a determinant of capsule expression. Upon analyses of NMB0065 mutations in meningococcal serogroups B, C and Y, we demonstrated a consistent phenotype in which the levels of total CPS produced by the mutants were similar to the wild type strains. Our results differ from the result of Geoffroy et al. [11], which reported that the serogroup C NMB0065, ctrE and ctrF transposon mutants produced < 10% of the wild-type capsule level. We have previously demonstrated that ctrE and ctrF mutants of the serogroup B strain NMB also express intracellular capsule at levels close to that of the wild type [9]. Thus, this discrepancy may be due to strain variation, or more likely, caused by differences in the degree of cell lysis, in which their study detected extracellular capsule with limited cell lysis.
The intracellular capsule expression of all NMB0065 mutants in different serogroups was consistent with their sensitivity to killing by normal human serum, increased surface hydrophobicity and large electron-translucent zones (lacunae) of accumulated intracellular CPS as revealed by immuno-gold electron microscopy. There are no discernible differences in the composition and the structures of the capsular polymers and the lipid anchor. Thus, NMB0065 was not involved in biosynthesis or lipidation of the polymer. In addition, DOC-PAGE and MALLS (multi-angle laser light scattering) analysis of the mutant and wild type CPSs (data not shown) did not reveal noticeable differences in the polymer length, suggesting that NMB0065 was not involved in the capping of polymer chain length. Thus, it is unlikely that NMB0065 participates in the assembly of capsular polymers, but perhaps contribute in linking the capsule biosynthesis complex with the CtrA-D transport apparatus in conjunction with CtrE and CtrF proteins. ctrE and ctrF are conserved in all meningococcal serogroups, while NMB0065 is only present amongst meningococci expressing sialic acid capsules (serogroups B, C, Y and W-135) and is absent in the non-sialic acid producing serogroups A or X meningococci. The exclusive linkage of NMB0065 and its homologues with sialic acid-containing CPS in meningococci suggests a possible functional role in ushering the assembled sialic polymers for translocation. We propose that NMB0065 be designated as CtrG to indicate its function as part of the CPS translocation machinery. Interestingly, the NMB0065 homologue of E. coli (neuE) encoded within the kps gene cluster is present in K1 and K92 strains that express polysialic acid capsule, but not in the strains producing non-sialic acid group II capsules.
While proteins of unknown function that exhibit slightly higher homology to CtrG were found in Streptomyces coelicolor and Synechococcus spp., NeuE of E. coli that expresses polysialic acid capsule is the best characterized homologue of CtrG [41]. However, several features distinguish CtrG and NeuE. First, CtrG is a 318-aa protein, while NeuE contains 411 residues (Accession no. AY937259, [26]). They share sequence similarity (25% identity and 45% similarity) over the central ~220 residues. The phoA fusion and subcellular fractionation analyses (Fig. 2) indicated that NMB0065 is a soluble cytoplasmic protein peripherally associated with the membrane, while NeuE is believed to associate with the inner membrane via a predicted transmembrane domain near the C terminus with similarity to a polyprenyl-binding motif [25, 26]. Pair-wise alignment of CtrG and NeuE (Fig. 1B) indicated that CtrG does not contain the corresponding C-terminal TM domain of NeuE. Andreishcheva and Vann [26] have shown that NeuE, together with KpsC and KpsS (meningococcal CtrE and CtrF homologues, respectively), was required for efficient de novo synthesis of polysialic acid capsule in the absence of an exogenous acceptor both in vitro and in vivo. Full-length intracellular lipidated polymers and a decreased polymerase activity were detected in a neuE mutant of K1 E. coli [34]. Thus, NeuE appears to be involved in both de novo biosynthesis and transport of assembled capsule in E. coli. The E. coli polysialyltransferase NeuS, a membrane bound enzyme catalyzes the elongation reaction in the absence of other kps gene products, but cannot initiate the polymerization of sialic acid [25, 42]. In contrast, the meningococcal polysialyitransferase SynD (SiaD) performs both functions [43], and has also been purified as an active soluble polymerase in the absence of membrane association or other capsule gene products [44]. Thus, the difference in the functional characteristics of NeuS and SynD appears to correlate with the phenotypic difference between the ctrG and neuE mutants. CtrG seems to only participate in meningococcal capsule surface expression.
In conclusion, we have determined the cellular location of CtrG and demonstrated that it is not required for the production of assembled meningococcal capsule polymers. CtrG engages the transmembrane export complex for proper surface expression of sialic acid capsular polysaccharides.
Acknowledgments
We thank Hong Yi at the Neurology Microscopy Facility, Emory University, for help in electron microscopy and S. K. Gudlavalleti at Laboratory of Bacterial Polysaccharides, Center for Biologics Evaluation and Research, Food and Drug Administration for multi-angle laser light scattering analysis. Antibodies were a gift from Wendell Zollinger, Department of the Army, Walter Reed Army Institute of Research Washington, DC. We thank B. Barquera for providing pBADphoA. We are grateful to Larry Martin and Soma Sannigrahi for technical assistance and Lane Pucko for administrative assistance. This work was supported by Public Health Service grant AI40247 from the National Institutes of Health to D.S.S. and in part by grant AI061031 from the National Institutes of Health to Y.T.
Abbreviations
- CPS
capsular polysaccharide
- DOC
deoxycholic acid
- NHS
normal human serum
- TM
transmembrane
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stephens DS, Greenwood B, Brandtzaeg P. Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet. 2007;369:2196–2210. doi: 10.1016/S0140-6736(07)61016-2. [DOI] [PubMed] [Google Scholar]
- 2.Rosenstein NE, Perkins BA, Stephens DS, Popovic T, Hughes JM. Meningococcal disease: recent progress and future challenges. N Engl J Med. 2001;344:1378–1388. doi: 10.1056/NEJM200105033441807. [DOI] [PubMed] [Google Scholar]
- 3.Liu TY, Gotschlich EC, Dunne FT, Jonssen EK. Studies on the meningococcal polysaccharides. II. Composition and chemical properties of the group B and group C polysaccharide. J Biol Chem. 1971;246:4703–4712. [PubMed] [Google Scholar]
- 4.Bhattacharjee AK, Jennings HJ, Kenny CP, Martin A, Smith IC. Structural determination of the polysaccharide antigens of Neisseria meningitidis serogroups Y, W-135, and BO1. Can J Biochem. 1976;54:1–8. doi: 10.1139/o76-001. [DOI] [PubMed] [Google Scholar]
- 5.Liu TY, Gotschlich EC, Jonssen EK, Wysocki JR. Studies on the meningococcal polysaccharides. I. Composition and chemical properties of the group A polysaccharide. J Biol Chem. 1971;246:2849–2858. [PubMed] [Google Scholar]
- 6.Bundle DR, Jennings HJ, Kenny CP. Studies on the group-specific polysaccharide of Neisseria meningitidis serogroup X and an improved procedure for its isolation. J Biol Chem. 1974;249:4797–4801. [PubMed] [Google Scholar]
- 7.Frosch M, Weisgerber C, Meyer TF. Molecular characterization and expression in Escherichia coli of the gene complex encoding the polysaccharide capsule of Neisseria meningitidis group B. Proc Natl Acad Sci U S A. 1989;86:1669–1673. doi: 10.1073/pnas.86.5.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swartley JS, Marfin AA, Edupuganti S, Liu LJ, Cieslak P, Perkins B, Wenger JD, Stephens DS. Capsule switching of Neisseria meningitidis. Proc Natl Acad Sci U S A. 1997;94:271–276. doi: 10.1073/pnas.94.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzeng YL, Datta AK, Strole CA, Lobritz MA, Carlson RW, Stephens DS. Translocation and surface expression of lipidated serogroup B capsular Polysaccharide in Neisseria meningitidis. Infect Immun. 2005;73:1491–1505. doi: 10.1128/IAI.73.3.1491-1505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun YH, Bakshi S, Chalmers R, Tang CM. Functional genomics of Neisseria meningitidis pathogenesis. Nat Med. 2000;6:1269–1273. doi: 10.1038/81380. [DOI] [PubMed] [Google Scholar]
- 11.Geoffroy MC, Floquet S, Metais A, Nassif X, Pelicic V. Large-scale analysis of the meningococcus genome by gene disruption: resistance to complement-mediated lysis. Genome Res. 2003;13:391–398. doi: 10.1101/gr.664303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menard R, Sansonetti PJ, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzeng YL, Zhou X, Bao S, Zhao S, Noble C, Stephens DS. Autoregulation of the MisR/MisS two-component signal transduction system in Neisseria meningitidis. J Bacteriol. 2006;188:5055–5065. doi: 10.1128/JB.00264-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tzeng YL, Datta A, Kolli VK, Carlson RW, Stephens DS. Endotoxin of Neisseria meningitidis composed only of intact lipid A: inactivation of the meningococcal 3-deoxy-D-manno-octulosonic acid transferase. J Bacteriol. 2002;184:2379–2388. doi: 10.1128/JB.184.9.2379-2388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melchers K, Schuhmacher A, Buhmann A, Weitzenegger T, Belin D, Grau S, Ehrmann M. Membrane topology of CadA homologous P-type ATPase of Helicobacter pylori as determined by expression of phoA fusions in Escherichia coli and the positive inside rule. Res Microbiol. 1999;150:507–520. doi: 10.1016/s0923-2508(99)00106-0. [DOI] [PubMed] [Google Scholar]
- 16.Manoil C. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 1991;34:61–75. doi: 10.1016/s0091-679x(08)61676-3. [DOI] [PubMed] [Google Scholar]
- 17.Clark VL, Campbell LA, Palermo DA, Evans TM, Klimpel KW. Induction and repression of outer membrane proteins by anaerobic growth of Neisseria gonorrhoeae. Infection & Immunity. 1987;55:1359–1364. doi: 10.1128/iai.55.6.1359-1364.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gudlavalleti SK, Datta AK, Tzeng YL, Noble C, Carlson RW, Stephens DS. The Neisseria meningitidis serogroup A capsular polysaccharide O-3 and O-4 acetyltransferase. J Biol Chem. 2004;279:42765–42773. doi: 10.1074/jbc.M313552200. [DOI] [PubMed] [Google Scholar]
- 19.Swartley JS, Liu LJ, Miller YK, Martin LE, Edupuganti S, Stephens DS. Characterization of the gene cassette required for biosynthesis of the (a1-->6)-linked N-acetyl-D-mannosamine-1-phosphate capsule of serogroup A Neisseria meningitidis. J Bacteriol. 1998;180:1533–1539. doi: 10.1128/jb.180.6.1533-1539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kahler CM, Martin LE, Shih GC, Rahman MM, Carlson RW, Stephens DS. The (a2-->8)-linked polysialic acid capsule and lipooligosaccharide structure both contribute to the ability of serogroup B Neisseria meningitidis to resist the bactericidal activity of normal human serum. Infect Immun. 1998;66:5939–5947. doi: 10.1128/iai.66.12.5939-5947.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gotschlich EC. Development of polysaccharide vaccines for the prevention of meningococcal diseases. Monogr Allergy. 1975;9:245–258. [PubMed] [Google Scholar]
- 22.Rahman MM, Guard-Petter J, Asokan K, Carlson RW. The structure of the capsular polysaccharide from a swarming strain of pathogenic Proteus vulgaris. Carbohydr Res. 1997;301:213–220. doi: 10.1016/s0008-6215(97)00093-1. [DOI] [PubMed] [Google Scholar]
- 23.Parkhill J, Achtman M, James KD, Bentley SD, Churcher C, Klee SR, Morelli G, Basham D, Brown D, Chillingworth T, Davies RM, Davis P, Devlin K, Feltwell T, Hamlin N, Holroyd S, Jagels K, Leather S, Moule S, Mungall K, Quail MA, Rajandream MA, Rutherford KM, Simmonds M, Skelton J, Whitehead S, Spratt BG, Barrell BG. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 24.Tzeng YL, Noble C, Stephens DS. Genetic basis for biosynthesis of the (alpha 1-->4)-linked N-acetyl-D-glucosamine 1-phosphate capsule of Neisseria meningitidis serogroup X. Infect Immun. 2003;71:6712–6720. doi: 10.1128/IAI.71.12.6712-6720.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steenbergen SM, Wrona TJ, Vimr ER. Functional analysis of the sialyltransferase complexes in Escherichia coli K1 and K92. J Bacteriol. 1992;174:1099–1108. doi: 10.1128/jb.174.4.1099-1108.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andreishcheva EN, Vann WF. Gene products required for de novo synthesis of polysialic acid in Escherichia coli K1. J Bacteriol. 2006;188:1786–1797. doi: 10.1128/JB.188.5.1786-1797.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steenbergen SM, Vimr ER. Biosynthesis of the Escherichia coli K1 group 2 polysialic acid capsule occurs within a protected cytoplasmic compartment. Mol Microbiol. 2008;68:1252–1267. doi: 10.1111/j.1365-2958.2008.06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melen K, Krogh A, von Heijne G. Reliability measures for membrane protein topology prediction algorithms. J Mol Biol. 2003;327:735–744. doi: 10.1016/s0022-2836(03)00182-7. [DOI] [PubMed] [Google Scholar]
- 29.Claros MG, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 30.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 31.Karlyshev AV, Linton D, Gregson NA, Lastovica AJ, Wren BW. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol Microbiol. 2000;35:529–541. doi: 10.1046/j.1365-2958.2000.01717.x. [DOI] [PubMed] [Google Scholar]
- 32.Kroncke KD, Boulnois G, Roberts I, Bitter-Suermann D, Golecki JR, Jann B, Jann K. Expression of the Escherichia coli K5 capsular antigen: immunoelectron microscopic and biochemical studies with recombinant E. coli. J Bacteriol. 1990;172:1085–1091. doi: 10.1128/jb.172.2.1085-1091.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cieslewicz M, Vimr E. Reduced polysialic acid capsule expression in Escherichia coli K1 mutants with chromosomal defects in kpsF. Mol Microbiol. 1997;26:237–249. doi: 10.1046/j.1365-2958.1997.5651942.x. [DOI] [PubMed] [Google Scholar]
- 34.Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev. 2004;68:132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moxon ER, Kroll JS. The role of bacterial polysaccharide capsules as virulence factors. Curr Top Microbio Immunol. 1990;150:65–85. doi: 10.1007/978-3-642-74694-9_4. [DOI] [PubMed] [Google Scholar]
- 36.Taylor CM, Roberts IS. Capsular polysaccharides and their role in virulence. Contrib Microbiol. 2005;12:55–66. doi: 10.1159/000081689. [DOI] [PubMed] [Google Scholar]
- 37.Stephens DS, Spellman PA, Swartley JS. Effect of the (a 2-->8)-linked polysialic acid capsule on adherence of Neisseria meningitidis to human mucosal cells. J Infect Dis. 1993;167:475–479. doi: 10.1093/infdis/167.2.475. [DOI] [PubMed] [Google Scholar]
- 38.Virji M, Makepeace K, Ferguson DJ, Achtman M, Moxon ER. Meningococcal Opa and Opc proteins: their role in colonization and invasion of human epithelial and endothelial cells. Mol Microbiol. 1993;10:499–510. doi: 10.1111/j.1365-2958.1993.tb00922.x. [DOI] [PubMed] [Google Scholar]
- 39.Salit IE, Morton G. Adherence of Neisseria meningitidis to human epithelial cells. Infect Immun. 1981;31:430–435. doi: 10.1128/iai.31.1.430-435.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stephens DS, McGee ZA. Attachment of Neisseria meningitidis to human mucosal surfaces: influence of pili and type of receptor cell. J Infect Dis. 1981;143:525–532. doi: 10.1093/infdis/143.4.525. [DOI] [PubMed] [Google Scholar]
- 41.Vimr ER, Steenbergen SM. Early molecular-recognition events in the synthesis and export of group 2 capsular polysaccharides. Microbiology. 2009;155:9–15. doi: 10.1099/mic.0.023564-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGowen MM, Vionnet J, Vann WF. Elongation of alternating alpha 2,8/2,9 polysialic acid by the Escherichia coli K92 polysialyltransferase. Glycobiology. 2001;11:613–620. doi: 10.1093/glycob/11.8.613. [DOI] [PubMed] [Google Scholar]
- 43.Edwards U, Muller A, Hammerschmidt S, Gerardy-Schahn R, Frosch M. Molecular analysis of the biosynthesis pathway of the alpha-2-->8 polysialic acid capsule by Neisseria meningitidis serogroup B. Mol Microbiol. 1994;14:141–149. doi: 10.1111/j.1365-2958.1994.tb01274.x. [DOI] [PubMed] [Google Scholar]
- 44.Freiberger F, Claus H, Gunzel A, Oltmann-Norden I, Vionnet J, Muhlenhoff M, Vogel U, Vann WF, Gerardy-Schahn R, Stummeyer K. Biochemical characterization of a Neisseria meningitidis polysialyltransferase reveals novel functional motifs in bacterial sialyltransferases. Mol Microbiol. 2007;65:1258–1275. doi: 10.1111/j.1365-2958.2007.05862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolfgang M, Lauer P, Park HS, Brossay L, Hebert J, Koomey M. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol. 1998;29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- 46.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 47.Stephens DS, Swartley JS, Kathariou S, Morse SA. Insertion of Tn916 in Neisseria meningitidis resulting in loss of group B capsular polysaccharide. Infect Immun. 1991;59:4097–4102. doi: 10.1128/iai.59.11.4097-4102.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tzeng YL, Datta A, Strole C, Kolli VS, Birck MR, Taylor WP, Carlson RW, Woodard RW, Stephens DS. KpsF is the arabinose-5-phosphate isomerase required for 3-deoxy-D-manno-octulosonic acid biosynthesis and for both lipooligosaccharide assembly and capsular polysaccharide expression in Neisseria meningitidis. J Biol Chem. 2002;277:24103–24113. doi: 10.1074/jbc.M200931200. [DOI] [PubMed] [Google Scholar]
- 49.Duffy EB, Barquera B. Membrane topology mapping of the Na+-pumping NADH: quinone oxidoreductase from Vibrio cholerae by PhoA-green fluorescent protein fusion analysis. J Bacteriol. 2006;188:8343–8351. doi: 10.1128/JB.01383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]