Abstract
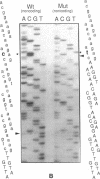
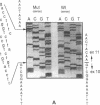
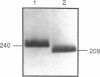
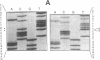
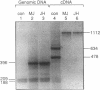
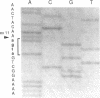
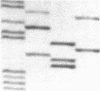
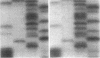
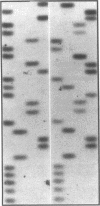
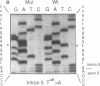
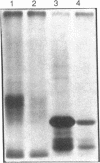
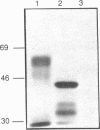
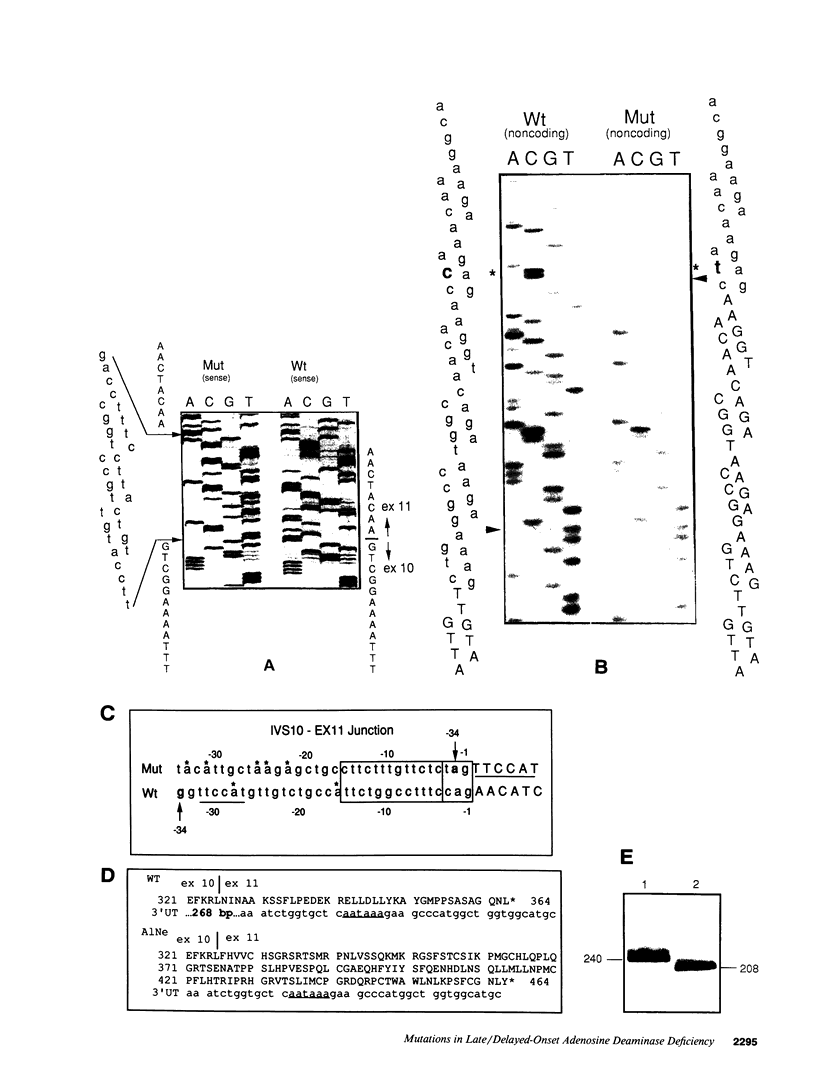
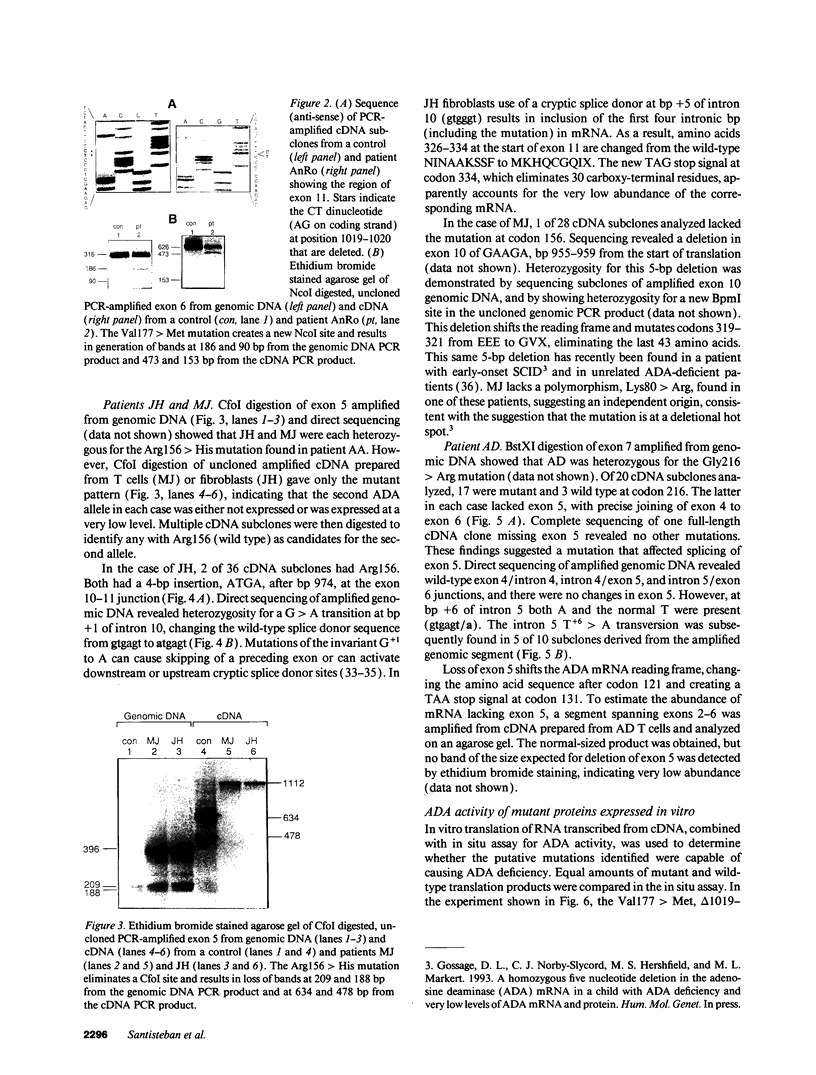
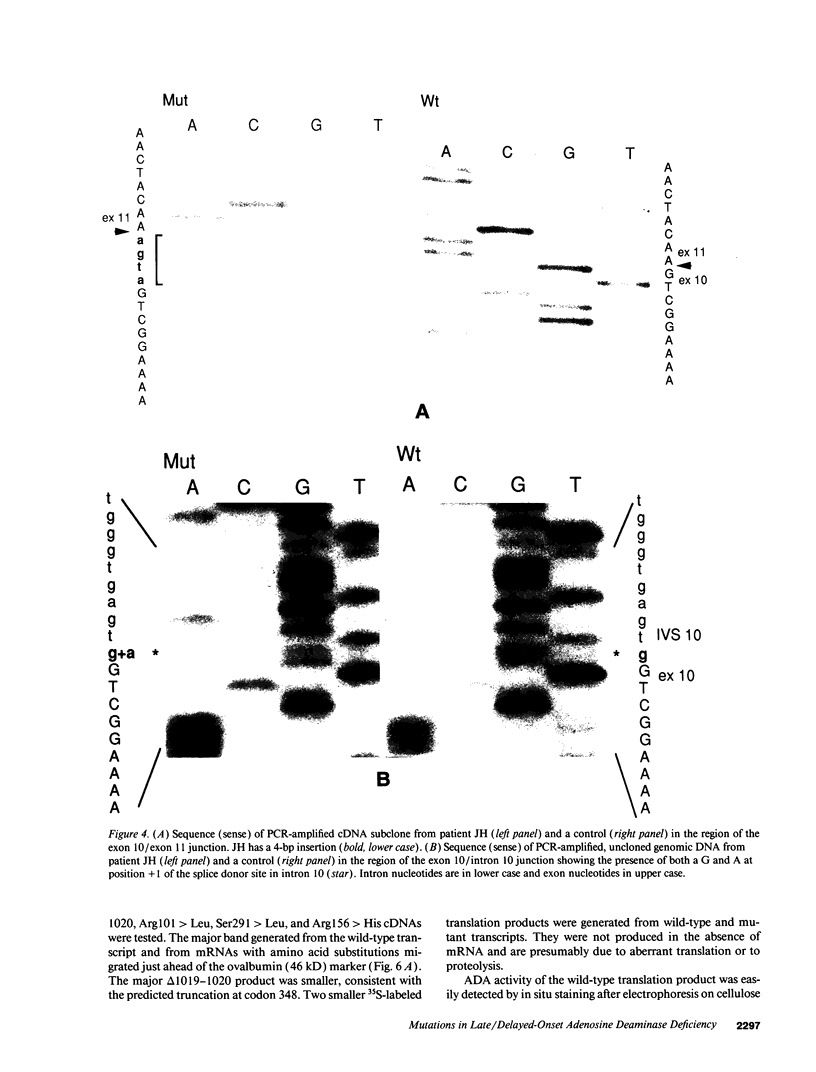
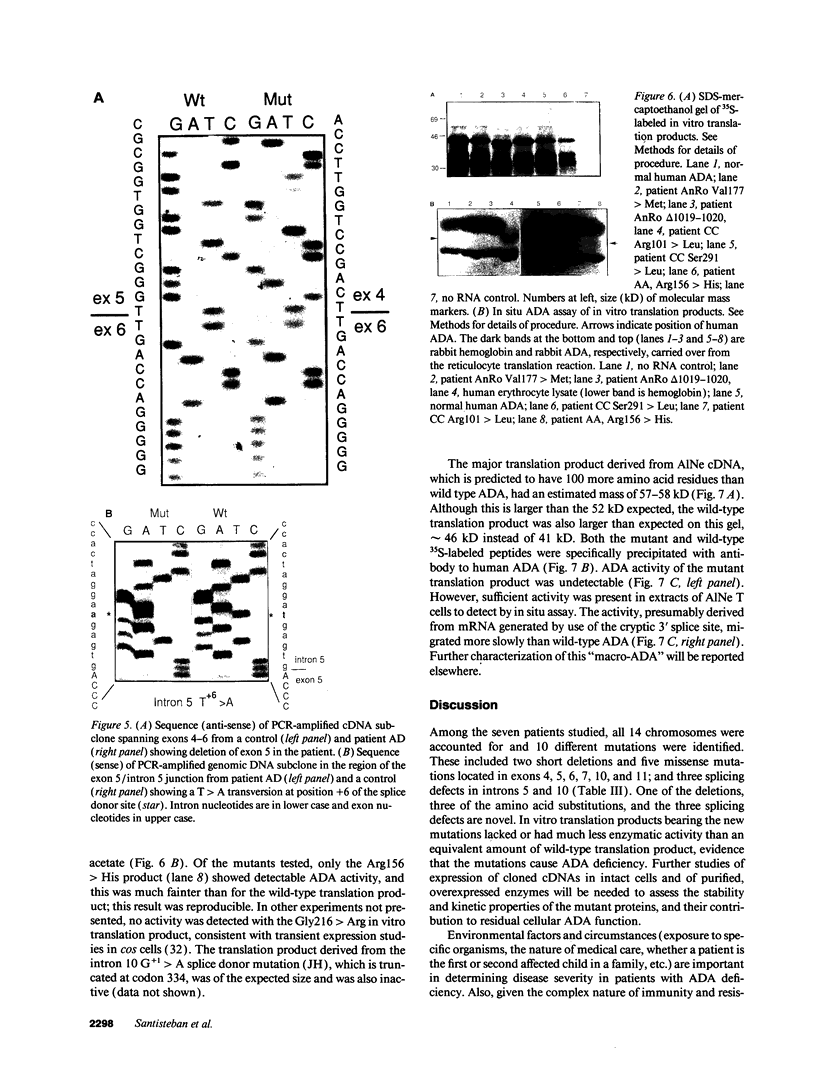
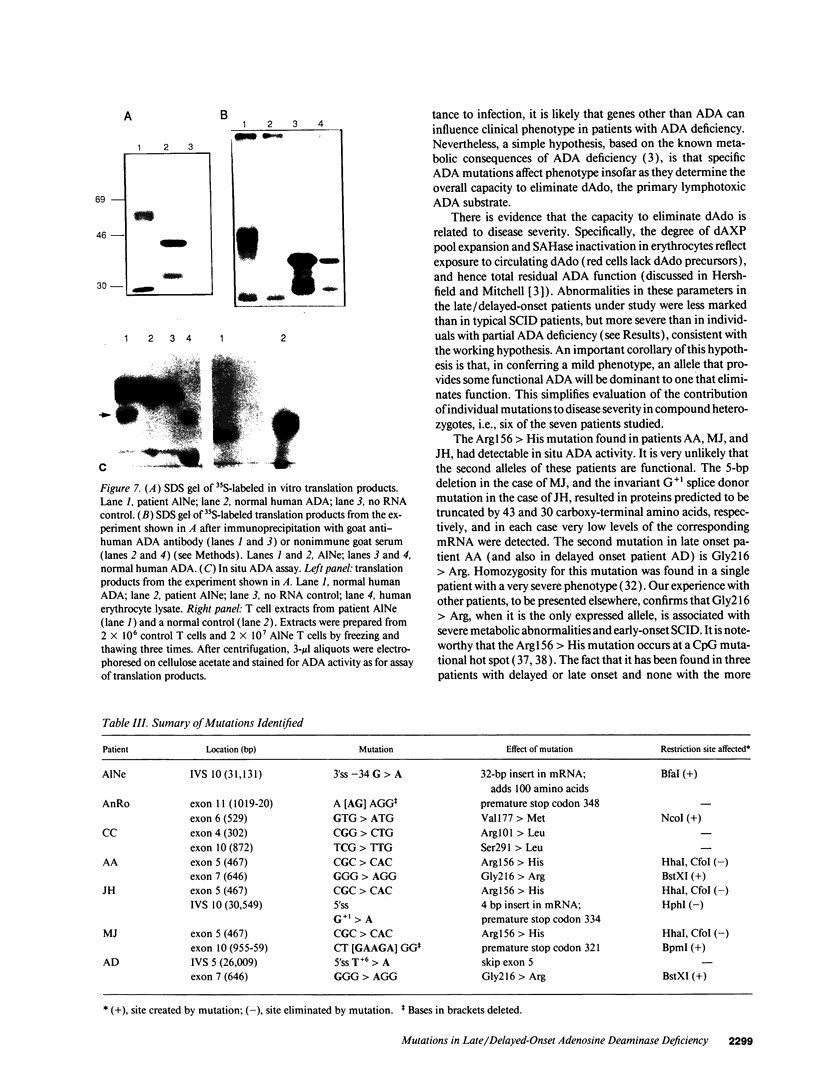
We examined the genetic basis for adenosine deaminase (ADA) deficiency in seven patients with late/delayed onset of immunodeficiency, an underdiagnosed and relatively unstudied condition. Deoxyadenosine-mediated metabolic abnormalities were less severe than in the usual, early-onset disorder. Six patients were compound heterozygotes; 7 of 10 mutations found were novel, including one deletion (delta 1019-1020), three missense (Arg156 > His, Arg101 > Leu, Val177 > Met), and three splicing defects (IVS 5, 5'ss T+6 > A; IVS 10, 5'ss G+1 > A; IVS 10, 3'ss G-34 > A). Four of the mutations generated stop signals at codons 131, 321, 334, and 348; transcripts of all but the last, due to delta 1019-1020, were severely reduced. delta 1019-1020 (like delta 955-959, found in one patient and apparently recurrent) is at a short deletional hot spot. Arg156 > His, the product of which had detectable activity, was found in three patients whose second alleles were unlikely to yield active ADA. The oldest patient diagnosed was homozygous for a single base change in intron 10, which activates a cryptic splice acceptor, resulting in a protein with 100 extra amino acids. We speculate that this "macro ADA," as well as the Arg156 > His, Arg101 > Leu, Ser291 > Leu, and delta 1019-1020 products, may contribute to mild phenotype. Tissue-specific variation in splicing efficiency may also ameliorate disease severity in patients with splicing mutations.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akeson A. L., Wiginton D. A., Dusing M. R., States J. C., Hutton J. J. Mutant human adenosine deaminase alleles and their expression by transfection into fibroblasts. J Biol Chem. 1988 Nov 5;263(31):16291–16296. [PubMed] [Google Scholar]
- Ammann A. J., Cowan M. J., Martin D. W., Wara D. W. Dipyridamole and intravenous deoxycytidine therapy in a patient with adenosine deaminase deficiency. Birth Defects Orig Artic Ser. 1983;19(3):117–120. [PubMed] [Google Scholar]
- Andrews L. G., Markert M. L. Exon skipping in purine nucleoside phosphorylase mRNA processing leading to severe immunodeficiency. J Biol Chem. 1992 Apr 15;267(11):7834–7838. [PubMed] [Google Scholar]
- Arredondo-Vega F. X., Kurtzberg J., Chaffee S., Santisteban I., Reisner E., Povey M. S., Hershfield M. S. Paradoxical expression of adenosine deaminase in T cells cultured from a patient with adenosine deaminase deficiency and combine immunodeficiency. J Clin Invest. 1990 Aug;86(2):444–452. doi: 10.1172/JCI114730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badley J. E., Bishop G. A., St John T., Frelinger J. A. A simple, rapid method for the purification of poly A+ RNA. Biotechniques. 1988 Feb;6(2):114–116. [PubMed] [Google Scholar]
- Baumann B., Potash M. J., Köhler G. Consequences of frameshift mutations at the immunoglobulin heavy chain locus of the mouse. EMBO J. 1985 Feb;4(2):351–359. doi: 10.1002/j.1460-2075.1985.tb03636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkvens T. M., Schoute F., van Ormondt H., Meera Khan P., van der Eb A. J. Adenosine deaminase gene expression is regulated posttranscriptionally in the nucleus. Nucleic Acids Res. 1988 Apr 25;16(8):3255–3268. doi: 10.1093/nar/16.8.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonthron D. T., Markham A. F., Ginsburg D., Orkin S. H. Identification of a point mutation in the adenosine deaminase gene responsible for immunodeficiency. J Clin Invest. 1985 Aug;76(2):894–897. doi: 10.1172/JCI112050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens R. P., Fenton W. A., Rosenberg L. R. Identification of RNA splicing errors resulting in human ornithine transcarbamylase deficiency. Am J Hum Genet. 1991 Jun;48(6):1105–1114. [PMC free article] [PubMed] [Google Scholar]
- Chang Z. Y., Nygaard P., Chinault A. C., Kellems R. E. Deduced amino acid sequence of Escherichia coli adenosine deaminase reveals evolutionarily conserved amino acid residues: implications for catalytic function. Biochemistry. 1991 Feb 26;30(8):2273–2280. doi: 10.1021/bi00222a033. [DOI] [PubMed] [Google Scholar]
- Cohen A., Hirschhorn R., Horowitz S. D., Rubinstein A., Polmar S. H., Hong R., Martin D. W., Jr Deoxyadenosine triphosphate as a potentially toxic metabolite in adenosine deaminase deficiency. Proc Natl Acad Sci U S A. 1978 Jan;75(1):472–476. doi: 10.1073/pnas.75.1.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M. S., Donofrio J., Hutton J. J., Hahn L., Daoud A., Lampkin B., Dyminski J. Identification and quantitation of adenine deoxynucleotides in erythrocytes of a patient with adenosine deaminase deficiency and severe combined immunodeficiency. J Biol Chem. 1978 Mar 10;253(5):1619–1626. [PubMed] [Google Scholar]
- Cooper D. N., Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet. 1988 Feb;78(2):151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H., Farabaugh P. J., Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978 Aug 24;274(5673):775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Cowan M. J., Wara D. W., Ammann A. J. Deoxycytidine therapy in two patients with adenosine deaminase deficiency and severe immunodeficiency disease. Clin Immunol Immunopathol. 1985 Oct;37(1):30–36. doi: 10.1016/0090-1229(85)90132-1. [DOI] [PubMed] [Google Scholar]
- Daar I. O., Maquat L. E. Premature translation termination mediates triosephosphate isomerase mRNA degradation. Mol Cell Biol. 1988 Feb;8(2):802–813. doi: 10.1128/mcb.8.2.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geffner M. E., Stiehm E. R., Stephure D., Cowan M. J. Probable autoimmune thyroid disease and combined immunodeficiency disease. Am J Dis Child. 1986 Nov;140(11):1194–1196. doi: 10.1001/archpedi.1986.02140250120047. [DOI] [PubMed] [Google Scholar]
- Girault D., Le Deist F., Debré M., Pérignon J. L., Herbelin C., Griscelli C., Sciudery D., Hershfield M., Fischer A. Traitement du déficit en adénosine désaminase par l'adénosine désaminase couplée au polyéthylène glycol. Arch Fr Pediatr. 1992 Apr;49(4):339–343. [PubMed] [Google Scholar]
- Goguel V., Rosbash M. Splice site choice and splicing efficiency are positively influenced by pre-mRNA intramolecular base pairing in yeast. Cell. 1993 Mar 26;72(6):893–901. doi: 10.1016/0092-8674(93)90578-e. [DOI] [PubMed] [Google Scholar]
- Grandchamp B., Picat C., de Rooij F., Beaumont C., Wilson P., Deybach J. C., Nordmann Y. A point mutation G----A in exon 12 of the porphobilinogen deaminase gene results in exon skipping and is responsible for acute intermittent porphyria. Nucleic Acids Res. 1989 Aug 25;17(16):6637–6649. doi: 10.1093/nar/17.16.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Hershfield M. S. Apparent suicide inactivation of human lymphoblast S-adenosylhomocysteine hydrolase by 2'-deoxyadenosine and adenine arabinoside. A basis for direct toxic effects of analogs of adenosine. J Biol Chem. 1979 Jan 10;254(1):22–25. [PubMed] [Google Scholar]
- Hershfield M. S., Buckley R. H., Greenberg M. L., Melton A. L., Schiff R., Hatem C., Kurtzberg J., Markert M. L., Kobayashi R. H., Kobayashi A. L. Treatment of adenosine deaminase deficiency with polyethylene glycol-modified adenosine deaminase. N Engl J Med. 1987 Mar 5;316(10):589–596. doi: 10.1056/NEJM198703053161005. [DOI] [PubMed] [Google Scholar]
- Hershfield M. S., Chaffee S., Sorensen R. U. Enzyme replacement therapy with polyethylene glycol-adenosine deaminase in adenosine deaminase deficiency: overview and case reports of three patients, including two now receiving gene therapy. Pediatr Res. 1993 Jan;33(1 Suppl):S42–S48. doi: 10.1203/00006450-199305001-00236. [DOI] [PubMed] [Google Scholar]
- Hershfield M. S., Kredich N. M., Ownby D. R., Ownby H., Buckley R. In vivo inactivation of erythrocyte S-adenosylhomocysteine hydrolase by 2'-deoxyadenosine in adenosine deaminase-deficient patients. J Clin Invest. 1979 Apr;63(4):807–811. doi: 10.1172/JCI109367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka Y., Palella T. D., O'Toole T. E., Tarlé S. A., Kelley W. N. Human adenine phosphoribosyltransferase. Identification of allelic mutations at the nucleotide level as a cause of complete deficiency of the enzyme. J Clin Invest. 1987 Nov;80(5):1409–1415. doi: 10.1172/JCI113219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn R. Adenosine deaminase deficiency. Immunodefic Rev. 1990;2(3):175–198. [PubMed] [Google Scholar]
- Hirschhorn R., Chakravarti V., Puck J., Douglas S. D. Homozygosity for a newly identified missense mutation in a patient with very severe combined immunodeficiency due to adenosine deaminase deficiency (ADA-SCID). Am J Hum Genet. 1991 Oct;49(4):878–885. [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn R., Ellenbogen A., Tzall S. Five missense mutations at the adenosine deaminase locus (ADA) detected by altered restriction fragments and their frequency in ADA--patients with severe combined immunodeficiency (ADA-SCID). Am J Med Genet. 1992 Jan 15;42(2):201–207. doi: 10.1002/ajmg.1320420213. [DOI] [PubMed] [Google Scholar]
- Hirschhorn R. Identification of two new missense mutations (R156C and S291L) in two ADA- SCID patients unusual for response to therapy with partial exchange transfusions. Hum Mutat. 1992;1(2):166–168. doi: 10.1002/humu.1380010214. [DOI] [PubMed] [Google Scholar]
- Hirschhorn R. Overview of biochemical abnormalities and molecular genetics of adenosine deaminase deficiency. Pediatr Res. 1993 Jan;33(1 Suppl):S35–S41. doi: 10.1203/00006450-199305001-00194. [DOI] [PubMed] [Google Scholar]
- Hirschhorn R., Vawter G. F., Kirkpatrick J. A., Jr, Rosen F. S. Adenosine deaminase deficiency: frequency and comparative pathology in autosomally recessive severe combined immunodeficiency. Clin Immunol Immunopathol. 1979 Sep;14(1):107–120. doi: 10.1016/0090-1229(79)90131-4. [DOI] [PubMed] [Google Scholar]
- Kishimoto T. K., O'Conner K., Springer T. A. Leukocyte adhesion deficiency. Aberrant splicing of a conserved integrin sequence causes a moderate deficiency phenotype. J Biol Chem. 1989 Feb 25;264(6):3588–3595. [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T., Ruskin B., Green M. R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984 Apr;36(4):993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Krawczak M., Cooper D. N. Gene deletions causing human genetic disease: mechanisms of mutagenesis and the role of the local DNA sequence environment. Hum Genet. 1991 Mar;86(5):425–441. doi: 10.1007/BF00194629. [DOI] [PubMed] [Google Scholar]
- Kuivaniemi H., Kontusaari S., Tromp G., Zhao M. J., Sabol C., Prockop D. J. Identical G+1 to A mutations in three different introns of the type III procollagen gene (COL3A1) produce different patterns of RNA splicing in three variants of Ehlers-Danlos syndrome. IV. An explanation for exon skipping some mutations and not others. J Biol Chem. 1990 Jul 15;265(20):12067–12074. [PubMed] [Google Scholar]
- Levy Y., Hershfield M. S., Fernandez-Mejia C., Polmar S. H., Scudiery D., Berger M., Sorensen R. U. Adenosine deaminase deficiency with late onset of recurrent infections: response to treatment with polyethylene glycol-modified adenosine deaminase. J Pediatr. 1988 Aug;113(2):312–317. doi: 10.1016/s0022-3476(88)80271-3. [DOI] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvit J., DiLella A. G., Brayton K., Ledley F. D., Robson K. J., Woo S. L. GT to AT transition at a splice donor site causes skipping of the preceding exon in phenylketonuria. Nucleic Acids Res. 1987 Jul 24;15(14):5613–5628. doi: 10.1093/nar/15.14.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashima Y., Murakami A., Weleber R. G., Kennaway N. G., Clarke L., Shiono T., Inana G. Nonsense-codon mutations of the ornithine aminotransferase gene with decreased levels of mutant mRNA in gyrate atrophy. Am J Hum Genet. 1992 Jul;51(1):81–91. [PMC free article] [PubMed] [Google Scholar]
- McInnes B., Potier M., Wakamatsu N., Melancon S. B., Klavins M. H., Tsuji S., Mahuran D. J. An unusual splicing mutation in the HEXB gene is associated with dramatically different phenotypes in patients from different racial backgrounds. J Clin Invest. 1992 Aug;90(2):306–314. doi: 10.1172/JCI115863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs H. D., Buckley R. H., Kobayashi R. H., Kobayashi A. L., Sorensen R. U., Douglas S. D., Hamilton B. L., Hershfield M. S. Antibody responses to bacteriophage phi X174 in patients with adenosine deaminase deficiency. Blood. 1992 Sep 1;80(5):1163–1171. [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983 Apr 14;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Treisman R., Proudfoot N. J., Shander M., Maniatis T. A single-base change at a splice site in a beta 0-thalassemic gene causes abnormal RNA splicing. Cell. 1982 Jul;29(3):903–911. doi: 10.1016/0092-8674(82)90452-4. [DOI] [PubMed] [Google Scholar]
- Uberti J., Lightbody J. J., Wolf J. W., Anderson J. A., Reid R. H., Johnson R. M. The effect of adenosine on mitogenesis of ADA-deficient lymphocytes. Clin Immunol Immunopathol. 1978 Aug;10(4):446–458. doi: 10.1016/0090-1229(78)90157-5. [DOI] [PubMed] [Google Scholar]
- Uberti J., Peterson W. D., Jr, Lightbody J. J., Johnson R. M. A phenotypically normal revertant of an adenosine deaminase-deficient lymphoblast cell line. J Immunol. 1983 Jun;130(6):2866–2870. [PubMed] [Google Scholar]
- Urlaub G., Mitchell P. J., Ciudad C. J., Chasin L. A. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989 Jul;9(7):2868–2880. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil D., Bernard M., Combates N., Wirtz M. K., Hollister D. W., Steinmann B., Ramirez F. Identification of a mutation that causes exon skipping during collagen pre-mRNA splicing in an Ehlers-Danlos syndrome variant. J Biol Chem. 1988 Jun 25;263(18):8561–8564. [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Wiginton D. A., Adrian G. S., Hutton J. J. Sequence of human adenosine deaminase cDNA including the coding region and a small intron. Nucleic Acids Res. 1984 Mar 12;12(5):2439–2446. doi: 10.1093/nar/12.5.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiginton D. A., Kaplan D. J., States J. C., Akeson A. L., Perme C. M., Bilyk I. J., Vaughn A. J., Lattier D. L., Hutton J. J. Complete sequence and structure of the gene for human adenosine deaminase. Biochemistry. 1986 Dec 16;25(25):8234–8244. doi: 10.1021/bi00373a017. [DOI] [PubMed] [Google Scholar]
- Wilson D. K., Rudolph F. B., Quiocho F. A. Atomic structure of adenosine deaminase complexed with a transition-state analog: understanding catalysis and immunodeficiency mutations. Science. 1991 May 31;252(5010):1278–1284. doi: 10.1126/science.1925539. [DOI] [PubMed] [Google Scholar]