Abstract
Viral mutations at Gag residue 242 and relevant viral polymorphisms were analyzed in a cohort of 42 individuals with primary HIV-1 subtype C infection using single-genome amplification/sequencing. In HLA-B*57/5801-negative subjects infected with 242N escape variant, reversion to Asn appeared at median (IQR) 103 (97;213) days post-seroconversion (p/s), and became dominant at 193 (170;215) days p/s. In subjects expressing HLA-B*57/5801 and infected with the wild type virus, the T242N escape appeared at 203 (196;231) days p/s, reached dominance at 277 (265;315) days p/s, and became complete at 323 (289;373) days p/s. HLA-B*57/5801-negative subjects infected with 242N escape variant did not show reduced viral load or increased CD4 count. The study highlights differential selection of T242N escape by HLA-B*57 and B*5801, and suggests the presence HLA-B*57/5801-mediated immune pressure is able to control replication of the wild type virus encoding Thr at Gag residue 242, but fails to suppress the T242N escape variant.
Keywords: HIV-1 subtype C, gag/Gag, mutation, 242, dynamics, timing, viral load, CD4+ T cell count, HLA-B*57, HLA-B*5801
Introduction
The HIV-1 Gag protein is one of the most functionally and structurally constrained proteins of HIV-1, and is a robust inducer of potent virus-specific immune responses. Potential association between T cell responses to Gag and better control of viral replication, lower viral set point, and more favorable disease prognosis (Betts et al., 2001; Boaz et al., 2002; Edwards et al., 2002; Geldmacher et al., 2007; Kiepiela et al., 2007; Masemola et al., 2004; Ndongala et al., 2009; Novitsky et al., 2003; Novitsky et al., 2006; Ramduth et al., 2005; Rolland et al., 2008; Serwanga et al., 2009; Zuniga et al., 2006) has led to a wide interest in HIV-1 Gag as an attractive immunogen for an HIV-1 vaccine design.
The MHC class I HLA alleles can affect the progression of HIV disease (O'Brien and Nelson, 2004). HIV-infected subjects expressing HLA-B*57 or -B*5801 have better clinical prognoses than HLA-B*57/5801-negative individuals (Altfeld et al., 2003; Goulder et al., 1996; Kaslow et al., 1996; Klein et al., 1998; Migueles et al., 2000), which is likely due to efficient processing and presentation of viral epitopes to virus-specific CD8+ T cells. In fact, the number of Gag escape mutations targeted by HLA-B-restricted CTL responses was associated with lower viral load in newly infected individuals (Goepfert et al., 2008).
The HIV-1 subtype B Gag p24 epitope TSTLQEQIGW, known as TW10 at Gag amino acid residues 240 to 249, was found to be one of the three most targeted CTL epitopes in individuals expressing MHC class I HLA-B*57 or -B*5801 within the first 6 months of infection (Altfeld et al., 2003). Kawashima et al. reported a strong correlation between frequency of escape mutation at Gag position 242 and HLA-B*57/5801 prevalence in the population (Kawashima et al., 2009). The HIV-1 subtype C version of TW10 epitope is TSTLQEQIAW (Los Alamos National Laboratory, 2009). In chronic HIV-1 infection, 74% of HLA-B*57/5801-positive individuals exhibit the T242N mutation (Martinez-Picado et al., 2006). In a series of in vitro experiments, the T242N mutation was associated with reduced viral replicative capacity (Martinez-Picado et al., 2006) providing a justification for reversion to the wild-type Thr at Gag residue 242 after transmission to individuals who do not express HLA-B*57/5801. The T242N substitution reduces viral fitness (Leslie et al., 2004; Martinez-Picado et al., 2006). In vitro experiments addressing the fitness cost of mutation at Gag residue 242 determined the viral relative replication capacity of T242N escape as 0.86 (Boutwell, Rowley, and Essex, 2009). It has been recently shown that HLA-B*57/5801-negative individuals infected with the 242N escape mutant have lower viral load and higher CD4+ T cell counts (Chopera et al., 2008).
However, the timing of in vivo Gag mutations including one of the best-studied HIV-1 epitopes, TW10, has been understudied. The limitation of cross-sectional studies to address timing of viral mutations is well recognized. For example, if time of infection is unknown or cannot be estimated, the prospective sampling during chronic HIV infection has limited ability to address the time the mutation occur in the course of HIV infection, and to compare or synchronize the timing data between studies. In contrast, studies focusing on primary HIV infection cohorts have power to analyze timing of mutations, but few have addressed the evolution of Gag mutations. For example, tracking viral sequence changes in Gag p24 in five acutely infected HLA-B*5703-positive individuals, Crawford et al. found that T242N escape occurred at a median of 13 months post-seroconversion (p/s) ranging from 4 to 18 months (Crawford et al., 2009). Besides the small sample size, systematic frequent longitudinal sampling is still a challenge even for primary HIV infection cohorts.
While it is known that in vivo N242T reversion might be rapid upon transmission, the time of N242T reverse mutation has been addressed sporadically within a relatively small number of cases, or infrequent sampling (Chopera et al., 2008; Crawford et al., 2009; Kawashima et al., 2009; Leslie et al., 2004; Thobakgale et al., 2009), or without the context of time of infection/seroconversion (Crawford et al., 2007). Leslie et al. reported vertical transmission of HIV-1 from an HLA-B57/5801-positive mother to HLA-B57/5801-negative children that resulted in N242T reversion from 5 to 9 months after transmission (Leslie et al., 2004). In a subject followed longitudinally, the N242T variants were not seen at day 43 p/s but were detected at day 818 p/s, and reached complete substitution by day 1,541 p/s (Leslie et al., 2004). Chopera et al. observed N242T reversions between six and 24 months post-infection in five of six individuals infected with the 242N variant (Chopera et al., 2008). In nine HLA-B*5703-negative individuals the reverse N242T mutations were detected at a median of 23 months p/s ranging from 7 to 55 months (Crawford et al., 2009). Recently Thobakgale et al. reported two cases of vertical transmission of 242N variant from HLA-B*5702-positive mother to HLA-B*5702-negative child followed by reversion to 242T between 773 and 819 days post delivery, and from HLA-B*5801-positive mother to HLA-B*5801-negative child without reversion up to 1,363 days post delivery (Thobakgale et al., 2009). Cumulatively, these studies provide an important step toward better understanding of the dynamics of viral mutational pathways in HIV infection. At the same time, the relatively vague time intervals of observed mutations highlight the importance of addressing the timing of appearance and fixation of in vivo HIV mutations.
In this study we focused on the timing patterns of viral mutations at Gag residue 242 within epitope TW10, and analyzed potential associations between Gag 242 mutations and viral load in primary HIV-1 subtype C infection utilizing a cohort of 42 individuals with estimated time of seroconversion and frequent sampling during primary infection.
Material and methods
Study subjects
A detailed characterization of the primary HIV-1C infection cohort from Botswana is provided elsewhere (Novitsky et al., 2009a; Novitsky et al., 2009b; Novitsky et al., 2009c; Novitsky et al., 2009d; Novitsky et al., 2008). Briefly, 8 acutely and 34 recently infected subjects with estimated time of seroconversion were enrolled and followed up prospectively for 500+ days thereafter. For acutely infected subjects the time of seroconversion was estimated as the midpoint between the last seronegative test and the first seropositive test (within a week in most cases). For recently infected subjects the time of seroconversion was estimated by Fiebig stage assignment (Fiebig et al., 2003) as described elsewhere (Novitsky et al., 2009b). For a time zero we have chosen to use the estimated time of seroconversion rather the estimated time of HIV infection because frequent sampling in this study allows reliable measurement of the time of seroconversion based on a series of laboratory tests, which can be more accurate than estimation the time of HIV infection. For comparison with other studies, the timing numbers in this study can be adjusted by adding 14 to 21 days, and will correspond to the estimated time of infection. The cohort included 9 males and 33 females with median age of 27 years (IQR 25–32, range 20–56) at enrollment. All subjects in the study were Botswana nationals, and all infections were HIV-1 subtype C (Novitsky et al., 2009a; Novitsky et al., 2009d). Viral load and CD4+ T cell counts were assessed prospectively during follow-up (Novitsky et al., 2009a; Novitsky et al., 2009d; Novitsky et al., 2008).
Viral sequences
Both viral RNA from plasma and cell-associated proviral DNA were used for amplification of viral sequences. The synthesized cDNA and proviral DNA templates were subjects for single-genome amplification by limiting dilutions (Liu et al., 1996; Palmer et al., 2005) with some modifications. Primers F2NST (Carr et al., 1999) (5′-GCG GAG GCT AGA AGG AGA GAG ATG G) and 1448L (5′-AGG GGT CGC TGC CAA AGA GTG ATT) were used in the first-round PCR. Primers gag-5U (5′-GTG CGA GAG CGT CAA TAT TAA GAG) and 1445L (5′-GGT CGC TGC CAA AGA GTG ATT) were used in the second-round PCR. Amplicons were purified by Exo-SAP (Dugan et al., 2002), and were sequenced directly at both strands on the ABI 3730 DNA Analyzer using BigDye technology. HYPERMUT v.2.0 (Rose and Korber, 2000) was used to identify and remove hypermutated sequences from analysis. The median analyzed viral quasispecies in Gag (IQR) was 58.5 (42.0; 73.0) per subject, and 11.1 (8.8; 13.6) viral sequences per time point per subject. Details on RNA and DNA sequences used in the study and their accession numbers are presented elsewhere (Novitsky et al., 2009c). The frequencies of translated amino acid sequences were analyzed using MargFreq (Ray, 2008) per time point per subject. The frequency of each amino acid residue was expressed as a fraction of 1 in the pool of viral quasispecies at a given time point. Evolution of amino acid frequencies was plotted over time in relation to the estimated time of seroconversion as time 0.
Multiplicity of transmitted viruses was tested by using the methodology described by Keele et al. (Keele et al., 2008), and reported elsewhere (Novitsky et al., 2009c). In summary, maximum achievable gag diversity of more than 1% and maximum achievable number of differences (Hamming distances) of 14 or more were used as a signature for transmission of multiple viral variants for analysis of gag quasispecies sequences within the first 100 days post seroconversion. In this study, a total of 30 out of 42 subjects (71%) were infected with single or closely related viral variants, while transmission of multiple viral variants was evident in 12 individuals (29%).
Extended HIV-1 subtype C consensus sequence
The analyzed amino acid frequency at studied Gag residues was compared with the extended HIV-1 subtype C consensus that was generated as described elsewhere (Novitsky et al., 2002). A total of 1,871 HIV-1 subtype C gag sequences with a length of at least 500 nt were retrieved from the Los Alamos HIV Database at http://www.hiv.lanl.gov (access date September 17, 2009). The set was comprised of sequences from Bangladesh (n=13), Botswana (198), Ethiopia (39), France (26), India (67), Israel (22), Kenya (16), Malawi (218), Soudi Arabia (20), South Africa (783), Tanzania (21), United Kingdom (23), and Zambia (344); fewer than ten sequences per country from Argentina (2), Australia (5), Brazil (7), Canada (7), Democratic Republic of Congo (7), Cameroon (1), China (2), Djibouti (1), Denmark (2), Spain (6), Gabon (2), Georgia (1), Nigeria (2), Russia (1), Rwanda (2), Senegal (8), Somalia (2), Uganda (3), USA (3), Uruguay (1), Yemen (1), and Zimbabwe (6); and one sequence with unknown origin. Frequencies of amino acids at each codon were expressed as percentages, and the low threshold of detection was 0.5%.
MHC class I HLA typing
High-resolution HLA typing was performed using AlleleSEQR HLA Sequencing-Based Typing kit (Celera, Alameda, CA) according to the manufacturer's instructions. Assignment of HLA alleles was implemented by Assign SBT ver. 3.5.1.42 (Conexio Genomics, Applecross, Australia). Polymorphisms outside the targeted exons that could not resolve heterozygote combinations are presented as HLA allele variants in brackets or as two-digit HLA typing results. Related to the study analyses, nine of 42 individuals expressed HLA-B*57/5801: one B*5702, three B*5703, and five B*5801.
Statistical analysis
Timing data were summarized with medians (interquartile range for 25% and 75%). Comparisons of continuous outcomes between two groups were based on Mann-Whitney rank sum tests. Ninety-five percent confidence intervals on shift parameter were based on inversions of the Mann-Whitney rank sum tests. In analyses of plasma RNA load, measurements originated during the first 50 days p/s were excluded from analysis due to potential effect of the viral load peak. Comparisons between groups were performed for time intervals 50–200, 201–300, 301–400, and 401–500 days p/s computed as mean values for each subject within the analyzed time intervals. All reported p-values were 2-sided and not adjusted for multiple testing. Confidence intervals for shift parameter were obtained using R, version 2.10.1 (The R Project for Statistical Computing, Vienna, Austria). All other statistical analyses were implemented using SigmaStat v.3.5. Plots were generated by SigmaPlot v.10 and refined in Adobe Illustrator CS4.
Results
To assess the in vivo dynamics of viral quasispecies in the context of Gag residue 242, the frequency and timing of Gag mutations were analyzed longitudinally in the cohort of 42 subjects with estimated time of seroconversion. Potential associations between frequency of viral mutations at Gag residue 242 and plasma RNA load or CD4+ T cell counts were analyzed in individuals expressing or not expressing HLA-B*57/5810.
In vivo escape and reversion at Gag residue 242
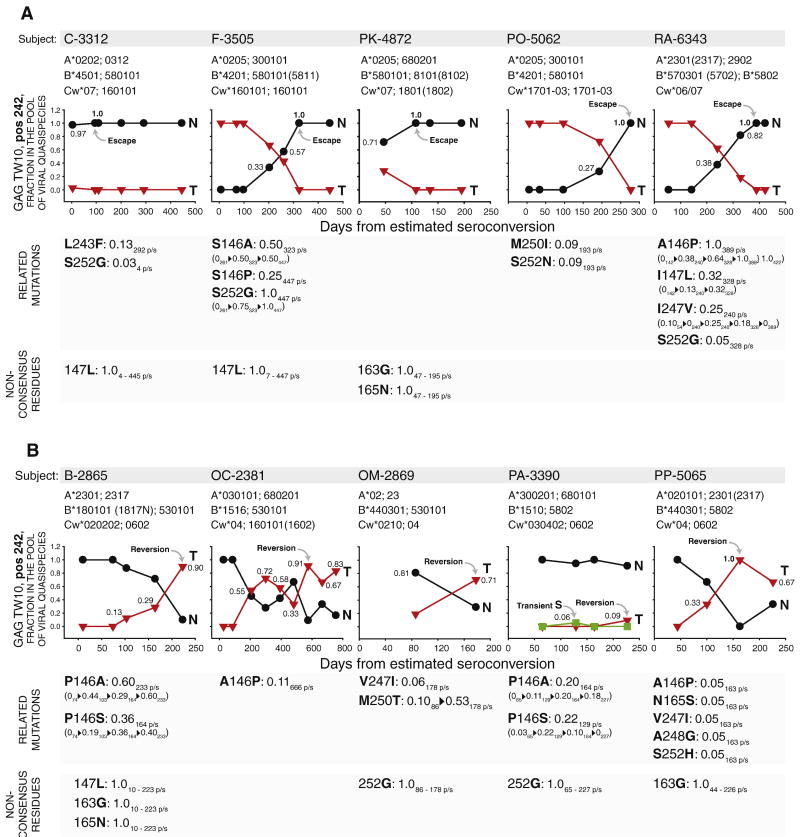
Changes in amino acid frequency at Gag residue 242 were detected in 12 out of 42 (28.6%) subjects. To evaluate the evolutionary patterns of mutational pathways, the observed frequency of viral quasispecies at Gag residue 242 were plotted over time with time zero as the estimated time of seroconversion (Fig. 1). The escape mutation from Thr to Asn was detected in five subjects including two acutely infected individuals, C-3312 and F-3505, expressing either HLA-B*57 (subject RA-6343) or B*580101 (four subjects; Fig. 1A). In three of those subjects, F-3505, PO-5062, and RA-6343, Thr was a single amino acid in the pool of viral quasispecies present at the time close to seroconversion, which was confirmed by 2 to 3 sampling time points before escape occurred. The transmission of a single viral variant in these subjects was confirmed by analysis of gag diversity and Hamming distances at the earliest time points, as presented elsewhere (Novitsky et al., 2009c). The escape from Thr to Asn was gradual, which was evident from increasing frequencies in the pool of viral quasispecies over time. In two other subjects expressing HLA-B*580101, C-3312 and PK-4872, a mixture of both Thr and Asn was detected at the earliest available time points, day 4 p/s (242T 95%CI: 0.92–1.02) and day 47 p/s (242T 95%CI: 0.37–1.05), respectively, but Thr was completely substituted by Asn at the next time point in both subjects (Fig. 1A). The T242N escape was complete (reached 100% in the pool of viral quasispecies) in all five subjects. It is likely that the coexistence of both Thr and Asn in the pool of viral quasispecies upon transmission to a new host can facilitate escape mutations in individuals expressing HLA-B*57/5801.
Figure 1.
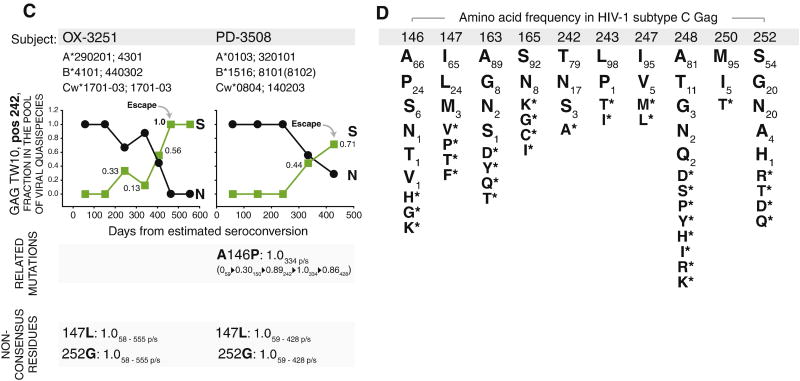
The dynamics of in vivo mutations at Gag residue 242 and related polymorphisms in 12 subjects infected with HIV-1 subtype C. Figures A, B, and C outline subject ID, MHC class I HLA profile, frequency (axis y) and kinetics (axis x) of viral quasispecies in the pool of viral quasispecies at Gag residue 242, and dynamics of related mutations below the corresponding graph, if any. Time is shown in days from estimated time of seroconversion. Numbers within the graphs outline frequency of closest amino acid at given time point in the pool of viral quasispecies (the frequency of the second amino acid equals 1 − frequency of the shown amino acid). The related polymorphisms were analyzed at Gag residues 146, 147, 163, 165, 243, 247, 248, 250, and 252. Presence of the related polymorphisms is shown as a stretch of changing amino acids at specified Gag residue (e.g., L243F delineates substitution of Leu by Phe at position 243) followed by observed frequency of emerging amino acid residue at specified time point in relation to the estimated seroconversion (e.g., L243F: 0.13 292 p/s stands for the 13% frequency of the emerging amino acid, Phe, at day 292 p/s, and, given the time scale at the 242-graph, represents a transient polymorphism observed at a single time point). In case of dynamic changes in the frequency, the observed related polymorphisms are presented as a stretch of amino acid frequencies at the specified time point (e.g., “0261▶0.50323▶0.50447” below “S146A: 0.50 323 p/s” delineates changing frequency of Ala at Gag residue 146 from 0 at day 261 p/s to 0.50 at day 323 p/s, and remaining at frequency 0.50 at day 447 p/s; frequency of “0.50 323 p/s” immediately following “S146A” outlines the highest-frequency first time observed at the specified time point). Non-consensus residues without detectable dynamics in the pool of viral quasispecies are outlined at the bottom of graphs A, B, and C (e.g., “147L: 1.0 4 – 445 p/s” delineates the presence of non-consensus Leu at Gag residue 147 (Iso corresponds to the consensus amino acid at position 147; see Fig. 1D) at frequency 100% over the follow-up period from day 4 p/s to day 445 p/s). A: Five HLA-B*57/5801-positive subjects with T242N escape mutations. B: Five HLA-B*57/5801-negative subjects with N242T reverse mutations. C: Two HLA-B*57/5801-negative subjects with N242S mutations. D: List of amino acid residues related to mutations at Gag residue 242 (amino acid positions are shaded by gray bar), and frequency of corresponding amino acid residues among HIV-1 subtype C (n=1,871; for details see Extended HIV-1 subtype C consensus sequence in the Material and methods section).
In contrast, five subjects who did not express HLA-B*57 or B*5801 demonstrated reversion from Asn to Thr or a mix of Thr/Asn (Fig. 1B). Analysis of multiplicity of HIV-1 infection provided evidence for a single transmitted viral variant in four of five subjects, B-2865, OC-2381, OM-2869, and PA-3390. Although Asn at amino acid position 242 was found in 10 out of 10 sequenced quasispecies in subject PP-5065 at day 44 p/s, levels of gag diversity and Hamming distances in this subject argued for transmission of multiple viral variants. Therefore, we cannot exclude a possibility that minor 242T variant was present in the pool of transmitted quasispecies in subject PP-5065 below the level of detection by methodology used in the current study. The gradual nature of the N242T reversion was similar to the pattern of T242N escape. The opposite patterns of mutations, escape and reversion, observed in subjects expressing or not expressing HLA-B*57/5801, respectively, further confirm a remarkably efficient restriction of the TW10 epitope by MHC class I HLA HLA-B*57 and B*5801. Interestingly, four of five HLA-B*57/5801-negative subjects did not reach complete amino acid substitution during reversion over the follow-up period, even by day 755 p/s, as seen in subject OC-2381 (Fig. 1B). A transient Ser was observed at day 129 p/s in subject PA-3390. Two subjects, PA-3390 and PP-5065, expressed HLA-B*5802, highlighting the inability of this MHC class I HLA molecule to restrict epitope TW10.
Viral mutation from Asn to Ser at Gag residue 242 that was described previously as a transient mutation (Crawford et al., 2007) was detected in two HLA-B*57/5801-negative subjects, OX-3251 and PD-3508 (Fig. 1C). These two subjects did not share any class I HLA allele (Fig. 1C), suggesting that some alternative mechanisms may be involved. Subject PD-3508 had Ala-to-Pro mutation at Gag residue 146 as one of only six Gag mutations occurring from 59 to 428 days p/s, while subject OX-3251 had as many as 47 mutations across Gag from 58 to 555 days p/s but none was among the list of polymorphisms related to mutations at Gag residue 242 (see below).
Gag polymorphisms relevant to mutations at residue 242
Mutations at Gag residue 242 are likely to be affected by other mutants within or flanking the TW10, ISW9, or KF11 epitopes (Brumme et al., 2008; Crawford et al., 2009; Crawford et al., 2007; Martinez-Picado et al., 2006; Matthews et al., 2009; Matthews et al., 2008). Therefore, subjects with detectable mutations at Gag residue 242 were analyzed for viral polymorphisms at Gag residues 146, 147, 163, 165, 243, 247, 248, 250, and 252. The amino acid frequencies in the HIV-1 subtype C Gag consensus at these positions are outlined in Figure 1D. Polymorphisms at specified Gag residues were detected in 10 of 12 analyzed cases. The Ser-to-Gly at residue 252 accompanied T242N escape in three of five cases (Fig. 1A). In addition, the Ser-to-Asn and Ser-to-His substitutions at position 252 were detected in one subjects with T242N escape, PO-5062 (Fig. 1A), and one subject with N242T reversion, PP-5065 (Fig. 1B), respectively. The heterogeneity of viral polymorphisms at Gag residue 252 was evident by a complete S252G substitution in subject F-3505, and transient appearance of Gly or Asn in three other T242N escape cases (Fig. 1A) and transient appearance of His in case of N242T reversion (Fig. 1B). The diverse polymorphisms at Gag residue 146 were observed in seven of 12 subjects accompanying four of five N242T reversions (Fig. 1B), two T242N escape (Fig. 1A), and one N242S mutation (Fig. 1C). A complete A146P mutation was detected in two subjects, RA-6343 (Fig. 1A) and PD-3508 (Fig. 1C). Other polymorphisms at Gag residue 146 were represented by the A146P escape (subjects OC-2381 and PP-5065; Fig. 1B), or P146A reversion (subjects B-2865 and PA-3390; Fig. 1B), or toggling between Pro and Ser (subjects F-3505, Fig. 1A; and B-2865 and OM-2869, Fig. 1B). Other observed polymorphisms at positions relevant to mutations at Gag residue 242 included transient escape mutations at I147L (subject RA-6343), L243F (C-3312), I247V (RA-6343), A248G (PP-5065), and M250I (PO-5062); dominant escape at M250T (OM-2869); and transient reversions at N165S and V247I (both in PP-5065). Except described polymorphisms related to mutations at Gag residue 242, non-consensus amino acid substitutions were detected in nine of 12 subjects (Fig. 1A, 1B, and 1C). The 147L, 163G, 165N, and 252G were the most common non-changing substitutions accompanying viral mutations at Gag residue 242. Two subjects with N242S mutations have similar profile of 147L and 252G (Fig. 1C), suggesting that the background of 147L and 252G might provide replicative advantage for the 242S variant as compared with the 242T wild type virus.
Timing of in vivo mutations at Gag residue 242
Timing constraints of the in vivo mutations at Gag residue 242 were assessed separately in subjects expressing or not expressing MHC class I HLA-B*57/5801 (Fig. 2). Appearance, dominance, and completeness occurred within relatively tight time intervals among subjects. Thus, in the three HLA-B*57/5801-positive subjects infected with the wild type Thr at Gag residue 242, the T242N escape appeared at 193, 203, and 240 days p/s, reached dominance at 261, 277, and 328 days p/s, and became complete at 277, 323, and 389 days p/s (Fig. 2A). In five HLA-B*57/5801-negative subjects infected with 242N variant, the 242T reverse mutation appeared at median (IQR) 103 (97;213) days p/s, became dominant at 193 (170;215) days p/s, but reached completeness only in one of five subjects, PP-5065, over the follow-up period (Fig. 2A). While the appearance time did not differ between reverse (n=5) and escape mutations (n=3; p=0.39), the time to reach dominance at Gag residue 242 appeared to be shorter for reverse mutations (p=0.057). Transmission of 242TN-mix to subjects expressing HLA-B*57/5801 resulted in fast 242N escape that reached completeness at 95 day p/s in subject C-3312 and at 108 day p/s in subject PK-4872, although a rapid escape before the time of sampling in the last case cannot be ruled out. No evidence for transmission of 242TN-mix to HLA-B*57/5801-negative subjects was found, but we cannot exclude this in subject OM-2869 due to late sampling.
Figure 2.
Timing of in vivo mutations at Gag residue 242. The time scale shows days from estimated seroconversion. In the box plots, the boundary of the box closer to zero indicates the 25th percentile, a solid line within the box marks the median value, and the boundary of the box farther from zero indicates the 75th percentile. A: Timing of escape mutation T242N in HLA-B*57/5801-positive subjects infected with the wild-type 242T. B: Timing of reverse mutation N242T in HLA-B*57/5801-negative subjects. Time of completeness for reverse mutation was not calculated because N242T reached completeness in only one of five cases.
Viral RNA load in HLA-B*57/5801-negative subjects
Subjects who do not express HLA-B*57/5801 were reported to have lower viral load upon transmission of 242N variant (Chopera et al., 2008) due to the apparent lower fitness of escape 242N variant. We tested whether viral RNA load differed between HLA-B*57/5801-negative subjects with (n=7; Figs. 1B and 1C)) and without (n=26 who did not express HLA-B*57/5801 and did not have mutations at Gag residue 242 in the study cohort)) transmitted Asn at Gag residue 242 over time (Fig. 3A). We found no evidence for reduced viral load in HLA-B*57/5801-negative subjects infected with 242N at any of the analyzed time intervals up to 500 days p/s (Fig. 3A). Next, we tested whether HLA-B*57/5801-negative subjects infected viral variant encoding Asn at Gag residue 242 have increased CD4+ T cell count (Fig. 3B). Similarly to viral load, no differences were detected in the levels of CD4+ T cell count between groups of HLA-B*57/5801-negative subjects with and without transmitted Asn.
Figure 3.
Median viral RNA load and CD4+ T cell counts in MHC class I HLA-B*57/5801-negative subjects are compared between groups with and without transmitted Asn at Gag residue 242. The upper error represents the 75th percentile. Comparisons between groups were based on Mann-Whitney rank sum test.
Measurements originated during the first 50 days p/s were excluded from analysis. A. Plasma viral RNA load over four time intervals: 50–200, 201–300, 301–400, and 401–500 days p/s. B. CD4+ T cell count over four time intervals: 50–200, 201–300, 301–400, and 401–500 days p/s.
Viral RNA load in HLA-B*57/5801-positive subjects
Not all subjects expressing HLA-B*57/5801 demonstrate Gag T242N escape mutation in primary HIV-1 subtype C infection. Four out of nine HLA-B*57/5801-positive subjects in this study maintained wild type Thr at Gag residue 242 without evidence for escape to Asn over the follow-up period. We tested whether viral RNA load differs in HLA-B*57/5801-positive subjects infected with the wild type virus with and without T242N escape mutation in the primary HIV-1 subtype C infection. Subjects without the detectable T242N escape mutation maintained significantly lower viral load than subjects with T242N escape up to 500 days p/s (Fig. 4A). Three out of four HLA-B*57/5801-positive individuals with wild type 242T expressed HLA-B*5703 or -B*5702, while four out of five HLA-B*57/5801-positive individuals with 242N escape expressed HLA-B*5801. Analysis of the CD4+ T cell counts between subjects with and without T242N mutation revealed a trend toward higher CD4 count in HLA-B*57/5801-positive subjects without T242N escape mutation at three out of four time intervals (Fig. 4B).
Figure 4.
Median viral load and CD4+ T cells in MHC class I HLA-B*57/5801-positive subjects with and without mutations at Gag residue 242. The upper error represents the 75th percentile. P-values were calculated using a Mann-Whitney rank sum test. Ninety-five percent confidence intervals on shift parameter were based on inversions of the Mann-Whitney rank sum tests (shown below the corresponding p-value for each time interval). Positive value of a shift parameter indicates that the values are greater in the group with mutations at Gag residue 242 than the group without. Measurements originated during the first 50 days p/s were excluded from analysis A. Plasma viral RNA load over four time intervals in relation to the estimated time of seroconversion: 50–200, 201–300, 301–400, and 401–500 days p/s. B. CD4+ T cell count over four time intervals in relation to the estimated time of seroconversion: 50–200, 201–300, 301–400, and 401–500 days p/s.
Discussion
This study addressed patterns and timing of the in vivo mutations at the key amino acid residue 242 within the Gag epitope TW10 in primary HIV-1 subtype C infection. Four major findings prompt discussion. First, the dynamics of viral mutations at Gag residue 242 are predictable in most cases based on the profile of transmitted virus and expression (or no expression) of MHC class I HLA-B*57/5801 by the host. Second, if the mutation occurs, the time of viral mutations at Gag residue 242 might be predicted. Third, HLA-B*57/5801-negative subjects infected with 242N virus did not have lower viral RNA load or higher CD4+ T cell counts when compared with HLA-B*57/5801-negative subjects infected with the wild type virus over the first year of HIV-1 subtype C infection, and therefore, our data do not support the previous report by Chopera et al. (Chopera et al., 2008). Fourth, we found that HLA-B*57/5801-positive subjects who maintain the wild type Thr at Gag residue 242 without escape to Asn during primary HIV-1 subtype C infection have significantly lower viral RNA load.
The dissecting of viral quasispecies sampled frequently soon after seroconversion revealed early dynamics of the in vivo profile of viral mutations at Gag residue 242 in primary HIV-1 subtype C infection. Consistent with the previous studies (Brumme et al., 2008; Chopera et al., 2008; Crawford et al., 2009; Crawford et al., 2007; Leslie et al., 2004; Martinez-Picado et al., 2006; Matthews et al., 2009; Matthews et al., 2008), expressing or not expressing HLA-B*57/5801 by an HIV-infected host is predictive of the direction of mutation, escape or reversion, at Gag residue 242. An escape from Thr to Asn was seen in HLA-B*57/5801-positive subjects, while reversion from Asn to Thr was observed in HLA-B*57/5801-negative individuals. Two HLA-B*57/5801-negative subjects in this study demonstrated mutation from Asn to Ser, raising the question why no reversion to Thr occurred. In both subjects Asn encoded by AAC was substituted by Ser encoded by AGC (transition), while in subjects reverting from Asn to Thr, there was a substitution from AAC to ACC (transversion) on the background of non-consensus 147L and 252G. Although transition is more likely than transversion, the overall frequency of Asn-to-Thr at Gag residue 242 is higher than frequency of Asn-to-Ser, which warrants further study addressing the mechanistic reason for these substitutions and potential compensatory role of the 147L and 252G background for N242S mutation.
In the case of transmission of a single viral variant, time of T242N escape was between 203 and 323 days p/s, while N242T reversion occurred between 103 and 193 days p/s. The reverse mutations at Gag residue 242 reached a state of dominance earlier than escape mutations (p=0.009). However, in contrast to escape mutations, the reverse mutations at Gag residue 242 did not reach complete amino acid substitution in four of five subjects over the follow-up period, which can be attributable to a relatively short follow-up period in some cases with N242T reversion.
Our data suggests differential timing of T242N escape mutation in HLA-B*57/5801-positive subjects depending on the profile of transmitted viral quasispecies. We found that it matters whether transmitted virus includes only a wild type Thr at Gag residue 242, or a mix of both Thr and Asn. Transmission of the wild type Thr led to appearance of T242N variants around 203 days p/s that advanced to compete T242N substitution by day 323 p/s. In contrast, transmission of 242TN-mix resulted in much faster T242N escape that reached completeness by day 115 p/s. Together with the timing patterns of viral mutations in HLA-B*57/5801-negative subjects, this finding provides important insight for modeling viral evolution on a population level, and generates a hypothesis that predicts increase of the 242N and 242TN-mix in the fraction of transmitted HIV-1 subtype C viruses in the population with substantial proportion of HLA-B*57/5801 individuals over time.
We found no difference between levels of viral RNA load or CD4 counts in HLA-B*57/5801-negative subjects infected with 242N escape variant or with the wild type virus. This is in contrast to the recent report by Chopera et al. (Chopera et al., 2008), which is likely due to some methodological differences and confounding biases in both studies, as illustrated below. Chopera et al. (Chopera et al., 2008) analyzed nine cases with transmitted A146X (X=P or S), and six of those also showed T242N. In our study, no cases of transmitted A146X without T242N were observed, and the heterogeneous patterns of mutations at Gag residue 146 included Ser-to-Ala, Ser-to-Pro, Pro-to-Ala, Pro-to-Ser, Ala-to-Pro, or no mutation (Fig. 1). It is also possible that early advantage for the host due to low fitness of transmitted 242N variant may result in low viral load later on in the course of disease, and if so, our study was not powered to address associations at later time points. Given the limitation of small sample size, further larger studies with longer follow-up periods are needed to resolve whether and when transmitted 242N is associated with lower viral RNA load in HLA-B*57/5801-negative subjects, if any.
HLA-B*57/5801-positive subjects maintaining the wild type Thr at Gag residue 242 had significantly lower viral RNA load than subjects with T242N escape. This finding suggests the HLA-B*57/5801-mediated immune pressure is able to control replication of the wild type virus encoding Thr at Gag position 242, but cannot suppress viral replication of the T242N escape variant. Although the 242N viral variant has a lower fitness in the in vitro assay, its replication capacity is higher than 242T in the presence of the in vivo immune pressure. Most of individuals expressing HLA-B*57 maintained wild type virus encoding Thr at Gag residue 242, while most of individuals expressing HLA-B*5801 developed T242N escape. This finding supports the notion that selection of HIV-1 escape mutants within TW10 epitope by subjects expressing HLA-B*57 and HLA-B*5801 differs with more frequent selection of T242N escape by individuals expressing HLA-B*5801, and can explain the observed lower HIV RNA load in subjects without T242N escape mutation.
Conclusions
The study analyzed the dynamics of in vivo mutations at Gag residue 242 and related polymorphisms during primary HIV-1 subtype C infection, and demonstrated predictive directions and timing constraints of reverse, N242T, and escape, T242N, mutations based on the host's MHC class I HLA-B profile and transmitted viral variant(s). No difference in viral load or CD4 counts was found between HLA-B*57/5801-negative subjects infected with the wild type virus encoding Thr and the mutant virus encoding Asn at Gag residue 242. Lack of the T242N escape in HLA-B*57/5801-positive subjects infected with the wild type virus was associated with better control of viral replication during the early phase of HIV-1 subtype C infection.
Acknowledgments
We are very grateful to all participants in the Tshedimoso study in Botswana. We thank Gaseboloke Mothowaeng, Florence Modise, S'khatele Molefhabangwe, and Sarah Masole for their dedication and outstanding work in the clinic and outreach, and Lemme Kebaabetswe and Raabya Rossenkhan for excellent lab support. We greatly appreciate the enthusiasm and strong commitment of Mary Fran McLane in achieving the overall study goals. We express thanks to Molly Pretorius Holme and Michael Roy for excellent support in communication with IRBs. Finally, we thank Lendsey Melton for outstanding editorial assistance. The primary HIV-1 subtype C infection study in Botswana, the Tshedimoso study, was supported and funded by NIH grant R01 AI057027.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altfeld M, Addo MM, Rosenberg ES, Hecht FM, Lee PK, Vogel M, Yu XG, Draenert R, Johnston MN, Strick D, Allen TM, Feeney ME, Kahn JO, Sekaly RP, Levy JA, Rockstroh JK, Goulder PJ, Walker BD. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS. 2003;17(18):2581–91. doi: 10.1097/00002030-200312050-00005. [DOI] [PubMed] [Google Scholar]
- Betts MR, Ambrozak DR, Douek DC, Bonhoeffer S, Brenchley JM, Casazza JP, Koup RA, Picker LJ. Analysis of Total Human Immunodeficiency Virus (HIV)-Specific CD4+ and CD8+ T-Cell Responses: Relationship to Viral Load in Untreated HIV Infection. J Virol. 2001;75(24):11983–11991. doi: 10.1128/JVI.75.24.11983-11991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boaz MJ, Waters A, Murad S, Easterbrook PJ, Vyakarnam A. Presence of HIV-1 Gag-Specific IFN-gamma(+)IL-2(+) and CD28(+)IL-2(+) CD4 T Cell Responses is Associated with Nonprogression in HIV-1 Infection. J Immunol. 2002;169(11):6376–85. doi: 10.4049/jimmunol.169.11.6376. [DOI] [PubMed] [Google Scholar]
- Boutwell CL, Rowley CF, Essex M. Reduced viral replication capacity of human immunodeficiency virus type 1 subtype C caused by cytotoxic-T-lymphocyte escape mutations in HLA-B57 epitopes of capsid protein. J Virol. 2009;83(6):2460–8. doi: 10.1128/JVI.01970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumme ZL, Tao I, Szeto S, Brumme CJ, Carlson JM, Chan D, Kadie C, Frahm N, Brander C, Walker B, Heckerman D, Harrigan PR. Human leukocyte antigen-specific polymorphisms in HIV-1 Gag and their association with viral load in chronic untreated infection. AIDS. 2008;22(11):1277–86. doi: 10.1097/QAD.0b013e3283021a8c. [DOI] [PubMed] [Google Scholar]
- Carr JK, Laukkanen T, Salminen MO, Albert J, Alaeus A, Kim B, Sanders-Buell E, Birx DL, McCutchan FE. Characterization of subtype A HIV-1 from Africa by full genome sequencing. AIDS. 1999;13(14):1819–26. doi: 10.1097/00002030-199910010-00003. [DOI] [PubMed] [Google Scholar]
- Chopera DR, Woodman Z, Mlisana K, Mlotshwa M, Martin DP, Seoighe C, Treurnicht F, de Rosa DA, Hide W, Karim SA, Gray CM, Williamson C. Transmission of HIV-1 CTL Escape Variants Provides HLA-Mismatched Recipients with a Survival Advantage. PLoS Pathogens. 2008;4(3):e1000033. doi: 10.1371/journal.ppat.1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford H, Lumm W, Leslie A, Schaefer M, Boeras D, Prado JG, Tang J, Farmer P, Ndung'u T, Lakhi S, Gilmour J, Goepfert P, Walker BD, Kaslow R, Mulenga J, Allen S, Goulder PJ, Hunter E. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J Exp Med. 2009;206(4):909–21. doi: 10.1084/jem.20081984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford H, Prado JG, Leslie A, Hue S, Honeyborne I, Reddy S, van der Stok M, Mncube Z, Brander C, Rousseau C, Mullins JI, Kaslow R, Goepfert P, Allen S, Hunter E, Mulenga J, Kiepiela P, Walker BD, Goulder PJ. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J Virol. 2007;81(15):8346–51. doi: 10.1128/JVI.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan KA, Lawrence HS, Hares DR, Fisher CL, Budowle B. An improved method for post-PCR purification for mtDNA sequence analysis. J Forensic Sci. 2002;47(4):811–8. [PubMed] [Google Scholar]
- Edwards BH, Bansal A, Sabbaj S, Bakari J, Mulligan MJ, Goepfert PA. Magnitude of Functional CD8+ T-Cell Responses to the Gag Protein of Human Immunodeficiency Virus Type 1 Correlates Inversely with Viral Load in Plasma. J Virol. 2002;76(5):2298–2305. doi: 10.1128/jvi.76.5.2298-2305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, Heldebrant C, Smith R, Conrad A, Kleinman SH, Busch MP. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17(13):1871–9. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- Geldmacher C, Currier JR, Herrmann E, Haule A, Kuta E, McCutchan F, Njovu L, Geis S, Hoffmann O, Maboko L, Williamson C, Birx D, Meyerhans A, Cox J, Hoelscher M. CD8 T-cell recognition of multiple epitopes within specific Gag regions is associated with maintenance of a low steady-state viremia in human immunodeficiency virus type 1-seropositive patients. J Virol. 2007;81(5):2440–8. doi: 10.1128/JVI.01847-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goepfert PA, Lumm W, Farmer P, Matthews P, Prendergast A, Carlson JM, Derdeyn CA, Tang J, Kaslow RA, Bansal A, Yusim K, Heckerman D, Mulenga J, Allen S, Goulder PJ, Hunter E. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J Exp Med. 2008;205(5):1009–17. doi: 10.1084/jem.20072457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder PJ, Bunce M, Krausa P, McIntyre K, Crowley S, Morgan B, Edwards A, Giangrande P, Phillips RE, McMichael AJ. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res Hum Retroviruses. 1996;12(18):1691–8. doi: 10.1089/aid.1996.12.1691. [DOI] [PubMed] [Google Scholar]
- Kaslow RA, Carrington M, Apple R, Park L, Munoz A, Saah AJ, Goedert JJ, Winkler C, O'Brien SJ, Rinaldo C, Detels R, Blattner W, Phair J, Erlich H, Mann DL. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2(4):405–11. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- Kawashima Y, Pfafferott K, Frater J, Matthews P, Payne R, Addo M, Gatanaga H, Fujiwara M, Hachiya A, Koizumi H, Kuse N, Oka S, Duda A, Prendergast A, Crawford H, Leslie A, Brumme Z, Brumme C, Allen T, Brander C, Kaslow R, Tang J, Hunter E, Allen S, Mulenga J, Branch S, Roach T, John M, Mallal S, Ogwu A, Shapiro R, Prado JG, Fidler S, Weber J, Pybus OG, Klenerman P, Ndung'u T, Phillips R, Heckerman D, Harrigan PR, Walker BD, Takiguchi M, Goulder P. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009 doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proceedings of the National Academy of Sciences. 2008;105(21):7552–7. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, Bishop K, van der Stok M, Nair K, Khan N, Crawford H, Payne R, Leslie A, Prado J, Prendergast A, Frater J, McCarthy N, Brander C, Learn GH, Nickle D, Rousseau C, Coovadia H, Mullins JI, Heckerman D, Walker BD, Goulder P. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13(1):46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- Klein MR, van der Burg SH, Hovenkamp E, Holwerda AM, Drijfhout JW, Melief CJ, Miedema F. Characterization of HLA-B57-restricted human immunodeficiency virus type 1 Gag- and RT-specific cytotoxic T lymphocyte responses. J Gen Virol. 1998;79(Pt 9):2191–201. doi: 10.1099/0022-1317-79-9-2191. [DOI] [PubMed] [Google Scholar]
- Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, Tang Y, Holmes EC, Allen T, Prado JG, Altfeld M, Brander C, Dixon C, Ramduth D, Jeena P, Thomas SA, St John A, Roach TA, Kupfer B, Luzzi G, Edwards A, Taylor G, Lyall H, Tudor-Williams G, Novelli V, Martinez-Picado J, Kiepiela P, Walker BD, Goulder PJ. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med. 2004;10(3):282–9. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- Liu SL, Rodrigo AG, Shankarappa R, Learn GH, Hsu L, Davidov O, Zhao LP, Mullins JI, Haynes BF, Pantaleo G, Fauci AS. HIV Quasispecies and Resampling. Science. 1996;273(5274):413c–417. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
- Los Alamos National Laboratory. HIV Sequence Databases and Compendia. 2009;2009 [Google Scholar]
- Martinez-Picado J, Prado JG, Fry EE, Pfafferott K, Leslie A, Chetty S, Thobakgale C, Honeyborne I, Crawford H, Matthews P, Pillay T, Rousseau C, Mullins JI, Brander C, Walker BD, Stuart DI, Kiepiela P, Goulder P. Fitness Cost of Escape Mutations in p24 Gag in Association with Control of Human Immunodeficiency Virus Type 1. J Virol. 2006;80(7):3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masemola A, Mashishi T, Khoury G, Mohube P, Mokgotho P, Vardas E, Colvin M, Zijenah L, Katzenstein D, Musonda R, Allen S, Kumwenda N, Taha T, Gray G, McIntyre J, Karim SA, Sheppard HW, Gray CM. Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. J Virol. 2004;78(7):3233–43. doi: 10.1128/JVI.78.7.3233-3243.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews PC, Leslie AJ, Katzourakis A, Crawford H, Payne R, Prendergast A, Power K, Kelleher AD, Klenerman P, Carlson J, Heckerman D, Ndung'u T, Walker BD, Allen TM, Pybus OG, Goulder PJ. HLA footprints on human immunodeficiency virus type 1 are associated with interclade polymorphisms and intraclade phylogenetic clustering. J Virol. 2009;83(9):4605–15. doi: 10.1128/JVI.02017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews PC, Prendergast A, Leslie A, Crawford H, Payne R, Rousseau C, Rolland M, Honeyborne I, Carlson J, Kadie C, Brander C, Bishop K, Mlotshwa N, Mullins JI, Coovadia H, Ndung'u T, Walker BD, Heckerman D, Goulder PJ. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J Virol. 2008;82(17):8548–59. doi: 10.1128/JVI.00580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, Hallahan CW, Selig SM, Schwartz D, Sullivan J, Connors M. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A. 2000;97(6):2709–14. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndongala ML, Peretz Y, Boulet S, Doroudchi M, Yassine-Diab B, Boulassel MR, Rouleau D, Tremblay C, Leblanc R, Routy JP, Sekaly RP, Bernard NF. HIV Gag p24 specific responses secreting IFN-gamma and/or IL-2 in treatment-naive individuals in acute infection early disease (AIED) are associated with low viral load. Clin Immunol. 2009 doi: 10.1016/j.clim.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Novitsky V, Gilbert P, Peter T, McLane MF, Gaolekwe S, Rybak N, Thior I, Ndung'u T, Marlink R, Lee TH, Essex M. Association between virus-specific T-cell responses and plasma viral load in HIV-1 subtype C infection. J Virol. 2003;77(2):882–890. doi: 10.1128/JVI.77.2.882-890.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Lagakos S, Herzig M, Bonney C, Kebaabetswe L, Rossenkhan R, Nkwe D, Margolin L, Musonda R, Moyo S, Woldegabriel E, van Widenfelt E, Makhema J, Essex M. Evolution of proviral gp120 over the first year of HIV-1 subtype C infection. NIHMSID # 79286. Virology. 2009a;383(1):47–59. doi: 10.1016/j.virol.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Smith UR, Gilbert P, McLane MF, Chigwedere P, Williamson C, Ndung'u T, Klein I, Chang SY, Peter T, Thior I, Foley BT, Gaolekwe S, Rybak N, Gaseitsiwe S, Vannberg F, Marlink R, Lee TH, Essex M. HIV-1 subtype C molecular phylogeny: consensus sequence for an AIDS vaccine design? J Virol. 2002;76(11):5435–5451. doi: 10.1128/JVI.76.11.5435-5451.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Wang R, Kebaabetswe L, Greenwald J, Rossenkhan R, Moyo S, Musonda R, Woldegabriel E, Lagakos S, Essex M. Better Control of Early Viral Replication Is Associated with Slower Rate of Elicited Antiviral Antibodies in the Detuned EIA during Primary HIV-1C Infection. J Acquir Immune Defic Syndr. 2009b;52:265–272. doi: 10.1097/QAI.0b013e3181ab6ef0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Wang R, Margolin L, Baca J, Kebaabetswe L, Rossenkhan R, Bonney C, Herzig M, Nkwe D, Moyo S, Musonda R, Woldegabriel E, van Widenfelt E, Makhema J, Lagakos S, Essex M. Timing constraints of in vivo gag mutations during primary HIV-1 subtype C infection. PLoS One. 2009c;4(11):e7727. doi: 10.1371/journal.pone.0007727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Woldegabriel E, Kebaabetswe L, Rossenkhan R, Mlotshwa B, Bonney C, Finucane M, Musonda R, Moyo S, Wester C, van Widenfelt E, Makhema J, Lagakos S, Essex M. Viral Load and CD4+ T Cell Dynamics in Primary HIV-1 Subtype C Infection. J Acquir Immune Defic Syndr. 2009d;50(1):65–76. doi: 10.1097/QAI.0b013e3181900141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Woldegabriel E, Wester C, McDonald E, Rossenkhan R, Ketunuti M, Makhema J, Seage GR, 3rd, Essex M. Identification of primary HIV-1C infection in Botswana. NIHMSID # 79283. AIDS Care. 2008;20(7):806–11. doi: 10.1080/09540120701694055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky VA, Gilbert PB, Shea K, McLane MF, Rybak N, Klein I, Thior I, Ndung'u T, Lee TH, Essex ME. Interactive association of proviral load and IFN-gamma-secreting T cell responses in HIV-1C infection. Virology. 2006;349(1):142–55. doi: 10.1016/j.virol.2006.02.006. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Nelson GW. Human genes that limit AIDS. Nat Genet. 2004;36(6):565–74. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- Palmer S, Kearney M, Maldarelli F, Halvas EK, Bixby CJ, Bazmi H, Rock D, Falloon J, Davey RT, Jr, Dewar RL, Metcalf JA, Hammer S, Mellors JW, Coffin JM. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol. 2005;43(1):406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramduth D, Chetty P, Mngquandaniso NC, Nene N, Harlow JD, Honeyborne I, Ntumba N, Gappoo S, Henry C, Jeena P, Addo MM, Altfeld M, Brander C, Day C, Coovadia H, Kiepiela P, Goulder P, Walker B. Differential immunogenicity of HIV-1 clade C proteins in eliciting CD8+ and CD4+ cell responses. J Infect Dis. 2005;192(9):1588–96. doi: 10.1086/496894. [DOI] [PubMed] [Google Scholar]
- Ray SC. MargFreq. 2008 ( http://sray.med.som.jhmi.edu/SCRoftware/MargFreq/). SCRoftware.
- Rolland M, Heckerman D, Deng W, Rousseau CM, Coovadia H, Bishop K, Goulder PJ, Walker BD, Brander C, Mullins JI. Broad and Gag-biased HIV-1 epitope repertoires are associated with lower viral loads. PLoS ONE. 2008;3(1):e1424. doi: 10.1371/journal.pone.0001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PP, Korber BT. Detecting hypermutations in viral sequences with an emphasis on G --> A hypermutation. Bioinformatics. 2000;16(4):400–1. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]
- Serwanga J, Shafer LA, Pimego E, Auma B, Watera C, Rowland S, Yirrell D, Pala P, Grosskurth H, Whitworth J, Gotch F, Kaleebu P. Host HLA B*allele-associated multi-clade Gag T-cell recognition correlates with slow HIV-1 disease progression in antiretroviral therapy-naive Ugandans. PLoS ONE. 2009;4(1):e4188. doi: 10.1371/journal.pone.0004188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thobakgale CF, Prendergast A, Crawford H, Mkhwanazi N, Ramduth D, Reddy S, Molina C, Mncube Z, Leslie A, Prado J, Chonco F, Mphatshwe W, Tudor-Williams G, Jeena P, Blanckenberg N, Dong K, Kiepiela P, Coovadia H, Ndung'u T, Walker BD, Goulder PJ. Impact of HLA in mother and child on disease progression of pediatric human immunodeficiency virus type 1 infection. J Virol. 2009;83(19):10234–44. doi: 10.1128/JVI.00921-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga R, Lucchetti A, Galvan P, Sanchez S, Sanchez C, Hernandez A, Sanchez H, Frahm N, Linde CH, Hewitt HS, Hildebrand W, Altfeld M, Allen TM, Walker BD, Korber BT, Leitner T, Sanchez J, Brander C. Relative Dominance of Gag p24-Specific Cytotoxic T Lymphocytes Is Associated with Human Immunodeficiency Virus Control. J Virol. 2006;80(6):3122–3125. doi: 10.1128/JVI.80.6.3122-3125.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]